Abstract
Niemann-Pick disease type C1 (NPC1) is a lysosomal disorder due to impaired intracellular cholesterol transport out of the endolysosomal compartment.. Marked heterogeneity has been observed in individuals with the same NPC1 genotype, thus suggesting a significant effect of modifier genes. Prior work demonstrated that decreased SOAT1 activity decreased disease severity in an NPC1 mouse model. Thus, we hypothesized that a polymorphism associated with decreased SOAT1 expression might influence the NPC1 phenotype. Phenotyping and genomic sequencing of 117 individuals with NPC1 was performed as part of a Natural History trial. Phenotyping included determination of disease severity and disease burden. Significant clinical heterogeneity is present in individuals homozygous for the NPC1I1061T variant and in siblings. Analysis of the SOAT1 polymorphism, rs1044925 (A>C), showed a significant association of the C-allele with earlier age of neurological onset. The C-allele may be associated with a higher Annualized Severity Index Score as well as increased frequency of liver disease and seizures. A polymorphism associated with decreased expression of SOAT1 appears to be a genetic modifier of the NPC1 phenotype. This finding is consistent with prior data showing decreased phenotypic severity in Npc1-/-:Soat1-/- mice and supports efforts to investigate the potential of SOAT1 inhibitors as a potential therapy for NPC1.
Keywords: Niemann-Pick disease, type C1, NPC1, sterol O-acyltransferase 1, SOAT1, ACAT1, genetic modifiers, neurodegeneration
1. Introduction
The phenotypes associated with genetic diseases often show incomplete penetrance and variable expressivity. For specific disorders, much of this phenotypic heterogeneity may be due to specific pathological variants in the causative gene that result in variable levels of residual protein function. However, phenotypic heterogeneity is also observed in individuals with the same genotype. An example of this would be incomplete penetrance and variable expressivity manifested in sibling pairs. Although environmental differences likely contribute, genetic modifiers also play a role. The concept of genetic modifiers contributing to human disease variability was initially proposed by Haldane in 1941 [1]. Genetic modifiers can either increase or decrease phenotypic severity; thus, their identification could help provide guardians and patients with prognostic information. In addition, identification of genetic modifiers that alleviate disease severity may provide insight into potential therapeutic approaches.
It is well-established that modifiers may significantly impact the phenotype of rare genetic disorders. Genomic databases consisting of healthy individuals have led to the identification of resilient individuals who, based on their genome sequence, would have been expected to manifest a severe childhood disorder [2,3]. Identification of genes that function as phenotypic modifiers for rare diseases has proven to be difficult, likely due to quantitatively defining the phenotype and statistical constraints due to the rarity of these diseases [4]. Underscoring the difficulty of identifying genetic modifiers, a 2020 bibliographic review by Rahit et al. [5] only identified 24 genes that appear to function as genetic modifiers for rare Mendelian disorders. In this paper, we demonstrate the use of a hypothesis-based approach to characterize a potential genetic modifier of Niemann-Pick disease, type C1.
Niemann-Pick disease, type C1 (NPC1, MIM 257220) is an autosomal recessive, inborn error of intracellular cholesterol transport due to impaired function of NPC1. A similar, but much rarer disorder, Niemann-Pick disease, type C2 (NPC2, MIM 607625), is caused by impaired function of NPC2. NPC2 is a small ~25–27 kDa luminal lysosomal protein and NPC1 is a large 142 kDa (calculated) transmembrane protein localized in the endolysosomal membrane. NPC1 and NPC2 are encoded by NPC1 on chromosome 18q11 and NPC2 on chromosome14q24. These two proteins, NPC1 and NPC2, act in concert to move cholesterol from the endolysosomal system and make it bioavailable to the cell. Cholesterol esters contained in lipoprotein particles, such as low-density lipoprotein, enter the cell via receptor-mediated endocytosis. The cholesterol esters are then de-esterified via lysosomal acid lipase (LAL). After removal of the fatty acid via LAL, the unesterified cholesterol is transported by NPC2 to the N-terminal domain of NPC1. NPC1 then facilitates the transit of cholesterol from the endolysosomal lumen to the cytoplasmic face of the endolysosomal membrane where it then can be distributed to other cellular membranes and become functionally bioavailable [6]. Thus, in either NPC1 or NPC2 disease, biallelic variants of either NPC1 or NPC2 lead to both endolysosomal storage of unesterified cholesterol and a corresponding deficiency of cellular bioavailable cholesterol. Increased expression of SREBP2 regulated genes, which increases both endogenous synthesis and exogenous cholesterol uptake, is evidence of the functional cholesterol deficiency, even though total cellular cholesterol may be increased [7].
NPC1 is an ultrarare disease with an incidence of the classical disease estimated to be in the order of 1/110,000 [8]. Classically, NPC1 was diagnosed via cellular staining with filipin, a fluorescent compound that specifically labels unesterified cholesterol; however, filipin staining of patient fibroblasts has been supplanted by plasma-based tests measuring levels of cholestane-3β,5α,6β-triol, N-(3β,5α,6β-trihydroxy-cholan-24-oyl)glycine or N-palmitoyl-O-phosphocholineserine/lysosphingomyelin-509 [9,10]. NPC1 can also be diagnosed by sequencing NPC1. Molecular diagnosis has been facilitated by inclusion of NPC1 on sign/symptom gene panels as well as exome/genome sequencing. It is hoped that improved diagnostic modalities will decrease the 4–5-year diagnostic delay that has been observed [10].
Infants with liver disease often present with cholestatic jaundice. Although this can be severe and lead to diagnosis of NPC1, it is often transient and resolves with time. Onset of the neurological signs and symptoms typically follows hepatic disease and has an insidious progression. The NPC1 neurological phenotype is characterized by progressive neurological dysfunction which includes supranuclear vertical gaze palsy, cerebellar ataxia and cognitive impairment. However, the NPC1 phenotype is heterogeneous both with respect to age of neurological onset and the specific sign/symptom complex manifested by individuals [8,11]. Clinically it has been recognized that the NPC1 phenotype can vary between siblings and individuals with the same NPC1 genotype. Case reports have recently been summarized by Las Heras et al. [12]. Clinical data also suggests that ApoE isotype can modify the NPC1 phenotype. Specifically, in a small case series, the ApoE4 isoform is associated with increased neuropathological findings [13] and earlier onset of neurological signs/symptoms [14]. Miglustat has been shown to be clinically effective in slowing neurological disease progression [15,16,17] and extending survival [18]. Although miglustat is approved for the treatment of NPC in most countries, it is only available off-label in the United States. Currently, there are no FDA-approved therapies for NPC1. Thus, identification of genes/metabolic processes that modify the NPC1 neurological phenotype could provide insight into novel therapeutic approaches.
Multiple studies using different experimental approaches with NPC1 models have provided evidence for genetic modifiers of NPC1. The Sturley research group, using an NPC1 yeast model, has identified 12 pathways and 13 genes in a genome-wide, conditional synthetic lethality screen [19]. These data, in combination with a high-throughput drug screening by Pipalia et al. [20], contributed to identification of histone deacetylase inhibition as a potential therapy for NPC1. High-throughput drug screens have shown that modulation of NPC1 activity by either protein stabilization or upregulation of NPC1 expression can decrease unesterified cholesterol storage. These include HDAC-inhibitor-mediated stabilization of variant NPC1 proteins [20] and alexidine-mediated increased NPC1 expression [21]. RNAi [22] and CRISPRi screens [23] identified TMEM97 and SNX13, respectively, as genes that modify unesterified cholesterol storage in cells deficient for NPC1 function. Genetic background in Npc1 mutant mice can significantly influence the severity of the NPC1 phenotype. Homozygous Npc1 mutant mice on a C57Bl/6J background have a more severe phenotype compared to the same mutant alleles on a Balb/cJ background [24,25,26]. Rodriguez-Gil et al. [25] performed a QTL analysis for lifespan between these two different strains, and found significant evidence for linkage to markers on mouse chromosomes 1 and 7. Similarly, Zhang and Erickson [27] observed evidence of a phenotypic modifier on mouse chromosome 19. Multiple studies have evaluated potential genetic modifiers in double-mutant mice. Disruption of genes encoding enzymes involved in glycosphingolipid synthesis, GM2/GD2 synthase [28] and GM3 synthase [29], decrease neuropathology in Npc1 mutant mice. Recognition that decreased glycosphingolipid synthesis was associated with phenotypic improvement contributed to the development of miglustat as a therapy for NPC1. Although neuroinflammation is a predominant neuropathological finding in NPC1, modulation of genes expressed as part of the immune response in general has had relatively limited impact on the survival of Npc1 mutant mice. Examples include disruption of Il6 [30], C1qa and C3 [31] and Ccl3 [32]. In contrast, a recent work suggests that modulation of either STING or IRF3 may have beneficial effects in NPC1 [33,34]. Decreased expression of tau (Mapt) and amyloid precursor protein (App) increased phenotypic severity [35,36]. Impairment of non-lysosomal glucosylceramidase (Gba2, [37]) also increased phenotypic severity in double-mutant mice. These preclinical data support the hypothesis that genetic modifiers may contribute to phenotypic heterogeneity in NPC1 disease.
Sterol O-acyltransferase 1 (SOAT1) and sterol o-acyltransferase 2 (SOAT2) catalyze the intracellular esterification of cholesterol. SOAT1 is also referred to as ACAT1; however, ACAT1 can also refer to the mitochondrial enzyme acetyl-CoA acetyltransferase. SOAT1 is ubiquitously expressed, and it is the isoform primarily expressed in the brain. SOAT2 is predominantly expressed by intestinal enterocytes and liver. Impaired function of either SOAT1 or SOAT2 appears to have a beneficial effect on the NPC1 phenotype in mouse models. Double-mutant Soat1-/-:Npc1-/- mice, relative to Soat+/+:Npc1-/- mice, have delayed development of the NPC1 disease phenotype, Purkinje neurons preservation and increased survival [38]. Consistent with its expression in liver, double-mutant Soat2-/-:Npc1-/- mice manifested an attenuated liver phenotype with decreased accumulation of unesterified cholesterol and decreased serum transaminase levels [39]. As noted above, although NPC1 is characterized by increased unesterified cholesterol storage, there is paradoxically a functional cellular cholesterol deficiency. Esterification of the bioavailable cholesterol by SOAT1 or SOAT2 may compound this deficiency. Thus, inhibition of intracellular cholesterol esterification may improve the cellular cholesterol deficiency and explain the decrease in phenotypic severity observed in both Soat1-/-:Npc1-/- and Soat2-/-:Npc1-/- mice in comparison to single-mutant Npc1-/- mice.
Several studies have evaluated associations between rs1044925, a single nucleotide polymorphism in the 3′-untranslated region of SOAT1 (C/A), with serum lipid levels [40,41], atherosclerotic disease [42] and Alzheimer’s disease/Dementia [43,44,45]. The rs1044925 A-allele is considered protective and associated with decreased SOAT1 mRNA expression [46]. Single-eQTL data also suggests that the rs1044925 A-allele is associated with decreased expression in multiple neuronal tissues (GTExPortal). Combining these observations with the data showing an attenuated neurological disease phenotype in double-mutant Soat1-/-:Npc1-/- mice, we hypothesized that the rs1044925 A-allele could be a genetic modifier of the NPC1 phenotype in individuals with NPC1.
In this paper we characterize the degree of phenotypic heterogeneity present in individuals with NPC1 with the same NPC1 genotype (NPC1I1061T/I1061T and siblings). The existence of genetic modifiers has been proposed based on the multiple case reports describing phenotypic heterogeneity in individuals with the same NPC1 genotype (reviewed in [12,47]; however, our data provide the first formal quantitative evidence for genetic modifiers in NPC1 disease. We also show that the SOAT1 rs1044925 polymorphism C-allele is associated with a more severe NPC1 phenotype. Specifically, the C-allele is associated with an earlier age of neurological onset.
2. Results
2.1. Evidence for Genetic Modifiers in Individuals with Niemann-Pick Disease, Type C1
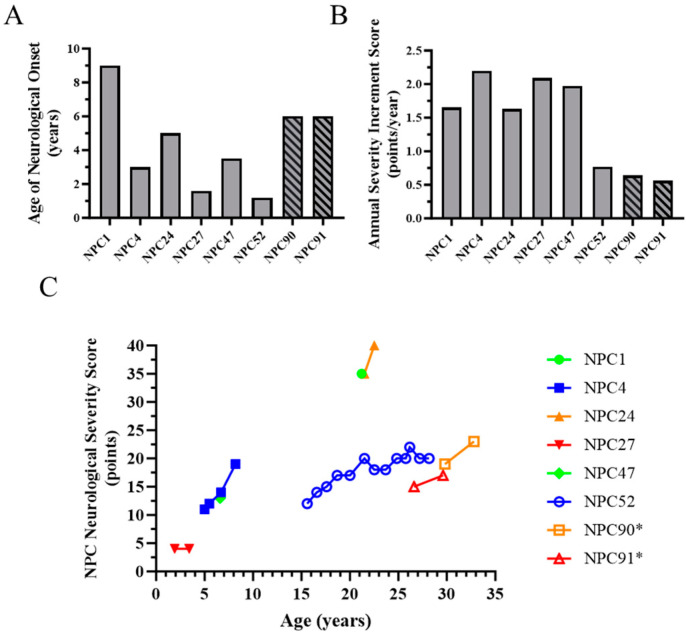
NPC1 neurological phenotypic heterogeneity can be characterized by age of neurological onset, the NPC Neurological Severity Scale (NPC NSS, [11]) and the Annualized Severity Index Score (ASIS, [48]). The NPC NSS is a Likert-like scale that assesses neurological severity in nine major and eight minor domains. Higher NPC NSS scores indicate increased disease burden. The full 17-domain NPC NSS and an abbreviated 5-domain (Ambulation, Fine Motor, Swallowing, Speech, Cognition) NPC NSS have been shown to ascertain clinically relevant aspects of the disease and measure clinically significant changes [49,50]. The NPC NSS can be used both retrospectively and prospectively to describe progression in individuals with NPC1. The ASIS age normalizes the NPC NSS. Decreased age of neurological onset and increased ASIS scores are indicative of a more severe NPC1 neurological phenotype. The most prevalent NPC1 pathological variant in individuals of Western European heritage is p.I1061T (c.3182T>C) [51]. Although individuals who are homozygous for NPC1I1061T have the same NPC1 genotype, they manifest significant phenotypic heterogeneity. In a natural history cohort followed at the National Institutes of Health Clinical Center, we evaluated eight individuals with a homozygous NPC1 p.I1061T genotype. Two of these individuals (NPC90 and NPC91) are siblings. Age of neurological onset varied from 1.2 to 9 years old in this cohort of individuals with NPC1I1061T/I1061T genotype (Figure 1A). Mean age of neurological onset was 4.4 ± 2.6 years. Similarly, baseline ASIS values varied from 0.56 to 2.20 points/year with a mean of 1.44 ± 0.68 points/year (Figure 1B). NPC NSS scores, a measure of increasing neurological disease burden and progression, are provided in Figure 1C. These data demonstrate the variable disease progression in individuals homozygous for the p.I1061T missense mutation.
Figure 1.
Phenotypic heterogeneity in NPC1 individuals homozygous for the p.I1061T (c.T3182C) variant. (A) Age of neurological onset in eight NPC1I1061T/I1061T individuals. NPC90 and NPC91 are full siblings. (B) Annual Severity Increment Scores in these same eight NPC1I1061T/I1061T individuals. (C) Longitudinal progression of the 17-domain NPC Neurological Severity Score in these eight NPC1I1061T/I1061T individuals. * siblings.
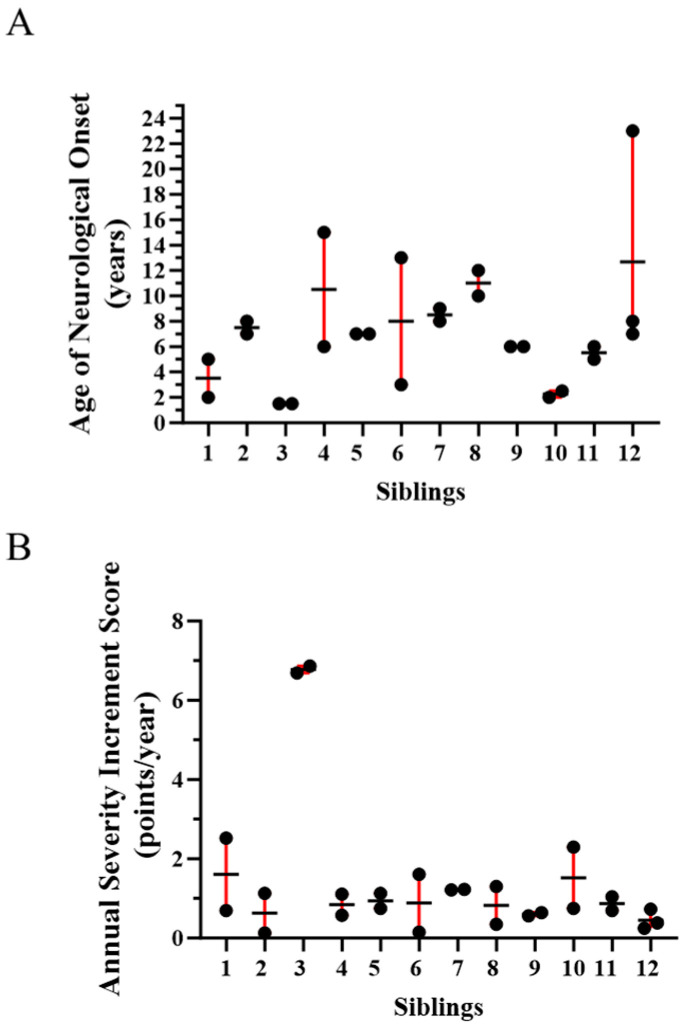
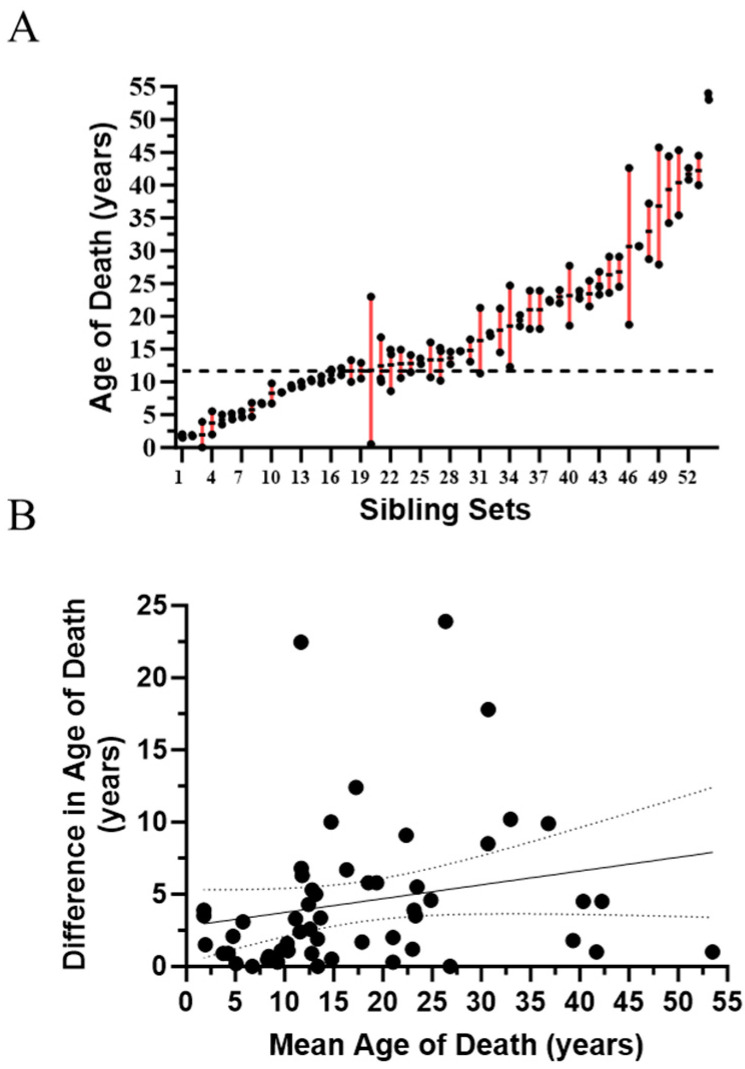
Our NPC1 natural history cohort includes 12 sibling sets manifesting NPC1 neurological symptoms. Genotypes for these sibling sets are provided in Supplementary Table S1. The “common” p.I1061T (c.3182T>C) allele accounted for approximately a third of the alleles in these siblings (8/24). A second pathological NPC1 allele has not been identified in one of the sibling pairs; however, the NPC1 diagnosis is supported by clinical and biochemical testing. Consistent with siblings sharing half of their genome, age of neurological onset was similar (≤1.25-fold difference) in 8/12 (67%). However, the difference in the age of neurological onset ranged from 2.5- to 4.3-fold in the remaining four sibling sets (Figure 2A). We also evaluated ASIS in these sibling sets (Figure 2B). Again, ASIS was similar (≤2-fold difference) in the majority (7/12, 58%), but varied 3.1- to 10.7-fold in the remaining five sibling pairs. We were also able to obtain age of death information on 54 sets of siblings (49 pairs, four triplets, one quadruplet) from the National Niemann Pick Disease Foundation. Age of death was relatively consistent in siblings where the mean age of death was under ~12 years of age (Figure 3A). However, in the remaining 34 sibling sets (mean age of death > 12 years), the difference in the range of age of death was ≥5 years in 16 (47%) and ≥10 years in 6 (18%). The variability appeared to increase after ~12 years of age, but there was no relationship (r2 = 0.05, p = 0.11) between difference in age of death and mean age of death for the siblings (Figure 3B). Although much of these data predate the use of miglustat for treatment of NPC1, earlier diagnosis and earlier interventions could have had a positive influence on the second child and thus confound the interpretation of these data. To explore this possibility, we determined the order of death for 34 sibling pairs where the cause of death was neurological and the difference in the age of death was more than one year. The first-born sibling died at a younger age in 19/34 (56%) cases. This was not significantly different than what one would expect if order of death was random (p = 0.63, Chi-square). Consistent with this result, survival curves show no appreciable difference for first or second born siblings (Supplementary Figure S1A). A male/female bias was not present; 47% of the siblings were male and mean age of death for males (18.3 ± 13.1 years) and females (16.5 ± 10.8 years) was similar (Supplementary Figure S1B). These data from siblings, in combination with the data from individuals homozygous for the p.I1061T variant, quantitatively support the conclusion of significant clinical phenotypic heterogeneity in NPC1 individuals irrespective of having the same NPC1 genotype.
Figure 2.
Phenotypic heterogeneity in twelve sets of siblings with NPC1. (A) Age of neurological onset in NPC1 siblings. Mean (horizontal bar) and range (red bar) are indicated. (B) Annual Severity Increment Scores in NPC1 siblings. Mean (horizontal bar) and range (red bar) are indicated.
Figure 3.
Variable age of death in 54 sets of siblings with NPC1. (A) Age of death is plotted for 54 sets of siblings. Range is indicated by the red bars. The horizontal dashed line is at 12 years of age. (B) Correlation (r2 = 0.05, p = 0.11) between mean age of death for the sibling sets and difference in the age of death. Dashed lines indicate the 95% confidence interval.
2.2. In Vitro Cellular Phenotype Heterogeneity
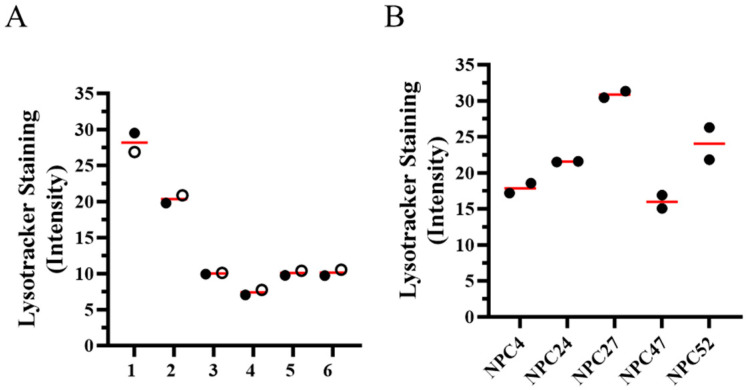
In addition to clinical phenotypic heterogeneity, we also looked to see if the NPC1 cellular phenotype was also heterogeneous. Lysotracker staining intensity is a measure of the intracellular acidic compartment volume and is typically increased in NPC1 fibroblasts and correlated with an age adjusted severity score [52]. Lysotracker staining intensity was also positively correlated with age of neurological onset in a panel of NPC1 fibroblasts [53]. From these prior data, we were able to extract lysotracker staining intensity values for cell lines from six sibling pairs (Figure 4A) and five homozygous p.I1061T cell lines (Figure 4B). Lysotracker staining intensity was similar in the six sibling pairs; however, mean lysotracker staining fold-change (stained vs. unstained) varied 1.9-fold (16.0 ± 1.3 to 30.9 ± 0.6) in cell lines homozygous for p.I1061T.
Figure 4.
Lysotracker staining intensity in fibroblasts from sibling pairs and NPC1I1061T/I1061T individuals. Lysotracker data was previously published [53] but repurposed for this analysis. (A) Lysotracker staining intensity in six sibling pairs. The mean value of two measurements is indicated for each sibling as either an open or filled circle. Sibling pairs 1, 5 and 6 correspond to sibling pairs 1, 5 and 6 in Figure 2. Sibling pair 3 corresponds to sibling pair 2 in Figure 3. (B) Lysotracker staining intensity in five individuals homozygous for the p.I1061T variant. Two measurements from each individual, with mean value indicated by the red bar, are included.
2.3. SOAT1 Polymorphism (rs1044925) Genotyping in Individuals with NPC1
SNP rs1044925 genotyping and phenotypic data were available from 117 individuals (234 alleles) with NPC1. The frequencies of the reference A- and C-alleles were 0.671 and 0.329, respectively. The genotype ratio of 50 AA, 57 AC and 10 CC did not differ significantly (p = 0.70, Chi-square) from the expected Mendelian ratio. The allele frequency in this cohort also did not differ significantly (p = 0.18, Chi-square) from the expected global frequency of 0.393 and 0.606 for C- and A-alleles, respectively (https://www.ncbi.nlm.nih.gov/snp/rs1044925, release version 20230706150541, accessed on 24 January 2024).
2.4. The SOAT1 rs1044925 C-Allele Is Associated with a More Severe NPC1 Phenotype
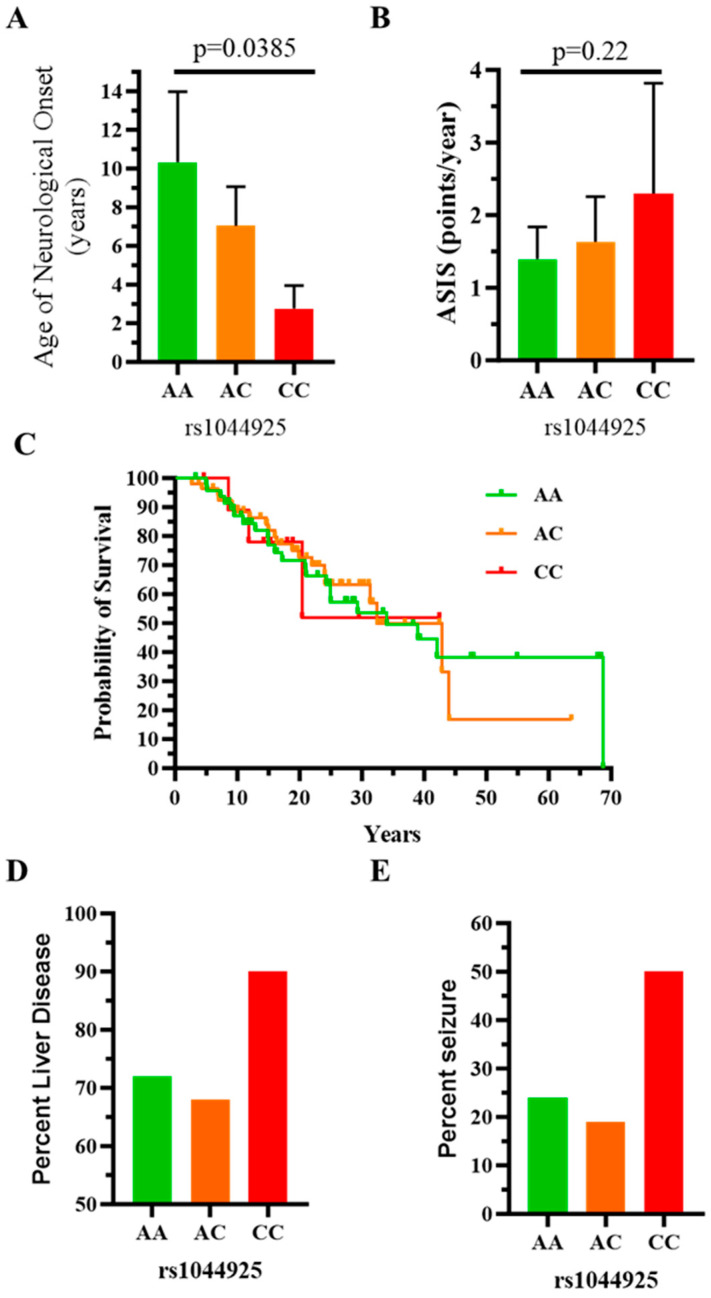
To determine if the SOAT1 rs1044925 polymorphism correlated with NPC1 phenotype, we stratified both age of neurological onset and baseline ASIS scores by rs1044925 genotype. The mean (95% confidence interval) age of neurological onset decreased from 10.3 (6.8, 14.0) to 7.0 (5.0, 9.1) to 2.8 (1.6, 4.0) years in individuals with AA, AC and CC genotypes, respectively (p = 0.0385, Kruskal-Wallis one-way ANOVA; Figure 5A). Although mean (95% confidence interval) ASIS values increased progressively from 1.4 (1.0, 1.8) to 1.6 (1.0, 2.3) to 2.3 (0.8, 3.8) points per year in individuals with AA, AC and AC genotypes, respectively, this was not significant (p = 0.22, Kruskal-Wallis one-way ANOVA; Figure 5B). Individual values for both age of neurological onset and ASIS are provided in Supplementary Table S2. We also explored survival in this cohort. No significant difference was observed between individuals with rs1044925 AA, AC and CC genotypes (Figure 5C). However, consistent with a more severe phenotype, the frequency of neonatal liver disease was higher in individuals with the CC genotype (90%) than in individuals with either the AA (72%) or AC (68%) genotype (Figure 5D). Similarly, the frequency of seizures was 2-fold higher in individuals with CC genotype (50%) than in individuals with either the AA (24%) or AC (19%) genotypes (Figure 5E). Genotype data for rs1044925 was available for 11 of the sibling sets. In 10/11 sibling sets, both siblings had the same rs1044925 genotype (AA n = 5 and AC n = 5). In sibling pair 6 (Figure 2), rs1044025 genotyping was AC and CC. Although one needs to be cautious about overinterpreting one example, the sibling with the CC genotype had an earlier age of neurological onset and a higher Annual Severity Increment Score. Taken in aggregate, these data are consistent with the conclusion that the SOAT1 rs1044925 allele is associated with a more severe NPC1 phenotype.
Figure 5.
The SOAT1 rs1044925 polymorphism C-allele is associated with a more severe NPC1 phenotype. (A) Age of neurological onset decreased significantly (p = 0.0385, Kruskal-Wallis one-way ANOVA) with the genotypic series of AA, AC and CC. (B) Although not significant (p = 0.22, Kruskal-Wallis one-way ANOVA), mean Annualized Severity Index Scores increased with the genotypic series of AA, AC and CC. (C) Survival curves for NPC1 individuals with the SOAT1 rs1044925 AA (green), AC (orange) and CC (red) genotypes. Frequency of liver disease (D) and seizures (E) was higher in individuals with SOAT1 rs1044925 CC genotype relative to individuals with either an AA or AC genotype. Mean and 95% confidence intervals are plotted in A and B.
3. Discussion
Phenotypic heterogeneity is a common observation in genetic disorders. Phenotypic heterogeneity can manifest both as incomplete penetrance and variable expressivity of the disease manifestations. Although environmental, microbiome and epigenetic differences clearly influence disease manifestations, variants genes, other than the disease causative gene, can play a major role. In NPC1, the NPC1 genotype and specifically residual NPC1 function is likely the primary determinate; however, significant preclinical and clinical data supports the hypothesis that genetic modifiers significantly influence disease severity in individuals with NPC1.
In this paper, we characterized and quantified the phenotypic heterogeneity in eight individuals who are homozygous for the common p.I1061T variant and twelve sibling pairs/sets who were evaluated at the NIH. Individuals homozygous for NPC1I10611T/I1061T showed marked phenotypic heterogeneity with respect to both measures of disease severity (Age of Neurological Onset and Annual Severity Increment Score) and neurological disease burden/progression (NPC NSS). Although the longitudinal data are limited, the progression slope of the NPC NSS in the five individuals where it could be estimated ranged from 0.6 to 5.0 points/year. This supports the conclusion that the NPC1 neurological phenotype is markedly different in individuals homozygous for the common p.I1061T variant.
Similar observations were made for siblings. In general, the age of neurological onset is similar in siblings; however, marked differences were observed in a third of the sibling pairs/sets. While, in general, ASIS values are similar in sibling pair/sets, substantially different ASIS values were observed in 5/12 (~42%). We were also able to obtain survival data on 54 sets of siblings. Age of death varied widely with the difference being ≥ten years in six and ≥five years in 17 sibling sets. Variability in the age of death increased in sibling sets where the mean age of death was greater than ~12 years. Environmental factors may vary in NPC1I10611T/I1061T individuals and thus potentially contribute to phenotypic heterogeneity. However, environmental differences contributing to phenotypic heterogeneity are less likely in siblings. This variability did not appear to be related to birth order, thus decreasing the likelihood that improved care in a second child contributed substantially to the heterogeneity in survival. Although phenotypic heterogeneity has previously been noted in individuals with the same NPC1 variants (reviewed in [12,47]), to our knowledge, the phenotypic data presented in this paper represent the largest sibling and homozygous p.I1061T cohorts quantitatively described to date, and these data clearly support the likelihood that genetic modifiers contribute to phenotypic heterogeneity in NPC1.
The effect of genetically inhibiting intracellular cholesterol esterification has been studied in both Soat1-/-:Npc1-/- [38] and Soat2-/-:Npc1-/- [39] mice. In both cases, it is hypothesized that inhibition of cholesterol esterification increases intracellular cholesterol bioavailability. Consistent with predominant liver expression of SOAT2, Soat2-/-:Npc1-/-: mice had decreased unesterified cholesterol storage in liver tissue and improved liver pathology. Similarly, consistent with expression in the central nervous system, Soat-/-:Npc1-/- mice, relative to Soat+/+:Npc1-/- mice, had delayed onset of neurological signs, improved neuropathology and significantly increased lifespans [38].
We hypothesized, based on the observation that the NPC1 phenotype is ameliorated in Soat1-/-:Npc1-/- mice, that functional variants in the SOAT1 gene could contribute to genetic heterogeneity in individuals with NPC1. To test this hypothesis, we correlated the genotype of a single nucleotide polymorphism, rs1044925 (A, C), with clinical parameters of disease severity in 117 individuals with NPC1. This single nucleotide polymorphism is encoded in the 3′-UTR of SOAT1. The A-allele, relative to the C-allele, is experimentally associated with decreased mRNA expression [46] and single-eQTL data specifically supports decreased expression in neuronal tissues (GTExPortal). Thus, we postulated that the A-allele would be associated with a less severe NPC1 phenotype. The data in this paper show that the C-allele is significantly associated with an earlier age of neurological onset with mean age of neurological onset decreasing from 10.3 to 2.8 years of age in individuals homozygous for the A- and C-alleles, respectively. Although not statistically significant, it is notable that the mean ASIS valued increase with the genotypic series of AA, AC and CC. This is consistent with the pattern one would expect of increased NPC neurological severity being associated with the C-allele. No difference in survival was noted, but again, consistent with increased phenotypic severity being associated with the C-allele, we observed an increased percentage of individuals with liver disease and seizures in those with CC genotype. This study does have limitations that temper the conclusion. Although the cohort of individuals with NPC1 being reported in this paper is large for NPC1, numbers are still very limited when trying to establish associations between genetic variants and phenotypic findings. This limitation is inherent to the study of rare diseases and compounded in rare diseases, such as NPC1, where there is significant phenotypic heterogeneity. It is also likely that other genetic modifiers, such as ApoE isotype, contribute to this phenotypic heterogeneity. These factors impact the ability to establish statistical significance. With these caveats established, the data we present is consistent with SOAT1 being a genetic modifier of the NPC1 phenotype, and specifically supports the idea that decreased expression of SOAT1 activity is beneficial in NPC1. This conclusion is both consistent with and substantiated by the observation of decreased phenotypic severity in Soat1-/-:Npc1-/- mice relative to Soat1+/+:Npc1-/- mice reported by Rogers et al. [38].
In conclusion, the data provided in this paper supports the hypothesis that genetic modifiers significantly contribute to phenotypic heterogeneity in NPC1 and specifically suggests that SOAT1 is a genetic modifier of the NPC1 phenotype. By extension, our data also support the hypothesis that inhibition of SOAT1 increases intracellular cholesterol bioavailability and may have a beneficial effect in NPC1. This latter observation provides support for developing inhibitors of SOAT1 as a potential therapeutic approach for the treatment of this lethal neurodegenerative disorder.
4. Materials and Methods
4.1. Study Participants and Ethical Approval
Individuals with NPC1 were enrolled in observational studies (NCT00344331 or NCT05588167) conducted at the National Institutes of Health in Bethesda, Maryland. Ethics approval for the conduct of this clinical trial was obtained from the Eunice Kennedy Shriver National Institute of Child Health and Human Development Institutional Review Board or the National Institutes of Health Intramural Institutional Review Board. Written consent for participation was provided by either a guardian or the participant. When appropriate and feasible, assent was obtained.
The diagnosis of NPC1 was obtained by appropriate clinical, biochemical or molecular testing. NPC1 genotype was obtained from clinical laboratory reports. In-depth phenotyping included medical record and history review as well as physical examination by individuals familiar with findings associated with NPC1 (NYF and FDP). Age of neurological onset was established by historical review. NPC Neurological Severity Scores were determined as previously described [11]. The full 17-domain scale was used for this study. Annual Severity Increment Scores were determined as previously described [48]. For analysis of age of neurological onset and Annual Severity Increment Scores, data from individuals who had not yet manifested NPC1 neurological symptoms was removed (n = 6). Historical data on age of death for siblings was provided by the National Niemann-Pick Disease Foundation (https://nnpdf.org/).
4.2. DNA Samples and Genomic Sequencing
Genomic DNA was obtained from either skin fibroblasts collected as part of a natural history study or from blood samples. Fibroblasts were cultured (37 °C, 5% CO2) in Dulbecco’s modified Eagle’s Medium (DMEM, ThermoFisher, Waltham, MA, USA), supplemented with 10% fetal bovine serum (FBS, Stemcell Technologies, Cambridge, MA, USA), 1X Pen/Strep and 2 mM Glutamax (ThermoFisher) in T-75 flasks. After reaching confluency, fibroblasts were isolated using 0.05% Trypsin, pelleted via low-speed centrifugation and stored at −80 °C until extracted. Genomic DNA was isolated from the fibroblast pellets using the DNeasy Blood and Tissue kit (Qiagen, Germantown, MD, USA). Whole blood samples were collected in EDTA tubes and stored at −80 °C. Genomic DNA was obtained from these samples using the QIAmp DNA Blood Midi kit (Qiagen). Quality and quantity of the DNA was determined by NanoDrop spectroscopy and the Qubit high-sensitivity dsDNA fluorescence dye binding assay kit (ThermoFisher).
Genomic sequencing was performed using the NIH Intramural Sequencing Center. Alignment was performed against human_g1k_v37 (HG19) and variants called using GAK (Broad Institute, Cambridge, MA, USA) using the “Best Practices” workflow [54], generating both single-sample VCF and GVCF files filtered as described. The presence of known SNPs was interrogated by evaluating single-sample VCF files at the given location. Average mapped coverage was ~40x and genotyping was performed for rs1044925.
4.3. Statistical Methods and Graphing
GraphPad Prism version 10.0.2 was used for the production of graphical images and statistical analysis.
Acknowledgments
We would like to acknowledge the invaluable contribution of the individuals with NPC1 and their guardians who have made the effort to participate in our clinical trials. We would like to thank the National Niemann-Pick disease foundation for the data on siblings who have died as a result of this devastating disease. We along with the NPC community owe a debt of gratitude to Bill Pavan. His commitment to advancing NPC science helped lay the foundation for this work. We would like to thank Cristin Davidson for reading and editing the manuscript. Finally, we would like to acknowledge the assistance of multiple students who assisted in pulling data from our database and supporting our clinical protocols. Most recently these include Hibaaq Mohamed and Cameron Padilla.
Supplementary Materials
The supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms25084217/s1.
Author Contributions
Conceptualization, N.Y.F., C.A.W., W.E.B. and F.D.P.; Methodology, N.Y.F., W.E.B. and F.D.P.; Formal Analysis, N.Y.F. and F.D.P.; Investigation, N.Y.F., D.A., K.M., J.I., J.L.R.-G., C.A.W. and N.X.C.; Resources, J.L.R.-G. and F.D.P.; Data Curation, N.Y.F., D.A., J.I. and N.X.C.; Writing—Original Draft Preparation, F.D.P.; Writing—Review and Editing, N.Y.F. and N.X.C.; Visualization, F.D.P.; Supervision, N.Y.F., D.A., C.A.W., N.X.C. and F.D.P.; Funding, W.E.B., and F.D.P. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Ethics approval for the conduct of this clinical trial was obtained from the Eunice Kennedy Shriver National Institute of Child Health and Human Development Institutional Review Board or the National Institutes of Health Intramural Institutional Review Board. Protocols were NCT00344331 (06-CH-0186) and NCT05588167 (001018). Initial approval dates were 14 August 2006 and 28 November 2022, respectively. Both protocols undergo yearly IRB review.
Informed Consent Statement
Written informed consent was obtained from all participants or their guardians. Assent was obtained if feasible (appropriate age and cognitive functioning).
Data Availability Statement
The underlying SNP genotyping, age of neurological onset and ASIS data are provided in Supplementary Table S2. Anonymized or coded clinical data is available for IRB approved research related to SLOS upon request. Genomic data has been deposited in dbGaP (phs003214.v1.p1).
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This study was supported by the intramural research program of NICHD NIH (ZIA HD008988 and ZIA HD008989, F.D.P.). This study was specifically supported by a grant from the Together Strong Foundation (W.E.B., W.J.P., F.D.P.). The NIH Natural History (N.Y.F., F.D.P.) study has been supported by the Ara Parseghian Medical Research Foundation.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Haldane J.B.S. The relative importance of principal and modifying genes in determining some human diseases. J. Genet. 1941;41:149–157. doi: 10.1007/BF02983018. [DOI] [Google Scholar]
- 2.Chen R., Shi L., Hakenberg J., Naughton B., Sklar P., Zhang J., Zhou H., Tian L., Prakash O., Lemire M., et al. Analysis of 589,306 genomes identifies individuals resilient to severe Mendelian childhood diseases. Nat. Biotechnol. 2016;34:531–538. doi: 10.1038/nbt.3514. [DOI] [PubMed] [Google Scholar]
- 3.Tarailo-Graovac M., Zhu J.Y.A., Matthews A., van Karnebeek C.D.M., Wasserman W.W. Assessment of the ExAC data set for the presence of individuals with pathogenic genotypes implicated in severe Mendelian pediatric disorders. Genet. Med. 2017;19:1300–1308. doi: 10.1038/gim.2017.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Genin E., Feingold J., Clerget-Darpoux F. Identifying modifier genes of monogenic disease: Strategies and difficulties. Hum. Genet. 2008;124:357–368. doi: 10.1007/s00439-008-0560-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rahit K., Tarailo-Graovac M. Genetic Modifiers and Rare Mendelian Disease. Genes. 2020;11:239. doi: 10.3390/genes11030239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cologna S.M., Rosenhouse-Dantsker A. Insights into the Molecular Mechanisms of Cholesterol Binding to the NPC1 and NPC2 Proteins. Adv. Exp. Med. Biol. 2019;1135:139–160. doi: 10.1007/978-3-030-14265-0_8. [DOI] [PubMed] [Google Scholar]
- 7.Xie C., Turley S.D., Pentchev P.G., Dietschy J.M. Cholesterol balance and metabolism in mice with loss of function of Niemann-Pick C protein. Am. J. Physiol. 1999;276:E336–E344. doi: 10.1152/ajpendo.1999.276.2.E336. [DOI] [PubMed] [Google Scholar]
- 8.Vanier M.T. Niemann-Pick disease type C. Orphanet J. Rare Dis. 2010;5:16. doi: 10.1186/1750-1172-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang X., Ory D.S. Advancing Diagnosis and Treatment of Niemann-Pick C disease through Biomarker Discovery. Explor. Neuroprotective Ther. 2021;1:146–158. doi: 10.37349/ent.2021.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patterson M.C., Clayton P., Gissen P., Anheim M., Bauer P., Bonnot O., Dardis A., Dionisi-Vici C., Klünemann H.H., Latour P., et al. Recommendations for the detection and diagnosis of Niemann-Pick disease type C: An update. Neurol. Clin. Pract. 2017;7:499–511. doi: 10.1212/CPJ.0000000000000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yanjanin N.M., Vélez J.I., Gropman A., King K., Bianconi S.E., Conley S.K., Brewer C.C., Solomon B., Pavan W.J., Arcos-Burgos M., et al. Linear clinical progression, independent of age of onset, in Niemann-Pick disease, type C. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2010;153B:132–140. doi: 10.1002/ajmg.b.30969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Las Heras M., Szenfeld B., Ballout R.A., Buratti E., Zanlungo S., Dardis A., Klein A.D. Understanding the phenotypic variability in Niemann-Pick disease type C (NPC): A need for precision medicine. NPJ Genom. Med. 2023;8:21. doi: 10.1038/s41525-023-00365-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saito Y., Suzuki K., Nanba E., Yamamoto T., Ohno K., Murayama S. Niemann-Pick type C disease: Accelerated neurofibrillary tangle formation and amyloid beta deposition associated with apolipoprotein E epsilon 4 homozygosity. Ann. Neurol. 2002;52:351–355. doi: 10.1002/ana.10266. [DOI] [PubMed] [Google Scholar]
- 14.Fu R., Yanjanin N.M., Elrick M.J., Ware C., Lieberman A.P., Porter F.D. Apolipoprotein E genotype and neurological disease onset in Niemann-Pick disease, type C1. Am. J. Med. Genet. A. 2012;158A:2775–2780. doi: 10.1002/ajmg.a.35395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Solomon B.I., Smith A.C., Sinaii N., Farhat N., King M.C., Machielse L., Porter F.D. Association of Miglustat With Swallowing Outcomes in Niemann-Pick Disease, Type C1. JAMA Neurol. 2020;77:1564–1568. doi: 10.1001/jamaneurol.2020.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patterson M.C., Mengel E., Vanier M.T., Moneuse P., Rosenberg D., Pineda M. Treatment outcomes following continuous miglustat therapy in patients with Niemann-Pick disease Type C: A final report of the NPC Registry. Orphanet J. Rare Dis. 2020;15:104. doi: 10.1186/s13023-020-01363-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pineda M., Walterfang M., Patterson M.C. Miglustat in Niemann-Pick disease type C patients: A review. Orphanet J. Rare Dis. 2018;13:140. doi: 10.1186/s13023-018-0844-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patterson M.C., Garver W.S., Giugliani R., Imrie J., Jahnova H., Meaney F.J., Nadjar Y., Vanier M.T., Moneuse P., Morand O., et al. Long-term survival outcomes of patients with Niemann-Pick disease type C receiving miglustat treatment: A large retrospective observational study. J. Inherit. Metab. Dis. 2020;43:1060–1069. doi: 10.1002/jimd.12245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Munkacsi A.B., Chen F.W., Brinkman M.A., Higaki K., Gutiérrez G.D., Chaudhari J., Layer J.V., Tong A., Bard M., Boone C., et al. An “exacerbate-reverse” strategy in yeast identifies histone deacetylase inhibition as a correction for cholesterol and sphingolipid transport defects in human Niemann-Pick type C disease. J. Biol. Chem. 2011;286:23842–23851. doi: 10.1074/jbc.M111.227645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pipalia N.H., Cosner C.C., Huang A., Chatterjee A., Bourbon P., Farley N., Helquist P., Wiest O., Maxfield F.R. Histone deacetylase inhibitor treatment dramatically reduces cholesterol accumulation in Niemann-Pick type C1 mutant human fibroblasts. Proc. Natl. Acad. Sci. USA. 2011;108:5620–5625. doi: 10.1073/pnas.1014890108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pugach E.K., Feltes M., Kaufman R.J., Ory D.S., Bang A.G. High-content screen for modifiers of Niemann-Pick type C disease in patient cells. Hum. Mol. Genet. 2018;27:2101–2112. doi: 10.1093/hmg/ddy117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartz F., Kern L., Erz D., Zhu M., Gilbert D., Meinhof T., Wirkner U., Erfle H., Muckenthaler M., Pepperkok R., et al. Identification of cholesterol-regulating genes by targeted RNAi screening. Cell Metab. 2009;10:63–75. doi: 10.1016/j.cmet.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 23.Lu A. Sorting (Nexin-13) out Novel Insights into Endolysosomal Cholesterol Export. Contact. 2022;5:25152564221114513. doi: 10.1177/25152564221114513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Praggastis M., Tortelli B., Zhang J., Fujiwara H., Sidhu R., Chacko A., Chen Z., Chung C., Lieberman A.P., Sikora J., et al. A murine Niemann-Pick C1 I1061T knock-in model recapitulates the pathological features of the most prevalent human disease allele. J. Neurosci. 2015;35:8091–8106. doi: 10.1523/JNEUROSCI.4173-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodriguez-Gil J.L., Watkins-Chow D.E., Baxter L.L., Elliot G., Harper U.L., Wincovitch S.M., Wedel J.C., Incao A.A., Huebecker M., Boehm F.J., et al. Genetic background modifies phenotypic severity and longevity in a mouse model of Niemann-Pick disease type C1. Dis. Model. Mech. 2020;13:dmm042614. doi: 10.1242/dmm.042614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parra J., Klein A.D., Castro J., Morales M.G., Mosqueira M., Valencia I., Cortés V., Rigotti A., Zanlungo S. Npc1 deficiency in the C57BL/6J genetic background enhances Niemann-Pick disease type C spleen pathology. Biochem. Biophys. Res. Commun. 2011;413:400–406. doi: 10.1016/j.bbrc.2011.08.096. [DOI] [PubMed] [Google Scholar]
- 27.Zhang J., Erickson R.P. A modifier of Niemann Pick C 1 maps to mouse chromosome 19. Mamm. Genome. 2000;11:69–71. doi: 10.1007/s003350010013. [DOI] [PubMed] [Google Scholar]
- 28.Gondre-Lewis M.C., McGlynn R., Walkley S.U. Cholesterol accumulation in NPC1-deficient neurons is ganglioside dependent. Curr. Biol. 2003;13:1324–1329. doi: 10.1016/S0960-9822(03)00531-1. [DOI] [PubMed] [Google Scholar]
- 29.Lee H., Lee J.K., Bae Y.C., Yang S.H., Okino N., Schuchman E.H., Yamashita T., Bae J.S., Jin H.K. Inhibition of GM3 synthase attenuates neuropathology of Niemann-Pick disease Type C. by affecting sphingolipid metabolism. Mol. Cells. 2014;37:161–171. doi: 10.14348/molcells.2014.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki M., Sugimoto Y., Ohsaki Y., Ueno M., Kato S., Kitamura Y., Hosokawa H., Davies J.P., Ioannou Y.A., Vanier M.T., et al. Endosomal accumulation of Toll-like receptor 4 causes constitutive secretion of cytokines and activation of signal transducers and activators of transcription in Niemann-Pick disease type C (NPC) fibroblasts: A potential basis for glial cell activation in the NPC brain. J. Neurosci. 2007;27:1879–1891. doi: 10.1523/JNEUROSCI.5282-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopez M.E., Klein A.D., Scott M.P. Complement is dispensable for neurodegeneration in Niemann-Pick disease type C. J. Neuroinflammation. 2012;9:216. doi: 10.1186/1742-2094-9-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lopez M.E., Klein A.D., Hong J., Dimbil U.J., Scott M.P. Neuronal and epithelial cell rescue resolves chronic systemic inflammation in the lipid storage disorder Niemann-Pick C. Hum. Mol. Genet. 2012;21:2946–2960. doi: 10.1093/hmg/dds126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chu T.T., Tu X., Yang K., Wu J., Repa J.J., Yan N. Tonic prime-boost of STING signalling mediates Niemann-Pick disease type C. Nature. 2021;596:570–575. doi: 10.1038/s41586-021-03762-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang A., Chen C., Mei C., Liu S., Xiang C., Fang W., Zhang F., Xu Y., Chen S., Zhang Q., et al. Innate immune sensing of lysosomal dysfunction drives multiple lysosomal storage disorders. Nat. Cell Biol. 2024;26:219–234. doi: 10.1038/s41556-023-01339-x. [DOI] [PubMed] [Google Scholar]
- 35.Nunes A., Pressey S.N., Cooper J.D., Soriano S. Loss of amyloid precursor protein in a mouse model of Niemann-Pick type C disease exacerbates its phenotype and disrupts tau homeostasis. Neurobiol. Dis. 2011;42:349–359. doi: 10.1016/j.nbd.2011.01.028. [DOI] [PubMed] [Google Scholar]
- 36.Pacheco C.D., Elrick M.J., Lieberman A.P. Tau deletion exacerbates the phenotype of Niemann-Pick type C mice and implicates autophagy in pathogenesis. Hum. Mol. Genet. 2009;18:956–965. doi: 10.1093/hmg/ddn423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marques A.R., Aten J., Ottenhoff R., van Roomen C.P., Herrera Moro D., Claessen N., Vinueza Veloz M.F., Zhou K., Lin Z., Mirzaian M., et al. Reducing GBA2 Activity Ameliorates Neuropathology in Niemann-Pick Type C Mice. PLoS ONE. 2015;10:e0135889. doi: 10.1371/journal.pone.0135889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rogers M.A., Chang C.C., Maue R.A., Melton E.M., Peden A.A., Garver W.S., Lee J., Schroen P., Huang M., Chang T.Y. Acat1/Soat1 knockout extends the mutant Npc1 mouse lifespan and ameliorates functional deficiencies in multiple organelles of mutant cells. Proc. Natl. Acad. Sci. USA. 2022;119:e2201646119. doi: 10.1073/pnas.2201646119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lopez A.M., Jones R.D., Repa J.J., Turley S.D. Niemann-Pick C1-deficient mice lacking sterol O-acyltransferase 2 have less hepatic cholesterol entrapment and improved liver function. Am. J. Physiol. Gastrointest. Liver Physiol. 2018;315:G454–G463. doi: 10.1152/ajpgi.00124.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu D.F., Yin R.X., Aung L.H.H., Hu X.J., Cao X.L., Miao L., Li Q., Yan T.T., Wu J.Z., Pan S.L. Polymorphism of rs1044925 in the acyl-CoA:cholesterol acyltransferase-1 gene and serum lipid levels in the Guangxi Bai Ku Yao and Han populations. Lipids Health Dis. 2010;9:139. doi: 10.1186/1476-511X-9-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu D.F., Yin R.X., Aung L.H.H., Li Q., Yan T.T., Zeng X.N., Huang K.K., Huang P., Wu J.Z., Pan S.L. Sex-specific association of ACAT-1 rs1044925 SNP and serum lipid levels in the hypercholesterolemic subjects. Lipids Health Dis. 2012;11:9. doi: 10.1186/1476-511X-11-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y.T., Wang Y.H., Ma Y.T., Fu Z.Y., Yang Y.N., Ma X., Li X.M., Adi D., Liu F., Chen B.D. ACAT-1 gene polymorphism is associated with increased susceptibility to coronary artery disease in Chinese Han population: A case-control study. Oncotarget. 2017;8:89055–89063. doi: 10.18632/oncotarget.21649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alavez-Rubio J.S., Martinez-Rodriguez N., Escobedo-de-la-Pena J., Garrido-Acosta O., Juarez-Cedillo T. Relationship Between Genetic Variants of ACAT1 and APOE with the Susceptibility to Dementia (SADEM Study) Mol. Neurobiol. 2021;58:905–912. doi: 10.1007/s12035-020-02162-3. [DOI] [PubMed] [Google Scholar]
- 44.Lämsä R., Helisalmi S., Herukka S.K., Tapiola T., Pirttilä T., Vepsäläinen S., Hiltunen M., Soininen H. Study on the association between SOAT1 polymorphisms, Alzheimer’s disease risk and the level of CSF biomarkers. Dement. Geriatr. Cogn. Disord. 2007;24:146–150. doi: 10.1159/000105164. [DOI] [PubMed] [Google Scholar]
- 45.Zhao F.G., Wang Y.H., Yang J.F., Ma Q.L., Tang Z., Dong X.M., Chan P. Association between acyl-coenzyme A: Cholesterol acyltransferase gene and risk for Alzheimer’s disease in Chinese. Neurosci. Lett. 2005;388:17–20. doi: 10.1016/j.neulet.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 46.Brunner F.S.M. Effects of a SOAT1 Haplotype on SOAT1 mRNA Expression and on the Risk for Sporadic Alzheimer’s Disease. Swill Federal Institute of Technology Zurich; Zürich, Switzerland: 2006. [Google Scholar]
- 47.Pfrieger F.W. The Niemann-Pick type diseases—A synopsis of inborn errors in sphingolipid and cholesterol metabolism. Prog. Lipid Res. 2023;90:101225. doi: 10.1016/j.plipres.2023.101225. [DOI] [PubMed] [Google Scholar]
- 48.Cortina-Borja M., Te Vruchte D., Mengel E., Amraoui Y., Imrie J., Jones S.A., i Dali C., Fineran P., Kirkegaard T., Runz H., et al. Annual severity increment score as a tool for stratifying patients with Niemann-Pick disease type C and for recruitment to clinical trials. Orphanet J. Rare Dis. 2018;13:143. doi: 10.1186/s13023-018-0880-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Evans W., Patterson M., Platt F., Guldberg C., Mathieson T., Pacey J. International consensus on clinical severity scale use in evaluating Niemann-Pick disease Type C in paediatric and adult patients: Results from a Delphi Study. Orphanet J. Rare Dis. 2021;16:482. doi: 10.1186/s13023-021-02115-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patterson M.C., Lloyd-Price L., Guldberg C., Doll H., Burbridge C., Chladek M., íDali C., Mengel E., Symonds T. Validation of the 5-domain Niemann-Pick type C Clinical Severity Scale. Orphanet J. Rare Dis. 2021;16:79. doi: 10.1186/s13023-021-01719-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Millat G., Marçais C., Rafi M.A., Yamamoto T., Morris J.A., Pentchev P.G., Ohno K., Wenger D.A., Vanier M.T. Niemann-Pick C1 disease: The I1061T substitution is a frequent mutant allele in patients of Western European descent and correlates with a classic juvenile phenotype. Am. J. Hum. Genet. 1999;65:1321–1329. doi: 10.1086/302626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Te Vruchte D., Speak A.O., Wallom K.L., Al Eisa N., Smith D.A., Hendriksz C.J., Simmons L., Lachmann R.H., Cousins A., Hartung R., et al. Relative acidic compartment volume as a lysosomal storage disorder-associated biomarker. J. Clin. Investig. 2014;124:1320–1328. doi: 10.1172/JCI72835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baxter L.L., Watkins-Chow D.E., Johnson N.L., Farhat N.Y., Platt F.M., Dale R.K., Porter F.D., Pavan W.J., Rodriguez-Gil J.L. Correlation of age of onset and clinical severity in Niemann-Pick disease type C1 with lysosomal abnormalities and gene expression. Sci. Rep. 2022;12:2162. doi: 10.1038/s41598-022-06112-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., Del Angel G., Rivas M.A., Hanna M., et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The underlying SNP genotyping, age of neurological onset and ASIS data are provided in Supplementary Table S2. Anonymized or coded clinical data is available for IRB approved research related to SLOS upon request. Genomic data has been deposited in dbGaP (phs003214.v1.p1).