Abstract
From a ferulic-acid-degrading Pseudomonas fluorescens strain (BF13), we have isolated a transposon mutant, which retained the ability to bioconvert ferulic acid into vanillic acid but lost the ability to further degrade the latter acid. The mutant, BF13-97, was very stable, and therefore it was suitable to be used as a biocatalyst for the preparative synthesis of vanillic acid from ferulic acid. By use of resting cells we determined the effect on the bioconversion rate of several parameters, such as the addition of nutritional factors, the concentration of the biomass, and the carbon source on which the biomass was grown. The optimal yield of vanillic acid was obtained with cells pregrown on M9 medium containing p-coumaric acid (0.1% [wt/vol]) as a sole carbon source and yeast extract (0.001% [wt/vol]) as a source of nutritional factors. Under these conditions, 1 mg (wet weight) of biomass produced 0.23 mg of vanillic acid per h. The genomic region of BF13-97 flanking the transposon's site of insertion was cloned and sequenced revealing two open reading frames of 1,062 (vanA) and 954 (vanB) bp, respectively. The van genes are organized in a cluster and encode the subunits of the vanillate-O-demethylase, which catalyzes the first step of the vanillate catabolism. Amino acid sequences deduced from vanA and vanB genes were shown to have high identity with known VanAs and VanBs from Pseudomonas and Acinetobacter spp. Highly conserved regions known to exist in class IA oxygenases were also found in the vanillate-O-demethylase components from P. fluorescens BF13. The terminal oxygenase VanA is characterized by a conserved Rieske-type [2Fe-2S]R ligand center. The reductase VanB contains a plant-type ferredoxin [2Fe-2S]Fd, flavin mononucleotide, and NAD-ribose binding domains which are located in its C-terminal and N-terminal halves, respectively. Transfer of wild-type vanAB genes to BF13-97 complemented this mutant, which recovered its ability to grow on either vanillic or ferulic acid.
Lignin-related aromatic acids, which contain phenylpropane (C6-C3) type structures (such as ferulic acid and related compounds), are abundant molecules that play important functions in plant cells, as antimicrobial compounds, signaling molecules, and phytoalexins (23). They commonly occur, free or in combined form, in fruits, vegetables, grains, beans, leaves, seeds, nuts, grasses, flowers, and other types of vegetation and can be easily extracted from some agriculture by-products (22). The catabolism of these compounds is an important aspect for the mineralization of plant wastes because they are released during the breakdown of lignin and cell wall materials by white-rot fungii. Moreover, there is a growing interest in the potential use of ferulic acid as feedstock for the biocatalytic conversion into other valuable molecules such as styrenes, polymers, epoxydes, alkylbenzenes, vanillin and vanillic acid derivatives, guaiacol, cathecol, and protocatechuic-acid-related cathecols (22).
We previously isolated a Pseudomonas fluorescens strain, named BF13, which utilized some phenylpropenoids (ferulic and m- and p-coumaric acids) as the sole carbon source (4). This strain, when degrading ferulic acid, transiently forms in the culture medium a certain amount of vanillic acid. The latter is a valuable product, used as a starting material in the chemical synthesis of oxygenated aromatic chemicals, such as vanillin, one of the most important flavor molecules (7, 22). We studied the possibility of enhancing the formation of vanillic acid during ferulic acid degradation by using suspensions of BF13 cells at high density. We demonstrated that the use of cells not adapted to ferulic acid improved the production of vanillic acid, which remained in the medium for a long period, facilitating its recovery (4). Moreover, we observed that this strain tolerated, better than others, high concentrations of ferulic acid and vanillic acid, which is a relevant property in view of a bioconversion process (unpublished data).
In this work our aim was to isolate a mutant of P. fluorescens BF13 which was unable to bioconvert vanillic acid into protocatechuic acid and use this mutant as a biocatalyst for the production of vanillic acid. The molecular characterization of this mutant allowed us to clone the genes from P. fluorescens BF13, which encode the terminal oxygenase (VanA) and the reductase (VanB) subunits of the vanillate-O-demethylase: the enzyme responsible for the demethylation of vanillate to protocatechuate. We will discuss the homology data between the deduced amino acid sequence of the vanillate-O-demethylase components from BF13 with those of already known VanAs and VanBs (5, 20, 32).
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Strains were grown at 37°C (Escherichia coli) or 30°C (P. fluorescens BF13) in Luria-Bertani (LB) medium (15) or mineral medium M9 (26) supplemented with carbon sources as indicated in the text. Ferulic, p-coumaric, protocatechuic, vanillic, and succinic acids were added to the medium at final concentrations of 0.1% (wt/vol). Glucose was added at final concentration of 0.5% (wt/vol). Solid media contained 1.5% (wt/vol) Difco (Detroit, Mich.) Bacto-Agar. BF13-97 was maintained at 4°C on LB agar containing kanamycin (50 mg/ml) or as frozen stocks at −20°C in LB containing 20% glycerol (Difco). Ampicillin, kanamycin, and tetracycline were used at final concentrations of 100, 50, and 25 μg/ml, respectively.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Reference or source |
|---|---|---|
| Strains | ||
| P. fluorescens BF13 | Wild type, ferulate positive, vanillate positive | 4 |
| P. fluorescens BF13-97 | BF13 derivative, ferulate negative, vanillate negative, Kmr (TnMod-OKm) | This study |
| E. coli JM109 | recA1 endA1 gyrA96 thi-1 hsdR17 (rk− mk+) supE44 relA1 λ− Δ(lac-proAB) [F′ traD36 proAB+ lacIqZΔM15] | 34 |
| E. coli S17-1 | recA harboring the tra genes of plasmid RP4 in the chromosome, proA thi-1 | 28 |
| Plasmids | ||
| pTZ18/19R | Apr | Pharmacia |
| pLAFR3 | Broad-host-range cosmid vector derived from IncP1 plasmid pRK290; RK2 replicon λcos+ lacZα; Mob+ Tra− Tcr | 3 |
| pRK2013 | Kmr; Tra+; ColE1 replicon | 24 |
| pGEM-T Easy | Apr | Promega |
| pGV1 | pGEM-T Easy containing PCR-amplified vanAB operon | This study |
| pCC1 | pLAFR3 with an EcoRI fragment from pGV1 containing vanAB operon | This study |
| pTnMod-OKm | Used as a source of TnMod-OKm; pMB1 replicon; Tn5 tnp; RP4 oriT; Kmr | 9 |
| pP1 | Kmr; 1.4-kb PstI fragment from P. fluorescens BF13-97 chromosome containing TnMod-OKm | This study |
| pB1 | Kmr; 1.3-kb BamHI fragment from P. fluorescens BF13-97 chromosome containing TnMod-OKm | This study |
| pE1 | Kmr; 6.4-kb EcoRI fragment from P. fluorescens BF13-97 chromosome containing TnMod-OKm | This study |
Isolation of insertion mutants.
The conjugatable suicide transposon TnMod-OKm was introduced into P. fluorescens BF13 by triparental mating using pRK2013 as described by Dennis and Zylstra (9). Kanamycin-resistant colonies were tested for the ability to utilize either ferulic acid or vanillic acid as a sole carbon source on M9 agar plates containing kanamycin (50 μg/ml). Growth was scored after 36 h at 30°C.
Cloning and DNA manipulations.
Genomic DNA from Pseudomonas strains was prepared by the procedure of Goldberg and Ohman (11). Standard protocols were used for DNA cloning and transformation and for plasmid DNA purification (26). Restriction endonuclease digestions and ligations with T4 ligase were done in accordance with the manufacturer's instructions (Life Technologies). The Concert extraction system (Life Technologies) was used for the recovery of DNA fragments from agarose gels. The vanA vanB operon was amplified by PCR from BF13 genomic DNA as a template with primers with the following sequences: 5′-GGCACCATTAACCATGATGTC-3′ for the upstream sequence and 5′-CTAGAGGTCCAGCACCAGCA-3′ for the downstream sequence. Amplification was performed with Taq polymerase (Qiagen) with an initial denaturation step of 2.0 min at 94°C, followed by 30 cycles of 1.0 min at 94°C (denaturation), 2.0 min at 60°C (annealing), and 2.0 min at 68°C (extension); this was followed by a 10-min final extension at 68°C. The fragment generated was purified by agarose gel electrophoresis and band extraction. It was then ligated into the pGEM-T Easy vector (Promega) to generate pGV1, removed as an EcoRI fragment, and directly ligated to pLAFR3 (3) to produce pCC1 (Table 1).
DNA sequencing and sequence data analysis.
DNA fragments carrying the vanA vanB genes were subcloned into pTZ18R or pTZ19R to prepare recombinant plasmids for sequencing. Nucleotide sequences were determined by MediGenomyx (Martinsried, Germany). M13 primers (universal and reverse) and additional specific primers were used to sequence both strands completely. Sequence analysis was done using the Sequence Analysis Software by the Genetics Computer Group (GCG Package version 8.1 [10]). Alignment of the sequences was done with the CLUSTAL W package (31), and searches for amino acid sequence similarities to entries in the databases were done by using FASTA (19) and BLAST (1). The potentially informative sites, i.e., those with two amino acid states shared by at least two sequences, were used for phylogenetic reconstruction. In our analysis, gaps were treated as missing data, and character state changes were weighted equally. Phylogenetic trees were reconstructed using the PAUP (30) “branch-and-bound” option. Bootstrap values were calculated from 1,000 replicates of branch-and-bound searches with the furthest addition sequence. In order to test the accuracy of the results, the phylogenetic reconstruction was also performed by the maximum-likelihood method using the PUZZLE (version 4.0 [29]) program (not shown). The topologies of the phylogenetic trees generated by the two methods were congruent and consistent.
Quantitative determination of vanillic acid by resting cells.
Preparation of cells and the quantitative determination of vanillic acid production were carried out as described previously (25). In brief, wet biomass obtained after centrifugation (6,000 × g at 4°C) was washed twice in saline phosphate buffer (4.2 mM Na2HPO4, 2.2 mM KH2PO4, 0.9 mM NaCl, 1.9 mM NH4Cl) before being resuspended in the same buffer. The reaction was performed in a 100-ml Erlenmeyer flask containing 20 ml of buffer supplemented with a sterile solution of ferulic acid and incubated on a rotary shaker at 30°C. During the span of 24 h 1-ml samples were withdrawn periodically and centrifuged at 15,000 × g (5). The supernatant was analyzed directly for substrate and intermediary metabolite by high-pressure liquid chromatography (HPLC).
HPLC analysis.
HPLC was performed on a Kontron (Milan, Italy) liquid chromatograph. Data acquisition and processing were controlled by the Kontron Data System 450 program (version 3.40) running on an IBM computer attached to a plotter (Kontron Plotter 800). The separations were carried out on a Supelcosil LC-18-S column (150 by 4.6 mm; Supelco, Inc., Bellefonte, Pa.) protected with a 1-cm guard cartridge (Phase Separations, Ltd.). Compounds were eluted with a gradient in acetonitrile-water (1% acetic acid) in which the concentration of acetonitrile was varied as follows over time: 0 min, 15%; 0 to 8 min, increase to 20%; 8 to 18 min, linear increase to 40%; 18 to 23 min, 40%; 23 to 26 min, increase to 95%; 26 to 30 min, 95%; and 30 to 33 min, decrease to 15%. The flow rate was 0.8 ml/min. Sample detection was achieved at dual wavelengths, 254 and 280 nm, and injection volumes were 10 μl. Compounds were identified and quantitated by comparison of the retention times and peak areas with those of authentic samples. They were eluted at the following retention times: ferulic acid, 10.3 min; and vanillic acid, 5.5 min. The identification of the vanillic acid was confirmed from the mass spectra of its trimethylsilylated derivative, which was carried out by the Organic Chemistry Unit of our Department. For vanillic acid, the mass spectrum has a molecular ion at position 312 (M+, 100%), a base peak at position 297 (M+, 98%), and major fragments at positions 282 (32%), 267 (56%), 253 (38%), 223 (65%), 193 (40%), 126 (21%), and 73 (81%).
Nucleotide sequence accession number.
The DNA sequences reported here have been submitted to GenBank under accession number AJ245887.
Chemicals.
All chemicals were of the highest purity commercially available and were purchased from Fluka (Buchs, Switzerland) and Carlo Erba (Milan, Italy). Luria Broth Base (Millers LB Broth Base) was from Gibco BRL (Life Technologies).
RESULTS
Isolation of a P. fluorescens BF13 mutant blocked in the demethylation of vanillic acid to protocatechuic acid.
P. fluorescens BF13 was found to be especially capable of transiently accumulating a significant amount of vanillic acid in the medium during ferulic acid degradation (4). By transposon mutagenesis, using the self-cloning minitransposon TnMod-Okm (9), we isolated one mutant of BF13 that was unable to further degrade vanillic acid. Three thousand TnMod mutants were screened for their ability to grow on either ferulic or vanillic acid, and three mutants were isolated. Two of these mutants (BF13-17 and BF13-125) could not grow on ferulic acid but retained their ability to grow on vanillic acid. Glucose-grown cells from these mutants were unable to dissimilate ferulic acid, which remained unchanged in the medium. We focused on the third mutant (BF13-97), which was unable to grow on either ferulic or vanillic acid but grew on protocatechuic acid. HPLC analysis of broth samples from a BF13-97 culture on minimal medium containing glucose, as the carbon source, and ferulic acid showed that the vanillic acid intermediate quantitatively accumulated in the medium without being further degraded. This indicated that the demethylation of vanillic acid to protocatechuic acid did not occur in the mutant.
The stability of the mutant was tested growing BF13-97 cells in LB medium without kanamycin for 20 generations, plating appropriate dilutions of the liquid culture on LB agar plates and, in addition, screening 103 independent clones for either antibiotic resistance or growth in M9 medium containing vanillic acid as a sole carbon source. All cells retained the kanamycin resistance and failed to grow on vanillic acid, suggesting that the mutation was very stable.
Cloning of the genes involved in the demethylation of vanillic acid to protocatechuic acid.
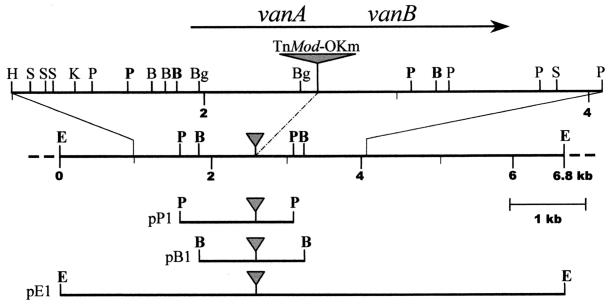
In order to clone the sequences flanking the TnMod's site of insertion, total genomic DNA from BF13-97 was digested with restriction enzymes that cut outside the TnMod, self-ligated under conditions favorable for intramolecular reactions, and used to transform E. coli JM109, selecting with the kanamycin resistance determinant encoded by TnMod-OKm. The genomic DNA was digested with either BamHI, EcoRI, or PstI, and three independent clones were obtained. As shown in Fig. 1, the BamHI (pB1) and PstI (pP1) clones were partially overlapping and were included in the larger EcoRI (pE1) clone.
FIG. 1.
Restriction map of the genomic region from P. fluorescens BF13-97 flanking the TnMod-OKm's site of insertion (indicated by a gray triangle). The region was partially sequenced, and a more detailed map of the sequenced portion is shown on the top. The arrow indicates the location and direction of transcription of the vanAB genes as deduced from the nucleotide sequence. The independent fragments carrying the TnMod, which were self-ligated and used to transform E. coli JM109, are shown at the bottom. The sites which delimited the ends of these fragments are indicated in boldface on the map. The names of the corresponding kanamycin-resistant plasmids are shown on the left. Restriction site designations: B, BamHI; Bg, BglI; E, EcoRI; H, HindIII; K, KpnI; P, PstI; S, SmaI.
On the basis of the restriction map of these clones, whose construction was facilitated by the presence of rare restriction sites near the inverted repeats of the minitransposon, we determined the physical map of the corresponding genomic region of BF13-97 (Fig. 1).
Sequencing of the region containing the vanillate-O-demethylase genes from P. fluorescens BF13.
The genomic regions flanking the TnMod's point of insertion were subcloned in the pTZ18R and pTZ19R vectors and sequenced. Sequence analysis of a 3,053-bp fragment overlapping this site revealed two open reading frames (ORFs) of 1,062 and 954 bp. The two ORFs, designated vanA and vanB, appear to be transcribed in the same direction (Fig. 1). They were separated by a 60-bp noncoding region, where we could not detect typical transcription termination or promoter sequences. The initiation codon of each ORF was preceded by a putative ribosome-binding site, suggesting that translation of these regions was possible. Moreover, a promoter-like sequence (TTGACA-N18-TTTAAT), which was preceded by an AT-rich region, was observed 41 bp upstream of the translation start codon of vanA. The organization of the vanA and vanB genes indicate that the two genes are clustered as an operon and are regulated by a common promoter located in front of the start codon of the vanA gene.
The G+C content of vanA (64.5%) and vanB (66.1%) was slightly higher than the general G+C content of the chromosome of P. fluorescens (59 to 62% [14]). This high value is due to the preferential usage of G and C at the wobble base position, as has been found for other Pseudomonas genes (33). The codon usage in vanA and vanB was very similar.
Sequence comparisons.
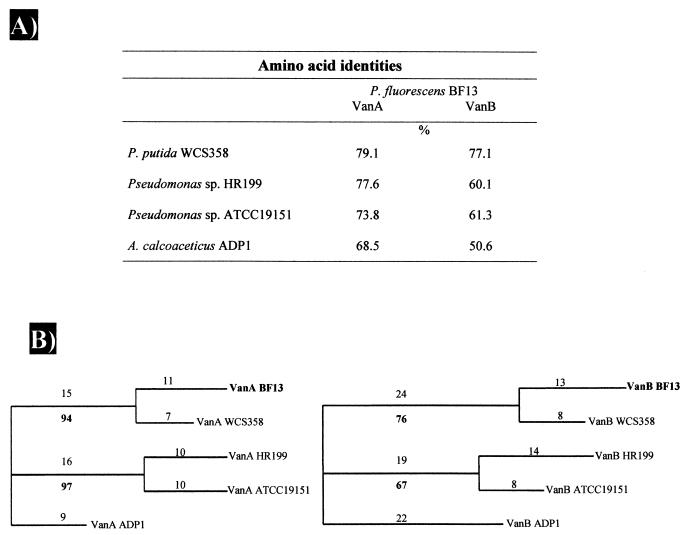
Sequence comparison analyses of the vanA and vanB gene products revealed significant similarities with the corresponding subunits of the vanillate-O-demethylases. Based on the homology data, vanA and vanB were identified as the cistrons for the terminal oxygenase subunit, VanA, and the reductase subunit, VanB, respectively. The VanA sequence from BF13 exhibited 68.5 to 79% identity with those of three VanAs from Pseudomonas sp. and one VanA from Acinetobacter sp. The deduced amino acid sequence of VanB showed significant identity, 50.6 to 77%, to those of the corresponding VanBs (Fig. 2A).
FIG. 2.
Sequence homologies (A) and phylogenetic trees (B) of terminal oxygenase VanA and reductase VanB components of vanillate-O-demethylase systems. The percent identity is based on pairwise alignment of BF13 gene products with corresponding protein of individual strains listed by using the LFASTA program. Trees were generated from potentially phylogenetically informative sequence data using the branch-and-bound option of the PAUP program. Sequences from A. calcoaceticus ADP1 strain were used as outgroups. The numbers above the branches indicate the number of changes within the lineage. The numbers below the branches are bootstrap values and represent the percentage of trees generated in 1,000 bootstrap replicates that show each grouping. No node was supported by a bootstrap value of <50%.
Complementation of BF13-97 mutant.
As previously shown, BF13-97 was deduced to be defective in the demethylation of vanillic acid into protocatechuic acid and contained a copy of TnMod-OKm inserted into the operon encoding the vanillate-O-demethylase subunits. It was, therefore, predictable that wild-type vanA vanB genes would be capable of complementing this mutant, restoring its ability to catabolize vanillic acid and utilize this compound as a sole carbon source. To test this hypothesis, the vanA vanB operon plus its putative promoter region were PCR amplified and cloned into the tetracycline-resistant vector pLAFR3 to give pCC1 (see Materials and Methods). E. coli S17.1 clones containing this recombinant cosmid were then mated with BF13-97, and kanamycin-tetracycline-resistant transconjugants were selected. The complemented mutant recovered its ability to grow on either vanillic acid or ferulic acid.
Bioconversion experiments.
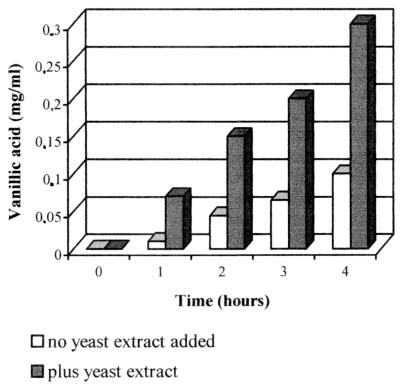
The potentialities of BF13-97 as biocatalyst for the production of vanillic acid were tested in bioconversion experiments carried out utilizing resting cells. Cells in M9 culture were harvested by centrifugation, washed, and suspended in saline-phosphate buffer and then incubated aerobically in the presence of 1 mg of ferulic acid per ml (as described in Materials and Methods). The decrease in the ferulic acid concentration and the increase of the vanillic acid catabolite in the medium were monitored by frequent sample analysis by HPLC. In preliminary experiments we observed that by adding nutritional factors to the M9 medium we could obtain a biomass which converted ferulic acid into vanillic acid threefold more rapidly (Fig. 3). This result was achieved by supplementing the medium with 0.001% (wt/vol) yeast extract. The addition of higher amounts (up to 0.01% [wt/vol]) of yeast extract failed to improve the bioconversion rate. Further experiments were, therefore, carried out by adding 0.001% (wt/vol) yeast extract to the medium on which the biomass was grown.
FIG. 3.
Vanillic acid production from BF13-97 cells pregrown with or without yeast extract as a source of nutritional factors. Biomass was grown in M9 medium containing glucose (0.5% [wt/vol]) as a carbon source and ferulic acid (0.1% [wt/vol]) as an inducer. Yeast extract was added at a concentration of 0.001%. Cells (1 mg [wet weight]/ml) were incubated in saline-phosphate buffer containing ferulic acid (1 mg/ml) as described in Materials and Methods. The data reported in the figure refer to the first 4 h of incubation.
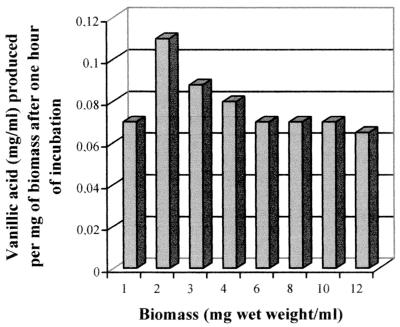
We next examined the effect on the bioconversion rate of the biomass concentrations. The experiments were performed using cells from ferulic-acid-induced cultures, which were grown on medium containing glucose as the carbon source. The optimal biomass concentration should be considered that which allows higher rates of bioconversion of ferulic acid into vanillic acid. In Fig. 4 the amounts (milligrams/milliliter) of vanillic acid per milligram (wet weight) of biomass produced after 1 h of incubation are shown. Better results were achieved with 2 mg (wet weight)/ml of biomass, at which concentration the maximal levels of vanillic acid (0.11 mg/ml) were obtained.
FIG. 4.
Effect of the biomass concentration on the formation of vanillic acid by resting cells of P. fluorescens BF13-97. Biomass was grown on M9 medium, containing glucose (0.5% [wt/vol]), ferulic acid (0.1% [wt/vol]), and yeast extract (0.001% [wt/vol]), and was incubated in the presence of 1 mg of ferulic acid per ml. The data, expressed as milligrams of vanillic acid per milliliter per milligram (wet weight) of biomass, refer to the first hour of incubation.
In experiments carried out with wild-type BF13 cells, we previously observed that the carbon source used for the cell growth affected the bioconversion rates (4). We analyzed whether similar effects were observed with the mutant cells. BF13-97 was grown on minimal medium containing glucose, succinate, or p-coumaric acid (a phenylpropenic compound structurally related to ferulic acid) as the carbon source, and the production of the enzymes involved in the first steps of the ferulic acid/p-coumaric degradative pathway was induced by using either ferulic or p-coumaric acid. These experiments showed that BF13-97 cells grown in the presence of p-coumaric acid were twice as active (0.23 mg of vanillate per ml per hour were produced by 1 mg [wet weight] of biomass) as ferulic acid-induced cells grown on glucose (0.11 mg/ml) and 60% more active than cells grown on succinate plus ferulic acid (0.14 mg/ml).
DISCUSSION
The use of P. fluorescens BF13 as a biocatalyst for the production of vanillic acid from ferulic acid was limited to batch processes because the ability of this strain to degrade vanillic acid could be impaired but not completely abolished by means of resting cells (4).
In order to develop a biocatalyst, which retained the ability of strain BF13 to bioconvert ferulic acid into vanillic acid rapidly but could also be used in continuous processes or in processes based on the use of immobilized cells, it was necessary to isolate a mutant unable to degrade vanillic acid.
Using transposon mutagenesis we have isolated a stable mutant, BF13-97, which had the desired properties. The mutation did not affect the velocity of ferulic acid consumption, which was similar to that of wild-type BF13 strain, but it blocked the demethylation of vanillic acid to protocatechuic acid. Furthermore, this mutant retained the ability to tolerate high concentrations of ferulate and vanillate in the medium. We used BF13-97 to determine the optimal conditions for the production of vanillic acid by means of resting cells. Better results were obtained in experiments with biomass from p-coumaric-grown cultures. Under this condition, 1 mg (wet weight) of biomass produced 0.23 mg of vanillic acid per h. It should be noted that, even with wild-type BF13, the optimal yield of vanillic acid was obtained by using p-coumaric-grown cells (4). The effect of p-coumaric acid is due to the ability of this compound to induce the production of enzymes involved in the first steps of ferulate catabolism, but not that of vanillate-O-demethylase, which catalyzes the first step of vanillate degradation (25).
From our experiments it came out that BF13-97 cells pregrown on nonaromatic sources of carbon (glucose or succinate), albeit in the presence of ferulic acid as an inducer, were less efficient than p-coumaric-grown cells in bioconverting ferulic acid into vanillic acid. In addition, we observed a slight inhibition effect on the bioconversion rate when glucose, rather than succinate, was used as a carbon source. This effect was also observed when cells were pregrown on medium with both glucose and p-coumaric acid, which was used as an inducer and as a carbon source as well (data not shown). All together these results suggested that nonaromatic carbon sources might have a possible regulatory role on the ferulic acid degradation by P. fluorescens BF13. This hypothesis should be further investigated. It is noteworthy that carbon source-dependent repression in many Pseudomonas species was previously reported to affect the catabolism of several aromatic carbon sources, such as ethylbenzene (8), styrene (17), and toluene (12), and the activity of protocatechuate 3,4-dioxygenase (35).
Another result that emphasized the influence of the physiological state of cells on the bioconversion rate was obtained in experiments with yeast extract. When added to the growth medium as a source of nutritional factors, yeast extract allowed us to obtain a biomass which more rapidly converted ferulic acid into vanillic acid (Fig. 3). It could be suggested that the ratio of carbon source to nutrient concentration played a key role in the metabolic flow. A stimulatory effect of yeast extract on the biodegradation of aromatics was reported for some chlorobenzoate-degrading bacteria (2). In that case it was speculated that yeast extract might contribute to a reduction of the environmental stress due to the presence of chlorobenzoates. This phenomenon should not be particularly relevant in our experimental conditions because a limited toxicity (10% growth inhibition) of ferulic acid on wild-type BF13 was detected only at a concentration fourfold higher than the one used in the present work (1 mg/ml [2]).
We also analyzed the relationship between the amount of biomass and the bioconversion rate, and in this way we could show that the yield of vanillic acid per milligram (wet weight) of biomass is not correlated to the increase of biomass concentration. The major effect observed when the biomass was varied from 2 to 12 mg (wet weight)/ml was a decrease in the velocity of vanillic acid formation (Fig. 4). We suppose that this effect could be partly due to the fact that at higher cell densities we have a higher O2 demand and probably, during the incubation, a lower O2 availability. In accordance with this hypothesis we observed that the bioconversion rate was drastically reduced when cells were incubated statically and in test tubes rather than in flasks (data not shown). Similar results were obtained with resting cells of Rhodotorula rubra by Huang et al. (13). These authors showed that the conversion of ferulic acid into vanillic acid by R. rubra IFO889 was affected by the incubation conditions and occurred efficiently only under aerobic conditions.
In the present work we also cloned and sequenced the genomic region from BF13-97 flanking the TnMod's site of insertion. Furthermore, by complementation experiments, we demonstrated that the insertional inactivation of the van operon was responsible for the specific block in the vanillate catabolism of BF13-97.
It is particularly interesting to find out that the entire gene organization of the vanA vanB region, as well as the homology of their products (summarized in Fig. 2A), is highly conserved among van operons. In contrast, the intergenic spacing between vanA and vanB is quite different: ranging from 0 in Pseudomonas sp. strain HR199 (20) to 57 bases in P. fluorescens BF13.
Several conserved regions that occur in amino acid sequences of known vanillate-O-demethylases and other class IA oxygenases were found in the sequences of VanA and VanB from P. fluorescens BF13. The consensus sequences for the Rieske-type [2Fe-2S] binding motif (21) and the mononuclear iron (II) coordination site (16) were present in the amino acids of the deduced VanA at positions 48-C-X-H-X16-C-X2-H-60 and 154-D-X2-H-X4-H-162, respectively. It is noteworthy that these motives are separated by 90 to 100 amino acids as in other oxygenases (6).
Moreover, in the C-terminal half of VanA three arginine residues (at positions 289, 331, and 332) are conserved that are postulated to be required for substrate binding in other class IA oxygenases (16).
Regions containing consensus amino acid sequences for the binding of flavin mononucleotide (48-RQYSL-52) and NAD-ribose (112-GGIGITPLSMAEQL-126) cofactors and chloroplast-type ferredoxin [2Fe-2S] (265-C-X4-C-X2-C-X29-C-303) were identified in the N-terminal and C-terminal halves of VanB, respectively. The same arrangement is conserved in the reductase components of class IA oxygenase systems, including vanillate-O-demethylase, phtalate 4,5-dioxygenase, and 3-chlorobenzoate 3,4-dioxygenase (16).
All of the VanA and VanB protein sequences available from the databases were compared with the corresponding gene products from P. fluorescens BF13. The phylogenetic trees that were obtained from these alignments were very similar (Fig. 2B). Both the VanA and the VanB families could be divided into three clusters. One cluster includes vanillate-O-demethylase components of P. fluorescens BF13 and P. putida WCS358, a second cluster comprises those of Pseudomonas sp. strain HR199 and Pseudomonas sp. strain ATCC 19151, while the Van proteins from A. calcoaceticus ADP1 fall into a third cluster.
The close evolutionary relationship of the gene cluster in BF13 and WCS358 was also confirmed by the noncoding DNA upstream of the start codon of vanA (Fig. 5). Compared with the nucleotide sequence of BF13 over the region 46 bases upstream, the corresponding sequence of WCS358 was 76% identical, while that of HR199 showed only 54% identity. In this region a BamHI cutting site is present (indicated in italics in Fig. 5), which is conserved in all three sequences. We noticed that this site is composed of a 16-base motif (5′-TCAATGGATCCATAAA-3′), which is directly repeated three times in the upstream sequence of BF13 vanA (Fig. 5).
FIG. 5.
Alignment of DNA sequence upstream of the start codon of BF13 vanA with corresponding upstream regions of P. putida WCS358 and Pseudomonas sp. strain HR199 vanA. Identical bases are in reverse type, and the conserved BamHI cutting site is indicated in italics. The 16-base motif containing the BamHI site, which is repeated three times in the upstream sequence of BF13 vanA, is shown below.
The significance of the presence of these repeats in P. fluorescens BF13 is unknown and needs to be investigated. It would also be interesting to know the regulatory mechanism of the vanillate-O-demethylase genes, which should provide more insight into the evolution of these genes among bacteria.
ACKNOWLEDGMENTS
This work was supported by a grant from Consiglio Nazionale delle Ricerche, applied project Biotechnology (PF 49).
We thank C. Leandro for technical assistance.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Armenante P M, Fava F, Kafkewitz D. Effect of yeast extract on growth kinetics during aerobic biodegradation of chlobenzoic acids. Biotechnol Bioeng. 1995;47:227–233. doi: 10.1002/bit.260470214. [DOI] [PubMed] [Google Scholar]
- 3.Bally M, Wretlind B, Lazdunski A. Protein secretion in Pseudomonas aeruginosa: molecular cloning and characterization of the xcp-1 gene. J Bacteriol. 1989;171:4342–4348. doi: 10.1128/jb.171.8.4342-4348.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barghini P, Montebove F, Ruzzi M, Schisser A. Optimal conditions for bioconversion of ferulic acid into vanillic acid by Pseudomonas fluorescens BF13 cells. Appl Microbiol Biothechnol. 1998;49:309–314. [Google Scholar]
- 5.Brunel F, Davison J. Cloning and sequencing of Pseudomonas genes encoding vanillate demethylase. J Bacteriol. 1988;170:4924–4930. doi: 10.1128/jb.170.10.4924-4930.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butler C S, Mason J R. Structure-function analysis of the bacterial aromatic ring-hydroxylating dioxygenases. Adv Microb Physiol. 1997;38:47–84. doi: 10.1016/s0065-2911(08)60155-1. [DOI] [PubMed] [Google Scholar]
- 7.Cheetham P. The use of biotransformation for the production of flavours and fragrances. Trends Biotechnol. 1993;11:478–488. [Google Scholar]
- 8.Corkery D M, O'Connor K E, Buckley C M, Dobson A D W. Ethylbenzene degradation by Pseudomonas fluorescens strain CA-4. FEMS Microbiol Lett. 1994;124:23–27. doi: 10.1111/j.1574-6968.1994.tb07256.x. [DOI] [PubMed] [Google Scholar]
- 9.Dennis J J, Zylstra G J. Plasposon: modular self-cloning minitransposon derivate for rapid genetic analysis of gram-negative bacterial genomes. Appl Environ Microbiol. 1998;64:2710–2715. doi: 10.1128/aem.64.7.2710-2715.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldberg J B, Ohman D E. Cloning and expression in Pseudomonas aeruginosa of a gene involved in production of alginate. J Bacteriol. 1984;158:1115–1121. doi: 10.1128/jb.158.3.1115-1121.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holten A, Marqués S, Möhler I, Jakubzik U, Timmis K N. Carbon source-dependent inhibition of xyl operon expression of the Pseudomonas putida TOL plasmid. J Bacteriol. 1994;176:1773–1776. doi: 10.1128/jb.176.6.1773-1776.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang Z, Dostal L, Rosazza J P N. Mechanisms of ferulic acid conversions to vanillic acid and guaiacol by Rhodotorula rubra. J Biol Chem. 1993;268:954–958. [PubMed] [Google Scholar]
- 14.Krieg N R, Holt J G. Bergey's manual of systematic bacteriology. Vol. 1. Baltimore, Md: The Williams and Wilkins Co.; 1984. [Google Scholar]
- 15.Miller J. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratories; 1972. [Google Scholar]
- 16.Nakatsu C, Straus N A, Campbell Wyndham R. The nucleotide sequence of the Tn5271 3-chlorobenzoate 3,4-dioxygenase genes (cbaAB) unites the class IA oxygenases in a single lineage. Microbiology. 1995;141:485–495. doi: 10.1099/13500872-141-2-485. [DOI] [PubMed] [Google Scholar]
- 17.O'Connor K, Buckley C M, Hartmans S, Dobson A D W. Possible regulatory role for nonaromatic carbon source in styrene degradation by Pseudomonas putida CA-3. Appl Environ Microbiol. 1995;61:544–548. doi: 10.1128/aem.61.2.544-548.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otaka E, Ooi T. Examination of protein sequence homologies. V. New perspectives on evolution between bacterial and chloroplast-type ferredoxins inferred from sequence evidence. J Mol Evol. 1989;29:246–254. doi: 10.1007/BF02100208. [DOI] [PubMed] [Google Scholar]
- 19.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Priefert, H., J. Rabenhorst, and A. Steinbuchel. 1997. Molecular characterization of genes of Pseudomonas sp. strain HR199 involved in bioconversion of vanillin to protocatechuate. 179:2595–2607. [DOI] [PMC free article] [PubMed]
- 21.Rieske J S, MacLennan D H, Coleman R. Isolation and properties of an iron-protein from the (reduced coenzyme Q)–cytochrome c reductase complex of the respiratory chain. Biochem Biophys Res Commun. 1964;15:338–344. [Google Scholar]
- 22.Rosazza J. Biocatalysis, microbiology and chemistry: the power of positive linking. ASM News. 1995;61:241–245. [Google Scholar]
- 23.Rosazza J, Huang Z, Dostal L, Rosseau B. Review: biocatalytic transformation of ferulic acid: an abundant aromatic natural product. J Ind Microbiol. 1995;15:457–471. doi: 10.1007/BF01570016. [DOI] [PubMed] [Google Scholar]
- 24.Ruvkun G B, Ausubel F M. A general method for site-directed mutagenesis in prokaryotes. Nature. 1981;289:85–88. doi: 10.1038/289085a0. [DOI] [PubMed] [Google Scholar]
- 25.Ruzzi M, Barghini P, Montebove F, Schiesser A P. Effect of the carbon source on the utilization of ferulic, m- and p-coumaric acids by a Pseudomonas fluorescens strain. Ann Microbiol. 1997;47:87–96. [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Shine J, Dalgarno L. Determinant of cistron specificity in bacterial ribosomes. Nature. 1975;254:34–38. doi: 10.1038/254034a0. [DOI] [PubMed] [Google Scholar]
- 28.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology. 1983;1:784–790. [Google Scholar]
- 29.Strimmer K, von Haeseler A. Quartet puzzling: a quartet maximum likelihood method for reconstructing tree topologies. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- 30.Swofford D L. PAUP: phylogenetic analysis using parsimony, version 3.1.1. Champaign, Ill: Illinois Natural History Survey; 1991. [Google Scholar]
- 31.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Venturi V, Zennaro F, Degrassi G, Okeke B C, Bruschi C V. Genetics of ferulic bioconversion to protocatechuit acid in plant-growth-promoting Pseudomonas putida WCS358. Microbiology. 1998;144:965–973. doi: 10.1099/00221287-144-4-965. [DOI] [PubMed] [Google Scholar]
- 33.West S E H, Iglewski B H. Codon usage in Pseudomonas aeruginosa. Nucleic Acids Res. 1988;16:9323–9335. doi: 10.1093/nar/16.19.9323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 35.Zylstra G J, Olsen R H, Ballou D P. Cloning, expression, and regulation of the Pseudomonas cepacia protocatechuate 3,4-dioxygenase genes. J Bacteriol. 1989;171:5907–5917. doi: 10.1128/jb.171.11.5907-5914.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]