Abstract
In vivo transduction of nondividing cells by human immunodeficiency virus type 1 (HIV-1)-based vectors results in transgene expression that is stable over several months. However, the use of HIV-1 vectors raises concerns about their safety. Here we describe a self-inactivating HIV-1 vector with a 400-nucleotide deletion in the 3′ long terminal repeat (LTR). The deletion, which includes the TATA box, abolished the LTR promoter activity but did not affect vector titers or transgene expression in vitro. The self-inactivating vector transduced neurons in vivo as efficiently as a vector with full-length LTRs. The inactivation design achieved in this work improves significantly the biosafety of HIV-derived vectors, as it reduces the likelihood that replication-competent retroviruses will originate in the vector producer and target cells, and hampers recombination with wild-type HIV in an infected host. Moreover, it improves the potential performance of the vector by removing LTR sequences previously associated with transcriptional interference and suppression in vivo and by allowing the construction of more-stringent tissue-specific or regulatable vectors.
Retroviral vectors are attractive tools for human gene therapy. First, they stably integrate into the chromosomes of their targets, a likely requisite for long-term expression. Second, they do not transfer viral genes, avoiding transduced cells that are destroyed by virus-specific cytotoxic T cells. Third, they have a relatively large cloning capacity, sufficient for most envisioned clinical situations. In addition to these characteristics, which are common to all retroviral vectors, vectors derived from lentiviruses offer one great advantage over their oncoretroviral counterparts: they can transduce nondividing cells, a crucial asset for genetically modifying tissues considered the main potential targets of gene therapy, such as the brain, the muscle, the liver, the lungs, and the hematopoietic system. Illustrating these properties, vectors derived from human immunodeficiency virus type 1 (HIV-1) allow for the efficient in vivo delivery, integration, and stable expression of transgenes into cells such as neurons, hepatocytes, and myocytes (2, 14, 17, 18). Although this opens exciting prospects for human gene therapy, the biosafety of HIV-based vectors requires a most careful evaluation, considering the pathogenicity of the parental virus.
Two components are involved in the making of a virus-based gene delivery system: first, the packaging elements, encompassing the structural proteins as well as the enzymes necessary to generate an infectious particle, and second, the vector itself, that is, the genetic material which will be transferred to the target cell. Biosafety safeguards, one goal of which is to prevent the emergence of replication-competent recombinants (RCRs), can be introduced in designing both of these components.
The packaging unit of the first generation of HIV-based vectors comprised all of the HIV-1 proteins except the envelope (18). A major step towards clinical acceptability was the subsequent demonstration that the fundamental properties of this system were left intact after deletion of four additional viral genes, encoding proteins proven or likely to represent crucial virulence factors: Vpr, Vif, Vpu, and Nef (31). More recent studies now indicate that the main transactivator of HIV, Tat, is also dispensable for generation of a fully efficient vector (7). What could be termed the third-generation packaging unit of HIV-1-based vectors thus conserves only three of the nine genes present in the genome of the parental virus: gag, pol, and rev. This eliminates the possibility that a wild-type virus will be reconstituted through recombination.
The system would be further improved if the transcriptional elements of HIV were removed from the vector. The modalities of reverse transcription, which generates both U3 regions of an integrated provirus from the 3′ end of the viral genome, facilitate this task by allowing the creation of so-called self-inactivating (SIN) vectors. Self-inactivation relies on the introduction of a deletion in the U3 region of the 3′ long terminal repeat (LTR) of the DNA used to produce the vector RNA. During reverse transcription, this deletion is transferred to the 5′ LTR of the proviral DNA. If enough sequence is eliminated to abolish the transcriptional activity of the LTR, the production of full-length vector RNA in transduced cells is abolished. This minimizes the risk that RCRs will emerge. Furthermore, it reduces the likelihood that cellular coding sequences located adjacent to the vector integration site will be aberrantly expressed, either due to the promoter activity of the 3′ LTR or through an enhancer effect. Finally, a potential transcriptional interference between the LTR and the internal promoter driving the transgene is prevented by the SIN design.
SIN vectors have been derived from murine leukemia virus (MLV) and spleen necrosis virus (SNV) (6, 12, 29, 30). Their development, however, has highlighted some of the difficulties inherent in this approach. The 3′ LTR is indeed involved in the polyadenylation of the viral RNA, a function that requires sequence elements often spread over U3, R, and U5. A U3 deletion conferring self-inactivation must eliminate as many of the transcriptionally important motifs from the LTR as possible while sparing the polyadenylation determinants. Because of overlaps between these two functional entities, most MLV-derived SIN vectors carry a deletion limited to the enhancer and as a consequence conserve significant transcriptional activity in their LTRs. One attempt to mutate the TATA box dramatically decreased the titers of the resulting vector, presumably because polyadenylation was rendered inefficient (29).
Studies on the regulation of HIV-1 polyadenylation have located the main cis-acting element governing the polyadenylation of the viral genomic RNA distal to the TATA box, just upstream of the R region of the LTR (5, 26, 27). This suggests that HIV-1-derived vectors may tolerate large U3 deletions and even a complete removal of the viral promoter without functional loss. Verifying this prediction, we report here on the successful development of HIV-based SIN vectors. Extensive U3 deletions, including one which removed the TATA box and resulted in an almost complete loss of LTR promoter activity, could be introduced without altering vector titers. Furthermore, none of the in vitro and in vivo properties of HIV-derived vectors were compromised by the SIN configuration.
MATERIALS AND METHODS
SIN plasmids. (i) pHR′SIN plasmids.
A KpnI-XbaI fragment containing the polypurine tract and the 3′ LTR was excised from a pHR′ plasmid and subcloned into the corresponding sites of pUC18. This plasmid was digested completely with EcoRV and partially with PvuII and self-ligated. A plasmid carrying a 400-nucleotide-long deletion of U3 was recovered. An XhoI linker was inserted in the EcoRI site of the deletion plasmid, and an XhoI-XbaI fragment was cloned back into the pHR′CMVlacZ plasmid digested with the corresponding enzymes. All other SIN-18 plasmids were obtained by substituting reporter genes (encoding luciferase, enhanced green fluorescence protein [GFP], and Neo) for lacZ. All reporter genes were swapped as BamHI-XhoI cassettes. The pHR′ vector plasmids used in this study differ from the plasmids originally described (17) by an XhoI-KpnI deletion removing 118 nucleotides from the Nef-coding sequence upstream of the polypurine tract and a deletion of 1,456 nucleotides of human sequence downstream of the 3′ LTR. This human sequence remained from the original cloning of the HXB2 proviral genome. The two deletions did not affect vector titers or transgene expression in dividing 293T cells.
(ii) pRLLSIN plasmids.
The construction of pRRL plasmids containing a chimeric 5′ LTR made of Rous sarcoma virus U3 and HIV-1 R/U5 regions is described elsewhere (7). pRRLPGK-GFPSIN-18, pRRLPGK-GFPSIN-36, pRRLPGK-GFPSIN-45, and pRRLPGK-GFPSIN-78 are vectors in which the 3′ LTR sequences from position −418 to −18, −36, −45, and −78, respectively, have been deleted from pRRLPGK-GFP.
pRRLPGK-GFPSIN-18 was generated by replacing the 590-bp EcoRI-AflII fragment from pRRLPGK-GFP with the 200-bp EcoRI-AflII fragment from pHR′CMVlacZSIN-18 in a four-part ligation with a 2.95-kb AflII fragment, a 2.8-kb AflII-BamHI fragment, and a 760-bp BamHI-EcoRI fragment from pRRLPGK-GFP.
pRRLPGK-GFPSIN-36 was derived from pRRLPGK-GFP by replacing the 493-bp BbsI-AlwNI fragment in the 3′ LTR with an oligonucleotide linker consisting of 5′-GATATGATCAGATC-3′ and 5′-CTGATCA-3′. The linker was ligated with a 540-bp AlwNI-AvrII fragment and a 6.1-kb AvrII-BbsI fragment from pRRLPGKGFP in a three-part ligation. pRRLPGK-GFPSIN-45 was generated similarly by using the oligonucleotides 5′-GATATGATCAGAGCCCTCAGATC-3′ and 5′-CTGAGGGCTCTGATCA-3′. The two oligonucleotides 5′-GATATGATCAGGAGGCGTGGCCTGGGCGGGACTGGGGAGTGGCG AGCCCTCAGATC-3′ and 5′-CTGAGGGCTCGCCACTCCCCAGTCCCGCCCAGGCCACGCCTCCTGATCA-3′ were used to generate pRRLPGK-GFPSIN-78.
Other plasmids.
The envelope plasmid pMD.G and the packaging plasmid pCMVΔR8.91 have been described previously (31).
Cells.
Dulbecco’s modified Eagle medium (Gibco) was supplemented with 10% fetal calf serum and a combination of penicillin-streptomycin and glutamine (Gibco). 293T, HeLa, HeLa-tat, 208 F, and NIH 3T3 cells were cultured in supplemented Dulbecco’s modified Eagle medium in a 10% CO2 atmosphere. SupT1 cells were cultured in RPMI 1640 medium (modified) (JRH Biosciences) supplemented with 10% fetal calf serum and 2 mM l-glutamine in a 5% CO2 atmosphere. Rat thyroid PC C13 cell lines immortalized with either E1A or v-Raf have been described previously (1). Primary human T lymphocytes were isolated and transduced as previously described (8). Gamma irradiation (8,000 rads) was delivered to cells in suspension as in previous studies (31) by a 3-min exposure to a 60Co source.
Northern blot analysis.
Total RNA was isolated from transduced HeLa cells by using RNAsol B as suggested by the manufacturer. RNA (10 to 20 μg) was separated on 1% agarose gel by using NorthernMax (Ambion) reagents and transferred to a Zetabind membrane by capillary transfer. A GFP-specific probe was 32P labelled by random priming.
Vector stock preparation.
Stocks were prepared as previously described (31) by transient cotransfection of three plasmids into 293T cells. The p24 concentration was determined by antigen immunoadsorbtion with a kit from the National Cancer Institute. Vector production and gene delivery were done in a biosafety level 2 environment. Vector-producing cells and transduced cells were fixed by a 30-min incubation in phosphate-buffered saline containing 4% paraformaldehyde before fluorescence-activated cell sorter (FACS) analysis on a Becton Dickinson FACScan.
In vitro transduction.
In vitro transduction experiments were done in six-well plates (Costar). Filtered vector-containing medium was added 24 h after the cells (2 × 105 cells/well) had been seeded and was left until cells were analyzed 48 to 60 h later. Typically, the following amounts of p24 were used: 0.1 ng for titration of lacZ vectors by X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining, 1 to 5 ng for luciferase assay, and 10 to 20 ng for β-galactosidase (β-Gal) enzyme assay and for titration of GFP vectors by FACS analysis. Multiplicities of infection can be estimated assuming that 1 ng of p24 corresponds to 1,000 to 5,000 transducing units (TU).
HIV-1 infection.
Vesicular stomatitis virus (VSV) G-pseudotyped HIV-1 particles were generated by transfection into 293T cells of the plasmid HXBH10ΔenvCAT, a Vpu-positive HIV-1 derivative with a deletion in env and a chloramphenicol acetyltransferase (CAT) gene in place of nef (a kind gift of H. Göttlinger, Dana-Farber Cancer Institute), and pMD.G. The conditioned medium was collected and filtered, and 50 ng of p24 antigen was used to infect overnight 106 SupT1 cells. Infected cells were assayed by p24 immunostaining or CAT assay to demonstrate similar extents of infection (not shown).
In vivo gene delivery.
Vector particles were concentrated from filtered supernatants by two rounds of centrifugation. Fisher 344 male rats weighing approximately 220 g were obtained from Harlan Sprague-Dawley and housed in accordance with published National Institutes of Health guidelines. All surgical procedures were performed with the rats under isofluorane gas anesthesia with aseptic instruments. Two microliters of lentivirus vector in phosphate-buffered saline was injected slowly (0.5 μl per min) into the striatum under stereotaxic guidance. One month after the injection, the animals were sacrificed and the brains were analyzed for GFP expression by immunocytochemistry. The primary anti-GFP antibody was purchased from Clontech and used at a 1:1,000 dilution. Biotinylated rabbit anti-goat secondary antibody, streptavidin conjugated to horseradish peroxidase, and the VIP chromagen kit were from Vector.
RESULTS
A 400-nucleotide-long deletion in the U3 region of the HIV-1 3′ LTR does not affect vector titers.
The upstream sequence element essential for polyadenylation of the HIV-1 genomic RNA has been mapped to a region situated between the TATA box and the beginning of the R region (26, 27). In contrast, all of the major determinants responsible for regulating the HIV-1 LTR promoter activity (including the so-called negative response element, the two NFκB and the NF-ATc binding sites, the three SP1 binding sites, and the TATA box) are located within the boundaries of a 400-nucleotide-long EcoRV-PvuII fragment which does not overlap with the upstream sequence element (Fig. 1). Based on this premise, this fragment was deleted from the 3′ LTR of the pHR′ CMVlacZ plasmid used to generate a β-Gal-expressing HIV-based vector. In the resulting pHR′CMVlacZSIN-18 construct, only 53 nucleotides were left in U3: 35 nucleotides upstream of the EcoRV site to preserve efficient recognition and processing by integrase and 18 nucleotides downstream of the PvuII site to govern polyadenylation. Transducing particles were produced by transient transfection of three plasmids into 293T cells as previously described (31): the multiply deleted packaging construct pCMVΔR8.91, which encodes Gag, Pol, Tat, and Rev; a plasmid expressing the surface glycoprotein (G) of VSV; and the vector DNA itself, in this case either the original pHR′CMVlacZ plasmid or its U3 deletion pHR′ CMVlacZSIN-18 version. The two vectors gave comparable titers as measured with 293T cells as targets: 1,476 ± 232 TU per ng of p24 capsid antigen for the SIN-18 vector and 1,544 ± 126 TU/ng of p24 for the control. The blue color following X-Gal treatment appeared already after 3 h in cells transduced with the SIN-18 vector, whereas cells transduced with full-length U3 vector scored positive only after 6 to 8 h. This suggested that LacZ expression was higher when the flanking LTRs were deleted. The β-Gal activity in cells transduced by the SIN-18 vector was indeed twice that found in cells containing the parental vector, with the number of transduced cells being equal (Fig. 2). A similar observation was made with a pair of full-length and SIN-18 luciferase-expressing vectors, although in this case the number of transduced cells could not be determined (not illustrated).
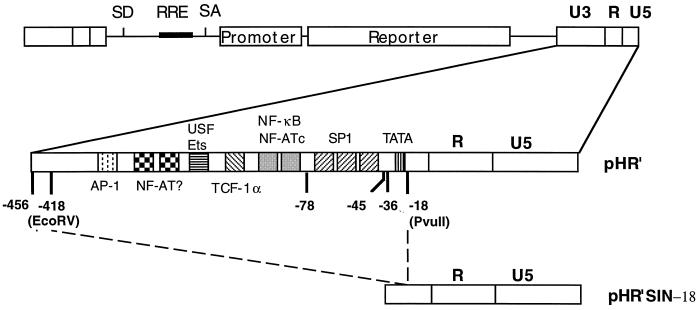
FIG. 1.
Structure of SIN HIV-derived vectors. A schematic representation of an HIV-1 vector with enlarged 3′ LTR to show the binding sites for differents transcription factors on U3 is shown (not to scale). Although the 3′ LTR is depicted, the nucleotide numbering refers to the cap site at the beginning of R as +1 as for a 5′ LTR. Position −418 is the 5′ limit of all deletions; positions −78, −45, −36, and −18 indicate the 3′ limits of the different deletions described in the text. The deletion generating the SIN-18 vector created a novel BglII site. Details on the nuclear factors binding U3 can be found in references 10, 15, and 22 and references therein. SD, splice donor; RRE, Rev-response element; SA, splice acceptor. The GenBank accession number for the wild-type 3′ LTR is M1991.
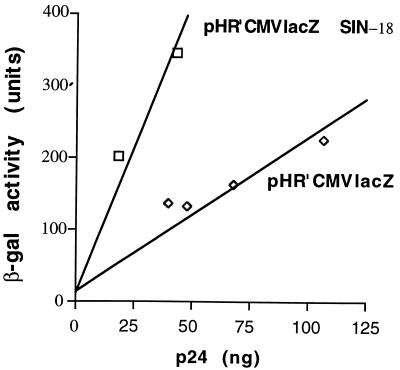
FIG. 2.
Expression of a lacZ transgene delivered by SIN or full-length LTR vectors. 293T cells were transduced with equal volumes (200 μl) of two HR′CMVlacZSIN-18 or four HR′CMV lacZ vector stocks. Titers (TU per nanogram of p24) were similar for all stocks. β-Gal activity (in arbitrary units) at 48 h postinfection is plotted against the amount of p24 in vector stocks. Cells transduced with SIN-18 vectors express more than twice as much β-Gal per nanogram of p24 than cells transduced with full-length LTR vectors.
Vectors with less extensive U3 deletions were also generated (Fig. 1). In the SIN-36 and SIN-45 vectors the TATA box is intact, while in the SIN-78 vector the TATA box and the three SP1 binding sites are preserved. All U3 deletion vectors had transducing abilities that were comparable to that of their full-length U3 parent in both HeLa cells and peripheral blood lymphocytes (Table 1).
TABLE 1.
Deletions in the U3 region of the HIV-1 LTR do not affect vector titersa
| Vector | Titer on HeLa cellsb (106 TU/ml) | Transduction of human PBLsc (%) |
|---|---|---|
| RRLPGK-GFP | 18 ± 2.4 | 50.75 ± 1.12 |
| RRLPGK-GFPSIN-18 | 14.5 ± 2.7 | 59.27 ± 0.90 |
| RRLPGK-GFPSIN-36 | 13 ± 2.1 | 55.86 ± 2.26 |
| RRLPGK-GFPSIN-45 | 12.7 ± 1.1 | 54.38 ± 1.16 |
| RRLPGK-GFPSIN-78 | 12.8 ± 1.2 | 55.59 ± 1.20 |
Fluorescent cells were scored by FACS analysis 6 days after transduction. Results are averages and standard deviations for duplicate determinations for an experiment representative of three performed.
The end point titer was determined by multiplying the number of fluorescent cells by the vector dilution. Samples were selected from the linear portion of the vector dose-response curve.
Percentage of fluorescent human peripheral blood lymphocytes (PBLs) after infection of 106 cells with 1 ml of vector-containing medium.
Unlike plasmids previously engineered to produce SNV-based SIN vectors (6), pHR′SIN plasmids have no polyadenylation signal downstream of the U3 deletion LTR to remedy a possible weakness of the RNA 3′ end processing. Thus, similar titers for SIN and regular vectors suggested that even the LTR with the most extensive U3 deletion had retained good polyadenylating activity (Table 1). Polyadenylation of the SIN-derived transcripts was also assumed to be efficient in target cells because of the good expression of the transgene in the SIN setting.
Transcriptional impact of U3 deletions.
As a first approach to determine the effects of the various U3 deletions on vector-derived RNA production in target cells, HeLa cells transduced with regular or SIN GFP-expressing vectors were subjected to Northern blot analyses, using a GFP probe capable of detecting transcripts produced from both the LTR and the phosphoglycerate kinase (PGK) internal promoter (Fig. 3). A small amount of spliced, LTR-derived RNA was detectable in HeLa cells transduced with the full-length U3 vector, despite the absence of Tat (lane 1). This residual LTR promoter activity was also observed with a deletion limited to sequences upstream of the SP1 sites, in the SIN-78 vector (lane 5). However, no LTR-driven transcript was detected with any of the SIN vectors lacking the SP1 binding sites (lanes 2 to 4).
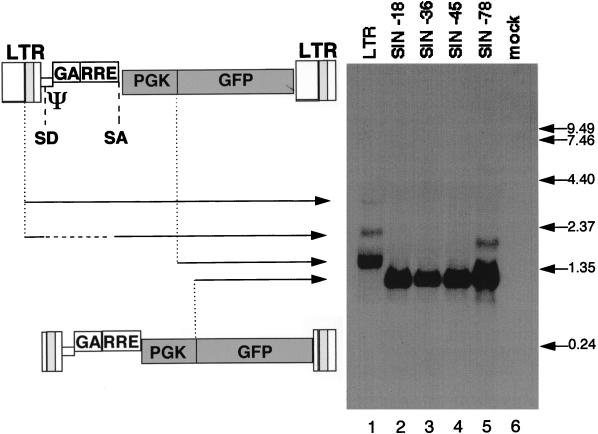
FIG. 3.
Northern blot analysis of vector-derived transcripts in transduced HeLa cells. Total RNA was extracted from HeLa cells transduced with an RRLPGK-GFP vector (lane 1) or with its SIN versions SIN-18 (lane 2), SIN-36 (lane 3), SIN-45 (lane 4), and SIN-78 (lane 5). In lane 1, three bands with the sizes expected for the LTR-derived transcripts (unspliced and spliced) and the PGK-derived transcripts are visible. As expected for HeLa cells, transcription was initiated much more frequently at the internal PGK promoter than at the 5′ HIV-1 LTR. In lanes 2 to 5, transcripts derived from SIN vectors are 340 to 400 nucleotides smaller than the corresponding transcripts in lane 1. RNA initiated at the HIV-1 LTR is detectable in lane 5 but not in lanes 2 to 4. Positions of molecular size markers (in kilobases) are indicated on the right. Ψ, encapsidation signal; SD, splice donor; SA, splice acceptor.
The high sensitivity of the luciferase activity assay and the strong stimulation of HIV-1 LTR promoter activity by Tat were exploited to analyze more accurately the transcriptional activities of the SIN vectors. For this, vectors containing the luciferase cDNA without an internal promoter, that is, those in which transgene expression is controlled exclusively by the HIV-1 5′ LTR, were used. Luciferase activity was measured in HeLa-tat, HeLa, 293T, and NIH 3T3 cells infected with normalized amounts of HR′luciferase or HR′luciferaseSIN-18 vectors (Table 2). With the full-length U3 vector, a strong production of luciferase was detected in HeLa-tat cells, while it was moderate in 293T cells and weak in HeLa and NIH 3T3 cells. The results obtained with 293T cells may reflect the presence in these cells of the adenovirus early protein E1A (24), which is known to stimulate HIV-1 LTR promoter activity (16). The U3 deletion present in HR′luciferaseSIN-18 resulted in a 350-fold reduction of luciferase activity in HeLa-tat and 293T cells. The very low levels of luciferase in HeLa-tat cells transduced with HR′luciferaseSIN-18 confirmed the transfer of the U3 deletion to the 5′ LTR and the minimal transcriptional activity of the U3 deletion LTR even in the presence of Tat. However, the SIN-18 vector still induced higher levels of luciferase in HeLa-tat cells than in HeLa cells. As the deletion abrogates transcription from the upstream LTR, this raised the possibility that the U3 deletion was repaired at a low but detectable frequency (see below) or that a promoter trap mechanism was enhanced by the presence of Tat in target cells. To investigate this point further, vectors expressing β-Gal without an internal promoter were used. In HeLa-tat cells, after normalization for p24 content of the inocula, the β-Gal-expressing HR′lacZ SIN-18 vector induced titers of 320 ± 11 TU/ml, compared with 4.1 × 105 ± 0.6 × 105 TU/ml for the control HR′LacZ vector. No positive cells were detected among HeLa cells exposed to 1 ml of either type of vector. The complete Tat dependence of β-Gal expression suggested that it resulted from U3 repair, although one could not completely exclude that Tat-mediated transcriptional activation enhanced promoter trapping. If transgene expression from the promoterless SIN-18 vector resulted entirely from U3 repair, then the frequency of this event, based on a comparison of the relative titers of the SIN-18 and wild-type vectors in HeLa-tat cells, was close to 1/1,000. It is important to recall, however, that in this case vector particles were generated by cotransfecting plasmids carrying a simian virus 40 (SV40) origin of replication in cells containing the SV40 large T antigen, a setting highly favorable for DNA recombination.
TABLE 2.
Promoter activity of the HIV-1 LTR with a U3 deletion
| Vector | Luciferase activity (RLU) in transduced cellsa
|
|||
|---|---|---|---|---|
| HeLa-tat | HeLa | 293T | NIH 3T3 | |
| HR′luciferaseSIN-18 | 245 ± 66 | 96 ± 12 | 34 ± 13 | 4.5 ± 0.6 |
| HR′luciferase | 87,553 ± 9,038 | 1,888 ± 272 | 11,613 ± 1,733 | 349 ± 71 |
| ΔU3/wild-type U3b | 0.0028 | 0.05 | 0.0029 | 0.012 |
Two HR′luciferaseSIN-18 and two HR′luciferase vector stocks were independently produced and used to transduce in parallel and in duplicate HeLa-tat, HeLa, 293T, or NIH 3T3 cells. Under the conditions used (2 ng of p24 with 2 × 105 cells), the multiplicity of infection can be estimated to be about 0.01. For each vector type the results for the two stocks were averaged. The given relative light unit (RLU) values are for 20 μl of a 200-μl protein extract. The luciferase activity in each protein extract was assayed in duplicate and averaged, and the background value for each cell type was subtracted. Results are means and standard deviations.
Ratio of the activities obtained with the two types of vectors.
The potential impact of interactions between the LTR and the internal promoter was probed by evaluating the effect of deleting U3 on the production of a luciferase reporter expressed from two different internal promoters (cytomegalovirus [CMV] and PGK), using various cell types as targets (Table 3). Because the GC-rich sequence of the mouse PGK promoter contains only three ATG triplets, LTR-derived RNAs can be translated and can contribute to the expression of transgenes delivered by HR′PGK vectors. In contrast, the AT-rich CMV promoter sequence contains 17 ATG triplets, which impair the translation of LTR-derived RNAs. Despite this difference, with both promoters the SIN-18/wild-type U3 ratio was 2 in 293T cells, suggesting that in these targets the presence of a full-length LTR interferes with transcription from the internal promoter. The adenovirus early gene E1A, which is expressed in 293T cells, appeared to be responsible for this phenomenon, because a similar SIN-18/wild-type U3 ratio was noted in 293 cells, excluding a role for the SV40 large T antigen, and in a rat thyroid cell line immortalized with E1A but not in one immortalized with v-Raf (data not shown). However, the level of LTR activity per se did not seem to be the key factor in inducing promoter interference, because HeLa and HeLa-tat cells transduced with HR′CMV-GFP vectors expressed the same level of GFP even though the HIV-1 LTR is 50 times more active in HeLa-tat cells than in HeLa cells. With a PGK internal promoter, a moderate but consistently positive effect of the U3 deletion on transgene expression was observed in 293T, HeLa, SupT1, and 3T3 cells (Table 3 and data not shown).
TABLE 3.
Relative expression of the luciferase gene delivered by U3 deletion and full-length LTR HIV-1 vectors
| Target cells | Relative expressiona using the following internal promoter:
|
||
|---|---|---|---|
| CMVb
|
PGKc (dividing cells) | ||
| Dividing cells | Gamma-irradiated cellsd | ||
| 293T | 2.39 | 2.10 | 2.2 |
| HeLa | 0.47 | 0.82 | 1.1e |
| F208 | 0.47 | NDf | 0.56 |
| 3T3 | 0.69 | 0.49 | 1.09e |
Luciferase activity (mean from two assays) was plotted against p24, and a linear regression was calculated for each type of vector by using Criketgraph as for Fig. 2. Values are the ratios of the slopes.
Stocks of HR′CMVluciferase and HR′CMVluciferaseSIN-18 vectors were prepared in quadruplicate and used to transduce in parallel and in duplicate all four cell types.
Stocks of HR′PGKluciferase and HR′PGKluciferaseSIN-18 were prepared in triplicate and used to transduce in parallel and in duplicate all four cell types.
Eight thousand rads was delivered by a 3-min exposure to a 60Co source.
Transductions by U3 deletion and full-length LTR vectors were not significantly different (by Student’s t test).
ND, not determined.
Pattern of activation of SIN vectors following HIV infection of transduced cells.
To investigate further the degree of transcriptional inactivation resulting from the various U3 deletions, human T lymphoid SupT1 cells stably transduced with wild-type or SIN PGK-GFP vectors were infected with envelope-defective, VSV G-pseudotyped HIV-1. Because the PGK promoter allows for the translational readthrough of LTR-derived transcripts, increases in GFP levels were used as a reflection of Tat-induced LTR activation. In cells containing the full-length U3 or the SIN-78 vector, GFP expression was stimulated following infection, while no such phenomenon was observed in cells transduced with the SIN-45, SIN-36, or SIN-18 vector (Fig. 4). Confirming the results obtained with HeLa-tat cells, these data indicate that also in an established T-cell line the HIV-1 LTR remains active despite the absence of all of the transcriptional elements located upstream of the SP1 binding sites and that the SIN-18 design abrogates this activity. Correspondingly, while full-length U3 and SIN-78 vectors could be rescued by HIV infection of transduced cells, recombine with the viral genome, and possibly be mobilized to new targets, these risks are theoretically alleviated by the use of the SIN-18 vector.
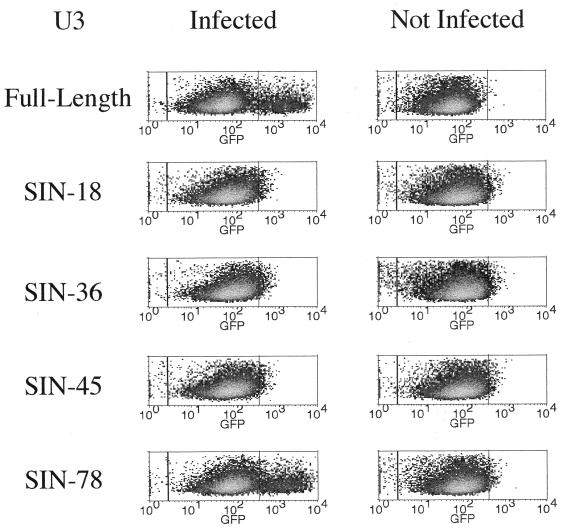
FIG. 4.
Activation pattern of HIV-1 vectors following infection of transduced SupT1 cells by HIV-1. Human lymphocytic SupT1 cells were transduced at a high multiplicity of infection by HIV-derived vectors carrying a PGK-GFP expression cassette and either a full-length LTR or the indicated U3 deletion construct. Six days later, the stably transduced cells were infected with VSV G-pseudotyped HIV-1 or were mock treated, and 48 h later they were analyzed by FACS for GFP fluorescence. Infection with HIV-1 strongly enhanced the expression of GFP in cells transduced by a vector with a full-length U3 LTR or the −78 deletion construct, while it had no effect on cells transduced with vectors having larger U3 deletions. The left and middle quadrants represent the fluorescence of cells not transduced and transduced by the GFP vector, respectively. The right quadrant includes cells with increased GFP expression upon infection by HIV-1. The increased expression of GFP indicates activation of vector transcription from the LTR and is due to translational readthrough of the PGK promoter sequence upstream of the GFP cDNA (see text). The increased in fluorescence intensity was 30-fold for cells transduced by the full-length LTR and 21-fold for those transduced by the SIN-78 vector. The HIV-1 had a deletion in the envelope gene and was thus limited to one round of infection. Similar patterns of Tat responsiveness were observed when HeLa-tat cells were transduced with the various vectors (not shown).
Efficient in vivo gene delivery by SIN vectors.
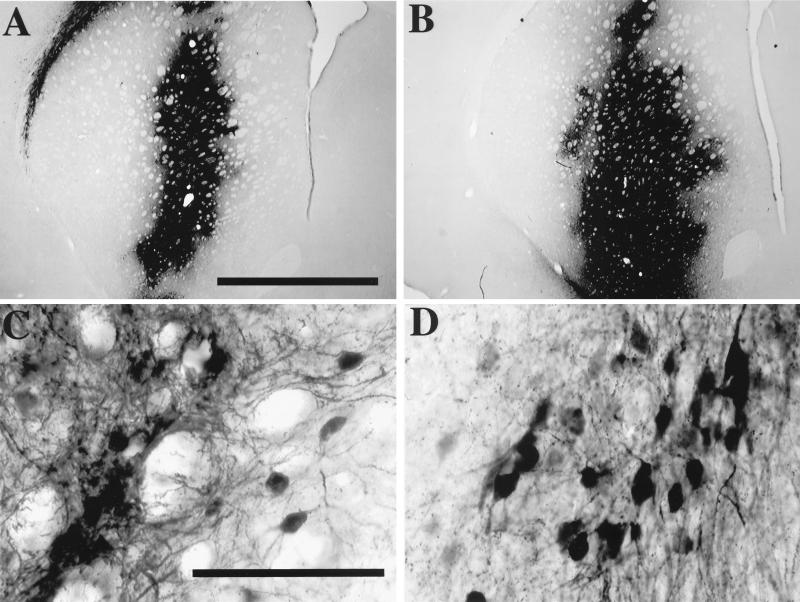
The results described above indicated that the inactivation design did not interfere with the transduction of cellular targets in vitro by HIV-derived vectors. The performance of SIN vectors was assayed in an in vivo delivery model that demands high efficiency of gene transfer and expression. Vectors carrying a PGK-GFP expression cassette with full-length U3 or U3 with the sequence from position −418 to −18 deleted were concentrated to high titers, matched for particle content by p24 antigen, and injected bilaterally in the neostriata of two groups of three adult rats. The animals were sacrificed after 1 month, and serial sections of the brain were analyzed for GFP expression by fluorescence (not illustrated) and immunostaining (Fig. 5). Both types of vector transduced neurons very efficiently: GFP-positive cells were detected at a very high density throughout most of the striata of all the injected animals. The level of transgene expression directed by the SIN-18 vector appeared to be even higher than that obtained with the wild-type vector. These results provide evidence that a SIN HIV-derived vector is an efficient vehicle for in vivo gene delivery.
FIG. 5.
In vivo transduction of GFP into neurons by SIN or full-length LTR vectors. HIV-1 vectors carrying a PGK-GFP expression cassette with the full-length U3 region (A and C) or the −18 deletion construct (B and D) were concentrated by ultracentrifugation and normalized for particle content prior to injection into the corpora striata of adult rats. One month after injection, brain sections were stained for immunoreactivity to the GFP protein. Both types of vectors transduced neurons very efficiently. The SIN vector often appeared to achieve a higher level of transgene expression. A representative section close to the injection site is shown for one of six injected striata per vector. Bars in panels A and C, 2 and 0.1 mm, respectively (magnifications are the same for panels B and D, respectively).
DISCUSSION
This study demonstrates that a large U3 deletion in the LTR of an HIV-1-based vector confers efficient self-inactivation without lowering the vector titer or impairing the expression of the transgene both in vitro and in vivo.
The HIV-1-based SIN vector presented here offers all of the previously claimed advantages of SIN retroviral vectors. First, the extensive U3 deletion of SIN-18 abolishes the viral promoter activity, thereby preventing the synthesis of full-length vector RNA in target cells. This results in minimizing the risk that a replication-competent retrovirus will emerge or that a cellular gene located immediately downstream of the 3′ LTR will be aberrantly expressed. Furthermore, the elimination of the LTR enhancer sequences in the SIN-18 design precludes the activation of a promoter located at a distance from the vector integration site. The so-called enhancer-less MLV vectors still have an active albeit attenuated viral promoter, and LTR-derived RNAs have been readily detected in transduced cells (11, 23). With such vectors, the spread of potential RCRs would not be limited to a single round of infection. Only the SNV-based vector developed by Olson et al. (20) and a chimeric MLV-based vector developed by Hawley et al. (12) are transcriptionally disabled to an extent comparable to that obtained with the HIV vector described here.
The SIN design also prevents potential interferences between the viral LTRs and the internal promoter, a phenomenon which can have profound implications in gene therapy. For instance, it was observed that the liver-specific albumin promoter loses its tissue specificity when flanked by MLV sequences (28). The mechanisms of promoter interference remain poorly understood. According to the classical view of promoter occlusion (4, 21), the presence of an active upstream viral LTR should decrease the activity of the internal promoter. Results presented by Yee et al. (29) and Chen et al. (3) initially gave credence to the promoter occlusion theory, but this conception was subsequently challenged by two well-documented studies. Taking advantage of the fact that the MLV LTR is transcriptionally competent in fibroblasts but not in embryonic stem cells, Soriano et al. have shown that the activity of different internal promoters is influenced by the sequence but not by the levels of transcriptional activity of the upstream LTR (23). Another study with an MLV vector in which the U3 region was replaced by a tetracycline-inducible promoter showed that activation of the chimeric LTR did not affect transcription from an internal promoter (13). In our system, the activation of the upstream HIV-1 LTR by Tat also failed to induce the occlusion of a downstream CMV promoter. Nevertheless, the comparison of various SIN and full-length HIV vectors revealed some promoter- and cell-specific differences in the degree of promoter interference, but in all cases the magnitude of these effects was minimal. While LTR-induced transcriptional inactivation of transgenes in vivo has not yet been described for HIV-based vectors, it may be relevant for the transduction of novel targets. The SIN design might help to avoid such an occurrence. Furthermore, the creation of tissue-specific and inducible vectors will be significantly facilitated by the availability of the SIN vector described here, which allows both the delivery of an internal expression cassette without flanking sequences that might influence its transcription and the swapping of novel enhancer-promoter sequences in the place of the deletion.
It is possible that the SIN HIV-1 vector described here underwent a repair of the U3 deletion at a maximal frequency of 1/1,000. This is much lower than that reported for the first generation of SNV- and avian leukosis virus-based U3 deletion vectors (9, 19). Moreover, it is likely that in our system the bulk of the repairing events occurred by recombination of the transfected plasmids. Documenting exactly what this frequency is in the current system is of little relevance, because only vectors produced from stable packaging cell lines will ultimately be considered for clinical use. When such cell lines become available for HIV vectors, it is likely that their SIN versions will exhibit the same low repair frequency as the newest generation of SNV-based SIN vectors (20).
The SIN design slightly increases the packaging capacity of HIV-based vectors by removing 400 bases of virus-derived sequence. Experiments performed with HIV-1 derivatives harboring the cDNAs of selectable markers in place of nef have demonstrated that viruses with a genome of more than 11 kb of RNA can maintain a full infectivity (25). In its current configuration, the SIN-18 vector contains approximately 1.7 kb of HIV-specific cis-acting sequence. Assuming that an internal promoter will occupy on average 500 bases, HIV-based vectors should be able to accommodate transgenes of at least 8.8 kb.
Finally, from a biosafety point of view, the newest generation of HIV-1-based vectors appears to be particularly reliable. Major improvements were brought to the original packaging system, first by deleting vif, vpr, vpu, env, and nef (31) and subsequently by removing tat and by expressing the gag-pol and rev genes from split genomes (7). Of note is that the strict Rev dependence of HIV-1 allows a distribution of the constituents of the vector-packaging system into more independent entities than is possible with MLV-based vectors. Here, we further demonstrate that a SIN HIV-based vector retains all of the properties of its full-length parent. When produced by packaging cell lines incorporating all of these safeguards, HIV-1-based vectors should meet the most stringent safety requirements for clinical applications.
ACKNOWLEDGMENTS
We thank H. Göttlinger for the CAT-expressing HIV-1 derivative and W. Haseltine and E. Terwilliger for HeLa-tat-III cells, obtained through the NIH AIDS Research and Reagent Reference Program.
This work was supported by a grant from the Swiss National Science Foundation and by a professorship from the Giorgi-Cavaglieri Foundation to D.T. R.Z. was the recipient of a fellowship from the Swiss National Science Foundation.
REFERENCES
- 1.Berlingieri M T, Manfioletti G, Santoro M, Bandiera A, Visconti R, Giancotti V, Fusco A. Inhibition of HMGI-C protein synthesis suppresses retrovirally induced neoplastic transformation of rat thyroid cells. Mol Cell Biol. 1995;15:1545–1553. doi: 10.1128/mcb.15.3.1545. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 2.Blömer U, Naldini L, Kafri T, Trono D, Verma I M, Gage F H. Highly efficient and sustained gene transfer in adult neurons with a lentivirus vector. J Virol. 1997;71:6641–6649. doi: 10.1128/jvi.71.9.6641-6649.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen B F, Hsieh C L, Chen D S, Hwang L H. Improved gene expression by a U3-based retroviral vector. Biochem Biophys Res Commun. 1992;184:330–337. doi: 10.1016/0006-291x(92)91197-x. [DOI] [PubMed] [Google Scholar]
- 4.Cullen B R, Lomedico P T, Ju G. Transcriptional interference in avian retroviruses—implications for the promoter insertion model of leukaemogenesis. Nature. 1984;307:241–245. doi: 10.1038/307241a0. [DOI] [PubMed] [Google Scholar]
- 5.DeZazzo J D, Kilpatrick J E, Imperiale M J. Involvement of long terminal repeat U3 sequences overlapping the transcription control region in human immunodeficiency virus type 1 mRNA 3′ end formation. Mol Cell Biol. 1991;11:1624–1630. doi: 10.1128/mcb.11.3.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dougherty J P, Temin H M. A promoter-less retroviral vector indicates that there are sequences in U3 required for 3′ RNA processing. Proc Natl Acad Sci USA. 1987;84:1197–1201. doi: 10.1073/pnas.84.5.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dull T, Zufferey R, Kelly M, Mandel R J, Nguyen M, Trono D, Naldini L. A third-generation lentivirus vector with a conditional packaging system. J Virol. 1998;72:8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finer M H, Dull T J, Quin L, Farson D, Roberts M R. Kat: a high efficiency retroviral transduction system for primary human T lymphocytes. Blood. 1994;83:43–50. [PubMed] [Google Scholar]
- 9.Flamant F, Aubert D, Legrand C, Cosset F L, Samarut J. Importance of 3′ non-coding sequences for efficient retrovirus-mediated gene transfer in avian cells revealed by self-inactivating vectors. J Gen Virol. 1993;74:39–46. doi: 10.1099/0022-1317-74-1-39. [DOI] [PubMed] [Google Scholar]
- 10.Giacca M, Gutierrez M I, Menzo S, D’Adda Di Fagagna F, Falaschi A. Human binding site for transcription factor USF/MLTF mimics the negative regulatory element of human immunodeficiency virus type 1. Virology. 1992;186:133–147. doi: 10.1016/0042-6822(92)90067-y. [DOI] [PubMed] [Google Scholar]
- 11.Hafenrichter D G, Wu X, Rettinger S D, Kennedy S C, Flye M W, Ponder K P. Quantitative evaluation of liver-specific promoters from retroviral vectors after in vivo transduction of hepatocytes. Blood. 1994;84:3394–3404. [PubMed] [Google Scholar]
- 12.Hawley R G, Covarrubias L, Hawley T, Mintz B. Handicapped retroviral vectors efficiently transduce foreign genes into hematopoietic stem cells. Proc Natl Acad Sci USA. 1987;84:2406–2410. doi: 10.1073/pnas.84.8.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hwang J J, Li L, Anderson W F. A conditional self-inactivating retrovirus vector that uses a tetracycline-responsive expression system. J Virol. 1997;71:7128–7131. doi: 10.1128/jvi.71.9.7128-7131.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kafri T, Blömer U, Peterson D A, Gage F H, Verma I M. Sustained expression of genes delivered directly into liver and muscle by lentiviral vectors. Nat Genet. 1997;17:314–317. doi: 10.1038/ng1197-314. [DOI] [PubMed] [Google Scholar]
- 15.Michael N L, D’Arcy L, Ehrenberg P K, Redfield R R. Naturally occuring genotypes of human immunodeficiency virus type 1 long terminal repeat display a wide range of basal and Tat-induced activities. J Virol. 1994;68:3163–3174. doi: 10.1128/jvi.68.5.3163-3174.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nabel G J, Rice S A, Knipe D M, Baltimore D. Alternative mechanisms for activation of human immunodeficiency virus enhancer in T cells. Science. 1988;239:1299–1302. doi: 10.1126/science.2830675. [DOI] [PubMed] [Google Scholar]
- 17.Naldini L, Blömer U, Gallay P, Ory D, Mulligan R, Gage F H, Verma I M, Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 18.Naldini L, Blömer U, Gage F H, Trono D, Verma I M. Efficient transfer, integration and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc Natl Acad Sci USA. 1996;93:11382–11388. doi: 10.1073/pnas.93.21.11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olson P, Temin H M, Dornburg R. Unusually high frequency of reconstitution of long terminal repeats in U3-minus retrovirus vectors by DNA recombination or gene conversion. J Virol. 1992;66:1336–1343. doi: 10.1128/jvi.66.3.1336-1343.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olson P, Nelson S, Dornburg R. Improved self-inactivating retroviral vector derived from spleen necrosis virus. J Virol. 1994;68:7060–7066. doi: 10.1128/jvi.68.11.7060-7066.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Proudfoot N J. Transcriptional interference and termination between duplicated alpha-globin gene constructs suggests a novel mechanism for gene regulation. Nature. 1986;322:562–565. doi: 10.1038/322562a0. [DOI] [PubMed] [Google Scholar]
- 22.Sieweke M H, Tekotte H, Jarosch U, Graf T. Cooperative interaction of Ets-1 with USF-1 required for HIV-1 enhancer activity in T cells. EMBO J. 1998;17:1728–1739. doi: 10.1093/emboj/17.6.1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soriano P, Friedrich G, Lawinger P. Promoter interactions in retrovirus vectors introduced into fibroblasts and embryonic stem cells. J Virol. 1991;65:2314–2319. doi: 10.1128/jvi.65.5.2314-2319.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spector D J. The pattern of integration of viral DNA sequences in the adenovirus 5-transformed human cell line 293. Virology. 1983;130:533–538. doi: 10.1016/0042-6822(83)90107-1. [DOI] [PubMed] [Google Scholar]
- 25.Trono D, Baltimore D. A human cell factor is essential for HIV-1 Rev action. EMBO J. 1990;9:4155–4160. doi: 10.1002/j.1460-2075.1990.tb07638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valsamakis A, Zeichner S, Carswell S, Alwine J C. The human immunodeficiency virus type 1 polyadenylation signal: a 3′ long terminal repeat element upstream of the AAUAAA necessary for efficient polyadenylation. Proc Natl Acad Sci USA. 1991;88:2108–2112. doi: 10.1073/pnas.88.6.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valsamakis A, Schek N, Alwine J C. Elements upstream of the AAUAAA within the human immunodeficiency virus polyadenylation signal are required for efficient polyadenylation in vitro. Mol Cell Biol. 1992;12:3699–3705. doi: 10.1128/mcb.12.9.3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu X, Holschen J, Kennedy S C, Ponder K P. Retroviral vector sequences may interact with some internal promoters and influence expression. Hum Gene Ther. 1996;7:159–171. doi: 10.1089/hum.1996.7.2-159. [DOI] [PubMed] [Google Scholar]
- 29.Yee J K, Moores J C, Jolly D J, Wolff J A, Respress J G, Friedmann T. Gene expression from transcriptionally disabled retroviral vectors. Proc Natl Acad Sci USA. 1987;84:5197–5201. doi: 10.1073/pnas.84.15.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu S F, von Ruden T, Kantoff P W, Graber C, Seiberg M, Ruther U, Anderson W F, Gilboa E. Self-inactivating retroviral vectors designed for transfer of whole genes into mammalian cells. Proc Natl Acad Sci USA. 1986;83:3194–3198. doi: 10.1073/pnas.83.10.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zufferey R, Nagy D, Mandel R J, Naldini L, Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol. 1997;15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]