Abstract
Recent interest in the use of porcine organs, tissues, and cells for xenotransplantation to humans has highlighted the need to characterize the properties of pig endogenous retroviruses (PERVs). Analysis of a variety of pig cells allowed us to isolate and identify three classes of infectious type C endogenous retrovirus (PERV-A, PERV-B, and PERV-C) which have distinct env genes but have highly homologous sequences in the rest of the genome. To study the properties of these env genes, expression plasmids for the three env genes were constructed and used to generate retrovirus vectors bearing corresponding Env proteins. Host range analyses by the vector transduction assay showed that PERV-A and PERV-B Envs have wider host ranges, including several human cell lines, compared with PERV-C Env, which infected only two pig cell lines and one human cell line. All PERVs could infect pig cells, indicating that the PERVs have a potential to replicate in pig transplants in immunosuppressed patients. Receptors for PERV-A and PERV-B were present on cells of some other species, including mink, rat, mouse, and dog, suggesting that such species may provide useful model systems to study infection and pathogenicity of PERV. In contrast, no vector transduction was observed on nonhuman primate cell lines, casting doubt on the utility of nonhuman primates as models for PERV zoonosis. Interference studies showed that the three PERV strains use receptors distinct from each other and from a number of other type C mammalian retroviruses.
Pig-to-human xenotransplantation has the potential to alleviate the shortage of allogeneic organs for transplantation (1, 25). In addition, it may also allow the development of novel therapies by providing unlimited supplies of cells and tissues (9, 11, 13, 18). Recently, substantial progress has been made in overcoming immunological barriers to cross-species transplantation (25, 27). At the same time, however, serious concerns that zoonotic infections might occur as a result of xenotransplantation have been expressed (1, 6, 30). Our report that an established pig cell line produces a porcine endogenous retrovirus (PERV) that can infect human cells fueled these concerns (23). Subsequently, the isolation of human tropic PERV from stimulated miniswine peripheral blood lymphocytes (38) has shown that normal pig cells can also produce potentially hazardous virus. PERVs may be difficult to eliminate from donor animals because multiple copies of PERV genomes are present in normal pig genomes (2, 16, 23). PERV infection may have serious impact on the health of not only transplant recipients but also the human population at large, if spread of an undetected infectious agent into the community were to take place (3, 31). To assess the risk posed by the PERVs for pig-to-human transplantation, a greater understanding of the properties of the PERVs is required.
Sequence analyses indicate that the infectious PERVs are closely related to one another in their gag and pol genes, with maximum amino acid divergence of around 5% (2, 16a, 23). The PERVs are members of the mammalian type C retrovirus genus showing closest homology to the gibbon ape leukemia virus (GALV) pol gene, with about 70% amino acid identity, and 60 to 70% identity to murine leukemia viruses (MLV). However, three distinct env genes have now been identified in PERV clones. Two of these env genes, PERV-A and PERV-B, were cloned from human 293 cells infected with PK15 virus (16). The third distinct class of PERV env gene, here designated PERV-C, was reported as a part of a full-length PERV genome isolated from miniature swine lymphocytes (PERV-MSL) and from a swine lymphoma (PERV-Tsukuba-1) (2, 32). The three types show marked differences in the VRA, VRB, and PRO regions of SU surface glycoprotein (2, 16). Differences in these regions determine the host range specificity of the different classes of MLV (4, 5). These observations suggest that the PERVs belong to three distinct classes with different host range specificities. To test this idea, the functions of the three types of PERV env gene were examined and correlated to production, infection, and replication of PERVs in cell culture. Recombinant retrovirus vectors bearing PERV Env proteins were developed and their host ranges, cell tropism, and interference with each other as well as with other type C retroviruses were examined. The results of these experiments are the subject of this report.
MATERIALS AND METHODS
Viruses and cell culture.
The origins of the MLV-A 1504, MLV-X NZB, feline endogenous retrovirus RD114, feline leukemia virus (FeLV)-B, and GALV SF virus strains have been described previously (29). A polytropic MLV AKR13 strain (MCF) and the FeLV-C Sarma strain were kind gifts from B. Chesebro and O. Jarrett, respectively. Recombinant, helper-free lacZ pseudotypes bearing Env proteins of MLV-A 4070A, RD114, and GALV SEATO (26) were employed. lacZ pseudotypes were rescued from TEL cells (human TE671 cells harboring MFGnlslacZ vector) by replication-competent MLV-X, FeLV-B, and FeLV-C or from ML cells (mink Mv-1-Lu cells with MFGnlslacZ) by MCF as described previously (8, 33, 34). Cynomolgus monkey skin fibroblast CYNOM-K1, chimpanzee skin fibroblast CP132, and baboon lymphoblast 26CB1 cells were obtained from the European Collection of Animal Cell Cultures. A pig aorta endothelial cell line, PAE (21), was kindly provided by C. J. Marshall. The other cell lines have been described in previous reports by researchers from the Institute of Cancer Research (8, 23, 29, 35). Two different batches of the human fibrosarcoma HT1080 cell line, HT1080-1 and HT1080-2, and its derivative, the FLYRD18 packaging cell line (8), were examined. Virus transmission was carried out by either cocultivation with irradiated producer cells or cell-free infection by using 0.45-μm-pore-size filtered cell supernatants (23). Titration of lacZ pseudotypes was performed in 48-well multiwell plates as described previously (34).
lacZ pseudotype viruses bearing PERV Envs.
The following clones were used: PERV-A, an env clone which differs from the reported consensus sequence (EMBL y12238) at three amino acids (N-to-S change at position 348, K-to-N change at 448, and R-to-G change at 583); PERV-B, an env clone which differs from the consensus sequence (EMBL y12239) at three amino acids (G-to-E change at 127, T-to-S change at 419, and S-to-P change at 595) (16); and PERV-C, a full-length cDNA clone (GenBank AF038600) (2). KasI-EcoRI fragments (corresponding to nucleotides [nt] 230 to 2312 from y12238, nt 940 to 3003 from y12239, and nt 5639 to 7636 from AF038600), which contain the env coding sequence except the first seven amino acids in the putative signal sequence, were subcloned into the BamHI-EcoRI cloning sites of BlueScript KS- (Stratagene) together with a synthetic double strand linker with BamHI and KasI sticky ends. This linker was formed by annealing positive-strand (5′-GATCCTCTAGACCACCATGCATCCCACGTTAAGCTG-3′) and negative-strand (5′-GCGCCAGCTTAACGTGGGATGCATGGTGGTCTAGAG-3′) oligonucleotides. It contains a new XbaI site and an improved Kozak sequence, CCACC, upstream of the sequences encoding the first eight amino acids of Env. Use of this oligonucleotide introduced an R-to-W change in PERV-A and PERV-C sequences at amino acid 7. The env coding sequences were excised by XbaI (in the synthetic oligonucleotide) and ClaI (in the BlueScript multiple cloning site downstream of the EcoRI site) and cloned into the FBMOSALF env expression plasmid (7) from which most of the Moloney MLV env coding sequence had been removed by digestion with XbaI and ClaI (the XbaI site is located 12 nt upstream of the env initiation codon; the ClaI site is found just upstream of the membrane spanning domain of TM), resulting in PERV Env expression plasmids. These plasmids were transfected into TELCeB6 cells, which express high amounts of MLV core particles incorporating an MFGnlslacZ vector (8). After selection of the transfectants with 50-μg/ml phleomycin, virus supernatant was harvested from the pooled phleomycin-resistant cell population, filtered (0.45-μm pore size), and used immediately in lacZ infection assays.
Reverse transcriptase (RT) assay and RT-PCR.
To assess RT activity, 500 μl of culture supernatant was filtered (0.45-μm pore size), pelleted at 355,000 × g for 15 min, and resuspended in 10 μl of 1% Nonidet P-40. Five microliters was then tested in a PCR-based method (28). PERV-A- and PERV-B-specific RT-PCR amplifications were carried out with the previously reported primer pairs PL170-PL171 and PL172-PL173 (16), respectively. For specific detection of PERV-C, a new primer pair, PL205 (5′-CTGACCTGGATTAGAACTGG-3′) plus PL206 (5′-ATGTTAGAGGATGGTCCTGG-3′), was designed. cDNA was synthesized with an RT-PCR kit (Stratagene) according to the manufacturer’s instructions. The PCR conditions were 92°C for 4 min (1 cycle), 94°C for 30 s, annealing (at 53°C for PERV-A and PERV-B and 58°C for PERV-C) for 45 s, 72°C for 30 s (30 cycles), and 72°C for 5 min (1 cycle).
RESULTS
Production and transmission of PERVs.
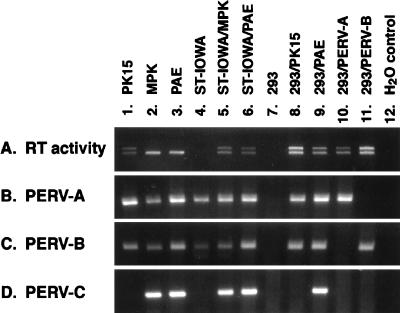
As a starting point for our analyses of the biological properties of the different classes of PERV we examined the expression and infectivity of endogenous retroviruses produced by a number of porcine cell lines. As we have previously reported (23), pig PK15 and MPK but not ST-IOWA cells produce infectious retroviruses with RT activity (Fig. 1, gel A). Viruses produced by PK15 can infect both ST-IOWA and human 293 cells, whereas viruses from MPK cells infect ST-IOWA but not 293 cells (23). A fourth cell line, PAE, also produces RT activity transmissible to both 293 and ST-IOWA cells.
FIG. 1.
Transmission of RT activity and PERV RNA expression. Cell supernatant was assayed for RT activity by a PCR-based method (gel A). RNA expression for PERV-A, PERV-B, and PERV-C was examined by RT-PCR (gels B, C, and D). ST-IOWA/MPK, ST-IOWA cells after cocultivation with MPK cells; ST-IOWA/PAE, ST-IOWA cells after cocultivation with PAE cells; 293/PK15, 293 cells infected with PK15 viral supernatant; 293/PAE, 293 cells after cocultivation with PAE cells.
RT-PCR assays specifically detecting expression of PERV-A or PERV-B showed that all four pig cell lines express PERV-A and PERV-B env genes regardless of infectious virus production and that both PERV-A and PERV-B were transmitted from PK15 and PAE cells to 293 cells (Fig. 1, gels B and C). In order to clarify if both PERV-A and PERV-B were infectious for 293 cells or if only one was infectious and the other was transmitted by phenotypic mixing, biological separation of the two virus strains was carried out. Individual wells of 48-well multiwell dishes plated with 10,000 293 cells were challenged with threefold serial dilutions of the supernatant of PK15 cells, cultured for 4 weeks, and then assayed for RT production and PERV-A RNA and PERV-B RNA expression. Five of 12 cultures challenged with 12 μl of supernatant and 2 of 5 cultures challenged with 4 μl of supernatant tested positive for RT, indicating that the composite 50% tissue culture infectious dose of PK15 supernatant was about 100/ml. By RT-PCR, one culture expressed only PERV-A, and not PERV-B, and was designated 293/PERV-A (Fig. 1, lane 10). Six cultures were found to express PERV-B, but not PERV-A, and one of them was named 293/PERV-B and used for further study (Fig. 1, lane 11). RT activity was transmissible from 293/PERV-A and 293/PERV-B to ST-IOWA cells and mink Mv-1-Lu cells by cocultivation (data not shown). These results demonstrate that both PERV-A and PERV-B can individually infect 293, ST-IOWA, and Mv-1-Lu cells.
A primer pair specific to PERV-C env was designed to examine the expression of PERV-C env (Fig. 1, gel D). MPK and PAE cells, but neither PK15 nor ST-IOWA cells, expressed PERV-C. PERV-C expression was transmitted to ST-IOWA by cocultivation with irradiated MPK or PAE cells. Because the virus from MPK cells infects ST-IOWA cells, but not 293 cells, we hypothesized that PERV-C Env is responsible for transmission of RT activity from MPK to ST-IOWA cells and that PERV-C has a more limited cell tropism than PERV-A and PERV-B. On the other hand, we did observe transmission of the PERV-C genome from PAE to 293 cells. However, this was accompanied by transmission of PERV-A and PERV-B (Fig. 1, lanes 9), making it more likely that transfer of PERV-C occurred as a result of phenotypic mixing rather than that PERV-C produced by PAE cells has a wider host range. This question will be addressed further below.
Generation of recombinant lacZ vectors bearing PERV Env proteins.
PERV env genes were subcloned in a FBSALF env expression plasmid (7) and transfected into TELCeB6 cells which produce an MLV-based vector core encoding a lacZ marker gene (8). Virus harvests from pooled populations of phleomycin-resistant transfectants were tested for transduction of the lacZ gene on 293 and ST-IOWA cells and their derivatives which had been infected with different PERVs. Table 1 shows the titers of recombinant lacZ pseudotypes for the PERV subtypes, lacZ(PERV) pseudotypes, together with RD114 [lacZ(RD114)] produced by a similar construct (26) as a control. A helper-positive lacZ pseudotype produced by PAE, lacZ(PAE), which was predicted to contain all three lacZ(PERV)s, was also tested.
TABLE 1.
Rescue and self-interference of lacZ pseudotypes by recombinant PERV envelopes
| Target cell (cell/virus) |
lacZ titer (U/ml)a
|
||||
|---|---|---|---|---|---|
| PERV-Ab | PERV-Bb | PERV-Cb | PAEc | RD114b | |
| 293 | 1,900 | 2,300 | <2 | 400 | 500,000 |
| 293/PERV-A | 2 | 1,600 | <2 | 36 | 600,000 |
| 203/PERV-B | 2,100 | <2 | <2 | 162 | 460,000 |
| 293/PK15 | <2 | <2 | <2 | 2 | 480,000 |
| 293/PAE | 2 | 4 | <2 | 2 | 650,000 |
| ST-IOWA | 35,000 | 940 | 430 | 7,300 | 250,000 |
| ST-IOWA/PERV-A | 560 | 460 | 560 | 2,600 | 190,000 |
| ST-IOWA/PERV-B | 36,000 | 110 | 410 | 7,100 | 180,000 |
| ST-IOWA/MPK | 9,600 | 204 | <2 | 130 | 84,000 |
| ST-IOWA/PK15 | 290 | 60 | 300 | 2,300 | 220,000 |
| ST-IOWA/PAE | 530 | <2 | <2 | 4 | 130,000 |
Representative titers of at least two independent experiments are shown.
Recombinant lacZ pseudotypes were produced by stable transfection of env expression plasmids for PERV-A, PERV-B, and PERV-C, and RD114 into TELCeB6 cells.
lacZ(PAE) was produced by transduction of PAE cells with helper-free lacZ(GALV).
lacZ(PERV-A) and lacZ(PERV-B) plated on both 293 and ST-IOWA cells, consistent with the tropism of replication-competent PERV-A and PERV-B produced by PK15 cells (see above). In contrast, lacZ(PERV-C) plated on ST-IOWA cells, but not 293 cells, consistent with the host range seen for PERV-C produced by MPK cells. The lacZ(PERV-A) titer was reproducibly higher on ST-IOWA cells than on 293 cells, whereas the lacZ(PERV-B) titer was higher on 293 cells than on ST-IOWA cells. All three lacZ(PERV) titers were relatively low compared with many other standard type C pseudotypes, including lacZ(RD114) (Tables 1 and 2), suggesting that PERV envelope-receptor systems may be less efficient for virus entry to cells than those of some other well-studied type C viruses. This property may be responsible for the relatively low titer of replication-competent viruses produced by PK15 cells (see above) (23).
TABLE 2.
Cross-interference between mammalian type C retroviruses
| Target cell (cell/virus) | Blocking of lacZ pseudotypesa
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PERV-A | PERV-B | PERV-C | MLV-A | MLV-X | MCF | RD114 | GALV | FeLV-B | FeLV-C | |
| ST-IOWA/PERV-A | ++ | − | − | NA | − | NA | − | − | − | − |
| ST-IOWA/PERV-B | − | + | − | NA | − | NA | − | − | − | − |
| ST-IOWA/MPK | +/− | − | ++ | NA | − | NA | − | − | − | − |
| 293/PERV-A | ++ | − | NA | − | − | − | − | − | − | − |
| 293/PERV-B | − | ++ | NA | − | − | − | − | − | − | − |
| 293/MLV-A | − | − | NA | ++ | − | − | − | − | − | − |
| 293/MLV-X | − | − | NA | − | ++ | ++ | − | − | − | − |
| 293/MCF | − | − | NA | − | − | ++ | − | − | − | − |
| 293/RD114 | − | − | NA | − | − | − | ++ | − | − | − |
| 293/GALV | − | − | NA | − | − | − | − | ++ | ++ | − |
| 293/FeLV-B | − | − | NA | − | − | − | − | ++ | ++ | − |
| 293/FeLV-C | − | − | NA | − | − | − | − | − | − | ++ |
−, >20% infectivity compared with that on uninfected cells; +, 2 to 20%; ++, <2%; +/−, occasionally <20%; NA, not applicable because these viruses had titers of <10 U/ml on uninfected cells. Two or more independent experiments were carried out.
PERV interference.
The lacZ pseudotype vectors were used to investigate the receptor specificities of PERV-A, PERV-B and PERV-C (Table 1). Preinfection of 293 cells with PERV-A and PERV-B interfered with lacZ(PERV-A) and lacZ(PERV-B), respectively. No cross-interference was seen, implying that these Envs recognize different receptors. As expected, both pseudotypes were blocked on 293 cells expressing both PERV-A and PERV-B after infection with supernatants of PK15 and PAE cells. lacZ(PAE) infection was effectively blocked on 293/PK15 and 293/PAE cells, but only partially blocked on 293/PERV-A and 293/PERV-B cells, indicating that PAE cells produce functional Env proteins of both PERV-A and PERV-B. Because PERV-C env was transmitted from PAE to 293 and ST-IOWA cells (Fig. 1), it is possible that lacZ(PAE) contains lacZ pseudotype particles bearing PERV-C Env proteins. However, it is unlikely that such PERV-C pseudotypes can infect 293 cells, because lacZ(PAE) was effectively blocked on 293/PK15 cells which produce PERV-A and PERV-B but do not contain PERV-C. This observation substantiates our hypothesis that PERV-C genomes can be transmitted to human cells by phenotypic mixing with PERV-A or PERV-B even if PERV-C itself is not infectious to human cells.
Self-interference of PERV-A was also observed on ST-IOWA cells, although that of PERV-B was modest (Table 1). lacZ(PERV-C) showed interference by viruses from MPK and PAE cells, but not by PERV-A, PERV-B, or their mixture (PK15 virus), consistent with the expression of PERV-C in these cells. No significant cross-interference among the three PERVs was observed except that lacZ(PERV-A) reproducibly plated on ST-IOWA/MPK at a reduced titer (10 to 30% of the titer on uninfected ST-IOWA) (Tables 1 and 2). It is possible that PERV-A env genes, which are not infectious but are capable of interfering with infectious PERV-A Env, were transmitted from MPK to ST-IOWA cells. ST-IOWA/PAE was refractory to all three lacZ(PERV) pseudotypes, consistent with the interpretation that PAE cells produce all three classes of PERV. Taken together, these data imply that PERV-A, PERV-B, and PERV-C use different receptors, thereby justifying placement of viruses carrying the three env genes into three different classes.
Cross-interference between PERVs and other type C retroviruses.
Human 293 cells chronically infected with different type C retroviruses were established. These cultures as well as PERV-infected 293 and ST-IOWA cells were tested for infection with lacZ vectors bearing different virus envelopes. Table 2 summarizes the relative infectivity on the preinfected cultures compared with that on uninfected parental cells. No cross-interference between PERVs and the other type C viruses tested was observed, while a full cross-interference between GALV and FeLV-B and a nonreciprocal interference between MLV-X and MCF were found, in accordance with the previous reports that these pairs of viruses each belong to a single receptor-interference group (17, 29, 36). PERV-A and PERV-B therefore appear to form separate, independent receptor interference groups on human cells. However, we reproducibly observed a reduction of titer of lacZ(PERV-A) on mink Mv-1-Lu cells preinfected with MCF (approximately 10% of the titer on uninfected Mv-1-Lu cells; data not shown), suggesting that MCF and PERV-A may share a receptor or that their receptors may be structurally or functionally related. However, Mv-1-Lu/MLV-X was fully sensitive to lacZ(PERV-A) and fully resistant to lacZ(MCF) (data not shown). Some human cells (e.g., TE671 and HeLa cells; see Table 3) were sensitive to lacZ(PERV-A) but not lacZ(MCF). These observations suggest that PERV-A does not belong to the MLV-X/MCF receptor group and that MCF and PERV-A do not have a common receptor. It therefore seems likely that the PERVs interact with receptors different from those used by other members of the mammalian C type retrovirus genus.
TABLE 3.
Host range and cell tropism of PERV-A, PERV-B, and PERV-C envelopes
| Species | Cell | Titer (U/ml) of lacZ pseudotypea
|
||
|---|---|---|---|---|
| PERV-A | PERV-B | PERV-C | ||
| Pig | PK15 | <4 | 2,100b | 1,550 |
| ST-IOWA | 30,000 | 940 | 3,700 | |
| PAE | 6 | <2 | <2 | |
| MPK | 164 | <4 | <4 | |
| Human | 293 | 12,000 | 3,800 | <2 |
| TE671 | 5,800 | 320 | <2 | |
| HeLa | 24,000 | 420 | <2 | |
| HT1080-1 | 82 | 290 | 890 | |
| HT1080-2 | 16 | 350 | 10 | |
| FLYRD18 | 48 | 25 | 12 | |
| HOS | <2 | <2 | <2 | |
| U87 | 830 | 120 | <2 | |
| MRC-5 | 150 | <2 | <2 | |
| SupT1 | <2 | 2 | <2 | |
| Raji | 2 | 200 | <2 | |
| Molt-4 | 2 | 2 | <2 | |
| Nonhuman primates | ||||
| African green monkey | Vero | <2 | <2 | <2 |
| COS-7 | <2 | <2 | <2 | |
| Rhesus | FRhK | <2 | <2 | <2 |
| Cynomolgus | CynomK1 | <2 | <2 | NT |
| Baboon | 26CB1 | <2 | <2 | <2 |
| Chimpanzee | CP132 | <2 | <2 | NT |
| Mink | Mv-1-Lu | 7,800 | 5,600 | <2 |
| Mouse | NIH 3T3 | <4 | 2,100 | <2 |
| M. dunni | <4 | 2,100 | <2 | |
| Rat | NRK | <4 | 9,600 | <2 |
| HSN | <4 | 12,400 | <2 | |
| Hamster | CHO | 32 | <2 | <2 |
| BHK | <2 | <2 | <2 | |
| Rabbit | SIRC | <4 | 1,380 | <2 |
| Bat | Tb-1-Lu | <2 | 14 | <2 |
| Dog | Th2CfS+L− | <2 | <2 | <2 |
| D17 | 760 | <2 | <2 | |
| Cat | CCCS+L− | 190 | <2 | <2 |
Averages of duplicate titers of lacZ pseudotypes are shown. Errors were less than 30% of the values. All cultures except hamster cells were sensitive to control pseudotypes, lacZ(RD114), lacZ(GALV), or lacZ(MLV-A). BHK and CHO cells could be transduced by lacZ(MLV-E) after tunicamycin treatment. NT, not tested.
PK15 cells were resistant to lacZ(PERV-B) in an earlier, preliminary experiment.
Host range and cell tropism of PERVs.
Thirty-two cell lines of various origins were screened for expression of PERV receptors by infection with lacZ pseudotypes (Table 3). Four pig cell lines were differentially susceptible to the three lacZ(PERV)s, presumably reflecting, at least in part, their endogenous expression of replication-competent PERVs which may interfere with lacZ pseudotypes. It therefore seems likely that many, if not all, pigs carry the receptor genes allowing entry of these three classes of PERV and that PERV may actively replicate in pigs at some stage of their life.
lacZ(PERV-A) and lacZ(PERV-B) plated on several human cell lines, in accordance with our previous report that PERV produced by PK15 cells could be transmitted to several human cell lines and primary cell strains (23). The titer of lacZ(PERV-A) on 293 cells in this experiment was about sixfold higher than that in the earlier titration, described in Table 1, for unknown reason. This variability suggests that the level of PERV-A receptor can vary in some circumstances. Indeed, another batch of 293 cells appeared 10- to 50-fold less sensitive to lacZ(PERV-A) than the batch used in this paper (data not shown). lacZ(PERV-C) appeared to be more restricted in human cells even though, perhaps surprisingly, it did plate with a significant titer on a batch (HT1080-1) of the human fibrosarcoma HT1080 cell line (Table 3). This cell line was an early passage used shortly after receipt from the ATCC. To confirm this result we examined a clonal derivative of this HT1080 cell batch, FLYRD18 (8), as well as another independent batch of HT1080 cells, HT1080-2. Both were also weakly sensitive to lacZ(PERV-C). HT1080-1 and FLYRD18 cells were examined by PCR and fluorescence-activated cell sorter analyses to confirm their species origin. While a human endogenous retrovirus element RTVL-H sequence was amplified from both cell lines, a sensitive PCR assay for pig mitochondrial cytochrome oxidase subunit II gene (22) was negative (data not shown), excluding the possibility of pig cell contamination. Anti-human CD59 and anti-human β-microglobulin antibodies stained almost the entire cell population of HT1080-1 and FLYRD18 cells, but they did not stain ST-IOWA cells at all (data not shown). Thus, although PERV-C has more limited titer, host range, and cell tropism than PERV-A and PERV-B, PERV-C Env may have the potential to allow virus entry to some human cells.
Among the other species, none of the nonhuman primate cell lines was sensitive to any lacZ(PERV) (Table 3), raising doubt as to the utility of primate animal models for study of PERV zoonosis. In contrast, mink Mv-1-Lu cells were sensitive to both lacZ(PERV-A) and lacZ(PERV-B), consistent with the observation that replication-competent PERV-A and PERV-B could be transmitted from 293/PERV-A and 293/PERV-B, respectively. lacZ(PERV-A) plated on hamster CHO, dog D17, and cat CCCS+L− cells, while lacZ(PERV-B) plated on mouse, rat, and rabbit cells, suggesting that these species could be used as models to study in vivo infection and pathogenesis by PERVs. Animal cells infected with PERVs may also be useful for raising antibodies against PERVs in their syngeneic or allogeneic animals. It is noteworthy that different cell lines from the same species (e.g., dog, hamster, and human cells) showed variable susceptibility to lacZ(PERV) pseudotypes, suggesting that PERV receptors are not ubiquitously expressed and/or are polymorphic in their PERV Env recognition. It is possible that posttranslational modifications, such as glycosylation, of receptor molecules may control sensitivity to PERVs, although tunicamycin treatment (20, 37) of human HeLa, HOS, HT1080, and 293 cells and hamster BHK and CHO cells did not alter their sensitivities to PERVs significantly (data not shown).
DISCUSSION
Sequence studies have identified PERVs carrying three different env genes. In this study, we have shown that the three env genes specify interactions with three different receptors, justifying the division of the PERV element into three functional classes with different host ranges. Whether additional classes of PERV element exist remains an open question. Hybridization studies indicate that there are up to 50 PERV proviruses in the pig genome (2, 16, 23). It is not yet clear what fraction of these proviruses encode intact retroviruses, though our studies with the various cell lines provide evidence that there must be at least one functional germ line copy in each class. These studies also indicate that some of these elements are likely to be defective, since PERV-A and PERV-B env sequences are expressed by ST-IOWA and MPK cells but these cells are permissive to infection by these viruses and do not make virus capable of infecting human cells. Indeed, expression of deleted proviruses in a number of porcine lines has been detected (2, 16a). Many pig lines have been reported to spontaneously express RT activity (24), and it is important to determine whether this represents the transcription of a limited subset of the endogenous elements. Genomic cloning and expression of full-length PERV genomes are under way in order to estimate how many potentially infectious proviruses are inherited by different pig herds.
PERV-A and PERV-B are capable of infecting a number of human cell lines. Consequently, they have the potential to infect the recipients of pig-to-human xenotransplants, although there has been no evidence for pig-to-human retrovirus infection despite close interspecies contacts (e.g., in farmers and abattoir workers). It would therefore be desirable to eliminate functional proviruses from transplant donor animals or to control expression of these classes in transplant tissues and organs. Toward this aim, genomic mapping and functional characterization of the PERV loci are required. The class-specific primers for PCR and the specific hybridization probes prepared with these primers will be very useful for this purpose.
PERV-C appears to be of lesser concern. Under some circumstances, an MLV vector bearing PERV-C Env could infect human HT1080 cells (Table 3). Moreover, PERV-C env was transmitted from pig PAE cells to human 293 cells in the presence of PERV-A and PERV-B, presumably by phenotypic mixing. Because it is probable that transmission and coreplication of PERV-C with PERV-A or PERV-B may enhance the chance of recombination between PERVs and human retrovirus and related sequences to give rise to more harmful viruses, absence of PERV-C would be desirable for donor animals. While all pig cell lines and primary tissues tested so far express both PERV-A and PERV-B RNAs, expression of PERV-C transcripts could not be detected in ST-IOWA or PK15 cells (Fig. 1) and only a small proportion of PBL samples from different individual Landrace × Duroc pigs were positive for PERV-C RT-PCR (23a). Breeding of pigs free of PERV-C may not prove as difficult as eliminating PERV-A and PERV-B.
All three PERV Envs recognized receptors on some pig cell lines, suggesting that they have potential to replicate in pigs as well as in transplanted pig tissues in immunosuppressed humans. Once production of infectious PERV has occurred, integrations may occur at novel chromosomal sites favoring higher-level provirus expression, thereby increasing the risk of infection of a transplant recipient’s cells. Also of concern might be the effect of PERV replication on transplanted organs and/or cells. No link between PERV production and illness in pigs has been clearly established, although PERV-C was originally cloned from a malignant lymphoma cell line (32). More-detailed etiological studies of PERV expression and replication in aged pigs would be valuable.
A variety of further investigations are required to assess the potential infectious disease risk associated with xenotransplantation (10, 19). Initial clinical trials of xenotransplantation should be limited in scope and involve careful screening of patients and their contacts for evidence of zoonotic infections. Again, the virus class-specific probes we have developed may be useful for this purpose. Screening of patients already exposed to live pig cells and tissues may also be informative (22). At the same time, model animal experiments could shed light on the processes of in vivo infection and pathogenesis of PERVs. It is noteworthy that so far no lacZ(PERV) has plated on nonhuman primate cell lines, though it should be noted that the titers of PERV-B and PERV-C pseudotypes were only moderate on permissive cells. Old-world primates provide useful animal model recipients for pig-to-human xenotransplantation, because like humans they produce natural antibodies against a major xenoantigen, the galactosyl(α1-3)galactosyl sugar epitope (12, 14, 15). However, the result that all primate cell lines tested lack receptors for PERVs (Table 3) suggests that primate models may not be suitable for the studies of PERV zoonosis. Further comparative studies on sensitivity to PERVs with primary cells of nonhuman primate species and humans may allow the identification of appropriate models. Alternatively, the use of small animals may prove fruitful; we have here identified a number of animal species whose cells express receptors for PERV-A and/or PERV-B, and these species may be useful for model studies of PERV infection and pathogenicity.
ACKNOWLEDGMENTS
This work was supported by the Medical Research Council, the Health and Safety Executive, and BioTransplant, Inc.
REFERENCES
- 1.Advisory Group on the Ethics of Xenotransplantation for the Department of Health. Animal tissue into humans. Norwich, England: Her Majesty’s Stationery Office; 1997. [Google Scholar]
- 2.Akiyoshi D E, Denaro M, Zhu H, Greenstein J L, Banerjee P T, Fishman J A. Identification of a full-length cDNA for an endogenous retrovirus of miniature swine. J Virol. 1998;72:4503–4507. doi: 10.1128/jvi.72.5.4503-4507.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allan J S. Xenotransplantation at a crossroads: prevention versus progress. Nat Med. 1996;2:18–21. doi: 10.1038/nm0196-18. [DOI] [PubMed] [Google Scholar]
- 4.Battini J L, Danos O, Heard J M. Receptor-binding domain of murine leukemia virus envelope glycoproteins. J Virol. 1995;69:713–719. doi: 10.1128/jvi.69.2.713-719.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Battini J L, Heard J M, Danos O. Receptor choice determinants in the envelope glycoproteins of amphotropic, xenotropic, and polytropic murine leukemia viruses. J Virol. 1992;66:1468–1475. doi: 10.1128/jvi.66.3.1468-1475.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butler D. Last chance to stop and think on risks of xenotransplants. Nature. 1998;391:320–324. doi: 10.1038/34749. [DOI] [PubMed] [Google Scholar]
- 7.Cosset F L, Morling F J, Takeuchi Y, Weiss R A, Collins M K, Russell S J. Retroviral retargeting by envelopes expressing an N-terminal binding domain. J Virol. 1995;69:6314–6322. doi: 10.1128/jvi.69.10.6314-6322.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cosset F L, Takeuchi Y, Battini J L, Weiss R A, Collins M K. High-titer packaging cells producing recombinant retroviruses resistant to human serum. J Virol. 1995;69:7430–7436. doi: 10.1128/jvi.69.12.7430-7436.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deacon T, Schumacher J, Dinsmore J, Thomas C, Palmer P, Kott S, Edge A, Penney D, Kassissieh S, Dempsey P, Isacson O. Histological evidence of fetal pig neural cell survival after transplantation into a patient with Parkinson’s disease. Nat Med. 1997;3:350–353. doi: 10.1038/nm0397-350. [DOI] [PubMed] [Google Scholar]
- 10.Fishman J A. Miniature swine as organ donors for man: strategies for prevention of xenotransplant-associated infections. Xenotransplantation. 1994;1:47–57. [Google Scholar]
- 11.Gage, F. H. 1998. Cell therapy. Nature 392(Suppl.):18–24. [PubMed]
- 12.Galili U, Swanson K. Gene sequences suggest inactivation of alpha-1,3-galactosyltransferase in catarrhines after the divergence of apes from monkeys. Proc Natl Acad Sci USA. 1991;88:7401–7404. doi: 10.1073/pnas.88.16.7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Groth C G, Korsgren O, Tibell A, Tollemar J, Moller E, Bolinder J, Ostman J, Reinholt F P, Hellerstrom C, Andersson A. Transplantation of porcine fetal pancreas to diabetic patients. Lancet. 1994;344:1402–1404. doi: 10.1016/s0140-6736(94)90570-3. [DOI] [PubMed] [Google Scholar]
- 14.Hamadeh R M, Jarvis G A, Galili U, Mandrell R E, Zhou P, Griffiss J M. Human natural anti-Gal IgG regulates alternative complement pathway activation on bacterial surfaces. J Clin Investig. 1992;89:1223–1235. doi: 10.1172/JCI115706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larsen R D, Rivera Marrero C A, Ernst L K, Cummings R D, Lowe J B. Frameshift and nonsense mutations in a human genomic sequence homologous to a murine UDP-Gal:beta-D-Gal(1,4)-D-GlcNAc alpha(1,3)-galactosyltransferase cDNA. J Biol Chem. 1990;265:7055–7061. [PubMed] [Google Scholar]
- 16.Le Tissier P, Stoye J P, Takeuchi Y, Patience C, Weiss R A. Two sets of human-tropic pig retrovirus. Nature. 1997;389:681–682. doi: 10.1038/39489. [DOI] [PubMed] [Google Scholar]
- 16a.Le Tissier, P., and J. P. Stoye. Unpublished data.
- 17.Loiler S A, DiFronzo N L, Holland C A. Gene transfer to human cells using retrovirus vectors produced by a new polytropic packaging cell line. J Virol. 1997;71:4825–4828. doi: 10.1128/jvi.71.6.4825-4828.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Makowa L, Cramer D V, Hoffman A, Breda M, Sher L, Eiras-Hreha G, Tuso P J, Yasunaga C, Cosenza C A, Wu G D, Chapman F A, Podesta L. The use of a pig liver xenograft for temporary support of a patient with fulminant hepatic failure. Transplantation. 1995;59:1654–1659. doi: 10.1097/00007890-199506270-00002. [DOI] [PubMed] [Google Scholar]
- 19.Michaels M G. Infectious concerns of cross-species transplantation: xenozoonoses. World J Surg. 1997;21:968–974. doi: 10.1007/s002689900335. [DOI] [PubMed] [Google Scholar]
- 20.Miller D G, Miller A D. Tunicamycin treatment of CHO cells abrogates multiple blocks to retrovirus infection, one of which is due to a secreted inhibitor. J Virol. 1992;66:78–84. doi: 10.1128/jvi.66.1.78-84.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyazono K, Okabe T, Urabe A, Takaku F, Heldin C H. Purification and properties of an endothelial cell growth factor from human platelets. J Biol Chem. 1987;262:4098–4103. [PubMed] [Google Scholar]
- 22.Patience C, Patton G S, Takeuchi Y, Weiss R A, McClure M O, Rydberg L, Breimer M E. No evidence of pig DNA or retroviral infection in patients with short-term extracorporeal connection to pig kidneys. Lancet. 1998;352:699–701. doi: 10.1016/S0140-6736(98)04369-4. [DOI] [PubMed] [Google Scholar]
- 23.Patience C, Takeuchi Y, Weiss R A. Infection of human cells by an endogenous retrovirus of pigs. Nat Med. 1997;3:282–286. doi: 10.1038/nm0397-282. [DOI] [PubMed] [Google Scholar]
- 23a.Patience, C. Unpublished data.
- 24.Phan Thanh L, Kaeffer B, Bottreau E. Porcine retrovirus: optimal conditions for its biochemical detection. Arch Virol. 1992;123:255–265. doi: 10.1007/BF01317262. [DOI] [PubMed] [Google Scholar]
- 25.Platt, J. L. 1998. New directions for organ transplantation. Nature 392(Suppl.):11–17. [DOI] [PubMed]
- 26.Porter C D, Collins M K, Tailor C S, Parkar M H, Cosset F L, Weiss R A, Takeuchi Y. Comparison of efficiency of infection of human gene therapy target cells via four different retroviral receptors. Hum Gene Ther. 1996;7:913–919. doi: 10.1089/hum.1996.7.8-913. [DOI] [PubMed] [Google Scholar]
- 27.Sachs D H. Transplantation. Curr Opin Immunol. 1996;8:671–673. doi: 10.1016/s0952-7915(96)80084-7. [DOI] [PubMed] [Google Scholar]
- 28.Silver J, Maudru T, Fujita K, Repaske R. An RT-PCR assay for the enzyme activity of reverse transcriptase capable of detecting single virions. Nucleic Acids Res. 1993;21:3593–3594. doi: 10.1093/nar/21.15.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sommerfelt M A, Weiss R A. Receptor interference groups of 20 retroviruses plating on human cells. Virology. 1990;176:58–69. doi: 10.1016/0042-6822(90)90230-o. [DOI] [PubMed] [Google Scholar]
- 30.Stoye J P, Coffin J M. The dangers of xenotransplantation. Nat Med. 1995;1:1100. doi: 10.1038/nm1195-1100a. [DOI] [PubMed] [Google Scholar]
- 31.Stoye, J. P., P. Le Tissier, Y. Takeuchi, C. Patience, and R. A. Weiss. Endogenous retroviruses: a potential problem for xenotransplantation? Ann. N. Y. Acad. Sci., in press. [DOI] [PubMed]
- 32.Suzuka I, Shimizu N, Sekiguchi K, Hoshino H, Kodama M, Shimotohno K. Molecular cloning of unintegrated closed circular DNA of porcine retrovirus. FEBS Lett. 1986;198:339–343. doi: 10.1016/0014-5793(86)80432-x. [DOI] [PubMed] [Google Scholar]
- 33.Tailor C S, Takeuchi Y, O’Hara B, Johann S V, Weiss R A, Collins M K. Mutation of amino acids within the gibbon ape leukemia virus (GALV) receptor differentially affects feline leukemia virus subgroup B, simian sarcoma-associated virus, and GALV infections. J Virol. 1993;67:6737–6741. doi: 10.1128/jvi.67.11.6737-6741.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takeuchi Y, Cosset F L, Lachmann P J, Okada H, Weiss R A, Collins M K. Type C retrovirus inactivation by human complement is determined by both the viral genome and the producer cell. J Virol. 1994;68:8001–8007. doi: 10.1128/jvi.68.12.8001-8007.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takeuchi Y, Simpson G, Vile R G, Weiss R A, Collins M K. Retroviral pseudotypes produced by rescue of a Moloney murine leukemia virus vector by C-type, but not D-type, retroviruses. Virology. 1992;186:792–794. doi: 10.1016/0042-6822(92)90049-u. [DOI] [PubMed] [Google Scholar]
- 36.Takeuchi Y, Vile R G, Simpson G, O’Hara B, Collins M K, Weiss R A. Feline leukemia virus subgroup B uses the same cell surface receptor as gibbon ape leukemia virus. J Virol. 1992;66:1219–1222. doi: 10.1128/jvi.66.2.1219-1222.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Talbot S J, Weiss R A, Schulz T F. Reduced glycosylation of human cell lines increases susceptibility to CD4-independent infection by human immunodeficiency virus type 2 (LAV-2/B) J Virol. 1995;69:3399–3406. doi: 10.1128/jvi.69.6.3399-3406.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilson C A, Wong S, Muller J, Davidson C E, Rose T M, Burd P. Type C retrovirus released from porcine primary peripheral blood mononuclear cells infects human cells. J Virol. 1998;72:3082–3087. doi: 10.1128/jvi.72.4.3082-3087.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]