Abstract
Rhizopus oryzae is used for industrial production of lactic acid, yet little is known about the genetics of this fungus. In this study I cloned two genes, ldhA and ldhB, which code for NAD+-dependent l-lactate dehydrogenases (LDH) (EC 1.1.1.27), from a lactic acid-producing strain of R. oryzae. These genes are similar to each other and exhibit more than 90% nucleotide sequence identity and they contain no introns. This is the first description of ldh genes in a fungus, and sequence comparisons revealed that these genes are distinct from previously isolated prokaryotic and eukaryotic ldh genes. Protein sequencing of the LDH isolated from R. oryzae during lactic acid production confirmed that ldhA codes for a 36-kDa protein that converts pyruvate to lactate. Production of LdhA was greatest when glucose was the carbon source, followed by xylose and trehalose; all of these sugars could be fermented to lactic acid. Transcripts from ldhB were not detected when R. oryzae was grown on any of these sugars but were present when R. oryzae was grown on glycerol, ethanol, and lactate. I hypothesize that ldhB encodes a second NAD+-dependent LDH that is capable of converting l-lactate to pyruvate and is produced by cultures grown on these nonfermentable substrates. Both ldhA and ldhB restored fermentative growth to Escherichia coli (ldhA pfl) mutants so that they grew anaerobically and produced lactic acid.
Global lactic acid production is estimated to be more than 100,000 tons per year, and approximately 75% of the lactic acid produced is used in the food industry as an acidulant for flavor or as an antimicrobial agent (26). More recent uses for lactic acid have been driven by ecological interest and include production of the nonchlorinated solvent ethyl lactate and the biodegradable plastic polylactic acid. Polylactic acid is a polymer whose properties are similar to those of polyolefins, and it could replace a significant portion of the polyethylene terephthalate-based polymers, which are produced at a rate of approximately 15 million tons per year worldwide (4, 14, 26). Lactic acid can be synthesized chemically, but such synthesis results in a mixture of d and l isomers. The products of microbiological fermentations depend on the organism used and also may include a mixture the two isomers or individual isomers in a stereospecific form. The desired stereospecificity of the product depends on the intended use; however, l-(+)-lactic acid is the form desired for most applications.
In 1936, Lockwood et al. found that in a chemically defined medium, Rhizopus oryzae was able to aerobically convert glucose to large amounts of l-(+)-lactic acid (18). Research on lactic acid production by Rhizopus spp. has continued primarily because of the ease of product purification and the ability of the fungus to utilize both complex carbohydrates and pentose sugars (35, 38). Production of lactic acid by Rhizopus cultures is often preferred to bacterial fermentations, because lactobacilli require that the growth medium be supplemented with complex components, such as yeast extract, which adds to the cost and complicates purification.
Rhizopus cells can produce more than 1.5 mol of lactic acid from 1 mol of glucose under aerobic conditions (19), and the remainder of the glucose is converted to mycelial mass, glycerol, fumarate, or ethanol. Previously, other authors have described (22, 24, 25, 39) the presence of two lactate dehydrogenase (LDH) enzymes in R. oryzae. An NAD+-dependent LDH (EC 1.1.1.27) that converts pyruvate to lactate but exhibits negligible activity for the reverse reaction is produced during early growth and synthesis of lactic acid. It is thought that after glucose is exhausted, a second LDH activity is involved in NAD+-independent oxidation of l-lactic acid to pyruvic acid.
In this study, my objective was to isolate and characterize the gene involved in synthesis of lactic acid by R. oryzae, so I focused on the NAD+-dependent conversion of pyruvate to lactate. In the process of cloning this gene, I identified a second gene that appears to encode another NAD+-dependent LDH that has not been described previously. This is the first description of ldh genes in a fungal species, and my results fill a significant void between information concerning higher eukaryotic (e.g., plant, mammalian, fish, etc.) ldh and prokaryotic ldh genes, which have been extensively studied.
MATERIALS AND METHODS
Preparation of genomic and cDNA libraries.
R. oryzae NRRL 395 DNA was purified by cetyltrimethylammonium bromide extraction (2), partially digested with Sau3A, and used to construct a genomic library in Lambda Zap Express (Stratagene, La Jolla, Calif.) as recommended by the manufacturer. To prepare the cDNA library, R. oryzae spores were germinated for 24 h with shaking at 30°C in RZ medium, which contained (per liter) 100 g of glucose, 2 g of (NH4)2SO4, 0.5 g of KH2PO4, 0.25 g of MgSO4, 2.2 mg of ZnSO4 · 7H2O, and 0.5 mg of MnCl2 · 4H2O. Calcium carbonate chips (Malinckrodt Baker, Paris, Ky.) were added to control the pH, and the preparation was incubated for an additional 8 h before the mycelium was harvested in order to isolate RNA by a hot phenol method (2). A cDNA library was prepared by following the manufacturer's instructions for a Lambda Zap unidirectional cDNA synthesis kit (Stratagene).
Isolation of LDH.
R. oryzae was grown under conditions similar to those used for cDNA library preparation. Mycelium was collected by filtration and was washed with ice-cold sodium phosphate buffer (100 mM, pH 7.0) before it was suspended in 1 volume of the same buffer containing glycerol (10%, vol/vol), dithiothreitol (0.15 mg/ml), phenylmethylsulfonyl fluoride (1 mM), pepstatin A (1 μg/ml), and leupeptin (0.5 μg/ml). The cell mass was broken by agitating it with 0.45-mm-diameter glass beads for 3 min at 4,000 rpm in a Braun (Frankfurt, Germany) homogenizer and was clarified by centrifugation at 12,000 × g for 30 min. The protein that precipitated between 35 and 55% ammonium sulfate saturation was collected and dissolved in BTP buffer (50 mM bis-Tris-propane [pH 6.8], 10% glycerol, 0.15 mg of dithiothreitol per ml). The crude protein was desalted and separated from high-molecular-weight polysaccharides by using a Sephadex G-150 column (2.5 by 30 cm) equilibrated in BTP buffer. Fractions containing activity were combined, concentrated with a Centriprep-20 concentrator (Amicon, Beverly, Mass.), and injected onto a SynChropak AX300 anion-exchange high-performance liquid chromatography (HPLC) column (SynChrom, Inc., Linden, Ind.) equilibrated in BTP buffer. The column was washed with BTP buffer until all unbound proteins were removed. LDH was eluted by using a linear 0 to 0.5 M NaCl gradient in the same buffer. Combined fractions were desalted and concentrated by washing in the Centriprep-20 concentrator. The protein was again resolved by anion-exchange chromatography by using a Bio-Gel TSK-DEAE-5-PW column (Bio-Rad, Hercules, Calif.) and the conditions described above. Pooled fractions were concentrated and separated by using a Sephacryl 300 column (1.5 by 115 cm) that was equilibrated in BTP buffer. The fractions that were collected were precipitated, resolved by denaturing polyacrylamide gel electrophoresis (PAGE), and transferred to a polyvinylidene difluoride membrane (6). The protein corresponding to the predominant semipure 36-kDa enzyme was eluted, digested, and sequenced by workers at the Protein/DNA Technology Center of Rockefeller University.
Analysis of LDH activity and zymograms.
LDH activity was assayed spectrophotometrically by measuring the first-order change in absorbance at 340 nm as a result of oxidation of NADH or reduction of NAD+. Reductive LDH activity (pyruvate → lactate) was determined by monitoring the decrease in absorbance of 175 μM NADH in 0.1 M bis-Tris-propane (pH 6.8), after the reaction was started by adding sodium pyruvate to a final concentration of 4 mM. LDH oxidative activity (lactate → pyruvate) was determined by measuring the increase in absorbance with 200 mM lithium lactate–0.1 M Tris-Cl (pH 8.0), after the reaction was started with 750 μM (final concentration) NAD+. The pH values for reaction buffers were chosen based on the results of preliminary experiments in which I determined the optimal conditions that resulted in the maximum change in absorbance. All protein concentrations were adjusted to ensure that the change in absorbance followed first-order kinetics for at least 3 to 5 min. All assays were performed in triplicate, and 1 U of enzyme activity was defined as the amount of activity necessary to convert 1 μmol of NADH to NAD+ per min or 1 μmol of NAD+ to NADH per min. Protein concentrations were determined with a Bio-Rad protein assay kit.
LDH zymograms were obtained by separating 10 μg of protein on a native 7.5% PAGE gel for 2 h at 4 W. LDH reductive activity was detected by soaking the gel for 1 h at 33°C in a solution containing 0.1 M bis-Tris-propane (pH 6.8), 1 mM NADH, and 25 mM sodium pyruvate. The gel was washed twice in water and then developed in a solution containing 0.1 M Tris-Cl (pH 8.0), 0.25 mg of nitroblue tetrazolium per ml, and 0.02 mg of phenazine methylsulfate per ml. NADH converted to NAD+ does not form a blue precipitate and leaves a clear zone on a gel. Oxidative reaction activity was detected by soaking the gel in a solution containing 0.1 M Tris-Cl (pH 8.0), 750 μM NAD+, 200 mM lithium lactate, 0.1 mg of nitroblue tetrazolium per ml, and 0.02 mg of phenazine methylsulfate per ml until bands resulting from the conversion of NAD+ to NADH were clearly visible. The intensities of bands in zymograms were determined with Kodak 1D Image Analysis software (Eastman Kodak, Rochester, N.Y.).
Isolation of ldh genes.
A method which involved an anchored PCR (2) performed with a degenerate primer was used to isolate an ldh gene fragment from R. oryzae. The degenerate primer 5′-(GC)(AT)(AG) TC(GAT) CC(GA) TG(CT) TCA CC-3′ was designed to preferentially anneal to a region encoding the consensus amino acid motif GEHGDS, which is involved in substrate binding and proton transfer for NAD+-dependent l-LDH enzymes (10, 36). This primer was used with M13 reverse primer to amplify the upstream region of ldh from the cDNA library. PCR amplification was performed as described by Ausubel et al. (2), except that the following program was used: 25 cycles consisting of 95°C for 45 s, 52°C for 45 s, and 72°C for 90 s. A 600-bp fragment was recovered and TA cloned into pCRII/TOPO (Invitrogen, Carlsbad, Calif.). Sequence analysis confirmed that the fragment represented a partial ldh gene. A probe was prepared from the gel-purified fragment and used to isolate hybridizing clones from both genomic and cDNA libraries.
Transcript differentiation and expression of ldhA and ldhB.
R. oryzae was grown in RZ medium containing 1.5% glycerol and 0.5% Trypticase peptone for 18 h with shaking at 30°C. The mycelium was then filtered and transferred to fresh RZ medium containing 1% (wt/vol) glucose, 1% (wt/vol) xylose, 1% (wt/vol) trehalose, 1% (wt/vol) Trypticase peptone, 1% (wt/vol) glycerol, 1% (wt/vol) ethanol, or 1% (wt/vol) sodium lactate. The preparations were incubated for an additional 5.5 h, and then RNA was isolated from the cultures. Northern analysis was performed by using 10 μg of RNA from each culture. Additionally, this RNA was used along with oligo(dT) primers to synthesize first-strand cDNA with the Superscript System as recommended by the manufacturer (Life Technologies, Gaithersburg, Md.). PCR amplification was performed as described above by using this cDNA as the template and primers specific for the ldhB sequence (5′-GCAGACGCAGCCAGTGTAAGCA-3′ and 5′-CAACGGCTGCCCACCAATC-3′).
In a similar expression study, R. oryzae was grown in RZ medium containing 2% glycerol for 72 h. The mycelium was harvested and transferred to fresh medium containing 1% glucose, 1% xylose, 1% trehalose, 1% glycerol, 1% ethanol, and 1% sodium lactate. The preparations were incubated for 16 h before both protein and RNA were isolated. The RNA was used to make cDNA which served as a template for amplification with PCR primers referred to as universal ldh primers (5′-ACACGCCCATCCGAGCAGG-3′ and 5′-GCACAGGCACCAATTCCATAAAAC-3′). These universal primers were designed to anneal to regions that are identical in both ldhA and ldhB genes. The amplified product was TA cloned, and 10 to 20 isolates from each sample preparation were sequenced to determine whether each ldh transcript was present. Protein samples were analyzed to determine whether LDH activity was present by using spectrophotometric methods and zymogram analysis.
Time course expression studies were performed at 32°C in 400 ml of RZ medium containing 10% glucose by using a bubble column apparatus (diameter, 4.5 cm; length, 37 cm) constructed in my laboratory. Spores were inoculated to a final concentration of 106 spores/ml, and sterile air was sparged from the bottom of the column at a rate of 0.4 liter/min. Ammonium hydroxide was added with a peristaltic pump to maintain the pH at 5.5. Samples were taken and used to determine lactic acid and glucose concentration. Additionally, mycelia were collected and used for protein and RNA isolation. Lactic acid and glucose contents were analyzed by HPLC by using an HPX-87H column (Bio-Rad Laboratories) and a model 410 differential refractometer (Waters, Milford, Mass.). An enzymatic lactate detection kit (product no. 735; limit of detection, 0.2 mM; Sigma Chemical Co., St. Louis, Mo.) was used when the concentration of lactic acid was below the limit of detection of HPLC methods.
Complementation of Escherichia coli.
E. coli DC1368(thr-1 leu-6 thi-6 lacY tonA22 rpsL ldhA::kan pfl::Cam) was provided by D. P. Clark (Southern Illinois University, Carbondale). This strain, which lacks a functional LdhA and pyruvate-formate lyase (Pfl), is not able to grow anaerobically because it cannot regenerate NAD+ fermentatively (20). Plasmid pBluescript II KS(−) (Stratagene) containing either a genomic 1.8-kb HindIII fragment containing ldhA or a 3.2-kb HindIII fragment containing ldhB was introduced into E. coli DC1368. Transformants were tested to determine whether they grew anaerobically in rich broth (3) supplemented with cysteine-HCl (50 mg/ml) and glucose (0.4%). Culture tubes were flushed with nitrogen gas and sealed with rubber stoppers. Transformants containing only the vector served as negative controls.
Molecular analyses.
Northern and Southern analyses were performed by using the Genius system (Boehringer Mannheim, Indianapolis, Ind.) as recommended by the manufacturer. RNase protection assays (RPA) were performed with RPA-III (Ambion, Austin, Tex.) by using conditions used for nondenaturing 5% PAGE. Gel-purified, full-length ldhA was used for random primed labeling with digoxigenin and as a probe for Southern analysis. A 263-bp region of ldhA between BamHI and EcoRV restriction sites was used to prepare digoxigenin-labeled RNA by T3 transcription. RNA-labeled probes were used for both RPA and Northern analyses.
Multiple protein sequence comparisons were performed as Clustal analyses by using Lasergene Megalign (DNA Star, Madison, Wis.). The aligned sequences were arranged by bootstrap analysis and tree construction by using TreeCon (33). Default program settings were used for all analyses.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the ldhA and ldhB sequences are AF226154 and AF226155, respectively.
RESULTS
Isolation of ldh genes.
A 600-bp fragment was amplified by using anchored PCR performed with a degenerate primer and the cDNA library as the template. This fragment appeared to be a novel ldh fragment when the data were analyzed by performing Blast comparisons (1). Using this partial ldh gene as a probe, I isolated seven overlapping clones from the cDNA library that differed by only 5 to 10 nucleotides at the 5′ end and by the location of polyadenylation. There were high degrees of amino acid sequence similarity between the predicted translation product of this gene, designated ldhA, and l-LDH proteins from other species. Comparisons between the genomic and cDNA sequences of ldhA showed that introns were not present in this gene. The 5′ ends of all of the cDNA fragments isolated were within 5 to 10 nucleotides of the upstream start codon. A total of 69 amino acids were sequenced from the amino terminus and three internal fragments of the purified protein. All of these amino acids were identical to amino acids in the 320-amino-acid sequence predicted on the basis of translation of ldhA.
I also isolated a second gene, designated ldhB, that was more than 90% identical to ldhA. The ldhB gene could not be isolated from the cDNA library that was prepared from mRNA that was isolated from R. oryzae which was actively producing lactic acid in a glucose-containing medium. By avoiding glucose-repressing conditions, ldhB fragments were recovered following PCR amplification of cDNA prepared from RNA obtained from glycerol-grown cultures. No introns were present in ldhB, which was surprising since ldhB genomic DNA and cDNA have a 40-bp insertion at position 929. This insertion contains a stop codon 94 bp upstream of the TGA codon present in both genes and results in a putative 302-amino-acid protein, LdhB. It is possible that unprocessed RNA served as the template for cDNA synthesis and PCR amplification, or contaminating DNA may have been present. If this 40-bp insertion were an intron, splicing of ldhB could restore sequence identity to the carboxyl ends of LdhA and LdhB. Hybridization of total genomic DNA to full-length ldhA resulted in detection of only ldhA and ldhB. The intensity of the ldhB signal was only slightly less than the intensity of the ldhA signal, but ldhB was easily detected and distinguished from ldhA by using restriction enzymes XhoI, XbaI, HindIII, and ClaI (data not shown).
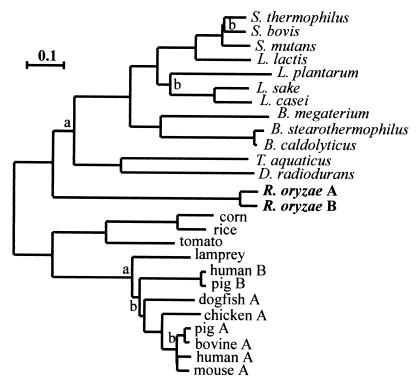
The deduced protein sequences were compared with the sequences of other NAD+-dependent LDH subunits (Fig. 1). Thermus aquaticus LdhA (23) exhibited the greatest similarity to R. oryzae LDH; the level of similarity was 42%, as calculated by pairwise Lipman-Pearson comparisons. However, this level of similarity may not be significant, since most of the other LDH protein sequences were 34 to 40% similar to the R. oryzae LDH sequence. Nucleotide comparisons resulted in even lower levels of similarity, which explained the difficulty which I encountered when I used heterologous probes at the beginning of this study. Because no fungal ldh genes had been described previously, I originally tried to make a probe from almost full-length Streptococcus bovis (37), Lactococcus lactis (17), and bovine (12) genes, but I did not detect any specific hybridization with either the cDNA or genomic libraries.
FIG. 1.
Relationship of LDH subunits from numerous hosts. A most parsimonious tree for 26 LDH amino acid sequences is shown. Levels of amino acid substitution are expressed as percentages (bar = 10%). Most of the nodes have levels of bootstrap support of 99 to 100%; the only exceptions are the nodes labeled a (78 to 80%) or b (44 to 60%). Data for Streptococcus thermophilus (13), Streptococcus bovis (37), Streptococcus mutans (5), Lactococcus lactis (17), Lactobacillus plantarum (30), Lactobacillus sake (accession no. U26688), Lactobacillus casei (15), Bacillus megaterium (34), Bacillus stearothermophilus (40), Bacillus caldolyticus (40), Thermus aquaticus (23), Deinococcus radiodurans (21), corn (9), rice (16), tomato (8), human A (32), human B (27), pigs A and B (31), bovine A (12), mouse A, chicken A (11), dogfish A (28), and lamprey (29) have been published previously.
Regulated expression of ldhA and ldhB.
The similarity between the deduced LdhA protein sequence and the purified NAD+-dependent LDH sequence was evidence that LdhA plays a role in lactic acid synthesis. It was not clear that ldhB was expressed or functional. The high degree of similarity between ldhA and ldhB made it difficult to differentiate the transcripts of the two genes. Northern analysis of glucose-grown cultures always resulted in detection of only one transcript, although it was unlikely that I could have distinguished between ldhA and ldhB. PCR amplification with the universal LDH primers always yielded ldhA DNA when the template was cDNA from cultures that were actively producing lactic acid in a glucose-containing medium.
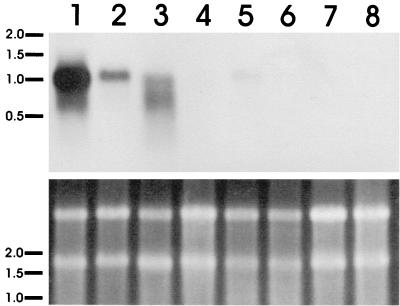
The Northern analysis was repeated with glycerol-peptone-grown mycelium that was transferred to media containing different carbon sources (Fig. 2). Glycerol-peptone was chosen as a carbon source for growing the mycelium because I previously determined that there was not enough fermentable sugar to support production of detectable levels of lactate (>0.2 mM). The amount of ldh transcript that accumulated was largest in the presence of sugars that could be fermented to lactic acid; the maximum amount of transcript was observed in the presence of glucose, followed by xylose and trehalose. The signals for the remaining cultures were almost undetectable, although 1.1-kb bands were observed if blots were exposed to film for an extended time. There appeared to be slight differences in transcript size, although it is possible that this was a result of variation in poly(A) RNA modification. PCR performed with ldhB primers and cDNA from this RNA resulted in amplification of a single 430-bp product for all samples except the samples grown in the presence of glucose, xylose, and trehalose (data not shown).
FIG. 2.
Northern analysis of ldh transcript accumulation after glycerol-peptone-grown R. oryzae was transferred to media containing new carbon sources and incubated for 5.5 h. Lane 1, glucose; lane 2, xylose; lane 3, trehalose; lane 4, peptone; lane 5, glycerol; lane 6, ethanol; lane 7, lactate; lane 8, glycerol-peptone. The gel used for transfer to the membrane is shown at the bottom. The locations of molecular weight standards (in kilobases) are indicated on the left. Due to the similarity of ldhA and ldhB, the labeled ldhA riboprobe detected both transcripts.
To obtain additional evidence that ldhB is expressed only in the presence of nonfermentable sugars, mycelium from a glycerol-grown culture was transferred to fresh RZ medium containing one of several different carbon sources. When universal ldh primers were used, only ldhA fragments were isolated from the amplified product obtained with cDNA made from the glucose-, xylose-, and trehalose-grown cultures. For cultures grown in the presence of glycerol, ethanol, and lactate, almost one-half of the cloned fragments examined were ldhB, suggesting that this gene is functional and apparently glucose repressed.
Enzymatic and zymogram analyses of crude protein extracts from the cultures revealed that reductive LDH activity (pyruvate → lactate) was present in glucose-, xylose-, and trehalose-grown cultures (Table 1). Areas of negative staining on the zymograms had an Rf of 0.18. The glucose-grown culture contained the most reductive LDH activity, while the cultures transferred to glycerol-, ethanol-, and lactate-containing media had the least. This contrasts with the results obtained for lactate-, ethanol-, and glycerol-grown cultures, all of which were positive (Rf, 0.44) for oxidative conversion of lactate to pyruvate. I hypothesized that this activity was due to the product of ldhB, although this hypothesis has not been confirmed yet. The results of my enzymatic analysis of oxidative activity did not correlate well with the results of the zymogram analysis and may reflect some LdhA oxidative activity.
TABLE 1.
LDH activity after glycerol-grown Rhizopus cultures were transferred to media containing new carbon sources
| Carbon source in growth mediuma | Reductive LDH activity (pyruvate → lactate)
|
Oxidative LDH activity (lactate → pyruvate)
|
||
|---|---|---|---|---|
| Sp act (mU/mg of protein)b | Net intensity of band on zymogramc | Sp act (mU/mg of protein)b | Net intensity of band on zymogramc | |
| Glucose | 900 | 330,000 | 56 | NDd |
| Xylose | 280 | 85,000 | 34 | ND |
| Trehalose | 250 | 64,000 | 24 | ND |
| Glycerol | 130 | ND | 9 | 68,000 |
| Ethanol | 37 | ND | 18 | 200,000 |
| Lactate | 96 | ND | 20 | 120,000 |
Cells were induced in the new media for 16 h at 30°C.
Determined by spectrophotometric analysis.
Sum of background-subtracted pixel values for the visible band.
ND, not detected.
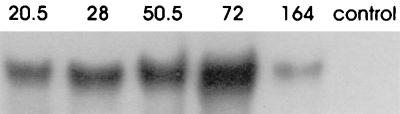
The time course study performed with the bubble column apparatus was not optimized for maximum production of lactic acid but was designed to reveal the differential expression of each gene. The majority of the glucose was depleted by 72 h, and production of additional lactic acid was minimal after this (Table 2). RPA showed that the greatest accumulation of ldh transcripts occurred at 72 h (Fig. 3), which correlated well with enzymatic analysis results (Table 3). Zymogram data confirmed that the maximum reductive LDH activity occurred at 72 h and revealed that NAD+-dependent oxidative LDH activity was present at 164 h. However, the ldh transcript present at 164 h, when the glucose was virtually depleted, was probably a combination of both transcripts. The RPA analysis was performed under conditions that allowed detection of both ldhA and ldhB.
TABLE 2.
Glucose and lactic acid concentrations during R. oryzae fermentation in RZ medium containing 10% glucose in a bubble column apparatus
| Time (h) | Glucose concn (%, wt/vol) | Lactic acid concn (%, wt/vol) |
|---|---|---|
| 20.5 | 9.5 | NDa |
| 28 | 9.0 | 0.18 |
| 50.5 | 5.8 | 2.2 |
| 72 | 1.1 | 6.0 |
| 164 | 0.0 | 6.0 |
ND, not detected.
FIG. 3.
RPA for ldh accumulation in a bubble column apparatus. RNA was purified at various times during fermentation and hybridized to the ldhA riboprobe for RPA analysis. The conditions used for the analysis were such that the probe protected both ldhA and ldhB. The control was yeast RNA hybridized to the probe. The numbers at the top indicate the times (in hours) that the RNA were obtained.
TABLE 3.
LDH activity during growth in a bubble column
| Time (h) | Reductive LDH sp act (pyruvate → lactate) (mU/mg of protein)a | Oxidative LDH sp act (lactate → pyruvate) (mU/mg of protein)a |
|---|---|---|
| 20.5 | 190 | NDb |
| 28 | 620 | ND |
| 50.5 | 520 | ND |
| 72 | 2,200 | 180 |
| 164 | 90 | 90 |
Determined by spectrophotometric analysis.
ND, not detected.
Complementation of E. coli.
Both ldhA and ldhB genes could restore the fermentative ability of E. coli DC1368 mutants so transformants grew anaerobically. All of the ldh transformants produced 10 to 25 mM lactic acid, and the orientation of the ldh gene in relation to the lacZ promoter had little effect on growth or lactate production. No growth or lactic acid production occurred under anaerobic conditions with transformants containing only the pBluescript plasmid.
DISCUSSION
This is the first description of ldh genes isolated from a fungus. Protein sequence comparisons with other LDH revealed the unique phylogeny of these genes. One of the genes, ldhA, encodes an NAD+-dependent l-LDH which is involved in lactic acid production by R. oryzae. There have been several previous reports that have described the role of this protein, although most of the previous work was performed with partially purified enzyme preparations because the short half-life of the enzyme prevented further purification (22, 24, 25). I overcame the instability of the enzyme by including proteinase inhibitors, dithiothreitol, and glycerol in my preparations. Yu and Hang (39) claimed that their enzyme was very stable in phosphate buffer, but it is difficult to interpret the results of these authors, since the total activity of their purified enzyme was only 4.6 U and the specific activity was 15.4 U/mg of protein.
My findings concerning expression of the reductive NAD+-dependent enzyme, LdhA, in glucose-containing medium correlate well with the findings of Pritchard (24, 25). Enzyme activity was greatest in the presence of high concentrations of glucose and quickly disappeared after the glucose was depleted. I also found that both activity and transcripts were present in media containing xylose or trehalose. These results were not surprising, since R. oryzae is known to produce lactic acid when it is grown in the presence of xylose (35) and I measured lactate accumulation when trehalose was used as the carbon source (data not shown). Even though I did not detect lactic acid production in cultures grown in media containing ethanol, glycerol, and lactate, reductive LDH activity was detected by spectrophotometric assays; however, it was not detected by zymogram analysis. The results of incorporating pyrazole, an inhibitor of alcohol dehydrogenase, into the spectrophotometric assay preparations led me to believe that the difference was probably a result of pyruvate decarboxylase and alcohol dehydrogenase activities that interfered with the spectrophotometric analysis (unpublished data). Thus, I think that LdhA activity is probably negligible or absent in the presence of gluconeogenic substrates. The small amount of ldhA transcripts may indicate that there are both transcriptional and translational control mechanisms.
The function of ldhB is not clear, although this gene appears to be glucose repressed and ldhB expression occurs concomitantly with the appearance of an NAD+-dependent LDH that can catalyze oxidization of lactate to pyruvate. I originally hypothesized that LdhB was involved in lactic acid production during anaerobic stress, in a manner similar to that in other eukaryotes that produce multiple LDH isozymes (7, 8), but I did not detect the ldhB transcript under anoxic conditions when glucose was present (unpublished data). This enzyme activity also was present when fermentation exhausted the available glucose. I believe that this LDH is distinct from an enzyme that was previously described as an NAD+-independent LDH and is also produced following the disappearance of glucose (24). This NAD+-independent enzyme catalyzed oxidation of lactate to pyruvate by using dichlorophenol indolephenol, ferricyanide, or cytochrome c, but not oxygen, as an electron acceptor and could not use either NAD+ or NADH as a cofactor. I have also detected this NAD+-independent activity but have not been able to demonstrate it by zymogram analysis. This NAD+-independent LDH could be a membrane-bound flavoprotein coupled to the respiratory chain and may be required for aerobic growth on lactate.
Although I detected oxidative NAD+-dependent LDH activity (lactate → pyruvate) for the protein associated with ldhB, this does not mean that the protein has an oxidative function in vivo, as both ldhA and ldhB restore fermentative growth to E. coli DC1368 pfl ldh mutants. The ability of ldhB to complement these mutants was not expected, since I did not find any reductive activity associated with LdhB. It is possible that my assays were not sensitive enough to detect this conversion and that LdhB can catalyze both oxidative and reductive reactions.
During fermentation, slight decreases in lactic acid production may occur when glucose is exhausted, and these decreases are followed by increases in lactic acid levels. Pritchard (24) attributed the decreases in lactic acid levels to the NAD+-independent LDH but was not able to explain the increases in lactate levels. I hypothesize that LdhB is responsible for this phenomenon, since it is expressed during this phase of growth and it can produce lactic acid in E. coli DC1368. I suggest that the source of carbon for the increases in lactic acid levels is a storage sugar, such as trehalose. Although I found only ldhA transcripts in trehalose-grown cultures, the high concentrations of sugar that I used may not be representative of the concentrations found in mycelia.
In summary, my results suggest that at least three different LDH are produced by R. oryzae. Two of these enzymes, LdhA and LdhB, require the cofactor NAD+, while the third enzyme is probably a mitochondrial NAD+-independent LDH that is used for oxidative utilization of lactate. The results of transcriptional and enzyme analyses show that ldhA is probably responsible for most of the lactic acid produced by this fungus. I have only begun to examine the molecular mechanisms that control transcription and translation of this gene, although it is known that the ldhA transcript is present even with nonfermentable carbon sources, such as glycerol and ethanol. My results suggest that the function of the second gene, ldhB, may be associated with conversion of storage sugars to lactic acid, but further work is required to substantiate this hypothesis.
ACKNOWLEDGMENT
I thank Kristina Glenzinski for exemplary and invaluable technical assistance.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1995. [Google Scholar]
- 3.Bunch P K, Mat-Jan F, Lee N, Clark D P. The ldhA gene encoding the fermentative lactate dehydrogenase of Escherichia coli. Microbiology. 1997;143:187–195. doi: 10.1099/00221287-143-1-187. [DOI] [PubMed] [Google Scholar]
- 4.Datta R, Tsai S P, Bonsignore P, Moon S H, Frank J R. Technological and economic potential of poly(lactic acid) and lactic acid derivatives. FEMS Microbiol Rev. 1995;16:221–231. [Google Scholar]
- 5.Duncan M J, Hillman J D. DNA sequence and in vitro mutagenesis of the gene encoding the fructose-1,6-diphosphate-dependent l-(+)-lactate dehydrogenase of Streptococcus mutans. Infect Immun. 1991;59:3930–3934. doi: 10.1128/iai.59.11.3930-3934.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernandez J, Andrews L, Mische S M. An improved procedure for enzymatic digestion of polyvinylidene difluoride-bound proteins for internal sequence analysis. Anal Biochem. 1994;214:112–117. doi: 10.1006/abio.1994.1148. [DOI] [PubMed] [Google Scholar]
- 7.Firth J D, Eberts B L, Ratcliffe P J. Hypoxic regulation of lactate dehydrogenase. J Biol Chem. 1995;270:21021–21027. doi: 10.1074/jbc.270.36.21021. [DOI] [PubMed] [Google Scholar]
- 8.Germain V, Raymond P, Ricard B. Differential expression of two tomato lactate dehydrogenase genes in response to oxygen deficit. Plant Mol Biol. 1997;35:711–721. doi: 10.1023/a:1005854002969. [DOI] [PubMed] [Google Scholar]
- 9.Good A G, Paetkau D H. Identification and characterization of a hypoxically induced maize lactate dehydrogenase gene. Plant Mol Biol. 1992;19:693–697. doi: 10.1007/BF00026795. [DOI] [PubMed] [Google Scholar]
- 10.Goward C R, Nicholls D J. Malate dehydrogenase: a model for structure, evolution, and catalysis. Protein Sci. 1994;3:1883–1888. doi: 10.1002/pro.5560031027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirota Y, Katsumata A, Takeya T. Nucleotide and deduced amino acid sequences of chicken lactate dehydrogenase-A. Nucleic Acids Res. 1990;18:6432. doi: 10.1093/nar/18.21.6432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishiguro N, Osame S, Kagiya R, Ichijo S, Shinagawa M. Primary structure of bovine lactate dehydrogenase-A and its synthesis in Escherichia coli. Gene. 1990;91:281–285. doi: 10.1016/0378-1119(90)90101-v. [DOI] [PubMed] [Google Scholar]
- 13.Ito Y, Sasaki T. Cloning and nucleotide sequencing of l-lactate dehydrogenase gene from Streptococcus thermophilus M-192. Biosci Biotechnol Biochem. 1994;58:1569–1573. doi: 10.1271/bbb.58.1569. [DOI] [PubMed] [Google Scholar]
- 14.Kascak J S, Kominek J, Roehr M. Lactic acid. In: Rehm H J, Reed G, Puhler A, Stadler P, editors. Biotechnology. Weinheim, Germany: VCH Press; 1996. pp. 293–306. [Google Scholar]
- 15.Kim S F, Baek S J, Pack M Y. Cloning and nucleotide sequence of the Lactobacillus casei lactate dehydrogenase gene. Appl Environ Microbiol. 1991;57:2413–2417. doi: 10.1128/aem.57.8.2413-2417.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li T, Takano T, Matsumura H, Takeda G. Molecular cloning of rice lactate dehydrogenase cDNA. Jpn J Breed. 1993;43:129–133. [Google Scholar]
- 17.Llanos R M, Hillier A J, Davidson B E. Cloning, nucleotide sequence, expression, and chromosomal location of ldh, the gene encoding l-(+)-lactate dehydrogenase, from Lactococcus lactis. J Bacteriol. 1992;174:6956–6964. doi: 10.1128/jb.174.21.6956-6964.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lockwood L B, Ward G E, May O E. The physiology of Rhizopus oryzae. J Agric Res. 1936;53:849–857. [Google Scholar]
- 19.Margulies M, Vishniac W. Dissimilation of glucose by the MX strain of Rhizopus. J Bacteriol. 1961;81:1–9. doi: 10.1128/jb.81.1.1-9.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mat-Jan F, Alam K Y, Clark D P. Mutants of Escherichia coli deficient in the fermentative lactate dehydrogenase. J Bacteriol. 1989;171:342–348. doi: 10.1128/jb.171.1.342-348.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Narumi I, Watanabe H. Sequence and analysis of the l-lactate dehydrogenase encoding gene of Deinococcus radiodurans, a suitable mesophilic counterpart for Thermus. Gene. 1996;172:117–119. doi: 10.1016/0378-1119(96)00108-4. [DOI] [PubMed] [Google Scholar]
- 22.Obayashi A, Yorifuji H, Yamagata T, Ijichi T, Kanie M. Respiration in organic acid forming molds. Part I. Purification of cytochrome c, coenzyme Q and l-lactic dehydrogenase from lactate forming Rhizopus oryzae. Agric Biol Chem. 1966;30:717–724. [Google Scholar]
- 23.Ono M, Matsuzawa H, Ohta T. Nucleotide sequence and characteristics of the gene for l-lactate dehydrogenase of Thermus aquaticus YT-1 and the deduced amino acid sequence of the enzyme. J Biochem. 1990;107:21–26. doi: 10.1093/oxfordjournals.jbchem.a123004. [DOI] [PubMed] [Google Scholar]
- 24.Pritchard G G. NAD+ independent l-lactate dehydrogenase from Rhizopus oryzae. Biochim Biophys Acta. 1971;250:25–34. doi: 10.1016/0005-2744(71)90116-1. [DOI] [PubMed] [Google Scholar]
- 25.Pritchard G G. Factors affecting the activity and synthesis of NAD dependent lactate dehydrogenase in Rhizopus oryzae. J Gen Microbiol. 1973;78:125–137. [Google Scholar]
- 26.Raber L R. Green chemistry here to stay. Chem Eng News. 1998;1998(July):25–26. [Google Scholar]
- 27.Sakai I, Sharief F S, Pan Y C, Li S S. The cDNA and protein sequence of human lactate dehydrogenase B. Biochem J. 1987;248:933–936. doi: 10.1042/bj2480933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stock D W, Powers D A. The cDNA sequence of the lactate dehydrogenase A of the spiny dogfish (Squalus acanthias): corrections to the amino acid sequence and analysis of the phylogeny of vertebrate lactate dehydrogenases. Mol Mar Biol Biotechnol. 1995;4:284–294. [PubMed] [Google Scholar]
- 29.Stock D W, Whitt G S. Evolutionary implications of the cDNA sequence of the single lactate dehydrogenase of a lamprey. Proc Natl Acad Sci USA. 1992;89:1799–1803. doi: 10.1073/pnas.89.5.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taguchi H, Ohta T. d-Lactate dehydrogenase is a member of the d-isomer specific 2-hydroxyacid dehydrogenase family. Cloning, sequencing, and expression in E. coli of the d-lactate dehydrogenase gene of Lactobacillus plantarum. J Biol Chem. 1991;266:12588–12594. [PubMed] [Google Scholar]
- 31.Tsuji S, Qureshi M A, Hou E W, Fitch W M, Li S S. Evolutionary relationship of lactate dehydrogenases (LDH) from mammals, birds, an amphibian, fish, barley, and bacteria: LDH cDNA sequence from Xenopus, pig, and rat. Proc Natl Acad Sci USA. 1994;91:9392–9396. doi: 10.1073/pnas.91.20.9392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsujibo H, Tiano H F, Li S S. Nucleotide sequences of the cDNA and an intronless pseudogene for human lactate dehydrogenase-A isozyme. Eur J Biochem. 1985;147:9–15. doi: 10.1111/j.1432-1033.1985.tb08711.x. [DOI] [PubMed] [Google Scholar]
- 33.Van de Peer Y. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Applic Biosci. 1994;10:569–570. doi: 10.1093/bioinformatics/10.5.569. [DOI] [PubMed] [Google Scholar]
- 34.Waldvogel S, Weber H, Zuber H. Nucleotide sequence of the l-lactate dehydrogenase gene from the mesophilic bacterium B. megaterium. Preparation and properties of a hybrid lactate dehydrogenase compromising moieties of the B. megaterium and B. sterothermophilus enzymes. Biol Chem. 1987;368:1391–1399. doi: 10.1515/bchm3.1987.368.2.1391. [DOI] [PubMed] [Google Scholar]
- 35.Wang C W, Lu Z, Tsao G T. Lactic acid production by pellet-form Rhizopus oryzae in a submerged system. Appl Biochem Biotechnol. 1995;51/52:57–71. [Google Scholar]
- 36.Wigley D B, Gamblin S J, Turkenburg J P, Dodson E J, Piontek K, Muirhead H, Holbrook J J. Structure of a ternary complex of an allosteric lactate dehydrogenase from Bacillus stearothermophilus at 2.5 Å resolution. J Mol Biol. 1992;223:317–335. doi: 10.1016/0022-2836(92)90733-z. [DOI] [PubMed] [Google Scholar]
- 37.Wyckoff H A, Chow J, Whitehead T R, Cotta M A. Cloning, sequence, and expression of the l-(+) lactate dehydrogenase of Streptococcus bovis. Curr Microbiol. 1997;34:367–373. doi: 10.1007/s002849900197. [DOI] [PubMed] [Google Scholar]
- 38.Yu R-C, Hang Y D. Kinetics of direct fermentation of agricultural commodities to l(+)-lactic acid by Rhizopus oryzae. Biotechnol Lett. 1989;11:597–600. [Google Scholar]
- 39.Yu R-C, Hang Y D. Purification and characterization of NAD-dependent lactate dehydrogenase from Rhizopus oryzae. Food Chem. 1991;41:219–225. [Google Scholar]
- 40.Zuelli F, Weber H, Zuber H. Nucleotide sequences of lactate dehydrogenase genes from the thermophilic bacteria Bacillus stearothermophilus, B. caldolyticus, and B. caldotenax. Biol Chem. 1987;368:1167–1177. doi: 10.1515/bchm3.1987.368.2.1167. [DOI] [PubMed] [Google Scholar]