Abstract
NS1, the major nonstructural protein of the parvovirus minute virus of mice, is a multifunctional phosphoprotein which is involved in cytotoxicity, transcriptional regulation, and initiation of viral DNA replication. For coordination of these various functions during virus propagation, NS1 has been proposed to be regulated by posttranslational modifications, in particular phosphorylation. Recent in vitro studies (J. P. F. Nüesch, R. Corbau, P. Tattersall, and J. Rommelaere, J. Virol. 72:8002–8012, 1998) provided evidence that distinct NS1 activities, notably the intrinsic helicase function, are modulated by the phosphorylation state of the protein. In order to study the dependence of the initiation of viral DNA replication on NS1 phosphorylation and to identify the protein kinases involved, we established an in vitro replication system that is devoid of endogenous protein kinases and is based on plasmid substrates containing the minimal left-end origins of replication. Cellular components necessary to drive NS1-dependent rolling-circle replication (RCR) were freed from endogenous serine/threonine protein kinases by affinity chromatography, and the eukaryotic DNA polymerases were replaced by the bacteriophage T4 DNA polymerase. While native NS1 (NS1P) supported RCR under these conditions, dephosphorylated NS1 (NS1O) was impaired. Using fractionated HeLa cell extracts, we identified two essential protein components which are able to phosphorylate NS1O, are enriched in protein kinase C (PKC), and, when present together, reactivate NS1O for replication. One of these components, containing atypical PKC, was sufficient to restore NS1O helicase activity. The requirement of NS1O reactivation for characteristic PKC cofactors such as Ca2+/phosphatidylserine or phorbol esters strongly suggests the involvement of this protein kinase family in regulation of NS1 replicative functions in vitro.
Minute virus of mice (MVM) is the prototype of the genus Parvovirus. Members of this genus consist of nonenveloped spherical particles of about 20 to 24 nm in diameter, comprising a linear single-strand DNA genome of approximately 5.1 kb. Parvovirus DNA encodes the two structural (VP) and at least four nonstructural (NS) polypeptides, of which the 83-kDa nuclear phosphoprotein NS1 is the only viral product necessary for viral DNA replication in all cell types (22, 54). Replication of the parvovirus genome involves the formation of monomeric and concatemeric duplex DNA intermediates. These replicative forms are produced by an unidirectional, single-strand copy mechanism (for a review, see reference 16) which resembles the rolling-circle replication (RCR) mechanism described for single-stranded DNA plasmids, bacteriophages, and geminiviruses (for a review, see reference 35). After conversion of the single-strand genome into a monomeric duplex, which is executed solely by cellular components (3), replication initiates at site-specific, single-strand nicks introduced by the viral NS1 protein into origin sequences located at either end of the genome (14, 17, 18). This cleavage reaction leaves NS1 covalently attached to the 5′ end at the nick site and generates a base-paired 3′ hydroxyl group which serves as a primer for DNA synthesis (8, 13, 19, 59).
The minimal origin sequence at the left-end telomere has been mapped and consists of approximately 50 bp located within the stem of the Y-shaped terminal structure (13). This sequence comprises binding sites for the cellular component PIF (parvovirus initiation factor) (9) and for NS1 (21) and an NS1 nick site (13). The NS1 binding and nick sites are separated by an AT-rich sequence, which most likely facilitates local unwinding during the nicking reaction. In the left-end hairpin structure of the genome, between the binding sites for PIF and NS1, there is a functionally important mismatched “bubble,” with a 5′-GAA-3′ triplet on one strand opposite a 5′-GA-3′ doublet on the other strand. When replication through the hairpin unfolds and copies the palindrome, a double-strand intermediate is generated, in which these tri- and dinucleotide sequences are located on either side of the axis of symmetry. Although the origin sequences of both arms are nearly identical, only the arm containing the GA dinucleotide serves as an active origin for NS1-mediated RCR, while the trinucleotide-containing counterpart remains silent (13).
Besides its key role as the initiator protein for viral DNA replication, NS1 is essential for several additional processes during the viral life cycle. In particular, the NS1 protein is a strong trans activator of the parvovirus P38 promoter that controls capsid gene expression (64). Furthermore, NS1 trans regulates nonparvovirus promoters (27, 70), and it exerts cytotoxic and/or cytostatic effects for which oncogene-transformed cells appear to be preferential targets (5, 7, 53). To account for the temporal coordination of these various functions during virus multiplication, NS1 has been proposed to be regulated by posttranslational modifications such as phosphorylation. NS1 was indeed found to be phosphorylated in infected cells (2, 11, 20, 51). Moreover, recent in vitro studies have shown that HeLa cell-derived native NS1 differs from its dephosphorylated counterpart in its capacity for distinct biochemical activities involved in viral DNA replication (60).
In order to study the effect of phosphorylation on NS1-driven initiation of DNA replication, we used a previously described RCR system, which is based on plasmid substrates containing the minimal left-end origins of replication (8, 13). This system was modified to deplete its protein components from endogenous kinases, allowing purified native NS1 (NS1P) to be compared with dephosphorylated NS1 (NS1O) with regard to their respective replication activities. In contrast to standard HeLa cell extracts, the kinase-free replication system was severely impaired in its ability to support RCR when supplied with NS1O as compared with NS1P. In reactivation experiments, the combination of two distinct protein fractions from HeLa cell extracts proved to be able to restore at least in part the replication activity of NS1O in the kinase-free system. This reactivation was dependent upon the presence of either acid lipids and Ca2+ or the phorbol ester 12-O-tetradecanoylphorbol-13-acetate (TPA). This dependence on cofactors, together with the capacity of both fractions to phosphorylate NS1O in vitro, strongly suggests that members of the protein kinase C (PKC) family are responsible for regulation of the NS1 replicative functions. Previous analyses of selected biochemical activities of native NS1P versus NS1O polypeptides have shown that the intrinsic helicase function of the viral product is strikingly dependent upon phosphorylation (60). One of the protein components necessary to rescue the replication activity of NS1O in the kinase-free system, which was enriched in atypical PKC, was also found to reactivate the helicase function of NS1O.
MATERIALS AND METHODS
Viruses and cells.
Recombinant vaccinia viruses were propagated in monolayer cultures of BSC-40 cells, collected, and purified over a sucrose cushion as described previously (41), except for the release of virus from infected cells, which was achieved by three cycles of freezing and thawing instead of sonication. Recombinant vaccinia viruses were constructed as previously described (57). The 293 cell line was adapted to suspension and grown in spinner bottles with Joklik’s medium supplemented with 10% fetal calf serum. HeLa-S3 cells were grown in spinner bottles in the presence of 5% fetal calf serum.
Production and purification of native and dephosphorylated NS1.
Wild-type NS1 and mutant NS1 were produced from recombinant vaccinia viruses in suspension cultures of HeLa-S3 cells and harvested at 18 h postinfection (57, 60). His-tagged NS1 present in nuclear extracts was dephosphorylated, or not, with calf intestine alkaline phosphatase and purified immediately on Ni2+-NTA agarose columns (60). NS1 preparations were analyzed by discontinuous sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and proteins were detected by Coomassie blue staining. All NS1 preparations were tested for activities in various in vitro assays.
Plasmids.
Plasmid pTMHis is a derivative of pTM-1 (52), which allows the expression of N-terminal His6-tagged proteins. The presence of a unique NcoI site allows the in-frame cloning of the gene of interest starting from its initiation codon. pTMHis was constructed by insertion of the annealed oligonucleotide pair 5′-CATGCACCATCACCACCATCACGCCATGGAATTC-3′ and 5′-GAATTCCATGGCGTGATGGTGGTGATGGTG-3′ into the NcoI- and SmaI-cleaved pTM-1 vector. The plasmid used to obtain recombinant vaccinia viruses expressing His-tagged PKCα was constructed by insertion of the full-length human PKCα-coding sequence (30) into NcoI- and EcoRI-cleaved pTMHis. Plasmids used as templates for in vitro replication assays were pL1-2TC and pL1-2GAA, containing the minimal active left-end MVM origin and the corresponding inactive origin, respectively (13). The bacterial expression plasmid pYT202am, containing the MVM sequence from nucleotide (nt) 225 to 534, served to produce peptides for rabbit immunization and generation of NS-specific antisera (23). pQE-PKCγ was constructed by insertion of the BamHI-to-SmaI fragment (nt 1332 to 1956) of human PKCγ cDNA (37) into pQE-30 (Qiagen). pQE-PKCζ was produced by insertion of the HindII-to-BamHI fragment (nt 981 to 1403) of human PKCζ cDNA (34) into pQE-32 (Qiagen).
Purification of peptides and production of antisera.
Peptides from pYT202amNS, pQE-PKCγ, or pQE-PKCζ were expressed overnight in the presence of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), extracted, and purified as described previously (23). Antisera were produced by multiple injections into rabbits. For Western blot analyses with anti-PKCγ (αPKCγ) or αPKCζ antibodies, IgGs were affinity purified on peptide columns by using the immunizing peptides (31).
Preparation of l-threonine and protamine affinity columns.
Affinity chromatography columns were prepared by coupling l-threonine or protamine sulfate with NIH-activated Hi-Trap columns (5 ml; Pharmacia) according to the manufacturer’s instructions. l-Threonine (Sigma) or protamine chloride (Sigma) was dissolved in coupling buffer (0.1 M NaCO3 [pH 8.3], 0.5 M NaCl) at 10 mg/ml and allowed to interact with the column material for 1 h at room temperature by recirculation.
Protein extraction and fractionation by column chromatography.
S100 extracts from 293S and HeLa cells were prepared and fractionated on phosphocellulose columns to obtain fractions P1, P2, and P3 as described previously (8, 68), except that P3 was eluted at 1 M NaCl (see Fig. 2A). To remove endogenous serine/threonine kinases, P1 from a 10-liter suspension culture of 293 cells was further purified on a 5-ml l-threonine affinity Hi-Trap column in buffer A (25 mM Tris [pH 7.5], 1 mM EDTA, 1 mM dithiothreitol [DTT], 174 μg of phenylmethylsulfonyl fluoride [PMSF] per ml, 10% glycerol) containing 150 mM NaCl. Individual flowthrough fractions were dialyzed against buffer B (20 mM HEPES [pH 7.5], 5 mM MgCl2, 5 mM KCl, 0.1 mM DTT, 17.4 μg of PMSF per ml, 10% glycerol, 20% sucrose) overnight at 4°C and stored in aliquots at −80°C. To determine the extent of purification, all fractions were tested for their ability to phosphorylate NS1O in in vitro kinase assays. P2-pol was obtained by P2 fractionation on DE52 columns. P2 from a 10-liter HeLa cell culture was loaded on a DE52 column (5 ml of resin/liter of original culture) in buffer A containing 50 mM NaCl. After thorough washing with the same buffer, P2-pol was eluted with buffer A containing 1 M NaCl. The eluate was dialyzed against buffer B and frozen in aliquots.
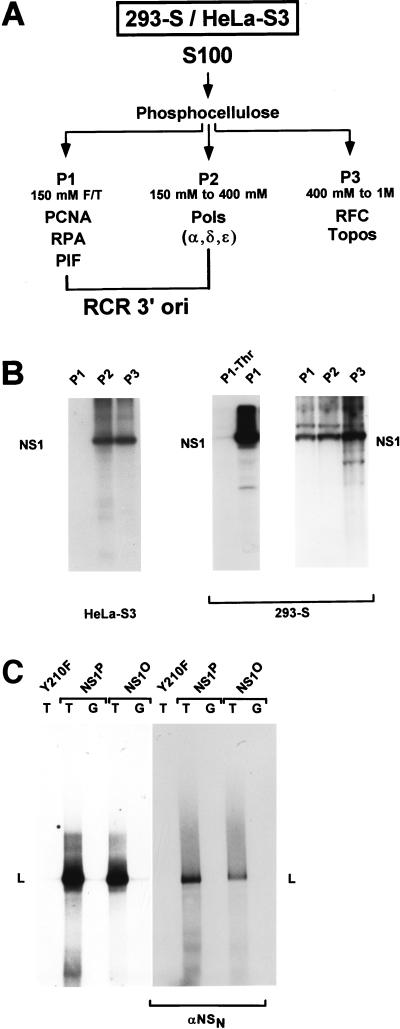
FIG. 2.
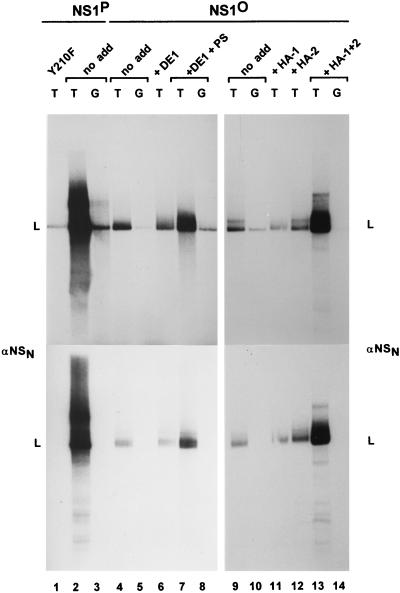
Fractionation of cell extracts by phosphocellulose chromatography. S100 replication extracts were prepared from 293-S or HeLa-S3 cells and fractionated on phosphocellulose columns as described by Tsurimoto and Stillman (68). (A) Scheme of replication extract fractionation into P1, P2, and P3, with the expected distribution of known replication factors (8, 55, 68). The NaCl concentrations used for elution are indicated. F/T, flowthrough; PCNA, proliferating cell nuclear antigen; RPA, replicator protein A (also designated RF-A or human single-stranded-DNA-binding protein); PIF, parvovirus initiation factor; Pols, eukaryotic DNA polymerases; RFC: replication factor C; Topos, eukaryotic topoisomerases. As indicated, P1 plus P2 have been shown to contain all cellular components necessary to support NS1-dependent RCR from the active left-end (3′) origin (ori) (8). (B) Detection of NS1O-phosphorylating protein kinases in the distinct phosphocellulose fractions. NS1O was incubated with the indicated fractions in the presence of [γ-32P]ATP for 30 min at 37°C and analyzed by SDS-PAGE after immunoprecipitation with αNSN (23). The migration of NS1 is indicated. P1-Thr corresponds to the P1 fraction from 293-S cells after purification on l-threonine affinity columns. (C) Comparison of NS1P and NS1O in RCR assays with P1-Thr derived from 293 cells plus P2-pol derived from HeLa cells, with plasmid templates containing the minimal active (T) or inactive (G) left-end origin. Since NS1O has been shown to be activated for helicase activity by members of the PKC family (60), the reactions were carried out in the presence of the PKC cofactors Ca2+ and PS. The replication-deficient NS1 mutant Y210F was used as a negative control. Linearized, labeled reaction products (L) were analyzed by 0.8% agarose gel electrophoresis, either directly after proteinase K treatment (left panel) or after immunoprecipitation with αNSN (right panel).
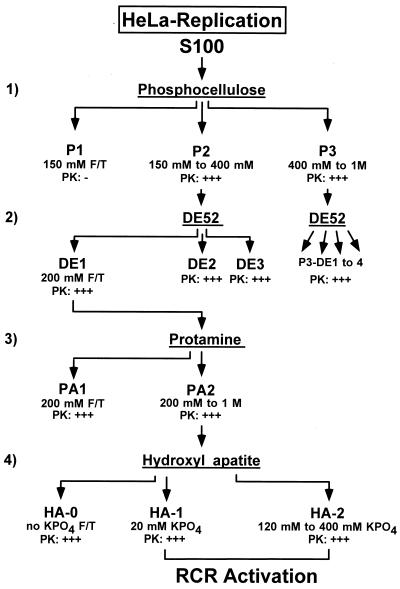
Protein kinases present in the HeLa cell-derived P2 fraction were further purified by consecutive anion-exchange, protamine affinity, and hydroxylapatite chromatographies (see Fig. 4). (i) P2 was adjusted to 200 mM NaCl and fractionated on a DE52 column (step 2). The flowthrough at 200 mM NaCl (DE-1) and the elution fractions DE-2 (200 to 500 mM NaCl) and DE-3 (500 mM to 1 M NaCl) were collected. All fractions were dialyzed against buffer B, frozen, and stored at −80°C. The protein kinases necessary to achieve extensive reactivation of NS1O in replication assays were found to be confined to fraction DE-1. (ii) PKC family members contained in DE-1 were further purified by protamine affinity chromatography (step 3), using a fast performance liquid chromatography (FPLC) system (Pharmacia). DE-1 (corresponding to a 6-liter culture) was loaded on a 5-ml protamine Hi-Trap column with a constant flow rate of 0.5 ml/min. After collection of the flowthrough (PA-1), the column was washed with buffer C (20 mM HEPES [pH 7.5], 1 mM EDTA, 0.1 mM DTT, 10% glycerol) containing 200 mM NaCl. The PKC-containing fraction PA-2 was then eluted with buffer C containing 1 M NaCl and the protease inhibitors PMSF (174 μg/ml), leupeptin (1 μg/ml), and aprotinin (1 μg/ml). PA-2 was dialyzed, adjusted to 50% glycerol, and stored in aliquots at −80°C. (iii) PKC isoforms present in fraction PA-2 were separated by FPLC on hydroxylapatite columns (step 4). PA-2 (corresponding to a 3-liter culture) was adjusted to 200 mM NaCl, loaded on a 5-ml hydroxylapatite column (Merck) with constant flux (0.5 ml/min), and washed with 30 ml of buffer C containing 50 mM NaCl. After collection of the flowthrough and wash, the HA-1 fraction was eluted from the column with buffer D (150 mM NaCl, 20 mM KPO4 [pH 7.5], 10% glycerol, and the protease inhibitors PMSF, leupeptin, and aprotinin). The protein peak was identified by UV monitoring (280 nm) and collected. HA-2 was then eluted with a linear gradient between buffer D and buffer E (150 mM NaCl, 0.5 M KPO4 [pH 7.5], 10% glycerol, and protease inhibitors) and consisted of pooled fractions recovered between 120 and 400 mM KPO4. All fractions were dialyzed against buffer B containing 50 mM NaCl overnight at 4°C, adjusted to 50% glycerol, and frozen in aliquots at −80°C.
FIG. 4.
Purification scheme used to identify NS1O-activating protein kinases. HeLa cell replication extracts were analyzed for protein kinases which are able to phosphorylate and activate NS1O for RCR in a kinase-free system. Chromatography steps 1 (phosphocellulose) and 2 (anion exchange) allow the bulk separation of protein kinases. Steps 3 (protamine) and 4 (hydroxylapatite) are performed to further purify members of the PKC family. Selective elution of the fractions under investigation from rows 1, 2, and 3 was achieved with the indicated NaCl concentrations, while the hydroxylapatite-bound components (row 4) were eluted in phosphate buffer. PK: − and PK: +++, undetectable and strong protein kinase activities, respectively, assayed in vitro with NS1O as a substrate. F/T, flowthrough.
Replication assays.
Replication assays were carried out as described previously (13) in the presence of optimized amounts of the various cell fractions, using approximately 0.2 μg of His-tagged vaccinia virus-produced NS1 (determined by Coomassie blue staining after SDS-PAGE). In the modified replication system, 3 U of T4 DNA polymerase (Boehringer Mannheim) was used instead of cellular polymerases. Each assay was carried out in a 20-μl total volume of 20 mM HEPES-KOH (pH 7.5)–5 mM MgCl2–5 mM KCl–1 mM DTT–0.05 mM each deoxynucleoside triphosphate–4 mM ATP–40 mM creatine phosphate–1 μg of phosphocreatine kinase–10 μCi of [α-32P]dATP (3,000 mCi/mmol)–20 ng of the appropriate DNA template. After incubation at 37°C for 2 h, the reaction was stopped by adding 60 μl of 20 mM Tris (pH 7.5)–10 mM EDTA–0.2% SDS and incubating the mixture at 70°C for at least 30 min. In order to quantify the extent of DNA replication, 3 μl of the terminated reaction mixture was spotted in duplicate on DE81 filters, washed extensively with 0.5 M Na2HPO4, and analyzed for incorporated radioactivity by scintillation counting. 32P-labeled replication products were also linearized with restriction endonuclease HindIII and analyzed by agarose gel electrophoresis, either directly after proteinase K digestion or after immunoprecipitation with αNSN antiserum (13). This antiserum was raised against the common N terminus of MVM NS proteins (23).
In vitro kinase reactions.
In vitro kinase reactions were performed as described previously (60), using various amounts of protein extracts, 100 ng of dephosphorylated NS1O, and 10 μCi of [γ-32P]ATP (3,000 mCi/mmol) in 20 μl of 20 mM HEPES-KOH (pH 7.5)–7 mM MgCl2–5 mM KCl–1 mM DTT. After incubation for 30 min at 37°C, the reactions were stopped by adding the same volume of 20 mM Tris (pH 7.5)–5 mM EDTA–0.2% SDS and heating for 30 min at 70°C. One-fifth of the reaction products were immunoprecipitated with αNSN antiserum, and in vitro-labeled NS1 was detected by 8% SDS–PAGE and autoradiography.
Helicase assays.
Helicase assays were performed as described previously (59, 60) with M13-VAR as a template. Reaction mixtures with 10 to 100 ng of purified NS1 were incubated for 40 min at 37°C. For reactivation experiments, titrated amounts of protein extracts were added to the reaction mixtures together with one or more of the following PKC cofactors: 2 mM Ca2+, 1 μg of l-α-phosphatidyl-l-serine (PS) per μl, or 5 nM TPA. None of these PKC cofactors alone had any influence on the helicase function of native NS1P, dephosphorylated NS1O, or mutant NS1 proteins used as negative controls.
Western blot analyses.
Protein extracts were fractionated by discontinuous 10% SDS–PAGE, blotted on nitrocellulose membranes, and revealed with rabbit antibodies directed against the most conserved domain of PKC (αPKCγ and αPKCζ), or with mouse antibodies specific for the atypical PKCι (Transduction Laboratories). The αPKCγ and αPKCζ polyclonal antibodies were affinity purified on peptide columns, used at 0.6 mg of IgG per ml, and revealed with 125I-labeled protein A (ICN; 0.2 μCi/ml). Mouse αPKCι antibodies were used at a 1:2,500 dilution, and bound antibodies were revealed with a 1:5,000 dilution of horseradish peroxidase-conjugated anti-mouse IgGs by using the ECL system (Amersham).
RESULTS
NS1 phosphorylation is required for RCR.
Previous investigations comparing biochemical activities of dephosphorylated NS1 (NS1O) and native NS1 (NS1P), both derived from HeLa cells, revealed modulation of site-specific binding to the left-end origin, site-specific nicking, and helicase and ATPase activities. In contrast, no significant difference between NS1O and NS1P could be observed in their respective capacities for dimer-bridge resolution in an in vitro replication assay with HeLa cell extracts (60). It was suggested that NS1O became phosphorylated by kinases present in cell extracts, leading us to develop a kinase-free replication system, as presented in this study.
Resolution of the left-end dimer-bridge junction is a complicated assay, probably requiring a number of so-far-unknown cellular components. In addition, this assay is rather insensitive, because each initiation event is associated with synthesis of only a short stretch of labeled DNA. This prompted us to use a more simple assay, which consists of the NS1-dependent initiation of RCR of nonpalindromic substrates carrying the MVM origins of replication (13), to study the effect of phosphorylation on NS1 replicative functions. When performed with standard replication extract, wild-type NS1, and a plasmid containing an active origin of replication, this assay leads to the synthesis of several kilobases of labeled DNA, which makes it far more sensitive than dimer-bridge resolution. In addition, in the absence of an active origin or without functional NS1, only marginal replication due to repair synthesis is observed, which facilitates the analysis of the modulation of NS1 activity (13).
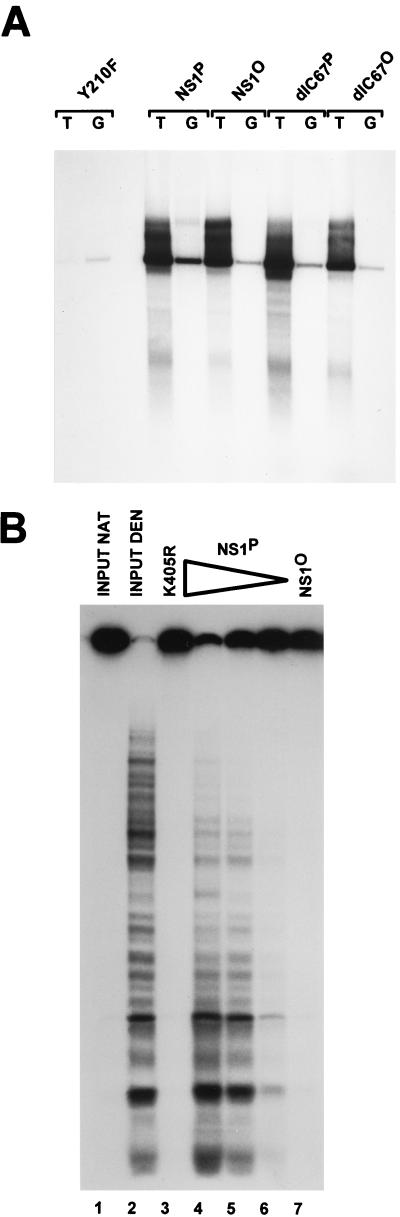
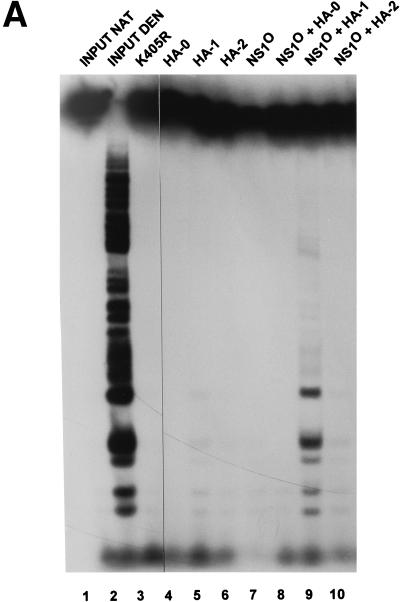
In a first step, NS1P was compared with NS1O in regard to the ability to support RCR from a parvovirus origin in standard HeLa cell extracts. Purified NS1P and NS1O were prepared as reported previously (60). Plasmids carrying the active (TC) or inactive (GAA) left-end origin of MVM DNA replication were used as substrates. The mutant NS1 derivative Y210F, which is impaired in site-specific nicking due to an amino acid substitution for the active-site tyrosine but which is proficient in helicase activity (59), served as a negative control for NS1 replicative function. As shown in Fig. 1A, NS1O was able to support RCR in crude HeLa cell extracts almost as efficiently as NS1P, confirming previously reported findings with cloned dimer-bridge as a substrate (60). Indeed, the replication activities of both wild-type NS1 and the replication-competent NS1 mutant dlC67 were reduced to only a small extent by dephosphorylation; i.e., NS1O sustained approximately 60% of the level of [32P]dATP incorporation into newly synthesized DNA as did NS1P (data not shown). To verify dephosphorylation, we also tested NS1O for its biochemical activities in absence of any additional proteins. The most striking difference between native and dephosphorylated NS1 has been described for the helicase function (60). As illustrated in Fig. 1B, NS1O helicase activity was indeed reduced more than 10-fold compared to that of the native polypeptide, in contrast to the significant activity of NS1O in replication assays. The helicase-deficient NS1 mutant K405R served as a negative control in these experiments.
FIG. 1.
Comparison of replicative functions of native NS1P and dephosphorylated NS1O. NS1 was expressed from recombinant vaccinia viruses in HeLa-S3 cells and harvested at 18 h postinfection by preparing nuclear extracts. Dephosphorylated NS1O was obtained by treatment with calf intestine alkaline phosphatase. His-tagged native NS1P and phosphatase-treated NS1O were purified from nuclear extracts by Ni2+-NTA affinity chromatography. (A) NS1P and NS1O were compared for their capacities to support RCR in standard HeLa cell replication extracts, using plasmids containing the left-end active (T) or inactive (G) origin as substrates, in the presence of [32P]dATP. The reaction products were linearized with HindIII and analyzed by 0.8% agarose gel electrophoresis. The linkage tyrosine mutant Y210F (59) served as a negative control. The dlC67 mutant is replication competent (60) and was also analyzed in its native (P) and dephosphorylated (O) forms. (B) NS1P (100, 30, and 10 ng [lanes 4 to 6, respectively]) and NS1O (100 ng [lane 7]) were compared for their intrinsic helicase activities, using M13-VAR template, for 40 min at 37°C in the presence of 2 mM ATP. The ATP-binding site mutant K405R (57) served as a negative control. Reaction products were analyzed by native 7% PAGE in the presence of 0.1% SDS. Lanes 1 and 2, native (NAT) and denatured (DEN) input DNA, respectively.
One possible explanation for the replication competence of NS1O, despite the lack of helicase function, might be the presence of protein kinases within the cell extracts used in the former but not in the latter assay, which would lead to NS1O rephosphorylation and consequent reactivation. To test this possibility, and eventually to establish RCR in absence of endogenous protein kinases, we first determined the presence of NS1O-phosphorylating protein kinases after fractionation of replication extracts on phosphocellulose columns, taking advantage of recent developments in the identification of host cell determinants of this reaction. As indicated in Fig. 2A, it has been shown that phosphocellulose fractions P1 and P2 derived from 293 cells are sufficient to support RCR of plasmids containing the left-end origin in the presence of wild-type NS1 (8). When HeLa cell extracts were fractionated in the same way, protein kinases phosphorylating NS1 were found to be confined to fractions 2 and 3, with no detectable activity in the P1 flowthrough (Fig. 2B). Yet, HeLa cell P1 failed to support significant replication in the presence of P2 and native NS1P (data not shown) and hence could not be used in subsequent experiments. Therefore, we prepared P1 from 293 cells as previously described (8), in order to obtain sufficient replication factors therein to drive RCR. In contrast to HeLa cell P1, 293 cell P1 significantly phosphorylated NS1O, although the bulk of kinase activity was still found in fractions P2 and P3 (Fig. 2B). To remove the residual endogenous serine/threonine kinases from 293 cell P1, this fraction was purified over an l-threonine affinity column, resulting in P1-Thr, which was essentially free of NS1O-phosphorylating activity in comparison with the original P1 material (Fig. 2B). This kinase-free P1-Thr from 293 cells was then combined with fraction P2 obtained from HeLa cells in order to supply the cellular components allowing NS1-mediated RCR. As seen in Fig. 2C, NS1P was able to trigger RCR from the active (TC) origin in the presence of P1-Thr and P2 fractions. This reaction was specific, since it occurred to only a small extent when the inactive (GAA) origin was used as a substrate. Furthermore, as seen previously with standard HeLa extracts, NS1O was also able to support RCR under these conditions, i.e., in the presence of the sole protein kinases present in HeLa cell P2. Under these conditions, NS1O achieved close to 50% of the RCR activity of NS1P (Fig. 2C). The inactivity of the replication-deficient NS1 mutant Y210F, used as a negative control, and the immunoprecipitation of labeled DNA products with NS1 antiserum confirmed the specificity of the RCR reaction (Fig. 2C). Together, these results indicate that the phosphocellulose P3 fraction, as well as the protein kinases therein, are dispensable for RCR initiated by NS1 at the left-end origin.
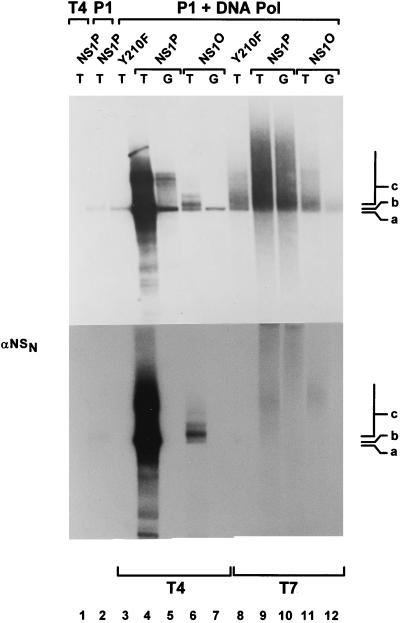
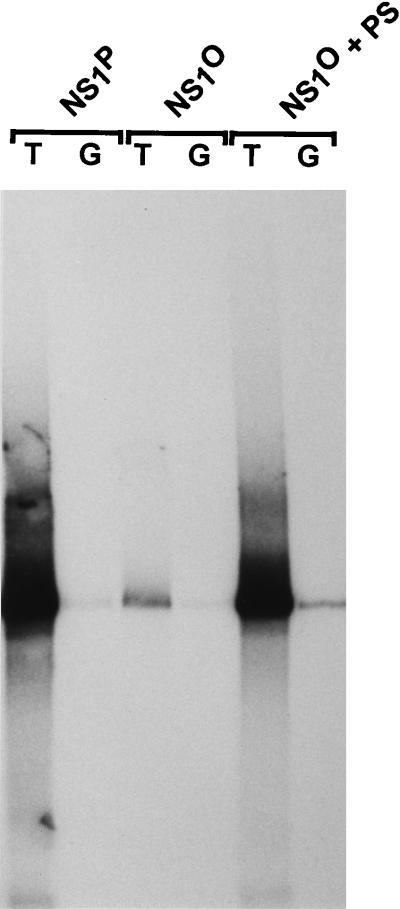
Fraction P2 is thought to provide the DNA polymerases necessary for NS1-dependent RCR at the left-end origin (8, 55, 68). In order to reconstitute a kinase-free system and supply DNA polymerases in absence of protein kinases, we decided to replace the whole fraction P2 by purified bacterial or bacteriophage polymerases. With native NS1P, no replication products were obtained when either P1 or P2 was omitted from the reaction (Fig. 3, lanes 1 and 2). On the other hand, DNA synthesis took place when the P1-Thr fraction was supplemented with Escherichia coli DNA polymerase I or the Klenow fragment thereof (data not shown) or with DNA polymerase from bacteriophage T7 or T4 (Fig. 3). In the presence of standard replication extracts, the active origin from pL1-2TC is recognized by NS1, allowing the establishment of a unidirectional single-strand replication fork which progresses around the circular plasmid (13). Initiation of replication is achieved by site- and strand-specific nicking performed by the NS1 protein, which remains covalently attached to the 5′ end of the nicked strand. In contrast to the active origin, the inactive origin in pL1-2GAA is not a substrate for nicking (8), and therefore no NS1-dependent replication occurs. We further analyzed the specificity of the RCR reactions taking place in the reconstituted, kinase-free system by using pL1-2GAA as an inactive substrate and the mutant Y210F as replication-deficient NS1 control. In addition, replication products were analyzed after immunoprecipitation with αNS antiserum. When E. coli Klenow fragment, DNA polymerase I (data not shown), or bacteriophage T7 polymerase (Fig. 3, lanes 8 to 12) was substituted for the eukaryotic polymerases, replication was found to occur irrespective of whether the templates contained an active (T) or inactive (G) origin. The lack of NS1-dependent initiation of the replication reactions driven by E. coli and phage T7 polymerases was also apparent from the failure of the αNS1 serum to immunoprecipitate labeled DNA products (Fig. 3, lower panel) and the significant DNA synthesis detected with the NS1 mutant Y210F (Fig. 3, lane 8). In contrast, as reported previously for simian virus 40 (SV40) DNA replication in vitro (69), T4 DNA polymerase could successfully substitute for the cellular DNA polymerases contained in P2 to give rise to a specific RCR reaction with plasmids containing the active left-end (TC) origin in the presence of P1 and NS1P (Fig. 3, lane 4). Indeed, only limited repair synthesis occurred with the substrate containing the inactive origin (Fig. 3, lane 5), while initiation was NS1 dependent, as apparent from the formation of αNS1-immunoprecipitatable replication products in the presence of wild-type NS1P but not Y210F (Fig. 3, lanes 3 and 4).
FIG. 3.
NS1-dependent RCR in a kinase-free in vitro system. Plasmids containing the active (T) or inactive (G) left-end origin were used as substrates to measure the capacity of NS1P and NS1O to support RCR in absence of endogenous protein kinases, in a system based on P1-Thr and bacteriophage DNA polymerases. T4 and T7, bacteriophage T4 and T7 DNA polymerases, respectively. The replication-deficient NS1 mutant Y210F was used as a negative control. Linearized 32P-labeled replication products were analyzed on 0.8% agarose gels, either directly after proteinase K digestion (top panel) or after immunoprecipitations with αNSN (bottom panel). The positions of the linearized plasmid (a) and slower-migrating products (b and c) are indicated.
Since we were able to obtain a specific RCR reaction by using a kinase-free P1-Thr and T4 DNA polymerase in the presence of native NS1P, we further investigated the requirements of this reaction for NS1 phosphorylation. As illustrated in Fig. 3, the patterns of labeled DNA products generated in the presence of NS1O and NS1P could be distinguished in three respects. (i) The overall level of DNA synthesis was much reduced when NS1 was dephosphorylated, which strongly suggests that phosphorylation modulates the capacity of NS1 for initiation and/or promoting RCR. (ii) Major products of the RCR reaction taking place in the presence of NS1P migrated more slowly than linearized plasmid DNA and can be ascribed to multiple rounds of plasmid template DNA copying, yielding circular molecules with single-stranded tails of various lengths (13). These intermediates, marked c in Fig. 3, were not formed efficiently in the presence of NS1O, although labeled DNA was detected in the plasmid length region, which argues for a role of NS1 phosphorylation in the strand displacement synthesis during RCR (and parvovirus DNA replication). This conclusion is in agreement with the previously reported main deficiency of NS1O in the helicase activity, which is thought to facilitate unwinding of the double-stranded template to allow the replication fork to proceed (Fig. 1B) (60). (iii) The predominantly labeled DNA product obtained with NS1O (marked b in Fig. 3) was slightly upshifted compared with the linearized plasmid (marked a). This mobility shift is expected for DNA molecules which underwent nicking and replication initiation, resulting in a short stretch of newly synthesized DNA in the absence of extensive strand displacement synthesis. This species b was not detected when replication-competent NS1P was used, suggesting that the lack of NS1 phosphorylation is associated with an impairment of growing-strand elongation in already initiated DNA molecules. It should also be stated that besides this modulation of the elongation step, the initiation of parvovirus DNA replication also appears to be stimulated by NS1 phosphorylation. Indeed, recently reported in vitro assays have shown that although it is proficient in site-specific nicking, NS1O is less efficient than NS1P for this function (60).
Reactivation of NS1O for RCR by members of the PKC family.
Since NS1O was able to support RCR in the presence of cell extracts containing endogenous protein kinases but distinguished itself from NS1P in its low capacity to achieve this reaction in the kinase-free in vitro replication system, we further attempted to reactive NS1O for RCR by providing exogenous cellular components. Previous investigations using commercially available PKC preparations indicated the involvement of this protein kinase family in regulation of NS1 helicase activity (60). Therefore, we performed the following reactivation experiments in the presence of Ca2+ and PS, which are known cofactors for PKC (44, 61). Indeed, under these conditions, NS1O replication activity could be stimulated to a significant extent by supplying the reaction mixture with limited amounts of whole HeLa cell replication extracts (data not shown). This result encouraged us to fractionate HeLa cell extracts in order to characterize the NS1O-activating components, and in particular to determine whether the rescue of NS1O replication activity cosegregated with NS1 phosphorylation by specific protein kinases as postulated.
Figure 4 gives the purification scheme for HeLa cell extracts used for these reactivation experiments. Individual fractions were tested for their ability to phosphorylate NS1O. In parallel, various protein concentrations were used to determine the capacity of individual or combined HeLa cell fractions for reactivating the RCR function of the underphosphorylated polypeptide. Native wild-type NS1P and the replication-deficient mutant Y210F served as positive and negative controls in these assays, respectively. The specificity of the reactions was also ascertained by using active (pL1-2TC) versus inactive (pL1-2GAA) origin-containing substrates and by immunoprecipitating NS1-bound replication products with αNSN antiserum. Chromatography steps 1 and 2 were designed to achieve a first bulk segregation of the multiple protein kinases which are able to phosphorylate NS1O. As illustrated in Fig. 2B and C, more than 50% of the NS1-targeted kinase activity segregated in phosphocellulose fraction P3 (step 1) and proved to be nonessential, since NS1O was almost as efficient as NS1P for RCR in the sole presence of proteins contained in P1-Thr and P2. Furthermore, neither the whole P3 fraction nor subfractions thereof were able to activate NS1O in the kinase-free replication system (data not shown). This result indicated that specific rather than random phosphorylation of NS1 was necessary for the replicative functions of the viral product. After further purification of P2 on anion-exchange columns (step 2), NS1O-stimulating activity was recovered in the low-affinity DE-1 fraction, eluting between 50 and 200 mM NaCl (Fig. 5, lanes 4 and 7). No additional stimulation resulted from the supply of kinases present in fractions eluting at higher salt concentrations, despite their ability to phosphorylate NS1O to a significant extent in vitro (data not shown).
FIG. 5.
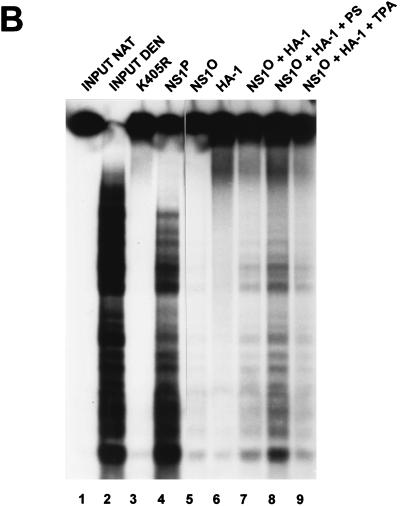
Activation of NS1O for RCR by fractionated HeLa cell extracts. NS1-dependent RCR of plasmids containing the left-end active (T) or inactive (G) origin was determined in a kinase-free in vitro system based on P1-Thr and T4 DNA polymerase. Activation of NS1O (lanes 4 to 14) was achieved by using the indicated protein components (see Fig. 4) in the presence or absence of protein kinase C cofactors. Only some of the protein-cofactor combinations that have been tested are illustrated, as most representative. Lanes 4 to 8 show the dependency of NS1O activation on PKC cofactors. PS, Ca2+ plus PS. Lanes 9 to 14 show the segregation of protein components allowing the reactivation of dephosphorylated NS1. The illustrated reactions were carried out in presence of the PKC activators Ca2+ and PS (HA-1) or TPA (HA-2 and HA-1 plus HA-2). It should be mentioned that TPA-stimulated HA-1 or Ca2++PS-stimulated HA-2 also failed to activate NS1O (data not shown), while the combined protein components (HA-1 plus HA-2) were able to rescue NS1O in the presence of Ca2++PS instead of TPA. The NS1 mutant Y210F served as a negative control (lane 1). Native NS1P was used as a phosphorylated, replication-competent standard (lanes 2 and 3). L, migration of the linearized plasmid. The lower panel presents reaction products after immunoprecipitation with αNSN.
The subsequent purification steps (steps 3 and 4) were applied to concentrate PKCs and to separate distinct isoforms of this protein kinase family. Protamine affinity columns are commonly used to purify PKC from bulk proteins, due to the high affinity of these kinases for basic protein substrates (61). On the other hand, at least some of the structurally related PKC isoforms can be separated on hydroxylapatite columns, based on their various requirements for Ca2+ ions and affinities to phosphate groups (32, 62). As expected, NS1O-activating components were retained on the protamine column and recovered in fraction PA-2 after high-salt elution (data not shown). A further concentration of these components was achieved by hydroxylapatite chromatography, yielding an inactive flowthrough and two bound fractions of low (HA-1) and high (HA-2) affinity (eluting at 20 and 120 to 400 mM KPO4, respectively). Though inactive on their own, the HA-1 and HA-2 fractions were able to reactivate NS1O when supplied in combination in RCR assays (Fig. 5, lanes 9 to 14; Table 1). The reaction stimulated by HA-1 and -2 was a specific NS1-dependent RCR process. Indeed, no significant DNA synthesis was observed when the inactive GAA origin was used as a substrate (Fig. 5, lane 14), and the reaction products obtained with the active (TC) origin were covalently attached to NS1 as shown by immunoprecipitation (Fig. 5, bottom panel, lane 13). To further evaluate the active components within fractions HA-1 and HA-2, we performed the reactivation experiments in the presence or absence of characteristic cofactors for PKCs (44, 61). As summarized in Table 1 and illustrated in Fig. 5 and 6, reactivation of NS1O in replication assays was strictly dependent upon addition of PS, Ca2++PS, TPA, or PS + TPA. This strongly argues for the involvement of members of the PKC family in modulation of NS1 replicative functions in vitro.
TABLE 1.
Dependence of the reactivation of NS1O replicative function on PKC cofactors
| Column fraction | Reactivation ofa:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| NS1O RCR activity
|
NS1O helicase activity
|
|||||||
| No co-factors | PS | TPA | Ca2+-PS | PS-TPA | No co-factors | PS | TPA | |
| HA-1 | − | − | ++ | − | + | −/+ | +++ | −/+ |
| HA-2 | − | − | − | − | − | − | − | − |
| HA-1 plus HA-2 | − | + | ++ | ++ | +++ | NDb | ND | ND |
The protein fraction-induced reactivation of NS1O functions in the presence of cofactors (PS, TPA, and Ca2+) is graded from − (no effect) to +++ (maximal stimulation). The following PKC isoforms are known to be typically activated by the cofactors: PS, atypical isoforms; TPA, classical and novel isoforms; no cofactors, proteolytically derived catalytic domain; Ca2+-PS, classical and atypical isoforms; PS-TPA, classical, atypical, and novel isoforms.
ND, not determined.
FIG. 6.
Effect of PKC cofactors on activation of the NS1O replicative functions in an entirely eukaryotic replication system. NS1P and NS1O were compared for RCR of plasmids containing the left-end active (T) or inactive (G) origin of replication by using P1-Thr plus P2-pol (containing endogenous DNA polymerases). NS1O activity was tested in either the absence or presence (+ PS) of 1.5 μg of PS. All reaction were performed in the presence of 2 mM CaCl2.
The reactivation experiments described above were carried out with a rather artificial replication system based on bacteriophage T4 DNA polymerase. To substantiate these data in the presence of eukaryotic polymerases, we took advantage of the requirement of NS1O activation for PKC cofactors. Unlike standard HeLa cell replication extracts, fractions P1-Thr (purified on phosphocellulose and l-Thr affinity columns) and P2-pol (purified on phosphocellulose and DE52 columns) are depleted of residual membrane structures which can serve as PKC activators (61). Assuming that the results obtained with the kinase-free reconstituted system can be extrapolated to mammalian DNA polymerases, these fractions should be supplemented with PKC activators in order to render NS1O competent for RCR. This prediction was tested by comparing NS1P and NS1O for RCR with P1-Thr plus P2-pol in the presence or absence of PKC cofactors. As shown in Fig. 6, the ability of NS1O (but not NS1P) to support RCR of pL1-2TC templates under these conditions was dependent upon addition of the PKC cofactors Ca2+ and PS, despite the fact that P2-pol contains, besides DNA polymerases, multiple protein kinases which do not require additional cofactors for activation. This experiment clearly demonstrates that dephosphorylated NS1O is not irreversibly inactivated with regard to its replication function, and it extends the above-mentioned results to suggest that phosphorylation by distinct protein kinases is required for NS1 activity in a purely eukaryotic DNA replication system.
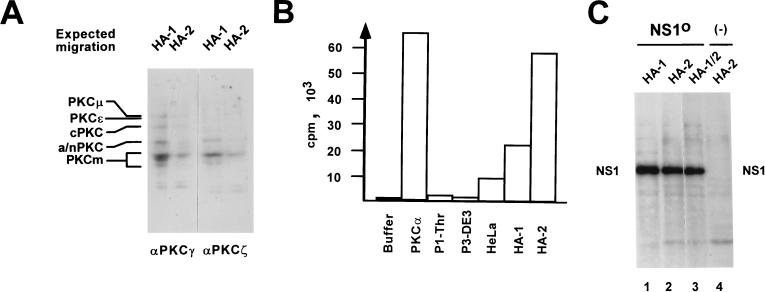
The purification profile and the cofactor requirements of the NS1O-reactivating components in replication assays strongly suggested PKC to be an essential kinase(s) for NS1 activation in vitro. In order to ascertain the presence of active PKC within the reactivating HeLa cell fractions, we produced polyclonal antibodies against the most-conserved regions of PKC, which are between amino acids 416 and 569 in PKCγ and between amino acids 310 and 444 in PKCζ, and performed Western blotting with these immune affinity-purified antisera. As illustrated in Fig. 7A, multiple proteins with the sizes of known PKC isoforms were indeed immunodetected within fractions HA-1 and HA-2, in addition to a major product of lower molecular weight corresponding to PKCm, the proteolytically cleaved catalytic domain of PKC. Moreover, as seen in Fig. 7B, both fractions HA-1 and HA-2 were enriched in PKC activity compared with crude HeLa cell extract or the negative controls P1-Thr (which does not contain significant protein kinase activity), and P3-DE3 (for which NS1O constitutes a target for in vitro phosphorylation but not replication reactivation [data not shown]), as determined with a commercially available PKC detection system (Amersham). Furthermore, by performing in vitro kinase assays in the presence of [γ-32P]ATP under conditions used in replication assays, NS1O was found to serve as a substrate for semipurified protein kinases contained in HA-1 and/or HA-2 (Fig. 7C). Surprisingly, overall NS1 phosphorylation was only slightly stimulated upon addition of PKC cofactors (data not shown). This may be because either additional kinases were still present within the HA-1 and -2 fractions or the cofactors target PKC on a specific phosphorylation site(s) within the NS1 polypeptide. Together, these results clearly demonstrated that cellular factors able to reactivate the replicative functions of NS1O copurified with highly active PKCs contained within both fractions HA-1 and HA-2. Furthermore, the requirement of HA-1 and -2-induced RCR reactivation for specific PKC cofactors substantiated the role of one or more PKC isoforms from HA-1 and/or HA-2 in the upmodulation of NS1 replicative functions.
FIG. 7.
Detection of PKC within fractions HA-1 and HA-2 from HeLa cell extracts. (A) Western blot analysis of HA-1 or HA-2 after 10% SDS–PAGE, using peptide affinity-purified polyclonal antisera that were raised against the most-conserved regions of classical PKCs (PKCγ, amino acids 416 to 569; PKCζ, amino acids 310 to 444) and revealed the various PKC isoforms by cross-reaction. The expected migration positions of known PKC isoforms are given on the left: cPKC, classical PKCα, -β, and -γ (approximately 80 kDa); a/nPKC, atypical PKCι/λ and -ζ (70 to 72 kDa) and novel PKCδ, -η, and -θ (72 to 78 kDa). PKCɛ (90 kDa) and PKCμ (115 kDa), higher-molecular-mass novel PKCs; PKCm (approximately 45 kDa), PKC catalytic domains produced by proteolytic cleavage. (B) PKC activity assays (Amersham) of fractions used for reactivation of NS1O in replication reactions. Buffer, negative control in the absence of added protein components; PKCα, 10 ng of His tag-purified recombinant PKCα produced by recombinant vaccinia virus expression in HeLa cells; P1-Thr, fraction containing no NS1O-targeted protein kinase activity (see Fig. 2B); P3-DE3: fraction which proved able to phosphorylate but failed to reactivate NS1O (see Fig. 4); HeLa, standard HeLa cell replication extract; HA-1 and HA-2, NS1O-activating fractions in RCR assays (see Fig. 5). The values are expressed as transferred 32P-labeled substrate per 10 ng (recombinant PKCα) or 10 μg (HeLa cell fractions) of total effector proteins. (C) In vitro phosphorylation of 1 μg of NS1O with protein fractions HA-1 and HA-2, used individually or in combination in the presence of PKC cofactors. The reactions were performed in the presence of [γ-32P]ATP for 30 min at 37°C, and radiolabeled NS1 was revealed by autoradiography after SDS-PAGE. In lane 4, the NS1O substrate was omitted. The migration of NS1 is indicated.
Stimulation of NS1O helicase activity by members of the PKC family.
The above-mentioned analyses of NS1-driven RCR in the kinase-free in vitro replication system indicated that a major NS1 phosphorylation-dependent step consisted of processive strand displacement synthesis (Fig. 3). This step of RCR (and also parvovirus DNA replication) is thought to involve the unwinding function of NS1. Consistently, NS1O was found to be heavily impaired for this biochemical activity in standard helicase assays (Fig. 1B) (60). In order to investigate whether the intrinsic helicase function of NS1 might be regulated by the same components as those identified with the in vitro RCR system, the HA- fractions were also tested for their ability to rescue NS1O in helicase assays. As illustrated in Fig. 8A, one of these fractions, HA-1, was able to stimulate the helicase function of NS1O to a significant extent, whereas HA-2 or the flowthrough (HA-0) had no detectable effect. None of the hydroxylapatite fractions exerted helicase activity by itself, even when tested in a 100-fold excess over the amount used to stimulate the NS1O helicase function (Fig. 8A, lanes 4 to 6), indicating that this stimulation resulted from the activation of NS1O rather than supply of a cellular helicase(s).
FIG. 8.
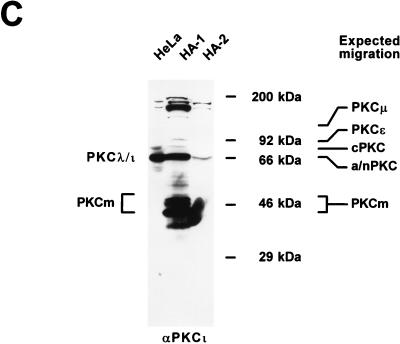
Reactivation of dephosphorylated NS1O for helicase activity. Helicase assays were performed as described for Fig. 1B, using M13-VAR as a template. (A) Hydroxylapatite column fractions HA-0, HA-1, and HA-2 were tested for their intrinsic helicase activities (lanes 4 to 6) and for activation of the NS1O helicase function in the presence of the PKC cofactor Ca2++PS (lanes 8 to 10). Lane 7 shows the helicase activity of unstimulated NS1O. The NS1 mutant K405R served as a negative control (lane 3). The reaction products were analyzed by native 7% PAGE in the presence of 0.1% SDS. Lanes 1 and 2, native (NAT) and denatured (DEN) input DNA, respectively. (B and C) Characterization of PKC isoforms present within the HA-1 fraction, which is able to reactivate NS1O for helicase activity. (B) Dependence of HA-1-induced reactivation of NS1O helicase function on defined PKC cofactors. Atypical PKCs (ι/λ and ζ) are stimulated by acid lipids (PS) but are insensitive to phorbol esters such as TPA, whereas novel and classical PKCs are only slightly activated by PS alone but respond strongly to TPA. Dephosphorylated NS1O was assayed for helicase activity either alone (lane 5) or in the presence of HA-1 with or without the indicated PKC cofactors (lanes 7 to 9). The NS1 mutant K405R (lane 3) and native NS1P (lane 4) served as negative and positive controls, respectively. The effect of HA-1 in the absence of NS1 is shown in lane 6. Lanes 1 and 2, native and denatured input DNA, respectively. (C) Immunodetection of atypical PKC in hydroxylapatite fractions. Equal protein amounts of HA-1 and HA-2 were analyzed by Western blotting with αPKCι antibodies (Transduction Laboratories). A HeLa cell replication extract served as a positive control. Size markers and expected migrations of different PKC isoforms are indicated on the right (for abbreviations, see the legend to Fig. 7A). The estimated migrations of PKCλ/ι and PKCm, which are recognized by αPKCι, are indicated on the left.
These assays were performed in the presence of Ca2++PS, i.e., under conditions which activate all members of the PKC family in vitro. PKCs have been subdivided into three subgroups according to their cofactor requirements. Classical (α, β, and γ) and novel (δ, ɛ, θ, η, and μ) PKC isoforms are stimulated through binding of phorbol esters (such as TPA) and only to a minor extent by binding of acid lipids (such as PS). Full activation of classical PKC is also achieved by Ca2+ in addition to PS, while novel PKCs do not contain a Ca2+-binding site. Members of the last group, designated atypical PKC isoforms (ι/λ and ζ), lack a calcium-binding domain and do not respond to TPA either, but they are stimulated by acid lipids. Finally, PKCm, the catalytic domain of PKC which derives from proteolytic cleavage, is constitutively active; i.e., its kinase activity is independent of cofactors (44, 61). In order to characterize the activating kinase(s) present in fraction HA-1, we further tested NS1O reactivation for helicase function in the presence of HA-1 together with selected PKC cofactors. As seen in Fig. 8B, the HA-1 fraction had a moderate, constitutive stimulatory effect in absence of cofactors (lane 7), which may be ascribed to PKCm and/or other cofactor-independent components. Interestingly, this activation was significantly enhanced upon supply of PS (lane 8), while TPA failed to increase the capacity of HA-1 to stimulate the helicase function of NS1O (lane 9). As summarized in Table 1, this PS responsiveness and TPA insensitivity of the HA-1-induced rescue of NS1O point to an atypical PKC(s) as the effector(s) mediating the dependence of NS1 helicase activity on phosphorylation. This conclusion was substantiated by Western blot analysis of fractions HA-1 and HA-2 with specific antibodies recognizing the atypical PKCι. As shown in Fig. 8C, the atypical PKCι segregated mainly to fraction HA-1 during the purification procedure, while HA-2, which was unable to activate NS1O in helicase assays, did not contain substantial amounts of this PKC isoform. The PS responsiveness of the NS1 helicase-stimulating factor(s) and its cosegregation with PKCι into HA-1 strongly suggested that atypical PKCs are involved in the upmodulation of the NS1 DNA-unwinding function.
DISCUSSION
NS1, the major nonstructural protein of MVM, is involved in multiple functions necessary for progeny virus production, ranging from DNA replication to promoter regulation and toxic action on the host cell (15). Such a variety of tasks is unlikely to be achievable by a single polypeptide and usually requires the multifunctional protein to interact with heterologous proteins (42), to self-assemble into higher-order oligomers (4), to associate with cofactors (61), and/or to become posttranslationally modified. The last possibility can provide an original polypeptide with functional heterogeneity through the addition of various molecule groups, catalyzed with high efficiency by enzymes present in rather small amounts, targeted on a large protein pool. All of the above-mentioned modes of regulation have been assigned to NS1, including interactions with cellular partner proteins (24, 36), oligomerization (58), and phosphorylation (20). Interestingly, NS1 oligomerization has been implicated in the control of NS1 replicative functions, in particular helicase activity (63). It is worth noting that NS1 self-assembly to produce higher-order oligomers is dependent on an intact nucleoside triphosphate-binding domain (58) and might thus be regulated by association with this cofactor. Furthermore, the ATP-bound form of NS1 was found to be most competent for site-specific DNA binding (21). Indeed, ATP binding and/or hydrolysis seems to be crucial for many NS1 functions, as apparent from the fact that mutagenesis of the ATP-binding domain abolishes all NS1 activities described so far (10, 21, 38, 39, 57–59). In turn, the NS1-associated ATP turnover was recently found to be controlled by phosphorylation, at least under in vitro conditions (60). Thus, the regulation of an activity which plays a pivotal role in NS1 functions may be traced back to the modification of NS1 through phosphorylation. NS1 dephosphorylation correlates with a reduction of NS1 ATPase activity in vitro, which is associated with an increase in the affinity of the viral product for its DNA recognition motif and with a decrease of helicase and site-specific nickase functions (60). This regulation might be physiologically relevant, since NS1 has been shown to be phosphorylated in vivo (2, 11, 20). The present work was carried out to further characterize the cellular protein kinases involved in the upmodulation of NS1 replicative functions.
In this study, using an in vitro replication system devoid of endogenous protein kinases, we obtained evidence that a central NS1 function necessary for progeny virus production, namely, replication initiated at the left-end origin, is upmodulated by phosphorylation of NS1 and that members of the PKC family participate in this stimulation. The phosphorylation dependence of NS1 is reminiscent of the regulation of large T antigen (LT), the initiator protein for SV40 DNA replication (71). LT, which is expressed in nondividing cells and is able to drive quiescent cells into S phase, becomes activated for replication by phosphorylation at T124 through cyclin-dependent kinases. This regulation results in a coordination between SV40 and host cell DNA replication (1, 47). As in the case of SV40, parvovirus DNA replication has been shown to be dependent on the S phase of host cells (15), hence the restriction of parvovirus multiplication in vivo to proliferating tissues (43). Yet, in contrast to the case for LT, NS1 production is limited during G0/G1 (25, 66), and parvoviruses fail to drive quiescent cells into S phase (15). Moreover, extracts derived from cells arrested in G0 are able to activate the replicative functions of dephosphorylated NS1 in vitro (56). Therefore, posttranslational modifications of NS1 do not account for the S phase dependency of parvovirus DNA replication. Another feature of parvoviruses is their striking oncotropism (65). This tropism could be mimicked in various cell culture systems, where the restrictions to parvovirus replication detected in normal parental cell lines were found to be at least partly overcome upon neoplastic transformation (12). In this context, it is interesting that PKC activators, such as phorbol esters, also exert strong effects on cell proliferation, differentiation, and, most intriguingly, tumor promotion (44, 61). Moreover, it has been reported that overexpression of PKCɛ, a novel PKC isoform which is activated by phorbol esters and has been detected in A9 cells (a natural host cell line permissive for MVMp [26]), leads to cell transformation in vitro (6, 49). These correlations of oncogenic transformation with changes in both PKC activity and cell permissiveness to parvovirus replication, together with the present evidence of a role of PKC in the regulation of the pivotal viral replicator protein NS1, raise the possibility that this regulation may contribute to the oncotropism of parvoviruses.
Native NS1P was able to initiate RCR, leading to extensive strand displacement synthesis in the absence of protein kinases. In contrast, NS1O had a restricted phenotype that was characterized by the formation of a distinct NS1-bound replication intermediate. This intermediate migrated in agarose gels as a molecule in which DNA synthesis was initiated but became arrested prior to strand displacement synthesis, suggesting that besides its role in the site-specific initiation of DNA replication, NS1 is also essential to drive the subsequent strand displacement reaction in a phosphorylation-dependent way. This is in agreement with previously reported findings that dephosphorylated NS1O is deficient in helicase activity (60), which would account for its inability to allow the replication fork to proceed during DNA synthesis. Therefore, regulation of the DNA-unwinding activity of NS1 might be of crucial importance to turn on replication. It should be stated, however, that the helicase deficiency of NS1O could be corrected by supplementing solely the HA-1 fraction, while RCR reactivation of NS1O required both protein fractions HA-1 and HA-2. Thus, more than one protein kinase seem to be necessary to switch NS1O on for DNA replication. The additional replicative functions of NS1 which are regulated by phosphorylation, besides helicase activity, are as yet undefined. One candidate is the nicking reaction, which is achieved by NS1O to a detectable, albeit reduced level compared to NS1P, both in the RCR assay and in an in vitro nicking assay performed in the absence of additional cellular components (60). However, this reduction could also be a consequence of the defect of NS1O in DNA unwinding, since the latter is thought to facilitate single-strand nicking (35, 67). Other potential phosphorylation-dependent NS1 replicative functions might be considered by analogy with SV40 LT. LT binds to the origin of replication in the absence of phosphorylation (45, 46, 50) and is able to interact with components of the basic replication machinery such as replicator protein A (48) or polymerase α primase (28, 29) to establish the replication complex by protein-protein interactions. It is tempting to speculate that NS1 may act in a similar way, since it is known that LT- and NS1-driven in vitro replications share specific requirements for eukaryotic DNA polymerases or the related T4 DNA polymerase (69) and for template DNA unwinding by the helicase activities of the respective viral proteins (71). While binding to target (ACCA) motifs on the viral DNA, NS1 fulfills functions involved not only in DNA replication but also in promoter regulation, raising the possibility that distinct NS1 phosphorylation events may regulate the interaction of the viral polypeptide with proteins from the basal replication and/or transcription machinery. NS1 was indeed demonstrated to interact specifically with general transcription factors (36, 40).
NS1 has been shown to be a target for phosphorylation by many protein kinases in vitro (2, 60). Indeed, most HeLa cell fractions tested in this study were found to exhibit NS1 phosphorylation activity. In contrast, only selected fractions of the original HeLa cell replication extract were able to activate NS1O for replication activity, pointing to the involvement of specific kinases and phosphorylation events in NS1 regulation. Based on the cofactor requirements for NS1O reactivation and the purification properties of effector kinases, we were able to assign, at least in part, the capacity for NS1 phosphorylation and reactivation in vitro to members of the PKC family. In particular, the NS1 unwinding function might be modulated by atypical PKCs, given its responsiveness to acid lipids but not to phorbol esters. As stated above, regulation of the NS1 replicative functions appears to be complex and to involve more than one protein kinase, as indicated by the fact that rescue of NS1O for RCR required at least two protein components that could be separated by hydroxylapatite chromatography. When combined, the active HA- fractions were able to rescue the RCR functions of NS1O to a significant extent in the presence of the phorbol ester TPA. Since TPA was unable to activate the PKC responsible for stimulating the helicase function of NS1, our results suggest that at least two PKCs may be involved in NS1 regulation. One of these kinases appears to be one of the classical or novel PKC isoforms (which respond to TPA) and controls an as-yet-undefined NS1 replicative function, while the other is a TPA-insensitive atypical PKC isoform that regulates the DNA-unwinding activity of NS1. The latter PKC may be activated by the former, thereby accounting for the sole requirement of TPA as a cofactor to rescue the RCR capacity of NS1O, in agreement with the fact that PKCs are themselves regulated by phosphorylation (33). Mapping of the NS1 phosphorylation sites involved in the modulation of replicative functions, as well as further characterization of the cellular protein kinases responsible for these modifications, should contribute to understanding the posttranslational regulation of NS1 activities.
ACKNOWLEDGMENTS
We are indebted to Bernard Moss (National Institutes of Health) for making pTM-1 plasmid and the vTF7-3 virus available and to Hubert Hug (Deutsches Krebsforschungzentrum) for providing full-length PKCα, PKCγ, and PKCζ cDNA clones. We are most grateful to Peter Tattersall and Susan Cotmore for sharing constructs used in our assays, stimulating discussions, and critical comments. We also thank Claudia Plotzky for technical assistance and Rainer Schmidt, Michael Gschwendt, and Jesper Christensen for helpful comments concerning fractionation of replication extracts and characterization of PKC.
This work was supported by the Commission of the European Communities and the German-Israeli Foundation for Scientific Research and Development. R.C. was supported in part by a fellowship from La Ligue Nationale Contre le Cancer.
REFERENCES
- 1.Adamczewsky J P, Gannon J V, Hunt T. Simian virus 40 large T antigen associates with cyclin A and p33-cdk2. J Virol. 1993;67:6551–6557. doi: 10.1128/jvi.67.11.6551-6557.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Astell C R, Liu Q, Harris C E, Brunstein J, Jindal H K, Tam P. Minute virus of mice cis-acting sequences required for genome replication and the role of the trans-acting viral protein, NS1. Prog Nucleic Acid Res Mol Biol. 1996;55:245–285. doi: 10.1016/s0079-6603(08)60196-8. [DOI] [PubMed] [Google Scholar]
- 3.Baldauf A, Willwand K, Mumtsidu E, Nüesch J P F, Rommelaere J. Formation of circular replicative form (RF) from single-stranded virion DNA and initiation of DNA replication at the RF 5′ telomere induced by the minute virus of mice nonstructural protein NS1. J Virol. 1997;71:971–980. doi: 10.1128/jvi.71.2.971-980.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradley M K, Griffin J D, Livingston D M. Relationship of oligomerization to enzymatic and DNA-binding properties of the SV40 large T antigen. Cell. 1982;28:125–134. doi: 10.1016/0092-8674(82)90382-8. [DOI] [PubMed] [Google Scholar]
- 5.Brandenburger A, Legendre D, Avalosse B, Rommelaere J. NS1 and NS2 proteins may act synergistically in the cytopathogenicity of parvovirus MVMp. Virology. 1990;174:576–584. doi: 10.1016/0042-6822(90)90110-d. [DOI] [PubMed] [Google Scholar]
- 6.Cacace A M, Guadagno S N, Krauss R S, Fabbro D, Weinstein I B. The epsilon isoform of protein kinase C is an oncogene when overexpressed in rat fibroblasts. Oncogene. 1993;8:2095–2104. [PubMed] [Google Scholar]
- 7.Caillet Fauquet P, Perros M, Brandenburger A, Spegelaere P, Rommelaere J. Programmed killing of human cells by means of an inducible clone of parvoviral genes encoding non-structural proteins. EMBO J. 1990;9:2989–2995. doi: 10.1002/j.1460-2075.1990.tb07491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christensen J, Cotmore S F, Tattersall P. A novel cellular site-specific DNA-binding protein cooperates with the viral NS1 polypeptide to initiate parvovirus DNA replication. J Virol. 1997;71:1405–1416. doi: 10.1128/jvi.71.2.1405-1416.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christensen J, Cotmore S F, Tattersall P. Parvovirus initiation factor (PIF): a novel human DNA binding factor which coordinately recognizes two ACGT motifs. J Virol. 1997;71:5733–5741. doi: 10.1128/jvi.71.8.5733-5741.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christensen J, Pederson M, Aasted B, Alexandersen S. Purification and characterization of the major nonstructural protein (NS1) of Aleutian mink disease parvovirus. J Virol. 1995;69:1802–1809. doi: 10.1128/jvi.69.3.1802-1809.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corbau R, Nüesch J P F, Salome N, Rommelaere J. Phosphorylation study of minute virus of mice NS1 protein, P20. VIIth International Parvovirus Workshop, Heidelberg, Germany. 1997. [Google Scholar]
- 12.Cornelis J J, Becquart P, Duponchel N, Salome N, Avalosse B L, Namba M, Rommelaere J. Transformation of human fibroblasts by ionizing radiation, a chemical carcinogen, or simian virus 40 correlates with an increase in susceptibility to the autonomous parvovirus H-1 and minute virus of mice. J Virol. 1988;62:1679–1686. doi: 10.1128/jvi.62.5.1679-1686.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cotmore S F, Tattersall P. An asymmetric nucleotide in the parvoviral 3′ hairpin directs segregation of a single active origin of DNA replication. EMBO J. 1994;13:4145–4152. doi: 10.1002/j.1460-2075.1994.tb06732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cotmore S F, Nüesch J P, Tattersall P. Asymmetric resolution of a parvovirus palindrome in vitro. J Virol. 1993;67:1579–1589. doi: 10.1128/jvi.67.3.1579-1589.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cotmore S F, Tattersall P. The autonomously replicating parvoviruses of vertebrates. Adv Virus Res. 1987;33:91–174. doi: 10.1016/s0065-3527(08)60317-6. [DOI] [PubMed] [Google Scholar]
- 16.Cotmore S F, Tattersall P. DNA replication in the autonomous parvoviruses. Semin Virol. 1995;6:271–281. [Google Scholar]
- 17.Cotmore S F, Nüesch J P, Tattersall P. In vitro excision and replication of 5′ telomeres of minute virus of mice DNA from cloned palindromic concatemer junctions. Virology. 1992;190:365–377. doi: 10.1016/0042-6822(92)91223-h. [DOI] [PubMed] [Google Scholar]
- 18.Cotmore S F, Tattersall P. In vivo resolution of circular plasmids containing concatemer junction fragments from minute virus of mice DNA and their subsequent replication as linear molecules. J Virol. 1992;66:420–431. doi: 10.1128/jvi.66.1.420-431.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cotmore S F, Tattersall P. The NS1 polypeptide of minute virus of mice is covalently attached to the 5′ termini of duplex replicative-form DNA and progeny single strands. J Virol. 1988;62:851–860. doi: 10.1128/jvi.62.3.851-860.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cotmore S F, Tattersall P. The NS1 polypeptide of the autonomous parvovirus MVM is a nuclear phosphoprotein. Virus Res. 1986;4:243–250. doi: 10.1016/0168-1702(86)90003-1. [DOI] [PubMed] [Google Scholar]
- 21.Cotmore S F, Christensen J, Nüesch J P, Tattersall P. The NS1 polypeptide of the murine parvovirus minute virus of mice binds to DNA sequences containing the motif [ACCA]2-3. J Virol. 1995;69:1652–1660. doi: 10.1128/jvi.69.3.1652-1660.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cotmore S F, D’Abramo A M, Carbonell L F, Bratton J, Tattersall P. The NS2 polypeptide of parvovirus MVM is required for capsid assembly in murine cells. Virology. 1997;231:267–289. doi: 10.1006/viro.1997.8545. [DOI] [PubMed] [Google Scholar]
- 23.Cotmore S F, Tattersall P. Organization of nonstructural genes of the autonomous parvovirus minute virus of mice. J Virol. 1986;58:724–732. doi: 10.1128/jvi.58.3.724-732.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cziepluch C, Kordes E, Poirey R, Grewenig A, Rommelaere J, Jauniaux J-C. Identification of a novel cellular TPR-containing protein, SGT, that interacts with the nonstructural protein NS1 of parvovirus H-1. J Virol. 1998;72:4149–4156. doi: 10.1128/jvi.72.5.4149-4156.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deleu L, Fuks F, Spitkovsky D, Horlein R, Faisst S, Rommelaere J. Opposite transcriptional effects of cyclic AMP-responsive elements on confluent or p27-kip-expressing cells versus serum-starved or growing cells. Mol Cell Biol. 1998;18:409–419. doi: 10.1128/mcb.18.1.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dettwiler, S. Unpublished observations.
- 27.Doerig C, Hirt B, Antonietti J P, Beard P. Nonstructural proteins of parvovirus B19 and minute virus of mice control transcription. J Virol. 1990;64:387–396. doi: 10.1128/jvi.64.1.387-396.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dornreiter I, Erdile L F, Gilbert I U, von Winkler W D, Kelly T J, Fanning E. Interaction of DNA polymerase alpha-primase with cellular replication protein A and SV40 T antigen. EMBO J. 1992;11:769–779. doi: 10.1002/j.1460-2075.1992.tb05110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dornreiter I, Hoss A, Arthur A K, Fanning E. SV40 T antigen binds directly to the catalytic subunit of DNA polymerase a. EMBO J. 1990;9:3329–3336. doi: 10.1002/j.1460-2075.1990.tb07533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finkenzeller G, Marme D, Hug H. Sequence of human protein kinase C a. Nucleic Acids Res. 1990;18:2183. doi: 10.1093/nar/18.8.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 32.Huang K-P, Huang F L. Purification and analysis of protein kinase C isozymes. In: Hunter T, Sefton B M, editors. Protein phosphorylation, part A. Vol. 200. San Diego, Calif: Academic Press, Inc.; 1991. pp. 241–252. [DOI] [PubMed] [Google Scholar]
- 33.Keranen L M, Dutil E M, Newton A C. Protein kinase C is regulated in vivo by three functionally distinct phosphorylations. Curr Biol. 1995;5:1394–1403. doi: 10.1016/s0960-9822(95)00277-6. [DOI] [PubMed] [Google Scholar]
- 34.Kochs G, Hummel R, Meyer D, Hug H, Marme D, Sarre T F. Activation and substrate specificity of the human protein kinase C a and z isoenzymes. Eur J Biochem. 1993;216:597–606. doi: 10.1111/j.1432-1033.1993.tb18179.x. [DOI] [PubMed] [Google Scholar]
- 35.Kornberg A, Baker T A. DNA replication. 2nd ed. New York, N.Y: W.H. Freeman & Co.; 1992. [Google Scholar]
- 36.Krady J K, Ward D C. Transcriptional activation by the parvoviral nonstructural protein NS-1 is mediated via a direct interaction with Sp1. Mol Cell Biol. 1995;15:524–533. doi: 10.1128/mcb.15.1.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuranami M, Powell C T, Hug H, Zeng Z, Cohen A M, Guillem J G. Differential expression of protein kinase C isoforms in human colorectal cancers. J Surg Res. 1995;58:233–239. doi: 10.1006/jsre.1995.1036. [DOI] [PubMed] [Google Scholar]
- 38.Legendre D, Rommelaere J. Terminal regions of the NS1 protein of the parvovirus minute virus of mice are involved in cytotoxicity and promoter trans-inhibition. J Virol. 1992;66:5705–5713. doi: 10.1128/jvi.66.10.5705-5713.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li X, Rhode S L., III Mutation of lysine 405 to serine in the parvovirus H-1 NS1 abolishes its functions for viral DNA replication, late promoter transactivation, and cytotoxicity. J Virol. 1990;64:4654–4660. doi: 10.1128/jvi.64.10.4654-4660.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lorson C, Pearson J, Burger L, Pintel D J. An Sp1-binding site and TATA element are sufficient to support full transactivation by proximally bound NS1 protein of minute virus of mice. Virology. 1998;240:326–337. doi: 10.1006/viro.1997.8940. [DOI] [PubMed] [Google Scholar]
- 41.Mackett M, Smith G L, Moss B. The construction and characterization of vaccinia virus recombinants expressing foreign genes. In: Glover D M, editor. DNA cloning: a practical approach. Oxford, United Kingdom: IRL Press; 1985. [Google Scholar]
- 42.Mantovani R, Li X-Y, Pessara U, van Huisjduijnen R H, Benoist C, Mathis D. Dominant negative analogs of NF-YA*. J Biol Chem. 1994;269:20340–20346. [PubMed] [Google Scholar]
- 43.Margolis G, Kilham L. Problems of human concern arising from animal models of intrauterine and neonatal infections due to viruses: a review. II. Pathogenic studies. Prog Med Virol. 1975;20:144–179. [PubMed] [Google Scholar]
- 44.Marks F, Gschwendt M. Protein kinase C. In: Marks F, editor. Protein phosphorylation. Weinheim, Germany: VCH Verlagsgesellschaft mbH; 1996. pp. 81–116. [Google Scholar]
- 45.McVey D, Ray S, Gluzman Y, Berger L, Wildeman A G, Marshak D R, Tegtmeyer P. cdc2 phosphorylation of threonine 124 activates the origin-unwinding functions of simian virus 40 T antigen. J Virol. 1993;67:5206–5215. doi: 10.1128/jvi.67.9.5206-5215.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McVey D, Woelker B, Tegtmeyer P. Mechanisms of simian virus 40 T-antigen activation by phosphorylation of threonine 124. J Virol. 1996;70:3887–3893. doi: 10.1128/jvi.70.6.3887-3893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McVey D, Brizuela L, Mohr I, Marshak D R, Gluzman Y. Phosphorylation of large tumor virus antigen by cdc2 stimulates SV40 DNA replication. Nature. 1989;341:503–507. doi: 10.1038/341503a0. [DOI] [PubMed] [Google Scholar]
- 48.Melendy T, Stillman B. An interaction between replication protein A and SV40 T antigen appears essential for primosome assembly during SV40 DNA replication. J Biol Chem. 1993;268:3389–3395. [PubMed] [Google Scholar]
- 49.Mischak H, Goodnight J A, Kolch W, Martiny-Baron G, Schaechtle C, Kazanietz M G, Blumberg P M, Pierce J H, Mushinski J F. Overexpression of protein kinase C-delta and -epsilon in NIH 3T3 cells induces opposite effects on growth, morphology, anchorage dependence and tumorigenicity. J Biol Chem. 1993;268:6090–6096. [PubMed] [Google Scholar]
- 50.Moarefi I F, Small D, Gilbert I, Hopfne M, Randall S K, Schneider C, Russo A A, Ramsperger U, Arthur A K, Stahl H, Kelly T J, Fanning E. Mutation of the cyclin-dependent kinase phosphorylation site in simian virus 40 (SV40) large T antigen specifically blocks SV40 origin DNA unwinding. J Virol. 1993;67:4992–5002. doi: 10.1128/jvi.67.8.4992-5002.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Molitor T W, Joo H S, Collett M S. Identification and characterization of a porcine parvovirus nonstructural polypeptide. J Virol. 1985;55:554–559. doi: 10.1128/jvi.55.3.554-559.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moss B, Elroy Stein O, Mizukami T, Alexander W A, Fuerst T R. Product review. New mammalian expression vectors. Nature. 1990;348:91–92. doi: 10.1038/348091a0. [DOI] [PubMed] [Google Scholar]
- 53.Mousset S, Ouadrhiri Y, Caillet Fauquet P, Rommelaere J. The cytotoxicity of the autonomous parvovirus minute virus of mice nonstructural proteins in FR3T3 rat cells depends on oncogene expression. J Virol. 1994;68:6446–6453. doi: 10.1128/jvi.68.10.6446-6453.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Naeger L K, Cater J, Pintel D J. The small nonstructural protein (NS2) of minute virus of mice is required for efficient DNA replication and infectious virus production in a cell-type-specific manner. J Virol. 1990;64:6166–6175. doi: 10.1128/jvi.64.12.6166-6175.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ni T-H, McDonald W F, Zolotukhin I, Melendy T, Waga S, Stillman B, Muzyczka N. Cellular proteins required for adeno-associated virus DNA replication in the absence of adenovirus coinfection. J Virol. 1998;72:2777–2787. doi: 10.1128/jvi.72.4.2777-2787.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nüesch, J. P. Unpublished observations.
- 57.Nüesch J P, Cotmore S F, Tattersall P. Expression of functional parvoviral NS1 from recombinant vaccinia virus: effects of mutations in the nucleotide-binding motif. Virology. 1992;191:406–416. doi: 10.1016/0042-6822(92)90202-z. [DOI] [PubMed] [Google Scholar]
- 58.Nüesch J P, Tattersall P. Nuclear targeting of the parvoviral replicator protein molecule NS1: evidence for self-association prior to nuclear transport. Virology. 1993;196:637–651. doi: 10.1006/viro.1993.1520. [DOI] [PubMed] [Google Scholar]
- 59.Nüesch J P, Cotmore S F, Tattersall P. Sequence motifs in the replicator protein of parvovirus MVM essential for nicking and covalent attachment to the viral origin: identification of the linking tyrosine. Virology. 1995;209:122–135. doi: 10.1006/viro.1995.1236. [DOI] [PubMed] [Google Scholar]
- 60.Nüesch J P F, Corbau R, Tattersall P, Rommelaere J. Biochemical activities of minute virus of mice nonstructural protein NS1 are modulated in vitro by the phosphorylation state of the polypeptide. J Virol. 1998;72:8002–8012. doi: 10.1128/jvi.72.10.8002-8012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parker P J, Dekker L V. Protein kinase C. New York, N.Y: Springer; 1997. [Google Scholar]
- 62.Parker P J, Marais R M. Purification of protein kinase C isotypes from bovine brain. In: Hunter B M S T, editor. Protein phosphorylation, part A. Vol. 200. San Diego, Calif: Academic Press, Inc.; 1991. pp. 234–241. [DOI] [PubMed] [Google Scholar]
- 63.Pujol A, Deleu L, Nüesch J P F, Cziepluch C, Jauniaux J-C, Rommelaere J. Inhibition of parvovirus minute virus of mice replication by a peptide involved in the oligomerization of the nonstructural protein NS1. J Virol. 1997;71:7397–7403. doi: 10.1128/jvi.71.10.7393-7403.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rhode S L I, Richard S M. Characterization of the trans-activation-responsive element of the parvovirus H-1 P38 promoter. J Virol. 1987;61:2807–2815. doi: 10.1128/jvi.61.9.2807-2815.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rommelaere J, Cornelis J J. Antineoplastic activity of parvoviruses. J Virol Methods. 1991;33:233–251. doi: 10.1016/0166-0934(91)90024-t. [DOI] [PubMed] [Google Scholar]
- 66.Schoborg R V, Pintel D J. Accumulation of MVM gene products is differentially regulated by transcription initiation. Virology. 1991;181:22–34. doi: 10.1016/0042-6822(91)90466-o. [DOI] [PubMed] [Google Scholar]
- 67.Snyder R O, Im D-S, Ni T, Xiao X, Samulsky R J, Muzyczka N. Features of the adeno-associated virus origin involved in substrate recognition by the viral Rep protein. J Virol. 1993;67:6096–6104. doi: 10.1128/jvi.67.10.6096-6104.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tsurimoto T, Stillman B. Replication factors required for SV40 DNA replication in vitro. I. DNA structure-specific recognition of a primer-template junction by eucaryotic DNA polymerases and their accessory proteins. J Biol Chem. 1991;266:1950–1960. [PubMed] [Google Scholar]
- 69.Tsurimoto T, Melendy T, Stillman B. Sequential initiation of lagging and leading strand synthesis by two different polymerase complexes at the SV40 DNA replication origin. Nature. 1990;346:534–539. doi: 10.1038/346534a0. [DOI] [PubMed] [Google Scholar]
- 70.Vanacker J-M, Corbau R, Adelmant G, Perros M, Laudet V, Rommelaere J. Transactivation of a cellular promoter by the NS1 protein of the parvovirus minute virus of mice through a putative hormone-responsive element. J Virol. 1996;70:2369–2377. doi: 10.1128/jvi.70.4.2369-2377.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weisshart K, Fanning E. Roles of phosphorylation in DNA replication. In: DePamphilis M L, editor. DNA replication in eucaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 295–330. [Google Scholar]