Abstract
Autographa californica nuclear polyhedrosis virus late and very late mRNAs are transcribed by an RNA polymerase consisting of four virus-encoded polypeptides: LEF-8, LEF-9, LEF-4, and p47. The 464-amino-acid LEF-4 subunit contains the signature motifs of GTP:RNA guanylyltransferases (capping enzymes). Here, we show that the purified recombinant LEF-4 protein catalyzes two reactions involved in RNA cap formation. LEF-4 is an RNA 5′-triphosphatase that hydrolyzes the γ phosphate of triphosphate-terminated RNA and a guanylyltransferase that reacts with GTP to form a covalent protein-guanylate adduct. The RNA triphosphatase activity depends absolutely on a divalent cation; the cofactor requirement is satisfied by either magnesium or manganese. LEF-4 also hydrolyzes ATP to ADP and Pi (Km = 43 μM ATP; Vmax = 30 s−1) and GTP to GDP and Pi. The LEF-4 nucleoside triphosphatase (NTPase) is activated by manganese or cobalt but not by magnesium. The RNA triphosphatase and NTPase activities of baculovirus LEF-4 resemble those of the vaccinia virus and Saccharomyces cerevisiae mRNA capping enzymes. We suggest that these proteins comprise a novel family of metal-dependent triphosphatases.
The m7GpppN cap structure of eukaryotic mRNA is formed cotranscriptionally by three enzymatic reactions: (i) the 5′ triphosphate end of the nascent RNA is hydrolyzed to a diphosphate by RNA triphosphatase; (ii) the diphosphate end is capped with GMP by GTP:RNA guanylyltransferase; and (iii) the GpppN cap is methylated by S-adenosylmethionine (AdoMet):RNA (guanine-N7) methyltransferase (3, 35). The mRNAs of most nuclear DNA viruses (e.g., papovaviruses, adenoviruses, and herpesviruses) are transcribed by RNA polymerase II, and their 5′ ends are modified by the host cell’s capping and methylating enzymes. However, vaccinia virus, which replicates entirely in the cytoplasm, encodes and encapsidates its own DNA-dependent RNA polymerase and mRNA capping apparatus. African swine fever virus, which has a cytoplasmic replication phase, also encodes and encapsidates an RNA polymerase and a capping enzyme. Chlorella virus PBCV-1 encodes a capping enzyme but appears not to encode its own RNA polymerase.
The triphosphatase, guanylyltransferase, and methyltransferase components of the capping apparatus are organized differently in viral, metazoan, and fungal systems. The vaccinia virus capping enzyme is a multifunctional protein that catalyzes all three steps in cap formation (40, 45). The triphosphatase, guanylyltransferase, and methyltransferase active sites are arranged in a modular fashion within a single 95-kDa polypeptide (7, 14, 22, 24, 26, 52). Metazoan species encode a two-component capping system consisting of a bifunctional triphosphatase-guanylyltransferase and a separate methyltransferase (16, 25, 41, 43, 47, 48, 53). The budding yeast Saccharomyces cerevisiae has a three-component system in which the triphosphatase, guanylyltransferase, and methyltransferase reactions are catalyzed by separate gene products (15, 23, 33, 42). The guanylyltransferase and methyltransferase domains are conserved between DNA viruses, fungi, and metazoans. In contrast, the triphosphatase components are structurally and mechanistically divergent.
Baculoviruses are large DNA viruses that replicate in the nuclei of insect cells. The prototypal member of this family is the Autographa californica nuclear polyhedrosis virus (AcNPV), which encodes ∼154 genes within a 134-kbp circular DNA genome (1). Baculovirus early mRNAs are synthesized by cellular RNA polymerase II (9, 18, 19). Late and very late genes are transcribed after the onset of viral DNA replication by a novel amanitin-resistant RNA polymerase that is induced in virus-infected cells (2, 9, 50). Baculovirus mRNAs isolated from infected cells at late times contain a 7-methylguanosine cap (5a). In order for late and very late mRNAs to be capped, the virus must either encode its own capping enzymes or enlist the cellular capping machinery.
Recent studies indicate that cellular RNA guanylyltransferases are targeted to nascent pre-mRNAs in transcription elongation complexes by binding to the phosphorylated form of the carboxyl-terminal domain (CTD) of the largest subunit of RNA polymerase II (5, 16, 25, 36, 53). The CTD consists of a series of tandem repeats of the heptad sequence Thr-Ser-Pro-Thr-Ser-Pro-Ser. The purified AcNPV RNA polymerase consists of four equimolar subunits encoded by the viral lef-8, lef-9, lef-4, and p47 genes (11). The LEF-8 and LEF-9 proteins include a short segment of homology to the two largest subunits of eukaryotic and prokaryotic DNA-dependent RNA polymerases (21, 29). None of the baculovirus RNA polymerase subunits has an element homologous to the RNA polymerase II CTD. Therefore, it seems unlikely that the cellular guanylyltransferase would be conscripted by the viral RNA polymerase.
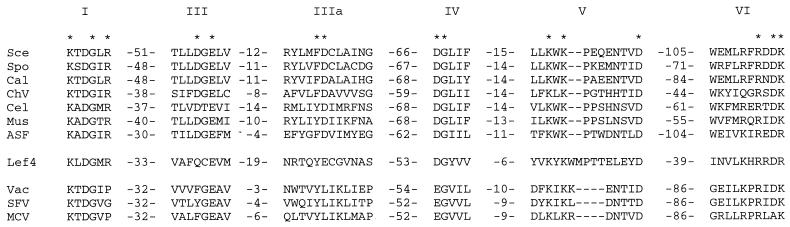
We noted that the 464-amino-acid LEF-4 subunit (28) contains the six signature motifs of the covalent nucleotidyltransferase superfamily, which includes the ATP-dependent DNA ligases and the GTP-dependent mRNA capping enzymes (39). The motifs in LEF-4 are arrayed in the same order as and with spacing similar to those of the RNA guanylyltransferase components of the poxvirus (31, 44), African swine fever virus (30), S. cerevisiae (33), Schizosaccharomyces pombe (38), Candida albicans (49), Chlorella virus (17), Caenorhabditis elegans (41, 48), and mammalian capping enzymes (25, 53) (Fig. 1). These enzymes react with GTP to form a covalent enzyme-GMP intermediate in which GMP is linked via a phosphoramidate (P-N) bond to the invariant lysine of motif I (KxDG). The GMP is then transferred to diphosphate-terminated RNA to form the GpppN cap.
FIG. 1.
Baculovirus LEF-4 contains the capping enzyme signature motifs. Six collinear sequence elements, designated motifs I, III, IIIa IV, V, and VI, are present in AcNPV LEF-4 and in cellular and viral capping enzymes. The amino acid sequences are aligned for the enzymes of S. cerevisiae (Sce), S. pombe (Spo), C. albicans (Cal), Chlorella virus PBCV-1 (ChV), mouse (Mus), African swine fever virus (ASF), AcNPV (Lef4), vaccinia virus (Vac), Shope fibroma virus (SFV), and molluscum contagiosum virus (MCV). The numbers of amino acid residues separating the motifs are indicated. The essential positions of the Sce enzyme motifs that have identical or similar functional groups in LEF-4 are denoted by asterisks.
The six nucleotidyltransferase motifs form a GTP binding pocket in the crystal structure of the Chlorella virus guanylyltransferase (13). Mutational analysis of the S. cerevisiae guanylyltransferase (Ceg1p) has shown that individual residues within the six motifs are essential for enzyme function (38, 48). The essential positions of the yeast guanylyltransferase that have identical or similar functional groups in LEF-4 are denoted by asterisks in Fig. 1. Based on intramotif conservation (e.g., Arg in motif I, Asp in motif IV, and Pro in motif V), LEF-4 appears more closely related to the guanylyltransferases from fungi, metazoans, Chlorella virus, and African swine fever virus than to the poxvirus enzymes.
The prediction from the protein sequence alignment is that LEF-4 is a virus-encoded capping enzyme. We tested this hypothesis by expressing LEF-4 in bacteria and characterizing the reactions catalyzed by the purified recombinant protein. We report that LEF-4 is a bifunctional enzyme with guanylyltransferase and 5′ triphosphatase activities. The substrate specificity and cofactor requirements of the LEF-4 triphosphatase are similar to those of the triphosphatase components of the vaccinia virus and yeast capping apparatus.
MATERIALS AND METHODS
Expression and purification of recombinant LEF-4.
The lef-4 open reading frame was amplified by PCR from a plasmid template containing a viral genomic DNA fragment (a gift of Linda Guarino). Oligonucleotide primers complementary to 5′ and 3′ ends of the gene were designed to introduce NdeI and BamHI restriction sites, respectively. The sequence of the 5′ sense primer was 5′-GTTGCCGTTATACATATGGACTACGGCGAT, and that of the 3′ antisense primer was 5′-CGGACTGCCCGTTGGATCCGCTTAACGTGC (restriction sites are underlined). PCR was carried out with Taq polymerase (Boehringer). The PCR product was digested with NdeI and BamHI and then inserted between the NdeI and BamHI sites of the T7-based expression plasmid pET16b (Novagen). The resulting plasmid, pET-LEF-4, was transformed into Escherichia coli BL21(DE3).
A 1-liter culture of E. coli BL21(DE3)/pET-LEF-4 was grown at 37°C in Luria-Bertani medium containing 0.1 mg of ampicillin per ml until the A600 reached 0.7. The culture was chilled for 30 min on ice, then adjusted to 2% ethanol, and incubated at 17°C for 15 h with continuous shaking. Cells were harvested by centrifugation, and the pellets were stored at −80°C. All subsequent procedures were performed at 4°C. Thawed bacteria were resuspended in 30 ml of buffer B (50 mM Tris HCl [pH 7.5], 150 mM NaCl, 10% sucrose) and then mixed with 10 ml of buffer B containing 0.8 mg of lysozyme per ml. The suspension was incubated on ice for 30 min, then adjusted to 0.1% Triton X-100, and incubated for an additional 30 min. The lysate was sonicated to reduce viscosity and then separated into soluble and insoluble fractions by centrifugation for 20 min at 18,000 rpm in a Sorvall SS34 rotor. The soluble fraction was mixed with 1.5 ml of Ni-nitrilotriacetic acid–agarose resin (Qiagen) that had been equilibrated with buffer C (20 mM Tris [pH 8.0], 300 mM NaCl, 10% glycerol, 0.1% Triton X-100), and the suspension was mixed by continuous rotation for 1 h. The Ni-agarose resin was recovered by centrifugation, and the supernatant was removed. The resin was resuspended in 20 ml of buffer C, and the slurry was poured into a column. The packed column was washed with 40 ml of buffer C and then eluted stepwise with 6-ml aliquots of buffer C containing 5, 10, 25, 50, 250, and 500 mM imidazole. The LEF-4 polypeptide at eluted at 250 mM imidazole, as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining. The 250 mM imidazole fraction was diluted 20-fold in buffer A (50 mM Tris HCl [pH 6.5], 2 mM dithiothreitol [DTT], 1 mM EDTA, 10% glycerol, 0.1% Triton X-100) and then applied to a 2-ml phosphocellulose column that had been equilibrated in buffer A. The column was eluted stepwise with buffer A containing 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, and 0.8 M NaCl. LEF-4 was recovered predominantly in the 0.3 and 0.4 M NaCl fraction (yielding 2 mg of LEF-4 protein). Protein concentrations were determined by the Bio-Rad dye binding assay with bovine serum albumin as the standard.
Sedimentation analysis.
An aliquot (0.2 ml; 50 μg of protein) of the 0.3 M NaCl phosphocellulose eluate fraction was applied to a 4.8-ml 15 to 30% glycerol gradient containing 0.3 NaCl in buffer A. The gradient was centrifuged for 19 h at 50,000 rpm in a Beckman SW50 rotor at 4°C. Fractions (0.15 ml) were collected from the bottom of the tube. Protein standards (catalase, bovine serum albumin, and cytochrome c) were sedimented in a parallel gradient.
RNA triphosphatase assay.
γ-32P-labeled triphosphate-terminated poly(A) was prepared as described previously (40). RNA triphosphatase reaction mixtures (10 μl) containing 50 mM Tris HCl (pH 7.5), 5 mM DTT, 10 pmol of [γ-32P]poly(A), MnCl2 or MgCl2 as specified, and enzyme were incubated for 15 min at 30°C. The reactions were halted by adding 1 μl of 1 M formic acid. Aliquots (5 μl) were spotted onto a polyethyleneimine (PEI)-cellulose thin-layer chromatography (TLC) plate. The TLC plate was developed with 0.75 M potassium phosphate (pH 4.3). 32Pi and [γ-32P]poly(A) were visualized by autoradiographic exposure of the plate. The extent of release of 32Pi from [γ-32P]poly(A) was quantitated by scanning the plate with a Fuji BAS1000 phosphorimager.
ATPase assay.
Reaction mixtures (20 μl) containing 50 mM Tris HCl (pH 7.5), 5 mM DTT, 1 mM MnCl2, 100 μM [γ-32P]ATP, and enzyme were incubated at 30°C for 15 min. The reaction was terminated by adding 1 μl of 1 M formic acid. Aliquots were spotted onto PEI-cellulose TLC plates, which were developed with 0.5 M LiCl–1 M formic acid. 32Pi and [γ-32P]ATP were visualized by autoradiographic exposure. 32Pi formation was quantitated by scanning the plate with a phosphorimager.
LEF-4–GMP complex formation.
Reaction mixtures (20 μl) containing 50 mM Tris HCl (pH 8.0), 5 mM DTT, 20 mM MgCl2, 1 mM [α-32P]GTP, and enzyme were incubated for 15 min at 30°C. The reaction was halted by adjusting the mixtures to 1% SDS. The samples were electrophoresed through a 10% polyacrylamide gel containing 0.1% SDS. Label transfer to the LEF-4 polypeptide was visualized by autoradiographic exposure of the dried gel and quantitated by scanning the gel with a phosphorimager.
Preparation of cap-labeled RNA.
Cap-labeled poly(A) [GpppA(pA)n; boldface denotes the site of labeling] was synthesized in a reaction mixture (80 μl) containing 50 mM Tris-HCl (pH 8.0), 1.25 mM MgCl2, 5 mM DTT, 200 pmol of triphosphate-terminated poly(A), 5 μM [α-32P]GTP, and 0.4 pmol of purified recombinant vaccinia virus capping enzyme (22). Methylated cap-labeled poly(A) [m7GpppA(pA)n] was synthesized in a similar reaction mixture supplemented with 50 μM AdoMet. The mixtures were incubated for 30 min at 37°C. Unincorporated GTP was removed by multiple rounds of trichloroacetic acid precipitation. The resuspended RNA was extracted with phenol-chloroform and recovered by ethanol precipitation.
RESULTS
Purification of AcNPV LEF-4 protein.
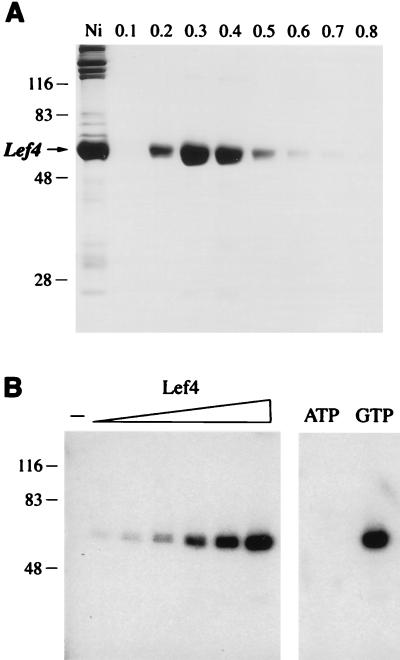
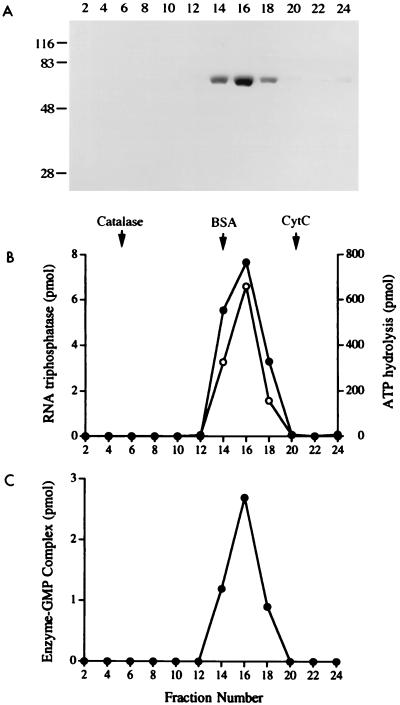
The lef-4 open reading frame was cloned into a T7 RNA polymerase-based vector so as to place the gene in frame with an N-terminal leader encoding a 21-amino-acid peptide with 10 tandem histidines. The expression plasmid was introduced into E. coli BL21(DE3), a strain that contains the T7 RNA polymerase gene. A novel 57-kDa polypeptide was detectable by SDS-PAGE in bacterial extracts (not shown). Initial purification of the His-tagged fusion protein was achieved by adsorption to Ni-agarose and elution with 250 mM imidazole; the eluate was highly enriched with respect to the 57-kDa LEF-4 polypeptide (Fig. 2A, lane Ni). This polypeptide was not present in the imidazole eluate when extracts of bacteria lacking the lef-4 expression plasmid were chromatographed in parallel (not shown). LEF-4 was purified further by adsorption to a column of phosphocellulose and step elution with 300 and 400 mM NaCl (Fig. 2A). The phosphocellulose preparation was nearly homogeneous with respect to the 57-kDa polypeptide, as judged by SDS-PAGE (Fig. 2A). Further characterization of recombinant LEF-4 was performed with the 0.3 M NaCl phosphocellulose eluate fraction.
FIG. 2.
Purification and guanylyltransferase activity of recombinant LEF-4. (A) Aliquots of the Ni-agarose 250 mM imidazole eluate fraction (Ni) and the phosphocellulose NaCl eluate fractions (NaCl concentrations indicated above the lanes) were analyzed by SDS-PAGE. A Coomassie blue-stained gel is shown. The positions and sizes (in kilodaltons) of marker proteins are indicated on the left. (B) Enzyme-guanylate complex formation. Left, reaction mixtures (20 μl) contained 20 mM MgCl2, 1 mM [α-32P]GTP, and increasing amounts of LEF-4 (0.16, 0.32, 0.63, 1.25, 2.5, or 5 pmol). LEF-4 was omitted from a control reaction (lane −). The reaction products were analyzed by SDS-PAGE. An autoradiogram of the gel is shown. The amount of [32P]GMP transferred to LEF-4 was determined by scanning the gel with a phosphorimager and is plotted in Fig. 3C as a function of input protein. Right, reaction mixtures contained 5 pmol of LEF-4 and either 1 mM [α-32P]ATP or 1 mM [α-32P]GTP.
Recombinant LEF-4 forms a covalent protein-GMP complex in vitro.
The mRNA guanylyltransferase reaction entails two sequential nucleotidyl transfer steps (39). In the first step, nucleophilic attack on the α phosphate of GTP by enzyme results in liberation of pyrophosphate and formation of a covalent enzyme-GMP intermediate. To assay guanylyltransferase activity of the expressed LEF-4 protein, we incubated the phosphocellulose fraction in the presence of 1 mM [α-32P]GTP and 20 mM MgCl2. This resulted in the formation of an SDS-stable nucleotidyl-protein adduct that migrated as a single 57-kDa species during SDS-PAGE (Fig. 2B, lane GTP). Labeling of this polypeptide was not detected when LEF-4 was incubated with 1 mM [α-32P]ATP (Fig. 2B, lane ATP). We conclude that the expressed LEF-4 protein is active in transguanylylation.
Characterization of the LEF-4 guanylyltransferase reaction.
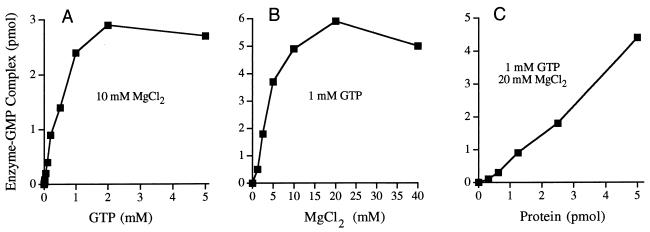
The yield of LEF-4–GMP complex in the presence of 10 mM magnesium was proportional to GMP concentration in the range of 0.01 to 1 mM and saturated at 2 mM GTP, with approximately 0.6 pmol of GMP bound per pmol of input LEF-4 (Fig. 3A). Half-saturation was attained at ∼0.5 mM GTP. Recombinant LEF-4 binds GTP with much lower affinity than other guanylyltransferases; e.g., enzyme-GMP formation by recombinant Chlorella virus, vaccinia virus, and mammalian guanylyltransferases saturates at 1 to 5 μM GTP (7, 16, 17). LEF-4–GMP formation in the presence of 1 mM GTP was strictly dependent on inclusion of magnesium chloride in the reaction mixture. The yield of LEF-4–GMP was proportional to magnesium concentration in the range of 1 to 10 mM and peaked at 20 mM (Fig. 3B). The protein concentration dependence of LEF-4–GMP formation in 1 mM GTP and 20 mM MgCl2 is shown in Fig. 2B and quantitated in Fig. 3C. In this experiment, 5 pmol of LEF-4 bound 4.5 pmol of GMP. We surmise that LEF-4 has a single site for covalent GMP binding and that most of the LEF-4 molecules in the enzyme preparation are in the unguanylylated form.
FIG. 3.
Characterization of the LEF-4 guanylyltransferase. (A) GTP dependence. Reaction mixtures (20 μl) contained 50 mM Tris-HCl (pH 8.0), 5 mM DTT, 10 mM MgCl2, 5 pmol of LEF-4, and either 0.01, 0.025, 0.05, 0.1, 0.2, 0.5, 1, 2, or 5 mM [α-32P]GTP. The amount of [32P]GMP transferred to LEF-4 is plotted as a function of GTP concentration. (B) Magnesium dependence. Reaction mixtures (20 μl) contained 1 mM [α-32P]GTP, 5 pmol of LEF-4, and MgCl2 as indicated. (C) LEF-4 titration. See the legend to Fig. 2B for details.
RNA 5′-triphosphatase activity of LEF-4.
The predicted active-site lysine nucleophile of the LEF-4 guanylyltransferase (residue Lys-255 in motif I) is located far from the N terminus of the polypeptide, just like in the vaccinia virus capping enzyme, in which the active-site residue is Lys-260 (6, 27). In the yeast and Chlorella virus guanylyltransferases, which are monofunctional and have no intrinsic RNA triphosphatase activity, the active site lysine is located much closer to the N terminus (at positions 67 to 82). This finding suggested that the LEF-4 N-terminal region might contribute an RNA triphosphatase function, as it does in the vaccinia virus enzyme (26, 51, 52).
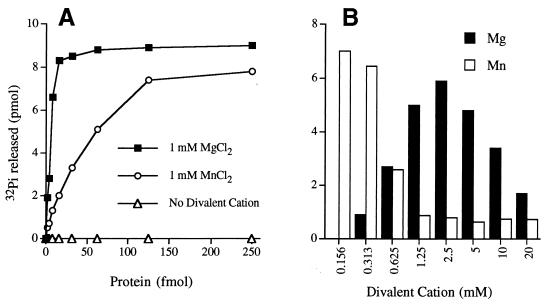
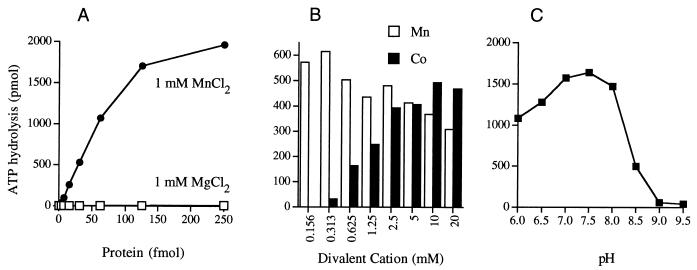
We found that purified recombinant LEF-4 is indeed an RNA triphosphatase. Activity was assayed by the liberation of 32Pi from 1 μM γ-32P-labeled triphosphate-terminated poly(A) in the presence of 1 mM magnesium chloride. The extent of γ phosphate hydrolysis during a 15-min incubation at 30°C was proportional to the amount of input protein (Fig. 4A). In the linear range of enzyme dependence, 950 fmol of 32Pi was released per fmol of LEF-4 (which translates to a turnover number of ∼1 s−1). Specific activity was lower when 1 mM manganese was substituted for 1 mM magnesium (Fig. 4A). No activity was detected in the absence of a divalent cation cofactor. A finer analysis of the divalent cation dependence of the LEF-4 RNA triphosphatase showed that magnesium and manganese were equally effective cofactors at their concentration optima, which were 0.16 to 0.31 mM for manganese and 2.5 mM for magnesium (Fig. 4B).
FIG. 4.
LEF-4 RNA triphosphatase activity. (A) Divalent cation dependence. Reaction mixtures (10 μl) contained 10 pmol of [γ-32P]poly(A), LEF-4 as specified, and either 1 mM MgCl2, 1 mM MnCl2, or no divalent cation. The extent of 32Pi release is plotted as function of input LEF-4. (B) Divalent cation dependence. Reaction mixtures contained 10 pmol of [γ-32P]poly(A), 5 fmol of LEF-4, and MgCl2 or MnCl2 at the concentrations specified.
Hydrolysis of nucleoside triphosphatases.
The activity of the LEF-4 triphosphatase component was not restricted to RNA 5′ ends. LEF-4 also hydrolyzed ATP. Remarkably, the ATPase of LEF-4 was specifically activated by manganese or cobalt but not by magnesium (Fig. 5). ATPase activity was assayed by the release of 32Pi from 100 μM [γ-32P]ATP in the presence of 1 mM manganese chloride. The extent of 32Pi release during a 15-min reaction at 30°C was proportional to the amount of input enzyme in the range of 0.2 to 6 nM LEF-4 (Fig. 5A). The substrate was hydrolyzed quantitatively at ≥12.5 nM enzyme (Fig. 5A). In the linear range of enzyme dependence, 17 pmol of 32Pi was released per fmol of LEF-4. There was no ATP hydrolysis when 1 mM magnesium was substituted for 1 mM manganese (Fig. 5A). Among other metals tested at a concentration of 1 mM only cobalt was an effective cofactor (Fig. 5B). Calcium, copper, and zinc failed to activate the ATPase (not shown). A finer analysis of the divalent cation dependence of the LEF-4 ATPase showed that manganese and cobalt were nearly equally effective cofactors at their concentration optima. Manganese supported activity over a broad range from 0.16 to 20 mM, whereas cobalt was most effective at 2.5 to 20 mM (Fig. 5B). LEF-4 ATPase activity with magnesium (optimal at 10 to 20 mM) was less than 2% of the manganese-dependent activity (not shown). Manganese- and cobalt-activated ATPase activities have also been reported for the vaccinia virus RNA triphosphatase (34, 40) and the S. cerevisiae RNA triphosphatase (15a). ATP hydrolysis by LEF-4 in 1 mM manganese was optimal at pH 7.0 to 8.0 and declined sharply at higher pH values (Fig. 5C).
FIG. 5.
Hydrolysis of ATP. (A) LEF-4 titration. Reaction mixtures (20 μl) contained 50 mM Tris-HCl (pH 7.5), 5 mM DTT, 100 μM [γ-32P]ATP, either 1 mM MgCl2 or 1 mM MnCl2, and LEF-4 as specified. The extent of 32Pi formation is plotted as function of input protein. (B) Divalent cation dependence. Reaction mixtures (20 μl) contained 50 mM Tris-HCl (pH 7.5), 100 μM [γ-32P]ATP, 50 fmol of LEF-4, and MnCl2 or CoCl2 as indicated. (C) pH dependence. Reaction mixtures (20 μl) contained 50 mM Tris buffer (pH as specified), 5 mM DTT, 1 mM MnCl2, 100 μM [γ-32P]ATP, and 50 fmol of LEF-4.
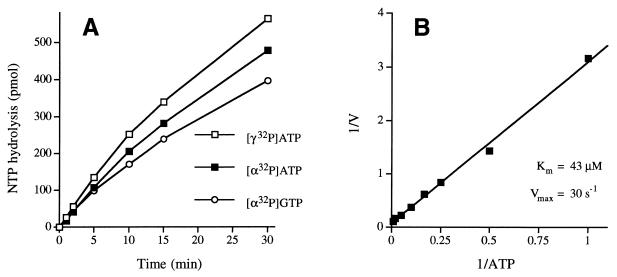
The rate of release of 32Pi from [γ-32P]ATP was nearly identical to the rate of conversion of [α-32P]ATP to [α-32P]ADP in a parallel reaction mixture containing the same concentration of LEF-4 (Fig. 6A). We detected no formation of [α-32P]AMP during the reaction. Hence, we conclude that LEF-4 catalyzes the hydrolysis of ATP to ADP and Pi. The rate of conversion of [α-32P]GTP to [α-32P]GDP was similar to the rate of ATP hydrolysis; we detected no formation of [α-32P]GMP during the reaction. LEF-4 activity on other NTP substrates was not tested.
FIG. 6.
Kinetics of NTP hydrolysis. (A) Reaction mixtures containing (per 20 μl) 1 mM MnCl2, 100 μM [γ-32P]ATP, [α-32P]ATP or [α-32P]GTP, and 30 fmol of LEF-4 were incubated at 30°C. Aliquots were withdrawn at the times specified and quenched immediately with formic acid. The reaction products were analyzed by PEI-cellulose TLC. The extent of 32Pi, [α-32P]ADP, or [α-32P]GDP formation (from 2,000 pmol of input ATP or GTP) is plotted as function of reaction time. (B) Steady-state kinetic parameters of ATP hydrolysis. Reaction mixtures (10 μl) containing 1 mM MnCl2, 8 fmol of LEF-4, and either 1, 2, 4, 6, 10, 20, 50, 100, or 200 μM [γ-32P]ATP were incubated at 30°C for 15 min. The extent of 32Pi formation (picomoles) was determined by TLC analysis of the reaction products. A double-reciprocal plot of the rate of 32Pi formation (min−1 = pmol of 32Pi formed/15) versus [ATP] is shown.
Kinetic parameters were determined by measuring the extent of 32Pi formation during a 15-min reaction as a function of input [γ-32P]ATP concentration in the range of 1 to 200 μM. From a double-reciprocal plot of the data (Fig. 6B), we calculated a Km of 43 μM ATP and a Vmax of 30 s−1.
Sedimentation analysis.
The native size of LEF-4 was gauged by sedimentation of the phosphocellulose protein fraction through a 15 to 30% glycerol gradient containing 0.3 M NaCl. The LEF-4 polypeptide sedimented as a single discrete peak coincident with the guanylyltransferase, RNA triphosphatase, and ATPase activity profiles (Fig. 7). An apparent sedimentation coefficient of 4.0S was calculated by comparison to marker proteins (catalase, 11.2S, 248 kDa; bovine serum albumin, 4.4S, 66 kDa; and cytochrome c, 1.9S, 13.4 kDa) that were centrifuged in a parallel gradient. We conclude that LEF-4 is a monomer.
FIG. 7.
Sedimentation analysis. A sample of the phosphocellulose preparation of LEF-4 was sedimented in a 15 to 30% glycerol gradient as described in Materials and Methods. Fractions were collected from the bottom of the tube. (A) Aliquots (20 μl) of the even-numbered gradient fractions were analyzed by SDS-PAGE. A Coomassie blue-stained gel is shown. The positions and sizes (in kilodaltons) of coelectrophoresed marker polypeptides are indicated at the left. (B) RNA triphosphatase reaction mixtures (10 μl) contained 1 mM MgCl2, 10 pmol of [γ-32P]poly(A), and 1 μl of a 1:100 dilution of the indicated glycerol gradient fractions. ATPase reaction mixtures (20 μl) contained 1 mM MnCl2, 100 μM [γ-32P]ATP, and 1 μl of a 1:30 dilution of the indicated gradient fraction. The RNA triphosphatase (left y axis; •) and ATPase (right y axis; ○) activity profiles are shown. The peaks of marker proteins catalase, bovine serum albumin (BSA), and cytochrome c (CytC), which were centrifuged in a parallel gradient, are indicated by arrows. (C) Guanylyltransferase reaction mixtures (20 μl) contained 20 mM MgCl2, 1 mM [α-32P]GTP, and 3 μl of the indicated gradient fraction.
Transfer of GMP from the RNA cap to LEF-4.
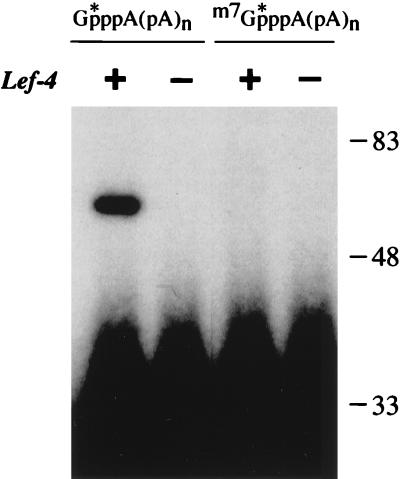
Although we presume that the LEF-4–[32P]GMP complex is an intermediate in cap synthesis, we were unable to detect formation of cap-labeled poly(A) when LEF-4 was incubated with 1 mM [α-32P]GTP, 10 mM MgCl2, and 5 μM triphosphate-terminated poly(A). The requirement by LEF-4 for very high GTP concentrations (and hence low GTP specific radioactivity) was a confounding technical factor. As alternative approach, we tested whether [32P]GMP could be transferred from the RNA cap to the LEF-4 protein via reversal of the capping reaction. [α-32P]GMP-labeled capped poly(A) was synthesized at high specific radioactivity, using the vaccinia virus capping enzyme, [α-32P]GTP, and triphosphate-terminated poly(A). Methylated cap-labeled poly(A) was synthesized in a parallel reaction containing AdoMet. The capped poly(A) products were recovered free of [α-32P]GTP by multiple rounds of precipitation with trichloroacetic acid and then with ethanol; the radiochemical purity of the capped RNA was confirmed by TLC chromatography (not shown). LEF-4 was incubated with cap-labeled poly(A) [GpppA(pA)n] in the presence of magnesium, and the reaction products were analyzed by SDS-PAGE. The cap-labeled poly(A) migrated near the bottom of the gel. Inclusion of LEF-4 in the reaction resulted in very low, but readily detectable, levels of label transfer to the 57-kDa LEF-4 polypeptide (Fig. 8). The inefficiency of the reverse capping reaction probably stems from the low affinity of LEF-4 for binding to the cap guanylate moiety (the concentration of cap-labeled ends in the reaction was ∼100 nM), just as it displays low affinity for binding to GTP. When LEF-4 was incubated with methylated cap-labeled poly(A) [m7GpppA(pA)n] in the presence of magnesium, no transfer of m7GMP from RNA to protein was detected (Fig. 8). Thus, cap methylation renders the LEF-4 guanylyltransferase reaction irreversible, as it does for Chlorella virus guanylyltransferase (17).
FIG. 8.
GMP transfer to LEF-4 from capped poly(A). Reaction mixtures (10 μl) containing 50 mM Tris HCl (pH 8.0), 5 mM DTT, 10 mM MgCl2, 5 pmol of LEF-4 (+ lanes), and 1 pmol of cap-labeled (denoted by the asterisk) poly(A) [GpppA(pA)n] or methylated cap-labeled poly(A) [m7GpppA(pA)n] were incubated for 20 min at 30°C. LEF-4 was omitted from control reaction mixtures (− lanes). The reactions were terminated by adding SDS to 1%, and the samples were analyzed by SDS-PAGE. An autoradiograph of the dried gel is shown. The positions and sizes (in kilodaltons) of marker proteins are denoted on the right.
DISCUSSION
LEF-4—a baculovirus mRNA capping enzyme.
The results of this study of AcNPV LEF-4, together with those of Guarino and colleagues (11, 11a), suggest that baculoviruses adopt a novel strategy to achieve targeting of cap formation to specific transcripts, i.e., by incorporating a capping enzyme, LEF-4, as an RNA polymerase subunit. This can be viewed as one extreme in a spectrum of capping enzyme-RNA polymerase interactions. The vaccinia virus and cellular guanylyltransferases are readily purified away from their cognate RNA polymerases; hence, they are not polymerase subunits. However, vaccinia virus capping enzyme forms a binary complex in solution with vaccinia virus RNA polymerase (12). This interaction appears to facilitate the capping of nascent RNA chains as soon as their 5′ ends are extruded from the RNA binding pocket on the elongating polymerase. Recent studies of yeast and metazoans indicate that cellular guanylyltransferases bind to the elongating form of RNA polymerase II via the phosphorylated CTD of the largest polymerase subunit (5, 16, 25, 53). This interaction may explain why only RNA polymerase II transcripts acquire a cap structure in vivo (36).
lef-4 is essential for baculovirus replication and for the expression of baculovirus late and very late genes in vivo (4, 28). Why is the lef-4 gene product essential? Are the RNA triphosphatase and/or guanylyltransferase activities of LEF-4 critical? Does LEF-4 play a structural role in assembly of the viral RNA polymerase? Is LEF-4 required for transcription per se? The vaccinia virus capping enzyme certainly plays a larger role in transcription beyond cap formation; it serves as a transcription termination factor during the synthesis of viral early mRNAs (37) and as an initiation factor during the transcription of intermediate genes (46). The termination function of vaccinia capping enzyme is unaffected by mutations that abrogate its catalytic activity in cap formation (22, 51).
The biochemical properties of recombinant LEF-4 are consistent with a role in catalyzing the first two steps of cap formation, whereby the LEF-4 triphosphatase would hydrolyze the phosphoanhydride bond between the β and γ phosphates of nascent baculovirus late and very late mRNAs, thus preparing them for capping by the LEF-4 guanylyltransferase. For this model to be plausible, the catalytic activities of LEF-4 must be robust enough to execute this function cotranscriptionally. Our data argue that this is the case for the RNA triphosphatase component. LEF-4 released one molecule of Pi from 1 μM triphosphate-terminated poly(A) per enzyme per second in the steady state. This is comparable to the steady-state turnover number of 1 to 2 s−1 for the mouse RNA triphosphatase domain on the same poly(A) substrate (16). The turnover number of LEF-4 is also quite close to the value of 0.5 to 0.8 s−1 reported for the vaccinia virus RNA triphosphatase (26).
The guanylyltransferase component of LEF-4 displays exceptionally low affinity for GTP in enzyme-GMP complex formation. (The guanylyltransferase reactions were performed in the presence of magnesium, which does not support the NTPase activity of LEF-4. Hence, the requirement for very high GTP concentrations is not attributable to a competing reaction that would deplete the substrate.) We were also unable to demonstrate the incorporation of labeled GMP into a cap structure. However, we did detect reversal of the RNA capping step. Transfer of GMP from the RNA cap to LEF-4 was extremely inefficient. We presume that recombinant LEF-4 binds as poorly to the cap guanylate as it does to GTP. These findings inject uncertainty into the presumption that LEF-4 is a self-contained capping enzyme.
An attractive hypothesis is that the guanylyltransferase activity of LEF-4 is more robust in the context of the native AcNPV RNA polymerase. There are two precedents in which capping activities are upregulated by protein-protein interactions. First, the very low basal cap methyltransferase activity of the vaccinia virus D1 protein is stimulated 50- to 100-fold by heterodimerization with the vaccinia virus D12 protein (14, 24). Second, the formation of enzyme-GMP complex by purified recombinant yeast guanylyltransferase (Ceg1p) is stimulated 10-fold by heterodimerization with the separately encoded yeast RNA triphosphatase Cet1p (15). Cet1p stimulates Ceg1p-GMP formation by increasing the affinity of the guanylyltransferase for GTP substrate (15). We speculate that interaction of LEF-4 with one or more of the other subunits of baculovirus RNA polymerase mass elicit a similar change in the substrate binding properties of LEF-4.
Addition of a cap guanylate to nascent baculovirus mRNAs by LEF-4 must be followed by guanine-N7 methylation in order to complete the capping reaction. Addition of the this methyl group renders the capping reaction irreversible. The guanine-N7 methyl group is also required for cap-dependent translation initiation. Our searches revealed no baculovirus homologues of the yeast and poxvirus RNA (guanine-7) methyltransferases. Thus, either baculoviruses encode a cap methyltransferase unrelated to the known enzymes or baculovirus late and very late mRNAs are N7-methylated by the host cell enzyme. The latter scenario has been suggested for Chlorella virus PBCV-1, which encodes its own guanylyltransferase but seems not to encode a cap-specific methyltransferase (17).
Evolution of the RNA triphosphatase component of the capping apparatus.
The identification and characterization of the LEF-4 triphosphatase, together with recent work on the vaccinia virus, metazoan, and yeast enzymes, underscores the existence of two mechanistically and structurally distinct classes of RNA triphosphatases: (i) the divalent cation-dependent triphosphatases exemplified by baculovirus LEF-4, vaccinia virus D1, and S. cerevisiae Cet1p and (ii) the divalent cation-independent RNA triphosphatases, e.g., the metazoan cellular enzymes and the 168-amino-acid baculovirus phosphatase BVP (10, 16, 32, 41), which contain the HCxAGxGR(S/T)G phosphate binding signature motif first described for the protein tyrosine phosphatases and dual-specificity protein phosphatases (8).
The metazoan RNA triphosphatases and BVP do not require a divalent cation cofactor for catalysis; indeed, they are inhibited by magnesium (10, 16, 41). Based on their structural similarity to the well-characterized protein phosphatases (8), it is posited that metazoan RNA triphosphatases execute a two-step ping-pong reaction in which (i) the γ phosphate of triphosphate-terminated RNA is transferred to a conserved cysteine nucleophile to form a phosphoenzyme intermediate and the diphosphate RNA product is expelled; and (ii) the phosphoenzyme is attacked by water to liberate Pi and expel the cysteine. Although a phosphoenzyme intermediate has not been demonstrated directly for metazoan RNA triphosphatases or BVP, mutations of the conserved active-site cysteine nucleophiles of mammalian and C. elegans RNA triphosphatase and BVP do abrogate enzyme activity (10, 15, 41). It is interesting that baculovirus replication still occurs in cultured insect cells when the BVP gene is deleted (20). The RNA triphosphatase function of BVP may be redundant to that of LEF-4 during AcNPV replication.
The LEF-4, vaccinia virus D1, and yeast Cet1p enzymes display remarkably similar biochemical characteristics in their hydrolysis of the β-γ phosphoanhydride linkage of RNA and NTP substrates. The activations of NTP hydrolysis by manganese and cobalt are the signature features of these enzymes (15a, 34, 40). Is there a common structural basis for metal-dependent catalysis? A database search with LEF-4 revealed extensive amino acid sequence identity to the LEF-4 equivalents of other baculovirus strains but no similarity to known NTPases or phosphatases. Moreover, computer-based comparisons reveal no sequence similarity between LEF-4 and either Cet1p or vaccinia virus D1 (exclusive of the D1 guanylyltransferase motifs). Yet, because we have already mapped essential catalytic residues within the vaccinia virus RNA triphosphatase (51), we have the capacity to screen by eye for conservation of essential side chains. We find that the metal-dependent RNA triphosphatases share three collinear sequence motifs (Fig. 9). These are present in yeast RNA triphosphatase Cet1p, in the triphosphatase-guanylyltransferase domains of the vaccinia virus, Shope fibroma virus, molluscum contagiosum virus, and African swine fever virus capping enzymes, and in baculovirus LEF-4. The residues that are essential for the RNA triphosphatase and ATPase activities of vaccinia virus capping enzyme (denoted by asterisks in Fig. 9) include four glutamates and an arginine. Alanine substitutions at any of these positions reduce phosphohydrolase specific activity by 2 to 3 orders of magnitude but have no impact on the guanylyltransferase activity of the vaccinia virus enzyme (51). All five residues essential for vaccinia virus triphosphatase activity are conserved.
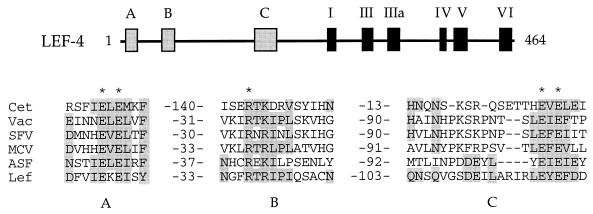
FIG. 9.
Conserved sequence elements of the metal-dependent RNA triphosphatases. Three conserved motifs, designated A, B, and C, in the RNA triphosphatases of S. cerevisiae (Cet), vaccinia virus (Vac), Shope fibroma virus (SFV), molluscum contagiosum virus (MCV), African swine fever virus (ASF), and AcNPV LEF-4 (Lef) are aligned in the figure. LEF-4 residues that are conserved in the other proteins are shaded. The numbers of amino acids separating the motifs are indicated. The five amino acids in the vaccinia virus capping enzyme that were found by mutational analysis to be essential for triphosphatase activity are denoted by asterisks. The locations of the three triphosphatase motifs and the six guanylyltransferase motifs within the LEF-4 polypeptide are illustrated.
The three triphosphatase motifs (designated A, B, and C in Fig. 9) are located N terminal to the six guanylyltransferase motifs of the LEF-4, poxvirus, and African swine fever virus enzymes. Motifs B and C are separated by 90 to 103 amino acids in the viral proteins, whereas the intervening segment is only 13 amino acids in yeast Cet1p. In vaccinia virus capping enzyme, the guanylyltransferase and triphosphatase functional domains overlap; e.g., the 90-amino-acid segment between motifs B and C contains residues that are essential for the guanylyltransferase but not for the triphosphatase (52). This may explain the longer motif B→motif C distance in the viral enzymes. The segment between motifs A and B is much longer in yeast Cet1p (140 amino acids) than in the viral proteins (30 to 37 amino acids). This region of Cet1p may well include a portion of the Ceg1p binding surface, a feature that would not apply to the viral triphosphatases.
We hypothesize that baculovirus LEF-4, yeast Cet1p, and vaccinia virus D1 comprise a novel family of metal-dependent RNA triphosphatases with a common active site. We speculate that the Arg in motif II contacts the negatively charged 5′ triphosphate moiety to promote attack by water on the γ phosphorus. The role of the essential glutamates may be to coordinate the divalent cation cofactor. An more extensive mutational analysis of the conserved motifs should illuminate the evolutionary connection between these enzymes.
REFERENCES
- 1.Ayres M D, Howard S C, Kuzio J, Lopez-Ferber M, Possee R D. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology. 1994;202:586–605. doi: 10.1006/viro.1994.1380. [DOI] [PubMed] [Google Scholar]
- 2.Beniya H, Funk C J, Rohrmann G F, Weaver R F. Purification of a virus-induced RNA polymerase from Autographa californica nuclear polyhedrosis virus-infected Spodoptera frugiperda cells that accurately initiates late and very late transcription in vitro. Virology. 1996;216:12–19. doi: 10.1006/viro.1996.0029. [DOI] [PubMed] [Google Scholar]
- 3.Bisaillon M, Lemay G. Viral and cellular enzymes involved in synthesis of mRNA cap structures. Virology. 1997;236:1–7. doi: 10.1006/viro.1997.8698. [DOI] [PubMed] [Google Scholar]
- 4.Carstens E B, Chan H, Yu H, Williams G V, Casselman R. Genetic analyses of temperature-sensitive mutations in baculovirus late expression genes. Virology. 1994;204:323–337. doi: 10.1006/viro.1994.1537. [DOI] [PubMed] [Google Scholar]
- 5.Cho E, Takagi T, Moore C R, Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxyl-terminal domain. Genes Dev. 1997;11:3319–3332. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Jun-Chuan Q, Weaver R F. Capping of viral RNA in cultured Spodoptera frugiperda cells infected with Autographa californica nuclear polyhedrosis virus. J Virol. 1982;43:234–240. doi: 10.1128/jvi.43.1.234-240.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cong P, Shuman S. Covalent catalysis in nucleotidyl transfer: a KTDG motif essential for enzyme-GMP complex formation by mRNA capping enzyme is conserved at the active sites of RNA and DNA ligases. J Biol Chem. 1993;268:7256–7260. [PubMed] [Google Scholar]
- 7.Cong P, Shuman S. Mutational analysis of mRNA capping enzyme identifies amino acids involved in GTP binding, enzyme-guanylate complex formation, and GMP transfer to RNA. Mol Cell Biol. 1995;15:6222–6231. doi: 10.1128/mcb.15.11.6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denu J M, Stuckey J A, Saper M A, Dixon J E. Form and function in protein dephosphorylation. Cell. 1996;87:361–364. doi: 10.1016/s0092-8674(00)81356-2. [DOI] [PubMed] [Google Scholar]
- 9.Fuchs L Y, Woods M S, Weaver R F. Viral transcription during Autographa californica nuclear polyhedrosis virus infection: a novel RNA polymerase induced in infected Spodoptera frugiperda cells. J Virol. 1983;67:3773–3776. doi: 10.1128/jvi.48.3.641-646.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gross C H, Shuman S. Characterization of a baculovirus-encoded RNA 5′-triphosphatase. J Virol. 1998;72:7057–7063. doi: 10.1128/jvi.72.9.7057-7063.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guarino L A, Xu B, Jin J, Dong W. A virus-encoded RNA polymerase purified from baculovirus-infected cells. J Virol. 1998;72:7985–7991. doi: 10.1128/jvi.72.10.7985-7991.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Guarino L A, Jin J, Dong W. Guanylyltransferase activity of the LEF-4 subunit of baculovirus RNA polymerase. J Virol. 1998;72:10003–10010. doi: 10.1128/jvi.72.12.10003-10010.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hagler J, Shuman S. A freeze-frame view of eukaryotic transcription during elongation and capping of nascent mRNA. Science. 1992;255:983–986. doi: 10.1126/science.1546295. [DOI] [PubMed] [Google Scholar]
- 13.Hakansson K, Doherty A J, Shuman S, Wigley D B. X-ray crystallography reveals a large conformational change during guanyl transfer by mRNA capping enzymes. Cell. 1997;89:545–553. doi: 10.1016/s0092-8674(00)80236-6. [DOI] [PubMed] [Google Scholar]
- 14.Higman M A, Christen L A, Niles E G. The mRNA (guanine-7-) methyltransferase domain of the vaccinia virus mRNA capping enzyme: expression in Escherichia coli and structural and kinetic comparison to the intact capping enzyme. J Biol Chem. 1994;269:14974–14981. [PubMed] [Google Scholar]
- 15.Ho C K, Schwer B, Shuman S. Genetic, physical, and functional interactions between the triphosphatase and guanylyltransferase components of the yeast mRNA capping apparatus. Mol Cell Biol. 1998;18:5189–5198. doi: 10.1128/mcb.18.9.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a.Ho, C. K., Y. Pei, and S. Shuman. Unpublished data.
- 16.Ho C K, Sriskanda V, McCracken S, Bentley D, Schwer B, Shuman S. The guanylyltransferase domain of mammalian mRNA capping enzyme binds to the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 1998;273:9577–9585. doi: 10.1074/jbc.273.16.9577. [DOI] [PubMed] [Google Scholar]
- 17.Ho C K, Van Etten J L, Shuman S. Expression and characterization of an RNA capping enzyme encoded by Chlorella virus PBCV-1. J Virol. 1996;70:6658–6664. doi: 10.1128/jvi.70.10.6658-6664.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoopes R R, Jr, Rohrmann G F. In vitro transcription of baculovirus immediate early genes: accurate initiation by nuclear extracts from both insect and human cells. Proc Natl Acad Sci USA. 1991;88:4513–4517. doi: 10.1073/pnas.88.10.4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huh N E, Weaver R F. Identifying the RNA polymerases that synthesize specific transcripts of the Autographa californica nuclear polyhedrosis virus. J Gen Virol. 1990;71:195–201. doi: 10.1099/0022-1317-71-1-195. [DOI] [PubMed] [Google Scholar]
- 20.Li Y, Miller L K. Properties of a baculovirus mutant defective in the protein phosphatase gene. J Virol. 1995;69:4533–4537. doi: 10.1128/jvi.69.7.4533-4537.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu A, Miller L K. Identification of three late expression factor genes within the 33.8- to 43.4-map-unit region of Autographa californica nuclear polyhedrosis virus. J Virol. 1994;68:6710–6718. doi: 10.1128/jvi.68.10.6710-6718.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo Y, Mao X, Deng L, Cong P, Shuman S. The D1 and D12 subunits are both essential for the transcription termination factor activity of vaccinia virus capping enzyme. J Virol. 1995;69:3852–3856. doi: 10.1128/jvi.69.6.3852-3856.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mao X, Schwer B, Shuman S. Yeast mRNA cap methyltransferase is a 50-kDa protein encoded by an essential gene. Mol Cell Biol. 1995;15:4167–4174. doi: 10.1128/mcb.15.8.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mao X, Shuman S. Intrinsic RNA (guanine-7) methyltransferase activity of the vaccinia virus capping enzyme D1 subunit is stimulated by the D12 subunit: identification of amino acid residues in the D1 protein required for subunit association and methyl group transfer. J Biol Chem. 1994;269:24472–24479. [PubMed] [Google Scholar]
- 25.McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Shuman S, Bentley D L. 5′ Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated C-terminal domain of RNA polymerase II. Genes Dev. 1997;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Myette J R, Niles E G. Domain structure of the vaccinia virus mRNA capping enzyme: expression in Escherichia coli of a subdomain possessing the RNA 5′ triphosphatase and guanylyltransferase activities and a kinetic comparison to the full-size enzyme. J Biol Chem. 1996;271:11936–11944. doi: 10.1074/jbc.271.20.11936. [DOI] [PubMed] [Google Scholar]
- 27.Niles E G, Christen L. Identification of the vaccinia virus mRNA guanylyltransferase active site lysine. J Biol Chem. 1993;268:24986–24989. [PubMed] [Google Scholar]
- 28.Passarelli A L, Miller L K. Identification of genes encoding late expression factors located between 56.0 and 65.4 map units of the Autographa californica nuclear polyhedrosis virus genome. Virology. 1993;197:704–714. doi: 10.1006/viro.1993.1646. [DOI] [PubMed] [Google Scholar]
- 29.Passarelli A L, Todd J W, Miller L K. A baculovirus gene involved in late gene expression predicts a large polypeptide with a conserved motif of RNA polymerases. J Virol. 1994;68:4673–4678. doi: 10.1128/jvi.68.7.4673-4678.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pena L, Yanez J, Revilla Y, Vinuela E, Salas M L. African swine fever virus guanylyltransferase. Virology. 1992;193:319–328. doi: 10.1006/viro.1993.1128. [DOI] [PubMed] [Google Scholar]
- 31.Senkevich T G, Koonin E V, Bugert J J, Darai G, Moss B. The genome of molluscum contagiosum virus: analysis and comparison with other poxviruses. Virology. 1997;233:19–42. doi: 10.1006/viro.1997.8607. [DOI] [PubMed] [Google Scholar]
- 32.Sheng Z, Charbonneau H. The baculovirus Autographa californica encodes a protein tyrosine phosphatase. J Biol Chem. 1993;268:4728–4733. [PubMed] [Google Scholar]
- 33.Shibagaki Y, Itoh N, Yamada H, Nagata S, Mizumoto K. mRNA capping enzyme: isolation and characterization of the gene encoding mRNA guanylyltransferase subunit from Saccharomyces cerevisiae. J Biol Chem. 1992;267:9521–9528. [PubMed] [Google Scholar]
- 34.Shuman S. Catalytic activity of vaccinia mRNA capping enzyme subunits coexpressed in Escherichia coli. J Biol Chem. 1990;265:11960–11966. [PubMed] [Google Scholar]
- 35.Shuman S. Capping enzyme in eukaryotic mRNA synthesis. Prog Nucleic Acid Res Mol Biol. 1995;50:101–129. doi: 10.1016/s0079-6603(08)60812-0. [DOI] [PubMed] [Google Scholar]
- 36.Shuman S. Origins of mRNA identity: capping enzymes bind to the phosphorylated C-terminal domain of RNA polymerase II. Proc Natl Acad Sci USA. 1997;94:12758–12760. doi: 10.1073/pnas.94.24.12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shuman S, Broyles S, Moss B. Purification and characterization of a transcription termination factor from vaccinia virions. J Biol Chem. 1987;262:12372–12380. [PubMed] [Google Scholar]
- 38.Shuman S, Liu Y, Schwer B. Covalent catalysis in nucleotidyl transfer reactions: essential motifs in Saccharomyces cerevisiae RNA capping enzyme are conserved in Schizosaccharomyces pombe and vaccinia capping enzymes and among DNA ligases. Proc Natl Acad Sci USA. 1994;91:12046–12050. doi: 10.1073/pnas.91.25.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shuman S, Schwer B. RNA capping enzyme and DNA ligase—a superfamily of covalent nucleotidyl transferases. Mol Microbiol. 1995;17:405–410. doi: 10.1111/j.1365-2958.1995.mmi_17030405.x. [DOI] [PubMed] [Google Scholar]
- 40.Shuman S, Surks M, Furneaux H, Hurwitz J. Purification and characterization of a GTP:pyrophosphate exchange activity from vaccinia virions. J Biol Chem. 1980;255:11588–11598. [PubMed] [Google Scholar]
- 41.Takagi T, Moore C R, Diehn F, Buratowski S. An RNA 5′-triphosphatase related to the protein tyrosine phosphatases. Cell. 1997;89:867–873. doi: 10.1016/s0092-8674(00)80272-x. [DOI] [PubMed] [Google Scholar]
- 42.Tsukamoto T, Shibagaki Y, Imajoh-Ohmi S, Murakoshi T, Suzuki M, Nakamura A, Gotoh H, Mizumoto K. Isolation and characterization of the yeast mRNA capping enzyme β subunit gene encoding RNA 5′-triphosphatase, which is essential for cell viability. Biochem Biophys Res Commun. 1997;239:116–122. doi: 10.1006/bbrc.1997.7439. [DOI] [PubMed] [Google Scholar]
- 43.Tsukamoto T, Shibagaki Y, Murakoshi T, Suzuki M, Nakamura A, Gotoh H, Mizumoto K. Cloning and characterization of two human cDNAs encoding the mRNA capping enzyme. Biochem Biophys Res Commun. 1998;243:101–108. doi: 10.1006/bbrc.1997.8038. [DOI] [PubMed] [Google Scholar]
- 44.Upton C, Stuart D, McFadden G. Identification and DNA sequence of the large subunit of the capping enzyme from Shope fibroma virus. Virology. 1991;183:773–777. doi: 10.1016/0042-6822(91)91009-6. [DOI] [PubMed] [Google Scholar]
- 45.Venkatesan S, Gershowitz A, Moss B. Modification of the 5′ end of mRNA: association of RNA triphosphatase with the RNA guanylyltransferase-RNA (guanine-7-) methyltransferase complex from vaccinia virus. J Biol Chem. 1980;255:903–908. [PubMed] [Google Scholar]
- 46.Vos J C, Sasker M, Stunnenberg H G. Vaccinia virus capping enzyme is a transcription initiation factor. EMBO J. 1991;10:2553–2558. doi: 10.1002/j.1460-2075.1991.tb07795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang S P, Shuman S. Structure-function analysis of the mRNA cap methyltransferase of Saccharomyces cerevisiae. J Biol Chem. 1997;272:14683–14689. doi: 10.1074/jbc.272.23.14683. [DOI] [PubMed] [Google Scholar]
- 48.Wang S P, Deng L, Ho C K, Shuman S. Phylogeny of mRNA capping enzymes. Proc Natl Acad Sci USA. 1997;94:9573–9578. doi: 10.1073/pnas.94.18.9573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamada-Okabe T, Shimmi O, Doi R, Mizumoto K, Arisawa M, Yamada-Okabe H. Isolation of the mRNA-capping enzyme and ferric-reductase-related genes from Candida albicans. Microbiology. 1996;142:2515–2523. doi: 10.1099/00221287-142-9-2515. [DOI] [PubMed] [Google Scholar]
- 50.Yang C L, Stetler D A, Weaver R F. Structural comparison of the Autographa californica nuclear polyhedrosis virus-induced RNA polymerase and the three nuclear RNA polymerases from the host, Spodoptera frugiperda. Virus Res. 1991;20:251–264. doi: 10.1016/0168-1702(91)90079-b. [DOI] [PubMed] [Google Scholar]
- 51.Yu L, Martins A, Deng L, Shuman S. Structure-function analysis of the triphosphatase component of vaccinia virus mRNA capping enzyme. J Virol. 1997;71:9837–9843. doi: 10.1128/jvi.71.12.9837-9843.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu L, Shuman S. Mutational analysis of the triphosphatase domain of vaccinia virus mRNA capping enzyme. J Virol. 1996;70:6162–6168. doi: 10.1128/jvi.70.9.6162-6168.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yue Z, Maldonado E, Pillutla R, Cho H, Reinberg D, Shatkin A J. Mammalian capping enzyme complements mutant S. cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA polymerase II. Proc Natl Acad Sci USA. 1997;94:12898–12903. doi: 10.1073/pnas.94.24.12898. [DOI] [PMC free article] [PubMed] [Google Scholar]