Abstract
To investigate the role of hepatitis C virus (HCV) quasispecies mutation in the pathogenesis of HCV infection, we analyzed changes in the genetic diversity of HCV genomes in 22 patients before and after liver transplantation by using heteroduplex mobility assay (HMA) technology. All patients were infected with HCV genotype 1 and developed high-titer posttransplant viremia. Each patient was classified according to the severity of posttransplant hepatitis, as assessed by standard biochemical and histological criteria. HCV quasispecies were characterized by HMA analysis of eight separate subgenomic regions of HCV, which collectively comprise 44% of the entire genome. The glycoprotein genes E1 and E2, as well as the nonstructural protein genes NS2 and NS3, had the greatest genetic divergence after liver transplantation (the change in the heteroduplex mobility ratio [HMR] ranged from 2.5 to 7.0%). In contrast, genes encoding the core, NS4, and NS5b proteins had the least amount of genetic divergence after liver transplantation (range, 0.3 to 1.2%). The E1/E2 region showed the greatest change in genetic diversity after liver transplantation, and the change in HMRs was 2.5- to 3.3-fold greater in patients with asymptomatic or moderate disease than in those with severe disease. The E1-5′ region of HCV quasispecies isolated from patients in the asymptomatic group had a significantly greater degree of diversification after liver transplantation than the same regions of HCV quasispecies isolated from patients in the severe disease group (P = 0.05). While changes in the genetic diversity of some nonstructural genes were also greater in asymptomatic patients or in patients with mild disease than in patients with severe disease, the results were not significant. Data from this cohort demonstrate that greater rates of HCV quasispecies diversification are associated with mild or moderate liver disease activity in this immunosuppressed population.
Hepatitis C virus (HCV), a member of the Flaviviridae family, is known to be a major causative agent of chronic liver diseases, including chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma (27). Chronic hepatitis C is now recognized as the leading indication for orthotopic liver transplantation in the United States, with nearly 100% of HCV-infected liver transplant recipients developing recurrent viremia after transplantation (4, 17, 22, 32, 44).
The HCV genome is a single-stranded, positive-sense RNA of about 9.5 kb, which exists as a viral quasispecies in infected humans (6, 25, 31, 41). HCV quasispecies are characterized by extensive genetic mutation in the hypervariable region 1 (HVR1) of the second envelope glycoprotein gene (E2) (25, 41). Mutation of this region of the genome is believed to be associated with viral persistence via immune escape mechanisms (15, 16, 29, 42). The role of evolution of HCV quasispecies in the development of chronic hepatitis C is currently unknown.
HCV-infected liver transplant recipients offer an opportunity to study the evolution of HCV quasispecies in a new host tissue and to assess the role of quasispecies diversification in the development of posttransplant hepatitis. In a previous study (23) of HCV-infected liver transplant recipients, we found that in the three patients who developed severe, recurrent hepatitis, quasispecies major variants present in pretransplant serum samples were efficiently propagated after liver transplantation and during acute and chronic posttransplant hepatitis. In contrast, in the two asymptomatic cases, we observed rapid depletion of pretransplant quasispecies major variants from posttransplant serum samples, followed by the emergence of quasispecies minor variants. These data suggested that the evolution of HCV quasispecies after liver transplantation may be related to posttransplant disease severity.
In order to extend our previous findings and to address the hypothesis that mutation of other HCV genes also correlates with severity of posttransplant hepatitis C, we analyzed the pre- and posttransplant HCV quasispecies in 22 HCV-infected liver transplant recipients. The 22 patients were selected based on two virological criteria: all patients were infected with HCV genotype 1 before and after liver transplantation, and all patients developed recurrent, high-titer HCV viremia within 30 days posttransplant. Additionally, at least five posttransplant liver biopsies were available in each case to evaluate the histopathologic course of posttransplant hepatitis C. Pre- and posttransplant HCV quasispecies were characterized by using the heteroduplex mobility assay (HMA) to analyze eight different regions of the viral genome.
MATERIALS AND METHODS
Patients and clinical monitoring.
The 22 patients were selected from a larger cohort of HCV-infected liver transplant recipients at the University of Washington Medical Center as described elsewhere (22, 39). In brief, all 22 patients had active HCV infection at the time of liver transplant as demonstrated by detection of HCV antibody and HCV viremia (see Table 1). HCV genotype was determined by restriction fragment length polymorphism analysis of the 5′ noncoding region (9). HCV RNA levels were determined by a combination of bDNA version 2.0 (Chiron Corporation, Emeryville, Calif.) and in-house quantitative PCR as described previously (20). The sensitivity limit of our PCR assay is less than 100 copies of HCV RNA per ml of serum (21, 24). All patients were monitored clinically by a common protocol in which the serum specimens were obtained immediately prior to liver transplantation and at regularly scheduled posttransplant intervals. The serum specimens were separated from whole blood within 2 h of venipuncture, aliquoted, and immediately stored at −70°C (22). Protocol liver biopsies were performed with informed consent at five time points during the posttransplant year, as described elsewhere (22).
TABLE 1.
Clinical and virological features of HCV infection after orthotopic liver transplantation
| Disease groupa and patient no. | HCV genotypeb | Post-Txc serum collection time (no. of mos) | HCV RNA titerd
|
|
|---|---|---|---|---|
| Pre-Tx | Post-Tx | |||
| Severe | ||||
| 1 | 1a | 25 | 6.5 | 7.7 |
| 2 | 1a | 24 | 6.5 | 7.9 |
| 3 | 1a | 17 | 6.7 | 7.7 |
| 4 | 1a | 13 | 5.8 | 8.0 |
| 5 | 1a | 12 | 7.2 | 7.4 |
| 6 | 1b | 14 | 4.0 | 7.1 |
| Group mean | 17.5 | 6.1 | 7.6 | |
| Moderate | ||||
| 7 | 1a | 13 | 4.0 | 6.8 |
| 8 | 1a | 12 | 4.0 | 5.5 |
| 9 | 1a | 12 | 6.0 | 7.8 |
| 10 | 1b | 14 | 6.4 | 7.2 |
| 11 | 1b | 25 | 6.0 | 6.9 |
| 12 | 1a | 12 | NT | 7.5 |
| 13 | 1b | 28 | 7.6 | 7.7 |
| 14 | 1a | 19 | 5.9 | 7.0 |
| 15 | 1a | 13 | 6.0 | 6.8 |
| Group mean | 16.4 | 5.7 | 7.0 | |
| Asymptomatic | ||||
| 16 | 1a | 12 | 5.3 | 7.3 |
| 17 | 1b | 15 | 5.6 | 8.5 |
| 18 | 1b | 14 | 6.4 | 6.8 |
| 19 | 1a | 14 | 5.5 | 6.6 |
| 20 | 1a | 24 | 4.7 | 9.5 |
| 21 | 1b | 6 | 6.8 | 7.1 |
| 22 | 1a | 19 | 5.6 | 7.4 |
| Group mean | 14.8 | 5.7 | 7.6 | |
| Total mean | 16.2 | 5.8 | 7.4 | |
See Materials and Methods for details on disease groups.
Classified by the method of Simmonds et al. (40).
Post-Tx, posttransplant.
Viral RNA titers are expressed as log transformed RNA equivalents per milliliter. Abbreviations: Pre-Tx and Post-Tx, pre- and posttransplant, respectively; NT, not tested.
Selection of patients for the current study was based on the following criteria. (i) All patients had HCV genotype 1 infections throughout the study period. (ii) None of the patients had evidence of clinically significant allograft rejection after liver transplantation. (iii) Patients 1 to 6 were selected because they developed severe posttransplant hepatitis, defined as biochemical and histological evidence of chronic active hepatitis in liver allografts within the first 6 months after liver transplantation, which progressed to bridging fibrosis or severe bridging necrosis in the allograft within the first year after transplantation. (iv) Patients 7 to 15 were selected because they developed moderately chronic active hepatitis within 6 months after liver transplantation, characterized by abnormal alanine aminotransferase (ALT) levels, lymphocytic infiltrations, and/or piecemeal necrosis without progression to bridging fibrosis or bridging necrosis during the follow-up period (range, 12 to 28 months). (v) Patients 16 to 22 were selected because they had asymptomatic HCV infections for up to 2 years after transplantation. Asymptomatic HCV infection was defined as the absence of histological evidence of liver injury in six consecutive liver biopsies performed 10 days, 21 days, 3 months, 6 months, 12 months, and 24 months after liver transplantation and by normal levels of ALT and other indices of hepatic function, which were monitored by a routine posttransplant protocol (22).
Amplification of multiple HCV genomic regions.
Eight different HCV genomic regions (see Fig. 1) were targeted for amplification by either single-round reverse transcription-PCR (RT-PCR) or nested RT-PCR. All primers used in the study are listed in Table 2. Primers not previously described were designed from nucleotide sequence alignments of published HCV genotype 1 genomic sequences. PCR conditions were optimized by varying the amount of MgCl2, annealing temperature, and extension time. Total RNA was extracted from 100 μl of serum by the single-step guanidinium method previously described (5) and resuspended in 10 μl of diethylpyrocarbonate-treated distilled water. RT-PCR was performed as described previously (43). The RNA was incubated at 70°C for 5 min and then reverse transcribed in a 25-μl reaction mixture containing 50 pmol of the external antisense primer, 3 mM MgCl2, 1 mmol of each deoxynucleoside triphosphate (dNTP), 1 mM dithiothreitol, 75 mM KCl, 5 mM Tris-HCl (pH 8.3), 20 U of RNase inhibitor (Pharmacia LKB, Piscataway, N.J.) and 130 U of Moloney murine leukemia virus reverse transcriptase (Gibco BRL, Gaithersburg, Md.). The mixture was incubated at 37°C for 60 min and then at 95°C for 5 min.
FIG. 1.
Map of the HCV genome showing the eight regions targeted for analysis by HMA. The name and length (in base pairs) of the regions are shown. See Table 2 for the sequences and positions of oligonucleotide primers. UTR, untranslated region.
TABLE 2.
Primers used for amplification of eight HCV genomic regions
| Region | Primer set | Polaritya | Positionb | Sequence | Reference |
|---|---|---|---|---|---|
| Core | External | S | 266–297 | 5′-GGGTCGCGAAAGGCCTTGTGGTACTGCCTGAT-3′ | 1 |
| AS | 838–809 | 5′-GTTGCATAGTTCACGCCGTCTTCCAGAACC-3′ | 1 | ||
| Nested | S | 276–305 | 5′-AGGCCTTGTGGTACTGCCTGATAGGGTGCT-3′ | 1 | |
| AS | 783–754 | 5′-CAGCGAATCCAAGAGGGGCGCCGACGAGCG-3′ | 1 | ||
| E1 and E2 | External | S | 802–841 | 5′-GCGTCCGGGTTCTGGAAGACGGCGTGAACTATGCAACAGG-3′ | 2 |
| AS | 1639–1600 | 5′-AGGCTTTCATTGCAGTTCAAGGCCGTGCTATTGATGTGCC-3′ | 2 | ||
| E1-5′ | Nested | S | 821–856 | 5′-AAGACGGCGTGAACTATGCAACAGGGAACCTTCCTGGTTG-3′ | 2 |
| AS | 1321–1295 | 5′-GACCAGTTCATCATCATATCCCATGCC-3′ | 2 | ||
| E1/E2 | Nested | S | 1295–1327 | 5′-GGCATGGGATATGATGATGAACTGGTCCCCTAC-3′ | |
| AS | 1626–1587 | 5′-AGTTCAAGGCCGTGCTATTGATGTGCCAACTGCCGTTGGT-3′ | |||
| NS2 | External | S | 2505–2527 | 5′-CTGTTCCTTCTGCTTGCAGACGC-3′ | 41 |
| AS | 3337–3318 | 5′-CCGTTGATGATGTCACCGCA-3′ | 41 | ||
| Nested | S | 2535–2555 | 5′-TGCTCCTGCTTGTGGATGATG-3′ | 41 | |
| AS | 3250–3228 | 5′-GCCACGGCCAGATCTCGCAAGCC-3′ | 41 | ||
| NS3-3′ | External | S | 4668–4687 | 5′-GGCTATACCGGCGACTTCGA-3′ | 3 |
| AS | 5290–5271 | 5′-GACATGCATGTCATGATGTA-3′ | 3 | ||
| NS4–5′ | External | S | 5471–5490 | 5′-GCTCAAGCCCCTCCCCCAT-3′ | |
| AS | 6040–6018 | 5′-AAAAGCCATCAATGAAGCAATG-3′ | |||
| Nested | S | 5595–5619 | 5′-CGCACCCAGTCACCAAATACATCA-3′ | ||
| AS | 6017–5997 | 5′-GGGTTACCAGGCAGCGTTGA-3′ | |||
| NS4-3′ | External | S | 5901–5921 | 5′-CCCGTCAGGCAGAGGTTATC-3′ | |
| AS | 6643–6621 | 5′-CCAGGTCTTAAAGTCGCTCAAC-3′ | |||
| Nested | S | 5933–5955 | 5′-CAGACCAACTGGCAAAAACTCG-3′ | ||
| AS | 6609–6587 | 5′-CAGTCCCAGATGTCCCTTAGCC-3′ | |||
| NS5b | External | S | 8245–8275 | 5′-TGGGGATCCCGTATGATACCCGCTGCTTTGA-3′ | 14 |
| AS | 8645–8616 | 5′-GGCGGAATTCCTGGTCATAGCCTCCGTGAA-3′ | 14 |
Abbreviations: S, sense; AS, antisense.
Positions (in nucleotides) according to the HCV1 sequence (7).
The first round of PCR was performed as follows: 10 μl of the cDNA was added to a 40-μl PCR mixture containing 50 pmol of the external, sense primer, 1.13 to 2.0 mM MgCl2, 23.5 mM Tris-HCl (pH 8.3), 35.5 mM KCl, and 1.5 U of Taq polymerase (Perkin-Elmer, Norwalk, Conn.). A “hot start,” nested PCR was then performed for some of the regions (see below and Table 2). In nested PCR, the bottom reaction mixture contained 2.0 to 3.0 mM MgCl2, 0.2 mmol of each dNTP, 10 mM Tris-HCl (pH 8.3), 15 mM KCl, and 50 pmol of each internal primer and was separated by a wax layer from the top reaction mixture containing 40 mM Tris-HCl (pH 8.3), 60 mM KCl, 1.5 U of Taq polymerase, and 2% of the first-round product. The PCRs were done in a Perkin-Elmer 9600 thermocycler, using 30 cycles with the following cycling parameters: template denaturation at 94°C for 30 s, primer annealing at 45 to 65°C for 20 s, and extension at 72°C for 30 to 60 s. A single final extension step was done at 72°C for 5 min to complete the amplification reaction. E1-5′ and E1/E2 genetic regions were both amplified from common PCR products using an external primer pair surrounding both regions. The NS3-3′ and NS5b regions were amplified by single-round PCR. Amplified products were analyzed on 1.0 to 2.0% agarose gels (38).
HMA.
HMA was performed as described previously with minor modifications (23, 43). Briefly, a universal probe of each genomic region was generated from HCV RNA present in one patient’s pretransplant serum. The amplified product was ligated into the pCR 2.1 vector and transformed into INVF′ cells (Invitrogen, San Diego, Calif.) according to the manufacturer’s instructions. Inserts were reamplified by PCR and purified by electroelution from agarose gels, and 20 ng of purified DNA was end labeled with T4 polynucleotide kinase (Gibco BRL) plus 100 μCi of [32P]ATP (Amersham, Arlington Heights, Ill.) to generate probes. Probes were then hybridized to heterogeneous (noncloned) PCR products derived from patient sera. Probe hybridized to itself (unlabeled) served as a marker for identification of homoduplexes. Since a universal probe (not patient specific) was used to hybridize with target HCV PCR products amplified from pre- and posttransplant sera, the sequences of all hybrids contain nucleotide differences relative to the probe sequence and the hybrids displayed retarded mobility in nondenaturing gels. These hybrids are referred to as heteroduplex bands. Since both strands of the probe were radiolabeled with 32P, multiple bands could be produced. However, the heteroduplex bands frequently yielded a single band, which is regarded as a doublet.
Calculation of changes in genetic diversity.
Changes in HCV quasispecies genetic diversity were calculated by comparing the changes in heteroduplex shift patterns before versus after liver transplantation for the various genomic regions under study. A previous study has demonstrated that the gel shift distance between homoduplex and heteroduplex bands (the average distance of multiple bands) is directly proportional to the number of nucleotide differences between the probe and target molecules (36). The heteroduplex mobility ratio (HMR) was calculated by dividing the distance in millimeters from the origin of the gel to the heteroduplex band(s) by the distance from the origin of the gel to the homoduplex control. In cases where both strands of the heteroduplex were clearly distinguishable, the average distance of each band was used to calculate the HMR (10, 43). To estimate the degree of genetic divergence within specific HCV genomic regions when comparing pre- versus posttransplant time points, the percent change in HMR was calculated as follows: percent change in HMR = {ABS [(HMRpost − HMRpre)/HMRpre]} × 100, where ABS is an absolute value and the pre and post subscripts are pre- and posttransplant, respectively. To assess sampling, cloning, and RT-PCR bias, parallel replication experiments were performed on undiluted or serially diluted specimens as described previously (23, 43); such experiments verified that our experimental methods yield highly reproducible results (data not shown).
Statistical analysis.
A two-sample paired Student’s t test was used to test whether the mean pretransplant viral RNA titers were different from the mean posttransplant viral RNA titers for all patients and for each disease group. A Mann-Whitney two-sample test was used to compare the percent change in HMR (pre- versus posttransplant) between the asymptomatic and moderate disease groups, the asymptomatic and severe disease groups, and the moderate disease and asymptomatic disease groups.
RESULTS
Clinical and virological features of HCV infection after liver transplantation.
Table 1 summarizes the clinical and virological features of HCV infection before and after liver transplantation for the 22 patients selected for the current study. All 22 patients had active HCV genotype 1 infections before and after liver transplantation; 15 had HCV subtype 1a infections, while 7 had HCV subtype 1b infections. The histopathological results of six consecutive liver biopsies obtained 10 days, 21 days, 3 months, 6 months, 12 months, and 24 months after liver transplantation were available for each patient and were used to place the patients in asymptomatic, moderate, or severe disease groups, as described in Materials and Methods.
HCV viral load was determined in pretransplant sera 1 or 2 days prior to liver transplantation, as described in Materials and Methods. Although patients in the severe disease group had the highest pretransplant viral RNA levels, the results were not significant (P > 0.5) (Table 1). Posttransplant sera were obtained at various time points after transplant, ranging from 6 to 28 months (mean, 16.2 months after transplant). Posttransplant HCV RNA titers were significantly higher than pretransplant titers for all patients regardless of disease status (mean viral load of 5.8 versus 7.4 log transformed RNA equivalents per ml for pretransplant versus posttransplant, respectively, P < 0.001). Finally, posttransplant viral loads were not significantly different between the three disease groups (Table 1), which is consistent with previous reports (22, 45).
Application of the HMA technique for multigene analysis.
We have previously demonstrated the utility of HMA as a technique for the longitudinal analysis of HCV quasispecies in liver transplant recipients (23) and in patients receiving interferon therapy (26, 36, 37). However, previous studies examined very restricted regions of the HCV genome (e.g., HVR1). Figure 1 depicts the eight HCV subgenomic regions targeted for analysis in the current study, which when taken together (4,190 nucleotides) make up 44.0% of the entire HCV nucleotide sequence. Table 2 presents a summary of oligonucleotide primers used for RT-PCR amplification as described in Materials and Methods.
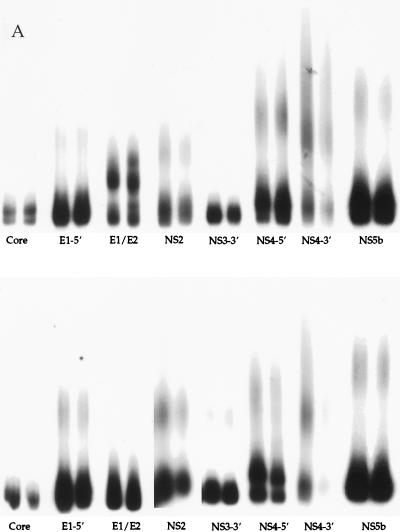
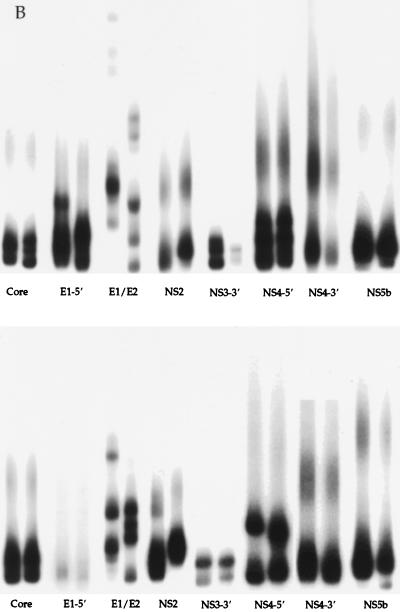
Representative results of the HMA multigene analysis technique are shown in Fig. 2. A universal probe of each HCV genomic region was produced from one liver transplant recipient infected with HCV genotype 1a and hybridized to heterogeneous amplification products of the corresponding region of each patient. Figure 2a depicts multigene analysis of HCV isolated from pre- and posttransplant sera from two patients who developed severe posttransplant hepatitis, characterized by bridging fibrosis, within the first 18 months after transplantation. For this experiment, multiple genetic regions isolated from pre- and posttransplant specimens were analyzed side by side after hybridization with radioactive probe. The pre- and posttransplant amplification products showed very similar gel shift patterns for all eight regions amplified, suggesting the patients were infected by a relatively stable and genetically homogeneous HCV quasispecies throughout the course of disease. Figure 2b illustrates multigene analysis of HCV genes amplified from pre- and posttransplant specimens from liver transplant recipients who developed moderate hepatitis. The HMA patterns for both patients in Fig. 2b (top and bottom) show changes after transplant in the E1/E2 and NS2 regions and show smaller differences in regions such as E1-5′ (upper panel), and NS4-5′ (lower panel). Figure 2c illustrates the HMA patterns for two patients with asymptomatic HCV infections. Again, major changes among the quasispecies populations can be seen after transplant, specifically in the E1/E2, NS2, NS4-5′, and NS4-3′ regions.
FIG. 2.
Representative multigene analysis of HCV quasispecies before and after liver transplantation for two representative patients (top and bottom panels) who developed either severe hepatitis (A), moderate hepatitis (B), or asymptomatic HCV infection (C) after transplantation (a total of six patients are shown). Pre- and posttransplant time points are run side by side for each HCV genomic region. A universal probe of each region was used for all six patients, as described in Materials and Methods. The absence of radioactive bands in lanes indicates the failure of PCR amplification for the specific region of interest (e.g., Fig. 2C, NS4-3′ and NS3-3′). See Fig. 1 for locations within the HCV genome.
Multigene analysis of HCV quasispecies after liver transplantation.
The previous section demonstrates the utility of the HMA technique for multigene analysis of HCV quasispecies in six representative patients. For the current study, 22 liver transplant recipients with HCV genotype 1 infection who fell into one of three disease categories were selected for multigene analysis by HMA. Figure 3 summarizes changes in viral quasispecies heterogeneity after liver transplantation for the entire cohort of 22 patients. The data are presented as mean percent change in HMR observed when comparing pretransplant quasispecies genes with posttransplant quasispecies genes, using probes as described in Materials and Methods. A higher value in mean percent change in HMR is indicative of a greater change in genetic heterogeneity after liver transplant.
FIG. 3.
HCV quasispecies genetic divergence after liver transplantation by HMA. Data are presented as mean percent changes in HMR (y axis) between pre- and posttransplant time points for each HCV genomic region (x axis). The data represent the average changes in HMR for the entire cohort. The number of patients (N) studied for each region is indicated above the bars.
Using the HMA technique, the most divergent region for the entire cohort after liver transplantation was the E1/E2 region, which contains the HVR1 (mean percent change in HMR of 7.04% after liver transplantation). The E1-5′, NS2, and NS3-3′ regions showed the next greatest divergence after liver transplantation, with mean percent changes in HMR of approximately 2.5, 2.9, and 3.3, respectively. The core, NS4-5′, NS4-3′, and NS5b regions showed the least amount of genetic divergence after liver transplantation, with a mean percent change in HMR of less than 1.5 for each region. Of the five nonstructural gene regions analyzed, the NS5b region showed the least genetic change (<0.2% change in HMR) after transplant. These results are in general agreement with previous studies of small numbers of patients using nucleotide sequence analysis on the mutation rates of specific regions of the HCV genome (34, 35).
Changes in HCV quasispecies heterogeneity according to posttransplant disease status.
The posttransplant disease classifications for the 22 patients in the current study are summarized in Table 1. Figure 4 summarizes the changes in HCV quasispecies heterogeneity after liver transplantation for the 22 patients according to posttransplant disease pattern. Changes in HCV structural genes are summarized in Fig. 4A, while changes in HCV nonstructural genes are summarized in Fig. 4B.
FIG. 4.
Genetic divergence of the HCV structural (A) and nonstructural (B) genes after liver transplantation according to disease group. Data were generated by HMA. The mean percent change in HMR between pre- and posttransplant time points is plotted along the y axis. See Materials and Methods and Table 1 for the classification of patients by disease severity.
In all three disease groups, minimal changes were observed within the core region in HCV quasispecies after liver transplantation, with mean percent changes in HMR ranging from 0.38 to 1.27. In contrast, the E1-5′ and E1/E2 regions of HCV quasispecies isolated from patients in the asymptomatic and moderate disease groups showed a greater genetic change after liver transplantation than the same regions isolated from HCV quasispecies in patients in the severe disease group. The E1-5′ region of the asymptomatic and moderate disease groups had average changes in HMR of 2.7 and 3.9% after liver transplantation, respectively, versus 0.3% for the disease group (P = 0.05) (Fig. 4A). The E1-E2 junction region showed the greatest degree of genetic divergence after liver transplantation for all three disease groups. However, HCV isolated from patients in the asymptomatic and moderate disease groups had, on average, a 2.5- to 3.3-fold-greater change in HMRs between pre- and posttransplant time points than HCV quasispecies isolated from patients in the severe disease group (Fig. 4A).
Figure 4B compares changes in HCV nonstructural genes after liver transplantation according to patient disease status. HCV quasispecies isolated from patients in the asymptomatic and moderate disease groups had, on average, at least a 1% greater change in HMR in the NS2, NS4-3′, and NS4-5′ regions than HCV quasispecies isolated from patients in the severe disease group. In the NS4-5′ region, patients in the asymptomatic group showed a mean percent change in HMR of 2.0 compared to 0.6 or 0.8 for patients in the moderate or severe disease group, respectively. Finally, all three disease groups showed a very similar degree of genetic change within the NS3-3′ region after transplant (3.2 to 3.5% change in HMR) (Fig. 4B).
In summary, these experiments indicate that in asymptomatic and moderate disease groups, the pretransplant HCV quasispecies underwent a greater degree of genetic divergence after liver transplantation than HCV quasispecies isolated from patients within the severe disease group. Multigene analysis by the HMA technique indicated that HCV quasispecies isolated from asymptomatic patients showed a greater percent change in HMR than those from patients in the severe disease group for all regions studied except NS3-3′. Likewise, HCV quasispecies isolated from the moderate disease group also had a greater percent change in HMR than the severe disease group for all regions studied except the core and NS3-3′ region. The mean percent changes in HMR of all the regions studied for each of the three disease groups are summarized in Fig. 5.
FIG. 5.
Multigene analysis of HCV quasispecies genetic divergence after liver transplantation, according to disease group. The y axis depicts the average percent change in HCV genetic divergence after liver transplantation for the eight genetic regions analyzed in the current study (Fig. 1). HMR were calculated as described in Materials and Methods.
DISCUSSION
To date, the vast majority of studies describing HCV quasispecies evolution in humans have focused on the most genetically diverse region of the HCV genome, namely, HVR1. In contrast, there have been very few longitudinal analyses of multiple HCV genes or entire HCV genomes in infected humans or chimpanzees. Previous studies tracking multiple HCV genomic regions over time have been restricted to evaluation of only one or a few infected individuals, and correlation with disease activity has been limited (30, 35). Therefore, one of the principal contributions of the present study is the systematic longitudinal analysis of eight individual HCV genomic regions (approximately 44% of the HCV genome) in a relatively large sample of patients (n = 22) with very thorough histopathological assessment of clinical disease status (at least five sequential posttransplant liver biopsies per patient). Thus, the present study represents a highly systematic approach to the important yet technically challenging question of the role of HCV quasispecies in the pathogenesis of liver disease in humans.
In a previous report, we described HCV quasispecies tracking patterns in five patients with genotype 1 infection, three of whom developed severe posttransplant hepatitis C and two of whom developed asymptomatic HCV viremia (23). Based on extensive analysis of HVR1, we concluded that pretransplant quasispecies major variants were efficiently propagated after liver transplantation in the patients with severe hepatitis, but not in the patients with asymptomatic infection. Rather, we detected the emergence of quasispecies minor variants in the latter group of patients. Our present findings support our previous observations in that HCV quasispecies isolated from patients in the severe disease group had significantly lower genetic divergence than HCV quasispecies isolated from patients with mild or asymptomatic infections of the same HCV genotype (genotype 1). Based on results from multigene analysis by HMA, we can now conclude that HCV genetic diversification in liver transplant recipients occurs in multiple regions of the HCV genome and is not restricted to HVR1. Furthermore, in nearly all regions studied, HCV diversification was greater in the asymptomatic or moderate disease group than in the severe disease group. For example, in the NS4-3′ region, diversification was 2.4-fold greater in asymptomatic patients than in patients with severe disease. However, the results reached statistical significance only for the envelope region, although it is likely that inclusion of more patients in each disease group would enhance the statistical significance of our findings for nonstructural genes. In our current study, the NS3 region was the only gene to show an equal degree of genetic divergence in patients from all three disease categories. While undersampling or specimen dropout could certainly account for these results, it is interesting to speculate that NS3 may be under different selective pressures than other HCV genes, especially since the NS3 gene product is a multifunctional enzyme with protease, nucleoside triphosphatase, and helicase activities (19).
From a mechanistic perspective, variation within the HCV genome is assumed to be caused by random mutation and selection of variants which are most fit to propagate in a given host. For example, in the immunocompetent host, antibodies directed against envelope gene products appear to play an important role in shaping quasispecies repertoires (29, 31, 42). However, there are likely to be many forces which influence the selection of variants in other regions of the HCV genome. In theory, the growth of specific HCV variants may be influenced by either positive selection (e.g., growth advantage based on translational or replicative fitness) or by negative selection (e.g., growth suppression based on immunological responses). Since most of the patients in the asymptomatic and moderate disease groups had changes in multiple regions of HCV genomes after transplantation, the results seem consistent with the hypothesis that a broadly directed pressure involving multiple HCV regions is protective to the host, even though viral replication continues at high levels.
Viral quasispecies selection by immunological mechanisms at the time of liver transplantation is one factor which requires careful study. Several studies have shown that the majority of HCV-infected liver transplant recipients remain anti-HCV antibody positive after transplantation (8, 13, 28). We are currently testing the hypothesis that the formation of antibody complexes with quasispecies major variants prior to transplant is associated with reduced viremia during the immediate posttransplant period and a more slowly progressive disease course. If such a mechanism were operational, it might argue for the use of pretransplant conditioning of HCV quasispecies, either by adoptive immunotherapy or perhaps treatment with agents such as interferon.
A second factor to be studied is the role of cellular immune responses in modulating disease activity after liver transplantation. In the nontransplant setting, rigorous cellular responses to HCV nonstructural antigens have been associated with viral clearance in acute hepatitis C (11, 12). Therefore, one might speculate that ineffective cellular immune responses contribute to the pathogenesis of hepatitis C in the transplant setting by allowing the propagation of more-cytopathic variants. One hypothesis to test is that specific HCV genes play a critical role in the development of posttransplant hepatitis. In asymptomatic patients, host pressures might prevent evolution of pathogenic quasispecies by direct selection of viral gene products with nontoxic biochemical properties. Thus, divergence of genes such as NS4 may reflect an attenuation of viral pathogenicity, allowing efficient replication but inefficient induction of hepatitis. An alternative hypothesis would be that viral determinants present in “pathogenic variants” are capable of suppressing critical host responses, allowing for persistence of such variants. In vitro experimentation and generation of infectious HCV molecular clones will be required to solve these questions in an unequivocal manner. We are in the process of expressing isolated HCV genes in human liver cell lines to investigate the interactions between viral and host proteins at the cellular level. The development of cell cultures that support HCV replication and animal models should also greatly facilitate meaningful classification of HCV phenotypes.
Genomic insertions and rearrangements of some members of the pestivirus genus have been directly associated with cytopathogenicity in cell culture and the development of severe host disease (reviewed in reference 33). Although not a primary objective of the current study, the multigene HMA technique provided an opportunity to scan a relatively large number of viral genomes for genetic insertions and rearrangements. While our preliminary analysis revealed no direct evidence of large genomic insertions or rearrangements, the occasional dropout during PCR amplification might be related to the presence of altered genomes. Importantly, Forns et al. (18) have recently described PCR and cloning bias in the generation of HCV protein expression constructs. Thus, although we cannot rule out the possibility that our current results are biased due to technical limitations, the results so far suggest HCV genomes are intact in patients with severe disease.
In summary, the current study demonstrates adaptation of the HMA technique for characterizing and tracking HCV quasispecies by analyzing multiple regions of the HCV genome in an immunosuppressed population of patients. This approach allowed a larger number of patients and a larger proportion of the HCV genome to be analyzed than in prior longitudinal studies of quasispecies diversity. We conclude that greater rates of HCV quasispecies diversification are associated with mild or moderate liver disease activity in this model, and we postulate that both host and viral factors play important roles in the pathogenesis of chronic hepatitis C in this population. Further research is necessary to determine the extent to which the observed results are due to a different type of immune response in asymptomatic patients compared with patients in the severe disease group, a replicative advantage of certain quasispecies populations in the new liver allografts of specific patient subgroups, and/or selective tropism of quasispecies variants for hepatic versus nonhepatic compartments. The techniques described herein should prove useful for monitoring HCV quasispecies during antiviral therapy, for systematic studies of HCV quasispecies transmission and tropism, and for optimizing tissue culture and animal infectivity models of hepatitis C.
ACKNOWLEDGMENTS
We thank Stephen Polyak, Larry Corey, and Martina Gerotto for helpful discussions and Corazon dela Rosa, Minjun Chung, Maureen Guajardo, and Anthony Marquardt for excellent technical support.
Funding for this research was provided by NIH grants U19 AI40032-02 and AI39049-02.
REFERENCES
- 1.Bukh J, Purcell R, Miller R. Importance of primer selection for the detection of hepatitis C virus RNA with the polymerase chain reaction assay. Proc Natl Acad Sci USA. 1992;89:187–191. doi: 10.1073/pnas.89.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bukh J, Purcell R H, Miller R H. Sequence analysis of the core gene of 14 hepatitis C virus genotypes. Proc Natl Acad Sci USA. 1994;91:8239–8243. doi: 10.1073/pnas.91.17.8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castillo I, Bartolome J, Quiroga J A, Carreno V. Comparison of several PCR procedures for detection of serum HCV-RNA using different regions of the HCV genome. J Virol Methods. 1992;38:71–79. doi: 10.1016/0166-0934(92)90170-i. [DOI] [PubMed] [Google Scholar]
- 4.Chazouilleres O, Kim M, Combs C, Ferrell L, Bacchetti P, Roberts J, Ascher N L, Neuwald P, Wilber J, Urdea M. Quantitation of hepatitis C virus RNA in liver transplant recipients. Gastroenterology. 1994;106:994–999. doi: 10.1016/0016-5085(94)90759-5. [DOI] [PubMed] [Google Scholar]
- 5.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 6.Choo Q L, Kuo G, Weiner A J, Overby L R, Bradley D W, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 7.Choo Q L, Richman K H, Han J H, Berger K, Lee C, Dong C, Gallegos C, Coit D, Medina S R, Barr P J, et al. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci USA. 1991;88:2451–2455. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crespo J, Carte B, Lozano J L, Casafont F, Rivero M, de la Cruz F, Pons-Romero F. Hepatitis C virus recurrence after liver transplantation: relationship to anti-HCV core IgM, genotype, and level of viremia. Am J Gastroenterol. 1997;92:1458–1462. [PubMed] [Google Scholar]
- 9.Davidson F, Simmonds P, Ferguson J C, Jarvis L M, Dow B C, Follett E A, Seed C R, Krusius T, Lin C, Medgyesi G A, et al. Survey of major genotypes and subtypes of hepatitis C virus using RFLP of sequences amplified from the 5′ non-coding region. J Gen Virol. 1995;76:1197–1204. doi: 10.1099/0022-1317-76-5-1197. [DOI] [PubMed] [Google Scholar]
- 10.Delwart E L, Shpaer E G, Louwagie J, McCutchan F E, Grez M, Rubsamen W H, Mullins J I. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science. 1993;262:1257–1261. doi: 10.1126/science.8235655. [DOI] [PubMed] [Google Scholar]
- 11.Diepolder H M, Gerlach J T, Zachoval R, Hoffmann R M, Jung M C, Wierenga E A, Scholz S, Santantonio T, Houghton M, Southwood S, Sette A, Pape G R. Immunodominant CD4+ T-cell epitope within nonstructural protein 3 in acute hepatitis C virus infection. J Virol. 1997;71:6011–6019. doi: 10.1128/jvi.71.8.6011-6019.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diepolder H M, Zachoval R, Hoffmann R M, Wierenga E A, Santantonio T, Jung M C, Eichenlaub D, Pape G R. Possible mechanism involving T-lymphocyte response to non-structural protein 3 in viral clearance in acute hepatitis C virus infection. Lancet. 1995;346:1006–1007. doi: 10.1016/s0140-6736(95)91691-1. [DOI] [PubMed] [Google Scholar]
- 13.Donegan E, Wright T L, Roberts J, Ascher N L, Lake J R, Neuwald P, Wilber J, Quan S, Kuramoto I K, Dinello R K, et al. Detection of hepatitis C after liver transplantation. Four serologic tests compared. Am J Clin Pathol. 1997;104:673–679. doi: 10.1093/ajcp/104.6.673. [DOI] [PubMed] [Google Scholar]
- 14.Enomoto N, Takada A, Nakao T, Date T. There are two major types of hepatitis C virus in Japan. Biochem Biophys Res Commun. 1990;170:1021–1025. doi: 10.1016/0006-291x(90)90494-8. [DOI] [PubMed] [Google Scholar]
- 15.Farci P, Alter H J, Wong D C, Miller R H, Govindarajan S, Engle R, Shapiro M, Purcell R H. Prevention of hepatitis C virus infection in chimpanzees after antibody-mediated in vitro neutralization. Proc Natl Acad Sci USA. 1994;91:7792–7796. doi: 10.1073/pnas.91.16.7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farci P, Shimoda A, Wong D, Cabezon T, De Gioannis D, Strazzera A, Shimizu Y, Shapiro M, Alter H J, Purcell R H. Prevention of hepatitis C virus infection in chimpanzees by hyperimmune serum against the hypervariable region 1 of the envelope 2 protein. Proc Natl Acad Sci USA. 1996;93:15394–15399. doi: 10.1073/pnas.93.26.15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.F′eray C, Gigou M, Samuel D, Paradis V, Wilber J, David M F, Urdea M, Reynes M, Br′echot C, Bismuth H. The course of hepatitis C virus infection after liver transplantation. Hepatology. 1994;20:1137–1143. doi: 10.1002/hep.1840200506. [DOI] [PubMed] [Google Scholar]
- 18.Forns X, Bukh J, Purcell R H, Emerson S U. How Escherichia coli can bias the results of molecular cloning: preferential selection of defective genomes of hepatitis C virus during the cloning procedure. Proc Natl Acad Sci USA. 1997;94:13909–13914. doi: 10.1073/pnas.94.25.13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallinari P, Brennan D, Nardi C, Brunetti M, Tomel L, Steinkuhler C, De Francesco R. Multiple enzymatic activities associated with recombinant NS3 protein hepatitis C virus. J Virol. 1998;72:6758–6769. doi: 10.1128/jvi.72.8.6758-6769.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gretch D, Corey L, Wilson J, dela Rosa C, Willson R, Carithers R, Jr, Busch M, Hart J, Sayers M, Han J. Assessment of hepatitis C virus RNA levels by quantitative competive RNA PCR: high titer viremia correlates with advanced stage of disease. J Infect Dis. 1994;169:1219–1225. doi: 10.1093/infdis/169.6.1219. [DOI] [PubMed] [Google Scholar]
- 21.Gretch D, dela Rosa C, Carithers R, Willson R, Williams B, Corey L. Assessment of hepatitis C viremia using molecular amplification technologies: correlations and clinical implications. Ann Int Med. 1995;123:321–329. doi: 10.7326/0003-4819-123-5-199509010-00001. [DOI] [PubMed] [Google Scholar]
- 22.Gretch D R, Bacchi C E, Corey L, Rosa C D, Lesniewski R R, Kowdley K, Gown A, Frank I, Perkins J D, Carithers R L., Jr Persistent hepatitis C virus infection following liver transplantation: clinical and virologic features. Hepatology. 1995;22:1–9. [PubMed] [Google Scholar]
- 23.Gretch D R, Polyak S J, Wilson J J, Carithers R L, Jr, Perkins J D, Corey L. Tracking hepatitis C virus quasispecies major and minor variants in symptomatic and asymptomatic liver transplant recipients. J Virol. 1996;70:7622–7631. doi: 10.1128/jvi.70.11.7622-7631.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gretch D R, Wilson J J, Carithers R L, Jr, dela Rosa C, Han J H, Corey L. Detection of hepatitis C virus RNA: comparison of one-stage polymerase chain reaction (PCR) with nested-set PCR. J Clin Microbiol. 1993;31:289–291. doi: 10.1128/jcm.31.2.289-291.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hijikata M, Kato N, Ootsuyama Y, Nakagawa M, Ohkoshi S, Shimotohno K. Hypervariable regions in the putative glycoprotein of hepatitis C virus. Biochem Biophys Res Commun. 1991;175:220–228. doi: 10.1016/s0006-291x(05)81223-9. [DOI] [PubMed] [Google Scholar]
- 26.Hofgartner W T, Polyak S J, Sullivan D G, Carithers R L, Jr, Gretch D R. Mutations in the NS5A gene of hepatitis c virus in North American patients infected with HCV genotype 1a or 1b. J Med Virol. 1997;53:118–126. doi: 10.1002/(sici)1096-9071(199710)53:2<118::aid-jmv3>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 27.Houghton M, Weiner A, Han J, Kuo G, Choo Q-L. Molecular biology of hepatitis C viruses: implications for diagnosis, development and control of viral disease. Hepatology. 1991;14:381–388. [PubMed] [Google Scholar]
- 28.Hsu H H, Wright T L, Tsao S C, Combs C, Donets M, Feinstone S M, Greenberg H B. Antibody response to hepatitis C virus infection after liver transplantation. Am J Gastroenterol. 1994;89:1169–1174. [PubMed] [Google Scholar]
- 29.Kato N, Sekiya H, Ootsuyama Y, Nakazawa T, Hijikata M, Ohkoshi S, Shimotohno K. Humoral immune response to hypervariable region 1 of the putative envelope glycoprotein (gp70) of hepatitis C virus. J Virol. 1993;67:3923–3930. doi: 10.1128/jvi.67.7.3923-3930.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kurosaki M, Enomoto N, Marumo F, Sato C. Rapid sequence variation of the hypervariable region of hepatitis C virus during the course of chronic infection. Hepatology. 1993;18:1293–1299. [PubMed] [Google Scholar]
- 31.Martell M, Esteban J I, Quer J, Genescà J, Weiner A, Esteban R, Guardia J, Gómez J. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. J Virol. 1992;66:3225–3229. doi: 10.1128/jvi.66.5.3225-3229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martell M, Esteban J I, Quer J, Vargas V, Esteban R, Guardia J, Gómez J. Dynamic behavior of hepatitis C virus quasispecies in patients undergoing orthotopic liver transplantation. J Virol. 1994;68:3425–3436. doi: 10.1128/jvi.68.5.3425-3436.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyers G, Thiel H-J. Molecular characterization of pestiviruses. Adv Vir Res. 1996;41:53–118. doi: 10.1016/s0065-3527(08)60734-4. [DOI] [PubMed] [Google Scholar]
- 34.Mizokami M, Gojobori T, Lau J Y N. Molecular evolutionary virology: its application to hepatitis C virus. Gastroenterology. 1994;107:1181–1182. doi: 10.1016/0016-5085(94)90245-3. [DOI] [PubMed] [Google Scholar]
- 35.Ogata N, Alter H J, Miller R H, Purcell R H. Nucleotide sequence and mutation rate of the H strain of hepatitis C virus. Proc Natl Acad Sci USA. 1991;88:3392–3396. doi: 10.1073/pnas.88.8.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Polyak S, Faulkner G, Carithers R, Jr, Corey L, Gretch D. Assessment of hepatitis C virus quasispecies heterogeneity by gel shift analysis: correlation with response to interferon therapy. J Infect Dis. 1997;175:1101–1107. doi: 10.1086/516448. [DOI] [PubMed] [Google Scholar]
- 37.Polyak S J, McArdle S, Liu S-L, Sullivan D G, Chung M, Hofgärtner W T, Carithers R L, Jr, McMahon B J, Mullins J I, Corey L, Gretch D R. Evolution of hepatitis C virus quasispecies in hypervariable region 1 and the putative interferon sensitivity-determining region during interferon therapy and natural infection. J Virol. 1998;72:4288–4296. doi: 10.1128/jvi.72.5.4288-4296.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1989. [Google Scholar]
- 39.Shuhart M C, Bronner M P, Gretch D R, Thomassen L V, Wartelle C F, Tateyama H, Emerson S S, Perkins J D, Carithers R L. Histological and clinical outcome after liver transplantation for hepatitis. Hepatology. 1997;26:1646–1652. doi: 10.1002/hep.510260638. [DOI] [PubMed] [Google Scholar]
- 40.Simmonds P, Holmes E C, Cha T A, Chan S W, McOmish F, Irvine B, Beall E, Yap P L, Kolberg J, Urdea M S. Classification of hepatitis C virus into six major genotypes and a series of subtypes by phylogenetic analysis of the NS-5 region. J Gen Virol. 1993;74:2391–2399. doi: 10.1099/0022-1317-74-11-2391. [DOI] [PubMed] [Google Scholar]
- 41.Weiner A J, Brauer M J, Rosenblatt J, Richman K H, Tung J, Crawford K, Bonino F, Saracco G, Choo Q L, Houghton M. Variable and hypervariable domains are found in the regions of HCV corresponding to the flavivirus envelope and NS1 proteins and the pestivirus envelope glycoproteins. Virology. 1991;180:842–848. doi: 10.1016/0042-6822(91)90104-j. [DOI] [PubMed] [Google Scholar]
- 42.Weiner A J, Geysen H M, Christopherson C, Hall J E, Mason T J, Saracco G, Bonino F, Crawford K, Marion C D, Crawford K A, Brunetto M, Barr P J, Miyamura T, McHutchinson J, Houghton M. Evidence for immune selection of hepatitis C virus (HCV) putative envelope glycoprotein variants: potential role in chronic HCV infections. Proc Natl Acad Sci USA. 1992;89:3468–3472. doi: 10.1073/pnas.89.8.3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson J J, Polyak S J, Day T D, Gretch D R. Characterization of simple and complex hepatitis C virus quasispecies by heteroduplex gel shift analysis: correlation with nucleotide sequencing. J Gen Virol. 1995;76:1763–1771. doi: 10.1099/0022-1317-76-7-1763. [DOI] [PubMed] [Google Scholar]
- 44.Wright T L, Donegan E, Hsu H H, Ferrell L, Lake J R, Kim M, Combs C, Fennessy S, Roberts J P, Ascher N L. Recurrent and acquired hepatitis C viral infection in liver transplant recipients. Gastroenterology. 1992;103:317–322. doi: 10.1016/0016-5085(92)91129-r. [DOI] [PubMed] [Google Scholar]
- 45.Zhou S, Terrault N A, Ferrell L, Hahn J A, Lau J Y, Simmonds P, Roberts J P, Lake J R, Ascher N L, Wright T L. Severity of liver disease in liver transplant recipients with hepatitis C virus infection: relationship to genotype and level of viremia. Hepatology. 1996;24:1041–1046. doi: 10.1002/hep.510240510. [DOI] [PubMed] [Google Scholar]