Abstract
The baculovirus Autographa californica nuclear polyhedrosis virus encodes a DNA-dependent RNA polymerase that is required for transcription of viral late genes. This polymerase is composed of four equimolar subunits, LEF-8, LEF-4, LEF-9, and p47. The LEF-4 subunit has guanylyltransferase activity, suggesting that baculoviruses may encode a full complement of capping enzymes. Here we show that LEF-4 is a bifunctional enzyme that hydrolyzes the gamma phosphates of triphosphate-terminated RNA and also hydrolyzes ATP and GTP to the respective diphosphate forms. Alanine substitution of five residues previously shown to be essential for vaccinia virus RNA triphosphatase activity inactivated the triphosphatase component of LEF-4 but not the guanylyltransferase domain. Conversely, mutation of the invariant lysine in the guanylyltransferase domain abolished the guanylyltransferase activity without affecting triphosphatase function. We also investigated the effects of substituting phenylalanine for leucine at position 105, a mutation that results in a virus that is temperature sensitive for late gene expression. We found that this mutation had no significant effect on the ATPase or guanylyltransferase activity of LEF-4 but resulted in a modest decrease in RNA triphosphatase activity.
Expression of baculovirus genes in infected cells is temporally regulated at the transcriptional level. Immediately after infection, only a subset of viral genes is expressed. These immediate-early genes are transcribed by host RNA polymerase II (pol II) and do not require the action of viral transcription factors (6, 11, 16, 31). Expression of IE1 during this stage allows for the transcription of the delayed-early class of genes which are also transcribed by pol II (10, 19, 43). IE1 binds to viral enhancer elements and presumably helps to recruit host pol II to the early promoters (9). Many delayed-early genes encode proteins required for viral DNA replication and late gene expression, and expression of these proteins sets the stage for late gene expression. The late and very late genes are transcribed by an RNA polymerase that is resistant to α-amanitin and is chromatographically distinct from the three host RNA polymerases (6, 42).
Recently we purified the virus-specific RNA polymerase and showed that it was composed of four subunits, all of which are virus encoded (13). This finding raises interesting questions regarding the mechanisms of posttranslational processing of late viral mRNAs. Both late and very late transcripts are capped at the 5′ ends with a typical 7-methylguanosine (m7G) structure (32). It has been shown in other systems that these mRNA caps are added cotranscriptionally when nascent RNAs are less than 30 nucleotides in length (1). Only mRNAs are capped, and this restriction is mediated by specific interactions of capping enzymes with their cognate RNA polymerases (4, 24). This model suggests that either the baculovirus RNA polymerase must interact with host capping enzymes or the baculoviruses must encode their own capping enzymes.
The accompanying report (12) provides a partial answer to this question with its demonstration that the LEF-4 subunit of Autographa californica nuclear polyhedrosis virus (AcNPV) RNA polymerase has guanylyltransferase activity. Furthermore, analysis of the LEF-4 amino acid sequence revealed the presence of a conserved KxDG motif and homology to five additional motifs that are common to viral and cellular guanylyltransferases, which strongly supports our enzymatic data. Guanylyltransferase is one of the three enzymatic functions required for the formation of a m7G cap on the 5′ ends of mRNAs. The other two enzymes needed are RNA triphosphatase, which removes the gamma phosphate from 5′ triphosphate termini, and RNA methyltransferase, which adds a methyl group to the N-7 position of the guanosine cap. Vaccinia virus encodes a multifunctional capping enzyme that catalyzes all three steps (15, 22, 23), which suggested to us that LEF-4 may also have additional functions. In support of this hypothesis, we noticed sequence similarities between the baculovirus LEF-4 proteins and residues previously shown to be essential for triphosphatase function of the vaccinia virus capping enzyme. Here we report that LEF-4 has RNA triphosphatase activity. Furthermore, we show that mutation of these conserved residues abrogates the triphosphatase function of LEF-4.
MATERIALS AND METHODS
Purification of RNA polymerase and in vitro transcription assays.
RNA polymerase was purified from Spodoptera frugiperda cells infected with vBAC-RNApol as previously described (13). In vitro transcription assays were performed as described by Xu et al. (41).
Construction of LEF4-intein clones.
The AcNPV genomic clone pHindIII-C was amplified by using Pfu I DNA polymerase. The 5′ primer (CTGCAGCCATGGACTACGGCGATTTTGTG) inserted an NcoI site (underlined) at the ATG codon of LEF-4. The 3′ primer (TTACCCGGGCACGATTCGGTCGCG) was designed to substitute a glycine residue for the C-terminal aspartate and added a SmaI site (underlined) at the C-terminal glycine. The PCR products were cloned into pBluescript and screened by restriction enzyme digestion. A clone containing LEF-4 was digested with NcoI and SmaI and ligated to pCYB4 (New England Biolabs) previously digested with NcoI and SmaI. This plasmid directs the synthesis of a fusion protein containing a self-cleavable affinity tag under control of the tac promoter. Recombinant DNAs were screened by restriction enzyme digestion, and a clone with the correct size insert was sequenced. Alanine substitution mutations in LEF-4 were constructed by using a QuikChange site-directed mutagenesis kit (Stratagene) as recommended by the manufacturer. Potential mutant clones were verified by DNA sequence analysis.
Purification of LEF-4 from bacteria.
Escherichia coli XL1-Blue cells containing pLEF4-intein were grown to late log phase, and expression of LEF-4 was induced by the addition of 0.4 mM 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside. After induction, cells were incubated at 20°C overnight. Cells were harvested by centrifugation, resuspended in buffer B (50 mM Tris [pH 7.9], 0.1 mM EDTA, 500 mM NaCl, 0.1% Triton X-100), lysed by sonication, and clarified by low-speed centrifugation. The resulting supernatant was loaded onto a 2-ml chitin column. The column was washed with 20 volumes buffer B plus 30 mM dithiothreitol (DTT) and allowed to sit overnight in the presence of 30 mM DTT. Cleaved LEF-4 was eluted with 3 column volumes of buffer B plus 30 mM DTT. Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) followed by staining with Coomassie brilliant blue showed a single protein that migrated in the expected position for LEF-4. LEF-4 was further purified by anion-exchange chromatography. Peak fractions were dialyzed against buffer A (50 mM Tris [pH 7.9], 0.1 mM EDTA, 1 mM DTT) containing 50 mM NaCl, and the protein was applied to a Mono Q HR 5/5 column (Pharmacia) previously equilibrated in the same buffer. The column was washed with 10 ml of loading buffer and then eluted with a 20-ml linear NaCl gradient from 50 to 500 mM. LEF-4 eluted from Mono Q in a single sharp peak at 230 mM NaCl. LEF-4 was quantitated by a Coomassie blue G250 binding assay performed as recommended by the manufacturer (Pierce).
Assay of LEF-4–GMP complex formation.
Standard reaction mixtures (25 μl) contained 50 mM Tris-HCl (pH 7.9), 1 to 5 mM MnCl2, 5 mM DTT, 5 μM [α-32P]GTP, and purified RNA polymerase or LEF-4 as indicated. Samples were incubated for 15 min at 30°C and then stopped by the addition of 1% SDS. Samples were boiled and electrophoresed through an SDS–8% polyacrylamide gel. Gels were fixed, dried, and exposed to film.
RNA triphosphatase assays.
RNA 5′-triphosphatase activity was assayed by the liberation of [32P]Pi from [γ-32P]GTP-terminated RNA. Template RNA was synthesized by in vitro transcription of pBluescript KS DNA previously digested with BssHII. The linearized template was transcribed with T3 RNA polymerase, using standard conditions for in vitro transcription in the presence of [γ-32P]GTP added at a final specific activity of 5,000 cpm/pmol. This produced a 150-nucleotide-long RNA transcript specifically labeled at the 5′ end. γ-32P-labeled RNA was purified from transcription reactions by three rounds of precipitation with 2.5 M LiCl, followed by standard ethanol precipitation. Standard RNA triphosphatase assay mixtures (20 μl) contained 50 mM Tris-HCl (pH 7.9), 5 mM DTT, 0.3 mM MnCl2, 50 mM KCl, 1 to 5 μM RNA, and enzyme as indicated. Reactions were incubated for 15 min at 30°C and then terminated by the addition of 1 M formic acid. Samples were applied to polyethylenimine (PEI)-cellulose thin-layer chromatography (TLC) plate (J. T. Baker) and chromatographed with 0.5 M KH2PO4 (pH 3.4). Reactions were quantitated by scanning the TLC plate in a FUJIX BAS PhosphorImager.
ATPase assays.
Standard reaction mixtures (20 μl) contained 50 mM Tris-HCl (pH 7.9), 5 mM DTT, 50 mM KCl, 0.3 mM MnCl2, 1 μM [γ-32P]ATP or GTP, and enzyme as indicated. Reactions were incubated 15 min at 30°C, and aliquots were spotted on PEI-cellulose plates. Reactions were quantitated by scanning the plates with a PhosphorImager.
RESULTS
RNA triphosphatase activity of RNA polymerase and purified LEF-4.
We have previously shown that the LEF-4 subunit of AcNPV RNA polymerase has guanylyltransferase activity, suggesting that LEF-4 is an RNA capping enzyme (12). Furthermore, analysis of the LEF-4 amino acid sequence revealed homology to six motifs that are shared among viral and cellular guanylyltransferases in the C-terminal half of the protein. Sequence analysis of the N-terminal portion of LEF-4 revealed homologies with the triphosphatase domains of capping enzymes in the poxviruses and African swine fever virus (Fig. 1). Two pairs of glutamic acid residues and a single arginine have been shown to be essential for the RNA triphosphatase and ATPase activities of vaccinia virus capping enzyme (45). These glutamates and arginines are present in AcNPV LEF-4 with a similar order and spacing as found in the vaccinia virus and African swine fever virus capping enzymes. Furthermore, these residues are conserved among all three of the baculovirus LEF-4 proteins that have been submitted to protein and nucleic acid databases. These similarities suggested to us that the LEF-4 subunit of baculovirus RNA polymerase may have RNA triphosphatase and ATPase activities in addition to guanylyltransferase function.
FIG. 1.
Sequence of the LEF-4 RNA triphosphatase domain. The amino acid sequence of the AcNPV LEF-4 from residues 1 to 194 is aligned with the LEF-4 sequences of Bombyx mori nuclear polyhedrosis virus (BmNPV) and Orgyia pseudotsugata nuclear polyhedrosis virus (OpNPV) and the N-terminal regions of the capping enzymes of vaccinia virus (VAC) and African swine fever virus (ASF). Gaps in the sequence alignment are denoted by hyphens; residues that are identical in at least four of the five sequences are shown in bold; residues shown to be essential for RNA triphosphatase activity in vaccinia virus are indicated by asterisks above the sequence (44). Leucine 105 (underlined) is substituted with a phenylalanine in the LEF-4 ts virus.
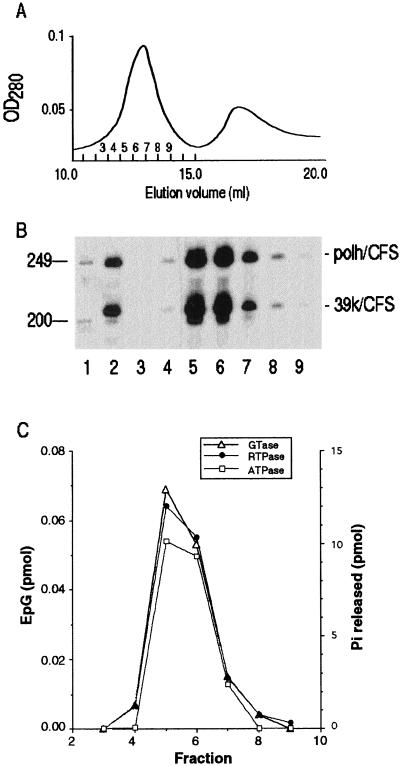
To test this hypothesis, purified AcNPV RNA polymerase was filtered through Superose 6 (Fig. 2A). As previously shown (12), a peak of guanylyltransferase activity coincided with the peak of absorbance at 280 nm and the peak of polymerase activity (Fig. 2B). To test for RNA triphosphatase activity, γ-32P-labeled RNA was prepared by in vitro transcription with T3 RNA polymerase in the presence of [γ-32P]GTP. This produces an RNA transcript that is singly labeled in the gamma phosphate position at the 5′ end of the RNA. RNA polymerase fractions were then incubated with 1 μM [γ-32P]RNA in the presence of 1 mM MnCl2. After incubation for 15 min at 30°C, free phosphate was separated from the RNA substrate by TLC on PEI-cellulose. As shown in Fig. 2B, RNA polymerase fractions catalyzed the hydrolysis of gamma phosphates from the RNA substrate. Furthermore, the extent of cleavage was proportional to input protein, and the peak of RNA triphosphatase activity exactly matched the peak of transcription activity. This finding indicates that RNA triphosphatase is an integral component of the baculovirus RNA polymerase.
FIG. 2.
RNA triphosphatase and ATPase activities of baculovirus RNA polymerase. (A) Gel filtration chromatography of RNA polymerase. RNA polymerase was filtered through Superose 6 as previously described (13). OD280, optical density at 280 nm. (B) In vitro transcription. Fractions corresponding to the peak of absorbance at 280 nm were assayed for in vitro transcription. The positions of molecular size markers are shown in kilodaltons on the left. The radiolabeled products corresponding to initiation from the polyhedrin (polh/CFS) and 39k (39k/CFS) promoters are shown on the right. (C) Guanyltransferase, RNA triphosphatase (RTPase), and ATPase assays. Guanylyltransferase mixtures (25 μl) contained 50 mM Tris HCl (pH 7.9), 5 mM DTT, 1 μM [α-32P]GTP, 1 mM MnCl2, and 2 μl of each fraction. Reactions were incubated at 30°C for 15 min, stopped by the addition of 1% SDS, and separated on an 8% polyacrylamide gel. RNA triphosphatase mixtures (10 μl) contained 50 mM Tris HCl (pH 7.9), 5 mM DTT, 1 mM MnCl2, 1 μM (5′ termini) γ-32P-labeled RNA, and 0.2 μl of each fraction. Reactions were incubated at 30°C for 15 min, stopped by the addition of 1 M formic acid, spotted on PEI-cellulose plates, and quantitated by scanning in a PhosphorImager. Picomoles of phosphate released is plotted on the right. ATPase mixtures (10 μl) contained 50 mM Tris HCl (pH 7.9), 5 mM DTT, 1 mM MnCl2, 1 μM [γ-32P]ATP, and 0.2 μl of each fraction. ▵, EpG; • and □, Pi released.
To examine the specificity of the triphosphatase activity, fractions were also incubated with [γ-32P]ATP, and the release of free phosphate was monitored by PEI chromatography. The peak of activity of gamma phosphate release exactly coincided with the three other enzymatic functions analyzed (Fig. 2B). Essentially identical results were obtained with [γ-32P]GTP as the substrate (data not shown). This finding suggests that the baculovirus enzyme, like the vaccinia virus capping enzyme, has nucleoside triphosphatase activity in addition to RNA triphosphatase activity.
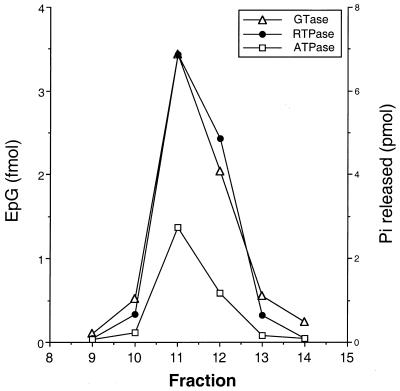
Triphosphatase activity of purified LEF-4.
To demonstrate that the RNA triphosphatase and ATPase activities were due to LEF-4 and not to other components of the RNA polymerase, purified LEF-4 was also assayed for both activities (Fig. 3). LEF-4 was overexpressed in baculovirus-infected cells and purified to near homogeneity by heparin-agarose and Mono Q chromatography as previously described (12). Purified LEF-4 was then filtered through a Superdex 200 gel exclusion column. Fractions corresponding to the peak of UV absorbance were assayed for guanylyltransferase activity, using the GMP label transfer assay. The same fractions were also active in the RNA triphosphatase and ATPase assays. The specific activities of the three enzymatic functions were nearly constant across the peak of protein. This result is consistent with our hypothesis that the triphosphatase and ATPase activities of RNA polymerase map to the LEF-4 subunit.
FIG. 3.
RNA triphosphatase and ATPase activities of LEF-4. LEF-4 was purified by recombinant baculovirus-infected cells and filtered through Superdex 200 as previously described (12). Fractions corresponding to the peak of absorbance at 280 nm were assayed for guanylyltransferase, RNA triphosphatase (RTPase), and ATPase activities as described in the legend to Fig. 2. Guanylyltransferase activity is plotted on the left; picomoles of phosphate released from gamma-labeled RNA or ATP is plotted on the right. ▵, EpG; • and □, Pi released.
Characterization of the RNA triphosphatase component of LEF-4.
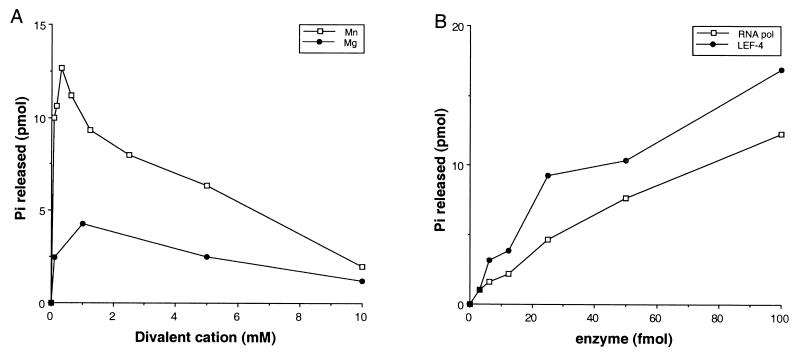
The RNA triphosphatase activity of LEF-4 was absolutely dependent on the addition of a divalent cation (Fig. 4A). Manganese was a more efficient cofactor than was magnesium at all concentrations tested. Activity peaked sharply at 0.3 mM and declined at higher concentrations. Magnesium was active over a broader range and was maximal at 1 mM, at which concentration the activity was 30% of that achieved with manganese. Essentially the same results were seen with RNA polymerase (data not shown).
FIG. 4.
Characterization of the RNA triphosphatase activity of LEF-4. (A) Cation requirements. Reaction mixtures contained 50 mM Tris HCl (pH 7.9), 5 mM DTT, 1 μM (5′ termini) γ-32P-labeled RNA, 0.1 pmol of LEF-4, and divalent cation as indicated in 20 μl. Reactions were incubated at 30°C for 15 min, spotted on PEI-cellulose plates, and quantitated by scanning in a PhosphorImager. Picomoles of phosphate released is plotted as a function of magnesium or manganese concentration. (B) Protein titration. Reaction mixtures (20 μl) contained 50 mM Tris HCl (pH 7.9), 5 mM DTT, 1 μM γ-32P-labeled RNA, 0.3 mM MnCl2, and AcNPV RNA polymerase or LEF-4 purified from baculovirus-infected insect cells as indicated. Picomoles of phosphate released is plotted as a function of input protein.
Under optimal conditions, the extent of gamma phosphate hydrolysis during a 15-min incubation was proportional to input protein with both sources of enzyme (Fig. 4B). In the linear range of enzyme concentration, the turnover number for RNA polymerase was 0.16 fmol/s/fmol of enzyme, while that of LEF-4 was 0.31 fmol/s/fmol.
Characterization of the nucleoside triphosphatase component of LEF-4.
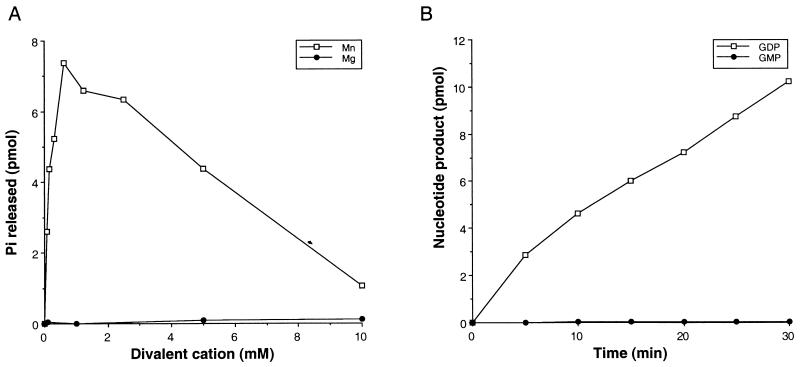
Hydrolysis of ATP was also absolutely dependent on the addition of a divalent cation (Fig. 5A). In the presence of manganese, ATPase activity increased linearly from 0.08 to 0.6 μM and then declined slowly at higher concentrations. Magnesium was a poor substitute for manganese, unlike the results observed for magnesium in the RNA triphosphatase assays. The ATPase activity at 10 mM magnesium was only 1% of the maximal activity gained with manganese.
FIG. 5.
Characterization of the nucleoside triphosphatase activity of LEF-4. (A) Cation requirements. Reaction mixtures contained 50 mM Tris HCl (pH 7.9), 5 mM DTT, 1 μM [α-32P]ATP, 0.2 pmol of LEF-4 purified from baculovirus-infected cells, and divalent cation as indicated in 20 μl. Reactions were incubated at 30°C for 15 min, spotted on PEI-cellulose plates, and quantitated by scanning in a PhosphorImager. Picomoles of phosphate released is plotted as a function of magnesium or manganese concentration. (B) Phosphate specificity. Reaction mixtures contained 50 mM Tris HCl (pH 7.9), 5 mM DTT, 1 μM [α-32P]GTP, 0.3 mM MnCl2 and 0.2 pmol of LEF-4 purified from baculovirus-infected cells in 20 μl. Picomoles of GDP or GMP produced is plotted as a function of input protein.
Specificity for the gamma phosphate was monitored by using [α-32P]GTP as the substrate (Fig. 5B). GDP accumulated linearly at a rate of 6.4 pmol/min/pmol of enzyme. GDP was slowly converted to GMP at a rate of 0.22 pmol/min/pmol of enzyme. Free phosphate was not detected during the 30-min time course.
Structure-function analysis of LEF-4.
To confirm that the RNA triphosphatase and ATPase activities observed were attributable to LEF-4, we expressed three mutated versions of LEF-4. Four glutamates and one arginine corresponding to residues previously shown to be essential for vaccinia virus triphosphatase activity (45) were changed to alanine. We also expressed a mutant protein in which the lysine residue of the guanylyltransferase KxDG motif was substituted with alanine. For ease of expression and purification, the mutant LEF-4 proteins were expressed in the E. coli expression system IMPACT I (New England Biolabs). This system produces fusion proteins with a self-cleavable intein tag linked to a chitin-binding domain. Cleavage of the intein tag produces a protein that is identical to the wild-type LEF-4 except for the substitution of a glycine residue for the C-terminal asparagine. This substitution does not affect the enzymatic activities of the protein, as all of our assays indicated that LEF-4 produced in E. coli was equivalent to native LEF-4 purified from baculovirus-infected cells (Fig. 6 and data not shown).
FIG. 6.
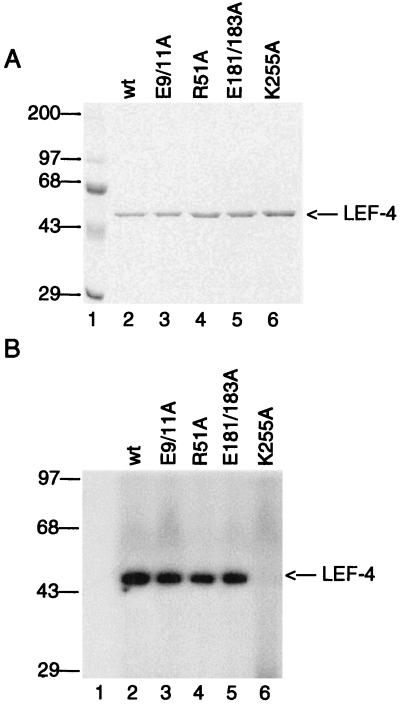
Purification and guanylyltransferase activities of LEF-4 mutants. (A) SDS-PAGE analysis of wild-type (wt) and mutant (E9/11A, R51A, K255A, and E181/183A) proteins purified from bacteria. Protein (1.35 μg per lane) purified by Mono Q chromatography was analyzed on SDS–8% polyacrylamide gels. The migration of prestained molecular markers is indicated on the left. (B) Guanylyltransferase assays. Guanylyltransferase activities were measured by formation of a covalent enzyme-GMP complex. Reaction mixtures (25 μl) contained 50 mM Tris (pH 7.9), 5 mM DTT, 5 mM MnCl2, 1 μM [32P]GTP, and 0.1 μg of the indicated protein. Reactions were stopped by the addition of 1% SDS and electrophoresed through an SDS–8% polyacrylamide gel. The positions of 14C-labeled protein markers are shown in kilodaltons on the left. The position of LEF-4 is indicated on the right.
After elution from the intein column, the LEF-4 protein samples were further purified by Mono Q chromatography. Figure 6 shows that the wild-type and mutant proteins expressed in E. coli were purified to single-band homogeneity. Guanylyltransferase assays were performed to ensure that the mutant proteins were correctly folded. The specific activities of the mutant and wild-type enzymes were determined within the linear range of enzymatic activity. With the wild-type enzyme, 0.5% of the available binding sites were radiolabeled, comparable to the level observed with native LEF-4 produced in baculovirus-infected cells (12). The activities of the triphosphatase mutants were equivalent to that of the wild-type protein, with the exception of that of E9/11A, which was 2.5-fold higher than the activity of the wild-type protein. As expected, the guanylyltransferase activity of the K255A mutant was undetectable. It has been established in other systems that nucleotidyltransferases form covalent intermediates with adenylate or guanylate linked to the lysine residue of the KxDG motif (5, 25). Thus, analysis of this mutation serves to demonstrate that the guanylyltransferase and triphosphatase activities are localized to separate domains.
RNA triphosphatase activities of the mutant and wild-type enzymes were assayed by release of 32Pi from 1 μM [γ-32P]RNA. The wild-type protein hydrolyzed 210 pmol of Pi per pmol of protein in a 15-min incubation. The specific activities of the mutant proteins relative to wild type are shown in Table 1. RNA triphosphatase activity was undetectable in the E181/183A mutant, reduced 100-fold in the E9/11A mutant, and reduced to 2.5% of control levels in the R51A mutant. ATPase activities were assayed by release of 32Pi from 1 μM [γ-32P]ATP. The wild-type enzyme liberated 108 pmol of Pi per mol of LEF-4 in 15 min at 30°C. The activity of all three mutants was reduced to less than 1% of that of the control. The K255A mutant retained both RNA triphosphatase and ATPase activities. These data are consistent with results obtained with corresponding mutations in the vaccinia virus capping enzyme (44).
TABLE 1.
Mutational effects of alanine substitutions on guanylyltransferase, RNA triphosphatase, and ATPase activities of LEF-4a
| Mutant | Sp act (% of wild-type activity)
|
||
|---|---|---|---|
| Guanylyltransferase | RTPase | ATPase | |
| E9/11A | 78 | 1.5 | 0.8 |
| R51A | 80 | 2.5 | <0.2 |
| E181/183A | 79 | <0.01 | <0.3 |
| K256A | <0.01 | 84.6 | 98.1 |
Wild-type and mutant proteins were purified by affinity and anion-exchange chromatography and then titrated for guanylyltransferase, RNA triphosphatase (RTPase), and ATPase activities as described in Materials and Methods. Specific activities were determined in the linear range of enzyme dependence. Two independent enzyme preparations were tested for each protein, and each preparation was tested in duplicate or triplicate. Values represent averages of all determinations.
Analysis of the L105F LEF-4 mutant.
The lef-4 gene was first described as the site of a mutation in a virus that was temperature sensitive (ts) for expression of late genes (28). The ts lesion in LEF-4 was mapped to a single-nucleotide transversion resulting in a substitution of phenylalanine for leucine at amino acid residue 105 (3), within the RNA triphosphatase domain (Fig. 1). This finding suggested the possibility that the defect in late gene expression may be due to incomplete hydrolysis of the gamma phosphates of late transcripts and therefore inefficient capping of late transcripts. One function of the m7G cap is to protect mRNAs from degradation. Therefore, a defect in capping could lead to rapid turnover of late mRNAs, producing the phenotype observed in this mutant.
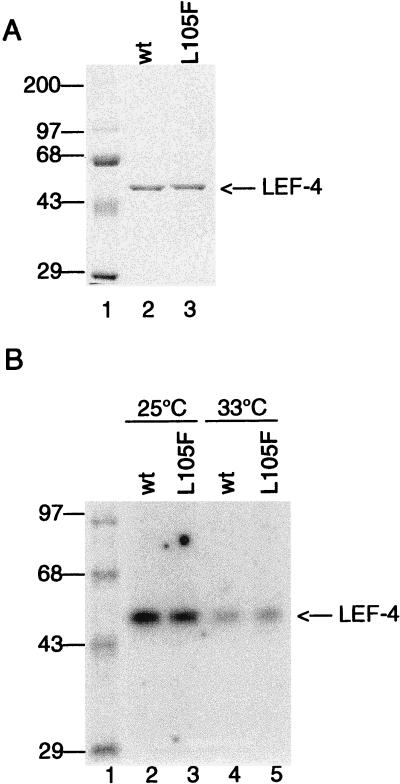
To characterize the L105F mutation, we expressed a mutant protein with this substitution as described above for the alanine substitutions. The protein was expressed at 20°C and tested for guanylyltransferase, RNA triphosphatase, and ATPase activities at 25 and 33°C. As shown in Fig. 7A, the purity of the L105F protein was commensurate with that of the wild-type protein. Guanylyltransferase activities were also equivalent to those of the wild-type protein at both permissive and nonpermissive temperatures. With both mutant and wild-type proteins, the activity was slightly higher at 25 than at 30°C, while the activity was reduced by half at the higher temperature (Fig. 7B). The wild-type protein bound GMP to an extent that occupied 0.7% of available sites at 25°C and only 0.2% at 33°C. The activity of the mutant protein was approximately 85% that of the wild type at both temperatures. These data indicate that substitution of the phenylalanine for leucine did not lead to temperature-dependent unfolding of LEF-4.
FIG. 7.
Characterization of the L105F protein. (A) SDS-PAGE analysis of wild-type (wt) and L105F mutant LEF-4 proteins purified from bacteria. Protein (1 μg per lane) purified by Mono Q chromatography was analyzed on SDS–8% polyacrylamide gels. The migration of prestained molecular markers is indicated in kilodaltons on the left. (B) Guanylyltransferase assays. Guanylyltransferase activities were measured by formation of a covalent enzyme-GMP complex. Reactions mixtures (25 μl) contained 50 mM Tris (pH 7.9), 5 mM DTT, 5 mM MnCl2, 1 μM [32P]GTP, and 0.1 μg of the indicated protein. Reactions were incubated for 15 min at 25°C (lanes 2 and 4) or 33°C (lanes 3 and 5), stopped by the addition of 1% SDS, and electrophoresed through an SDS–8% polyacrylamide gel. The positions of 14C-labeled protein markers are shown in kilodaltons on the left. The migration of LEF-4 is indicated on the right.
The RNA triphosphatase and ATPase activities of L105F were determined by protein titration at the permissive and nonpermissive temperatures. Both activities were indistinguishable from that of the wild-type protein at the permissive temperature (Table 2). At the nonpermissive temperature, the ATPase activity of the ts mutant was equivalent to wild-type activity but the RNA triphosphatase activity was reduced to 74% of the wild-type level. Mutational analyses of the vaccinia virus capping enzyme indicate that the RNA triphosphatase and ATPase activities map to the same active site (25, 44), and the data presented in Table 1 suggest that both activities of LEF-4 also colocalize. Therefore, it is possible that this residue maps to an RNA binding site rather than to the catalytic site. However, this effect is minimal and unlikely to account for the ts phenotype observed for the LEF-4 mutant virus.
TABLE 2.
Specific activities of L105F at the permissive and nonpermissive temperaturesa
| Enzyme assay | Sp act (% of wild-type activity)
|
|
|---|---|---|
| 25°C | 33°C | |
| Guanylyltransferase | 85 | 83 |
| RNA triphosphatase | 98 | 74 |
| ATPase | 103 | 93 |
Wild-type LEF-4 and L105F were purified by affinity and anion-exchange chromatography and then titrated for guanylyltransferase, RNA triphosphatase, and ATPase activities as described in Materials and Methods. Specific activities were determined in the linear range of enzyme dependence. Two independent enzyme preparations were tested for each protein, and each preparation was tested in duplicate or triplicate. Values represent averages of all determinations.
Thermal stability of L105F.
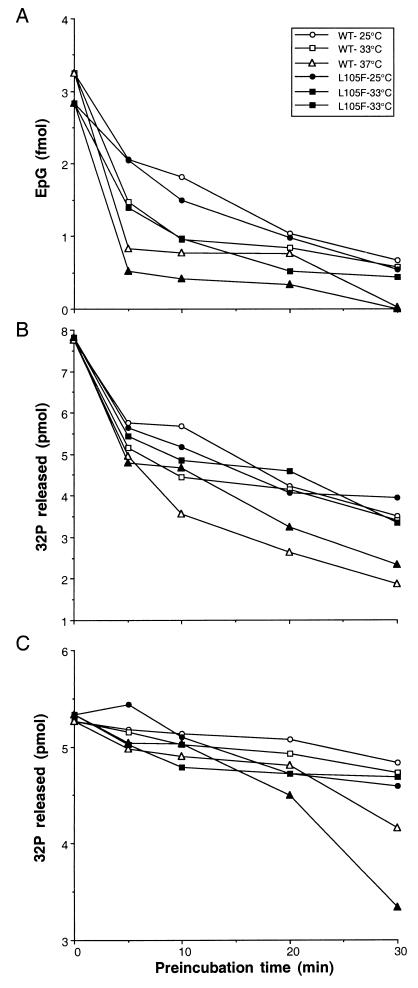
To further investigate the activities of L105F at the nonpermissive temperature, thermal inactivation profiles for the three enzymatic activities were produced (Fig. 8). Wild-type and L105F proteins were incubated at the permissive (25°C) or nonpermissive (33°C) temperature for defined lengths of time and then tested for guanylyltransferase and triphosphatase activities at the nonpermissive temperature. In addition, we analyzed the effect of preincubation at 37°C, which is higher than the nonpermissive temperature in vivo.
FIG. 8.
Thermal stability of LEF-4 L105F. (A) Guanylyltransferase assays. Guanylyltransferase activities were measured by formation of a covalent enzyme-GMP complex. Reactions mixtures (25 μl) contained 50 mM Tris (pH 7.9), 5 mM DTT, 5 mM MnCl2, 5 μM [32P]GTP, and 0.01 μM wild-type (WT) or L105F protein. After incubation in the absence of GTP for the indicated times at 25, 33, or 37°C, the samples shifted to 25°C, GTP was added, and the samples were incubated for 15 min. Reactions were quantitated by scanning of SDS-protein gels in a PhosphorImager. EpG, LEF-4. (B) ATPase assays. Reaction mixtures (20 μl) contained 50 mM Tris HCl (pH 7.9), 5 mM DTT, 1 μM [γ-32P]ATP, 0.3 mM MnCl2 and 0.01 μM of wild-type or L105F protein. (C) RNA triphosphatase assays. Reaction mixtures (20 μl) contained 50 mM Tris HCl (pH 7.9), 5 mM DTT, 1 μM γ-32P-labeled RNA, 0.3 mM MnCl2, and 5 nM of wild-type or L105F protein. Samples for RNA triphosphatase and ATPase activities were incubated for the indicated amounts of time at 25, 33, or 37°C in the absence of substrate; then the samples were shifted to 25°C, RNA or ATP was added, and the samples were incubated for 15 min. Reactions were quantitated by scanning TLC plates in a PhosphorImager. Each point represents the average of triplicate reactions.
The temperature inactivation profile for the guanylyltransferase activities indicated that both wild-type and mutant enzymes were sensitive to incubation at elevated temperatures (Fig. 8A). After 30 min at 37°C, the activity of the wild-type enzyme was reduced to 1% of control levels (no preincubation). Incubation for 30 min at 33 or at 25°C reduced activity to approximately 20% of control levels. At 25 and 33°C, the inactivation profiles of the L105F mutant mirrored those of the wild type, and the values averaged 87% of wild-type values. At 37°C, the difference between the two proteins was greater. The activity of L105F was averaged 50% of that of the wild type after incubation at 37°C.
ATPase profiles were similar for the wild type and L105F. The activities of both enzymes in ATPase assays decreased approximately 50% after 30 min of preincubation at 25°C, 56% at 33°C, and 74% at 37°C. RNA triphosphatase activities showed the highest level of thermal stability for both proteins. The inactivation profiles for L105F and the wild type were indistinguishable at 25 and 33°C, with both enzymes losing only 10% of their activities after 30 min of preincubation. L105F was somewhat more sensitive than the wild type to incubation at 37°C and retained only 60% of its activity after 30 min, while the wild-type enzyme retained 78% of its activity.
DISCUSSION
Eukaryotic viruses that replicate in the cytosol have evolved diverse strategies to ensure that their mRNAs are capped (2). RNA viruses either steal their caps from cellular mRNAs, encode their own capping enzymes, or circumvent the need for a cap by including elements in their RNAs that promote cap-independent translation. Poxviruses and African swine fever virus encode their own capping and methylating enzymes, while most other DNA viruses replicate in the nucleus and rely on host capping enzymes. Here we present data showing that baculoviruses have provided yet another solution to the capping problem. They replicate in the nucleus yet still encode at least two of the enzymes required for cap formation. Moreover, these enzymatic functions are incorporated into the baculovirus RNA polymerase.
In the accompanying report, we present data suggesting that LEF-4 is a guanylyltransferase (12). Analysis of the LEF-4 amino acid sequence revealed homology to five collinear motifs that are shared among viral and cellular guanylyltransferases (40), confirming the enzymatic data. These motifs are localized to the C-terminal half of the protein. Here we show that the N-terminal portion of the molecule shows local sequence conservation with the triphosphatase domain of vaccinia virus capping enzyme. Five residues in the vaccinia enzyme were previously shown to be required for ATPase and RNA triphosphatase activities (45). These amino acids are conserved among all of the baculovirus LEF-4 proteins in the same order and with similar spacing, suggesting that LEF-4 was also a triphosphatase.
We demonstrated that both RNA 5′-triphosphatase and ATPase activities copurified with the guanylyltransferase and transcription activities of AcNPV RNA polymerase. The specific activities of the four enzymes were essentially linear across the peak of protein, strongly suggesting that all four enzymes were contained within the same protein complex. Moreover, we showed that the RNA triphosphatase and ATPase activities coeluted, with the peak of LEF-4 protein when overexpressed as a single subunit. Further confirmation was provided by mutagenesis data showing that substitution of alanine at critical residues abrogated the triphosphatase and ATPase activities of LEF-4.
RNA 5′-triphosphatase is required for the hydrolysis of gamma phosphates from primary transcripts which have triphosphate termini. The resultant diphosphate-terminated RNAs serve as acceptors for the guanylyltransferase reaction in the production of GpppN caps on the ends of RNAs. Although we have not demonstrated that LEF-4 is capable of transferring GMP to diphosphate-terminated RNAs, it seems reasonable to propose that the function of LEF-4 is to cap RNAs transcribed by the baculovirus RNA polymerase.
Gross and Shuman (8) have recently demonstrated that AcNPV encodes another protein with RNA 5′-triphosphatase function. This protein, protein tyrosine phosphatase (PTPase), was originally identified on the basis of sequence homology with protein tyrosine/serine phosphatases (18, 36), and it was shown to be active in the hydrolysis of phosphotyrosine- and phosphoserine-containing substrates. However, the identification of a Caenorhabditis elegans RNA 5′-triphosphatase with a protein tyrosine/serine signature motif suggests that RNA may be the preferred substrate for some members of this family (37). Indeed, Gross and Shuman showed that PTPase hydrolyzed RNA substrates at a rate several orders of magnitude higher than that reported for protein substrates. Thus, it seems likely that this protein functions as an RNA 5′-triphosphatase and not a protein phosphatase. However, ptpase is not essential for viral replication and is not required for the transient expression of late and viral late genes, while lef-4 is required for both transient expression and productive viral infection (3, 20, 38). Furthermore, LEF-4 is part of the RNA polymerase complex and so is optimally positioned to serve as the primary protein involved in hydrolysis of primary termini of baculovirus late mRNAs.
The function of the ATPase activity of LEF-4 is unknown, as is the role of the ATP hydrolysis of the vaccinia virus capping enzyme. Vaccinia virus capping enzyme is also required for transcription termination, and early experiments suggested that the ATPase activity of vaccinia virus capping enzyme was required for transcription termination (14). However, recent alanine mutagenesis experiments showed that capping enzymes which are deficient in ATP hydrolysis are still active in promoting transcription termination (45). As yet, nothing is known regarding the mechanisms of transcription termination in the baculovirus system. Development of a termination assay will be essential in determining whether LEF-4 is involved in termination as well as capping of viral RNAs.
The ts mutation previously mapped to the lef-4 gene results in a substitution of phenylalanine for leucine at position 105 (3). The phenotype of this mutant is consistent with a defect in late gene expression, as it is characterized by decreased numbers of virions and decreased levels of late gene expression. L105 is located in the triphosphatase domain; therefore, we considered the possibility that the ts phenotype was due to a defect in mRNA capping. The mRNA cap plays essential roles in mRNA stability and initiation of translation. Thus, it seemed possible that production of uncapped RNAs could result in decreased translation of viral structural proteins due to rapid turnover of late RNAs. Bombyx mori and Orgyia pseudotsugata nuclear polyhedrosis viruses have leucine or valine at this position; thus, there is some sequence conservation among the baculoviruses at L105. To address this question, we produced a mutant LEF-4 protein with a phenylalanine at residue 105. The enzymatic activities of wild-type protein and L105F were compared by protein titration experiments at permissive and nonpermissive temperatures. We found that there were no significant differences in the guanylyltransferase or ATPase activities of the mutant protein relative to the wild-type protein at either the permissive or nonpermissive temperature. There was a slight, but reproducible, decrease (74%) in the RNA triphosphatase activity of the mutant relative to the wild-type protein at the nonpermissive temperature. The specific effect on RNA triphosphatase activity suggests that L105 may be part of the RNA binding site. RNA triphosphatase and ATPase activities map to the same active site; therefore, L105 is probably not part of the catalytic site, as the mutation affected only one of the activities.
More extensive thermal inactivation profiles showed that the thermal stability of L105F was equivalent to that of the wild type at permissive and nonpermissive temperatures. However, the guanylyltransferase and RNA triphosphatase activities of L105F were more sensitive than wild-type activities to prolonged incubation at 37°C, although the ATPase activities were equivalent at this temperature. Thus, differences were only seen at temperatures which are not physiologically relevant.
Further experimentation will be required to determine whether the phenotype of the L105F mutant is due to a defect in mRNA capping. A decrease in RNA triphosphatase activity is unlikely to result in a ts phenotype. It is well established that in the vaccinia virus system the rate of gamma phosphate hydrolysis greatly exceeds the rate of cap formation (39). RNAs with triphosphate ends are efficiently capped by the vaccinia virus capping enzyme (45), but it is unlikely that the resulting tetraphosphate caps would be efficiently translated. Therefore, significant differences in the activities of the two enzymes are required to ensure that guanylyltransferase never encounters a triphosphate end, thus preventing the formation of tetraphosphate cap structures. We cannot directly measure the relative rates of RNA triphosphatase and capping in LEF-4, because the enzyme is not active in the transfer of GMP to RNA. However, it seems reasonable to assume that the baculovirus enzyme is also much more active in hydrolysis than in guanylyltransferase. And if this is true, a 74% decrease in triphosphatase probably would not significantly alter the formation of triphosphate caps. Furthermore, even if 74% of the late RNAs were not capped or were incorrectly capped, the result would probably not be a lethal phenotype but instead merely a slight reduction in the synthesis of late viral proteins.
Comparison of the sequences surrounding L105 with sequences of the poxvirus capping enzymes or African swine fever virus capping enzyme did not reveal any sequence similarity in this region. This finding is consistent with our conclusions that L105 is not part of the active site of the enzyme and raises the possibility that substitution of phenylalanine, which has a large aromatic ring, destabilizes the interactions between the RNA polymerase subunits. Development of a reconstitution system for the baculovirus RNA polymerase should help to answer this question.
ACKNOWLEDGMENT
This research was supported by grant MCB 95-06233 from the National Science Foundation.
REFERENCES
- 1.Babich A, Nevins J R, Darnell J E. Early capping of transcripts from the adenovirus major late transcription unit. Nature. 1980;287:246–248. doi: 10.1038/287246a0. [DOI] [PubMed] [Google Scholar]
- 2.Bisaillon M, Lemay G. Viral and cellular enzymes involved in synthesis of mRNA cap structure. Virology. 1997;236:1–7. doi: 10.1006/viro.1997.8698. [DOI] [PubMed] [Google Scholar]
- 3.Carstens E B, Chan H, Yu H, Williams G V, Casselman R. Genetic analysis of temperature-sensitive mutations in baculovirus late expression factors. Virology. 1994;204:323–337. doi: 10.1006/viro.1994.1537. [DOI] [PubMed] [Google Scholar]
- 4.Cho E-J, Takagi T, Moore C R, Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cong P, Shuman S. Covalent catalysis in nucleotidyl transfer: a KTDG motif essential for enzyme-GMP complex formation by mRNA capping enzyme is conserved at the active sites of RNA and DNA ligases. J Biol Chem. 1993;268:7256–7260. [PubMed] [Google Scholar]
- 6.Fuchs L Y, Woods M S, Weaver R F. Viral transcription during Autographa californica nuclear polyhedrosis virus infection: a novel RNA polymerase induced in infected Spodoptera frugiperda cells. J Virol. 1983;48:641–646. doi: 10.1128/jvi.48.3.641-646.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glocker B, Hoopes R R, Rohrmann G F. In vitro transcription from baculovirus late gene promoter: accurate mRNA initiation by nuclear extracts prepared from infected Spodoptera frugiperda cells. J Virol. 1993;67:3771–3776. doi: 10.1128/jvi.67.7.3771-3776.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gross C H, Shuman S. Characterization of a baculovirus-encoded RNA 5′-triphosphatase. J Virol. 1998;72:7057–7063. doi: 10.1128/jvi.72.9.7057-7063.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8a.Gross C H, Shuman S. RNA 5′-triphosphatase, nucleoside triphosphatase, and guanylyltransferase activities of the baculovirus LEF-4 protein. J Virol. 1998;72:10020–10028. doi: 10.1128/jvi.72.12.10020-10028.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guarino L A, Dong W. Transient expression of an enhancer-binding protein in insect cells transfected with the Autographa californica nuclear polyhedrosis virus IE1 gene. J Virol. 1991;65:3676–3680. doi: 10.1128/jvi.65.7.3676-3680.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guarino L A, Smith M. Regulation of delayed-early gene transcription of dual TATA boxes. J Virol. 1992;66:3722–3739. doi: 10.1128/jvi.66.6.3733-3739.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guarino L A, Summers M D. Functional mapping of a trans-activating gene required for expression of a baculovirus delayed-early gene. J Virol. 1986;57:563–571. doi: 10.1128/jvi.57.2.563-571.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guarino L A, Jin J, Dong W. Guanylyltransferase activity of the LEF-4 subunit of baculovirus RNA polymerase. J Virol. 1998;72:10003–10010. doi: 10.1128/jvi.72.12.10003-10010.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guarino L A, Xu B, Jin J, Dong W. A virus-encoded RNA polymerase purified from baculovirus-infected cells. J Virol. 1998;72:7985–7991. doi: 10.1128/jvi.72.10.7985-7991.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hagler J, Luo Y, Shuman S. Mechanism of factor-dependent transcription termination by vaccinia RNA polymerase: kinetic coupling and requirement for ATP hydrolysis. J Biol Chem. 1994;269:10050–10060. [PubMed] [Google Scholar]
- 15.Higman M A, Christen L A, Niles E G. The mRNA (guanine-7-) methyltransferase domain of the vaccinia virus mRNA capping enzyme: expression in E. coli and structural and kinetic comparison to the intact capping enzyme. J Biol Chem. 1994;267:14974–14981. [PubMed] [Google Scholar]
- 16.Hoopes R R, Jr, Rohrmann G F. In vitro transcription of baculovirus immediate early genes: accurate initiation by nuclear extracts from both insect and human cells. Proc Natl Acad Sci USA. 1991;88:4513–4517. doi: 10.1073/pnas.88.10.4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huh N E, Weaver R F. Identifying the RNA polymerases that synthesize specific transcripts of the Autographa californica nuclear polyhedrosis virus. J Gen Virol. 1990;71:195–201. doi: 10.1099/0022-1317-71-1-195. [DOI] [PubMed] [Google Scholar]
- 18.Kim D, Weaver R F. Transcription mapping and functional analysis of the protein tyrosine/serine phosphatase (PTPase) gene of the Autographa californica nuclear polyhedrosis virus. Virology. 1993;195:587–596. doi: 10.1006/viro.1993.1410. [DOI] [PubMed] [Google Scholar]
- 19.Kogan P H, Blissard G W. A baculovirus gp64 early promoter is activated by host transcription factor binding to CACGTG and GATA elements. J Virol. 1994;68:813–822. doi: 10.1128/jvi.68.2.813-822.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Miller L K. Properties of a baculovirus mutant defective in the protein phosphatase gene. J Virol. 1995;69:4533–4537. doi: 10.1128/jvi.69.7.4533-4537.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mans R M, Knebel-Morsdorf D. In vitro transcription of pe38/polyhedrin hybrid promoters reveals sequences essential for recognition by the baculovirus-induced RNA polymerase and for the strength of very late viral promoters. J Virol. 1998;72:2991–2998. doi: 10.1128/jvi.72.4.2991-2998.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mao X, Shuman S. Intrinsic RNA (guanine-7) methyltransferase activity of the vaccinia virus capping enzyme D1 subunit is stimulated by the D12 subunit: identification of amino acid residues in the D1 protein required for subunit association and methyl group transfer. J Biol Chem. 1994;269:24472–24479. [PubMed] [Google Scholar]
- 23.Martin S A, Paoletti E, Moss B. Purification of mRNA guanylyltransferase and mRNA (guanine-7-) methyltransferase from vaccinia virions. J Biol Chem. 1975;250:9322–9329. [PubMed] [Google Scholar]
- 24.McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Shuman S, Bentley D L Amgen EST Program. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Myette J, Niles E G. Characterization of the vaccinia virus RNA 5′ triphosphatase and nucleoside phosphohydrolase activities: demonstration that both activities are carried out at the same active site. J Biol Chem. 1996;271:11936–11944. doi: 10.1074/jbc.271.20.11945. [DOI] [PubMed] [Google Scholar]
- 26.Miller L K, editor. The baculoviruses. New York, N.Y: Plenum Press; 1997. [Google Scholar]
- 27.Niles E G, Christen L. Identification of the vaccinia virus mRNA guanylyltransferase active site lysine. J Biol Chem. 1993;268:24968–24989. [PubMed] [Google Scholar]
- 28.Partington S, Yu H, Lu A, Carstens E B. Isolation of temperature-sensitive mutants of Autographa californica nuclear polyhedrosis virus: phenotypic characterization of mutants defective in very late gene expression. Virology. 1990;175:91–102. doi: 10.1016/0042-6822(90)90189-x. [DOI] [PubMed] [Google Scholar]
- 29.Passarelli A L, Miller L K. Identification of genes encoding late expression factors located between 56.0 and 65.4 map units of the Autographa californica nuclear polyhedrosis virus genome. Virology. 1993;197:704–714. doi: 10.1006/viro.1993.1646. [DOI] [PubMed] [Google Scholar]
- 30.Passarelli A L, Todd J W, Miller L K. A baculovirus gene involved in late gene expression predicts a large polypeptide with a conserved motif of RNA polymerases. J Virol. 1994;68:4673–4678. doi: 10.1128/jvi.68.7.4673-4678.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pullen S S, Friesen P D. Early transcription of the ie1 transregulator gene of Autographa californica nuclear polyhedrosis virus is regulated by DNA sequences within its 5′ noncoding leader region. J Virol. 1995;69:156–165. doi: 10.1128/jvi.69.1.156-165.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qin J-C, Weaver R F. Capping of viral RNA in cultured Spodoptera frugiperda cells infected with Autographa californica nuclear polyhedrosis virus. J Virol. 1982;43:234–240. doi: 10.1128/jvi.43.1.234-240.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rankin C B, Ooi B G, Miller L K. Eight base pairs encompassing the transcriptional start point are the major determinant for baculovirus polyhedrin expression. Gene. 1988;70:39–49. doi: 10.1016/0378-1119(88)90102-3. [DOI] [PubMed] [Google Scholar]
- 34.Rohrmann G F. Polyhedrin structure. J Gen Virol. 1986;67:1499–1508. doi: 10.1099/0022-1317-67-8-1499. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Sheng Z, Charbonneau H. The baculovirus Autographa californica encodes a protein tyrosine phosphatase. J Biol Chem. 1993;268:4728–4733. [PubMed] [Google Scholar]
- 37.Takagi T, Moore C R, Diehn F, Buratowski S. An RNA 5′ triphosphatase related to the protein tyrosine phosphatases. Cell. 1997;89:867–873. doi: 10.1016/s0092-8674(00)80272-x. [DOI] [PubMed] [Google Scholar]
- 38.Todd J W, Passarelli A L, Miller L K. Eighteen baculovirus genes, including lef-11, p35, 39k, and p47, support late gene expression. J Virol. 1995;69:968–974. doi: 10.1128/jvi.69.2.968-974.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Venkatesan S, Gershowitz A, Moss B. Modification of the 5′ end of mRNA: association of RNA triphosphatase with the RNA guanyltransferase-RNA (guanine-7-) methyltransferase complex from vaccinia virus. J Biol Chem. 1980;255:903–908. [PubMed] [Google Scholar]
- 40.Wang S P, Deng L, Ho C K, Shuman S. Phylogeny of mRNA capping enzymes. Proc Natl Acad Sci USA. 1997;94:9573–9578. doi: 10.1073/pnas.94.18.9573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu B, Yoo S, Guarino L A. Differential transcription of baculovirus late and very late promoters: fractionation of nuclear extracts by phosphocellulose chromatography. J Virol. 1995;69:2912–2917. doi: 10.1128/jvi.69.5.2912-2917.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang C L, Stetler D A, Weaver R F. Structural comparison of the Autographa californica nuclear polyhedrosis virus-induced RNA polymerases from the host, Spodoptera frugiperda. Virus Res. 1991;20:251–264. doi: 10.1016/0168-1702(91)90079-b. [DOI] [PubMed] [Google Scholar]
- 43.Yoo S, Guarino L A. The Autographa californica nuclear polyhedrosis virus ie2 gene encodes a transcriptional regulator. Virology. 1994;202:746–753. doi: 10.1006/viro.1994.1396. [DOI] [PubMed] [Google Scholar]
- 44.Yu L, Shuman S. Mutational analysis of the triphosphatase domain of vaccinia virus mRNA capping enzyme. J Virol. 1996;70:6162–6168. doi: 10.1128/jvi.70.9.6162-6168.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu L, Martins A, Deng L, Shuman S. Structure-function analysis of the triphosphatase component of vaccinia virus mRNA capping enzyme. J Virol. 1997;71:9837–9843. doi: 10.1128/jvi.71.12.9837-9843.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]