Abstract
We report on the functional cloning of a hitherto unknown member of the immunoglobulin (Ig) superfamily selected for its ability to confer susceptibility to herpes simplex virus (HSV) infection on a highly resistant cell line (J1.1-2 cells), derived by exposure of BHKtk− cells to a recombinant HSV-1 expressing tumor necrosis factor alpha (TNF-α). The sequence of herpesvirus Ig-like receptor (HIgR) predicts a transmembrane protein with an ectodomain consisting of three cysteine-bracketed domains, one V-like and two C-like. HIgR shares its ectodomain with and appears to be an alternative splice variant of the previously described protein PRR-1 (poliovirus receptor-related protein). Both HIgR and PRR-1 conferred on J1.1-2 cells susceptibility to HSV-1, HSV-2, and bovine herpesvirus 1. The viral ligand of HIgR and PRR-1 is glycoprotein D, a constituent of the virion envelope long known to mediate viral entry into cells through interaction with cellular receptor molecules. Recently, PRR-1, renamed HveC (herpesvirus entry mediator C), and the related PRR-2, renamed HveB, were reported to mediate the entry of HSV-1, HSV-2, and bovine herpesvirus 1, and the homologous poliovirus receptor was reported to mediate the entry of pseudorabies virus (R. J. Geraghty, C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and P. G. Spear, Science 280:1618–1620, 1998; M. S. Warner, R. J. Geraghty, W. M. Martinez, R. I. Montgomery, J. C. Whitbeck, R. Xu, R. J. Eisenberg, G. H. Cohen, and P. G. Spear, Virology 246:179–189, 1998). Here we further show that HIgR or PRR-1 proteins detected by using a monoclonal antibody to PRR-1 are widely distributed among human cell lines susceptible to HSV infection and commonly used for HSV studies. The monoclonal antibody neutralized virion infectivity in cells transfected with HIgR or PRR-1 cDNA, as well as in the human cell lines, indicating a direct interaction of virions with the receptor molecule, and preliminarily mapping this function to the ectodomain of HIgR and PRR-1. Northern blot analysis showed that HIgR or PRR-1 mRNAs were expressed in human tissues, with the highest expression being detected in nervous system samples. HIgR adds a novel member to the cluster of Ig superfamily members able to mediate the entry of alphaherpesviruses into cells. The wide distribution of HIgR or PRR-1 proteins among human cell lines susceptible to HSV infection, coupled with the neutralizing activity of the antibody in the same cells, provides direct demonstration of the actual use of this cluster of molecules as HSV-1 and HSV-2 entry receptors in human cell lines. The high level of expression in samples from nervous system makes the use of these proteins in human tissues very likely. This cluster of molecules may therefore be considered to constitute bona fide receptors for HSV-1 and HSV-2.
Following attachment of herpes simplex virus (HSV) to cells mediated by the interaction of two virion glycoproteins, gC and gB, with cell surface glycosaminoglycans (9; for a review, see reference 46), entry of the capsid into the cytoplasm occurs via a pH-independent fusion of the virion envelope with plasma membranes and involves at least four glycoproteins, gB, gD, and the heterodimer gH/gL (6, 15, 27, 41). The involvement of cellular receptor proteins binding gD rests on numerous lines of evidence. First, stable expression of gD in cell lines prevents infection (1, 7, 21). Incubation of gD-expressing cells with antibodies to gD releases the block (4, 8). Viral unrestricted mutants able to overcome the gD-mediated block carry mutations in gD (4, 8, 11). This suggested that expression of gD blocked infection by sequestering a cellular receptor required for HSV entry (21). Studies on unrestricted mutants carrying different mutations in gD led to the further suggestion that multiple forms of gD-binding cellular receptors may exist (4, 40). The notion that different gD-expressing alphaherpesviruses—HSV, pseudorabies (PRV), and bovine herpesvirus 1 (BHV-1)—may use common receptors for entry in some cell types rested on the observation that cells expressing gD of one of the viruses could restrict infection by the homologous as well as the heterologous viruses (10, 23, 38). Finally, anti-idiotypic antibodies mimicking gD bind to cell surfaces of commonly used cell lines and block virus infectivity (19), and cells susceptible to HSV infection bind gD in a saturable manner (20). Similar evidence implicating cellular cognate proteins does not exist for gB, gH, or gL. Studies with the resistant CHO cells led to the identification of herpesvirus entry mediator (HVEM, now HveA) (33), a novel member of the tumor necrosis factor-nerve growth factor (TNF/NGF) receptor family present primarily in activated T lymphocytes, which mediates efficient entry of some HSV-1 strains into resistant transfected cell lines.
We report the identification of a novel member of the immunoglobulin (Ig) superfamily that confers susceptibility to HSV infection by mediating entry of the virus into cells. Its identification was permitted by use of a cellular clone (J1.1-2 cells) highly resistant to HSV entry, derived by exposure of BHKtk− cells to a recombinant HSV expressing tumor necrosis factor alpha (TNF-α). From a human cDNA expression library, we selected a clone encoding a protein whose ectodomain is almost identical to that of poliovirus receptor related-1 protein (PRR-1) (28) and appears to be a splice variant isoform of it. The herpesvirus Ig-like receptor (HIgR) and PRR-1 each confers susceptibility to HSV-1, HSV-2, and BHV-1. Key properties of HIgR and PRR-1 that allow them to be considered bona fide receptors for HSV-1 and HSV-2 entry are their ability to bind glycoprotein D, their wide distribution among commonly used human cell lines susceptible to HSV infection, neutralization of virion infectivity by monoclonal antibody to the ectodomain of the HIgR/PRR-1 isoforms, and expression of the mRNA in human tissues, with the highest level of expression being detected in nervous system tissues.
MATERIALS AND METHODS
Cells, viruses, and plasmids.
All cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 5% fetal calf serum. The wild-type viruses HSV-1(F), HSV-2(G), and HSV-1(MP) were described previously (14, 18, 24). HSV (Sc-16, ANG, KOS, and HFEM), BHV-1, and PRV were described previously (17, 43, 49). U21, U10, and U30 carry mutations in the gD gene which overcome the gD-mediated block to infection (4, 8). R8996, a recombinant carrying a TNF-α gene under the control of the epidermal growth factor receptor I promoter in place of the γI34.5 gene and R8102, a recombinant carrying a lacZ gene fused to α27 and inserted between the UL3 and UL4 genes, were gifts from B. Roizman and will be described elsewhere. Viruses were grown and subjected to titer determination by plaque assay in Vero cells. Infectivity of R8102 was assayed by light microscopy of cells expressing β-galactosidase (33). Cells expressing PRR-1 were obtained by transfection of pLX1.12 containing PRR-1 cDNA cloned in the BamHI site of pLXSN. Stable transformants of J1.1-2 cells expressing HIgR or PRR-1 were obtained by G418 neomycin selection of pCF18- or pLX1.12-transfected cells.
Antibodies.
HD-1 (anti-gD; Goodwin Institute, Plantation, Fla.) (37) and 52S (anti-gH; American Type Culture Collection) (45) were used for J1.1-2 cell derivation; LP1 (anti-αTIF) (30), rabbit anti-gM (2), and 1240 to BHV-1 gB were used for immunostaining of infected cells; and monoclonal antibody (MAb) R1.302 to PRR-1 (29) was used to detect expression of HIgR and PRR-1 proteins and for infectivity neutralization assays. Alkaline phosphatase anti-rabbit and biotin-avidin anti-mouse antibodies (ABC-Kit) were from Sigma and Vector Laboratories, respectively.
Isolation of HIgR cDNA.
Screening of a HeLa cDNA library cloned unidirectionally in pcDNA3.1 (Invitrogen) was done as described previously (33). Briefly, the library was plated onto 100 plates and colonies were grown, harvested, and pooled into groups of 10 plates each. The pools were grown in liquid broth. Plasmid DNA was extracted and purified with Qiagen columns (Qiagen Companies; M-Medical, Florence, Italy). J1.1-2 cells in 25-cm2 flasks were transfected by means of LipofectAMINE (Gibco Laboratories, Milan, Italy) (2.5 μg of DNA in 20 μl of LipofectAMINE). After 30 h, they were exposed to R8102 (5 PFU/cell) for 16 h and assayed for β-galactosidase. In each experiment, replicate cultures were transfected with a plasmid carrying lacZ cloned in pcDNA3.1 or with pBEC10 (HveA) followed by infection with R8102 and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) staining. The cDNA pools that yielded the largest number of infected cells were subjected to repeated subdivisions. The final plasmid pCF18 was sequenced by the Department of Biological and Technological Research, Primm, Milan, San Raffaele Biomedical Scientific Park, Italy.
Reverse transcription-PCR (RT-PCR).
Total RNA was extracted with RNAzoldB (TEL-TEST, Friendswood, Tex.) and subjected to exhaustive DNase digestion, which was checked by lack of amplification of β-actin (47). RNA (2 μg) was retrotranscribed with avian myeloblastosis virus reverse transcriptase (Invitrogen cDNA cycle kit), as assessed by amplification of β-actin mRNA. HIgR, PRR-1, and HIgR plus PRR-1 sequences were cumulatively amplified with primers annealing to the 5′ portion of their mRNAs (5′-GTGAG GCTTT CCTTG AAGCG GTCCA TGTGG-3′ and 5′-GAGGG TACCC AGGCA GTGCT TCGAG-3′). Primers specific for the 3′ portions of HIgR or PRR-1 mRNAs were as follows: 5′-CTTCT GGGAG AGGGG CTGGT C-3′ and 5′-GTGGC CGTGT TCCTC ATCCT A-3′ (HIgR), and 5′-GCATC CTGCT GGTGT TGATT GTGGT-3′ and 5′-GTCTG AGTCG TCGGG GTACT GCAGG-3′ (PRR-1). HveA sequences were amplified with specific primers (5′-TGGGT ATGGT GGTTT CTCTC AGGG-3′ and 5′-CGTGA ATGAG GGTAT TGTCT CCTCC-3′). The PCR conditions for amplification of HIgR, PRR-1, and HIgR plus PRR-1 sequences were 10 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min, and then 35 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 1 min. The PCR conditions for amplification of HveA sequences were 35 cycles of 94°C for 1 min, 56°C for 1 min, and 72°C for 1 min (with a 3-s increase at each new cycle). PCR conditions for amplification of β-actin sequences were 45 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min. As a routine, samples were initially denatured for 4 min at 94°C.
Immunofluorescence and FACS analysis.
For immunofluorescence, cells grown in glass coverslips were fixed with acetone and reacted with MAb R1.302 (1:100) followed by biotinylated anti-mouse antibodies (Vectastain ABC kit) and Extravidin-tetramethylrhodamine-5-isothiocyanate (TRITC) (Sigma). Fluorescence-activated cell sorter (FACS) analysis was performed as described previously (29) and analyzed in a FACSscan flow cytometer.
Infectivity neutralization assays.
Cells grown in 96-well trays were preincubated with the indicated amounts of antibodies, either purified IgGs or ascites fluid, in 25 μl of medium for 2 h at 4°C. The appropriate amount of R8102 in 7.5 μl was then added for a further 90 min at 4°C. The viral inoculum was removed, and the cells were rinsed twice, overlaid with medium containing the same concentration of antibodies as present during the preabsorption, shifted to 37°C, and incubated for 16 h. β-Galactosidase activity was assayed as described previously (33). The optical density was read in a Bio-Rad enzyme-linked immunosorbent assay reader. For each antibody concentration, triplicate experiments were run. Data represent the average of at least two experiments. The 100% value represents data obtained with infected cells not exposed to antibodies. Infectivity neutralization was detected irrespective of whether purified IgGs or ascites fluids were used.
Northern blot analysis.
Human multiple-tissue Northern (MTN) membranes (Clontech) were hybridized with the BamHI fragment from pLX1.12 labeled with [32P]dCTP, as specified by the manufacturer. The membranes were also probed with a β-actin probe to verify the hybridation conditions.
Binding of gD to J1.1-2 cells expressing HIgR.
J1.1-2 cells in glass coverslips were transfected with pCF18, fixed with methanol 30 h later, incubated with biotinylated recombinant gD-1(Δ290–299t) (0.1 μg in 30 μl/coverslip) (36) for 1 h at room temperature followed by Extravidin-TRITC (Sigma) (1:100 in phosphate-buffered saline containing 20% fetal calf serum) for 1 h at room temperature. For biotinylation, 7 μg of gD was incubated with 1 μg of Immunopure sulfo-NHS-biotin (Pierce) in 50 mM sodium bicarbonate buffer (pH 8.5) for 2 h on ice.
RESULTS
Derivation of the J1.1-2 cell line, which is highly resistant to HSV infection.
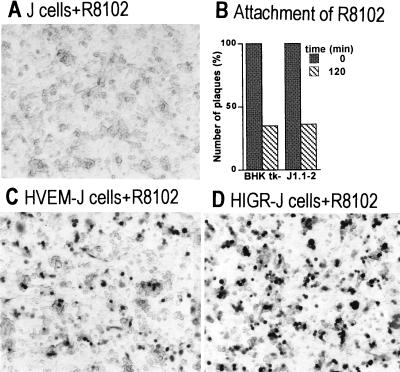
Of the numerous attempts to obtain a cell line resistant to infection by exposing different cell lines to a variety of HSV strains, the one that yielded the most useful cell line involved exposure of BHKtk− cells to recombinant HSV R8996, which secretes TNF-α in the medium. The few cells surviving a 10-PFU/cell infection were grown and infected two more times with the same virus at 100 PFU/cell. Some cells in the culture showed a persistent cytopathic effect, which was eliminated by cultivation of the cells for 20 days in the presence of HD-1 ascites fluid (1:250), a potent neutralizing monoclonal antibody to gD (37), and for an additional 20 days in the presence of a mixture of HD-1 and 52S (45), a neutralizing antibody to gH. The cells were cloned by single-cell plating with medium supplemented with 20% fetal calf serum and 20% conditioned medium from BHKtk− cells. Twelve clones were assayed for resistance to infection with R8102, which carries a lacZ gene fused to α-27, and infection was monitored by X-Gal staining. All clones were resistant to infection. In clone J1.1-2, no more than an average of 10 cells per monolayer (3 × 106 cells) exhibited evidence of infection after exposure to 5 PFU/cell (Fig. 1A). The J1.1-2 cells have been propagated for about 1 year without exhibiting a change in phenotype.
FIG. 1.
Characterization of J1.1-2 cells and identification of HIgR. (A) Micrograph of J1.1-2 (J) cells infected with the recombinant virus R8102 and stained for β-galactosidase to locate infected cells. No infected cell is visible. (B) Comparison of the extent of R8102 attachment to J1.1-2 and BHKtk− cells. Virus was allowed to attach to cells at 4°C; the virus present in the inoculum at zero time and that remaining unattached after 120 min were measured by a plaque assay in triplicate. (C and D) Micrographs of HveA(HVEM)-transfected (C) and HIgR-transfected (D) J1.1-2 cells infected with R8102 (5PFU/cell). X-Gal staining identified cells rendered susceptible by transfection.
Resistance in clone J1.1-2 is at the entry level.
In HSV-infected cells, α gene expression does not require prior viral protein synthesis. In R8102, the lacZ gene was driven by the α27 promoter. Thus, lack of lacZ expression indicated that the block to infection with R8102 in J1.1-2 cells preceded α gene expression. Additional evidence that block is at the entry level was as follows. (i) At 4°C, HSV attaches to but does not penetrate cells. We compared the extent of virus attachment to J1.1-2 and BHKtk− cells by measuring the amount of virus remaining unattached after 120 min of absorption at 4°C and found no difference (Fig. 1B). (ii) In the second experiment, J1.1-2 cells were transfected with the mediator of HSV entry, HVEM(HveA), and infected with R8102. The cells became infectable (Fig. 1C).
Selection of a human cDNA clone that confers susceptibility to J1.1-2.
J1.1-2 cells were transfected with DNA pools from a HeLa cDNA library, exposed to R8102 30 h after transfection, and monitored for β-galactosidase activity. The protocol described in Materials and Methods led to the selection of clone pCF18, which conferred susceptibility to all the transfected cells in the culture (Fig. 1D).
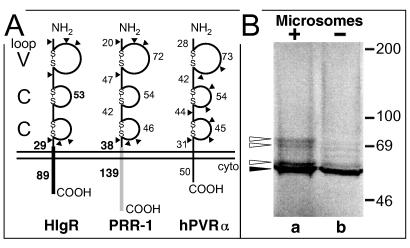
The approximately 2,500-bp insert in pCF18 contained an open reading frame predicted to encode a 458-amino-acid protein (designated HIgR) of 50.7 kDa (Fig. 2). The predicted protein had structural elements typical of type 1 membrane-bound proteins, an N-terminal signal sequence with cleavage at Ser30, and a hydrophobic transmembrane region at residues 346 to 369. The ectodomain carried, in addition to seven potential N-glycosylation sites, six cysteine residues and consensus Ig motifs which identified three domains, one V-like and two C-like, of 72, 53, and 46 residues, respectively (Fig. 2) (a representation of the predicted structure is shown in Fig. 3A). These features defined the protein encoded by pCF18 as belonging to the Ig superfamily. The sequence was identical up to nucleotide 1002 to that of PRR-1 cDNA (28), except for the absence of three nonconsecutive cytosines, which modified the amino acid sequence between residues 194 and 204 (Fig. 2). Divergence between HIgR and PRR-1 started at nucleotide 1002 immediately after a splice donor consensus sequence. Since alternative splicing is very frequent among Ig superfamily transcripts, e.g., in poliovirus receptor (PVR) (25) and PRR-2 (13), it seems highly probable that the cDNA encoded by pCF18 represents a previously unknown, alternative splice variant of PRR-1. HIgR has a shorter C-terminal cytoplasmic tail than PRR-1 (Fig. 2 and 3A). This is similar to PRR-2α and PRR-2δ, which share the ectodomain and differ in the size and sequence of the cytoplasmic tail (13).
FIG. 2.
Alignment of the HIgR and PRR-1 amino acid sequences by the ALIGN program, and relevant features of the proteins. The N-terminal signal sequences and transmembrane sequences are boxed, as are the six conserved cysteines involved in the formation of Ig domains. The arrow indicates the serine used for cleavage of the signal sequence. Potential N-glycosylation sites are underlined. The vertical line indicates the position of the splice site. The shaded box highlights the small divergent tract.
FIG. 3.
(A) Comparative schematic representation of the molecular structure of HIgR, PRR-1, and PVR. The V and C domains formed by cysteine bonds typical of this cluster of Ig superfamily members are shown, along with the number of residues in each region. The regions differing between HIgR and PRR-1, i.e., the transmembrane portion, the segment preceding it, and the cytoplasmic (cyto) tail, are in solid black for HIgR and shaded for PRR-1; the numbers of residues forming these latter regions are shown in bold type. Triangles indicate predicted N-glycosylation sites. (B) Electrophoretic mobility of HIgR synthesized by in vitro transcription-translation with 28.6 μCi of [35S]methionine (Radiochemical Center, Amersham, England) in the absence (lane b) or presence (lane a) of microsomes (Promega kit). Proteins were separated by denaturing electrophoresis in 8.5% polyacrylamide gels. The dried gel was analysed with a Bio-Rad PhosphoImager. The solid arrowhead points to unglycosylated precursor; the open arrowheads point to glycosylated species. The electrophoretic mobility of radioactive markers is shown on the right.
In vitro transcription-translation of pCF18 yielded a protein with an apparent mass of 57 kDa (Fig. 3B). The apparent molecular mass increased to 70 kDa in the presence of microsomes and showed discrete intermediate bands, consistent with glycosylation of some of the predicted sites.
HIgR confers susceptibility to a wide range of HSV-1 and HSV-2 strains.
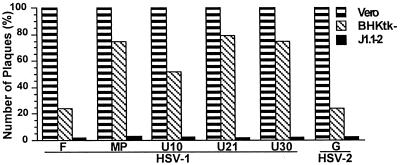
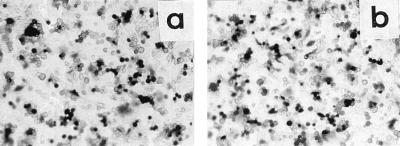
Preliminary experiments established that J1.1-2 cells were simultaneously resistant to all HSV-1 and HSV-2 strains tested. These included the wild-type strains HSV-1(F), HSV-1(KOS), HSV-1(SC-16), and HSV-2(G); the two syncytial strains HSV-1(MP) and HSV-1(HFEM) with mutations in gK (39) and gB (12, 42), which induce polykaryocyte formation of infected cells; four strains with mutations in gD which overcome gD-mediated block to infection, three of which (U21, U30, and U10) were selected in our laboratory (4, 8); and ANG, a clinical isolate (11, 43). Lack of plaque formation (Fig. 4 shows representative examples) or immunostaining of monolayers (Fig. 5a shows an example) were used to assess resistance to infection.
FIG. 4.
Relative plating efficiency of HSV-1 and HSV-2 strains in Vero, BHKtk−, and J1.1-2 cells. Plaques in Vero and BHKtk− cells were scored by Giemsa staining, and those in J1.1-2 cells were scored by immunostaining.
FIG. 5.
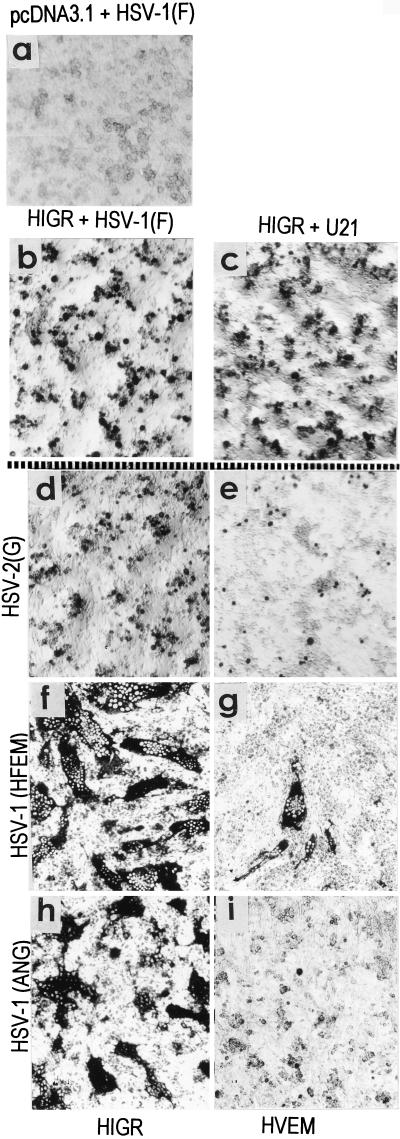
Effect of HIgR expression on the susceptibility of J1.1-2 cells to the indicated HSV-1 and HSV-2 strains. A gallery of representative micrographs of cells transfected with pcDNA3.1 vector alone (a), HIgR (b to d, f, and h), or HveA(HVEM) (e, g, and i) is shown. Infected cells were detected by immunostaining.
To determine the range of HSV strains to which HIgR conferred susceptibility, J1.1-2 cells transfected with pCF18 or pcDNA3.1 vector were exposed to the above virus strains. Infected cells were readily detected with each virus tested (Fig. 5b to d, f, and h). Cells infected with aggregating strains were single or formed small aggregates (Fig. 5 b to d). Cells infected with syncytial strains HSV-1(MP), HSV-1(HFEM), or HSV-1(ANG) formed syncytial plaques (Fig. 5f and h). The characteristic of J1.1-2 cells of resistance to a wide range of HSV strains differentiates J1.1-2 from CHO cells, which show moderate resistance to HSV-2 and to the syncytial strain MP. Furthermore, transfection of CHO cells with HveA increased their susceptibility to HSV-2 and to syncytial strain MP to a low level and did not induce susceptibility to the HSV gD mutants able to overcome the gD-mediated block (33). In contrast to HveA, transfection of J1.1-2 cells with HIgR conferred high sensitivity to all HSV-1 and HSV-2 strains tested. We also noticed that HIgR was much more efficient than HveA(HVEM) in conferring on J1.1-2 cells susceptibility to infection with HSV-2 and the syncytial strains MP, HFEM, and ANG (compare Fig. 5d to e, Fig. 5f to g, and Fig. 5h to i).
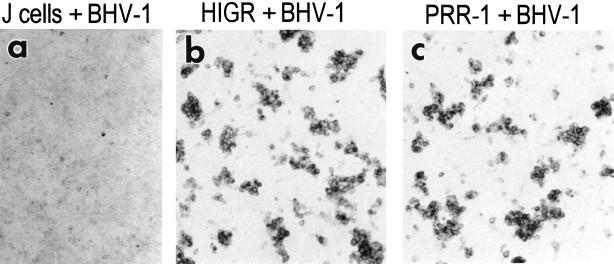
PRR-1 confers susceptibility on J1.1-2 cells.
J1.1-2 cells were transfected with pLX1.12 containing pPRR-1 cDNA and 30 h later were exposed to R8102. PRR-1 was as effective as HIgR in conferring susceptibility to HSV infection (Fig. 6). In a subsequent experiment, J1.1-2 cells were transfected with pLX1.12 and infected with all the HSV-1 and HSV-2 strains tested above. For all of them, transfection with PRR-1 was as effective in conferring susceptibility to J1.1-2 cells as HIgR was (data not shown). This result indicates that either isoform, HIgR or PRR-1, can function to mediate the entry of HSV into transfected J1.1-2 cells. The effect of PRR-1 in conferring on J1.1-2 cells susceptibility to a wide spectrum of HSV strains is similar to the effect of PRR-1(HveC) in conferring susceptibility on CHO cells, reported recently by Geraghty et al. (16).
FIG. 6.
Transfection of J1.1-2 cells with PRR-1 cDNA confers susceptibility to R8102. Micrographs of J1.1-2 cells transfected with pLX1.12 (PRR-1) (a), or pCF18 (HIgR) (b), infected with R8102, and stained with X-Gal are shown.
Transfection with HIgR or PRR-1 confers susceptibility to BHV-1.
As mentioned in the introduction, studies on restriction mediated by gD homologs encoded by HSV, BHV-1, and PRV suggest the possibility that the three alphaherpesviruses use common receptors in some cell lines (10, 23, 38). As shown in Fig. 7a, J1.1-2 cells were not susceptible to BHV-1 infection. However, susceptibility was restored by transfection with HIgR or PRR-1 (Fig. 7b and c). Somewhat to our surprise, J1.1-2 cells were susceptible to PRV infection; therefore, the effect of HIgR or PRR-1 expression on this virus could not be investigated. With respect to BHV-1, the results indicate that although HIgR and PRR-1 do not represent the natural receptors used by BHV-1 in bovine cells and in its natural host, the human HIgR or PRR-1 can be used promiscuously by BHV-1 to enter the cells. With respect to PRV, the results indicate that in J1.1-2 cells this virus finds a receptor, which cannot be utilized by HSV-1, HSV-2, or BHV-1.
FIG. 7.
Transfection of J1.1-2 cells with HIgR or PRR-1 confers susceptibility to BHV-1 infection. Micrographs of J1.1-2 cells transfected with pCF18 (HIgR) (b), pLX1.12 (PRR-1) (c), or pcDNA3.1 (a), infected with BHV-1, and immunostained with MAb 1240 to glycoprotein B are shown.
HIgR or PRR-1 mRNAs are present in human cell lines.
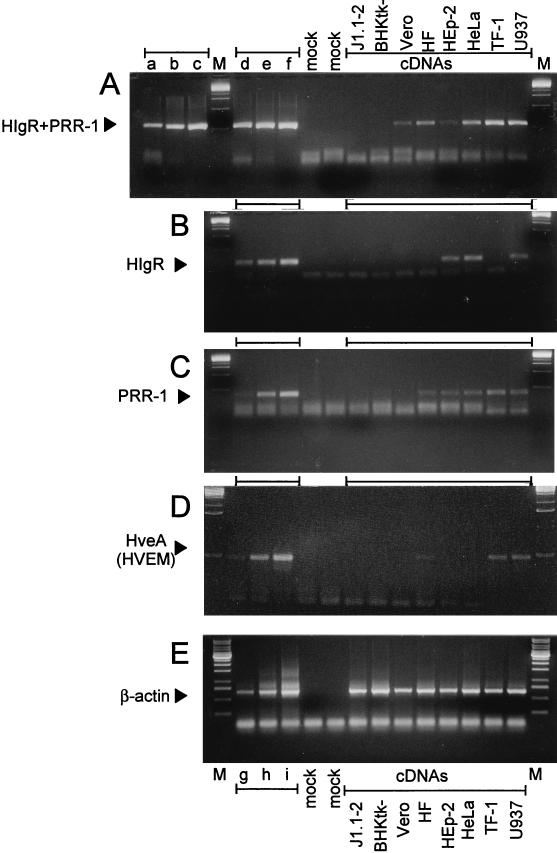
A central question is whether the gene encoding HIgR or PRR-1 is expressed in cells susceptible to HSV-1 infection. A preliminary assay for the presence of HIgR or PRR-1 mRNAs was performed by RT-PCR of RNA extracted from a number of cell lines susceptible to HVS infection and from J1.1-2 cells. Three sets of primers were used. The first set amplified the 5′ portion of the mRNAs in common between the two isoforms and therefore cumulatively amplified sequences of HIgR and PRR-1 (HIgR+PRR-1) (Fig. 8A). The other two sets of primers annealed to the 3′ portions of the mRNAs and were therefore specific either for HIgR or for PRR-1 (Fig. 8B and C). All cells of human origin, susceptible to HSV infection, i.e., HeLa, HEp-2, MRC human fibroblasts, TF-1, and U937, contained mRNAs for HIgR or PRR-1, or both, indicating that mRNAs for these molecules are widely expressed in human cells. Vero cells of simian origin also contained sequences amplified with the primers designed on the basis of the human gene sequence, suggesting the possible existence of a simian homolog. BHKtk− cells, which are susceptible to HSV infection but of hamster origin, did not. This probably reflects the specificity of the primers for the human gene and stringency of PCR conditions. The results with human cell lines are consistent with and extend previous data (16).
FIG. 8.
Agarose gel electrophoresis of RT-PCR-amplified sequences specific for HIgR plus PRR-1 (HIgR + PRR-1) (A), HIgR (B), PRR-1 (C), HveA(HVEM) (D), and β-actin (E). Sequences were amplified from cDNAs of the indicated cells (A to E) and from appropriate plasmids (A to D) or from cellular DNA (E). Each panel shows the intensity of amplified bands for increasing amounts of target sequences, as follows. (A) Lanes a to c and d to f contain 50, 500, and 5,000 copies of pLX1.12 (PRR-1) and pCF18 (HIgR), respectively. (B) Lanes d to f contain 5, 50, and 500 copies of pCF18. (C) Lanes d to f contain 50, 500, and 5,000 copies of pLX1.12. (D) Lanes d to f contain 50, 500, and 5,000 copies of pBEC10 for HveA. (E) Lanes g to i contain 0.1, 1, or 10 ng of DNA from human T lymphocytes. Lane M, molecular weight markers. Lanes mock, negative reaction controls lacking DNA but containing all other reagents. The sizes of the amplified products were 269, 119, 184, 233, and 674 bp for HIgR+PRR-1, HIgR, PRR-1, HveA, and β-actin, respectively. HF, MRC human fibroblasts.
The same cells were tested for presence of HveA mRNA by RT-PCR with specific primers. Only some cells were positive (Fig. 8D), confirming that HveA expression is limited to a few cell types (33). As a control, β-actin mRNA could be amplified from all cell lines tested (Fig. 8E). With respect to the mechanism of action of HIgR, the inference from this experiment is that HIgR enables HSV to enter J1.1-2 cells without the need for cooperation by HveA.
Distribution of HIgR or PRR-1 proteins in human cell lines.
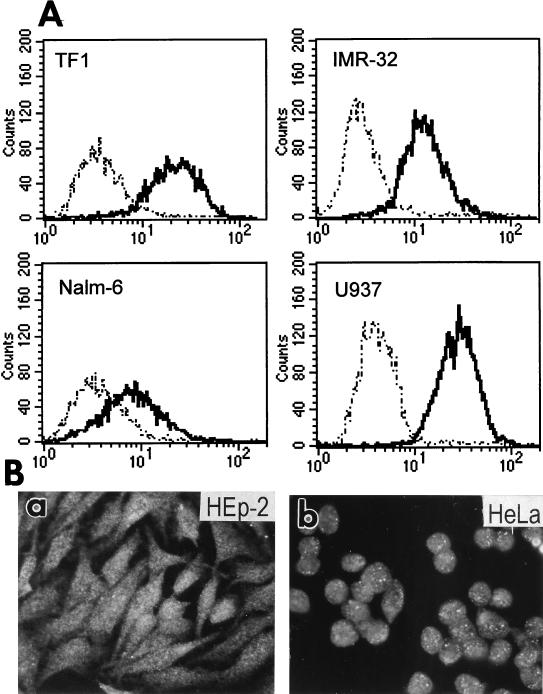
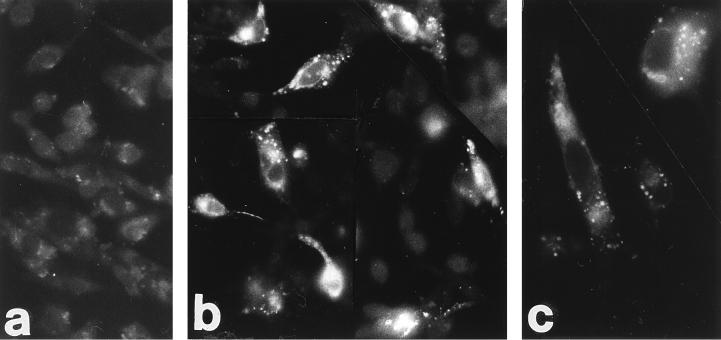
To ascertain if HIgR and PRR-1 are the authentic receptors used by HSV in human cell lines, it was crucial to detect the expression of these molecules at the protein level. We used a MAb to PRR-1 previously derived in one of our laboratories, designated MAb R1.302 (29). Since MAb R1.302 reacted with cells expressing HIgR (results not shown), the epitope recognized by MAb R1.302 must reside in the ectodomain of the two molecules. The distribution of HIgR and/or PRR-1 proteins was next investigated by flow cytometry or by indirect immunofluorescence and found to be positive on numerous human cell lines, i.e., HEp-2 and HeLa (epithelial cell carcinoma), TF-1 (hematopoietic progenitor), IMR-32 and Lan5 (neuroblastoma), Nalm-6 (lymphoid, precursor B), and 5637 and T24 (epithelial bladder carcinoma) (representative examples shown in Fig. 9). Previously, reactivity to MAb R1.302 was detected in additional human cells lines, including U937 (myeloid) (Fig. 9), Raji and Daudi (Burkitt’s lymphoma), RPMI 8866 and 0467 (B cells), RPMI 8266 and U266 (plasmacytoid), and CEM (T cells) (29). Together, these data indicate that HIgR and/or PRR-1 proteins are widely represented in human cell cultures of different lineages, including epidermal, neuronal, myeloid, and lymphoid.
FIG. 9.
Expression of HIgR and PRR-1 in human cell lines. (A) FACS analysis of TF-1, IMR-32, Nalm-6, and U937 cells. (B). Micrographs of HEp-2 (a) and HeLa (b) cells stained by indirect immunofluorescence with MAb R1.302 followed by biotinylated anti-mouse antibodies and Extravidin-TRITC.
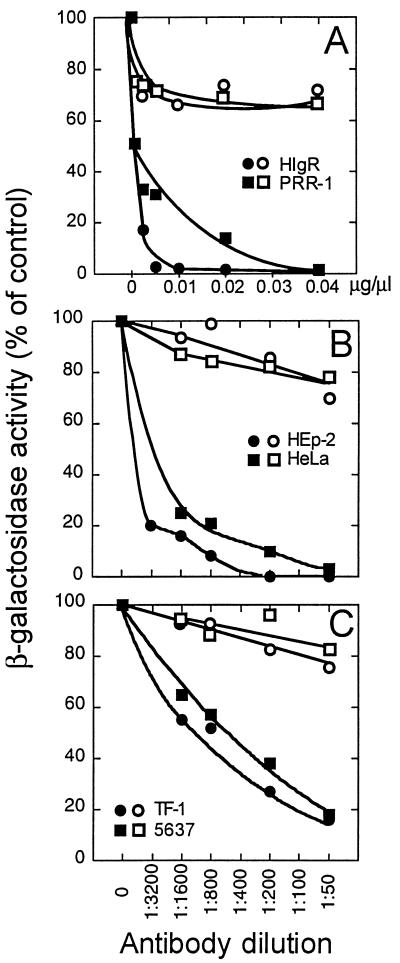
Neutralization of HIgR- and PRR-1-mediated HSV infectivity by anti-PRR-1 antibody.
In J1.1-2 cells stably transformed with pCF18 (HIgR) or pLX1.12 (PRR-1), exposure to increasing concentrations of MAb R1.302 before infection inhibited R8102 infectivity in a dose-dependent manner (Fig. 10A). Mouse IgGs had only a minor inhibitory effect. To ascertain if the HIgR or PRR-1 molecules detected in human cell lines are actually used as receptors by HSV, we carried out infectivity neutralization experiments on representative cell lines. The results in Fig. 10B and C show that in HEp-2, HeLa, TF-1, and 5637 cells, HSV infectivity was reduced in a dose-dependent fashion by MAb R1.302 whereas an unrelated MAb had only minimal effects. From these data, we can draw three conclusions. First, the experiment provides compelling evidence that susceptibility to infection in HIgR- and PRR-1-expressing J1.1-2 cells is dependent upon a direct interaction of virions with these molecules. Second, infectivity neutralization in human cell lines, coupled with protein expression, demonstrates that these molecules are actually used as receptors for entry. Third, since the portion of the molecule shared by HIgR and PRR-1 is the ectodomain (Fig. 2), the functional domain of HIgR or PRR-1 used to mediate HSV entry maps to the ectodomain of the molecules.
FIG. 10.
Neutralization of R8102 infectivity by MAb R1.302 to PRR-1. (A) Stable transformants of J1.1-2 cells expressing HIgR or PRR-1 in 96-well plates were preincubated with the indicated dilutions (micrograms per microliters) of IgGs from MAb R1.302 to PRR-1 (solid symbols) or from purified mouse IgGs (open symbols) for 2 h at 4°C. R8102 was added to the antibody-containing medium and allowed to absorb for further 90 min at 4°C. (B) Infectivity neutralization in HEp-2 and HeLa cells with MAb R1.302 ascites fluid (solid symbols) or unrelated MAb (open symbols). (C) Infectivity neutralization in TF-1 and 5637 cells with MAb R1.302, ascites fluid (solid symbols) or unrelated MAb (open symbols).
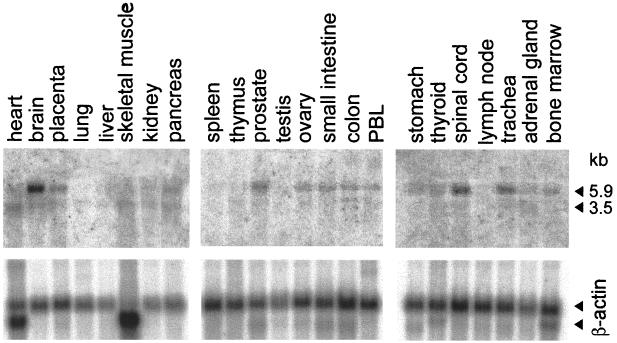
Distribution of HIgR or PRR-1 mRNAs in human tissues.
In this experiment, the expression of HIgR and/or PRR-1 in human tissues was assessed by Northern blot analysis carried out on human multiple-tissue Northern blot membranes from Clontech. The probe consisted of the BamHI fragment of pLX1.12 comprising the entire PRR-1 cDNA, and it hybridized to both HIgR and PRR-1 mRNAs. The results in Fig. 11 show that two bands of 5.9 and 3.5 kbp were detectable in several tissues. The highest level of expression was detected in samples from the nervous tissue, brain, and spinal cord, followed by the trachea, prostate, and pancreas.
FIG. 11.
Northern blot analysis of HIgR plus PRR-1 mRNA expression in human tissues. (A) Phosphoimages obtained with multiple-tissue Northern blot membranes hybridized with the BamHI fragment of pLX1.12 containing the entire PRR-1 cDNA. The probe did not distinguish between HIgR and PRR-1 mRNAs. Two bands, of 5.9 and 3.5 kbp, were detected. The highest level of expression occurred in brain and spinal cord tissues. (B) The same membranes hybridized with a β-actin probe.
Cells expressing HIgR bind gD.
J1.1-2 cells transfected with pCF18 or pcDNA3.1 were reacted with a biotinylated recombinant soluble form of gD [gD-1(Δ290–299t)] (36) followed by TRITC-conjugated Extravidin. The results in Fig. 12b and c show that a portion of the cells in the culture bound gD, based on fluorescence labeling. The number of labeled cells was consistent with a transient-expression assay. Fluorescence localized mainly to vesicles in the cytoplasm, as expected for a membrane-bound protein. Cells transfected with plasmid alone (Fig. 12a) or pCF18-transfected cells treated with Extravidin in the absence of gD (results not shown) were not labeled. Overlapping results were obtained with pLX1.12-transfected J1.1-2 cells (results not shown).
FIG. 12.
Binding of gD to HIgR-expressing J1.1-2 cells. Fluorescence micrographs of J1.1-2 cells transfected with pCF18 (b and c) or pcDNA3.1 (a) are shown. Cells fixed with methanol were incubated with biotinylated recombinant gD-1(Δ290–299t) followed by Extravidin-TRITC.
DISCUSSION
The key discovery reported here is that a novel member of the Ig superfamily, HIgR, which shares the ectodomain with PRR-1, and PRR-1 itself act as bona fide receptors for HSV-1 and HSV-2 entry into human cells.
The discovery of HIgR was made possible by use of the J1.1-2 cell line, which is highly resistant to entry of HSV-1, HSV-2, and BHV-1; this cell line was selected by repeated exposures of BHKtk− cells to a recombinant HSV strain expressing TNF-α. Screening of a human cDNA expression library for clones that restored susceptibility to J1.1-2 cells led to the isolation of a human cDNA clone which encodes a novel transmembrane protein with features typical of the Ig superfamily and an overall molecular organization essentially overlapping that of PVR (31), i.e., a V-like and two C-like domains bracketed by conserved cysteines. Its predicted amino acid sequence is almost identical, until amino acid 335, to that of PRR-1 (28). HIgR and PRR-1 diverge after a consensus sequence for a splice donor. Alternative splicing is a very generalized phenomenon in the Ig superfamily and contributes to the generation of Ig diversity. For example, PVR mRNAs originate from a 20-kb transcript, consist of eight exons, and, because of alternative splicing, yield at least four different transmembrane and secreted forms of the receptor (25). Similarly, the hPRR-2 homolog of PRR-1 exists in two forms, α and δ (13), which have an identical 5′ sequence and diverge in the 3′ portion (1,041 of 1,413 bp identical). On this basis, it seems likely that HIgR originates by alternative splicing of PRR-1 transcript.
While the manuscript was being readied for publication, it was reported that PRR-1, renamed HveC (for herpesvirus entry mediator C), mediates the entry of HSV-1, HSV-2, BHV-1, and PRV; that PVR itself can mediate the entry of PRV (16); and that PRR-2, renamed HveB (for herpesvirus entry mediator B), mediates the entry of HSV-1 gD-mutants and HSV-2 but not wild-type HSV-1 strains (48).
The relevant properties of HIgR and PRR-1(HveC) as mediators of HSV entry into human cells are as follows. (i) HIgR and PRR-1 permit the entry of all wild-type and mutant HSV-1 and HSV-2 strains tested, including mutants which induce the fusion of cells and mutants which overcome the gD-mediated restriction to infection. This property differentiates HIgR and PRR-1 (16) from HveA, which shows a narrower viral spectrum (33).
(ii) HIgR and PRR-1 permit HSV entry into J1.1-2 cells independently of each other and of HveA.
(iii) Cells expressing HIgR or PRR-1 bind gD, as expected from numerous studies pointing to gD as the virion component engaged in virus entry through interaction with cellular receptor molecules.
(iv) The HIgR and/or PRR-1 isoforms are highly distributed among human cell lines susceptible to HSV infection and commonly used for HSV studies, such as HEp-2, HeLa, human fibroblasts, U937, and numerous other human cell lines of different origin, such as IMR-32 and Lan5 (neuroblastoma), TF-1 (hematopoietic progenitor), 5637 and T24 (bladder carcinoma), Nalm-6 (lymphoid, precursor B), and plasmacytoid cells and T lymphocytes (29). mRNA analysis showed that some of these cell lines express one or the other isoform while some express both.
(v) A MAb to PRR-1 neutralizes HSV-1 infectivity in HIgR- or PRR-1-transformed cells as well as in human cell lines. This provides unambiguous evidence of the actual usage of these molecules as HSV-1 receptors in human cell lines. It also shows that viral entry mediated by HIgR or PRR-1 occurs through a direct interaction of the receptor molecules with virions.
(vi) HIgR and PRR-1 are two isoforms sharing the ectodomain. Thus, the functional domain that mediates alphaherpesvirus entry must reside in the ectodomain of the molecules.
(vii) HIgR and PRR-1 mRNAs are expressed in human tissues; the highest level of expression is detected in samples from the nervous system.
HIgR adds a novel member to the cluster of Ig homologs, which includes PVR, hPRR-1, hPRR-2α and δ, and homologous proteins of simian, murine, and rat origin (3, 13, 25, 26, 28, 31, 34). They share three characteristics: (i) a common molecular structure defined by the six conserved cysteins (Fig. 3A), (ii) the ability to originate multiple isoforms by alternative splicing, and (iii) the ability of the human members to mediate the entry of some alphaherpesviruses. The discovery of a novel member of the cluster suggests the possibility that the actual number of members capable of acting as receptors for herpesviruses is even larger than that known to date. It will be interesting to determine whether animal isoforms of the cluster can mediate HSV entry into animal cells, providing a basis for the broad host range of HSV in cell cultures and animals of different species.
In the past, proteins proposed as mediators of HSV entry were subsequently brought into question because of failure to comply with authenticity criteria. For basic fibroblast receptor (22), transfection in resistant cells did not enhance virus entry (32, 35, 44). For mannose-6-phosphate receptors, cell mutants defective in the expression of these proteins were infectable by HSV (5). Here we show that HIgR and PRR-1 fulfill criteria that allow them to be considered bona fide receptors for HSV-1 and HSV-2. First, these proteins are present in a variety of human cells susceptible to HSV infection, as detected by reactivity with MAbs. Second, the neutralizing activity of the antibody in the same cells which express the HIgR or PRR-1 proteins provides unambiguous evidence for the actual usage of these receptors in human cell lines. Third, since the virus must infect neurons to establish latency, the finding that mRNAs for HIgR or PRR-1 are expressed in human tissues, with the highest expression being detected in nervous-system samples, makes infection in humans feasible.
The present work confirms and extends the report on PRR-1(HveC) (16) by mapping to the ectodomain of HIgR and PRR-1 the functional domain mediating alphaherpesvirus entry into cells, by showing the actual distribution of HIgR and PRR-1 at the protein level in human cell cultures, by providing evidence for actual usage of the receptors in these same cells, and by indicating possible usage of receptors in humans in the pathway of neuron infection by HSV.
ACKNOWLEDGMENTS
We thank our colleagues Sara Di Gaeta, Margherita De Lillo, Mustapha Aoubala, and José Adelaide for generous help, and we are indebted to Elisabetta Romagnoli for assistance in derivation of the resistant cell lines and to Angela Algeri for invaluable suggestions. We thank P. G. Spear, Chicago, for the gift of pBEC10; G. Cohen and R. Eisenberg, Philadelphia, for recombinant gD; B. Roizman, Chicago, for gifts of R8102, R8996, and anti-gM antibodies; T. Minson, Cambridge, for LP1 antibody; M. Ackerman, Zurich, for MAb antibody 1240 to BHV-1 gB; and T. Mettenleiter, Insel Riems, for MAb B16-C8 to PRV.
The work at the University of Bologna was supported by grants from Target Project in Biotechnology; AIDS Project from Istituto Superiore di Sanità; BIOMED2 BMH4 CT95 1016 grant from UE; MURST 40%, University of Bologna 60%; and the pluriannual plan. The work at INSERM Marseille was supported by INSERM and grants from the Association de le Recherche pour le Cancer (ARC).
REFERENCES
- 1.Arsenakis M, Campadelli Fiume G, Roizman B. Regulation of glycoprotein D synthesis: does alpha 4, the major regulatory protein of herpes simplex virus 1, regulate late genes both positively and negatively? J Virol. 1988;62:148–158. doi: 10.1128/jvi.62.1.148-158.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baines J D, Roizman B. The UL10 gene of herpes simplex virus 1 encodes a novel viral glycoprotein, gM, which is present in the virion and in the plasma membrane of infected cells. J Virol. 1993;67:1441–52. doi: 10.1128/jvi.67.3.1441-1452.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonaldo M F, Lennon G, Soares M B. Normalization and subtraction: two approaches to facilitate gene discovery. Genome Res. 1996;6:791–806. doi: 10.1101/gr.6.9.791. [DOI] [PubMed] [Google Scholar]
- 4.Brandimarti R, Huang T, Roizman B, Campadelli Fiume G. Mapping of herpes simplex virus 1 genes with mutations which overcome host restrictions to infection. Proc Natl Acad Sci USA. 1994;91:5406–5410. doi: 10.1073/pnas.91.12.5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunetti C R, Burke R L, Hoflack B, Ludwig T, Dingwell K S, Johnson D C. Role of mannose-6-phosphate receptors in herpes simplex virus entry into cells and cell-to-cell transmission. J Virol. 1995;69:3517–3528. doi: 10.1128/jvi.69.6.3517-3528.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai W H, Gu B, Person S. Role of glycoprotein B of herpes simplex virus type 1 in viral entry and cell fusion. J Virol. 1988;62:2596–2604. doi: 10.1128/jvi.62.8.2596-2604.1988. . (Erratum, 62:4438.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campadelli-Fiume G, Arsenakis M, Farabegoli F, Roizman B. Entry of herpes simplex virus 1 in BJ cells that constitutively express viral glycoprotein D is by endocytosis and results in degradation of the virus. J Virol. 1988;62:159–167. doi: 10.1128/jvi.62.1.159-167.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campadelli-Fiume G, Qi S, Avitabile E, Foà-Tomasi L, Brandimarti R, Roizman B. Glycoprotein D of herpes simplex virus encodes a domain which precludes penetration of cells expressing the glycoprotein by superinfecting herpes simplex virus. J Virol. 1990;64:6070–6079. doi: 10.1128/jvi.64.12.6070-6079.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campadelli-Fiume G, Stirpe D, Boscaro A, Avitabile E, Foà-Tomasi L, Barker D, Roizman B. Glycoprotein C-dependent attachment of herpes simplex virus to susceptible cells leading to productive infection. Virology. 1990;178:213–222. doi: 10.1016/0042-6822(90)90396-9. [DOI] [PubMed] [Google Scholar]
- 10.Chase C C, Lohff C, Letchworth G J D. Resistance and susceptibility of bovine cells expressing herpesviral glycoprotein D homologs to herpesviral infections. Virology. 1993;194:365–369. doi: 10.1006/viro.1993.1269. [DOI] [PubMed] [Google Scholar]
- 11.Dean H J, Terhune S S, Shieh M T, Susmarski N, Spear P G. Single amino acid substitutions in gD of herpes simplex virus 1 confer resistance to gD-mediated interference and cause cell-type-dependent alterations in infectivity. Virology. 1994;199:67–80. doi: 10.1006/viro.1994.1098. [DOI] [PubMed] [Google Scholar]
- 12.De Luca N, Bzik D J, Bond V C, Person S, Snipes W. Nucleotide sequences of herpes simplex virus type 1 (HSV-1) affecting virus entry, cell fusion, and production of glycoprotein gB (VP7) Virology. 1982;122:411–423. doi: 10.1016/0042-6822(82)90240-9. [DOI] [PubMed] [Google Scholar]
- 13.Eberlé F, Dubreuil P, Mattei M G, Devilard E, Lopez M. The human PRR2 gene, related to the human poliovirus receptor gene (PVR), is the true homolog of the murine MPH gene. Gene. 1995;159:267–272. doi: 10.1016/0378-1119(95)00180-e. [DOI] [PubMed] [Google Scholar]
- 14.Ejercito P M, Kieff E D, Roizman B. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J Gen Virol. 1968;2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 15.Forrester A, Farrell H, Wilkinson G, Kaye J, Davis Poynter N, Minson T. Construction and properties of a mutant of herpes simplex virus type 1 with glycoprotein H coding sequences deleted. J Virol. 1992;66:341–348. doi: 10.1128/jvi.66.1.341-348.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geraghty R J, Krummenacher C, Cohen G H, Eisenberg R J, Spear P G. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science. 1998;280:1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 17.Hill T J, Field H J, Blyth W A. Acute and recurrent infection with herpes simplex virus in the mouse: a model for studying latency and recurrent disease. J Gen Virol. 1975;28:341–353. doi: 10.1099/0022-1317-28-3-341. [DOI] [PubMed] [Google Scholar]
- 18.Hoggan M D, Roizman B. The isolation and properties of a variant of herpes simplex producing multinucleated giant cells in monolayer cultures in the presence of antibody. Am J Hyg. 1959;70:208–219. doi: 10.1093/oxfordjournals.aje.a120071. [DOI] [PubMed] [Google Scholar]
- 19.Huang T, Campadelli Fiume G. Anti-idiotypic antibodies mimicking glycoprotein D of herpes simplex virus identify a cellular protein required for virus spread from cell to cell and virus-induced polykaryocytosis. Proc Natl Acad Sci USA. 1996;93:1836–1840. doi: 10.1073/pnas.93.5.1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson D C, Burke R L, Gregory T. Soluble forms of herpes simplex virus glycoprotein D bind to a limited number of cell surface receptors and inhibit virus entry into cells. J Virol. 1990;64:2569–2576. doi: 10.1128/jvi.64.6.2569-2576.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson R M, Spear P G. Herpes simplex virus glycoprotein D mediates interference with herpes simplex virus infection. J Virol. 1989;63:819–827. doi: 10.1128/jvi.63.2.819-827.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaner R J, Baird A, Mansukhani A, Basilico C, Summers B D, Florkiewicz R Z, Hajjar D P. Fibroblast growth factor receptor is a portal of cellular entry for herpes simplex virus type 1. Science. 1990;248:1410–1413. doi: 10.1126/science.2162560. [DOI] [PubMed] [Google Scholar]
- 23.Karger A, Mettenleiter T C. Glycoproteins gIII and gp50 play dominant roles in the biphasic attachment of pseudorabies virus. Virology. 1993;194:654–664. doi: 10.1006/viro.1993.1305. [DOI] [PubMed] [Google Scholar]
- 24.Kieff E D, Bachenheimer S L, Roizman B. Size, composition, and structure of the deoxyribonucleic acid of herpes simplex virus subtypes 1 and 2. J Virol. 1971;8:125–132. doi: 10.1128/jvi.8.2.125-132.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koike S, Horie H, Ise I, Okitsu A, Yoshida M, Iizuka N, Takeuchi K, Takegami T, Nomoto A. The poliovirus receptor protein is produced both as membrane-bound and secreted forms. EMBO J. 1990;9:3217–3224. doi: 10.1002/j.1460-2075.1990.tb07520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koike S, Ise I, Sato Y, Yonekawa H, Gotoh O, Nomoto A. A second gene for the African green monkey poliovirus receptor that has no putative N-glycosylation site in the functional N-terminal immunoglobulin-like domain. J Virol. 1992;66:7059–7066. doi: 10.1128/jvi.66.12.7059-7066.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ligas M W, Johnson D C. A herpes simplex virus mutant in which glycoprotein D sequences are replaced by β-galactosidase sequences binds to but is unable to penetrate into cells. J Virol. 1988;62:1486–1494. doi: 10.1128/jvi.62.5.1486-1494.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopez M, Eberlé F, Mattei M G, Gabert J, Birg F, Bardin F, Maroc C, Dubreuil P. Complementary DNA characterization and chromosomal localization of a human gene related to the poliovirus receptor-encoding gene. Gene. 1995;155:261–265. doi: 10.1016/0378-1119(94)00842-g. [DOI] [PubMed] [Google Scholar]
- 29.Lopez M, Jordier F, Bardin F, Coulombel L, Chabannon C, Dubreuil P. Identification of a new class of IgG superfamily antigens expressed in hemopoiesis. In: Kishomoto T, Kikutani H, von dem Borne A, Mason D Y, Miyasaka M, Moretta A, Okumura K, Shaw S, Springer T A, Sugumura K, Zola H, editors. Leukocyte typing IV. White cell differentiation antigens. New York, N.Y: Garland Publishing, Inc.; 1997. pp. 1081–1083. [Google Scholar]
- 30.McLean C, Buckmaster A, Hancock D, Buchan A, Fuller A, Minson A. Monoclonal antibodies to three non-glycosylated antigens of herpes simplex virus type 2. J Gen Virol. 1982;63:297–305. doi: 10.1099/0022-1317-63-2-297. [DOI] [PubMed] [Google Scholar]
- 31.Mendelsohn C L, Wimmer E, Racaniello V R. Cellular receptor for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell. 1989;56:855–865. doi: 10.1016/0092-8674(89)90690-9. [DOI] [PubMed] [Google Scholar]
- 32.Mirda D P, Navarro D, Paz P, Lee P L, Pereira L, Williams L T. The fibroblast growth factor receptor is not required for herpes simplex virus type 1 infection. J Virol. 1992;66:448–457. doi: 10.1128/jvi.66.1.448-457.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Montgomery R I, Warner M S, Lum B J, Spear P G. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87:427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 34.Morrison M E, Racaniello V R. Molecular cloning and expression of a murine homolog of the human poliovirus receptor gene. J Virol. 1992;66:2807–2813. doi: 10.1128/jvi.66.5.2807-2813.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muggeridge M I, Cohen G H, Eisenberg R J. Herpes simplex virus infection can occur without involvement of the fibroblast growth factor receptor. J Virol. 1992;66:824–830. doi: 10.1128/jvi.66.2.824-830.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nicola A V, Willis S H, Naidoo N N, Eisenberg R J, Cohen G H. Structure-function analysis of soluble forms of herpes simplex virus glycoprotein D. J Virol. 1996;70:3815–3822. doi: 10.1128/jvi.70.6.3815-3822.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pereira L, Klassen T, Baringer J R. Type-common and type-specific monoclonal antibody to herpes simplex virus type 1. Infect Immun. 1980;29:724–732. doi: 10.1128/iai.29.2.724-732.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petrovskis E A, Meyer A L, Post L E. Reduced yield of infectious pseudorabies virus and herpes simplex virus from cell lines producing viral glycoprotein gp50. J Virol. 1988;62:2196–2199. doi: 10.1128/jvi.62.6.2196-2199.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pogue-Geile K L, Spear P G. The single base pair substitution responsible for the Syn phenotype of herpes simplex virus type 1, strain MP. Virology. 1987;157:67–74. doi: 10.1016/0042-6822(87)90314-x. [DOI] [PubMed] [Google Scholar]
- 40.Roller R J, Roizman B. A herpes simplex virus 1 US11-expressing cell line is resistant to herpes simplex virus infection at a step in viral entry mediated by glycoprotein D. J Virol. 1994;68:2830–2839. doi: 10.1128/jvi.68.5.2830-2839.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roop C, Hutchinson L, Johnson D C. A mutant herpes simplex virus type 1 unable to express glycoprotein L cannot enter cells, and its particles lack glycoprotein H. J Virol. 1993;67:2285–2297. doi: 10.1128/jvi.67.4.2285-2297.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruyechan W T, Morse L S, Knipe D M, Roizman B. Molecular genetics of herpes simplex virus. II. Mapping of the major viral glycoproteins and of the genetic loci specifying the social behavior of infected cells. J Virol. 1979;29:677–697. doi: 10.1128/jvi.29.2.677-697.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schröder C H, Stegmann B, Lauppe H F, Kaerner H C. An unusual defective genotype derived from herpes simplex virus strain ANG. Intervirology. 1975;6:270–284. doi: 10.1159/000149481. [DOI] [PubMed] [Google Scholar]
- 44.Shieh M T, Spear P G. Fibroblast growth factor receptor: does it have a role in the binding of herpes simplex virus? Science. 1991;253:208–210. doi: 10.1126/science.1649495. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 45.Showalter S D, Zweig M, Hampar B. Monoclonal antibodies to herpes simplex virus type 1 proteins, including the immediate-early protein ICP 4. Infect Immun. 1981;34:684–692. doi: 10.1128/iai.34.3.684-692.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spear P G. Entry of alphaherpesviruses into cells. Semin Virol. 1993;4:167–80. [Google Scholar]
- 47.Walther W, Stein U, Eder C. RNA analysis using miniprep RNA in reverse transcription PCR. BioTechnique. 1994;17:674–675. [PubMed] [Google Scholar]
- 48.Warner M S, Geraghty R J, Martinez W M, Montgomery R I, Whitbeck J C, Xu R, Eisenberg R J, Cohen G H, Spear P G. A cell surface protein with herpesvirus entry activity (HveB) confers susceptibility to infection by mutants of herpes simplex virus type 1, herpes simplex virus type 2, and pseudorabies virus. Virology. 1998;246:179–189. doi: 10.1006/viro.1998.9218. [DOI] [PubMed] [Google Scholar]
- 49.Wildy P, Russel W C, Horne R W. The morphology of Herpes virus. Virology. 1960;12:204–222. doi: 10.1016/0042-6822(60)90195-1. [DOI] [PubMed] [Google Scholar]