Abstract
The entry of retroviruses into cells depends on receptor recognition by the viral envelope surface subunit SU followed by membrane fusion, which is thought to be mediated by a fusion peptide located at the amino terminus of the envelope transmembrane subunit TM. Several fusion determinants have been previously identified in murine leukemia virus (MLV) envelopes, but their functional interrelationships as well as the processes involved in fusion activation upon retroviral receptor recognition remain unelucidated. Despite both structural and functional similarities of their envelope glycoproteins, ecotropic and amphotropic MLVs display two different postbinding properties: (i) while amphotropic MLVs fuse the cells at neutral pH, penetration of ecotropic MLVs is relatively acid pH dependent and (ii) ecotropic envelopes are more efficient than amphotropic envelopes in inducing cell-to-cell fusion and syncytium formation. By exploiting the latter characteristic in the analysis of chimeras of ecotropic and amphotropic MLV envelopes, we show here that substitution of the ecotropic MLV proline-rich region (PRR), located in the SU between the amino-terminal receptor binding domain and the TM-interacting SU carboxy-terminal domains, is sufficient to revert the amphotropic low-fusogenic phenotype into a high-fusogenic one. Furthermore, we have identified potential β-turns in the PRR that control the stability of SU-TM associations as well as the thresholds required to trigger either cell-to-cell or virus-to-cell fusion. These data, demonstrating that the PRR functions as a signal which induces envelope conformational changes leading to fusion, have enabled us to derive envelopes which can infect cells harboring low levels of available amphotropic receptors.
Retroviruses have a common organization of their envelope glycoproteins, which consist of trimers of two subunits derived from a single protein precursor: a surface subunit, SU, harboring the determinants that interact with the cell surface receptor(s) and a transmembrane subunit, TM, whose functions include anchorage of the trimer complex in the viral membrane and promotion of the membrane fusion that follows interaction of the viral particle with the retroviral receptor (22). It is generally agreed that the fusion process of enveloped viruses is initiated by conformational rearrangements of the viral envelope glycoproteins. These rearrangements follow binding to the viral receptor, resulting in the exposure of domains more directly involved in fusion (54). The molecular mechanisms responsible for these structural changes are best understood in the case of entry of orthomyxoviruses. Thus, structural rearrangements of the influenza virus hemagglutinin are triggered by the acidic environment of the vesicles in which the virions have been endocytosed after their attachment to sialic acid residues harbored by cell surface glycoproteins (45). In the case of retroviruses, both pH-dependent and -independent viral entry has been described (31). Although conformational rearrangements of retroviral envelope glycoproteins are thought to be required for fusion (53), the precise determinants and steps involved in the putative conformational changes that follow interaction of retroviral envelopes with their receptors remain unelucidated. An understanding of these processes will greatly facilitate our ability to modulate retroviral infections as well as retrovirus-mediated gene targeting (11). Indeed, retrovirus-based gene transfer strategies utilize vectors pseudotyped with the amphotropic murine leukemia retrovirus (MLV) envelope because of the presence of the amphotropic receptor on human cells. Optimizing virus-cell fusion by engineering the amphotropic envelope will be highly desirable for several gene transfer applications.
Fusion determinants identified thus far in MLVs include (i) a fusion peptide located at the amino terminus of the TM subunit identified by sequence analogy to bona fide fusion peptides of other enveloped viruses (23) and (ii) several fusion-influencing determinants located at both the amino terminus of the SU subunit (4) and the carboxy terminus of the TM subunit (40, 43). The nature of the retroviral receptor eventually recognized by the envelope also seems to influence the fusogenic activity since ecotropic MLV (38) or amphotropic MLV chimeras harboring the ecotropic receptor binding domain (41) are much more fusogenic than other MLV strains when tested in cell-to-cell fusion assays. We show here that proline-rich regions (PRR) of MLV, located between the SU amino-terminal receptor binding domain and the TM-interacting SU carboxy-terminal domains, mediate envelope conformational changes and fusion activation. Furthermore, we identified potential β-turns in the PRR that determine both the stability of the SU-TM association as well as the thresholds necessary to trigger cell-to-cell and virus-to-cell fusion. Based on these results, we describe for the first time modified amphotropic envelopes with an enhanced virus-to-cell fusion and which allow efficient infection of cells with decreased levels of amphotropic receptor.
MATERIALS AND METHODS
Cell lines.
The TELCeB6 cell line (12) was derived from the TELac2 line after transfection and clonal selection of a Moloney murine leukemia virus (MoMLV)-based expression plasmid to produce Gag and Pol proteins. The TELac2 cells were originally derived from the TE671 human rhabdomyosarcoma cells (ATCC CRL8805) to express the nlsLacZ reporter retroviral vector (46). Production of infectious retroviral particles by TELCeB6 cells depends on newly introduced envelope expression vectors. Cerd9 and Cear13 cells (26) (kind gift of D. Kabat) are derived from CHO (Chinese hamster ovary) cells (ATCC CCL-61) and express either ecotropic MLV receptors alone or both ecotropic and amphotropic receptors, respectively. Cerd9, Cear13, and CHO cells were grown in Dulbecco modified Eagle medium (Life Technologies) supplemented with 10% fetal bovine serum and proline (Life Technologies). XC-A-ST cells were derived from XC rat sarcoma cells (ATCC CCL-165) after transfection with the pA-ST plasmid expressing the amino-terminal receptor binding domain of the amphotropic envelope glycoprotein (7). Expression of this amphotropic domain led to decreased availability of endogenous amphotropic receptors in selected XC-A-ST clones and poor infectibility of these cells by amphotropic envelope-pseudotyped retroviral vectors.
Construction of envelope expression vectors.
Plasmids FBASALF and FBMOSALF encoding the MLV-4070A amphotropic (noted as A) and MoMLV ecotropic (noted as MO) envelope glycoproteins, respectively, have been described elsewhere (10) and were used as backbones for construction and expression of envelope mutants. The FBASALF plasmid was modified to produce a highly cell-to-cell fusogenic form of the amphotropic glycoprotein, designated ARless envelope, by introducing a stop codon before the first amino acid of the intracytoplasmic p2-R peptide as previously described (43). Chimeric envelope glycoproteins in which BD, PRO, C, or TM envelope domains were swapped individually (Fig. 1) or in combinations (see Fig. 7) were generated by using allelic restriction sites that were already present or introduced by oligonucleotide site-directed mutagenesis (details and sequences available upon request) and cloned in the FBASALF envelope expression vector. For amphotropic and ecotropic glycoproteins, respectively, the boundaries of the various domains were defined as M31 to V237 and A34 to L262 for BD; G238 to P297 and G263 to A308 for PRO (G266 to A319 for Friend MLV [Fr-MLV]); G298 to R458 and G309 to R469 for C; and E459 to P654 and E470 to P665 for TM. Residues are numbered starting from the initiation methionine deduced from the amino acid sequences of the 4070A amphotropic MLV (34), the Moloney MLV (44) and the C57 strain of Fr-MLV (25) envelope glycoproteins. Substitution or deletion mutations were introduced in the PRR of the amphotropic 4070A-MLV by PCR-mediated mutagenesis (oligonucleotide sequences available upon request) and mutant glycoproteins were expressed from FBASALF-derived expression plasmids. The amino acid sequences of these mutants are shown below (see Fig. 4).
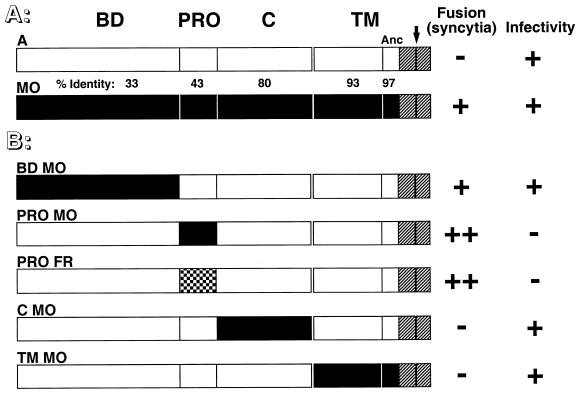
FIG. 1.
Schematic representation of envelope chimeras and their fusion properties. White and black boxes represent domains derived from amphotropic MLV-4070A envelope (A) and ecotropic Moloney-MLV (MO) or Fr-MLV (FR) envelope, respectively. The intracytoplasmic sequences, identical for both MLV classes, are shown as hatched boxes. (A) Domain organization of parental envelope. BD, amino-terminal receptor binding domain; PRO, proline-rich region; C, SU carboxy-terminal domain; TM, transmembrane subunit. Separation between ectodomain and anchor domain (Anc) of the TM subunit is indicated by the thin vertical black bar. The percentage of identical amino acids between each domain is indicated. The black arrow over the 4070A-MLV TM subunit marks the location of a premature stop codon introduced immediately before the R peptide to generate the cell-to-cell fusogenic ARless amphotropic envelope. (B) Chimeric envelope in which single domains were swapped. A summary of fusion and infection properties is shown to the right of the schematic representations. Cell-to-cell fusion activity was determined after transfection of the corresponding envelope expression vector in TELac2 cells and cocultivation with XC or XC-A-ST cells and is indicated as follows: −, absence of syncytia; +, presence of syncytia in XC cells; ++, presence of syncytia for chimeras with the amphotropic BD in both XC and X-A-ST cells (see detailed results in Table 1). Infectivity was tested by using supernatants harvested from stably transfected TELCeB6 packaging cells on XC cells and is indicated as follows: −, titers of less than 102 IU/ml; +, titers of greater than 106 IU/ml.
FIG. 7.
Schematic representation of envelope chimeras where multiple amphotropic and ecotropic domains were swapped. Domains derived from amphotropic MLV-4070A (white boxes) and ecotropic Moloney-MLV (MO) (black boxes) and sequences common to both MLV types (hatched boxes) are shown.
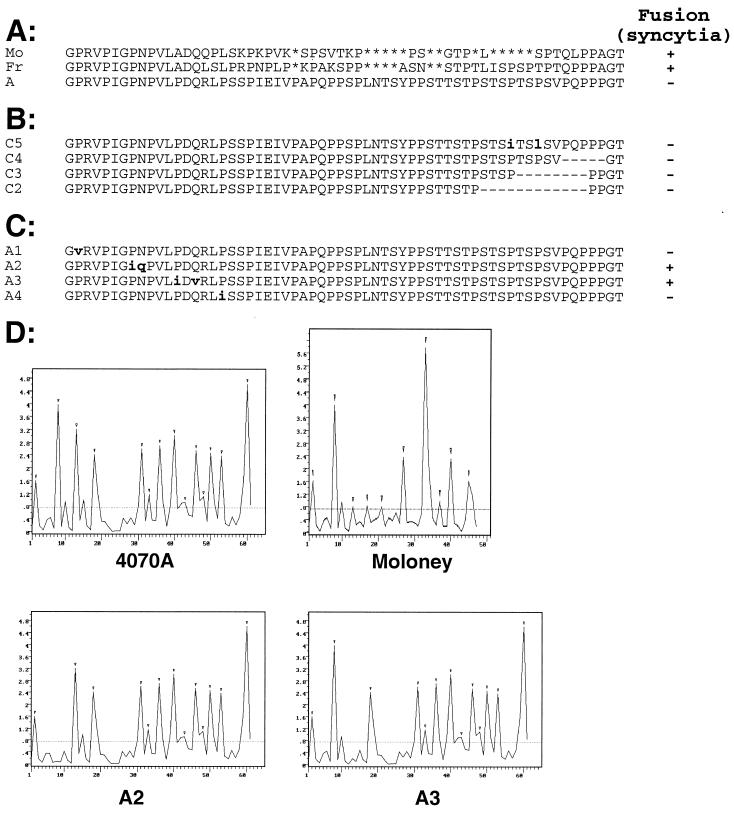
FIG. 4.
Fusion and β-turn profiles of 4070A-MLV PRR mutants. Two types of PRR mutants with either single amino acid substitutions (boldface and lowercase) or larger deletions (dashes) were derived from the 4070A-MLV PRR. The level of cell-to-cell fusion on XC cells observed with each parental or mutant envelope sequence is indicated as follows: −, absence of syncytia; +, presence of syncytia. (A) Alignment of the PRR amino acid sequences of parental ecotropic MoMLV (Mo) and Fr-MLV (Fr) envelope and amphotropic 4070A-MLV (A) envelope. Gaps introduced to optimize alignment are indicated by asterisks. (B) Sequences of 4070A-MLV mutants in the carboxy-terminal region of PRR. (C) Sequences of 4070A-MLV PRR mutants in the amino-terminal region of PRR. A second amino acid was substituted in the A2 and A3 mutants to avoid introduction of potential α-helices or β-strands not present in the parental MLV PRR (data not shown). (D) β-Turn probability profiles determined by the Chou-Fasman secondary structure prediction method (9) for parental 4070A-MLV (4070A) and MoMLV envelope PRRs and for 4070A-MLV PRR mutants with the second (A2) or third (A3) β-turn deleted. The y-axis values represent the probability P (of a turn) × 10−4 at each peptide residue (x axis). The small inverted triangles shown above the peaks show the β-turn accepted by the Chou-Fasman algorithm. The β-turn analysis was performed by using the 6.26 release of the PC/Gene software package (IntelliGenetics).
Transfections and infection assays.
Envelope glycoprotein expression plasmids were transfected by calcium phosphate precipitation into TELCeB6 or TELac2 cells as reported elsewhere (10). Virus-containing supernatants were collected after an overnight production from confluent env-transfected TELCeB6 cells and used for infection assays as described previously (10). Virus-containing supernatants were collected after an overnight production from freshly confluent env-transfected TELCeB6 cells in regular medium. Target cells were seeded in 24-well plates at a density of 5 × 104 cells per well. Viral supernatant dilutions containing 5 μg of Polybrene per ml were added, and cells were incubated for 3 to 5 h at 37°C. Viral supernatant was then removed, and cells were incubated in regular medium for 48 h. 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) staining and viral titer determination were performed as previously described (10) and expressed as LacZ infectious units (IU)/ml.
Antibodies.
Anti-gp70 (Quality Biotech Inc., Camden, N.J.) a goat antiserum raised against the Rausher leukemia virus gp70, was used diluted 1/2,000 for Western blots. Anti-SU, a rat monoclonal antibody 83A25 (17) cell culture supernatant against MLV SU, was used undiluted for fluorescence-activated cell sorting (FACS) analysis. Anti-TM, a mouse monoclonal antibody 372 (ATCC CRL-1893) (8) cell culture supernatant against MLV TM, was used undiluted for FACS analysis. Anti-CA (Quality Biotech Inc.), a goat antiserum raised against the Rausher leukemia virus p30 capsid protein (CA), was used diluted 1/10,000 for Western blots.
Immunoblots.
Virus samples from env-transfected TELCeB6 cells were prepared as previously described (10). Cell membrane preparations of env-transfected cells were processed as described elsewhere (2). Briefly, about 5 × 107 cells were harvested by EDTA treatment, washed two times in phosphate-buffered saline (PBS) and were suspended in 2 ml of ice-cold hypotonic lysis solution (10 mM Tris [pH 7.4], 2 mM MgCl2, 1 mM CaCl2) containing 1 mM phenylmethylsulfonyl fluoride. After centrifugation at 1,000 × g (4°C), the microsome-containing supernatant was kept and the pellet was relysed under the same conditions. Both supernatants were combined and ultracentrifuged at 100,000 × g for 30 min at 4°C in a precooled 70.1 Ti rotor (38,000 rpm). After slow deceleration, supernatant was discarded and excess fluid was wiped out from tubes. Pellets were then resuspended in 10 mM Tris (pH 7.4) (100 μl) resulting in suspension of membrane fragments which were further solubilized in 0.1% sodium dodecyl sulfate (SDS) and frozen at −70°C. Samples (30 μg for crude cell lysates and membrane preparations, and 20 μl for purified viruses and envelope producer cell supernatants) were mixed 5:1 (vol/vol) in a 375 mM Tris-HCl (pH 6.8) buffer containing 6% SDS, 30% β-mercaptoethanol, 10% glycerol, and 0.06% bromophenol blue, boiled for 3 min, and then run on SDS–10% acrylamide gels. After protein transfer onto nitrocellulose filters, immunostaining was performed in Tris base saline (pH 7.4) with 5% milk powder and 0.1% Tween 20. The blots were probed with the relevant antibody and developed with horseradish peroxidase-conjugated immunoglobulins raised against the species of each primary antibody (DAKO) and an enhanced chemiluminescence kit (Amersham Life Science).
Binding assays.
Target cells were washed in PBS and detached by a 10-min incubation at 37°C with 0.02% EDTA in PBS. Cells were washed in PBA (PBS with 2% fetal calf serum and 0.1% sodium azide). A total of 5 × 105 cells were incubated with virus supernatant for 45 min at 37°C in the presence of Polybrene (5 μg/ml). Cells were then washed with PBA and were incubated with the anti-SU antibody or the anti-TM antibodies for 45 min at 4°C. Cells were washed twice with PBA and incubated with either anti-rat or anti-mouse fluorescein isothiocyanate (FITC)-conjugated antibodies (DAKO), and 5 min before the two final washes in PBA, cells were counterstained with 20 μg of propidium iodide per ml. Fluorescence of living cells was analyzed with a fluorescence-activated cell sorter (FACSCalibur; Beckton Dickinson).
Cell-to-cell fusion assays.
Transfected cells were detached, counted, and reseeded at the same concentration (3 × 105 cells/well) in six-well plates. Fresh indicator cells (106 cells per well) were then added to the transfected cells and were cocultivated for 24 h. The coculture was stained by adding the May-Grunwald and Giemsa solutions (MERCK) according to the manufacturer’s recommendations.
RESULTS
Structural domains shared by the amphotropic 4070A (MLV-4070A) and ecotropic MoMLV envelopes include the following (Fig. 1A): (i) a ca. 200-amino-acid (aa) amino-terminal receptor binding domain (6), named BD domain, of known structure (18), which recognizes either PiT-2 amphotropic receptors (32, 51) present in most species including humans or the mCAT-1 ecotropic receptor (1) functionally expressed in murine and rat cells (42), respectively; (ii) the PRR, ranging between 45 to 59 aa and identified as PRO in our chimeric constructs (50); (iii) the carboxy-terminal C sequence of SU, approximately 160 aa and involved in SU-TM interactions (39); (iv) the 134-aa TM ectodomain which harbors the putative fusion peptide at its amino terminus (23); (v) the 32-aa cytoplasmic tail containing the small carboxy-terminal p2-R peptide whose late cleavage in virions increases envelope fusogenicity (40, 43). Whereas the ecotropic and amphotropic amino-terminal BD and PRR share only 33 and 43% identical residues, respectively, all other domains have more than 80% aa identity (see Fig. 1A). Most of these domains have been shown to contain regions which are involved in postbinding entry functions (4, 15, 16, 23, 33, 40, 43).
PRR modulates cell-to-cell fusion by MLV envelopes.
Ecotropic MLV envelope glycoproteins are more potent than amphotropic ones in inducing formation of syncytia in cell-to-cell fusion assays (38) (Fig. 2). To identify the region(s) responsible for the higher fusogenicity of ecotropic MLV envelopes, we generated a series of chimeric envelopes in which BD, PRO, C, and TM ecotropic domains were swapped within the amphotropic background envelope (Fig. 1B). The resulting envelopes are henceforth identified according to the substituted ecotropic domain(s). For example, PROMO and PROFR designate chimeric MLV-4070A-derived envelope glycoproteins which harbor the MoMLV and Fr-MLV PRR, respectively, whereas the CMO chimera contains the MoMLV SU carboxy-terminal domain (Fig. 1B).
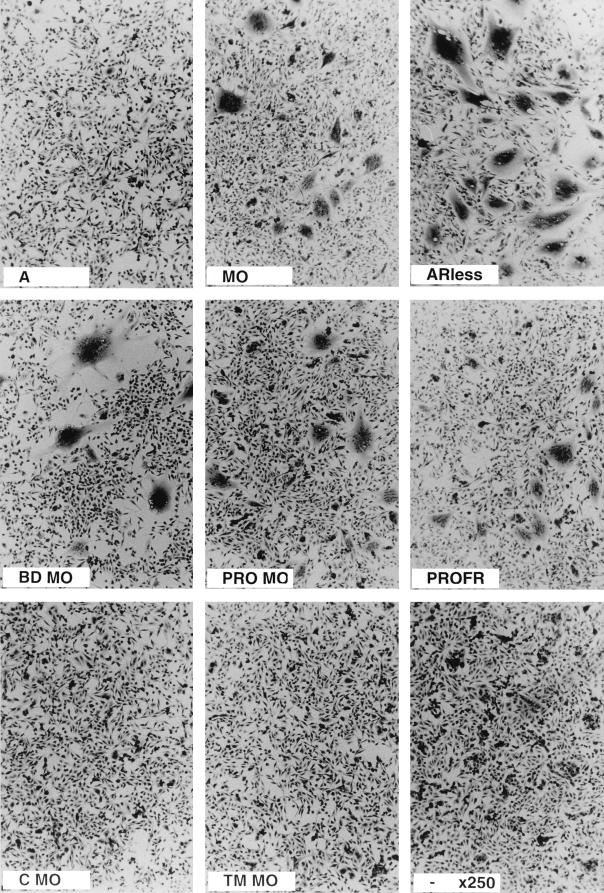
FIG. 2.
XC cell fusion assays of retroviral envelope mutants with single swapped domains. TELac2 cells were transfected with different envelope expression vectors before cocultivation for 24 h with XC indicator cells as described in the legend to Fig. 1. Magnification, ×250.
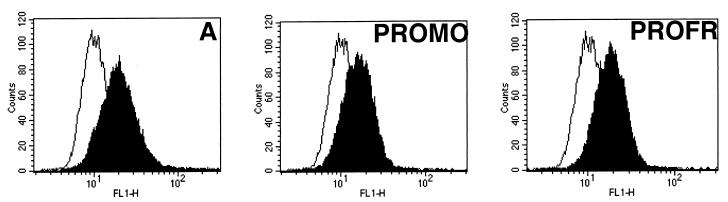
Cell-to-cell fusion was monitored by syncytium formation upon 24-h cocultivation of different indicator cells with cell lines expressing or not expressing MLV Gag-Pol core particles and transfected with the retroviral envelope to be tested. Two dramatically different cell-to-cell fusion phenotypes were observed (Fig. 2). Strong syncytium-inducing envelopes, similar to ecotropic MoMLV, included BDMO, PROMO, and PROFR, whereas CMO and TMMO were weak syncytium-inducing envelopes similar to MLV-4070A (Fig. 1 and 2). Syncytium formation was not affected by the presence (data not shown) or absence of MLV Gag-Pol core particles in the envelope-presenting cells (Fig. 2 and Table 1), demonstrating that fusion measured in this assay occurred by cell-to-cell contacts rather than by virus-to-cell interactions. The high fusogenicity observed with BDMO is in agreement with previous reports describing increased fusogenicity associated with recognition of the ecotropic receptor (41). However, we also observed highly efficient cell-to-cell fusion in envelopes lacking the ecotropic BD, such as the PROMO and PROFR chimeric envelopes which harbored the amphotropic BD (Fig. 1 and 2 and Table 1). Increased cell-to-cell fusion with the latter envelopes was observed with all indicator cell types tested, including human cells which lack the ecotropic receptor (data not shown). However, the most dramatic effect was observed with XC rat sarcoma cells (Fig. 2 and Table 1). Since similar or slightly weaker cell surface expression was detected for the hyperfusogenic PRR-mutated PROMO and PROFR chimeric envelopes compared to that of the parental MLV-4070A envelopes (Fig. 3), these data therefore indicated that differences in syncytium formation between parental and chimeric amphotropic envelopes were directly associated with a specific feature contained in the ecotropic PRO region which could sensitize the envelope fusion activity when inserted in an amphotropic background.
TABLE 1.
Effects of cell surface envelope on cell-to-cell fusion
| Cell surface envelope | Fusion indexa on cells:
|
|
|---|---|---|
| XC | XC-A-ST | |
| Noneb | 0.02 ± 0.01 | 0.02 ± 0 |
| MOc | 15 ± 2 | 14 ± 1 |
| Ad | 0.2 ± 0.1 | 0.05 ± 0.02 |
| ARlesse | 74 ± 4 | 0.85 ± 0.03 |
| PROMOc | 17 ± 3 | 16 ± 3 |
| PROFRc | 18 ± 3 | 23 ± 6 |
| A2c | 13 ± 1 | 17 ± 1 |
| A3c | 14 ± 1 | 17 ± 3 |
The fusion index is defined as the percentage of (N − S)/T, where N is the number of nuclei in the syncytia, S is the number of syncytia, and T is the total number of nuclei counted (3).
Mock-transfected cells.
Syncytia with more than 20 nuclei.
Small syncytia with less than four nuclei.
Syncytia with more than 40 nuclei in XC cells and less than 10 nuclei in XC-A-ST cells.
FIG. 3.
Cell surface expression of mutant envelopes with enhanced fusion activity. TELac2 cells transfected with the indicated envelope expression vectors shown in Fig. 1 were stained (black area) or not (white area) with 83A25 anti-SU rat monoclonal antibodies and analyzed by FACS analysis.
Limited proteolysis of MLV SU leads to cleavage at both ends of the PRR (28), suggesting that the PRR constitutes a separate domain of the SU which folds as a rigid structure. As described for feline leukemia viruses (20) and as suggested for the related MLV-4070A (50), the regular repetitions of proline-induced β-turns in MLV PRRs (Fig. 4D) might fold as polyproline β-turn helices (30). The Chou-Fasman structural analysis (9) shown in Fig. 4D shows the probability of β-turns in the PRRs of MLV-4070A and MoMLV and reveals some differences in the number and arrangement of their respective predicted β-turns (Fig. 4A and D). Hence, we sought to directly evaluate the role of these potential secondary structures in fusion by mutagenesis analyses.
The C2, C3, C4, and C5 point or deletion mutants designed to remove one or more of the seven carboxy-terminal β-turns of the amphotropic MLV-4070A PRR (Fig. 4B) were monitored for cell-to-cell fusion and were found to be as poorly fusogenic as the parental MLV-4070A envelope (Fig. 4). Proline-induced β-turns in the MLV-4070A PRR were less interwoven at the amino terminus than at the carboxy terminus (Fig. 4D), and thus each of the four potential MLV-4070A amino-terminal β-turns were inactivated individually by substitution of a valine or isoleucine, resulting in mutants A1, A2, A3, and A4 (Fig. 4C). Increased syncytium formation was not observed for A1 and A4 envelopes in which either the first or fourth amino-terminal β-turn, respectively, was mutated while maintaining a regular repetition of three potential contiguous β-turns (Fig. 4). In contrast, the A2 and A3 envelopes in which these contiguous β-turns were interrupted (Fig. 4C and D) were highly fusogenic (Fig. 4 and Table 1). These results indicated the critical importance of contiguous β-turns in mediating cell-to-cell fusion.
PRR determinants controlling cell-to-cell and virus-to-cell fusion thresholds.
As inferred from the results of syncytium assays, MLV-4070A glycoproteins harboring mutations in the PRR, such as PROMO, PROFR, A2, and A3, appeared more readily fusogenic in cell-to-cell fusion assays and thus seemed more reactive than wild-type amphotropic envelopes. They may thus require fewer PiT-2 amphotropic receptors to trigger their cell-to-cell fusogenicity. To test the relationship between increased fusogenicity and requirements for PiT-2 receptor molecules, we compared cell-to-cell fusion to either XC or XC-A-ST cells. In the latter, constitutive expression of an interfering amphotropic BD reduced the number of available functional PiT-2 receptors as demonstrated by the reduced capacities of either PROMO or MLV-4070A envelope glycoproteins to bind XC-A-ST cells compared to that of parental XC cells (see Fig. 5A). As expected, this resulted in an inhibition of fusion of XC-A-ST cells through cell-to-cell contacts by both the parental MLV-4070A envelope glycoprotein and the cytoplasmic tail-truncated ARless amphotropic envelope known to exert higher cell-to-cell fusion properties (Table 1). However, strong cell-to-cell fusion of XC-A-ST cells was still observed with the PRR-mutated PROMO, PROFR, A2, and A3 mutants (Table 1). The increased cell-to-cell fusion of the latter envelope mutants seemed to remain amphotropic receptor dependent, since cell-to-cell fusion of PiT-2-negative CHO cells with these envelopes was observed only upon de novo PiT-2 expression (data not shown). Altogether, these results suggested that mutations in the PRR β-turns facilitated cell-to-cell fusion via recognition of amphotropic PiT-2 receptors.
FIG. 5.
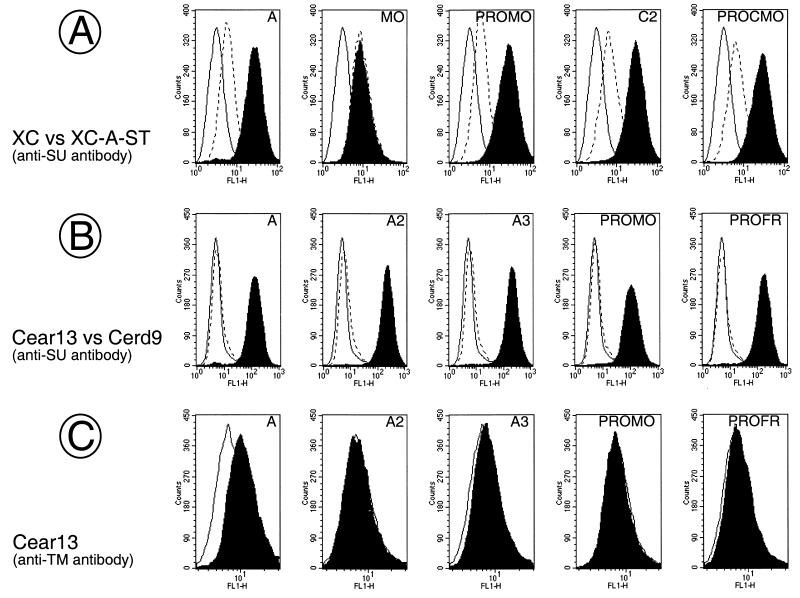
Binding assays of mutant envelopes with enhanced fusion activity. Binding assays were performed with supernatants of TELCeB6 cells transfected with the indicated envelope expression vectors depicted in Fig. 1, 4, and 7 on PiT-2-expressing XC cells and Cear13 cells (black area), on PiT-2-interfering XC-A-ST cells (broken lines, panel A) or on PiT-2-negative Cerd9 cells (broken lines, panel B), as indicated. The background fluorescence was provided by incubating XC or Cear13 cells with supernatant of nontransfected TELCeB6 cells (solid lines). The background fluorescence on XC-A-ST cells and on Cerd9 cells (not shown) was the same as that on XC cells and Cear13 cells, respectively. Incubated cells were stained with the indicated antienvelope antibodies. The envelope glycoprotein content of the different samples was normalized by immunoblotting of viral supernatant (A and B) or viral pellet (C).
We next examined the infectivity of cell-free virions harboring either parental, chimeric, or mutant envelopes. For the sake of clarity, infection resulting from Env-dependent viral entry is referred to here as virus-to-cell fusion. However, it should be noted that fusion, which can occur either directly at the level of the cell surface membrane or indirectly after internalization in endosomes (31, 33), requires other steps such as receptor binding and/or receptor internalization (14, 24). Surprisingly, the highly cell-to-cell fusogenic PROMO and PROFR chimeras and A2 and A3 point mutants yielded undetectable or significantly reduced titers on the cell lines tested, including rat, mouse, human, and PiT-2-transfected CHO hamster cells (Table 2 and data not shown). In contrast, virions pseudotyped with the remaining chimeric and mutant envelopes yielded similar titers ranging from 106 to 107 LacZ IU per ml on all cells tested, similar to both parental ecotropic and amphotropic virus titers (Table 2 and data not shown). All envelopes harboring the amphotropic BD were able to infect PiT-2-transfected CHO cells but not the parental CHO cells lacking PiT-2 (data not shown), thus indicating that PiT-2 was required for virion entry. Titration assays were then performed on the interfering XC-A-ST cells to assess whether modification of the PRR allowed virus-to-cell fusion when fewer PiT-2 receptors were available. Infectivity of parental amphotropic-pseudotyped virions was reduced by more than 1,000-fold in the interfering XC-A-ST cells, whereas almost all PRR infectious mutants were significantly more resistant to interference (Table 2). For example, infectivity of viruses carrying C2, C3, and C4 envelope glycoproteins with deletions in the carboxy-terminal end of the PRR was decreased by only approximately 10-fold, with titers remaining greater than 105 LacZ IU per ml (Table 2). These data indicated that although amphotropic envelopes carrying mutations at the carboxy terminus of the PRR did not exhibit increased infectivity on cells that had the wild-type number of PiT-2 receptors, they were more efficient than wild-type amphotropic virus in mediating virus entry in cells harboring few of available PiT-2 receptors.
TABLE 2.
Effects of virion-coating envelopes on viral infectiona
| Virion-coating envelope | Infectivityb on cells:
|
Level of interferencec | |
|---|---|---|---|
| XC | XC-A-ST | ||
| MO | 7.1 × 106 | 6.1 × 106 | 1 |
| A | 1 × 107 | 6.4 × 103 | 1,369 |
| PROMO | 2.2 × 101 | 1 × 101 | NA |
| PROFR | <1 × 101 | <1 × 101 | NA |
| A1 | 8.4 × 106 | 2.3 × 103 | 3,138 |
| A2 | <1 × 101 | <1 × 101 | NA |
| A3 | 7.8 × 101 | 3 × 101 | NA |
| A4 | 5.7 × 106 | 8.1 × 103 | 605 |
| C2 | 7.6 × 106 | 5.7 × 105 | 11 |
| C3 | 5.4 × 106 | 4.7 × 105 | 10 |
| C4 | 2.4 × 106 | 1.7 × 105 | 12 |
| C5 | 9.4 × 106 | 2.7 × 104 | 299 |
Results are from one representative experiment (of six experiments).
Infectivity is expressed as the number of LacZ IU per milliliter of viral supernatant.
Interference levels were calculated according to the following equation: (titer on XC cells/titer on XC-A-ST cells) × (MO titer on XC-A-ST cells/MO titer on XC cells). NA, not applicable.
The increased efficiency of these mutant envelopes in a virus-to-cell infection assay was not due to detectable differences in their processing, maturation, and virion incorporation properties (data not shown). Although we could not formally exclude the possibility that increased virus-to-cell fusion of PRR-mutated amphotropic envelopes on XC-A-ST cells was due to their interaction with an alternative receptor or coreceptor not recognized by the parental envelope, we found that parental and mutant envelopes bound XC-A-ST cells with the same efficiency (see below and Fig. 5A). Thus, even though carboxy-terminal PRR mutants did not show increased fusogenicity in cell-to-cell fusion assays (Fig. 4), they were more efficient in the virus-to-cell fusion assay (Table 2). As cell-to-cell fusion was affected only by mutations in the amino-terminal β-turns while virus-to-cell fusion was increased by mutations in the carboxy-terminal β-turns, our data indicate that these two regions of the PRR differentially modulate cell-to-cell membrane fusion and viral entry, most likely through interaction with PiT-2 receptors.
Cooperation between PRR and other SU domains in envelope stability and fusogenicity.
The ability of A2, A3, PROMO and PROFR mutants to drive cell-to-cell fusion but not virus-to-cell fusion (Tables 1 and 2) raised the possibility that changes introduced in these envelopes prevented interactions with the amphotropic PiT-2 receptor. To address this question, we performed binding assays by incubating supernatants of the various pseudotyped retroviruses on XC and XC-A-ST cells (Fig. 5A) as well as on PiT-2 amphotropic receptor-negative and -positive (PiT-2-transfected) Cerd9 and Cear13 CHO-derived cells, respectively (Fig. 5B and C). Cells were then stained with an anti-SU antibody in order to assess binding of the SU subunit to the receptor (10) or with an anti-TM antibody which allows recognition of envelope anchored to a viral particle (50). Assays performed with anti-SU antibodies showed that in comparison to the parental amphotropic envelope glycoproteins, binding specificity and affinity for PiT-2 were not significantly altered for the A2, A3, PROMO, PROFR, and the other mutant envelopes since they could bind with similar efficiencies to Cear13, but not Cerd9, cells (Fig. 5B). Similarly, no differences in binding could be found on XC cells between the mutant and wild-type amphotropic envelopes (Fig. 5A). Furthermore, endogenous expression of an interfering amphotropic BD in XC-A-ST cells decreased binding of all PRR-mutated and parental amphotropic envelopes with a similar efficiency (Fig. 5A). Additionally, staining with an anti-TM antibody revealed binding of virions harboring wild-type amphotropic envelope glycoproteins. However, no binding of viral particles generated with the A2, A3, PROMO, and PROFR envelopes was observed (Fig. 5C). These data indicated that although the SU of the latter mutants could fully recognize the PiT-2 receptor, it was not stably associated to virions, which probably explains their poor infectivity.
To assess whether our inability to detect an association between the SU and virions was due to an unstable SU-TM association in these mutants, immunoblotting was performed with cell membrane preparations, cell culture supernatants, and virions obtained from cells transfected with parental and mutant A2, A3, PROMO, and PROFR envelopes (Fig. 6). Two bands, corresponding to the unprocessed envelope precursor (PR) and the mature SU product, were detected in immunoblots of cell membrane preparations incubated with an anti-SU antibody (Fig. 6). In the case of the A2 and A3 point mutants, the migration positions of the two bands were the same as that of the parental amphotropic envelope glycoprotein. However, due to differences in the sizes and glycosylations of their respective PRRs (Fig. 4), the bands observed with the PROMO and PROFR chimeric envelope precursors exhibited faster mobilities, corresponding to expected molecular mass decreases of 15 and 6 kDa, respectively (Fig. 6). Although similar expression levels of the precursors were observed for all envelopes, as indicated by equivalent PR band intensities (Fig. 6) and pulse-chase labeling experiments (data not shown), the SU product was poorly detected in extracts from cells harboring A2, A3, PROMO, and PROFR mutants compared to parental amphotropic envelopes. This was due both to a less efficient precursor to SU maturation and to decreased stability of the latter mutant envelope glycoproteins compared to that of the wild-type amphotropic envelope (Fig. 6). Indeed, instability of these mutant chimeras was demonstrated by increased SU levels, indicative of shedding, in the culture medium and by low SU levels on the virions and cell membranes (Fig. 6). Thus, these data show the critical role of the potential amino-terminal MLV-4070A PRR second and third β-turns in SU-TM association and envelope conformation and suggest that the poor infectivity of the A2, A3, PROMO, and PROFR envelopes was due to increased SU shedding. In the A2, A3, PROMO, and PROFR mutants, increased SU shedding and decreased infectivity was concomitant with increased cell-to-cell fusion (Table 1). It is expected that increased shedding, resulting from changes in SU-TM interactions, leads to decreased virus-to-cell fusion due to altered interactions between virions and cell surface receptors. In contrast, SU shedding is not likely to significantly alter receptor interaction by cell surface-associated envelope, since SU-containing envelopes are continuously produced within the cell. Interestingly, in this cell context, a more unstable SU-TM interaction allowed a more efficient cell-to-cell fusion (Table 1).
FIG. 6.
Instability of mutant envelopes with enhanced cell-to-cell fusion activity. Cells were transfected with the indicated envelope expression vectors, and expression of both SU and envelope precursor (PR) was monitored in cell membrane preparations, cell culture supernatants, and viral pellets by immunoblotting with an anti-SU antiserum (Quality Biotech Inc.). Equivalent loading of viral pellets (bottom panel) was demonstrated by immunoblotting with an anti-capsid (CA) antiserum (Quality Biotech Inc.). The positions of PR, SU, and CA proteins are shown for MLV-4070A.
Increased envelope instability for A2, A3, PROMO, and PROFR envelopes suggested that a disruption of interactions between the PRR and other envelope regions occurred in these mutants and influenced both SU-TM interactions and fusogenicity. This is compatible with the finding that proline-rich sequences are involved in protein-protein interactions (55). Therefore, instability of the A2 and A3 mutants might be due to loss of envelope domain interactions critical for stability of the glycoprotein complex. Similarly, instability of the PROMO and PROFR chimeras might be due to nonoptimal interactions between the MoMLV PRR and the adjacent heterologous MLV-4070A domains. It is interesting to note that with the exception of the BD and PRR domains which have only 33 and 43% aa homology, respectively, all other ecotropic and amphotropic envelope domains are over 80% aa identical (Fig. 1A). Whereas BD hypervariability is linked to differences in the respective cognate receptors of these retroviruses, PRR hypervariability in MLVs and other mammalian type C retroviruses (25) might reflect the role of PRRs in adapting or facilitating interactions between the various adjacent domains of their envelopes.
To directly address the potential cooperation of PRR with distinct envelope domains, we generated chimeric glycoproteins in which the MoMLV PRR was associated with other MoMLV envelope domains in the context of an MLV-4070A background (Fig. 7). The combination of ecotropic PRR with the upstream ecotropic BD, as in BDPROMO, or in association with downstream ecotropic C and/or TM regions, as in PROCMO and PROCTMMO, led to increased envelope stability. This was demonstrated by the strong SU association to virions as monitored by immunoblotting (Table 3) and virion binding assays performed with anti-TM antibodies (data not shown). This observed increase in envelope stability provides evidence for a direct role of the PRR in stabilization of the envelope complex by allowing structural interactions between SU and TM proximal domains. This is in agreement with previous reports describing an influence of the MoMLV PRR on the conformation of the receptor binding domain (5, 6), stability of the SU-TM association (21, 57) and interactions between the amino- and carboxy-terminal domains of the MLV SU (35).
TABLE 3.
Characterization of chimeric envelope glycoproteins
| Envelopea | SU density on virionsb | Cell-cell fusionc | Infectivityd on cells:
|
Level of interferencee | |
|---|---|---|---|---|---|
| XC | XC-A-ST | ||||
| MO | ++ | + | 8 × 106 | 8 × 106 | 1 |
| A | ++ | − | 6 × 106 | 5.1 × 104 | 118 |
| PROMO | − | + | 4 × 101 | <1 × 101 | NA |
| BDPROMO | ++ | + | 2 × 106 | 2 × 106 | 1 |
| PROCMO | + | − | 2.2 × 106 | 2.7 × 105 | 8 |
| PROCTMMO | ++ | − | 2.3 × 106 | 4.5 × 104 | 50 |
Expression levels of SU glycoproteins on virion pellets as assessed by Western blot analysis. The ++, +, and − symbols indicate no difference, 1- to 10-fold-less SU expression, and less than 10-fold SU expression, respectively, compared to that of parental envelopes, respectively.
Fusion assays were performed by cocultivation of cells expressing the indicated envelope construct with XC cells. The presence (+) and absence (−) of syncytia are indicated.
Average titers of three experiments are shown. The standard errors did not exceed 30% of the titer values. Infectivity was expressed as LacZ IU per milliliter of viral supernatant.
Interference levels were calculated according to the following equation: (titer on XC cells/titer on XC-A-ST cells) × (MO titer on XC-A-ST cells/MO titer on XC cells). NA, not applicable.
With the exception of the BDPROMO chimera, whose high cell-to-cell fusion was most likely due to its ability to interact with the ecotropic receptor, the remaining chimeras demonstrated low cell-to-cell fusion concomitant to their increased stability (Table 3). These data therefore indicate a functional relationship between stability of the envelope complex and cell-to-cell fusion and suggest that the MLV PRR may control envelope conformational changes leading to fusion.
Additionally, as observed with the C2, C3, and C4 amphotropic mutant envelopes, viruses pseudotyped with the chimeric envelopes were highly infectious with titers of more than 106 LacZ IU per ml on XC cells (Table 3) and other cell types (data not shown). Specifically, titers obtained on XC-A-ST cells which have decreased levels of available amphotropic receptor were significantly higher for viruses harboring the PROCMO and PROCTMMO envelopes than for those carrying the parental amphotropic envelope (Table 3). Thus, in the two chimeras containing both ecotropic PRO and C domains with the amphotropic BD, the combination was sufficient both to prevent cell-to-cell fusion by maintaining envelope stability and to increase virus-to-cell fusogenicity.
DISCUSSION
Our results indicate that the PRR controls the transition of MuLV envelope glycoproteins from nonfusogenic to fusogenic conformations by controlling both the stability of the envelope complex and the thresholds required to trigger envelope-driven cell-to-cell fusion or virus-to-cell fusion. We have identified critical residues in the PRR that regulate these two functions. This is the first report of a retrovirus determinant in SU which passes on the fusion signal to the TM.
Modifications in the PRR of the amphotropic MLV envelope result in two different phenotypes: (i) high cell-to-cell fusion activity associated with decreased envelope stability and SU shedding and (ii) weak syncytium formation but increased virus-to-cell fusion associated with stability of the envelope glycoprotein complex. These two different phenotypes raise the possibility of a relationship between envelope (in)stability and cell-to-cell fusogenicity. Although others have previously noted that the requirements for cell-to-cell and virus-to-cell fusion differ (21, 36, 56–58), in this report we demonstrate a clear dissociation between the two phenomenons.
MLV PRR stabilizes envelope conformation.
Previous studies of chimeric MLV envelope glycoproteins have shown that although the MLV PRR is not directly involved in receptor recognition, it has an influence on the conformation of the receptor binding domain for certain strains of MLVs (5, 6). Other reports have also revealed the existence of an highly complex series of interactions between the different domains of MLV SUs, particularly between the N and C termini of mink cell focus-forming virus–MLV SU (35). Data in this report are in agreement with their results and furthermore suggest that one of the roles played by the PRR during retroviral infection is to stabilize a particular shape of the envelope glycoprotein, most probably by allowing structural interactions between protein domains which are proximal in the prefusogenic conformation of the envelope complex. A reason for the hypervariability of MLV PRR may therefore be that it provides to the glycoprotein complex a short adapter that accomodates subtle structural differences of protein domains between the different types of MLV envelopes, perhaps in relation with differential postbinding requirements. Indeed, while the PROMO envelope is not stable, the insertion of homologous MoMLV SU C-terminal (C and/or TM) or N-terminal (BD) domains confers stability to the chimeric glycoproteins (Table 3). The MLV TM ectodomain contains a leucine zipper that allows trimerization of the envelope complex (19). In addition, other subdomains of the MLV SU glycoprotein also contribute to the assembly and stability of the oligomer (48). Our data indicate that the MLV PRR may contain such determinants or, alternatively, may dictate a conformation of the glycoprotein that reveals other points of interaction in the envelope complex. In agreement with the results of others suggesting that the disruption of MoMLV PRR by linker insertion or mutagenesis led to instability of the envelope complex and to SU-TM dissociation (21, 57), in this study we identify amino acids (P245/N246 and P250/Q252) that are critical for the MLV-4070A envelope stability.
PRR structure-function relationship.
PRRs are among the most hypervariable regions found in the SU glycoproteins of MLVs and other mammalian type C retroviruses (25). However, the conservation of the sequence GPR(V/I)PIGPNP(I/L) at the amino termini of MLV PRRs suggests an important role for this subdomain. Indeed, recent findings of our laboratory indirectly revealed a particular property of the MLV PRR amino terminus. In this previous study (50), either the amino-terminal end or the whole amphotropic PRR were able to regulate the cooperation of two receptor binding domains between which it was inserted in a chimeric envelope glycoprotein. It seems likely that in the context of the wild-type envelope glycoprotein, the PRR is also able to regulate the necessary cooperation between the receptor binding domain and the fusion domain during virus entry.
The PRRs of mammalian type C retroviruses are likely to fold as regular and stable secondary structures. Limited proteolysis of MLV SU led to cleavage at both ends of the PRR (28) and suggested that it forms a separate domain of the glycoprotein which folds as a rigid structure. The regular arrangement of β-turns induced by the majority of the proline residues in type C mammalian retroviruses PRRs (MLV-4070A and MoMLV PRRs [Fig. 4D]) is compatible with their folding as polyproline β-turn helices (30). A recent report using synthetic peptide fragments derived from the feline leukemia virus A proline-rich region has shown that its PRR folds as a polyproline β-turn helix, a particularly ordered and stable structure which can self-assemble into complex ordered multimers (20). Moreover, the unusual properties of polyproline β-turn helices (49) may explain how the PRR might relay a fusion trigger following receptor binding. Indeed, a small deformation or movement induced by receptor interaction might be transmitted to the C-terminal fusion domain due to a major property of β-turn polyproline helices, development of elastomeric forces. This might trigger envelope fusion both by destabilizing the quaternary structure of the envelope complex and by unmasking SU C-terminal or TM epitopes required for membrane fusion. According to protein structure predictions, it seems possible that compared to its amphotropic counterpart, the MoMLV PRR is a more reactive polyproline β-turn helix (Fig. 4D). This may therefore explain the increased cell-to-cell or virus-to-cell fusion properties of amphotropic envelopes carrying a MoMLV PRR (Fig. 1 and 7 and Tables 1 and 3). Similarly, the tailoring of the MLV-4070A envelope, by disruption or removal of PRR β-turns (Fig. 4 and Tables 1 and 2), is also likely to increase its reactivity, thus providing such envelope mutants with their enhanced fusion phenotype.
MLV PRR plays a role in the initial fusion events.
Our data suggest that the highly fusogenic chimeras (PROMO, PROFR, A2, and A3) could induce cell-to-cell fusion as a result of their unstability and their propensity to shed from the envelope glycoprotein complex. This possibility would therefore rely on a very simple retroviral fusion mechanism whereby SU shedding is the primary cause of activation of the late steps of fusion and directly activates the membrane fusion properties of the TM subunit. However, although human immunodeficiency virus type 1 (HIV-1) TM expressed alone has been proposed to induce the formation of syncytia (37), which has been contested by others (29), it is difficult to draw up a direct relationship between shedding and fusion triggering. Indeed, while they were as fusogenic as PROMO and A2 envelopes, the PROFR and A3 chimeras seemed slightly more stable than the latter (Table 1 and Fig. 6). In addition, several unstable mutant MLV envelope glycoproteins have been described in the literature (21, 27, 57), but to our knowledge, this phenotype has never been correlated with an increased fusion activity. Moreover, envelope glycoproteins containing such constitutively active TM glycoproteins would be very toxic for the cells and would have prevented their stable expression. It is therefore likely that, similarly to HIV-1 (47), SU shedding is an indirect reflect of the fusion reaction and is a final consequence of conformational changes that occur in the envelope complex during the fusion pathway. Thus, the two critical functions of the PRR are most probably first, to induce a stable conformation of the SU which is required to control its fusogenic activity, and second, to facilitate structural rearrangements of the envelope complex following receptor binding. The envelope chimeras containing the structural modifications of the PRR that we describe here display a lower activation threshold for fusion and probably require less interaction with retroviral receptors to trigger membrane fusion. Thus, MLV SU PRR is most likely a fusion regulator rather than a positive fusion determinant, and it can be proposed that the MLV PRR regulates the transition between two conformations of the envelope glycoprotein: pre- and postreceptor binding. Our results are therefore consistent with a model of fusion in which interaction of the glycoprotein trimer with the retroviral receptor induces rearrangements of the envelope complex and, ultimately, SU shedding, a process which is controlled or at least facilitated by the PRR, leading to the late steps of membrane fusion and to recruitment of the membrane fusion properties of the TM subunit.
Applications to gene transfer technologies.
Envelope glycoproteins that mediate efficient virus entry even at very low PiT-2 receptor density will be of interest for certain gene therapy applications. Here, we describe several amphotropic envelopes with mutations in the carboxy terminus of the PRR which require a lower threshold to trigger virus-to-cell fusion. Upon infection of cells with low levels of amphotropic receptors, these mutants exhibit 100-fold-higher titers than those of the parental retroviruses. Low transduction efficiency of human target cells, hematopoietic cell progenitors for example, with retroviral vectors has been a recurring theme in human gene therapy trials and is thought to be due in part to a low density of PiT-2 receptors (13, 52). Packaging of vectors harboring the novel envelope glycoproteins reported here may allow for more efficient gene delivery to human cells with low levels of amphotropic receptors.
Numerous studies in several laboratories have aimed to retarget the tropism of type C retroviruses (11). Although the retargeting of retrovirus binding has generally been easily achieved via N-terminal extensions on MLV envelope glycoproteins with different ligand types, such as cytokines and single-chain antibodies, such envelope chimeras display an intrinsically low fusogenicity and hence are poorly infectious (10, 50). Results in this report may therefore provide a basis to engineer the fusion activity of retroviral vectors carrying intact or retargeted amphotropic MLV envelopes.
ACKNOWLEDGMENTS
We thank Edwige Delahaye for expert technical assistance and Naomi Taylor for critical reading of the manuscript.
This work was supported by Agence Nationale pour la Recherche contre le SIDA (ANRS), Association pour la Recherche contre le Cancer (ARC), Association Française de Lutte contre la Mucoviscidose (AFLM), Centre National de la Recherche Scientifique (CNRS), and Institut National de la Santé et de la Recherche Médicale (INSERM). M.S. is supported by INSERM and grants from CNRS (ATIPE), Fondation pour la Recherche Médicale (FRM) (Jeune Équipe) and ARC. S.J.R. is supported by the Medical Research Council (MRC).
REFERENCES
- 1.Albritton L M, Tseng L, Scadden D, Cunningham J M. A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell. 1989;57:659–666. doi: 10.1016/0092-8674(89)90134-7. [DOI] [PubMed] [Google Scholar]
- 2.Alterman L A, Crispe I N, Kinnon C. Characterization of the murine heat-stable antigen: an hematolymphoid differentiation antigen defined by the J11d, M1/69 and B2A2 antibodies. Eur J Immunol. 1990;20:1597–1602. doi: 10.1002/eji.1830200728. [DOI] [PubMed] [Google Scholar]
- 3.Andersen K B. A domain of murine retrovirus surface protein gp70 mediates cell fusion, as shown in a novel SC-1 cell fusion system. J Virol. 1994;68:3175–3182. doi: 10.1128/jvi.68.5.3175-3182.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bae Y, Kingsman S M, Kingsman A J. Functional dissection of the Moloney murine leukemia virus envelope protein gp70. J Virol. 1997;71:2092–2099. doi: 10.1128/jvi.71.3.2092-2099.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Battini J L, Danos O, Heard J M. Receptor-binding domain of murine leukemia virus envelope glycoproteins. J Virol. 1995;69:713–719. doi: 10.1128/jvi.69.2.713-719.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Battini J L, Heard J M, Danos O. Receptor choice determinants in the envelope glycoproteins of amphotropic, xenotropic, and polytropic murine leukemia viruses. J Virol. 1992;66:1468–1475. doi: 10.1128/jvi.66.3.1468-1475.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Battini J L, Rodrigues P, Müller R, Danos O, Heard J-M. Receptor-binding properties of a purified fragment of the 4070A amphotropic murine leukemia virus envelope glycoprotein. J Virol. 1996;70:4387–4393. doi: 10.1128/jvi.70.7.4387-4393.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chesebro B, Wehrly K, Cloyd M, Britt W, Portis J, Collins J, Nishio J. Characterization of mouse monoclonal antibodies specific for Friend murine leukemia virus-induced erythroleukemia cells: Friend-specific and FMR-specific antigens. Virology. 1981;112:131–144. doi: 10.1016/0042-6822(81)90619-x. [DOI] [PubMed] [Google Scholar]
- 9.Chou P Y, Fasman G D. Prediction of the secondary structure of proteins from their amino acid sequence. Annu Rev Biochem. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- 10.Cosset F-L, Morling F J, Takeuchi Y, Weiss R A, Collins M K L, Russell S J. Retroviral retargeting by envelopes expressing an N-terminal binding domain. J Virol. 1995;69:6314–6322. doi: 10.1128/jvi.69.10.6314-6322.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cosset F-L, Russell S J. Targeting retrovirus entry. Gene Ther. 1996;3:946–956. [PubMed] [Google Scholar]
- 12.Cosset F-L, Takeuchi Y, Battini J L, Weiss R A, Collins M K L. High-titer packaging cells producing recombinant retroviruses resistant to human serum. J Virol. 1995;69:7430–7436. doi: 10.1128/jvi.69.12.7430-7436.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crooks G M, Kohn D B. Growth factors increase amphotropic retrovirus binding to human CD34+ bone marrow progenitor cells. Blood. 1993;82:3290–3297. [PubMed] [Google Scholar]
- 14.Cudmore S, Reckmann I, Way M. Viral manipulations of the actin cytoskeleton. Trends Microbiol. 1997;5:142–148. doi: 10.1016/S0966-842X(97)01011-1. [DOI] [PubMed] [Google Scholar]
- 15.Denesvre C, Carrington C, Corbin A, Takeuchi Y, Cosset F-L, Schulz T, Sitbon M, Sonigo P. TM domain swapping of murine leukemia virus envelopes confers different infectious abilities despite similar incorporation into virions. J Virol. 1996;70:4380–4386. doi: 10.1128/jvi.70.7.4380-4386.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denesvre C, Sonigo P, Corbin A, Ellerbrok H, Sitbon M. Influence of transmembrane domains on the fusogenic abilities of human and murine leukemia retrovirus envelopes. J Virol. 1995;69:4149–4157. doi: 10.1128/jvi.69.7.4149-4157.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evans L H, Morrison R P, Malik F G, Portis J, Britt W J. A neutralizable epitope common to the envelope glycoproteins of ecotropic, polytropic, xenotropic, and amphotropic murine leukemia viruses. J Virol. 1990;64:6176–6183. doi: 10.1128/jvi.64.12.6176-6183.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fass D, Davey R A, Hamson C A, Kim P S, Cunningham J M, Berger J M. Structure of a murine leukemia virus receptor-binding glycoprotein at 2.0 angstrom resolution. Science. 1997;277:1662–1666. doi: 10.1126/science.277.5332.1662. [DOI] [PubMed] [Google Scholar]
- 19.Fass D, Harrison S C, Kim P S. Retrovirus envelope domain at 1.7 A resolution. Nat Struct Biol. 1996;3:465–469. doi: 10.1038/nsb0596-465. [DOI] [PubMed] [Google Scholar]
- 20.Fontenot J D, Tjandra N, Ho C, Andrews P C, Montelaro R C. Structure and self assembly of a retrovirus (FeLV) proline rich neutralization domain. J Biomol Struct Dyn. 1994;11:821–837. doi: 10.1080/07391102.1994.10508035. [DOI] [PubMed] [Google Scholar]
- 21.Gray K D, Roth M J. Mutational analysis of the envelope gene of Moloney murine leukemia virus. J Virol. 1993;67:3489–3496. doi: 10.1128/jvi.67.6.3489-3496.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hunter E, Swanstrom R. Retrovirus envelope glycoproteins. Curr Top Microbiol Immunol. 1990;157:187–253. doi: 10.1007/978-3-642-75218-6_7. [DOI] [PubMed] [Google Scholar]
- 23.Jones J S, Risser R. Cell fusion induced by the murine leukemia virus envelope glycoprotein. J Virol. 1993;67:67–74. doi: 10.1128/jvi.67.1.67-74.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kizhatil K, Albritton L M. Requirements for different components of the host cell cytoskeleton distinguish ecotropic murine leukemia virus entry via endocytosis from entry via surface fusion. J Virol. 1997;71:7145–7156. doi: 10.1128/jvi.71.10.7145-7156.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koch W, Hunsmann G, Friedrich R. Nucleotide sequence of the envelope gene of Friend murine leukemia virus. J Virol. 1983;45:1–9. doi: 10.1128/jvi.45.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozak S L, Siess D C, Kavanaugh M P, Miller A D, Kabat D. The envelope glycoprotein of an amphotropic murine retrovirus binds specifically to the cellular receptor/phosphate transporter of susceptible species. J Virol. 1995;69:3433–3440. doi: 10.1128/jvi.69.6.3433-3440.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Z, Pinter A, Kayman S C. The critical N-linked glycan of murine leukemia virus envelope protein promotes both folding of the C-terminal domains of the precursor polyprotein and stability of the postcleavage envelope complex. J Virol. 1997;71:7012–7019. doi: 10.1128/jvi.71.9.7012-7019.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Linder M, Wenzel V, Linder D, Stirm S. Structural elements in glycoprotein 70 from polytropic Friend mink cell focus-inducing virus and glycoprotein 71 from ecotropic Friend murine leukemia virus, as defined by disulfide-bonding pattern and limited proteolysis. J Virol. 1994;68:5133–5141. doi: 10.1128/jvi.68.8.5133-5141.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marcon L, Sodroski J. gp120-independent fusion mediated by the human immunodeficiency virus type 1 gp41 envelope glycoprotein: a reassessment. J Virol. 1994;68:1977–1982. doi: 10.1128/jvi.68.3.1977-1982.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsushima N, Creutz C E, Kretsinger R H. Polyproline, beta-turn helices. Novel secondary structures proposed for the tandem repeats within rhodopsin, synaptophysin, synexin, gliadin, RNA polymerase II, hordein, and gluten. Proteins. 1990;7:125–155. doi: 10.1002/prot.340070204. [DOI] [PubMed] [Google Scholar]
- 31.McClure M O, Sommerfelt M A, Marsh M, Weiss R A. The pH independence of mammalian retrovirus infection. J Gen Virol. 1990;71:767–773. doi: 10.1099/0022-1317-71-4-767. [DOI] [PubMed] [Google Scholar]
- 32.Miller D G, Edwards R H, Miller A D. Cloning of the cellular receptor for amphotropic murine retroviruses reveals homology to that for gibbon ape leukemia virus. Proc Natl Acad Sci USA. 1994;91:78–82. doi: 10.1073/pnas.91.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nussbaum O, Roop A, Anderson W F. Sequences determining the pH dependence of viral entry are distinct from the host range-determining region of the murine ecotropic and amphotropic retrovirus envelope proteins. J Virol. 1993;67:7402–7405. doi: 10.1128/jvi.67.12.7402-7405.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ott D, Friedrich R, Rein A. Sequence analysis of amphotropic and 10A1 murine leukemia virus: close relationship to mink cell focus-forming viruses. J Virol. 1990;64:757–766. doi: 10.1128/jvi.64.2.757-766.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ott D, Rein A. Basis for receptor specificity of nonecotropic murine leukemia virus surface glycoprotein gp70. J Virol. 1992;66:4632–4638. doi: 10.1128/jvi.66.8.4632-4638.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park B H, Matuschke B, Lavi E, Gaulton G N. A point mutation in the env gene of a murine leukemia virus induces syncytium formation and neurologic disease. J Virol. 1994;68:7516–7524. doi: 10.1128/jvi.68.11.7516-7524.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perez L G, O’Donnell M A, Stephens E B. The transmembrane glycoprotein of human immunodeficiency virus type 1 induces syncytium formation in the absence of the receptor binding glycoprotein. J Virol. 1992;66:4134–4143. doi: 10.1128/jvi.66.7.4134-4143.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pinter A, Chen T-E, Lowy A, Cortez N G, Siligari S. Ecotropic murine leukemia virus-induced fusion of murine cells. J Virol. 1986;57:1048–1054. doi: 10.1128/jvi.57.3.1048-1054.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pinter A, Kopelman R, Li Z, Kayman S C, Sanders D A. Localization of the labile disulfide bond between SU and TM of the murine leukemia virus envelope protein complex to a highly conserved CWLC motif in SU that resembles the active-site sequence of thiol-disulfide exchange enzymes. J Virol. 1997;71:8073–8077. doi: 10.1128/jvi.71.10.8073-8077.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ragheb J A, Anderson W F. pH-independent murine leukemia virus ecotropic envelope-mediated cell fusion: implications for the role of the R peptide and p12E TM in viral entry. J Virol. 1994;68:3220–3231. doi: 10.1128/jvi.68.5.3220-3231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ragheb J A, Yu H, Hofmann T, Anderson W F. The amphotropic and ecotropic murine leukemia virus envelope TM subunits are equivalent mediators of direct membrane fusion: implications for the role of the ecotropic envelope and receptor in syncytium formation and viral entry. J Virol. 1995;69:7205–7215. doi: 10.1128/jvi.69.11.7205-7215.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rein A. Interference grouping of murine leukemia viruses: a distinct receptor for the MCF-recombinant viruses in mouse cells. Virology. 1982;120:251–257. doi: 10.1016/0042-6822(82)90024-1. [DOI] [PubMed] [Google Scholar]
- 43.Rein A, Mirro J, Haynes J G, Ernst S M, Nagashima K. Function of the cytoplasmic domain of a retroviral transmembrane protein: p15E-p2E cleavage activates the membrane fusion capability of the murine leukemia virus Env protein. J Virol. 1994;68:1773–1781. doi: 10.1128/jvi.68.3.1773-1781.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shinnick T M, Lerner R A, Sutcliffe J G. Nucleotide sequence of Moloney murine leukemia virus. Nature (London) 1981;293:543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- 45.Skehel J J, Bayley P M, Brown E B, Martin S R, Waterfield M D, White J M, Wilson I A, Wiley D C. Changes in the conformation of influenza virus hemagglutinin at the pH optimum of virus-mediated membrane fusion. Proc Natl Acad Sci USA. 1982;79:968–972. doi: 10.1073/pnas.79.4.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takeuchi Y, Cosset F L, Lachmann P J, Okada H, Weiss R A, Collins M K L. Type C retrovirus inactivation by human complement is determined by both the viral genome and producer cell. J Virol. 1994;68:8001–8007. doi: 10.1128/jvi.68.12.8001-8007.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thali M, Furman C, Helseth E, Repke H, Sodroski J. Lack of correlation between soluble CD4-induced shedding of the human immunodeficiency virus type 1 exterior envelope glycoprotein and subsequent membrane fusion events. J Virol. 1992;66:5516–5524. doi: 10.1128/jvi.66.9.5516-5524.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tucker S P, Srinivas R V, Compans R W. Molecular domains involved in oligomerization of the Friend murine leukemia virus envelope glycoprotein. Virology. 1991;185:710–720. doi: 10.1016/0042-6822(91)90542-j. [DOI] [PubMed] [Google Scholar]
- 49.Urry D W. Entropic elastic processes in protein mechanisms. I. Elastic structure due to an inverse temperature transition and elasticity due to internal chain dynamics. J Protein Chem. 1988;7:1–34. doi: 10.1007/BF01025411. [DOI] [PubMed] [Google Scholar]
- 50.Valsesia-Wittmann S, Morling F J, Hatziioannou T, Russell S J, Cosset F-L. Receptor co-operation in retrovirus entry: recruitment of an auxiliary entry mechanism after retargeted binding. EMBO J. 1997;16:1214–1223. doi: 10.1093/emboj/16.6.1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.VanZeijl M, Johann S V, Cross E, Cunningham J, Eddy R, Shows T B, O’Hara B. An amphotropic virus receptor is a second member of the gibbon ape leukemia virus receptor family. Proc Natl Acad Sci USA. 1994;91:1168–1172. doi: 10.1073/pnas.91.3.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.von Laer D, Thomsen S, Vogt B, Donath M, Kruppa J, Rein A, Ostertag W, Stocking C. Entry of amphotropic and 10A1 pseudotyped murine retroviruses is restricted in hematopoietic stem cell lines. J Virol. 1998;72:1424–1430. doi: 10.1128/jvi.72.2.1424-1430.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weiss R A, Tailor C S. Retrovirus receptors. Cell. 1995;82:531–533. doi: 10.1016/0092-8674(95)90024-1. [DOI] [PubMed] [Google Scholar]
- 54.White J M. Membrane fusion. Science. 1992;258:917–924. doi: 10.1126/science.1439803. [DOI] [PubMed] [Google Scholar]
- 55.Williamson M P. The structure and function of proline-rich regions in proteins. Biochem J. 1994;297:249–260. doi: 10.1042/bj2970249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilson C A, Marsh J W, Eiden M V. The requirements for viral entry differ from those for virally induced syncytium formation in NIH 3T3/DTras cells exposed to Moloney murine leukemia virus. J Virol. 1992;66:7262–7269. doi: 10.1128/jvi.66.12.7262-7269.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu B W, Cannon P M, Gordon E M, Hall F L, Anderson W F. Characterization of the proline-rich region of murine leukemia virus envelope protein. J Virol. 1998;72:5383–5391. doi: 10.1128/jvi.72.7.5383-5391.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu N L, Cannon P M, Chen D, Anderson W F. Mutational analysis of the fusion peptide of Moloney murine leukemia virus transmembrane protein p15E. J Virol. 1998;72:1632–1639. doi: 10.1128/jvi.72.2.1632-1639.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]