Abstract
This study investigated the association between apolipoprotein E (APOE) gene polymorphisms (rs429358 and rs7412) and primary angle-closure glaucoma (PACG) and pseudoexfoliation glaucoma (PXG) in a Saudi cohort. Genotyping of 437 DNA samples (251 controls, 92 PACG, 94 PXG) was conducted using PCR-based Sanger sequencing. The results showed no significant differences in the allele and genotype frequencies of rs429358 and rs7412 between the PACG/PXG cases and controls. Haplotype analysis revealed ε3 as predominant, followed by ε4 and ε2 alleles, with no significant variance in PACG/PXG. However, APOE genotype analysis indicated a significant association between ε2-carriers and PACG (odds ratio = 4.82, 95% CI 1.52–15.26, p = 0.007), whereas no notable association was observed with PXG. Logistic regression confirmed ε2-carriers as a significant predictor for PACG (p = 0.008), while age emerged as significant for PXG (p < 0.001). These findings suggest a potential role of ε2-carriers in PACG risk within the Saudi cohort. Further validation and larger-scale investigations are essential to elucidate the precise role of APOE in PACG pathogenesis and progression.
Keywords: apolipoprotein E, angle-closure, genetics, glaucoma, polymorphisms, pseudoexfoliation, rs429358, rs7412, Saudi
1. Introduction
Glaucoma encompasses a spectrum of multifactorial ocular disorders marked by gradual deterioration of retinal ganglion cells (RGCs), optic nerve damage, and visual field impairment, often culminating in irreversible blindness if left untreated [1]. Among the subtypes of glaucoma, primary angle-closure glaucoma (PACG) and pseudoexfoliation glaucoma (PXG) exhibit distinct etiologies and clinical presentations [2,3]. PACG is characterized by the narrowing or closure of the drainage angle in the eye, often due to a shallow anterior chamber or a forward-positioned lens. This closure leads to increased intraocular pressure (IOP) and consequent optic nerve impairment [2]. On the other hand, PXG represents a form of secondary open-angle glaucoma, marked by the deposition of pseudoexfoliation material within various ocular tissues, such as the lens capsule, trabecular meshwork, and iris. This material obstructs aqueous drainage from the eye, resulting in increased IOP and optic nerve damage [3]. PACG and PXG significantly contribute to visual impairment and blindness globally, highlighting the importance of early detection and timely intervention to preserve vision and enhance patients’ quality of life [4].
Substantial evidence indicates a genetic predisposition to glaucoma, including PACG and PXG [5,6]. Given glaucoma’s complex and heterogeneous nature, elucidating the genetic factors contributing to PACG and PXG holds significant promise for advancing our understanding of their pathogenesis. This understanding is crucial not only for unraveling the pathogenesis but also for identifying early diagnostic markers, therapeutic interventions, and personalized management strategies [5,6,7].
Apolipoprotein E (APOE), a polymorphic gene located on chromosome 19q13.2, encodes a protein crucial for lipid metabolism and transport within the central nervous system and various ocular tissues [8,9]. APOE has also been detected in the pseudoexfoliative material [10]. Two common polymorphisms, rs429358 (T>C) at codon 112 and rs7412 (C>T) at codon 158, in the APOE gene give rise to three predominant APOE alleles in humans: ε2, ε3, and ε4. Each isoform differs subtly in amino acid composition at positions 112 and 158, resulting in distinct functional properties and disease-risk profiles [11]. APOE ε3, the most prevalent isoform in approximately 77% of the population, features a cysteine at position 112 and an arginine at position 158, and is considered neutral. In contrast, APOE ε4, characterized by arginine at both positions, is reported to increase the risk of atherosclerosis [12], Alzheimer’s disease, and other neurodegenerative conditions [13,14]. On the other hand, APOE ε2, with cysteine residues at both critical positions, is linked to a lower risk of Alzheimer’s but a heightened risk of type III hyperlipoproteinemia [15] and age-related macular degeneration [16,17].
Additionally, rare variants like APOE3-R136S (APOE3-Christchurch), APOE3-V236E (APOE3-Jacksonville), and APOE4-R251G [18,19,20] are thought to protect against Alzheimer’s disease. Beyond this, the APOE gene has garnered considerable attention in the field of neurodegenerative diseases, including those implicated in glaucomatous optic neuropathy [16,21,22,23]. Investigations exploring the association between the two common APOE polymorphisms and glaucoma susceptibility have yielded conflicting findings across different populations, including Saudi Arabia [24,25,26,27,28]. However, the precise role of APOE variants in PACG and PXG pathogenesis still needs to be elucidated, particularly within ethnically diverse populations such as those of the Saudi Arabian Peninsula.
Saudi Arabia, characterized by a high prevalence of consanguineous marriages and a distinctive genetic profile, offers a valuable setting for genetic studies on complex diseases such as glaucoma [29]. Investigating the APOE genotype distribution and its correlation with PACG and PXG in this population could provide crucial insights into the genetic determinants of these conditions, potentially uncovering novel biomarkers and therapeutic targets.
Based on this background, the present study aims to explore the genetic association of APOE polymorphisms (rs429358 and rs7412) in a PACG and PXG cohort of Saudi origin. Through genotyping analysis of these polymorphisms in PACG and PXG patients and ethnically matched controls, we seek to unravel potential associations between APOE genetic variants and glaucoma subtypes within this population.
2. Results
2.1. Demographic Characteristics of Study Cohort
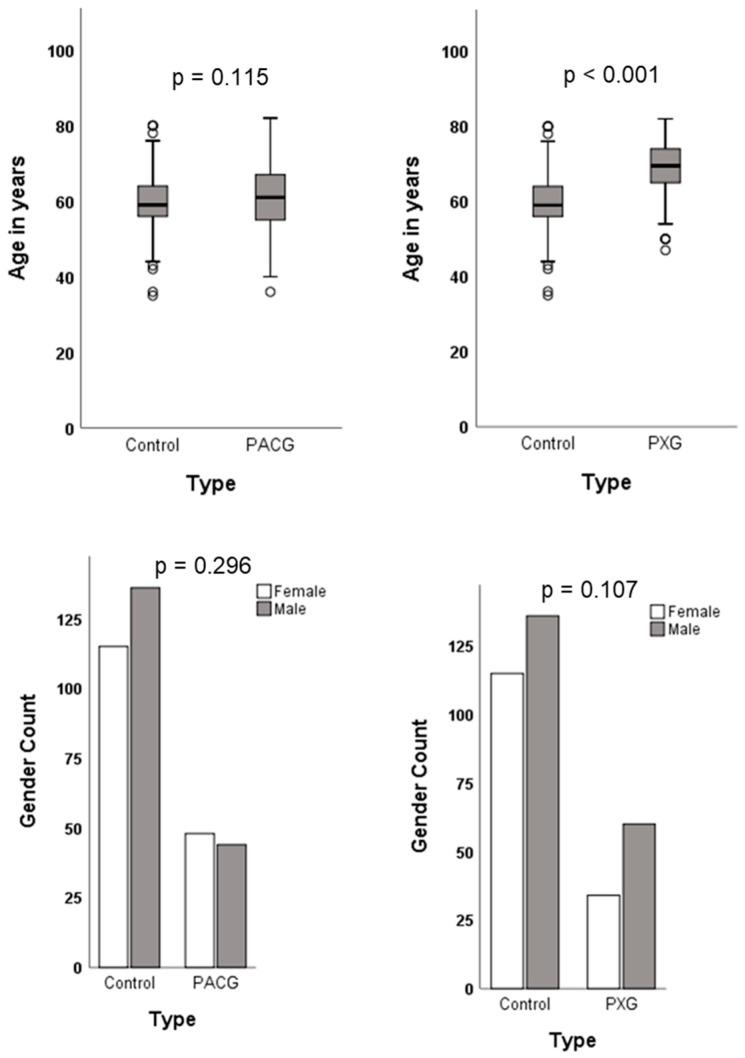
The demographic characteristics of the patient and control groups are illustrated in Figure 1. The mean ages of the study cohort were 59.7 (±7.0) years for the controls, 60.8 (±8.7) years for the PACG patients, and 68.8 (±7.7) years for the PXG patients. In the control group, there were 136 (54%) males and 115 (46%) females, while in the PACG group, there were 44 (48%) males and 48 (52%) females. Among the PXG patients, 60 (64%) were males and 34 (36%) were females. Age and gender distributions did not significantly differ between the PACG patients and controls. However, the PXG patients were significantly older than the controls (p < 0.001), with no significant difference in gender distribution.
Figure 1.
Demographic data of study cohort.
2.2. Association Analysis of rs429358 and rs7412 in the APOE Gene
We analyzed individual polymorphisms rs429358 and rs7412 in the APOE gene to determine their association with PACG and PXG. The polymorphisms showed no significant deviation from the Hardy–Weinberg equilibrium [30] (Table 1). The minor allele frequencies (MAF) of rs429358 and rs7412 were 0.10 and 0.03 in the controls, 0.11 and 0.05 in PACG, and 0.06 and 0.04 in PXG, respectively. There was no significant difference in MAF distribution between PACG and PXG compared to the controls (Table 1).
Table 1.
Minor allele frequency distribution of APOE polymorphisms.
| SNP ID | rs429358 | rs7412 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Type | Minor Allele | MAF | OR (95% CI) | p | HWE p | Minor Allele | MAF | OR (95% CI) | p | HWE p |
| Controls | C | 0.10 | Reference | - | 0.710 | T | 0.03 | Reference | - | 0.170 |
| PACG | C | 0.11 | 1.21 (0.70–2.1) | 0.475 | 0.086 | T | 0.05 | 1.79 (0.76–4.21) | 0.175 | 0.190 |
| PXG | C | 0.06 | 0.65 (0.33–1.24) | 0.187 | 1.000 | T | 0.04 | 1.34 (0.53–3.40) | 0.527 | 0.110 |
MAF—minor allele frequency, OR—odds ratio, 95% CI—95% confidence interval, HWE—Hardy–Weinberg Equilibrium, C—cytosine, T—thymine.
Genotype associations of APOE polymorphisms with PACG and PXG were examined using different genetic models. However, none of the polymorphisms showed significant associations (Tables S1 and S2). While rs429358 exhibited a moderately significant association with PACG in the recessive model (p = 0.044), this significance did not survive Bonferroni’s correction for multiple testing (0.05/2 = 0.025). Furthermore, this association lost significance after adjusting for age and gender (p = 0.063) (Table S1).
2.3. APOE Haplotype Association with PACG and PXG
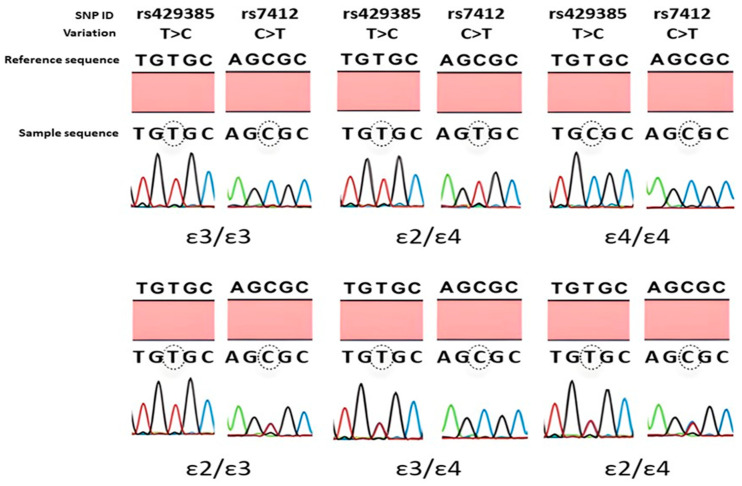
The haplotypes of the investigated polymorphisms in the APOE gene correspond to different APOE alleles (ε3, ε2, ε4) and genotypes (ε3/ε3, ε2/ε2, ε2/ε3, ε2/ε4, ε3/ε4, ε4/ε4). These genotypes were identified depending on the presence of T>C and C>T nucleotides at rs429358 and rs7412, respectively. The representative sequencing results of the identified APOE genotypes are presented in Figure 2.
Figure 2.
Representative sequencing results of APOE genotypes. The genotype calling was based on rs429358 (T>C) and rs7412 (C>T) polymorphisms. The circled nucleotide indicates the position of the nucleotide change compared to the reference sequence. Homozygous ε3/ε3 shows T/T and C/C, ε2/ε2 has T/T and T/T, ε4/ε4 has C/C and C/C, at the circled positions for rs429358 and rs7412, respectively. Likewise, heterozygous ε2/ε3 shows T/T and C/T, ε3/ε4 has T/C and C/C, ε2/ε4 has T/C and C/T, at the circled positions for rs429358 and rs7412, respectively.
This study explored the haplotype association of APOE alleles and genotypes with PACG and PXG, as summarized in Table 2 and Table 3. In the controls, APOE ε3 was the most common allele with a frequency of 87.6%, followed by ε4 (9.6%) and ε2 (2.8%). Similar trends were observed in the PACG and PXG patients. None of the allele distributions were significantly associated with PACG and PXG (Table 2 and Table 3). However, the distribution of the six different APOE genotypes was significant in PACG (Pearson chi-square = 16.36, df = 5, p = 0.006). The ε2/ε3 heterozygotes were found to increase the risk of PACG by over 5-fold, which was statistically significant (p = 0.009). Additionally, ε2-carriers had a significant 4.8-fold increased risk of PACG (p = 0.007) (Table 2). No significant associations were observed in the PXG patient group (Table 3).
Table 2.
Haplotype analysis of APOE polymorphisms according to APOE alleles and genotypes in primary angle-closure glaucoma (PACG).
| APOE | Controls n (%) |
PACG n (%) |
OR (95% CI) | p |
|---|---|---|---|---|
| Alleles | ||||
| ε3 | 440 (87.6) | 154 (83.7) | 1.00 | - |
| ε2 | 14 (2.8) | 9 (4.9) | 1.83 (0.78–4.32) | 0.225 |
| ε4 | 48 (9.6) | 21 (11.4) | 1.25 (0.72–2.15) | 0.470 |
| Genotypes a | ||||
| ε3/ε3 | 199 (79.3) | 66 (71.7) | 1.00 | - |
| ε2/ε2 | 1 (0.4) | 1 (1.1) | 3.01 (0.18–48.89) | 0.999 |
| ε2/ε3 | 4 (1.6) | 7 (7.6) | 5.27 (1.49–18.60) | 0.009 |
| ε2/ε4 | 8 (3.2) | 0 (0) | - | 0.205 |
| ε3/ε4 | 38 (15.1) | 15 (16.3) | 1.19 (0.61–2.30) | 0.730 |
| ε4/ε4 | 1 (0.4) | 3 (3.2) | 9.04 (0.92–88.45) | 0.053 |
| ε3/ε3 vs. All | 52 (20.7) | 26 (28.2) | 1.50 (0.87–2.60) | 0.147 |
| Carrier b | ||||
| ε3/ε3 | 199 (81.9) | 66 (71.7) | 1.00 | - |
| ε*2 c | 5 (2.0) | 8 (8.7) | 4.82 (1.52–15.26) | 0.007 |
| ε*4 d | 39 (16.0) | 18 (19.5) | 1.39 (0.74–2.60) | 0.320 |
OR—odds ratio, 95% CI—95% confidence interval. a Overall Pearson chi-square = 16.36, df = 5, p = 0.006. b ε2/ε4 were excluded from either ε*2 or ε*4 group. c Includes ε2/ε2 and ε2/ε3. d Includes ε4/ε4 and ε3/ε4.
Table 3.
Haplotype analysis of APOE polymorphisms according to APOE alleles and genotypes in pseudoexfoliation glaucoma (PXG).
| APOE | Controls n (%) |
PXG n (%) |
OR (95% CI) | p |
|---|---|---|---|---|
| Alleles | ||||
| ε3 | 440 (87.6) | 169 (89.9) | 1.00 | - |
| ε2 | 14 (2.8) | 7 (3.7) | 1.30 (0.51–3.28) | 0.557 |
| ε4 | 48 (9.6) | 12 (6.4) | 0.65 (0.33–1.25) | 0.197 |
| Genotypes a | ||||
| ε3/ε3 | 199 (79.3) | 77 (81.9) | 1.00 | - |
| ε2/ε2 | 1 (0.4) | 1 (1.1) | 2.58 (0.16–41.8) | 0.999 |
| ε2/ε3 | 4 (1.6) | 4 (4.2) | 2.80 (0.63–10.60) | 0.230 |
| ε2/ε4 | 8 (3.2) | 1 (1.1) | 0.32 (0.04–2.62) | 0.452 |
| ε3/ε4 | 38 (15.1) | 11 (11.7) | 0.75 (0.36–1.53) | 0.488 |
| ε4/ε4 | 1 (0.4) | 0 (0) | - | 0.999 |
| ε3/ε3 vs. All | 52 (20.7) | 17 (18.0) | 0.85 (0.46–1.55) | 0.583 |
| Carrier b | ||||
| ε3/ε3 | 199 (81.9) | 77 (82.8) | 1.00 | - |
| ε*2 c | 5 (2.0) | 5 (5.4) | 2.58 (0.72–9.17) | 0.156 |
| ε*4 d | 39 (16.0) | 11 (11.8) | 0.72 (0.35–1.49) | 0.489 |
OR—odds ratio, 95% CI—95% confidence interval. a Overall Pearson chi-square = 4.80, df = 5, p = 0.441. b ε2/ε4 were excluded from either ε*2 or ε*4 carrier group. c Includes ε2/ε2 and ε2/ε3. d Includes ε4/ε4 and ε3/ε4.
2.4. Logistic Regression Analysis of Risk Factors on Glaucoma Outcome
We further investigated the effects of risk factors such as age, gender, and APOE genotypes (ε3/ε3, ε2-, ε4-carriers) on the outcome of glaucoma (PACG and PXG) using logistic regression analysis. The analysis revealed statistically significant effects of APOE genotypes (p = 0.024) and ε2-carriers (p = 0.008) in the PACG patients. In the PXG patients, age emerged as a significant predictor (p < 0.001), with no significant effect observed for APOE genotypes (Table 4). When examining individual APOE variants (rs429358 and rs7412), age, and gender in relation to the risk of developing PACG or PXG, none of these variables showed a significant impact, except for age in the PXG patient group (p < 0.001). The effects of polymorphism were assessed using both co-dominant and dominant models (Table S3).
Table 4.
Binary logistic regression analysis of APOE genotypes effect in PACG and PXG.
| Group Variables |
B a | SE | Wald | OR (95% CI) | p |
|---|---|---|---|---|---|
| PACG | |||||
| Age | 0.023 | 0.017 | 1.812 | 1.02 (0.99–1.05) | 0.178 |
| Sex | −0.285 | 0.250 | 1.292 | 0.75 (0.46–1.23) | 0.256 |
| APOE genotypes | 7.487 | 0.024 | |||
| ε2-carriers b | 1.556 | 0.590 | 6.953 | 4.74 (1.49–15.06) | 0.008 |
| ε4-carriers c | 0.324 | 0.320 | 1.023 | 1.38 (0.74–2.59) | 0.312 |
| PXG | |||||
| Age | 0.163 | 0.021 | 61.389 | 1.18 (1.130–1.22) | 0.000 |
| Sex | 0.202 | 0.294 | 0.475 | 1.22 (0.69–2.17) | 0.490 |
| APOE genotypes | 1.342 | 0.511 | |||
| ε2-carriers b | 0.388 | 0.778 | 0.249 | 1.47 (0.32–6.77) | 0.618 |
| ε4-carriers c | −0.438 | 0.436 | 1.010 | 0.64 (0.27–1.51) | 0.315 |
OR—odds ratio, 95% CI—95% confidence interval. a B is the estimated coefficient, with standard error, SE. b Without ε4. c Without ε2.
2.5. Association between APOE Genotypes and Clinical Parameters of Glaucoma
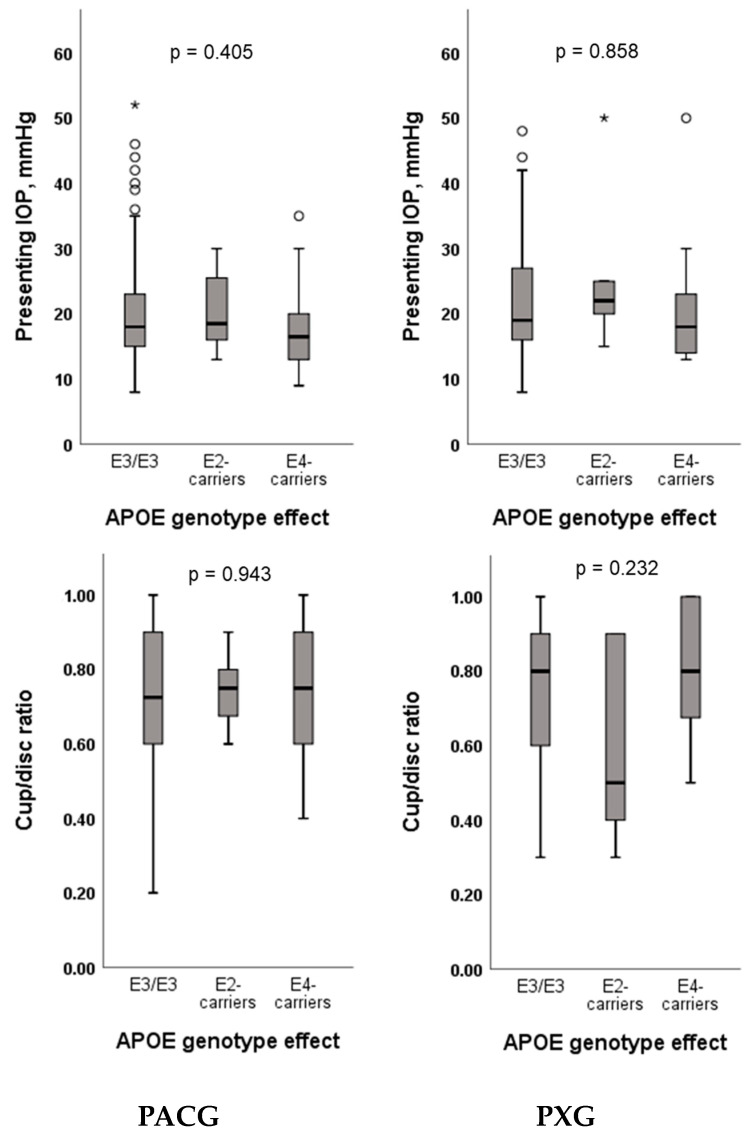
This study examined whether APOE genotypes correlate with clinical parameters of glaucoma, such as IOP and cup/disc ratio in both the PACG and PXG patients. However, no significant effects of APOE genotypes (ε3/ε3, ε2-, ε4-carriers) on IOP or cup/disc ratio were observed (Figure 3). Similarly, individual analysis of APOE polymorphisms (rs429358 and rs7412) did not show any significant association with IOP or cup/disc ratio (Figures S1 and S2).
Figure 3.
Association analysis of IOP and cup/disc ratio with APOE genotypes in primary angle-closure glaucoma (PACG) and pseudoexfoliation glaucoma (PXG) patients. * Outlier.
3. Discussion
The genetic variants of APOE (ε2, ε3, and ε4) are associated with the risk of developing several human diseases [31]. Understanding the function and impact of APOE in human health and disease remains a significant focus of research in neurology, cardiology, and ophthalmology [31,32]. The precise influence of genetic factors and polymorphisms in the complex polygenic nature of glaucoma among patients of Saudi Arabian descent remains poorly understood. Herein, we present findings indicating a positive association between the APOE ε2-carriers and PACG, but no association in PXG, in a Saudi cohort.
As illustrated in Table 5, the rs429358 and rs7412 polymorphisms in the APOE gene responsible for a Cys/Arg interchange and their haplotypes give rise to three major allelic APOE variants ε2, ε3, and ε4 [33]. The frequency distribution of these three major alleles varies worldwide (Table 6) [34,35]. While studies on non-human primates suggest the ε4 allele as the ancestral variant [36], modern human populations predominantly exhibit the ε3 variant, with frequencies ranging from 0.968 in Indians to 0.486 in Papuans [34,35]. The ε4 allele ranks as the second most common, with notably high frequencies observed among Pygmy populations in Central Africa (0.407), Khoisan populations in Southern Africa (0.370), Oceanians (including Papuans at 0.368 and Australian Aborigines at 0.260), and the European Saami people (0.310) [34]. In contrast, the ε2 allele is less common, ranging from rare to absent in Native Americans, Siberians, and Mongolians, but relatively more prevalent among Swedish (0.119), sub-Saharan African (0.116), Malay (0.140), and Papuan (0.145) populations [34,35,37]. Our study similarly reflects this global pattern, showing APOE ε3 as the most common allele (0.876), followed by ε4 (0.096) and ε2 (0.028). Generally, allele frequencies can vary significantly across populations due to factors such as genetic drift, migration, and natural selection [34,35,38]. Notably, our investigation found no association between the allele frequencies of rs429358, rs7412, and APOE haplotype (ε2, ε3, and ε4) and PACG/PXG compared to the controls.
Table 5.
Haplotypes associated with APOE allele and their frequency distribution in different populations.
| APOE Alleles | ε3 | ε2 | ε4 |
|---|---|---|---|
| Haplotype | rs429358-T rs7412-C |
rs429358-T rs7412-T |
rs429358-C rs7412-C |
| Residue combination | 112-Cys 158-Arg |
112-Cys 158-Cys |
112-Arg 158-Arg |
| Ethnicity | Allele frequency a | ||
| Europeans | 0.640–0.900 | 0.044–0.120 | 0.052–0.310 |
| Asians | 0.620–0.870 | 0.020–0.140 | 0.071–0.240 |
| Africans | 0.536–0.850 | 0.031–0.116 | 0.085–0.407 |
| Native Americans | 0.720–0.911 | 0.0–0.014 | 0.089–0.280 |
| Oceanians | 0.486–0.740 | 0.0–0.145 | 0.260–0.368 |
| Our study (Saudi Arabians) b | 0.876 | 0.028 | 0.096 |
Table 6.
Primer used for PCR amplification and Sanger sequencing of APOE genotypes.
| Primer Type | APOE Primer Sequences (5′–3′) | Thermal Cycling Conditions |
|---|---|---|
| Forward | a GACCATGAAGGAGTTGAAGGCCTAC | Initial denaturation—95 °C for 15 min Cycling—95 °C—1 min, 59 °C—30 s, 72 °C—1 min for 35 cycles Final extension—72 °C—10 min |
| Reverse | b GATGGCGCTGAGGCCGCGCT |
a TGTAAAACGACGGCCAGT and b CAGGAAACAGCTATGACC M13 sequences were tagged at the 5′ end of the PCR primers and used for Sanger sequencing as described in Methods.
The statistical evidence supporting a causal association between APOE variants and glaucoma remains less robust. Numerous studies with inconsistent findings have explored the association between APOE alleles/genotypes and adult-onset primary open-angle glaucoma (POAG) in different populations. In Japanese OAG patients, the ε3 allele increased OAG risk, the ε2 allele reduced risk, and the ε4 allele was linked to lower IOP [39]. Conversely, a smaller study in Saudi-origin POAG patients found a significant association with the ε4 allele [27], but our own study in a larger Saudi cohort contradicted this [28]. In Massachusetts and Canadian studies, the ε4 allele showed protective effects in POAG [24,40]. By contrast, in Brazilian POAG cases, the ε2 allele was associated with increased risk [41], while the ε4 allele was linked to neuroretinal thinning in normal-tension glaucoma (NTG) [42]. Conflicting results persist across diverse ethnic groups, including European [43], Chinese [44], Japanese [45], Turkish [46], and in meta-analyses [25,47] reflecting population-specific differences.
On the other hand, few studies have explored the association of APOE with PACG and PXG. A study in Saudi PACG patients found no association with APOE alleles and genotypes [27], and similar results were reported in large cohorts of German and Italian PXG patients [48]. Another study in Greek patients reported no APOE association in pseudoexfoliation syndrome (PXS)/PXG but found an increased risk of POAG in APOE ε2-carriers [49]. In a Turkish cohort, APOE ε2-carriers were at significantly increased risk of PXS [50]. However, this finding was not replicated in another Turkish study [51]. A recent Finnish study found that the APOE ε4 allele protects against POAG and NTG but not against PXG [32].
In our Saudi cohort, APOE ε2-carriers were found to be at significantly increased risk of PACG. However, similar to our earlier findings in a POAG cohort [28], no association of APOE variants was observed in PXG. A previous study has shown that ε2-carriers had significantly lower IOP than non-ε2-carriers in PXS patients [52]. While ε2-carriers in our study exhibited notably lower cup/disc ratios compared to ε3/ε3 and ε4-carriers in the PXG patients, however, no significant associations were found between APOE genotypes and clinical markers, such as IOP and cup/disc ratio, in the PACG and PXG patients. These observations support the hypothesis proposed by previous studies that APOE may be involved in modulating RGC degeneration via an IOP-independent mechanism(s) [24,32,53].
There are several mechanisms through which APOE could potentially play a role in the pathogenesis of glaucoma. APOE is produced by astrocytes, neurons, retinal Müller cells, and macrophages [9,54], and variations in the binding properties of APOE isoforms across different cell types can have significant functional consequences at both the cellular and molecular levels [55]. Different APOE isoforms have been demonstrated to confer differing levels of risk associated with glaucoma. Animal experiments suggest that APOE gene deletion (APOE−/−) and the ε4 isoform may reduce the risk of RGC loss in glaucoma by inhibiting kainic acid receptor signaling, modulating microglial activation, and reducing galectin-3 expression [21,24]. Conversely, the presence of the ε3 isoform and overall APOE gene expression (APOE+/+) may increase the risk of RGC death in glaucoma by promoting microglial phenotypic changes and upregulating galectin-3 [22]. However, definitive evidence regarding the beneficial or detrimental impact of the ε2 allele in glaucoma is lacking in the current literature. Nonetheless, ε2 is reported to be associated with the highest APOE protein levels [56]. Therefore, it can be speculated that ε2 allele might increase the risk of PACG, as observed in our study, through any of the aforementioned mechanisms.
Moreover, the involvement of APOE in lipid metabolism, complement system regulation, neuroinflammation, blood–brain barrier integrity, oxidative stress, mitochondrial function, and angiogenesis contributing to Alzheimer’s or age-related macular degeneration pathogenesis [13,16,31] suggests multifaceted mechanisms through which APOE may contribute to the pathogenesis of PACG. Interestingly, new findings have revealed a role for APOE in regulating microRNA-controlled cellular signaling in cells of the immune system and vascular wall, suggesting a role of APOE in intercellular communication [57]. The presence of a similar mechanism in PACG cannot be ruled out. However, whether APOE genotypes influence these functions remains to be investigated.
Overall, the association of the APOE ε2 genotype with PACG requires a comprehensive understanding of the potential biological mechanisms related to aqueous humor dynamics, vascular factors, genetic interactions, comparative analysis with other ocular diseases, and consideration of population-specific factors. Further research into these aspects is essential for elucidating the role of APOE in PACG and its potential clinical implications. By contrast, the absence of this association in PXG may be attributed to differences in the underlying disease pathophysiology, genetic heterogeneity, environmental influences, or limitations in sample size and statistical power.
To conclude, our results show, for the first time, a positive association of APOE ε2-carriers in PACG, indicating the potential implication of ε2 in elevating the risk of PACG within the Saudi cohort. This observation suggests a possible role for APOE genetic variants in the pathogenesis of PACG, adding to our understanding of the genetic underpinnings of this complex ocular disorder among individuals of Saudi Arabian ancestry. However, the results require a cautious interpretation since this study is limited by sample size, especially in the subgroup analysis. Therefore, further validation incorporating larger population-based cohorts and molecular and functional studies are warranted to elucidate the underlying mechanisms and factors contributing to these observed associations.
4. Materials and Methods
4.1. Study Design, Ethics Approval, and Participant Characteristics
We conducted a retrospective and exploratory case-control study, sanctioned by the Institutional Review Board Ethics Committee at the College of Medicine of King Saud University as per the principles of the Declaration of Helsinki guidelines for human research. Participants were recruited at King Abdulaziz University Hospital in Riyadh, Saudi Arabia, as described elsewhere [58].
Briefly, PACG patients (n = 92) exhibited clinical signs of anatomically closed angles, elevated intraocular pressure (IOP) (≥21 mmHg), optic disc damage with a cup/disc ratio of at least 0.7, and visual field defects. PXG patients (n = 94) demonstrated the presence of exfoliation material along the pupil margins or anterior lens capsule, glaucomatous optic nerve damage, and elevated IOP. Exclusion criteria encompassed secondary glaucoma types, optic neuropathies not associated with glaucoma, corticosteroid use, ocular trauma, inadequate fundus visualization, or refusal to participate. Healthy age- and gender- matched controls (n = 251), aged ≥ 40 years, exhibited normal IOP, open angles on gonioscopy, healthy optic discs, and lacked a family history of glaucoma.
4.2. Genotyping rs429358 and rs7412 Polymorphisms in the APOE Gene
Peripheral EDTA blood samples were utilized for DNA extraction, followed by PCR amplification and Sanger sequencing to identify the rs429358 (T>C) and rs7412 (C>T) variants of the APOE gene, as previously described [28]. The primers used for PCR amplification, Sanger sequencing, and the cycling conditions are outlined in Table 6. In brief, DNA samples were PCR amplified, followed by purification using the QIAquick PCR Purification Kit (Qiagen, Hilden, Germany) and sequencing with M13 primers using the BigDye Terminator V3.1 Cycle Sequencing kit (Applied Biosystems, Foster City, CA, USA). Subsequently, sequencing analysis was performed on the ABI 3730 XL sequencer (Applied Biosystems), and nucleotide variations and APOE genotypes were determined using CLC Sequence Viewer 6.0 (Qiagen), in comparison to the APOE reference sequence (NG_007084.2).
4.3. Statistical Analysis
Statistical analyses were performed using SPSS version 25 (IBM Inc., Chicago, IL, USA) and SNPStats online software version 1.0. A significance threshold of p < 0.05 was applied, with Bonferroni’s correction for multiple testing (p = 0.05/2 = 0.025) where appropriate. Data normality was assessed using the Kolmogorov–Smirnov test. Continuous variables were analyzed by the Mann–Whitney U test and the Kruskal–Wallis test for two-group and three-group comparisons, respectively. The categorical variables and deviation from Hardy–Weinberg equilibrium were assessed using chi-square and Fisher’s exact tests, as applicable [30]. The impact of multiple factors, including age, sex, and genotypes, on the disease outcome was evaluated using binary logistic regression analysis.
Acknowledgments
The authors would like to extend their appreciation to Al-Sheikh Ibrahim Al-Sultan for his valuable support. We also thank Abdulrahman Al-Mosa for his clinical assistance during the study.
Supplementary Materials
The supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms25084571/s1.
Author Contributions
A.A.K.: conceptualization, investigation, methodology, analysis, writing draft, review and editing, and supervision; T.A.A. and T.S.: investigation, methodology, data curation, manuscript review, and editing; T.K., R.R. and G.P.L.: data curation, data interpretation, re-sources, manuscript review, and editing; A.A.A., E.A., E.A.O. and F.A.A.: resources, data curation, manuscript review, and editing; S.A.A.-O.: concept, resources, funding acquisition, project administration, manuscript review, and editing. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and was approved by the Institutional Review Board Committee of the College of Medicine, King Saud University (protocol number #08–657).
Informed Consent Statement
Informed consent was obtained from all individual participants included in the study.
Data Availability Statement
The data supporting the conclusions of this article are all presented within the report.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manu-script, or in the decision to publish the results.
Funding Statement
This work was supported by King Saud University through the Vice Deanship of Scientific Research Chair and Glaucoma Research Chair in Ophthalmology (GRC-2024).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Fernandez-Albarral J.A., Ramirez A.I., de Hoz R., Matamoros J.A., Salobrar-Garcia E., Elvira-Hurtado L., Lopez-Cuenca I., Sanchez-Puebla L., Salazar J.J., Ramirez J.M. Glaucoma: From pathogenic mechanisms to retinal glial cell response to damage. Front. Cell Neurosci. 2024;18:1354569. doi: 10.3389/fncel.2024.1354569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun X., Dai Y., Chen Y., Yu D.Y., Cringle S.J., Chen J., Kong X., Wang X., Jiang C. Primary angle closure glaucoma: What we know and what we don’t know. Prog. Retin. Eye Res. 2017;57:26–45. doi: 10.1016/j.preteyeres.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Miglior S., Bertuzzi F. Exfoliative glaucoma: New evidence in the pathogenesis and treatment. Prog. Brain Res. 2015;221:233–241. doi: 10.1016/bs.pbr.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 4.Lee S.S., Mackey D.A. Glaucoma—Risk factors and current challenges in the diagnosis of a leading cause of visual impairment. Maturitas. 2022;163:15–22. doi: 10.1016/j.maturitas.2022.05.002. [DOI] [PubMed] [Google Scholar]
- 5.Kondkar A.A. Updates on genes and genetic mechanisms implicated in primary angle-closure glaucoma. Appl. Clin. Genet. 2021;14:89–112. doi: 10.2147/TACG.S274884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schlotzer-Schrehardt U. Genetics and genomics of pseudoexfoliation syndrome/glaucoma. Middle East Afr. J. Ophthalmol. 2011;18:30–36. doi: 10.4103/0974-9233.75882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaushik M., Tiwari P., Dada T., Dada R. Beyond the optic nerve: Genetics, diagnosis, and promising therapies for glaucoma. Gene. 2024;894:147983. doi: 10.1016/j.gene.2023.147983. [DOI] [PubMed] [Google Scholar]
- 8.Yang L.G., March Z.M., Stephenson R.A., Narayan P.S. Apolipoprotein e in lipid metabolism and neurodegenerative disease. Trends Endocrinol. Metab. 2023;34:430–445. doi: 10.1016/j.tem.2023.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson D.H., Ozaki S., Nealon M., Neitz J., Mullins R.F., Hageman G.S., Johnson L.V. Local cellular sources of apolipoprotein e in the human retina and retinal pigmented epithelium: Implications for the process of drusen formation. Am. J. Ophthalmol. 2001;131:767–781. doi: 10.1016/S0002-9394(00)00961-2. [DOI] [PubMed] [Google Scholar]
- 10.Sharma S., Chataway T., Burdon K.P., Jonavicius L., Klebe S., Hewitt A.W., Mills R.A., Craig J.E. Identification of loxl1 protein and apolipoprotein e as components of surgically isolated pseudoexfoliation material by direct mass spectrometry. Exp. Eye Res. 2009;89:479–485. doi: 10.1016/j.exer.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 11.Weisgraber K.H., Rall S.C., Jr., Mahley R.W. Human e apoprotein heterogeneity. Cysteine-arginine interchanges in the amino acid sequence of the apo-e isoforms. J. Biol. Chem. 1981;256:9077–9083. doi: 10.1016/S0021-9258(19)52510-8. [DOI] [PubMed] [Google Scholar]
- 12.Davignon J., Gregg R.E., Sing C.F. Apolipoprotein e polymorphism and atherosclerosis. Arteriosclerosis. 1988;8:1–21. doi: 10.1161/01.ATV.8.1.1. [DOI] [PubMed] [Google Scholar]
- 13.Raulin A.C., Doss S.V., Trottier Z.A., Ikezu T.C., Bu G., Liu C.C. Apoe in Alzheimer’s disease: Pathophysiology and therapeutic strategies. Mol. Neurodegener. 2022;17:72. doi: 10.1186/s13024-022-00574-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernandez-Calle R., Konings S.C., Frontinan-Rubio J., Garcia-Revilla J., Camprubi-Ferrer L., Svensson M., Martinson I., Boza-Serrano A., Venero J.L., Nielsen H.M., et al. Apoe in the bullseye of neurodegenerative diseases: Impact of the apoe genotype in Alzheimer’s disease pathology and brain diseases. Mol. Neurodegener. 2022;17:62. doi: 10.1186/s13024-022-00566-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rall S.C., Jr., Weisgraber K.H., Innerarity T.L., Mahley R.W. Structural basis for receptor binding heterogeneity of apolipoprotein e from type iii hyperlipoproteinemic subjects. Proc. Natl. Acad. Sci. USA. 1982;79:4696–4700. doi: 10.1073/pnas.79.15.4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu M.L., Quinn J., Xue K. Interactions between apolipoprotein e metabolism and retinal inflammation in age-related macular degeneration. Life. 2021;11:635. doi: 10.3390/life11070635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen L., Hoffmann T.J., Melles R.B., Sakoda L.C., Kvale M.N., Banda Y., Schaefer C., Risch N., Jorgenson E. Differences in the genetic susceptibility to age-related macular degeneration clinical subtypes. Investig. Ophthalmol. Vis. Sci. 2015;56:4290–4299. doi: 10.1167/iovs.15-16533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sepulveda-Falla D., Sanchez J.S., Almeida M.C., Boassa D., Acosta-Uribe J., Vila-Castelar C., Ramirez-Gomez L., Baena A., Aguillon D., Villalba-Moreno N.D., et al. Distinct tau neuropathology and cellular profiles of an apoe3 christchurch homozygote protected against autosomal dominant Alzheimer’s dementia. Acta Neuropathol. 2022;144:589–601. doi: 10.1007/s00401-022-02467-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu C.C., Murray M.E., Li X., Zhao N., Wang N., Heckman M.G., Shue F., Martens Y., Li Y., Raulin A.C., et al. Apoe3-jacksonville (v236e) variant reduces self-aggregation and risk of dementia. Sci. Transl. Med. 2021;13:eabc9375. doi: 10.1126/scitranslmed.abc9375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le Guen Y., Belloy M.E., Grenier-Boley B., de Rojas I., Castillo-Morales A., Jansen I., Nicolas A., Bellenguez C., Dalmasso C., Kucukali F., et al. Association of rare apoe missense variants v236e and r251g with risk of alzheimer disease. JAMA Neurol. 2022;79:652–663. doi: 10.1001/jamaneurol.2022.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yi W., Lv D., Sun Y., Mu J., Lu X. Role of apoe in glaucoma. Biochem. Biophys. Res. Commun. 2024;694:149414. doi: 10.1016/j.bbrc.2023.149414. [DOI] [PubMed] [Google Scholar]
- 22.Margeta M.A., Yin Z., Madore C., Pitts K.M., Letcher S.M., Tang J., Jiang S., Gauthier C.D., Silveira S.R., Schroeder C.M., et al. Apolipoprotein e4 impairs the response of neurodegenerative retinal microglia and prevents neuronal loss in glaucoma. Immunity. 2022;55:1627–1644.e7. doi: 10.1016/j.immuni.2022.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Omodaka K., Nishiguchi K.M., Yasuda M., Tanaka Y., Sato K., Nakamura O., Maruyama K., Nakazawa T. Neuroprotective effect against axonal damage-induced retinal ganglion cell death in apolipoprotein e-deficient mice through the suppression of kainate receptor signaling. Brain Res. 2014;1586:203–212. doi: 10.1016/j.brainres.2014.08.053. [DOI] [PubMed] [Google Scholar]
- 24.Margeta M.A., Letcher S.M., Igo R.P., Jr., Cooke Bailey J.N., Pasquale L.R., Haines J.L., Butovsky O., Wiggs J.L. Association of apoe with primary open-angle glaucoma suggests a protective effect for apoe epsilon4. Investig. Ophthalmol. Vis. Sci. 2020;61:3. doi: 10.1167/iovs.61.8.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang W., Zhou M., Huang W., Chen S., Zhang X. Lack of association of apolipoprotein e (apo e) epsilon2/epsilon3/epsilon4 polymorphisms with primary open-angle glaucoma: A meta-analysis from 1916 cases and 1756 controls. PLoS ONE. 2013;8:e72644. doi: 10.1371/journal.pone.0072644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y., Zhou Y.F., Zhao B.Y., Gu Z.Y., Li S.L. Apolipoprotein e gene epsilon4epsilon4 is associated with elevated risk of primary open angle glaucoma in asians: A meta-analysis. BMC Med. Genet. 2014;15:60. doi: 10.1186/1471-2350-15-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Al-Dabbagh N.M., Al-Dohayan N., Arfin M., Tariq M. Apolipoprotein e polymorphisms and primary glaucoma in saudis. Mol. Vis. 2009;15:912–919. [PMC free article] [PubMed] [Google Scholar]
- 28.Kondkar A.A., Sultan T., Azad T.A., Khatlani T., Alshehri A.A., Osman E.A., Lobo G.P., Almobarak F.A., Al-Obeidan S.A. Common variants rs429358 and rs7412 in apoe gene are not associated with poag in a saudi cohort. Biology. 2024;13:62. doi: 10.3390/biology13010062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alfares A., Alfadhel M., Wani T., Alsahli S., Alluhaydan I., Al Mutairi F., Alothaim A., Albalwi M., Al Subaie L., Alturki S., et al. A multicenter clinical exome study in unselected cohorts from a consanguineous population of saudi arabia demonstrated a high diagnostic yield. Mol. Genet. Metab. 2017;121:91–95. doi: 10.1016/j.ymgme.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 30.Namipashaki A., Razaghi-Moghadam Z., Ansari-Pour N. The essentiality of reporting hardy-weinberg equilibrium calculations in population-based genetic association studies. Cell J. 2015;17:187–192. doi: 10.22074/cellj.2016.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alagarsamy J., Jaeschke A., Hui D.Y. Apolipoprotein e in cardiometabolic and neurological health and diseases. Int. J. Mol. Sci. 2022;23:9892. doi: 10.3390/ijms23179892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liuska P.J., Ramo J.T., Lemmela S., Kaarniranta K., Uusitalo H., Lahtela E., Daly M.J., Harju M., Palotie A., Turunen J.A. Association of apoe haplotypes with common age-related ocular diseases in 412,171 individuals. Investig. Ophthalmol. Vis. Sci. 2023;64:33. doi: 10.1167/iovs.64.14.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahley R.W., Rall S.C., Jr. Apolipoprotein e: Far more than a lipid transport protein. Annu. Rev. Genom. Hum. Genet. 2000;1:507–537. doi: 10.1146/annurev.genom.1.1.507. [DOI] [PubMed] [Google Scholar]
- 34.Corbo R.M., Scacchi R. Apolipoprotein e (apoe) allele distribution in the world. Is apoe*4 a t’hrifty’ allele? Ann. Hum. Genet. 1999;63:301–310. doi: 10.1046/j.1469-1809.1999.6340301.x. [DOI] [PubMed] [Google Scholar]
- 35.Singh P.P., Singh M., Mastana S.S. Apoe distribution in world populations with new data from india and the uk. Ann. Hum. Biol. 2006;33:279–308. doi: 10.1080/03014460600594513. [DOI] [PubMed] [Google Scholar]
- 36.Fullerton S.M., Clark A.G., Weiss K.M., Nickerson D.A., Taylor S.L., Stengard J.H., Salomaa V., Vartiainen E., Perola M., Boerwinkle E., et al. Apolipoprotein e variation at the sequence haplotype level: Implications for the origin and maintenance of a major human polymorphism. Am. J. Hum. Genet. 2000;67:881–900. doi: 10.1086/303070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Egert S., Rimbach G., Huebbe P. Apoe genotype: From geographic distribution to function and responsiveness to dietary factors. Proc. Nutr. Soc. 2012;71:410–424. doi: 10.1017/S0029665112000249. [DOI] [PubMed] [Google Scholar]
- 38.Huebbe P., Rimbach G. Evolution of human apolipoprotein e (apoe) isoforms: Gene structure, protein function and interaction with dietary factors. Ageing Res. Rev. 2017;37:146–161. doi: 10.1016/j.arr.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 39.Mabuchi F., Tang S., Ando D., Yamakita M., Wang J., Kashiwagi K., Yamagata Z., Iijima H., Tsukahara S. The apolipoprotein e gene polymorphism is associated with open angle glaucoma in the japanese population. Mol. Vis. 2005;11:609–612. [PubMed] [Google Scholar]
- 40.Freeman E.E., Bastasic J., Grant A., Leung G., Li G., Buhrmann R., Roy-Gagnon M.H. Inverse association of apoe epsilon4 and glaucoma modified by systemic hypertension: The canadian longitudinal study on aging. Investig. Ophthalmol. Vis. Sci. 2022;63:9. doi: 10.1167/iovs.63.13.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Occhiutto M.L., de Melo M.B., Cabral de Vasconcellos J.P., Rodrigues T.A.R., Bajano F.F., Costa F.F., Costa V.P. “Association of apoe gene polymorphisms with primary open angle glaucoma in brazilian patients”. Ophthalmic Genet. 2021;42:53–61. doi: 10.1080/13816810.2020.1849314. [DOI] [PubMed] [Google Scholar]
- 42.Mullany S., Marshall H., Diaz-Torres S., Berry E.C., Schmidt J.M., Thomson D., Qassim A., To M.S., Dimasi D., Kuot A., et al. The apoe e4 allele is associated with faster rates of neuroretinal thinning in a prospective cohort study of suspect and early glaucoma. Ophthalmol. Sci. 2022;2:100159. doi: 10.1016/j.xops.2022.100159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mullany S., Diaz-Torres S., Schmidt J.M., Thomson D., Qassim A., Marshall H.N., Knight L.S.W., Berry E.C., Kolovos A., Dimasi D., et al. No strong association between the apolipoprotein e e4 allele and glaucoma: A multicohort study. Ophthalmol. Sci. 2023;3:100287. doi: 10.1016/j.xops.2023.100287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jia L.Y., Tam P.O., Chiang S.W., Ding N., Chen L.J., Yam G.H., Pang C.P., Wang N.L. Multiple gene polymorphisms analysis revealed a different profile of genetic polymorphisms of primary open-angle glaucoma in northern chinese. Mol. Vis. 2009;15:89–98. [PMC free article] [PubMed] [Google Scholar]
- 45.Tamura H., Kawakami H., Kanamoto T., Kato T., Yokoyama T., Sasaki K., Izumi Y., Matsumoto M., Mishima H.K. High frequency of open-angle glaucoma in japanese patients with Alzheimer’s disease. J. Neurol. Sci. 2006;246:79–83. doi: 10.1016/j.jns.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 46.Saglar E., Yucel D., Bozkurt B., Ozgul R.K., Irkec M., Ogus A. Association of polymorphisms in apoe, p53, and p21 with primary open-angle glaucoma in turkish patients. Mol. Vis. 2009;15:1270–1276. [PMC free article] [PubMed] [Google Scholar]
- 47.Song Q., Chen P., Liu Q. Role of the apoe epsilon2/epsilon3/epsilon4 polymorphism in the development of primary open-angle glaucoma: Evidence from a comprehensive meta-analysis. PLoS ONE. 2013;8:e82347. doi: 10.1371/journal.pone.0082347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krumbiegel M., Pasutto F., Mardin C.Y., Weisschuh N., Paoli D., Gramer E., Weber B.H., Kruse F.E., Schlotzer-Schrehardt U., Reis A. Apolipoprotein e genotypes in pseudoexfoliation syndrome and pseudoexfoliation glaucoma. J. Glaucoma. 2010;19:561–565. doi: 10.1097/IJG.0b013e3181ca76c4. [DOI] [PubMed] [Google Scholar]
- 49.Chiras D., Tzika K., Kokotas H., Oliveira S.C., Grigoriadou M., Kastania A., Dima K., Stefaniotou M., Aspiotis M., Petersen M.B., et al. Development of novel loxl1 genotyping method and evaluation of loxl1, apoe and mthfr polymorphisms in exfoliation syndrome/glaucoma in a greek population. Mol. Vis. 2013;19:1006–1016. [PMC free article] [PubMed] [Google Scholar]
- 50.Yilmaz A., Tamer L., Ates N.A., Camdeviren H., Degirmenci U. Effects of apolipoprotein e genotypes on the development of exfoliation syndrome. Exp. Eye Res. 2005;80:871–875. doi: 10.1016/j.exer.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 51.Yaylacioglu Tuncay F., Aktas Z., Ergun M.A., Ergun S.G., Hasanreisoglu M., Hasanreisoglu B. Association of polymorphisms in apoe and loxl1 with pseudoexfoliation syndrome and pseudoexfoliation glaucoma in a turkish population. Ophthalmic Genet. 2017;38:95–97. doi: 10.3109/13816810.2015.1126617. [DOI] [PubMed] [Google Scholar]
- 52.Ritland J.S., Utheim T.P., Utheim O.A., Espeseth T., Lydersen S., Semb S.O., Rootwelt H., Elsas T. Effects of apoe and chrna4 genotypes on retinal nerve fibre layer thickness at the optic disc and on risk for developing exfoliation syndrome. Acta Ophthalmol. Scand. 2007;85:257–261. doi: 10.1111/j.1600-0420.2006.00824.x. [DOI] [PubMed] [Google Scholar]
- 53.Lam C.Y., Fan B.J., Wang D.Y., Tam P.O., Yung Tham C.C., Leung D.Y., Ping Fan D.S., Chiu Lam D.S., Pang C.P. Association of apolipoprotein e polymorphisms with normal tension glaucoma in a chinese population. J. Glaucoma. 2006;15:218–222. doi: 10.1097/01.ijg.0000212217.19804.a7. [DOI] [PubMed] [Google Scholar]
- 54.Amaratunga A., Abraham C.R., Edwards R.B., Sandell J.H., Schreiber B.M., Fine R.E. Apolipoprotein e is synthesized in the retina by muller glial cells, secreted into the vitreous, and rapidly transported into the optic nerve by retinal ganglion cells. J. Biol. Chem. 1996;271:5628–5632. doi: 10.1074/jbc.271.10.5628. [DOI] [PubMed] [Google Scholar]
- 55.Guillaume D., Bertrand P., Dea D., Davignon J., Poirier J. Apolipoprotein e and low-density lipoprotein binding and internalization in primary cultures of rat astrocytes: Isoform-specific alterations. J. Neurochem. 1996;66:2410–2418. doi: 10.1046/j.1471-4159.1996.66062410.x. [DOI] [PubMed] [Google Scholar]
- 56.Riddell D.R., Zhou H., Atchison K., Warwick H.K., Atkinson P.J., Jefferson J., Xu L., Aschmies S., Kirksey Y., Hu Y., et al. Impact of apolipoprotein e (apoe) polymorphism on brain apoe levels. J. Neurosci. 2008;28:11445–11453. doi: 10.1523/JNEUROSCI.1972-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bouchareychas L., Raffai R.L. Apolipoprotein e and atherosclerosis: From lipoprotein metabolism to microrna control of inflammation. J. Cardiovasc. Dev. Dis. 2018;5:30. doi: 10.3390/jcdd5020030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kondkar A.A., Sultan T., Azad T.A., Osman E.A., Almobarak F.A., Lobo G.P., Al-Obeidan S.A. Evaluation of abca1 and fndc3b gene polymorphisms associated with pseudoexfoliation glaucoma and primary angle-closure glaucoma in a saudi cohort. Front. Genet. 2022;13:877174. doi: 10.3389/fgene.2022.877174. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the conclusions of this article are all presented within the report.