Abstract
Carbapenems are last-resort antibiotics used to treat multidrug-resistant bacterial infections. Resistance to carbapenems has been designated as an urgent threat and is increasing in healthcare settings. However, little is still known about the distribution and characteristics of carbapenem-resistant bacteria (CRB) outside of healthcare settings. Here, we surveyed the distribution of CRB in ten diverse freshwater and seawater environments in California, U.S., ranging from San Luis Obispo County to San Bernardino County, combining both direct isolation and enrichment approaches to increase the diversity of isolated CRB. From the locations surveyed, we selected 30 CRB for further characterization. These isolates were identified as members of the genera Aeromonas, Enterobacter, Enterococcus, Paenibacillus, Pseudomonas, Sphingobacterium, and Stenotrophomonas. These isolates were resistant to carbapenems, other β-lactams, and often to other antibiotics (tetracycline, gentamicin, or ciprofloxacin). We also found that nine isolates belonging to the genera Aeromonas, Enterobacter (blaIMI-2), and Stenotrophomonas (blaL1) produced carbapenemases. Overall, our findings indicate that sampling different types of aquatic environments and combining different isolation approaches increase the diversity of the environmental CRB obtained. Moreover, our study supports the increasingly recognized role of natural water systems as an underappreciated reservoir of bacteria resistant to carbapenems and other antibiotics, including bacteria carrying carbapenemase genes.
Keywords: carbapenems, carbapenem-resistant bacteria, carbapenemase, Aeromonas, Pseudomonas, Enterobacter, Enterococcus, Sphingobacterium, Paenibacillus
1. Introduction
Carbapenems are broad-spectrum, last-resort, β-lactam antibiotics used to treat multidrug-resistant infections [1,2,3,4]. They are used primarily to treat infectious bacteria resistant to other β-lactam antibiotics because carbapenems are resistant to hydrolysis by common β-lactamases [1,2,3,5]. Incidents related to carbapenem-resistant bacteria (CRB) have rapidly risen since the emergence of carbapenem-resistant Enterobacteriaceae (CRE) in the 1990s [6,7]. In 1990, there were almost no reported cases of CRE [8]. However, as of 2017, carbapenemase-producing CRE were found in every state of the U.S. [9].
Alongside decreased permeability and increased efflux, one of the most common forms of resistance to carbapenems is through the production of unique β-lactamases called carbapenemases, which are enzymes capable of degrading carbapenems [1,6]. Carbapenemases are found in Ambler classes A, B, and D, and are divided into two families based on their active site [10]. Class A carbapenemases contain a serine in their active site, while class B carbapenemases are metallo-enzymes that contain zinc ions [10]. Class D are also serine-based enzymes, but they are historically distinguished due to their ability to rapidly hydrolyze oxacillin [10,11]. Because carbapenems were originally found to resist hydrolysis by β-lactamases, the emergence of carbapenemases is of significant concern [10,12]. This problem is exacerbated by the fact that genes encoding for these enzymes are often found on genetic mobile elements such as conjugative plasmids [10], which favors their spread.
CRB pose a significant public health challenge. These bacteria are primarily associated with infections acquired in healthcare settings, and their prevalence in such environments is increasing [7,13,14,15,16,17,18,19,20,21,22,23,24,25,26]. In addition to CRE, which have been classified as an urgent threat by the Centers for Disease Control and Prevention (CDC), Acinetobacter baumannii and Pseudomonas aeruginosa strains that exhibit resistance to multiple antibiotics, including carbapenems, have been identified by the CDC as serious threats, often with limited treatment options [27]. However, despite their significant impact on public health and their increasing prevalence in healthcare settings, there is a notable lack of knowledge about the distribution of CRB and carbapenemase genes in the environment, particularly in the United States. Previous efforts to identify these bacteria have primarily focused on healthcare facilities or closely associated areas such as hospital wastewater [28,29,30,31,32]. However, recent research from Europe, Africa, and Asia has revealed the presence of carbapenem-resistant bacteria and genes in various environmental samples, including freshwater [33,34,35,36,37,38,39,40]. In the US, only two studies have examined CRB in water environments. First, a study conducted between 1999 and 2001 found CRB in seven out of sixteen rivers in the Midwest [41,42]. More recently, a study from our group examined the distribution and characteristics of CRB in ponds and lakes in the Los Angeles, CA area [43]. This study, together with a second study from our group investigating CRB in Southern California soils [44], has revealed that CRB and carbapenemase genes are more widespread than previously thought. However, further studies are still needed to better understand the spread, diversity, and characteristics of CRB in the environment in the US.
Here, we have investigated the frequency, distribution, and characteristics of CRB in freshwater and seawater aquatic environments in the broader region of the Central Coast and Southern California, using different isolation approaches to increase the diversity of the environmental CRB recovered. Overall, we have found CRB in all tested aquatic environments, although with variable abundance. We have also identified 30 CRB isolates, characterized their antibiotic resistance profiles, and found that all Gram-negative isolates were resistant to at least one non-carbapenem antibiotic, including seven isolates that were resistant to all but one of the antibiotics tested. We have also identified nine isolates that produce carbapenemases.
2. Materials and Methods
2.1. Collection of Water Samples and Isolation of Carbapenem-Resistant Bacteria (CRB)
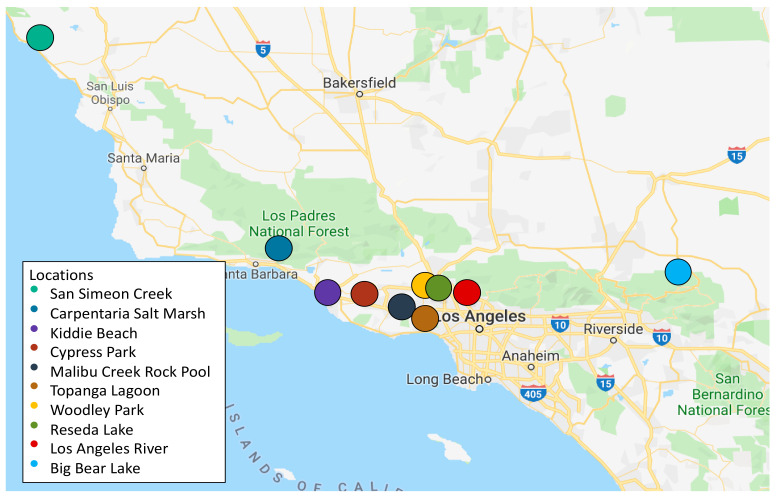
Between April 2018 and March 2019, we collected thirty water samples from ten different locations (Figure 1) to determine the abundance of CRB in these samples and isolate CRB for further characterization. Of the ten locations selected, seven had not been previously studied and three had been studied in our earlier experiments using a different isolation approach [43] and were resampled for comparison. The 10 locations surveyed here included a larger geographical area than our previous study and stretched from San Simeon, California to Big Bear Lake, California. Locations were selected considering their overall sanitation, proximity to residential areas, and/or proximity to agriculture. For example, Kiddie Beach in Oxnard, California is located in Ventura County not far from the Port Hueneme Naval Base. This particular beach is part of a sub-watershed for the western portion of the city of Oxnard, which includes the Oxnard west drain, residential runoff from both housing and the harbor, and runoff from the naval base [45]. In the years prior to sampling, the beach failed health and sanitation checks due to an overabundance of bacteria, as well as high levels of toxic metals such as zinc and lead [45,46]. While the outbreak has been controlled, bacterial levels were on the rise again in March 2018 [47], making it a prime sampling location. Likewise, all other locations were sampled based on their environmental health reports and history of pollution.
Figure 1.
Map of the locations sampled in this study for CRB.
For each location, four liters of surface water were collected in sterile vessels that were instantly closed and then transported to the lab immediately for testing under aseptic conditions. From each sample, 100 μL of water were directly plated into MacConkey agar (Fisher Scientific, Hampton, NH, USA) to determine the total count of Gram-negative bacteria, or MacConkey agar supplemented with 2 μg/mL meropenem (Ark Pharm, Inc., Arlington Heights, IL, USA) (MAC + M2) to determine the count of carbapenem-resistant Gram-negative bacteria. In addition, another 10 μL of water were spot-plated in MacConkey and MAC + M2 media after undergoing serial dilutions from 100 to 10−4 to determine total and CRB counts in samples with large bacterial counts. The plates were then grown at 37 °C for 18–24 h in aerobic conditions. Meropenem was the carbapenem used for the selection of CRB due to its greater effectiveness against Gram-negative bacteria [1], and because these bacteria are the main concern in healthcare settings. The concentration of meropenem used was the equivalent of “Intermediate” (or half the concentration for Resistant) of the Clinical and Laboratory Standards Institute (CLSI) minimum inhibitory concentration (MIC) break-point (4 μg/mL) for Enterobacteriaceae [48]. This concentration was selected to maximize the isolation of environmental CRB, especially when using a growth medium that is highly selective such as the MacConkey medium. It was also half of the concentration used in our previous study [43].
After initial collection and plating, the rest of the water sample was split into two-liter aliquots, each of which was filtered using a Stericup and Steritop vacuum-driven disposable bottle-top filter with a size of 0.22 μm, in order to concentrate the bacteria present in each aliquot. One filter was plated directly onto MacConkey agar supplemented with meropenem at 2 μg/mL and the other filter was placed into a BLCVM9 broth supplemented with 2 μg/mL of meropenem. Both the broth and the plate were grown at 37 °C for 24 h in aerobic conditions. BLCVM9 broth was developed for this study and contains 0.15% bile salts, 1% lactose, 0.001% crystal violet, and 1× M9 salts (Fisher Scientific). It was designed to primarily enrich enteric and Gram-negative bacteria while limiting the growth of Stenotrophomonas maltophilia. This bacterium is a non-lactose-fermenter and was over-represented in our previous study [43] because it is an abundant environmental opportunistic pathogen that is usually carbapenem-resistant [49]. After enrichment in BLCVM9 broth, 10 μL of culture were transferred to a MAC + M2 plate, and another 10 μL to a Mueller–Hinton agar plate supplemented with 2 μg/mL of meropenem and 40 μg/mL of X-gal (5-bromo-4-chloro-3-indolyl-β-D-glucopyranoside; to identify lactose-fermenters) (MH + X-gal + M2) to be struck for isolation. Representative colonies were restruck on MAC + M2 or MH + X-gal + M2, respectively, to confirm them as CRB, prioritizing lactose fermenters (colonies that were pink on MAC + M2 or blue on MH + X-gal + M2) when identified.
2.2. Selection, Identification, and Characterization of CRB Isolates
Among all isolated colonies obtained for each sample and isolation approach, we selected for further characterization only representative colonies that were phenotypically distinct to avoid selecting duplicates of the same isolate type from the same sample. Therefore, the number of selected isolates of each genus is not proportionally representative of their abundance in each sample. Overall, we selected a total of 30 distinct CRB from the 10 samples analyzed using these criteria.
The 30 CRB isolates selected for further characterization were first identified by Gram staining and PCR amplification of their 16S rRNA genes, followed by Sanger sequencing and BLAST analysis as previously described by Harmon et al. [43]. The oxidase test was performed as previously described [43] to further distinguish between closely related S. maltophilia, which is oxidase negative, and Pseudomonas species, most of which are oxidase positive [50]. After initial identification by 16S rRNA gene sequencing and BLAST analysis, the 16S sequences obtained were further analyzed by constructing phylogenetic trees based on the genus of the isolate, as determined by BLAST [51], and other sequences for the same genus found in GenBank (https://www.ncbi.nlm.nih.gov/genbank/ accessed on 13 April 2024). All of the sequences were aligned using ClustalW (https://www.genome.jp/tools-bin/clustalw accessed on 13 April 2024) [52] pairwise alignment first, and then ClustalW multiple alignment, both with the parameters set to a Gap open penalty of 15 and a Gap extension penalty of 6.66, and with the Weight Matrix set to IUB. The phylogenetic trees were constructed using Mega7 software (v7.0.26) [53], the Neighbor-Joining method, and the Jukes–Cantor statistical method, using 500 Bootstraps per tree.
The antibiotic susceptibility profile for each CRB isolate was determined using the Kirby–Bauer method [54] and the reference strain E. coli ATCC 25922 as a quality control, as we have previously described [43], and using three replicates per isolate and antibiotic. Gram-negative bacteria were tested using meropenem (10 μg), imipenem (10 μg), amoxicillin/clavulanic acid (20 μg/10 μg), cefotaxime (30 μg), ciprofloxacin (5 μg), gentamicin (10 μg), and tetracycline (30 μg) disks. The disks tested for Gram-positive bacteria included carbapenems: meropenem (10 μg), imipenem (10 μg), ertapenem (10 μg), and doripenem (10 μg); other β-lactams: amoxicillin/clavulanic acid (20 μg/10 μg); ampicillin (10 μg); vancomycin (30 μg); ciprofloxacin (5 μg), and tetracycline (30 μg) disks. For the vancomycin test, plates were incubated for a complete 24 h as recommended by the CLSI [48]. All antibiotic disks were purchased from Becton Dickinson (Franklin Lakes, NJ, USA). To determine whether isolates were susceptible, intermediate, or resistant to an antibiotic, we compared the average zone of inhibition diameter measurements for each isolate with the CLSI zone diameter breakpoint values [48] when available, or the European Committee on Antimicrobial Susceptibility Testing (EUCAST) values [55] as an alternative. For taxa in which the zone diameter breakpoints were not provided by CLSI or EUCAST, we used the CLSI Enterobacteriaceae breakpoint values [48].
2.3. Identification of Carbapenemase-Producing CRB Isolates
To identify carbapenemase-producing CRB isolates, we utilized the CarbaNP assay as previously described [43,44] to detect carbapenem hydrolysis. Briefly, we performed the CarbaNP assay [56,57,58] following the CLSI guidelines [48] and using 6 mg/mL of meropenem. CarbaNP-positive isolates were then confirmed using the Carbapenem Inactivation Method (mCIM) [59] in accordance with CLSI guidelines [48]. For the CarbaNP test, isolates positive for carbapenemase production appear as yellow because hydrolysis of meropenem lowers the pH and changes the color of the phenol red indicator [56,57,58]. Confirmation of carbapenemase production using the mCIM method involved observing a zone of inhibition of between 6 and 15 mm for E. coli ATCC 25922 when grown in the presence of a meropenem disk previously incubated for 4 h with the isolate being tested.
For CRB isolates confirmed as carbapenemase producers by the mCIM method, we used the EDTA-Carbapenem Inactivation Method (eCIM) to determine if the detected carbapenemase was a metallo-β-lactamase [48]. This method is similar to the mCIM, with the caveat of adding EDTA to the isolate-meropenem disk co-culture prior to the 4 h incubation to chelate metal ions and inactivate metallo-β-lactamases. As a result, metallo-β-lactamase carbapenemase producers no longer inactivate meropenem. Confirmation of metallo-β-lactamase production involved observing a zone of inhibition that increased by more than 5 mm compared to that obtained in the mCIM test.
2.4. Identification of Carbapenemase Genes
PCR detection of the IMI-2 carbapenemase gene (blaIMI-2) in carbapenemase-producing Enterobacter isolates was performed using the primers and program described by Harmon et al. [43]. PCR detection of the L1 carbapenemase gene (blaL1) in carbapenemase-producing Stenotrophomonas sp. isolates was performed using the primers and conditions described by Henriques et al. [40] to amplify blaL1 as previously described [43]. For each PCR, DNA-grade water was used as a non-template control, E. coli BW25113 as a negative control, and strains with each bla gene as positive controls as previously described [43].
Aeromonas veronii has been found to be resistant to carbapenems by producing the CphA, ImiS, or VIM-2 metallo-β-lactamases [33,60,61,62,63,64,65,66]. Thus, the following specific primers were used to detect the genes for these carbapenemases in our A. veronii CRB isolates. For blaCphA, the cphA Forward (5′-GGA TGA AGT GTG GAT TGG CCG-3′) and cphA Reverse (5′-TTA TGA CTG GGG TGC GGC-3′), which amplify 752 of 765 bp of the blaCphA gene (X57102), were designed. For blaImiS, the primers imiS Forward (5′-ATG ATG AAG GGT TGG ATA AAG T-3′) and imiS Reverse (5′-TTA TGA TTG TGA AGC CGC CT-3’) were designed to amplify 786 out of 922 bp of the blaImiS gene (NG_050415). For blaVIM-2, the primers designed by Belotti et al. [67] vim-2 Forward (5′-GAT GGT GTT TGG TCG CAT A-3′) and vim-2 Reverse (5′-CGA ATG CGC AGC ACC AG-3′) were used to amplify 390 out of the 801 bp of the blaVIM-2 gene. The PCR reaction mixes had a total volume of 50 μL per isolate, consisting of DreamTaq buffer, 0.2 mM dNTPs, 0.5 μM of each primer, 1.25 units of DreamTaq Polymerase, and 5 μL of template DNA (1 colony resuspended in 50 μL of DNA-grade water). The thermocycler programs used were: (1) for blaCphA, one cycle of 95 °C for 5 min; 35 cycles of 95 °C for 30 s, 58 °C for 40 s, and 72 °C for 50 s; one cycle of 72 °C for 7 min, and finally 4 °C for infinite; (2) for blaImiS, one cycle of 95 °C for 5 min; 35 cycles of 95 °C for 30 s, 53 °C for 30 s, and 72 °C for 60 s; one cycle of 72 °C for 10 min; and finally 4 °C for infinite; (3) for blaVIM-2, one cycle of 95 °C for 10 min; 35 cycles of 95 °C for 30 s, 52 °C for 40 s, and 72 °C for 50 s; then 72 °C for 5 min; and 4 °C for infinite time.
All carbapenemase gene PCR products were visualized by electrophoresis in a 1% agarose gel supplemented with 10 μL of 1:10,000 ethidium bromide prior to their submission to Laragen Inc. (Culver City, CA, USA) for Sanger sequencing. The sequences obtained were analyzed using LALIGN (SIB ExPASy: https://embnet.vital-it.ch/software/LALIGN_form.html accessed on 13 April 2024) to compare and align our sequences to reference sequences blaL1 (GenBank Accession number NG_047502) and blaIMI-2 (GenBank Accession number DQ173429).
3. Results and Discussion
3.1. Distribution, Abundance, and Isolation of Carbapenem-Resistant Bacteria (CRB) in Diverse Aquatic Environments
The rise of CRB in healthcare settings during the past 30 years is a global threat [6,7,13,14,15,16,17,18,19,20,21,22,23,24,25,26]. However, the role that the environment plays as a reservoir for CRB is not entirely understood, especially in the United States. In a previous study, we have identified CRB in a limited-scope survey of ponds and lakes in the Los Angeles (CA) area [43]. Therefore, our first goal here was to expand this survey to include a larger geographical area in South and Central California (ranging from San Simeon Creek in the north to Big Bear Lake in the east), and other types of aquatic environments such as rivers, marshes, estuaries, and beaches (Figure 1 and Table 1). The 10 locations sampled were selected because of their proximity to the community around them, long-standing history with pollution, or a history of bacterial outbreaks. Overall, we detected CRB in all aquatic environments tested, although with a low abundance (<10 to 20 CFU/mL), except for the Carpinteria Salt Marsh (5.2 × 103 CFU/mL, which is similar to the total bacterial counts obtained for this sample; Table 1). We speculate that the Thomas fire in December 2017, followed by a rain mudslide in January 2018, contributed to the higher total and CRB counts in this location. For the rest of the sampled locations, the low abundance of CRB found in this study in most samples is comparable to those in our previous study [43], and those previously found in Portuguese rivers [33].
Table 1.
Summary of locations sampled in this study and their total bacterial and CRB counts.
| Sample | Location | Collection Date (D/M/Y) | Location Type | Coordinates | Total Bacteria (CFU/mL) | CRB (CFU/mL) |
|---|---|---|---|---|---|---|
| C | Carpinteria Salt Marsh a | 24/4/18 | Marine estuary | 34.401244, 119.5401337 | 5.1 × 103 | 5.2 × 103 |
| K | Kiddie Beach b | 7/5/18 | Beach harbor | 34.1601144, 119.2235778 | ≤10 | ≤10 |
| LAR | LA River c | 4/6/18 | Urban river | 34.156572, 118.291143 | 2.2 × 103 | ≤10 |
| CP | Cypress Park d | 18/6/18 | Natural creek | 34.167243, 118.962229 | 5.2 × 102 | ≤10 |
| SS | San Simeon Creek e | 24/6/18 | Coastal wetlands | 35.5971832, 121.1218039 | 1 × 102 | ≤10 |
| TL | Topanga Lagoon f | 23/7/18 | Natural pool | 34.038536, 118.583085 | 7 × 102 | ≤10 |
| BBL | Big Bear Lake g | 21/8/18 | Natural freshwater lake | 34.245278, 116.917086 | 8 × 101 | ≤10 |
| WL | Woodley Lake h | 18/3/19 | Reclaimed water | 34.175326, 118.472829 | 3.1 × 103 | 10 |
| RL | Reseda Lake i | 18/3/19 | Artificial lake | 34.188714, 118.534383 | 2.4 × 102 | 20 |
| MRP | Malibu Rock Pool j | 28/3/19 | Natural pool | 34.096555, 118.729879 | 1.1 × 102 | ≤10 |
a Estuary fed by the Franklin, Santa Monica, and San Simeon creeks. b Kiddie Beach (Oxnard marina) receives residential and military runoff water. c The Los Angeles River runs from Simi Valley, CA to Long Beach, CA, bisecting the city. The riverbed alternates between cement and natural dirt. Our sample was collected at Glendale Narrows, which is an area of restoration where cement was removed to expose the natural riverbed. d Our sample was collected at Arroyo Conejo Creek, which spans the entire Conejo Valley and is the only watershed for the entire valley. The area is highly residential. e The creek is a natural wetland that receives agricultural and residential runoff. f Lagoon that forms behind a beach berm after seasonal rains and is fed by the Topanga Creek. g Natural reservoir for the San Bernardino Mountains, it is filled solely by snow runoff. h Wildlife wetland and bird sanctuary that is filled with reclaimed water from the Tillman Water Reclamation Plant (DCTWRP). i Asphalt lined urban lake with potable water that is treated with an algicide. j Natural pool filled with rain runoff from the Santa Monica Mountains.
Our second goal was to gain a deeper understanding of the diversity of CRB present in these environments. In our previous study, we had used MacConkey with 4 μg/mL meropenem media as our primary isolation media, and found that all CRB isolates were Gram-negatives, especially Stenotrophomonas sp. (63% of the isolates) and Pseudomonas sp. (22% of the isolates), followed by Enterobacter sp., Aeromonas sp., and Cupriavidus sp. isolates [43]. In the present study, we employed MacConkey agar with 2 μg/mL meropenem, a less stringent concentration of carbapenem, and added a second isolation approach based on first enriching CRB using BLCVM9 broth supplemented with 2 μg/mL of meropenem prior to streaking samples in both MacConkey with 2 μg/mL meropenem and Mueller–Hinton with 2 μg/mL meropenem. Using this approach, only three isolates (10%) out of the thirty CRB characterized were Stenotrophomonas sp., and new CRB types were identified, including Gram-positives (Enterococcus sp. and Paenibaciullus sp.) and Sphingobacterium sp. isolates (Figure 2, Figure 3, Figure 4, Figure 5, Figure 6, Figure 7 and Figure 8). These results indicate that, besides sampling different types of aquatic environments, the additional isolation approaches employed in this study may have increased the diversity of CRB obtained by limiting Stenotrophomonas sp. that potentially outgrew and/or outcompeted other bacteria in our previous study. This interpretation is strongly supported by the fact that seven out of the eight Gram-positive CRB isolates characterized in this study were obtained using our novel enrichment approach.
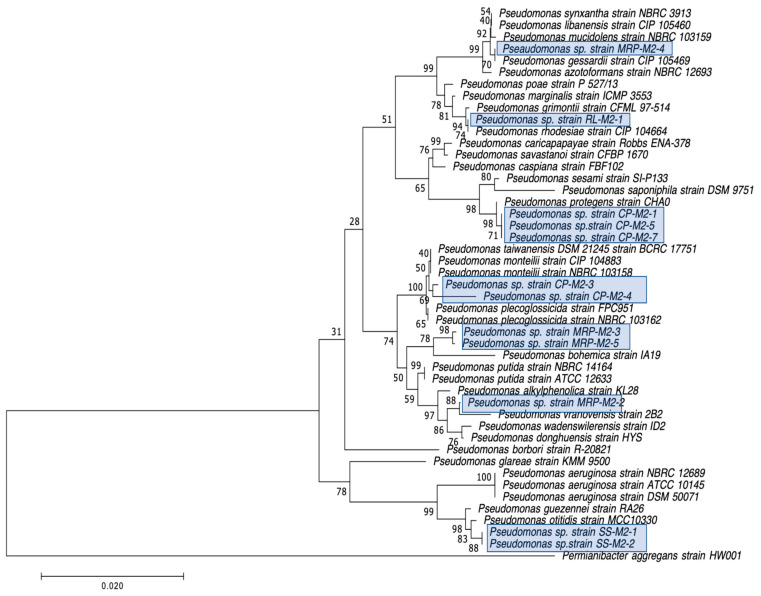
Figure 2.
Phylogenetic tree showing the relatedness between the 16S rRNA gene sequences of the carbapenem-resistant Pseudomonas sp. isolates obtained in this study (indicated in blue boxes) and those from Pseudomonas sp. isolates from previous studies obtained from GenBank. The scale bar at the bottom of the tree represents the number of nucleotide substitutions per site.
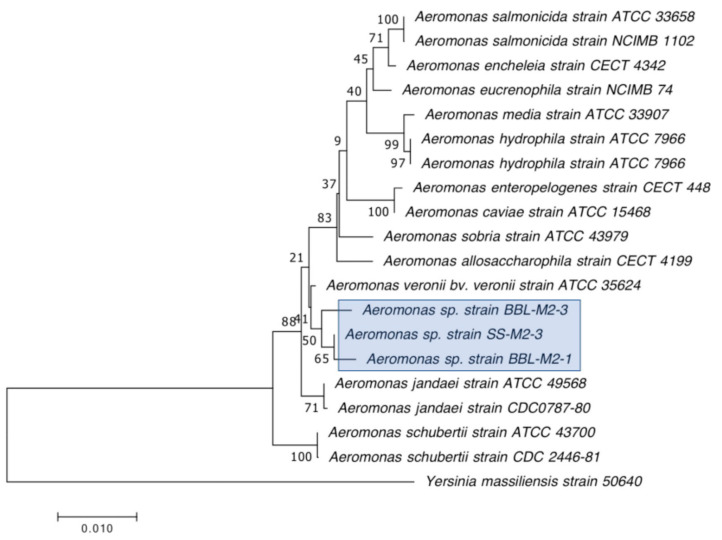
Figure 3.
Phylogenetic tree showing the relatedness between the 16S rRNA gene sequences of the carbapenem-resistant Aeromonas sp. isolates obtained in this study (indicated in blue boxes) and those from Aeromonas sp. isolates from previous studies obtained from GenBank. The scale bar at the bottom of the tree represents the number of nucleotide substitutions per site.
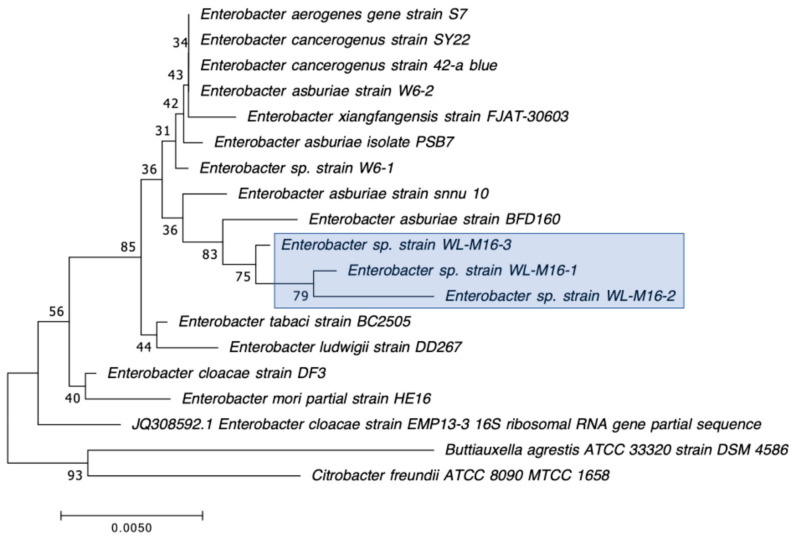
Figure 4.
Phylogenetic tree showing the relatedness between the 16S rRNA gene sequences of the carbapenem-resistant Enterobacter sp. isolates obtained in this study (indicated in blue boxes) and those from Enterobacter sp. isolates from previous studies obtained from GenBank. The scale bar at the bottom of the tree represents the number of nucleotide substitutions per site.
Figure 5.
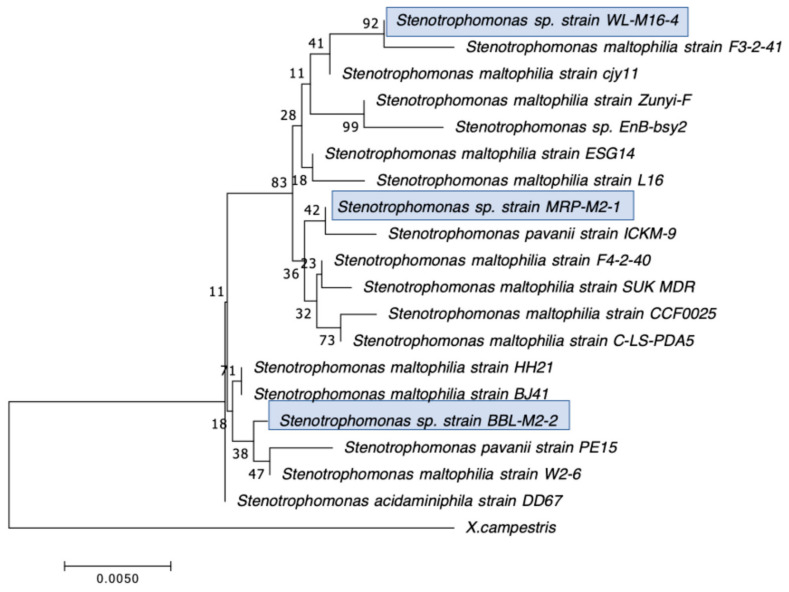
Phylogenetic tree showing the relatedness between the 16S rRNA gene sequences of the carbapenem-resistant Stenotrophomonas sp. isolates obtained in this study (indicated in blue boxes) and those from Stenotrophomonas sp. isolates from previous studies obtained from GenBank. The scale bar at the bottom of the tree represents the number of nucleotide substitutions per site.
Figure 6.
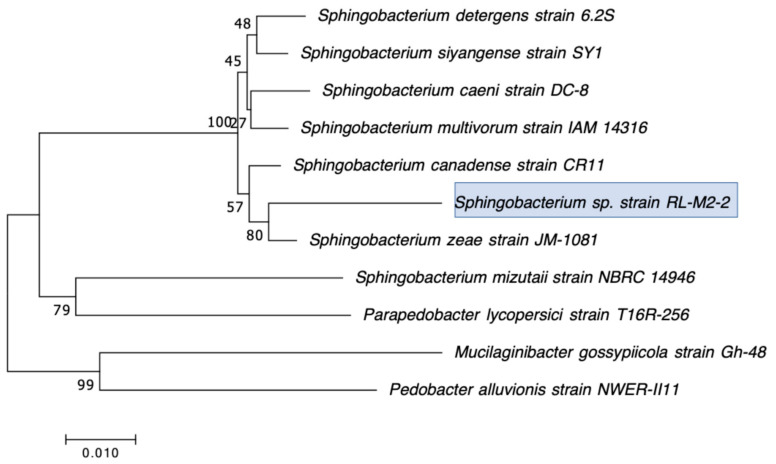
Phylogenetic tree showing the relatedness between the 16S rRNA gene sequences of the carbapenem-resistant Sphingobacterium sp. isolates obtained in this study (indicated in blue boxes) and those from Sphingobacterium sp. isolates from previous studies obtained from GenBank. The scale bar at the bottom of the tree represents the number of nucleotide substitutions per site.
Figure 7.
Phylogenetic tree showing the relatedness between the 16S rRNA gene sequences of the carbapenem-resistant Enterococcus sp. isolates obtained in this study (indicated in blue boxes) and those from Enterococcus sp. isolates previous studies obtained from GenBank. The scale bar at the bottom of the tree represents the number of nucleotide substitutions per site.
Figure 8.
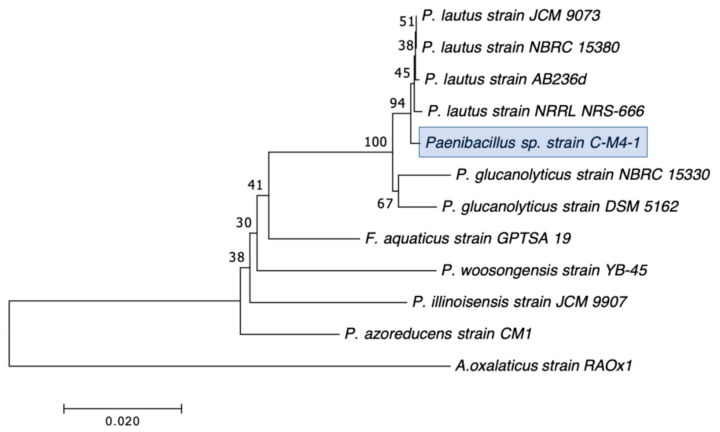
Phylogenetic tree showing the relatedness between the 16S rRNA gene sequences of the carbapenem-resistant Paenibacillus sp. isolates obtained in this study (indicated in blue boxes) and those from Paenibacillus sp. isolates previous studies obtained from GenBank. The scale bar at the bottom of the tree represents the number of nucleotide substitutions per site.
We preliminarily identified our 30 selected CRB isolates as three Aeromonas veronii, three Enterobacter asburiae, five Pseudomonas spp., one Pseudomonas plecoglossicida, three Pseudomonas putida, one Pseudomonas fluorescens, one Pseudomonas rhodesiae, one Sphingobacterium siyangensis, three Stenotrophomonas maltophilia, two Enterococcus lactis, two Enterococcus hirae, three Enterococcus faecium, and one Paenibacillus lautus (Figure 2, Figure 3, Figure 4, Figure 5, Figure 6, Figure 7 and Figure 8). Pseudomomas sp. isolates were the most abundant and widespread, comprising 12 (40%) of the 30 isolates. Prevalence of Pseudomonas sp. is expected from aquatic environments because Pseudomonas spp. are ubiquitous in the environment, being abundant in water, soil, and on plants [68]. Stenotrophomonas sp. is also widespread around the world, being isolated from numerous water sources [49]. Carbapenem-resistant E. asburiae have been previously found in aquatic environments, including one river in Portugal [33], four rivers in the midwestern US [41], and also in our previous study in a sample from Woodley Lake in Los Angeles, CA [43]. Other Gram-negatives identified in the present study included Aeromonas veronii and Sphingobacterium syangensis. Aeromonas veronii is a common aquatic bacterium that can exhibit intrinsic resistance to carbapenems [61]. In contrast, Sphingobacterium sp. resistant to carbapenems have not been previously reported in water environments in the U.S. to our knowledge. Sphingobacterium syangensis is isolated predominantly from soil [69], and members of the genus Sphingobacterium have been associated with pulmonary infections in cystic fibrosis patients [70]. Different species of Sphingobacterium sp. have been shown to possess varying levels of resistance to β-lactams (including carbapenems) and aminoglycosides [70].
Regarding the Gram-positive CRB isolated in this study, seven out of eight were Enterococcus sp., and one isolate was preliminarily identified as Paenibacillus lautus (Figure 7 and Figure 8). Interestingly, no Gram-positives were identified in our previous water study [43], although we found Enterococcus species in our survey for CRB in soil [44], which usually has a larger bacterial load. Our findings in this study suggest that the broader type of ecosystems sampled here, and especially the addition of an enrichment step (seven out of the eight Gram-positive CRB were obtained from the sub-samples subject to enrichment), contributed to the better isolation of Gram-positive CRB. Out of these, Enterococcus faecium (three isolates), all from Kiddie Beach in Oxnard, CA, was the most abundant. E. faecium is a human commensal organism associated with the gut microbiota [71], and thus its presence might indicate recent fecal contamination in this location. However, enterococci are also commonly found in marine environments, especially on sandy beaches due to their ability to form biofilms [71]. In recent years, enterococci, especially Enterococcus faecium, have been associated with nosocomial infections, including urinary tract infections, endocarditis, and bacteremia [72]. Thus, the isolation of carbapenem-resistant E. faecium in Kiddie Beach is a public health concern. We also found two Enterococcus hirae and two Enterococcus lactis. Interestingly, E. hirae has the ability to reduce copper toxicity in the environment, which is essential given the widespread use of copper in agricultural and urban settings [73]. E. lactis is associated with the food industry because it produces other lactic organic acids that act as biological preservatives [74]. However, Enterococcus lactis is not generally associated with environmental isolation, especially in aquatic environments [74]. Lastly, Paenibacillus lautus has been associated with both environmental and nosocomial isolation [75,76]. In the environment, P. lautus is considered an opportunistic pathogen isolated from the gut microbiota of ticks that can be transmitted to humans through tick bites, causing bacteremia [75]. In clinical settings, P. lautus was isolated from blood and abscesses in a hospital in Madrid, suggesting that it can be a nosocomial opportunistic pathogen [76].
3.2. Characterization of the Antibiotic Susceptibility Profile of Carbapenem-Resistant Bacteria (CRB) from Diverse Aquatic Environments
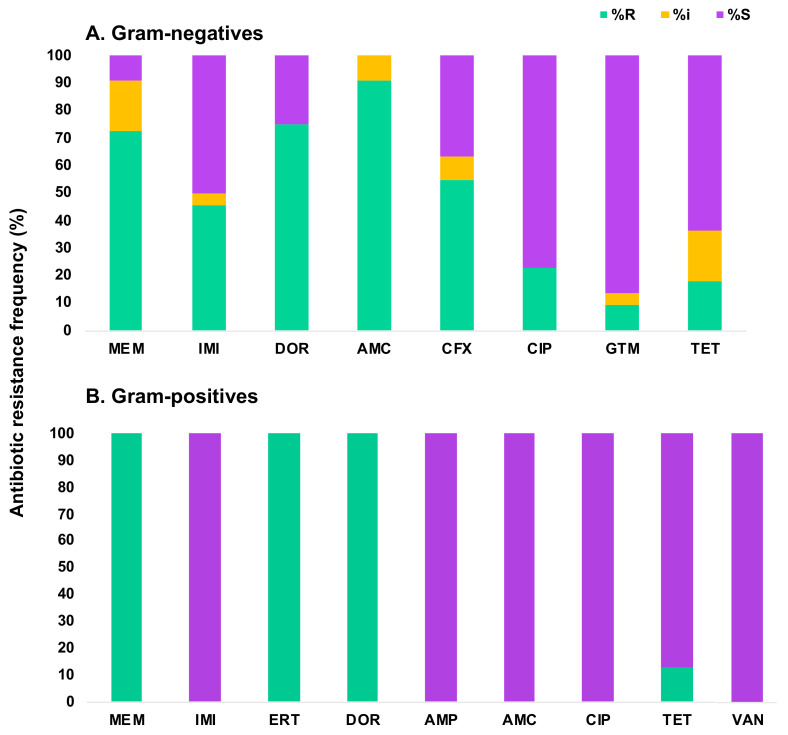
We next characterized the antibiotic susceptibility profiles of the 30 CRB isolates identified in this study (Table 2 and Table 3; Figure 9). Gram-negative bacteria testing included two carbapenems (meropenem and imipenem) and five non-carbapenem antibiotics (amoxicillin plus clavulanic acid, cefotaxime, ciprofloxacin, gentamicin, and tetracycline). These antibiotics were selected because they are commonly used to treat Gram-negative pathogens [48]. Overall, 20 out of the 22 selected Gram-negative CRB from all sampled locations were resistant or intermediate to at least one carbapenem, whereas two Aeromonas sp. isolates had zones of inhibition that were slightly above the intermediate threshold using the Enterobacteriaceae cutoffs for the carbapenems tested, and thus were scored as sensitive to carbapenems. However, both isolates would have been scored as intermediate for meropenem if we had used the Pseudomonas sp. EUCAST cut-off values [55]. Of the Gram-negative isolates characterized, 72.7% were resistant and 18.2% were intermediate to meropenem. In contrast, 45.5% were resistant to imipenem, predominately from the genera Stenotrophomonas and Enterobacter, and 4.5% were intermediate. Lastly, doripenem was tested against all Pseudomonas sp. isolates because it has shown potent inhibitory effects against P. aeruginosa [77]. We found that 75% of our Pseudomonas sp. isolates were resistant to this carbapenem. Of the non-carbapenem antibiotics, 90.9% of isolates showed resistance to amoxicillin plus clavulanic acid, and 9.1% were intermediate. Cefotaxime resistance was found in 54.5% of isolates, and 9.1% were intermediate. The isolates resistant to cefotaxime were identified as either Pseudomonas or Stenotrophomonas species. Only five isolates (all Pseudomonas sp., representing 22.7% of the tested Gram-negative CRB) were resistant to ciprofloxacin. For gentamicin, only 13.6% of the Gram-negative isolates tested, all of them Stenotrophomonas sp., were resistant or intermediate. Finally, 36.4% of the Gram-negatives CRB tested, all belonging to either the Pseudomonas or Stenotrophomonas genera, were resistant or intermediate to tetracycline (Table 2; Figure 9A). Interestingly, about one-third of the Gram-negative CRB characterized displayed a broad and concerning multidrug-resistant phenotype, being resistant or intermediate to all but one of the antibiotics tested.
Table 2.
Gram-negative CRB water isolates identified and characterized in this study.
| Isolate a | Closest Species Identified by BLAST of 16S rRNA Gene | CarbaNP c | CP d | Inhibition Zone (Diameter in mm) b | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MEM | IMI | DOR | AMC | CFX | CIP | GTM | TET | ||||
| CP-M2-1 | Pseudomonas sp. 18 | − | N/D | 0 | 16.7 | 16.7 | 0 | 5 | 22.6 | 18.3 | 6 |
| CP-M2-3 | Pseudomonas plecoglossicida | − | N/D | 5.3 | 27 | 19 | 0 | 10 | 25.3 | 18.3 | 14 |
| CP-M2-4 | Pseudomonas putida | − | N/D | 0 | 26 | 14.5 | 0 | 4.3 | 22.7 | 21.3 | 12 |
| CP-M2-5 | Pseudomonas spp. GC04 | − | N/D | 0 | 28.3 | 20.8 | 0 | 3.7 | 26 | 26 | 12.3 |
| CP-M2-7 | Pseudomonas sp. CHZYR63 | − | N/D | 0 | 19 | 16.3 | 0 | 0 | 23.3 | 17 | 7 |
| SS-M2-1 | Pseudomonas sp. J4AJ | − | N/D | 18.7 | 24.7 | 20.6 | 10 | 25.3 | 38.8 | 24.7 | 25.3 |
| SS-M2-2 | Pseudomonas sp. P7 | − | N/D | 9.7 | 27.3 | 18.4 | 5.7 | 25.3 | 33 | 23.7 | 16 |
| SS-M2-3 | Aeromonas veronii | + | UNK | 7 | 8 | N/D | 8 | 35 | 39 | 25 | 25.3 |
| BBL-M2-1 | Aeromonas veronii | + | UNK | 24.7 | 20.7 | N/D | 15 | 36.7 | 36.7 | 20.3 | 27.3 |
| BBL-M2-2 | Stenotrophomonas maltophilia | + | bla L1 | 0 | 0 | N/D | 0 | 0 | 26.7 | 0 | 11.7 |
| BBL-M2-3 | Aeromonas veronii | + | UNK | 23.5 | 24.7 | N/D | 9 | 34 | 31.8 | 17.8 | 27 |
| MRP-M2-1 | Stenotrophomonas maltophilia | + | bla L1 | 0 | 0 | N/D | 0 | 0 | 26 | 7.3 | 10.7 |
| MRP-M2-2 | Pseudomonas sp. W15Feb40A | − | N/D | 19.8 | 30.7 | 28.5 | 3.3 | 15.3 | 24.7 | 22.8 | 16 |
| MRP-M2-3 | Pseudomonas putida | − | N/D | 17 | 29.7 | 27.3 | 0 | 15 | 31.3 | 23.3 | 19.3 |
| MRP-M2-4 | Pseudomonas fluorescens | − | N/D | 19.7 | 21 | 21 | 0 | 4.7 | 35.7 | 32 | 34 |
| MRP-M2-5 | Pseudomonas putida | − | N/D | 12 | 25.3 | 22 | 0 | 13 | 28.7 | 23.7 | 17.3 |
| RL-M2-1 | Pseudomonas rhodesiae | − | N/D | 0 | 13.5 | 13 | 0 | 3 | 36 | 30 | 21.7 |
| RL-M2-2 | Sphingobacterium siyangensis | − | N/D | 21.2 | 15.8 | N/D | 13.7 | 26 | 29.7 | 15 | 24.5 |
| WL-M2-1 | Enterobacter asburiae | + | bla IMI-2 | 2.2 | 0 | N/D | 0 | 32 | 35.7 | 21.3 | 23.3 |
| WL-M2-2 | Enterobacter asburiae | + | bla IMI-2 | 2.2 | 0 | N/D | 0 | 32.3 | 35.7 | 21.3 | 23.3 |
| WL-M2-3 | Enterobacter asburiae | + | bla IMI-2 | 2.2 | 0 | N/D | 0 | 31.7 | 36 | 21.7 | 24 |
| WL-M2-4 | Stenotrophomonas maltophilia | + | bla L1 | 0 | 0 | N/D | 0 | 0 | 26.7 | 13.3 | 10.8 |
a Isolate number: in regular font, isolates obtained directly using MacConkey-meropenem medium; in bold, isolates enriched in BLCVM9-meropenem medium before streaking in MacConkey-meropenem medium; in bold and italics, isolates enriched in BLCVM9-meropenem medium before streaking in Mueller–Hinton-meropenem medium. The initial(s) before the first dash in the isolate designation indicates the isolation location (Table 1). b Resistant (green), Intermediate (yellow), Susceptible (purple), based on the CLSI [48] or EUCAST [55] (for Pseudomonas spp.) breakpoint values of the diameters of the zone of inhibitions of each antibiotic. For genera that did not have diameter breakpoint values by CLSI or EUCAST, the Enterobacteriaceae cutoff values were used. N/D indicates that it was not tested. MEM (meropenem), IMI (imipenem), DOR (doripenem), AMC (amoxicillin plus clavulanic acid), CFX (cefotaxime), CIP (Ciprofloxacin), GTM (Gentamicin), TET (Tetracycline). c “−” and “+” indicate a negative or positive result, respectively, for the CarbaNP test. d Carbapenemase gene identified by PCR and sequencing. UNK (unknown) indicates lack of detection of a carbapenemase gene by PCR using blacphA, blaImiS, or blaVIM-2 primers.
Table 3.
Gram-positive CRB water isolates identified and characterized in this study.
| Isolate a | Closest Species Identified by BLAST of 16S rRNA Gene | CarbaNP c | Inhibition Zone (Diameter in mm) b | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MEM | IMI | ERT | DOR | AMP | AMC | CIP | TET | VAN | |||
| C-M4-1 | Paenibacillus lautus | − | 5 | 25 | 0 | 12.3 | 23.3 | 26.7 | 22.3 | 34.3 | 26.3 |
| K-M2-1 | Enterococcus lactis | − | 2.7 | 23.7 | 2.3 | 11.7 | 25.7 | 31.3 | 23.6 | 30 | 24.3 |
| K-M2-2 | Enterococcus faecium | − | 2 | 24.3 | 0 | 9.6 | 21 | 29 | 24 | 29.3 | 24.8 |
| K-M2-3 | Enterococcus faecium | − | 4.7 | 22.7 | 4.7 | 9.7 | 21.7 | 27.3 | 23.3 | 27.7 | 24 |
| K-M2-4 | Enterococcus faecium | − | 0 | 20.3 | 0 | 6.7 | 19.3 | 27.3 | 23.3 | 30.7 | 25.3 |
| LAR-M2-1 | Enterococcus hirae | − | 5.5 | 24.7 | 8.3 | 10.7 | 22 | 27 | 24 | 26 | 22.5 |
| CP-M2-6 | Enterococcus hirae | − | 11 | 28 | 13 | 16.2 | 29.3 | 32 | 23 | 2.8 | 22.3 |
| RL-M2-3 | Enterococcus lactis | − | 14 | 27 | 11.7 | 11.3 | 25.7 | 27.3 | 25 | 30.3 | 25.3 |
a Isolate number: in regular font, isolate obtained directly on MacConkey-meropenem medium; in bold and italics, isolates enriched in BLCVM9-meropenem medium before streaking in Mueller–Hinton-meropenem medium. Intials(s) before the first dash indicates the isolation location (Table 1). b Resistant (green), Susceptible (purple), based on the CLSI [48] and EUCAST zone of inhibition diameter breakpoint values [55]. For genera that did not have diameter data in the CLSI manual, Enterobacteriaceae cut-offs were used. MEM (meropenem), IMI (imipenem), ERT (ertapenem), DOR (doripenem), AMP (ampicillin), AMC (amoxicillin plus clavulanic acid), CIP (ciprofloxacin), TET (tetracycline), VAN (vancomycin). c “−” indicate a negative result for the CarbaNP test.
Figure 9.
Antibiotic resistance frequency of Gram-negative (A) and Gram-positive (B) water isolates from this study. Green (% Resistant), Yellow (% Intermediate), Purple (% Susceptible). Carbapenems: meropenem (MEM), imipenem (IMI), doripenem (DOR), and ertapenem (ERT). Other β-lactams: amoxicillin-clavulanic acid (AMC), cefotaxime (CFX), and ampicillin (AMP). Other antibiotics: ciprofloxacin (CIP), gentamicin (GTM), tetracycline (TET), and vancomycin (VAN).
All eight Gram-positive CRB isolates were tested for resistance to carbapenems and other antibiotics commonly used to treat Gram-positive pathogens [48]. These antibiotics included four carbapenems (meropenem, imipenem, ertapenem, and doripenem), and four more non-carbapenem antibiotics (amoxicillin plus clavulanic acid, ciprofloxacin, tetracycline, and vancomycin). Overall, 100% of these isolates were resistant to three out of the four carbapenems tested (meropenem, ertapenem, and doripenem) but susceptible to imipenem (which is common for Gram-positives [78]). Interestingly, all isolates were susceptible to the non-carbapenem antibiotics tested except for one Enterococcus hirae isolate from Cypress Park, CA that was resistant to tetracycline (Table 3; Figure 9B).
Overall, these findings indicate that aquatic environments are an important reservoir of bacteria resistant to carbapenems and other antibiotics, especially Gram-negative CRB resistant to a broad spectrum of β-lactams. Of special concern was the finding that about one-third of the Gram-negative CRB characterized were resistant to nearly all antibiotics tested, given that some of them are opportunistic pathogens or could serve as reservoirs in the transmission of antibiotic resistance determinants.
3.3. Detection of Carbapenemase Production and Carbapenemase Genes in CRB Isolates
After characterizing them, we used the CarbaNP assay to determine whether carbapenemases contributed to the carbapenem-resistance phenotype of the 30 selected CRB isolates. Of them, nine were CarbaNP positive: three Enterobacter asburiae, three Stenotrophomonas maltophilia, and three Aeromonas veronii (Table 2 and Table 4). Using PCR and sequencing to identify their carbapenemase genes, we found that the E. asburiae and S. maltophilia isolates carried the blaIMI-2 and blaL1 carbapenemase genes, respectively. These two carbapenemase genes have previously been found in E. asburiae plasmids or in the chromosome of S. maltophilia isolates, respectively [42,79]. The high identity (>98%) between the blaIMI-2 variants identified here and the blaIMI-2 reference sequence (DQ173429) is similar to that found for E. asburiae isolates in our previous study [43]. For the blaL1 gene, the percent DNA identity between the blaL1 variants of the isolates from this study and the reference sequence (NG_047502) was 87.7% to 88.8% (96–97% protein similarity), in accordance with our previous study [43] and the blaL1 variability previously found in Stenotrophomonas sp. from healthcare settings [79].
Table 4.
Detection of Carbapenemases Production by CarbaNP, mCIM, and eCIM assays, and identification of carbapenemase genes by PCR and Sequencing.
| Isolate | Closest Species Identified by 16S rRNA Gene | CarbaNP | mCIM | eCIM | Carbapenemase Gene (% DNA Identity) | Carbapenemase (% Amino Acid Similarity) |
|---|---|---|---|---|---|---|
| SS-M2-3 | Aeromonas veronii | + | + | + | Unknown | Unknown |
| BBL-M2-1 | Aeromonas veronii | + | + | + | Unknown | Unknown |
| BBL-M2-2 | Stenotrophomonas maltophilia | + | N/D | N/D | blaL1 (88.6%) | L1 (96.9%) |
| BBL-M2-3 | Aeromonas veronii | + | + | + | Unknown | Unknown |
| MRP-M2-1 | Stenotrophomonas maltophilia | + | N/D | N/D | blaL1 (87.7%) | L1 (96.0%) |
| WL-M16-1 | Enterobacter asburiae | + | N/D | N/D | blaIMI-2 (99.2%) | IMI-2 (98.6%) |
| WL-M16-2 | Enterobacter asburiae | + | N/D | N/D | blaIMI-2 (98.3%) | IMI-2 (97.7%) |
| WL-M16-3 | Enterobacter asburiae | + | N/D | N/D | blaIMI-2 (99.8%) | IMI-2 (100%) |
| WL-M16-4 | Stenotrophomonas maltophilia | + | N/D | N/D | blaL1 (88.2%) | L1 (97.0%) |
Note: Unknown indicates the inability to identify the carbapenemase gene using targeted PCR with blaCphA-, blaImiS-, or blaVIM-2-specific primers. N/D: Not Determined (for carbapenemases directly identified by PCR and sequencing). % DNA identify or amino acid similarity compared to the reference sequences blaL1 (NG_047502) and blaIMI-2 (DQ173429). “+” Indicates a positive result for that test.
Interestingly, all three Aeromonas veronii isolates were positive for carbapenemase production using the CarbaNP test, despite two of them being susceptible to carbapenems according to the Enterobacteriaceae cut-offs (but intermediate to meropenem according to the Pseudomonas spp. cut-off) (Table 2 and Table 4). Moreover, prior studies have shown that carbapenemase-producing Aeromonas do not always appear as carbapenem-resistant under standard antibiotic susceptibility testing [61]. Thus, the carbapenemase activity of these three Aeromonas sp. isolates was further studied using the mCIM test. This test is recommended by the CLSI to confirm carbapenemase activity because the CarbaNP assay can yield false positives [59]. All three Aeromonas sp. isolates were confirmed as carbapenemase-positive using the mCIM test. Next, using the eCIM test [48], we found that their carbapenemases were Class B metallo-β-lactamases (Table 4). Because metallo-carbapenemase-producing A. veronii typically produce the chromosomal CphA, ImiS, or VIM-2 carbapenemases [61], we used PCR to test for the presence of the blaCphA, blaImiS, or blaVIM-2 genes in our isolates. Interestingly, all three isolates tested negative for these genes, including the highly carbapenem-resistant A. veronii SS-M2-3 isolate. These findings indicate that these A. veronii isolates either may carry poorly conserved variants of these genes, or different, potentially undiscovered carbapenemase genes, which we will investigate in future studies.
4. Conclusions
Carbapenem-resistant bacteria (CRB) and carbapenemase genes represent a major and increasing public health challenge. However, little is known about their distribution and diversity in the environment, especially in the U.S. This study contributes to addressing this gap in knowledge by sampling diverse freshwater and seawater aquatic environments and combining both direct isolation and enrichment approaches to determine the abundance, distribution, and diversity of CRB in California, U.S. Overall, we found a low abundance of CRB in the ten locations sampled, except for the Carpinteria Salt Marsh, which had been affected by a fire followed by rains and a mudslide. Identification and characterization of 30 selected CRB from the aquatic environments sampled revealed a greater diversity of both Gram-negative and Gram-positive CRB genera, compared to a prior study focused only on freshwater environments and not including CRB enrichment. The CRB isolated in the present study belonged to the genera Aeromonas, Enterobacter, Enterococcus, Paenibacillus, Pseudomonas, Sphingobacterium, and Stenotrophomonas. Interestingly, we found that all Gram-negative CRB characterized in this study were also resistant or intermediate to at least one non-carbapenem antibiotic, especially other β-lactams, and that one-third of them were resistant to nearly all antibiotics tested, which is of great concern. Finally, we found that nine Aeromonas sp., Enterobacter sp. (blaIMI-2), and Stenotrophomonas sp. (blaL1) isolates were carbapenemase producers. Overall, these findings expand our understanding of the role of natural water environments as important and often underappreciated reservoirs of bacteria resistant to carbapenems and other antibiotics, including carbapenemase-producing bacteria.
Acknowledgments
A.M. was directly supported by the CSUN MS tuition waiver.
Author Contributions
Conceptualization, A.M. and C.R.; formal analysis, A.M., M.L.E., D.E.H. and C.R.; investigation, A.M., M.L.E., D.E.H. and C.R.; writing—original draft preparation, A.M. and C.R.; writing—review and editing, D.E.H. and C.R.; supervision, C.R.; project administration, C.R.; funding acquisition, C.R. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
All 16S rRNA gene sequences obtained in this study have been deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank/) under the following accession numbers: MT790713–MT790742.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This research was funded by the California State University Northridge (CSUN) start-up funds and the CSUN Pandemic Recovery Grant for Research, Creative and Scholarly Activity grant to C. Ruiz.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Papp-Wallace K.M., Endimiani A., Taracila M.A., Bonomo R.A. Carbapenems: Past, present, and future. Antimicrob. Agents Chemother. 2011;55:4943–4960. doi: 10.1128/AAC.00296-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradley J.S., Garau J., Lode H., Rolston K.V., Wilson S.E., Quinn J.P. Carbapenems in clinical practice: A guide to their use in serious infection. Int. J. Antimicrob. Agents. 1999;11:93–100. doi: 10.1016/S0924-8579(98)00094-6. [DOI] [PubMed] [Google Scholar]
- 3.Vardakas K.Z., Tansarli G.S., Rafailidis P.I., Falagas M.E. Carbapenems versus alternative antibiotics for the treatment of bacteraemia due to Enterobacteriaceae producing extended-spectrum β-lactamases: A systematic review and meta-analysis. J. Antimicrob. Chemother. 2012;67:2793–2803. doi: 10.1093/jac/dks301. [DOI] [PubMed] [Google Scholar]
- 4.Williams P.C.M., Jones M., Snelling T.L., Duguid R., Moore N., Dickson B., Wu Y., Saunders J., Wijeratne P., Douangnouvong A., et al. Coverage gaps in empiric antibiotic regimens used to treat serious bacterial infections in neonates and children in Southeast Asia and the Pacific. Lancet Reg. Health Southeast Asia. 2024;22:100291. doi: 10.1016/j.lansea.2023.100291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin S.I., Kaye K.M. Beta-lactam antibiotics: Newer formulations and newer agents. Infect. Dis. Clin. N. Am. 2004;18:603–619. doi: 10.1016/j.idc.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 6.Nordmann P., Dortet L., Poirel L. Carbapenem resistance in Enterobacteriaceae: Here is the storm! Trends Mol. Med. 2012;18:263–272. doi: 10.1016/j.molmed.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 7.Jean S.S., Harnod D., Hsueh P.R. Global Threat of Carbapenem-Resistant Gram-Negative Bacteria. Front. Cell Infect. Microbiol. 2022;12:823684. doi: 10.3389/fcimb.2022.823684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martirosov D.M., Lodise T.P. Emerging trends in epidemiology and management of infections caused by carbapenem-resistant Enterobacteriaceae. Diagn. Microbiol. Infect. Dis. 2016;85:266–275. doi: 10.1016/j.diagmicrobio.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 9.Livorsi D.J., Chorazy M.L., Schweizer M.L., Balkenende E.C., Blevins A.E., Nair R., Samore M.H., Nelson R.E., Khader K., Perencevich E.N. A systematic review of the epidemiology of carbapenem-resistant Enterobacteriaceae in the United States. Antimicrob. Resist. Infect. Control. 2018;7:55. doi: 10.1186/s13756-018-0346-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Queenan A.M., Bush K. Carbapenemases: The versatile beta-lactamases. Clin. Microbiol. Rev. 2007;20:440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisher J.F., Meroueh S.O., Mobashery S. Bacterial resistance to beta-lactam antibiotics: Compelling opportunism, compelling opportunity. Chem. Rev. 2005;105:395–424. doi: 10.1021/cr030102i. [DOI] [PubMed] [Google Scholar]
- 12.Walther-Rasmussen J., Hoiby N. Class A carbapenemases. J. Antimicrob. Chemother. 2007;60:470–482. doi: 10.1093/jac/dkm226. [DOI] [PubMed] [Google Scholar]
- 13.Prabaker K., Weinstein R.A. Trends in antimicrobial resistance in intensive care units in the United States. Curr. Opin. Crit. Care. 2011;17:472–479. doi: 10.1097/MCC.0b013e32834a4b03. [DOI] [PubMed] [Google Scholar]
- 14.Rhomberg P.R., Jones R.N. Summary trends for the Meropenem Yearly Susceptibility Test Information Collection Program: A 10-year experience in the United States (1999–2008) Diagn. Microbiol. Infect. Dis. 2009;65:414–426. doi: 10.1016/j.diagmicrobio.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 15.Gupta N., Limbago B.M., Patel J.B., Kallen A.J. Carbapenem-resistant Enterobacteriaceae: Epidemiology and prevention. Clin. Infect. Dis. 2011;53:60–67. doi: 10.1093/cid/cir202. [DOI] [PubMed] [Google Scholar]
- 16.Guh A.Y., Bulens S.N., Mu Y., Jacob J.T., Reno J., Scott J., Wilson L.E., Vaeth E., Lynfield R., Shaw K.M., et al. Epidemiology of Carbapenem-Resistant Enterobacteriaceae in 7 US Communities, 2012–2013. JAMA. 2015;314:1479–1487. doi: 10.1001/jama.2015.12480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Duijn P.J., Dautzenberg M.J., Oostdijk E.A. Recent trends in antibiotic resistance in European ICUs. Curr. Opin. Crit. Care. 2011;17:658–665. doi: 10.1097/MCC.0b013e32834c9d87. [DOI] [PubMed] [Google Scholar]
- 18.Rodríguez-Martínez J.M., Poirel L., Nordmann P. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2009;53:4783–4788. doi: 10.1128/AAC.00574-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Correa A., Montealegre M.C., Mojica M.F., Maya J.J., Rojas L.J., De La Cadena E.P., Ruiz S.J., Recalde M., Rosso F., Quinn J.P., et al. First report of a Pseudomonas aeruginosa isolate coharboring KPC and VIM carbapenemases. Antimicrob. Agents Chemother. 2012;56:5422–5423. doi: 10.1128/AAC.00695-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cuzon G., Naas T., Villegas M.V., Correa A., Quinn J.P., Nordmann P. Wide dissemination of Pseudomonas aeruginosa producing beta-lactamase blaKPC-2 gene in Colombia. Antimicrob. Agents Chemother. 2011;55:5350–5353. doi: 10.1128/AAC.00297-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rizek C., Fu L., Dos Santos L.C., Leite G., Ramos J., Rossi F., Guimaraes T., Levin A.S., Costa S.F. Characterization of carbapenem-resistant Pseudomonas aeruginosa clinical isolates, carrying multiple genes coding for this antibiotic resistance. Ann. Clin. Microbiol. Antimicrob. 2014;13:43. doi: 10.1186/s12941-014-0043-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khuntayaporn P., Montakantikul P., Mootsikapun P., Thamlikitkul V., Chomnawang M.T. Prevalence and genotypic relatedness of carbapenem resistance among multidrug-resistant P. aeruginosa in tertiary hospitals across Thailand. Ann. Clin. Microbiol. Antimicrob. 2012;11:25. doi: 10.1186/1476-0711-11-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kallen A.J., Hidron A.I., Patel J., Srinivasan A. Multidrug resistance among gram-negative pathogens that caused healthcare-associated infections reported to the National Healthcare Safety Network, 2006–2008. Infect. Control Hosp. Epidemiol. 2010;31:528–531. doi: 10.1086/652152. [DOI] [PubMed] [Google Scholar]
- 24.Queenan A.M., Pillar C.M., Deane J., Sahm D.F., Lynch A.S., Flamm R.K., Peterson J., Davies T.A. Multidrug resistance among Acinetobacter spp. in the USA and activity profile of key agents: Results from CAPITAL Surveillance 2010. Diagn. Microbiol. Infect. Dis. 2012;73:267–270. doi: 10.1016/j.diagmicrobio.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 25.Viehman J.A., Nguyen M.H., Doi Y. Treatment options for carbapenem-resistant and extensively drug-resistant Acinetobacter baumannii infections. Drugs. 2014;74:1315–1333. doi: 10.1007/s40265-014-0267-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Centers for Disease Control and Prevention Carbapenem-Resistant Enterobacteriaceae in Healthcare Settings. [(accessed on 10 April 2024)]; Available online: https://www.cdc.gov/hai/organisms/cre/index.html.
- 27.Centers for Disease Control and Prevention . Antibiotic Resistance Threats in the United States, 2019. Centers for Disease Control and Prevention; Atlanta, GA, USA: 2019. [DOI] [Google Scholar]
- 28.Chagas T.P., Seki L.M., da Silva D.M., Asensi M.D. Occurrence of KPC-2-producing Klebsiella pneumoniae strains in hospital wastewater. J. Hosp. Infect. 2011;77:281. doi: 10.1016/j.jhin.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 29.White L., Hopkins K.L., Meunier D., Perry C.L., Pike R., Wilkinson P., Pickup R.W., Cheesbrough J., Woodford N. Carbapenemase-producing Enterobacteriaceae in hospital wastewater: A reservoir that may be unrelated to clinical isolates. J. Hosp. Infect. 2016;93:145–151. doi: 10.1016/j.jhin.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 30.Nasri E., Subirats J., Sanchez-Melsio A., Mansour H.B., Borrego C.M., Balcazar J.L. Abundance of carbapenemase genes (blaKPC, blaNDM and blaOXA-48) in wastewater effluents from Tunisian hospitals. Environ. Pollut. 2017;229:371–374. doi: 10.1016/j.envpol.2017.05.095. [DOI] [PubMed] [Google Scholar]
- 31.Nordmann P., Poirel L. Epidemiology and Diagnostics of Carbapenem Resistance in Gram-negative Bacteria. Clin. Infect. Dis. 2019;69:S521–S528. doi: 10.1093/cid/ciz824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kehl K., Schallenberg A., Szekat C., Albert C., Sib E., Exner M., Zacharias N., Schreiber C., Parcina M., Bierbaum G. Dissemination of carbapenem resistant bacteria from hospital wastewater into the environment. Sci. Total Environ. 2022;806:151339. doi: 10.1016/j.scitotenv.2021.151339. [DOI] [PubMed] [Google Scholar]
- 33.Tacão M., Correia A., Henriques I.S. Low Prevalence of Carbapenem-Resistant Bacteria in River Water: Resistance Is Mostly Related to Intrinsic Mechanisms. Microb. Drug Resist. 2015;21:497–506. doi: 10.1089/mdr.2015.0072. [DOI] [PubMed] [Google Scholar]
- 34.Girlich D., Poirel L., Nordmann P. Novel ambler class A carbapenem-hydrolyzing beta-lactamase from a Pseudomonas fluorescens isolate from the Seine River, Paris, France. Antimicrob. Agents Chemother. 2010;54:328–332. doi: 10.1128/AAC.00961-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poirel L., Barbosa-Vasconcelos A., Simões R.R., Da Costa P.M., Liu W., Nordmann P. Environmental KPC-producing Escherichia coli isolates in Portugal. Antimicrob. Agents Chemother. 2012;56:1662–1663. doi: 10.1128/AAC.05850-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Potron A., Poirel L., Bussy F., Nordmann P. Occurrence of the carbapenem-hydrolyzing beta-lactamase gene blaOXA-48 in the environment in Morocco. Antimicrob. Agents Chemother. 2011;55:5413–5414. doi: 10.1128/AAC.05120-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Isozumi R., Yoshimatsu K., Yamashiro T., Hasebe F., Nguyen B.M., Ngo T.C., Yasuda S.P., Koma T., Shimizu K., Arikawa J. bla(NDM-1)-positive Klebsiella pneumoniae from environment, Vietnam. Emerg. Infect. Dis. 2012;18:1383–1385. doi: 10.3201/eid1808.111816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zurfluh K., Hachler H., Nuesch-Inderbinen M., Stephan R. Characteristics of extended-spectrum beta-lactamase- and carbapenemase-producing Enterobacteriaceae Isolates from rivers and lakes in Switzerland. Appl. Environ. Microbiol. 2013;79:3021–3026. doi: 10.1128/AEM.00054-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Di D.Y., Jang J., Unno T., Hur H.G. Emergence of Klebsiella variicola positive for NDM-9, a variant of New Delhi metallo-beta-lactamase, in an urban river in South Korea. J. Antimicrob. Chemother. 2017;72:1063–1067. doi: 10.1093/jac/dkw547. [DOI] [PubMed] [Google Scholar]
- 40.Henriques I.S., Araujo S., Azevedo J.S., Alves M.S., Chouchani C., Pereira A., Correia A. Prevalence and diversity of carbapenem-resistant bacteria in untreated drinking water in Portugal. Microb. Drug Resist. 2012;18:531–537. doi: 10.1089/mdr.2012.0029. [DOI] [PubMed] [Google Scholar]
- 41.Ash R.J., Mauck B., Morgan M. Antibiotic resistance of gram-negative bacteria in rivers, United States. Emerg. Infect. Dis. 2002;8:713–716. doi: 10.3201/eid0807.010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aubron C., Poirel L., Ash R.J., Nordmann P. Carbapenemase-producing Enterobacteriaceae, U.S. rivers. Emerg. Infect. Dis. 2005;11:260–264. doi: 10.3201/eid1102.030684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harmon D.E., Miranda O.A., McCarley A., Eshaghian M., Carlson N., Ruiz C. Prevalence and characterization of carbapenem-resistant bacteria in water bodies in the Los Angeles-Southern California area. Microbiologyopen. 2019;8:e00692. doi: 10.1002/mbo3.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lopez N.V., Farsar C.J., Harmon D.E., Ruiz C. Urban and agricultural soils in Southern California are a reservoir of carbapenem-resistant bacteria. Microbiologyopen. 2020;9:1247–1263. doi: 10.1002/mbo3.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harbor Beaches of Ventura County (Kiddie Beach and Hobie Beach) Bacteria Total Maximum Daily Load. California State Water Resources Control Board; Sacramento, CA, USA: 2007. [Google Scholar]
- 46.Geosyntec . Bacteria Total Maximum Daily Load Compliance Report. GeoSyntec Consultants; Portland, OR, USA: 2016. [Google Scholar]
- 47.Staff Bacterial Levels Up at Six Ventura County Beaches; Public Warned to Avoid Ocean Water. VC Star. 2018. [(accessed on 13 April 2024)]. Available online: https://www.vcstar.com/story/news/2018/03/28/bacteria-levels-up-six-ventura-county-beaches-public-warned-avoid-ocean-water/467287002/
- 48.Clinical and Laboratory Standards Institute . Performance Standards for Antimicrobial Susceptibility Testing. 28th ed. CLSI Supplement M100; Wayne, PA, USA: 2018. [Google Scholar]
- 49.Denton M., Kerr K.G. Microbiological and clinical aspects of infection associated with Stenotrophomonas maltophilia. Clin. Microbiol. Rev. 1998;11:57–80. doi: 10.1128/CMR.11.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Holt J.G. Bergey’s Manual of Determinative Bacteriology. 9th ed. LWW; Baltimore, MD, USA: 1994. [Google Scholar]
- 51.Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumar S., Stecher G., Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hudzicki J. Kirby-Bauer Disk Diffusion Susceptibility Test Protocol. Am. Soc. Microbiol. 2009;15:1–23. [Google Scholar]
- 55.The European Committee on Antimicrobial Susceptibility Testing . Breakpoint Tables for Interpretation of MICs and Zone Diameters. The European Committee on Antimicrobial Susceptibility Testing; Växjö, Sweden: 2018. [Google Scholar]
- 56.Dortet L., Poirel L., Nordmann P. Rapid identification of carbapenemase types in Enterobacteriaceae and Pseudomonas spp. by using a biochemical test. Antimicrob. Agents Chemother. 2012;56:6437–6440. doi: 10.1128/AAC.01395-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dortet L., Poirel L., Nordmann P. Rapid detection of carbapenemase-producing Pseudomonas spp. J. Clin. Microbiol. 2012;50:3773–3776. doi: 10.1128/JCM.01597-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nordmann P., Poirel L., Dortet L. Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 2012;18:1503–1507. doi: 10.3201/eid1809.120355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pierce V.M., Simner P.J., Lonsway D.R., Roe-Carpenter D.E., Johnson J.K., Brasso W.B., Bobenchik A.M., Lockett Z.C., Charnot-Katsikas A., Ferraro M.J., et al. Modified Carbapenem Inactivation Method for Phenotypic Detection of Carbapenemase Production among Enterobacteriaceae. J. Clin. Microbiol. 2017;55:2321–2333. doi: 10.1128/JCM.00193-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bottoni C., Marcoccia F., Compagnoni C., Colapietro M., Sabatini A., Celenza G., Segatore B., Maturo M.G., Amicosante G., Perilli M. Identification of New Natural CphA Metallo-beta-Lactamases CphA4 and CphA5 in Aeromonas veronii and Aeromonas hydrophila Isolates from Municipal Sewage in Central Italy. Antimicrob. Agents Chemother. 2015;59:4990–4993. doi: 10.1128/AAC.00628-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen P.L., Ko W.C., Wu C.J. Complexity of β-lactamases among clinical Aeromonas isolates and its clinical implications. J. Microbiol. Immunol. Infect. 2012;45:398–403. doi: 10.1016/j.jmii.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 62.Drk S., Puljko A., Dzelalija M., Udikovic-Kolic N. Characterization of Third Generation Cephalosporin- and Carbapenem-Resistant Aeromonas Isolates from Municipal and Hospital Wastewater. Antibiotics. 2023;12:513. doi: 10.3390/antibiotics12030513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Libisch B., Giske C.G., Kovacs B., Toth T.G., Fuzi M. Identification of the first VIM metallo-beta-lactamase-producing multiresistant Aeromonas hydrophila strain. J. Clin. Microbiol. 2008;46:1878–1880. doi: 10.1128/JCM.00047-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Massidda O., Rossolini G.M., Satta G. The Aeromonas hydrophila cphA gene: Molecular heterogeneity among class B metallo-beta-lactamases. J. Bacteriol. 1991;173:4611–4617. doi: 10.1128/jb.173.15.4611-4617.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rossolini G.M., Walsh T., Amicosante G. The Aeromonas metallo-beta-lactamases: Genetics, enzymology, and contribution to drug resistance. Microb. Drug Resist. 1996;2:245–252. doi: 10.1089/mdr.1996.2.245. [DOI] [PubMed] [Google Scholar]
- 66.Walsh T.R., Neville W.A., Haran M.H., Tolson D., Payne D.J., Bateson J.H., MacGowan A.P., Bennett P.M. Nucleotide and amino acid sequences of the metallo-beta-lactamase, ImiS, from Aeromonas veronii bv. sobria. Antimicrob. Agents Chemother. 1998;42:436–439. doi: 10.1128/AAC.42.2.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Belotti P.T., Thabet L., Laffargue A., Andre C., Coulange-Mayonnove L., Arpin C., Messadi A., M’Zali F., Quentin C., Dubois V. Description of an original integron encompassing blaVIM-2, qnrVC1 and genes encoding bacterial group II intron proteins in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2015;70:2237–2240. doi: 10.1093/jac/dkv103. [DOI] [PubMed] [Google Scholar]
- 68.Romling U., Fiedler B., Bosshammer J., Grothues D., Greipel J., von der Hardt H., Tummler B. Epidemiology of chronic Pseudomonas aeruginosa infections in cystic fibrosis. J. Infect. Dis. 1994;170:1616–1621. doi: 10.1093/infdis/170.6.1616. [DOI] [PubMed] [Google Scholar]
- 69.Burgos-Diaz C., Pons R., Espuny M.J., Aranda F.J., Teruel J.A., Manresa A., Ortiz A., Marques A.M. Isolation and partial characterization of a biosurfactant mixture produced by Sphingobacterium sp. isolated from soil. J. Colloid. Interface Sci. 2011;361:195–204. doi: 10.1016/j.jcis.2011.05.054. [DOI] [PubMed] [Google Scholar]
- 70.Lambiase A., Rossano F., Del Pezzo M., Raia V., Sepe A., de Gregorio F., Catania M.R. Sphingobacterium respiratory tract infection in patients with cystic fibrosis. BMC Res. Notes. 2009;2:262. doi: 10.1186/1756-0500-2-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Byappanahalli M.N., Nevers M.B., Korajkic A., Staley Z.R., Harwood V.J. Enterococci in the environment. Microbiol. Mol. Biol. Rev. 2012;76:685–706. doi: 10.1128/MMBR.00023-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moellering R.C. Emergence of Enterococcus as a significant pathogen. Clin. Infect. Dis. 1992;14:1173–1176. doi: 10.1093/clinids/14.6.1173. [DOI] [PubMed] [Google Scholar]
- 73.Solioz M., Stoyanov J.V. Copper homeostasis in Enterococcus hirae. FEMS Microbiol. Rev. 2003;27:183–195. doi: 10.1016/S0168-6445(03)00053-6. [DOI] [PubMed] [Google Scholar]
- 74.Morandi S., Silvetti T., Brasca M. Biotechnological and safety characterization of Enterococcus lactis, a recently described species of dairy origin. Antonie Van. Leeuwenhoek. 2013;103:239–249. doi: 10.1007/s10482-012-9806-z. [DOI] [PubMed] [Google Scholar]
- 75.Loong S.K., Ishak S.N., Lim F.S., Khoo J.J., Tan S.N., Freddy-Jalin E.-J., Mohd-Taib F.S., Abubakar S. Paenibacillus lautus, an opportunistic bacterial pathogen, isolated from Ixodes granulatus Supino (Acari: Ixodidae) collected from a Müller’s giant Sunda rat (Sundamys muelleri) Syst. Appl. Acarol. 2018;23:597–602. doi: 10.11158/saa.23.4.2. [DOI] [Google Scholar]
- 76.Saez-Nieto J.A., Medina-Pascual M.J., Carrasco G., Garrido N., Fernandez-Torres M.A., Villalon P., Valdezate S. Paenibacillus spp. isolated from human and environmental samples in Spain: Detection of 11 new species. New Microbes New Infect. 2017;19:19–27. doi: 10.1016/j.nmni.2017.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mushtaq S., Ge Y., Livermore D.M. Doripenem versus Pseudomonas aeruginosa in vitro: Activity against characterized isolates, mutants, and transconjugants and resistance selection potential. Antimicrob. Agents Chemother. 2004;48:3086–3092. doi: 10.1128/AAC.48.8.3086-3092.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bassetti M., Nicolini L., Esposito S., Righi E., Viscoli C. Current status of newer carbapenems. Curr. Med. Chem. 2009;16:564–575. doi: 10.2174/092986709787458498. [DOI] [PubMed] [Google Scholar]
- 79.Avison M.B., Higgins C.S., von Heldreich C.J., Bennett P.M., Walsh T.R. Plasmid location and molecular heterogeneity of the L1 and L2 beta-lactamase genes of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 2001;45:413–419. doi: 10.1128/AAC.45.2.413-419.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All 16S rRNA gene sequences obtained in this study have been deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank/) under the following accession numbers: MT790713–MT790742.