Abstract
Cellular zinc (Zn2+) homeostasis is essential to human health and is under tight regulations. Human zinc transporter 1 (hZnT1) is a plasma membrane–localized Zn2+ exporter belonging to the ZnT family, and its functional aberration is associated with multiple diseases. Here, we show that hZnT1 works as a Zn2+/Ca2+ exchanger. We determine the structure of hZnT1 using cryo–electron microscopy (cryo-EM) single particle analysis. hZnT1 adopts a homodimeric structure, and each subunit contains a transmembrane domain consisting of six transmembrane segments, a cytosolic domain, and an extracellular domain. The transmembrane region displays an outward-facing conformation. On the basis of structural and functional analysis, we propose a model for the hZnT1-mediated Zn2+/Ca2+ exchange. Together, these results facilitate our understanding of the biological functions of hZnT1 and provide a basis for further investigations of the ZnT family transporters.
Human ZnT1 functions as a Zn2+/Ca2+ exchanger, and a cryo-EM structure of ZnT1 has been determined.
INTRODUCTION
As an essential trace metal ion in human, zinc is critical to numerous fundamental life processes by playing roles in maintaining the normal structures and functions of a large variety of proteins or cell signaling regulations (1, 2). Zinc homeostasis is under tight regulations of several factors, including the nonspecific metal-binding proteins named metallothioneins (MTs), which serve as an intracellular buffer, and two intramembrane zinc transporter families, the Zrt- and Irt-like protein (ZIP) family that imports zinc from the extracellular space or subcellular organelles into the cytoplasm and the zinc transporter (ZnT) family that exports zinc from the cytoplasm to the extracellular milieu or organelles (3, 4). Ten members have been identified in the human ZnT family, namely, ZnT1-ZnT10 (5). Of all these ZnTs, human ZnT1 (hZnT1) is first member identified and predominantly functions in the plasma membrane, thus playing a pivotal role in cellular zinc homeostasis (6–9). Homozygous ZnT1 gene knockout leads to embryonic lethal in mice (10), and aberrant hZnT1 expressions or functions have been found to be involved in multiple pathophysiological processes including Alzheimer’s disease (AD), cancer, obesity, and diabetes (5). Specially, expression levels of hZnT1 were reported to be substantially elevated in vulnerable brain regions of individuals with early AD or late AD but decreased in individuals in the preclinical stage of AD or with mild cognitive impairment, suggesting a close relationship between hZnT1 and AD onset (11–13). Up-regulation of hZnT1 expression level was also observed in several cancer types, such as prostate cancer, and in obesity (14–17). Thus, targeting hZnT1 may be a promising way of diagnosis and treatment of related diseases.
Although ubiquitously expressed and important to zinc homeostasis, the transport and energy coupling mechanisms of hZnT1 remain debatable. Bacterial homologs such as YiiP and certain hZnT members such as hZnT5 and hZnT8 have been shown to be Zn2+/H+ antiporters, which use the proton motive force to drive Zn2+ export (18–22). Earlier studies using human embryonic kidney 293T (HEK293T) cell–based fluorescent imaging and liposome-based coupled proton transport assay with the C-terminal truncated hZnT1 protein purified from Saccharomyces cerevisiae also suggested that hZnT1 functions as a Zn2+/H+ exchanger, and the export process is sodium independent but calcium sensitive (23, 24). A notable observation is that in the cell-based assay, no Zn2+ efflux was observed when calcium is absent in the extracellular medium, which was interpreted that Ca2+ had a stimulatory role to hZnT1’s transport activity (24). Another study using neuronal cell–based fluorescence measurements showed that a rise of pH (from 7.4 to 8.0) had little effect to hZnT1-mediated Zn2+ efflux (25). Instead, the transport is Ca2+ dependent, and Ca2+ influx was coupled to Zn2+ efflux, suggesting that hZnT1 is a Zn2+/Ca2+ exchanger in neuronal cells (25). Thus, the energy coupling mechanism of Zn2+ efflux by hZnT1 awaits further characterizations, especially through the in vitro analysis, which minimizes the influence of other cellular factors.
To solve the controversy of the energy coupler of Zn2+ export and unveil the transport mechanisms of hZnT1, in this study, we try to purify the full-length hZnT1 and determine its coupling mechanisms using both cell-based in vivo and proteoliposome-based in vitro assay systems. Our results support that hZnT1 functions as a Zn2+/Ca2+ exchanger. The structure of hZnT1 is determined using cryo–electron microscopy (cryo-EM) single particle analysis, providing a structural basis for further understanding of its transport mechanism.
RESULTS
Zn2+/Ca2+ exchange activity of hZnT1
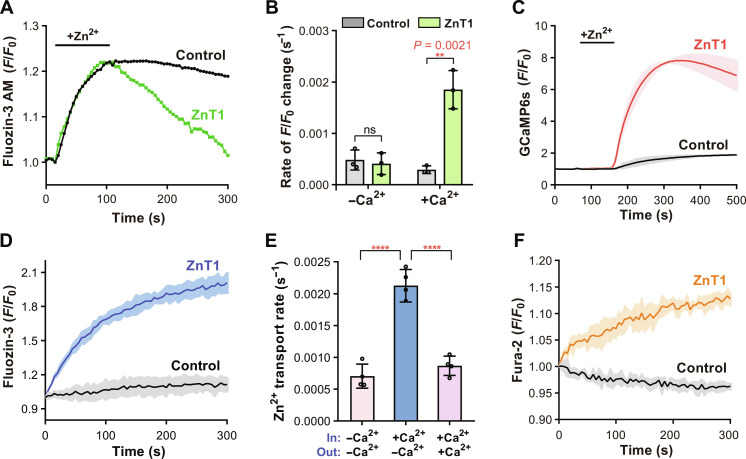
We first examined the Zn2+ efflux mediated by hZnT1 in the HEK293T cells. Cells were first loaded with the cell-permeable zinc-specific fluorescent indicator FluoZin-3 AM and then loaded with Zn2+ by incubation in the Ringer’s solution plus the ionophore zinc pyrithione and ZnCl2. After transfer into the Zn2+-free Ringer solution, rapid decrease in the fluorescence was observed in cells transfected with the wild-type (WT) hZnT1, but not in the control cells transfected with empty vector, confirming the hZnT1-mediated Zn2+ efflux (Fig. 1A). We tested the effects of proton gradients to the Zn2+ efflux by changing the extracellular buffer pH from 6.5 to 8.5. As the results showed, no significant differences were identified in the initial rate of fluorescence changes among different pH groups (fig. S1A), consistent with previous observations (25) and indicating that the efflux of Zn2+ is independent of proton gradients. The Ringer’s solution used as the extracellular assay buffer contains 1.8 mM CaCl2. If CaCl2 was removed from the Ringer’s solution, Zn2+ transport was impaired as indicated by the initial fluorescence change identified in cells transfected with the WT hZnT1 compared to the empty vector as a control (Fig. 1B), the same as previously reported (24, 25), suggesting that the presence of extracellular Ca2+ is critical to Zn2+ efflux. To find out whether Ca2+ is a transport activator or substrate of hZnT1, we monitored the intracellular Ca2+ level by coexpressing the specific Ca2+ fluorescent reporter GCaMP6s in HEK293T cells (26). After transfer into the Ringer’s solution, a rapid increase in the GCaMP6s fluorescence was observed in the WT hZnT1 cells, suggesting an elevation in the cytosolic Ca2+ accompanying the Zn2+ efflux (Fig. 1C). All above results obtained in the HEK293T cells are consistent with those reported in the neuronal cells (25), strongly supporting the notion that hZnT1 is a Zn2+/Ca2+ exchanger, instead of a Zn2+/H+ antiporter such as the other members of the ZnT family.
Fig. 1. hZnT1 functions as a Zn2+/Ca2+ exchanger.
(A) Zn2+ efflux by hZnT1 in HEK293T cells. Representative traces of the fluorescence are shown for cells on one coverslip. (B) Initial rates of Zn2+ efflux in HEK293T cells in absence or presence of Ca2+, respectively. Each data point represents the initial rate calculated from the mean of fluorescence changes of at least thirty cells from one coverslip and was repeated three times with three coverslips. (C) Ca2+ influx by hZnT1 in HEK293T cells. Data are obtained with at least thirty cells from one coverslip and repeated three times with three coverslips. (D) Zn2+ efflux by hZnT1 in the proteoliposome-based transport assay. (E) Initial Zn2+ transport rates of hZnT1 determined with the proteoliposomes-based assay. Presence or absence of Ca2+ in assay buffer (out) or inside proteoliposomes (in) is indicated. Data are obtained from four independent experiments. (F) Ca2+ transport by hZnT1 measured in the proteoliposome-based assay. For (C), (D), and (F), fluorescence traces are shown as solid line (mean) with shaded areas (SD) from at least three biological repeats. For (B) and (E), significances were determined using a two-tailed unpaired t test. ns, not significant. ****P < 0.0001. Data are means ± SD.
To confirm these findings and rule out possible interference factors in the whole cell, we tried to set up an in vitro assay system using reconstituted proteoliposomes. The full-length hZnT1 protein was expressed and purified using the transient expression system in suspension HEK293F cells. The liposomes were reconstituted with the membrane-impermeable fluorescent indicator FluoZin-3 and Ca2+ loaded inside, and Zn2+ transport into the liposomes was monitored by measuring fluorescence change after adding the liposomes into the Zn2+-containing extramembrane assay buffer. As the results showed, proteoliposomes loaded with the purified WT hZnT1 protein had a rapid increase in the fluorescence after adding into the assay buffer, while no such changes were observed in the protein-free liposomes as the negative control (Fig. 1D). By titrating the Zn2+ concentrations and determining the initial fluorescence change rate of hZnT1 (27, 28), we determined the Km (Michaelis constant) of Zn2+ concentration to be 0.38 ± 0.14 μM (fig. S1B). Notably, when Ca2+ was removed from the proteoliposomes and extramembrane assay buffer, we still observed Zn2+ transport into the liposomes mediated by ZnT1 (fig. S1C), suggesting that ZnT1 can also transport Zn2+ in the presence of a Zn2+ gradient alone. This is similar to the ZnT1 homologs including YiiP and hZnT8, which can also mediate Zn2+ efflux in the absence of a proton gradient (22, 27, 29). However, the transport rate was reduced compared to that in the presence of a transmembrane Ca2+ gradient (Fig. 1E and fig. S1C). Besides, if the proteoliposomes were added into the assay buffer with the same concentration of Ca2+ as the inside of proteoliposomes, then the transport rate was also reduced, confirming that a cross-membrane Ca2+ gradient is essential for hZnT1 to drive Zn2+ efflux (Fig. 1E and fig. S1C). To confirm that Ca2+ is transported by hZnT1, we preloaded the liposomes with Ca2+ and then added the liposomes into the assay buffer containing Zn2+ and the Ca2+ indicator Fura-2 to monitor the Ca2+ efflux. As the results showed, a notable increase in Fura-2 fluorescence was observed in the WT hZnT1-loaded proteoliposomes, further proving a Ca2+ transport associated with the Zn2+ (Fig. 1F). We also generated two double mutants at the predicted metal ion binding site of hZnT1 within in the transmembrane domain (TMD), H43A/H251A and D47A/D255A, and tested their Ca2+ transport activities. Similar to the empty liposomes as the negative control, the H43A/H251A or D47A/D255A mutant had no increase in the Fura-2 fluorescent signal (fig. S1D). Besides Fura-2, we also purified the Ca2+ fluorescent reporter GCaMP6s and loaded GCaMP6s together with Zn2+ into the liposomes to monitor the Ca2+ influx in the presence of a transmembrane Zn2+ gradient. A similar increase in GCaMP6s fluorescence signal was observed in the hZnT1-loaded proteoliposomes (fig. S1E). Together, these in vitro results support the notion that hZnT1 is a Zn2+/Ca2+ exchanger, rather than a proton gradient–driven Zn2+ transporter.
Structure determination of hZnT1
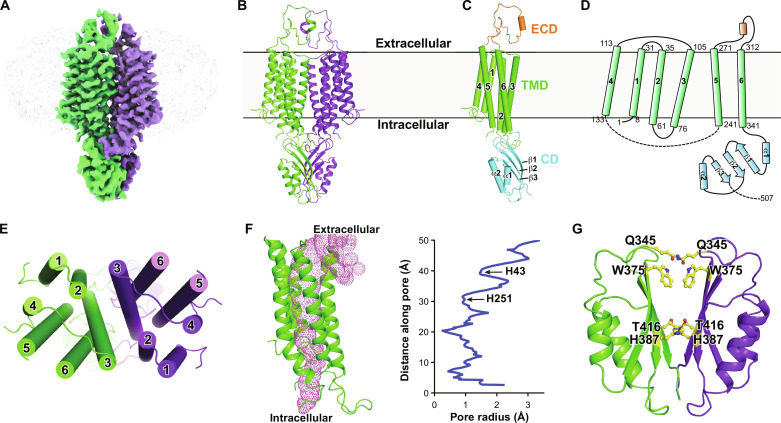
High-resolution structures have been reported for several Zn2+ exporters from different species, including the bacterial Zn2+/H+ transporter homolog YiiP in Shewanella oneidensis (SoYiiP) or Escherichia coli (EcYiiP), the hZnT1 orthologs hZnT7 and hZnT8, and the ZnT8 homolog in Xenopus tropicalis (XtZnT8) (27, 28, 30–35). Unlike the plasma membrane localization of hZnT1, hZnT7 and hZnT8 are localized in the Golgi apparatus and insulin secretory granules of pancreatic β cells, respectively (28, 34). hZnT1 shares relatively low sequence identities with either hZnT7 or hZnT8, about 27% each (fig. S2). We tried to determine its structure using cryo-EM single particle analysis with the purified protein in detergent glyco-diosgenin (GDN) from HEK293F cells (fig. S3, A and B). The two-dimensional (2D) classification results of the collected particles showed good protein features from different views (fig. S3C). However, we failed to get a high-resolution 3D electron microscopy (EM) map for structure modeling after extensive trials due to possible structural flexibilities and dynamics of hZnT1. The overall resolution only reached about 6.0 Å, insufficient for residue assignment (fig. S3, D and E). Nonetheless, a main-chain structure model for the transmembrane region was built, which shows the organization of the transmembrane helices (fig. S3F). To stabilize the protein and facilitate structure determination, two mutations at the predicted metal ion binding site within in the TMD, D47N and D255N, were introduced into hZnT1, a similar strategy used to get a high-resolution structure of hZnT8 (34). The protein expression and behavior were unaffected by the mutation (fig. S4A). We proceeded to determine its structure using cryo-EM with the protein purified in detergent GDN. After data collection and processing, an EM map with an overall resolution of 3.4 Å was lastly obtained as assessed by the gold standard Fourier shell correlation (FSC) criterion, allowing faithful model building and residue assignment (Fig. 2A and figs. S4, B to G; S5; and S6). Three hundred twenty-five of the total 507 amino acids were modeled (Fig. 2B). A large cytosolic loop between transmembrane helix 4 (TM4) and TM5 with about 100 amino acids (141 to 237) and the C-terminal about 85 amino acids (423 to 507) were missing in the structural model due to vague EM densities, suggesting large flexibilities of these segments.
Fig. 2. Overall architecture of hZnT1.
(A) EM density for the hZnT1-D47N/D255N double mutant (hZnT1-DM) at 3.4-Å resolution. Densities corresponding to the two hZnT1 protomers are colored green and purple, respectively. The surrounding micelle is shown in light gray. (B) Cartoon representation of the dimeric hZnT1-DM structure. (C) A side view of the hZnT1-DM protomer structure, which is organized into the TMD, CD, and ECD. (D) Topological diagram of hZnT1. The starting and ending residue numbers of each transmembrane helix are labeled aside. (E) A top view of the TMD of hZnT1-DM structure. (F) Ion accessibility analysis of the hZnT1 structure, as calculated using HOLE2 (49). The pore cavity is shown as magenta mesh. Calculated pore radius along the transmembrane region is shown. (G) Overview of the CD of hZnT1-DM. Residues in the dimer interface forming either hydrophobic interaction or hydrogen bonding are shown in sticks.
Overall architecture of hZnT1
The structure of hZnT1-D47N/D255N double mutant (named hZnT1-DM hereafter) exhibits a homodimeric architecture, consistent with previous biochemical and structural observations of hZnT7, hZnT8, or their homologs (28, 30, 34, 36). Each hZnT1 protomer can be divided into three parts: an extracellular domain (ECD), a TMD, and a cytosolic domain (CD) (Fig. 2, C and D). The ECD of hZnT1 is formed by a linker between TM5 and TM6 (residues 272 to 311). Particularly, this linker is unconserved and missing in all other hZnTs or the bacterial homologs with determined structures (fig. S2, A and B). Thus, the ECD region represents a unique structure feature for hZnT1. Residues 291 to 297 of this linker form a short α helix and mediate additional intersubunit contacts. If we truncate the ECD region of hZnT1 and replace it with the shorter linker between TM5 and TM6 of hZnT8 (hZnT1-ΔECD), then the protein expression was unaffected by the truncation. However, the Zn2+ transport activity of the ΔECD mutant was markedly decreased compared to the WT hZnT1 as measured in the proteoliposome-based assay (fig. S7A), indicating that the ECD not only plays a structural role in the dimer formation but also has an important role to the metal transport. The TMD of hZnT1 consists of six transmembrane segments (TMs), with both N and C terminus located in the cytoplasmic side (Fig. 2, C and D). TM2 and TM3 from each protomer form extensive hydrophobic interactions, mediating the dimer formation in the TMD (Fig. 2E and fig. S7B). TM2, TM3, TM5, and TM6 form a cavity that is solvent accessible from the extracellular side but completely sealed from the cytoplasmic side, as revealed by the pore radius analysis results (Fig. 2F), suggesting that the captured hZnT1 structure is in an outward-facing state. Notably, the TMD in the low-resolution WT hZnT1 structure merges well with that in the hZnT1-DM structure (fig. S3G). The CD of hZnT1 consists of two short α helices (CDα1 and CDα2) and three β sheets (CDβ1 to CDβ3) (Fig. 2, C, D, and G). The β sheets of one protomer closely packs against that of another protomer through both hydrophobic interactions and hydrogen bonding and interacts with the TMD regions such as the TM2-TM3 linker of another protomer, thus contributing to the dimer formation (Fig. 2G and fig. S7C).
Structures have been determined for both the WT hZnT8 and the TMD Zn2+ binding site mutant (hZnT8-DM) (34). The WT hZnT8 in the presence of Zn2+ and the hZnT8-DM mutant in the absence of Zn2+ exhibit an outward-facing conformation for both protomers, while the WT hZnT8 in the absence of Zn2+ exhibits heterogeneous conformations, with one protomer in the inward-facing state and the other in the outward-facing state (34). Structure alignment of the homodimeric structures between hZnT1-DM and hZnT8 either in the outward-facing state or heterogeneous state reveals large variations in the overall structure, with a root mean square deviation (RMSD) of 4.73 Å (507 Cα atoms aligned) and 4.87 Å (463 Cα atoms aligned), respectively (fig. S8, A and B). Unlike the V-shaped structure of the TMDs in hZnT8, which are largely separated at the extracellular side, the TMD from each protomer of hZnT1-DM shifts toward each other, making more extensive contacts. However, the observed structural variations in the CD dimer between hZnT1 and hZnT8 are mainly due to a rigid-body movement, as they merge well if we align the CDs alone, with a RMSD of 1.10 Å (104 Cα atoms aligned) (fig. S8C). Alignment of the hZnT1-DM monomer structure with the hZnT8 monomer either in the outward-facing state or inward-facing state reveals an RMSD of 2.96 Å (228 Cα atoms aligned) and 4.23 Å (225 Cα atoms aligned), respectively, further supporting the notion that the determined structure of hZnT1-DM is in the outward-facing state (fig. S8, D and F). This is further supported by the notion that the TMDs alone of hZnT1-DM and the outward-facing state of hZnT8-DM superpose well with an RMSD of 1.99 Å (188 Cα atoms aligned), especially for TM3 and TM6 (fig. S8E). However, dramatic changes are observed for the superposition of the TMDs of hZnT1-DM and the inward-facing state of hZnT8-WT. While TM3 and TM6 align well, TM1-TM2 and TM4-TM5 undergo large rotational movements (fig. S8G). The structural comparisons with hZnT8 indicates that hZnT1 may undergo similar rearrangements in the TMD during metal transport and that TM3 and TM6 remain largely unmoved, while TM1-TM2 and TM4-TM5 undergo marked rotational movement to expose the metal binding site to the extracellular side or the cytoplasm. The interdomain flexibilities between the two protomers have also been reported in ZnT1 homologs including YiiPs and hZnT7 (27, 28, 31, 32) and have been suggested to be critical during the Zn2+ transport.
Cryo-EM structures have also been reported recently for the hZnT7 in multiple states, including the Zn2+-unbound or Zn2+-bound states with the dimers both in the outward-facing state or one protomer in the inward-facing state while the other in the outward-facing state (28), providing a wealth of structural insights into the Zn2+ binding and transport process. Structural alignments of hZnT1 with hZnT7 also reveal that our determined hZnT1 structure aligns well with the Zn2+-unbound, outward-facing state of hZnT7 (fig. S9A), with an RMSD of 2.16 Å (476 Cα atoms aligned). Similar TM rearrangements were observed when aligning the TMD of the hZnT1-DM monomer and that of hZnT7 either in the outward-facing state (fig. S9B) or inward-facing states (fig. S9C), showing that TM1-TM2 and TM4-TM5 undergo large rotational movements during state transitions.
Metal ion binding sites of hZnT1
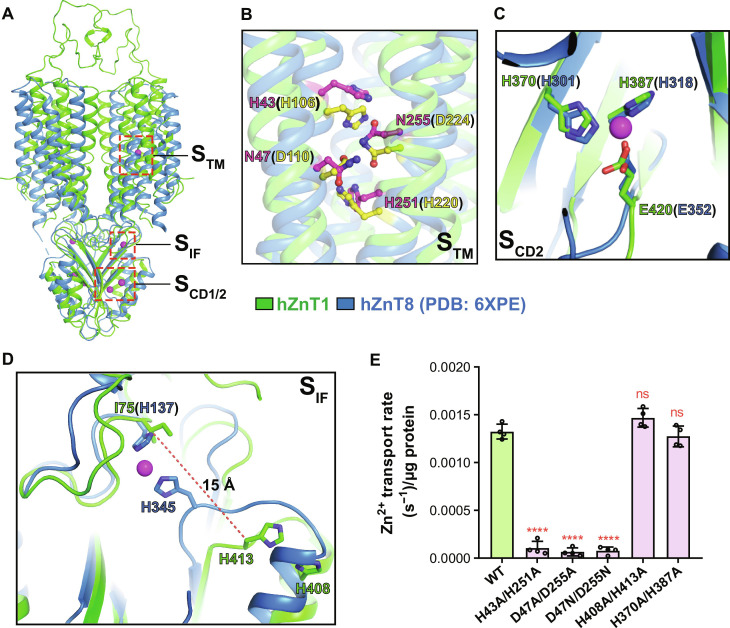
Several conserved Zn2+ binding sites have been proposed in the Zn2+-bound structures of bacterial YiiPs and hZnT8, including one site in the TMD (STM), one site in the TMD-CD interface (SIF), and two sites in the CD (SCD1 and SCD2) (Fig. 3A) (20, 31, 33). The metal binding site in the TMD is well preserved among hZnTs and YiiPs and is formed by a Histidine-Aspartate-Histidine-Aspartate (HDHD) or Aspartate-Aspartate-Histidine-Aspartate (DDHD) motif (fig. S2A) (34). In hZnT1, the corresponding residues are H43, D47, H251, and D255 (Fig. 3B). The H43A/H251A, D47A/D255A double mutant, or D47N/D255N mutant used for structure determinations lost the Zn2+ efflux activities as shown by the proteoliposome-based transport assays (Fig. 3E and fig. S10). Because two of the coordinating residues (D47 and D255) were mutated to asparagine to stabilize the protein conformation, no metal ion density was present in this site in the EM map of hZnT1-DM mutant. Besides, the four residues adopt quite similar configurations to the hZnT8-DM mutant structure or hZnT7 structures in the outward-facing state, suggesting a conserved metal binding profile in the transmembrane region (Fig. 3B and fig. S9D).
Fig. 3. Metal binding sites of hZnT1.
(A) Overall view of the putative Zn2+ binding sites in the hZnT1-DM structure (green) aligned with the WT hZnT8 structure [Protein Data Bank (PDB) code: 6XPE; blue]. (B) Superposition of the putative STM Zn2+ binding sites between hZnT1 (green) and WT hZnT8 (blue). Residues forming the binding site in WT hZnT1 are shown in sticks with carbon atoms colored magenta and in hZnT8 colored yellow. (C) Superposition of the putative SCD2 binding sites between hZnT1 (green) and hZnT8 (blue). (D) Superposition of the putative SIF binding sites between hZnT1 (green) and hZnT8 (blue). (E) Initial Zn2+ transport rates per migrogram protein for the WT hZnT1 and mutants in the STM, SIF, and SCD2 Zn2+ binding sites, as measured by the proteoliposome-based Zn2+ transport assay using FluoZin-3. Independent experiments were repeated four times for each construct. Data are means ± SD. Significances were determined using one-way analysis of variance (ANOVA) with Dunnett’s multiple comparisons test. ****P < 0.0001; ns, for WT versus mutants.
An N-terminal Histidine-Cysteine-Histidine (HCH) motif is involved in the formation of both Zn2+ binding sites in the CD of ZnT8, SCD1 and SCD2, especially SCD1 (34). However, in ZnT1, the N-terminal sequence is much shorter and the HCH motif is missing, thus unlikely to form a Zn2+ binding site as SCD1 (fig. S2A). Besides, the C terminus, which is involved in the formation of SCD1 in hZnT8, is unmodeled in the hZnT1-DM structure due to vague EM densities. Three metal-coordinating residues in SCD2 are invariant between hZnT1 (H370, H387, and E420) and hZnT8 (H301, H318, and E352) (Fig. 3C) and adopt almost identical configurations in the structures. Because no Zn2+ was added to the protein sample for structure determination, no clear EM density was observed for Zn2+ ion in this site. We examined the function of this site in hZnT1 by mutating both H370 and H387 to alanine and tested its Zn2+ transport activity. The result showed that the Zn2+ transport activity as indicated by the initial fluorescence change was unaffected by the double mutation in the proteoliposome-based assay system (Fig. 3E and fig. S10). We also tested the protein thermal stability of the WT hZnT1 and the H370/H387 mutant either in the absence or presence of Zn2+ using a microscale fluorescent–based assay (37). The result showed little difference between the WT and mutant proteins under either condition (fig. S11, A and B).
The SIF of hZnT8 is formed by two histidine residues, H137 in the TM2-TM3 linker and H345 in the CDα2-CDβ3 linker (Fig. 3D) (34). H345 is conserved in hZnT8 and hZnT1 (corresponding to H413 in hZnT1). However, H137 is substituted by an isoleucine residue, I75 in hZnT1 (Fig. 3D). In the hZnT1-DM structure without Zn2+, I75 and H413 are largely separated, with a distance of 15 Å between the Cβ atoms, much larger than the distance between H137 and H345 in the hZnT8-DM structure in the absence or presence of Zn2+ (Fig. 3D). Instead, H413 is much closer to H408 located in CDα2 (Fig. 3D). On the basis of sequence and structure variations, it is unlikely for I75 and H413 in hZnT1 to form a metal binding site as in hZnT8 or the bacterial YiiPs. In hZnT8, mutation of H137 and H345 leads to severely reduced Zn2+ transport rate, and this site is proposed to help recruit Zn2+ ions and facilitate the transport against its concentration gradient (34). For hZnT1, we mutated the two adjacent histidine residues (H408 and H413) to alanine and tested its Zn2+ transport activity using the proteoliposome-based assay. As the result showed, there was no significant difference in Zn2+ transport rate between the WT hZnT1 and the H408A/H413A mutant, suggesting a less conserved function (Fig. 3E and fig. S10). The protein thermal stability was also similar for the WT hZnT1 and the H413A mutant either in the absence or presence of Zn2+ (fig. S11, A and B). Together, of all the putative Zn2+ binding sites in hZnT1 including STM, SIF, and SCD, only the STM is the most critical to the Zn2+ transport.
Both as divalent metal ions, the coordination of Ca2+ and Zn2+ by proteins can vary in the binding geometries and residue preferences that Zn2+ typically forms a tetrahedral geometry with cysteine and/or histidine residues, while Ca2+ is coordinated in a pentagonal bipyramidal geometry with surrounding residues or water molecules, and the bond lengths and angles can also be different (38, 39). Thus, the location of the Ca2+ binding site in hZnT1 and whether Ca2+ binds to the same site in the TMD (STM) as Zn2+ await further investigations.
Metal translocation pathway in the TMD of hZnT1
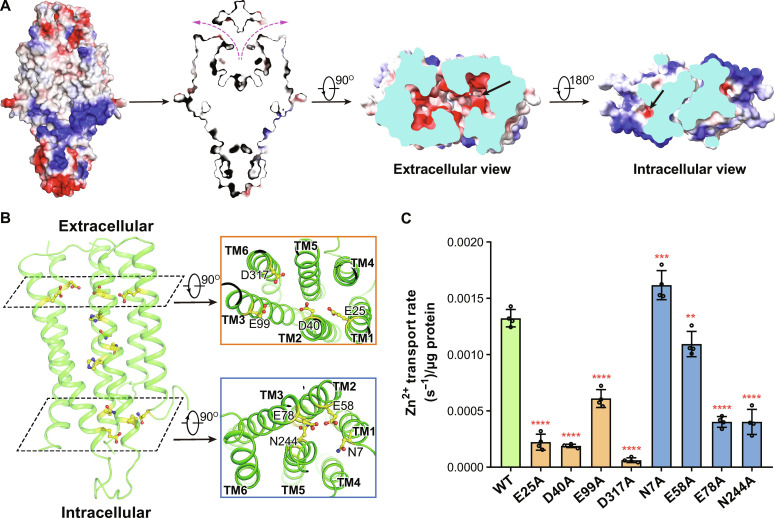
As aforementioned, the determined hZnT1-DM structure exhibits an outward-facing conformation with a large cavity formed on the extracellular side, which may be the path for Zn2+ release and Ca2+ binding. The cavity is strongly negative in electrostatic surface potential (Fig. 4A), contributed by mainly four negatively charged residues, including E25 in TM1, D40 in TM2, E99 in TM3, and D317 in TM6 (Fig. 4B). Notably, these residues are almost on the same layer above the STM, with the side chains pointing toward the extracellular cavity (Fig. 4B). We checked the functions of these negatively charged residues using point mutagenesis and the transport assays. Notable reductions in Zn2+ transport rate were detected for the E25A, D40A, and D317A mutants compared to the WT hZnT1 while reduced by about 50% for the E99A mutant (Fig. 4C and fig. S10). These results suggest that these negatively charged residues on the extracellular side are essential to the metal translocation of hZnT1, possibly through promoting either the metal binding/release or the conformational changes.
Fig. 4. Metal translocation pathway of hZnT1.
(A) Side and section views of the surface electrostatic potential of the hZnT1-DM structure. (B) The zinc binding site and two layers of acidic or polar residues along the metal permeation pathway in the TMD of hZnT1. Zoom-in views are shown for the acidic residues close to the extracellular side (orange box) and the polar/acidic residues on the intracellular side (cyan box). (C) Initial Zn2+ transport rates per microgram protein for the WT hZnT1 and alanine mutants of residues along the metal permeation pathway, as measured by the proteoliposome-based Zn2+ transport assay using FluoZin-3. Independent experiments were repeated four times for each construct. Data are means ± SD. Significances were determined using one-way ANOVA with Dunnet’s multiple comparisons test. **P = 0.0039, ***P = 0.0002, and ****P < 0.0001 for WT versus mutants.
On the intracellular side of the metal permeation pathway, there are four polar or negatively charged residues contributing to a negative electrostatic surface potential, including N7 in TM1, E58 in TM2, E78 in TM3, and N244 in TM5 (Fig. 4, A and B). In hZnT7 and hZnT8, there are also multiple acidic residues located on the intracellular side of the TMD (fig. S2A), and they are exposed to a cytosolic cavity in the inward-facing conformation of hZnT7 (28, 34). It has been proposed that these residues may act as a Zn2+ recruiting gate along the permeation pathway (28). For these residues in hZnT1, when mutated to alanine, the Zn2+ transport activity remained unaffected for N7 and E58A mutants while was reduced by more than 50% for E78 or N244, suggesting essential roles of these two residues in Zn2+ transport (Fig. 4C and fig. S10).
DISCUSSION
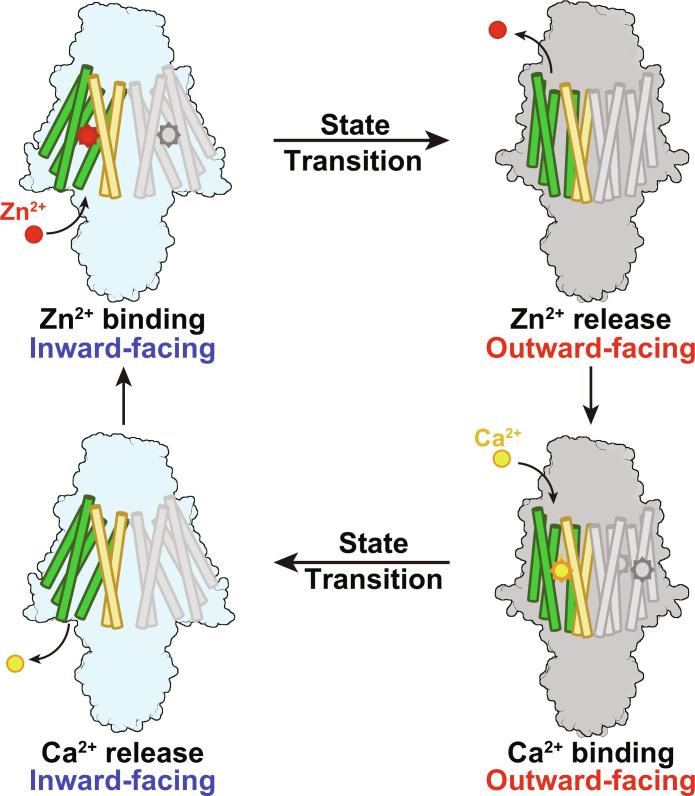
In this study, we show that instead of using the proton gradients as the transport energy source by other hZnT members with identified functions, hZnT1 mediates Zn2+ efflux with the downstream Ca2+ gradient. It is supported by both the cell-based and proteoliposome-based transport assay results in our studies and also consistent with previous reports by Gottesman et al. (25). Notably, even in the reports by Shusterman et al. (24), the calcium-dependent Zn2+ efflux by hZnT1 was also noticed but interpreted that calcium functions as a transport activator instead of a substrate. The Ca2+-coupled Zn2+ transport is meaningful, considering that hZnT1 is present in the plasma membrane where the cross-membrane calcium concentration is basically different and large for most cell types, while other hZnT family members are mainly present in acidic vesicles, such as hZnT5 in the Golgi apparatus and hZnT8 in the insulin secretory granules where the cross-membrane proton concentration varies markedly. Because the four residues forming this Zn2+ binding site are highly conserved in hZnT1, hZnT5, hZnT7, hZnT8, and the bacterial YiiPs (fig. S3A) and also adopt similar configurations in the hZnT1, hZnT7, and hZnT8 structures, the unique Ca2+ gradient–driven metal transport by hZnT1 cannot be explained by sequence or structural variations of these residues. The molecular basis and key residues, possibly lying along or near the metal translocation path, leading to the different energy sources of hZnT1-mediated Zn2+ transport, await further characterizations. In the structure of hZnT1, an ECD is identified, which mediates further dimer interface formation and is essential to the Zn2+ efflux as shown by the transport assay result (fig. S7A). The role of this domain remains to be investigated. On the basis of the functional and structural characterizations of hZnT1, together with the structural analysis of hZnT7, hZnT8, and the bacterial or eukaryotic homologs, we proposed a transport model for hZnT1-mediated Zn2+/Ca2+ exchange (Fig. 5). hZnT1 undergoes at least two different states during a transport cycle, the inward-facing state and the outward-facing state. Similar to hZnT7 and hZnT8, during the state transitions, TM3 and TM6 remain largely unchanged, forming a scaffold domain, while TM1, TM2, TM4, and TM5 undergo large rotational movement to alternatingly expose the metal binding site to the extracellular space or cytoplasm, forming a transport domain (Fig. 5). In the inward-facing state, Zn2+ binds to hZnT1, and the transporter undergoes a conformational change to the outward-facing state. Because the extracellular Ca2+ concentration is high, it binds to hZnT1. After Ca2+ binding, the transporter undergoes another conformational change and turns into the inward-facing state. Because the intracellular Ca2+ concentration is low, Ca2+ is released, and Zn2+ binds to the transporter to start another round of transport (Fig. 5). To further unveil the metal recognition and transport mechanism by hZnT1, structures of the hZnT1 under different conformational states or in complex with either Zn2+ or Ca2+ await to be determined. Nonetheless, the structural and functional characterizations of hZnT1 reported here facilitate our understanding of the Zn2+ efflux and set up a framework for further studies of the functional and energy-coupling variations among hZnT family transporters.
Fig. 5. Cartoon model for hZnT1-mediated Zn2+/Ca2+ exchange.
hZnT1 undergoes alternating inward-facing and outward-facing transitions to mediate metal transport. In the inward-facing state, Ca2+ is released to the cytosol and Zn2+ binds to hZnT1. In the outward-facing state, Zn2+ is released to the extracellular space and Ca2+ binds to hZnT1. During state transitions, TM3 and TM6 (colored orange) remain almost unchanged, while the other TMs (TM1, TM2, TM4, and TM5, colored green) undergo large rotational movement.
MATERIALS AND METHODS
Protein expression and purification
The cDNA sequence of the full-length WT hZnT1, which is publicly available at Uniprot (www.uniprot.org) with an accession code of Q9Y6M5, or hZnT1 variants was subcloned into a pCAG vector (Invitrogen) with a C-terminal Flag tag followed by a His10 tag. The suspension-cultured HEK293F cells (Sino Biological Inc.) were transfected with plasmids when reaching a density of 2.5 × 106 cells/ml. For 1 liter of cell culture, 1.5 mg of plasmids was premixed with 4 mg of linear polyethylenimines (Polysciences) in 45 ml of fresh medium for 15 min. Then, the mixture was added into the cell followed by a 30-min incubation. After 20 hours, 10 mM sodium butyrate were added to the cells cultured at 30°C. After 48-hour transfection, cells were collected by centrifugation at 3361g for 10 min. Cell pellet was resuspended in buffer containing 25 mM Hepes (pH 7.4) and 150 mM NaCl supplemented with 1.5% (w/v) n-dodecyl-β-d-maltoside (DDM; Anatrace), 0.3% (w/v) cholesteryl hemisuccinate (Sigma-Aldrich), 5 mM MgCl2, 5 mM adenosine 5´-triphosphate, 1 mM phenylmethylsulfonyl fluoride (PMSF), aprotinin (1.3 μg/ml), and pepstatin A (0.7 μg/ml) and then incubated for 2 hours at 4°C by gentle agitation. After detergent extraction, the supernatant was collected by centrifugation at 25,200g for 1 hour and incubated with the anti-FLAG M2 affinity gel (Sigma-Aldrich) at 4°C for 45 min. The resin was washed with 30 ml of buffer containing 25 mM Hepes (pH 7.4), 150 mM NaCl, and 0.02% (w/v) GDN (Anatrace). Protein was eluted with the wash buffer plus FLAG peptides (200 μg/ml), which was then concentrated with a 50-kDa cutoff Centricon (Millipore). Last, the sample was subjected to size exclusion chromatography using the Superose-6 Increase 10/300 column (Cytiva) in the buffer containing 25 mM Hepes (pH 7.4), 150 mM NaCl, and 0.01% (w/v) GDN (Anatrace). Peak fractions were collected and concentrated using a 50-kDa cutoff Centricon (Millipore) to 6 mg/ml for the cryo-EM analysis.
For the microscale fluorescent thermal stability assay, the WT hZnT1 and its mutants were purified in the presence of 0.5 mM EDTA during the membrane extraction and FLAG column affinity purification steps to remove endogenous Ca2+. The elution fractions were concentrated using a 50-kDa cutoff Centricon (Millipore) and purified on the Superose-6 Increase 10/300 column (Cytiva) in the buffer containing 25 mM Hepes (pH 7.4), 150 mM NaCl, and 0.01% (w/v) GDN (Anatrace). The peak fractions were concentrated to 0.1 mM using a 50-kDa cutoff Centricon for further analysis.
Protein expression and purification of GCaMP6s
The cDNA sequence of GCaMP6s (GenBank accession code: BCS79978.1) was cloned into the pET21b vector (Invitrogen) with the C-terminal His6 tag. E. coli strain BL21 (DE3) was used to express GCaMP6s. Cells were cultured in LB medium at 37°C to an OD600 of 1.0 and induced with 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 18°C overnight. Cells were collected by centrifugation at 3361g for 10 min, and the cell pellet was resuspended in buffer containing 25 mM tris-HCl (pH 8.0) and 150 mM NaCl supplemented with 1 mM PMSF. The cell suspension was lysed by sonication, and the insoluble fraction was removed by centrifugation at 25,200g for 1 hour. The supernatant was incubated with Ni-NTA beads (QIAGEN) at 4°C for 1 hour. The resin was washed with 40 ml of buffer containing 25 mM tris-HCl (pH 8.0), 150 mM NaCl, and 25 mM imidazole. The protein then was eluted with 6 ml of buffer containing 25 mM tris-HCl (pH 8.0), 150 mM NaCl, 300 mM imidazole, and further concentrated with a 10-kDa cutoff Centricon to 1.5 ml. A 20 mM EDTA was added into the eluent to remove metal ions. After an on-ice incubation of 40 min, the sample was applied to the Superdex 200 Increase 10/300 column (Cytiva). Peak fractions were collected and concentrated to 5 mg/ml.
Plasmid DNA constructs for the cell-based transport assays
The full-length, WT hZnT1 sequence was cloned into a pCAG vector (Invitrogen) with the C-terminal Flag tag and His10 tag. Sequence for the red, monomeric fluorescent protein mCherry (GenBank accession code: UFQ89828.1) was inserted between ZnT1 and the C-terminal Flag tag and His10 tag for the identification of successfully transfected cells. All hZnT1 variants and the empty vector plasmid were generated with the C-terminal mCherry, Flag, and His10 tag in the same way. The hZnT1-ΔECD mutation is generated by replacing the ECD region (residues 272 to 316) with the corresponding sequence of hZnT8 (residues 241 to 247). The GCaMP6s sequence was cloned into the pCAG vector with the C-terminal Flag and His10 tag.
Cell culture and transfection for the transport assay
HEK293T cells (Invitrogen) were cultured in Dulbecco’s modified Eagle’s medium/high glucose medium (Servicebio) supplemented with 10% fetal bovine serum (ExCell Bio.) and 1% penicillin-streptomycin (Sangon Biotech) and grown at 37°C with 5% CO2. Cells were cultured in 35-mm petri dishes, with three poly-d-lysine–coated (0.1 mg/ml) 12-mm glass coverslips placed at the bottom. Until reaching a confluency of about 50 to 60%, cells were transfected using jetOPTIMUS DNA transfection reagent (Polyplus-transfection), according to the manufacturer’s instructions.
HEK293T cell–based transport assay
Twenty-four hours after transfection, the glass coverslips with HEK293T cells loaded were washed with the Ringer’s solution containing 120 mM NaCl, 0.8 mM MgCl2, 5.4 mM KCl, 1.8 mM CaCl2, 20 mM Hepes (pH 7.4), and 15 mM glucose. To monitor Zn2+ efflux, cells were loaded with 1 μM fluorescent zinc indicator FluoZin-3 AM (Thermo Fisher Scientific) and incubated in the wash solution at room temperature for 30 min in dark. Transfected cells were imaged with a 488-nm excitation filter and an emission of 525-nm long-pass filter on an inverted microscope. The image capturing system is consist of a DG-5 wavelength switcher (Sutter Instrument), an ORCA-Flash4.0 LT+ complementary metal oxide-semiconductor (CMOS) camera (Hamamatsu), and a Ti2 microscope (Nikon). To measure Zn2+ efflux, cells loaded with FluoZin-3 AM were first incubated in Ringer’s solution (pH 7.4) and, after a baseline period, loaded with Zn2+ in the same solution plus 1 μM Zn2+ and 2 μM zinc pyrithione for 100 s. Then, cells were constantly superfused by gravity with Ringer’s solution at the indicated pH [Hepes (pH 7.4), MES (pH 6.5), or tris-HCl (pH 8.5)] to remove the Zn2+ pyrithione, and the fluorescent signal of the intracellular zinc was recorded during a time course of 250 s. The initial Zn2+ efflux rate was calculated as the average rate of fluorescence change upon washing with Zn2+-free Ringer’s solution within 100 s after the maximal signal. For Ca2+ influx measurements, the plasmids containing GCaMP6s and hZnT1 or the empty vector, respectively, were cotransfected into HEK293T cells with a 1:1 mass ratio. Cells were first incubated in Ca2+-free Ringer’s solution (pH 7.4) and then superfused with Ringer’s solution plus 5 mM Ca2+. The GCaMP6s fluorescence was excited at 488 nm and imaged with a 525-nm long-pass filter. For each experiment, at least 30 cells per coverslip were imaged and recorded, and all measurements were repeated at least three times with independent coverslips.
Proteoliposome-based zinc transport assay
For proteoliposome preparations, soybean lipids were resuspended to a final concentration of 20 mg/ml in the buffer containing 25 mM Hepes (pH 7.4), 100 mM KCl, and 1.8 mM CaCl2. The lipids were quick-frozen in liquid nitrogen and slowly thawed at room temperature for 10 times and then extruded through a 400-nm filter (Whatman) for 31 cycles. The liposome was then treated with 1% DDM for 1 hour at 4°C. The purified hZnT1 protein was loaded to liposome with a 1:100 protein:lipid (w/w) ratio, respectively, and incubated for 1 hour at 4°C. For detergent removal, the liposome was incubated overnight with Bio-Beads SM2 (400 mg/ml; Bio-Rad). After the beads were removed, 50 μM Fluozin-3 were added to the liposome. The mixtures were sonicated for 30 s and then frozen and thawed at room temperature. This step was repeated twice. After again sonicated for 30 s, the mixtures were extruded through a 400-nm filter for 21 cycles. To remove unincorporated FluoZin-3, the liposome was passed through a PD-10 desalting column (GE Healthcare) pre-equilibrated with the identical buffer. The eluted fractions were centrifuged at 160,000g for 1 hour at 4°C. After centrifugation, the liposomes were washed with ice-cold buffer [25 mM Hepes (pH 7.4) and 100 mM KCl] for three times to remove the remaining buffer and resuspended in the buffer containing 25 mM Hepes (pH 7.4) and 100 mM KCl to a final concentration of 100 mg/ml. All transport assays were performed at room temperature. For each experiment, following a baseline period, 1 μl of liposome was loaded into the assay buffer [25 mM Hepes (pH 7.4), 100 mM KCl, and 0.5 μM ZnCl2]. The Zn2+ transport activity of ZnT1 in proteoliposomes was monitored by the fluorescence change of FluoZin-3 (Thermo Fisher Scientific), a membrane-impermeable Zn2+ sensitive fluorescent indicator, using a Synergy H1 Multimode Reader (BioTek), with excitation and emission at 490 and at 525 nm, respectively, until a stable value was obtained. All transport experiments were repeated at least three times.
To measure Ca2+ efflux, after detergent removal, the liposomes were frozen and thawed at room temperature for eight cycles, followed by an extrusion through a 400-nm filter, and centrifuged at 160,000g for 1 hour. After centrifugation, the liposomes were washed with ice-cold buffer [25 mM Hepes (pH 7.4) and 100 mM KCl] for three times to remove the remaining buffer and resuspended in the buffer containing 25 mM Hepes (pH 7.4) and 100 mM KCl to a final concentration of 100 mg/ml. For each experiment, following a baseline period, 1 μl of liposome was loaded into the assay buffer [25 mM Hepes (pH 7.4) and 100 mM KCl] supplemented with various concentrations of ZnCl2 (0 to 2.5 μM) and 1 μM Fura-2 (Thermo Fisher Scientific). Fluorescence was excited at 340 and 380 nm and monitored at 510 nm. To measure the Ca2+ transport using GCaMP6s, during liposome preparation, purified GCaMP6s protein was added to the lipid resuspension buffer [25 mM Hepes (pH 7.4), 100 mM KCl, and 1 μM ZnCl2] with a 1:100 mass ratio versus soybean lipids. The fluorescence of GCaMP6s indicator was excited at 485 nm and monitored at 520 nm. The other steps are the same as described above, except that the assay buffer contains 1.8 mM CaCl2.
To test the effect of Ca2+ gradients to Zn2+ influx, the liposomes were prepared in buffer with or without CaCl2. For the liposomes without CaCl2 inside, soybean lipids were resuspended in buffer containing 25 mM Hepes (pH 7.4) and 100 mM KCl. For the measurements of zinc influx in the presence of outside CaCl2, 1 μl of liposome was added into the assay buffer with 25 mM Hepes (pH 7.4), 100 mM KCl, 1.8 mM CaCl2, and 0.5 μM ZnCl2. The other steps were the same as described above.
To normalize the time-dependent fluorescence from each measurement (FP), 4% octyl-β-d-glucloside (OG; Anatrace) in the assay buffer containing 64 mM ZnCl2 was mixed with proteoliposome for 40 min in dark to determine the maximal fluorescence (FPmax). The fluorescence of protein-free liposome (FL) was measured the same way as treated with 4% OG and 64 mM ZnCl2 to determine the background signal and normalize with the maximal fluorescence signal (FLmax). Zn2+ transport activity was then calculated using the equation (FP/FPmax) − (FL/FLmax) over time, and the initial rate was calculated by linear regression with in the first 40 s of liposome or proteoliposome, as described previously (27, 28, 32).
Microscale fluorescent thermal stability assay
N-[4-(7-diethylamino-4-methyl-3-coumarinyl) phenyl] maleimide (CPM; Sigma-Aldrich) was used for the thermal stability profiling of WT ZnT1 and the variants. CPM dye was dissolved in dimethyl sulfoxide at 4 mg/ml and diluted 1:40 in dilution buffer containing 25 mM Hepes (pH 7.4), 150 mM NaCl, and 0.025% DDM before use. Purified protein (5 μg) was incubated with 3 μl of diluted CPM while protected from light to reduce photobleaching. To determine the effect of Zn2+ on protein stability, purified proteins were mixed with 100 μM ZnCl2 and incubated with the diluted CPM buffer. The reaction mixture (20 μl in total) was added to a 384-well plate and heated in a controlled manner with a ramp rate of 3°C/min in a LightCycler 480 (Roche) real-time polymerase chain reaction instrument, ranging from room temperature to 99°C. The excitation wavelength was set at 440 nm, and the emission wavelength was set at 488 nm. Fluorescence intensity was continuously measured.
Sample preparation and cryo-EM data acquisition
For cryo-EM sample preparation, 4 μl of the purified WT hZnT1 protein or the hZnT1-DM mutant was applied to the glow-discharged holey carbon grids (Quantifoil Au R1.2/1.3, 300 mesh). The grid was blotted using Vitrobot Mark IV (Themo Fisher Scientific) operated at 8°C and 100% humidity with a blotting time of 3 s. Micrographs were acquired on a Titan Krios (FEI) electron microscope operating at 300 kV, equipped with the BioQuantum energy filter and a K3 direct electron detector (Gatan). Image stacks were recorded using E Pluribus Unum (EPU) software in the super-resolution mode with defocus values varied from −1.5 to −2.3 μm and an exposure time of 3 s and dose-fractionated to 32 frames with a total dose of 50 e−/Å2.
Image processing
A flowchart for the hZnT1-DM data processing is presented in fig. S5. Motion correction and dose weighting were performed using the RELION 3.1 implementation of MotionCor2 (40, 41). Defocus values were estimated by CTFFIND4 (42). After manually checked, 7509 micrographs were selected for particle picking. A total of 7,167,969 particles were automatically picked using cryoSPARC (v.3.2.0) (43). After 2D classification, 1,296,489 good particles were selected. These particles were classified into five classes in ab initio reconstruction, and the best class was selected for nonuniform refinement. After nonuniform refinement in the C2 symmetry, a map at 4.1 Å was obtained with 813,401 particles. Further classifications were performed with an ab initio reconstruction into five classes. A total of 594,368 good particles were selected from two classes, which were subjected to nonuniform refinement in the C2 symmetry, yielding a map with an overall resolution of 3.7 Å. Further ab initio reconstructions, heterogeneous refinement and nonuniform refinement using cryoSPARC were carried out to improve the resolution. Last, a map with an overall resolution of 3.4 Å with 550,472 particles was achieved. The overall resolution was estimated with the gold standard FSC at a 0.143 criterion with a high-resolution noise substitution method (44, 45). Local resolution variations were estimated using ResMap (46).
Model building and refinement
The 3.4 Å map for the hZnT1-DM was used for de novo model building by COOT (47). Bulky residues such as Phe, Tyr, Trp, and Arg were used to guide the sequence assignment, and the chemical properties of amino acids were considered to facilitate model building. Structure refinements were carried out by PHENIX in real space (48). Overfitting of the model was monitored by refining the model in one of the two independent maps from the gold standard refinement approach and testing the refined model against the other map. Statistics of the 3D reconstruction and model refinement can be found in table S1. In the final structure model of the hZnT1-DM, 325 of the total 507 amino acids were faithfully built and assigned. Ninety-seven amino acids (141 to 237) between the TM4 and TM5 and the last 85 amino acids in the C terminus were unmodeled due to vague EM densities.
Statistical analysis
Data were analyzed and prepared with GraphPad Prism 8.0 software. Data were considered significant if P < 0.05 with the unpaired two-tailed Student’s t test or the one-way analysis of variance (ANOVA) with Dunnett’s or Tukey’s multiple comparison test. The results are presented as means ± SD unless otherwise indicated.
Acknowledgments
We thank the Cryo-EM Center of the University of Science and Technology of China (USTC) for the EM facility support. We are grateful to all staff members for the technical support on cryo-EM data collection. We thank C. Cang and L. Liu for the technical and instrument supports on the transport assays.
Funding: This work was supported by grants from the Ministry of Science and Technology of the People’s Republic of China (National Key R&D Program, 2020YFA0509302 to L.S.), the Fundamental Research Funds for the Central Universities (WK9100000031 to L.S.), and the USTC Research Funds of the Double First-Class Initiative (YD9100002004 to L.S.). L.S. is supported by an Outstanding Young Scholar Award from the Qiu Shi Science and Technologies Foundation and a Young Scholar Award from the Cyrus Tang Foundation.
Author contributions: L.S. and X.L. conceived the project. X.W., X.L., and L.S. designed the experiments. C.S. and B.H. performed most of the molecular cloning, protein purification, transport assay, and structure determination work. C.S., B.H., and Y.G. performed the cryo-EM data collection. X.W., X.L., and L.S. wrote the manuscript. All authors contributed to data analysis, manuscript preparation, and revision.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The 3D cryo-EM density map of hZnT1-D47N/D255N double mutant has been deposited in the Electron Microscopy Data Bank under the accession number EMD-35949. Coordinates for the structure model have been deposited in the PDB under the accession code 8J2G.
Supplementary Materials
This PDF file includes:
Figs S1 to S11
Table S1
REFERENCES AND NOTES
- 1.Fukada T., Yamasaki S., Nishida K., Murakami M., Hirano T., Zinc homeostasis and signaling in health and diseases: Zinc signaling. J. Biol. Inorg. Chem. 16, 1123–1134 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bafaro E., Liu Y. T., Xu Y., Dempski R. E., The emerging role of zinc transporters in cellular homeostasis and cancer. Signal Transduct. Target. Ther. 2, 17029 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hara T., Takeda T., Takagishi T., Fukue K., Kambe T., Fukada T., Physiological roles of zinc transporters: Molecular and genetic importance in zinc homeostasis. J. Physiol. Sci. 67, 283–301 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cotrim C. A., Jarrott R. J., Martin J. L., Drew D., A structural overview of the zinc transporters in the cation diffusion facilitator family. Acta Crystallogr. D Struct. Biol. 75, 357–367 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang L. P., Tepaamorndech S., The SLC30 family of zinc transporters—A review of current understanding of their biological and pathophysiological roles. Mol. Aspects Med. 34, 548–560 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Coronado V. A., Naji M. S., Palmiter R. D., Cox D. W., Cloning and characterization of human zinc transporter 1 (ZNT1). Am. J. Hum. Genet. 73, 346–346 (2003). [Google Scholar]
- 7.Palmiter R. D., Findley S. D., Cloning and functional-characterization of a mammalian zinc transporter that confers resistance to zinc. EMBO J. 14, 639–649 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palmiter R. D., Huang L. P., Efflux and compartmentalization of zinc by members of the SLC30 family of solute carriers. Pflugers Arch. 447, 744–751 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Nishito Y., Kambe T., Zinc transporter 1 (ZNT1) expression on the cell surface is elaborately controlled by cellular zinc levels. J. Biol. Chem. 294, 15686–15697 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andrews G. K., Wang H. B., Dey S. K., Palmiter R. D., Mouse zinc transporter 1 gene provides an essential function during early embryonic development. Genesis 40, 74–81 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Lyubartseva G., Smith J. L., Markesbery W. R., Lovell M. A., Alterations of zinc transporter proteins ZnT-1, ZnT-4 and ZnT-6 in preclinical Alzheimer's disease brain. Brain Pathol. 20, 343–350 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lovell M. A., Smith J. L., Xiong S. L., Markesbery W. R., Alterations in zinc transporter protein-1 (ZnT-1) in the brain of subjects with mild cognitive impairment, early, and late-stage Alzheimer's disease. Neurotox. Res. 7, 265–271 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Zhang L. H., Wang X., Stoltenberg M., Danscher G., Huang L., Wang Z.-Y., Abundant expression of zinc transporters in the amyloid plaques of Alzheimer's disease brain. Brain Res. Bull. 77, 55–60 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Lehvy A. I., Horev G., Golan Y., Glaser F., Shammai Y., Assaraf Y. G., Alterations in ZnT1 expression and function lead to impaired intracellular zinc homeostasis in cancer. Cell Death Discov. 5, 144 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh C. K., Malas K. M., Tydrick C., Siddiqui I. A., Iczkowski K. A., Ahmad N., Analysis of zinc-exporters expression in prostate cancer. Sci. Rep. 6, 36772 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jing H. W., Kong C. Z., Liu T., Zeng Y., Zhang Z., Zinc transporter 1 (ZnT1) is overexpressed in bladder cancer and promotes the proliferation and invasion of bladder cancer BIU87 cells. Int. J. Clin. Exp. Med. 11, 5323–5331 (2018). [Google Scholar]
- 17.Maxel T., Svendsen P. F., Smidt K., Lauridsen J. K., Brock B., Pederser S. B., Rungby J., Larsen A., Expression patterns and correlations with metabolic markers of zinc transporters ZIP14 and ZNT1 in obesity and polycystic ovary syndrome. Front. Endocrinol. 8, 38 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grass G., Otto M., Fricke B., Haney C. J., Rensing C., Nies D. H., Munkelt D., FieF (YiiP) from Escherichia coli mediates decreased cellular accumulation of iron and relieves iron stress. Arch. Microbiol. 183, 9–18 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Ohana E., Hoch E., Keasar C., Kambe T., Yifrach O., Hershfinkel M., Sekler I., Identification of the Zn2+ binding site and mode of operation of a mammalian Zn2+ transporter. J. Biol. Chem. 284, 17677–17686 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoch E., Lin W., Chai J., Hershfinkel M., Fu D., Sekler I., Histidine pairing at the metal transport site of mammalian ZnT transporters controls Zn2+ over Cd2+ selectivity. Proc. Natl. Acad. Sci. U.S.A. 109, 7202–7207 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gupta S., Chai J., Cheng J., D'Mello R., Chance M. R., Fu D., Visualizing the kinetic power stroke that drives proton-coupled zinc(II) transport. Nature 512, 101–104 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daniels M. J., Jagielnicki M., Yeager M., Structure/function analysis of human ZnT8 (SLC30A8): A diabetes risk factor and zinc transporter. Curr. Res. Struct. Biol. 2, 144–155 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cotrim C. A., Jarrott R. J., Whitten A. E., Choudhury H. G., Drew D., Martin J. L., Heterologous expression and biochemical characterization of the human zinc transporter 1 (ZnT1) and its soluble C-terminal domain. Front. Chem. 9, 667803 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shusterman E., Beharier O., Shiri L., Zarivach R., Etzion Y., Campbell C. R., Lee I. H., Okabayashi K., Dinudom A., Cook D. I., Katz A., Moran A., ZnT-1 extrudes zinc from mammalian cells functioning as a Zn2+/H+ exchanger. Metallomics 6, 1656–1663 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Gottesman N., Asraf H., Bogdanovic M., Sekler I., Tzounopoulos T., Aizenman E., Hershfinkel M., ZnT1 is a neuronal Zn2+/Ca2+ exchanger. Cell Calcium 101, 102505 (2022). [DOI] [PubMed] [Google Scholar]
- 26.Chen T. W., Wardill T. J., Sun Y., Pulver S. R., Renninger S. L., Baohan A., Schreiter E. R., Kerr R. A., Orger M. B., Jayaraman V., Looger L. L., Svoboda K., Kim D. S., Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lopez-Redondo M. L., Coudray N., Zhang Z., Alexopoulos J., Stokes D. L., Structural basis for the alternating access mechanism of the cation diffusion facilitator YiiP. Proc. Natl. Acad. Sci. U.S.A. 115, 3042–3047 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bui H. B., Watanabe S., Nomura N., Liu K., Uemura T., Inoue M., Tsutsumi A., Fujita H., Kinoshita K., Kato Y., Iwata S., Kikkawa M., Inaba K., Cryo-EM structures of human zinc transporter ZnT7 reveal the mechanism of Zn2+ uptake into the Golgi apparatus. Nat. Commun. 14, 4770 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei Y. N., Fu D., Binding and transport of metal ions at the dimer interface of the Escherichia coli metal transporter YiiP. J. Biol. Chem. 281, 23492–23502 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Lu M., Fu D., Structure of the zinc transporter YiiP. Science 317, 1746–1748 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Lu M., Chai J., Fu D., Structural basis for autoregulation of the zinc transporter YiiP. Nat. Struct. Mol. Biol. 16, 1063–1067 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coudray N., Valvo S., Hu M., Lasala R., Kim C., Vink M., Zhou M., Provasi D., Filizola M., Tao J., Fang J., Penczek P. A., Ubarretxena-Belandia I., Stokes D. L., Inward-facing conformation of the zinc transporter YiiP revealed by cryoelectron microscopy. Proc. Natl. Acad. Sci. U.S.A. 110, 2140–2145 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lopez-Redondo M., Fan S., Koide A., Koide S., Beckstein O., Stokes D. L., Zinc binding alters the conformational dynamics and drives the transport cycle of the cation diffusion facilitator YiiP. J. Gen. Physiol. 153, e202112873 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xue J., Xie T., Zeng W., Jiang Y., Bai X. C., Cryo-EM structures of human ZnT8 in both outward- and inward-facing conformations. eLife 9, e58823 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang S. F., Fu C., Luo Y., Xie Q., Xu T., Sun Z., Su Z., Zhou X., Cryo-EM structure of a eukaryotic zinc transporter at a low pH suggests its Zn2+-releasing mechanism. J. Struct. Biol. 215, 107926 (2023). [DOI] [PubMed] [Google Scholar]
- 36.Lasry I., Seo Y. A., Ityel H., Shalva N., Pode-Shakked B., Glaser F., Berman B., Berezovsky I., Goncearenco A., Klar A., Levy J., Anikster Y., Kelleher S. L., Assaraf Y. G., A dominant negative heterozygous G87R mutation in the zinc transporter, ZnT-2 (SLC30A2), results in transient neonatal zinc deficiency. J. Biol. Chem. 287, 29348–29361 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kotov V., Bartels K., Veith K., Josts I., Subhramanyam U. K. T., Günther C., Labahn J., Marlovits T. C., Moraes I., Tidow H., Löw C., Garcia-Alai M. M., High-throughput stability screening for detergent-solubilized membrane proteins. Sci. Rep. 9, 10379 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katz A. K., Glusker J. P., Beebe S. A., Bock C. W., Calcium ion coordination: A comparison with that of beryllium, magnesium, and zinc. J. Am. Chem. Soc. 118, 5752–5763 (1996). [Google Scholar]
- 39.Torrance J. W., MacArthur M. W., Thornton J. M., Evolution of binding sites for zinc and calcium ions playing structural roles. Proteins 71, 813–830 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Zheng S. Q., Palovcak E., Armache J. P., Verba K. A., Cheng Y., Agard D. A., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zivanov J., Nakane T., Forsberg B. O., Kimanius D., Hagen W. J., Lindahl E., Scheres S. H., New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Rosenthal P. B., Henderson R., Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Chen S., McMullan G., Faruqi A. R., Murshudov G. N., Short J. M., Scheres S. H., Henderson R., High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy 135, 24–35 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kucukelbir A., Sigworth F. J., Tagare H. D., Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smart O. S., Neduvelil J. G., Wang X., Wallace B. A., Sansom M. S., HOLE: A program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360 (1996). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs S1 to S11
Table S1