Abstract
Purpose
Several studies have investigated the association between anorexia nervosa and polymorphisms of genes regulating serotonin neurotransmission, with a focus on the rs6311 polymorphism of 5-HTR2A. However, inconsistent results of these studies and conflicting conclusions of existing meta-analyses complicate the understanding of a possible association. We have updated these results and evaluated the involvement of other serotonin receptor gene polymorphisms in anorexia nervosa.
Methods
Adhering to PRISMA guidelines, we have searched studies on anorexia nervosa and serotonin-regulating genes published from 1997 to 2022, selected those concerning receptor genes and meta-analyzed the results from twenty candidate gene studies on the 5-HTR2A rs6311 polymorphism and the 5-HTR2C rs6318 polymorphism.
Results
Present analyses reveal an association for the 5-HTR2A rs6311 polymorphism, with G and A alleles, across eighteen studies (2049 patients, 2877 controls; A vs. G allele, Odds Ratio = 1.24; 95% Confidence Interval = 1.06–1.47; p = 0.009). However, after geographic subgrouping, an association emerged only in a Southern European area, involving five studies (722 patients, 773 controls; A vs. G allele, Odds Ratio = 1.82; 95% Confidence Interval = 1.41–2.37; p < 0.00001). No association was observed for the 5-HTR2C rs6318 polymorphism across three studies.
Conclusions
To date, the involvement in the pathophysiology of anorexia nervosa of the 5-HTR2A rs6311 polymorphism appears limited to a specific genetic and/or environmental context, while that of the 5-HTR2C rs6318 polymorphism seems excluded. Genome-wide association studies and epigenetic studies will likely offer deeper insights of genetic and environmental factors possibly contributing to the disorder.
Level of evidence
III Evidence obtained from well-designed cohort or case–control analytic studies.
Clinical trial registration PROSPERO registration number: CRD42021246122.
Keywords: Anorexia nervosa, Candidate gene studies, Serotonin, 5-HT2A, 5-HTR2A, 5-HT2C, 5-HTR2C
Introduction
Eating disorders (EDs), including anorexia nervosa (AN), bulimia nervosa (BN) and binge eating disorder (BED), are potential life-threatening multidimensional syndromes classified in the Diagnostic and statistical manual of mental disorders (DSM-5-TR, [1]). Although their pathogenesis is still unclear, a widely accepted bio-psycho-social model implicates psychological, familial, socio-cultural, and genetic factors. A significant genetic influence is suggested by family, twin, and molecular genetics studies [2–8]. An individual susceptibility to Eds is also acknowledged in the psychodynamic framework, coupled to the importance of family functioning in their onset and maintenance, especially in adolescents [9–12].
AN, the most thoroughly investigated ED, predominantly affects females with a lifetime prevalence of 0.2–3% [13–15]. It is characterized by a morbid fear of gaining weight that leads to food avoidance [13] and is associated with factors such as cognitive impairments [16] including poor information processing, rigid thinking patters and attentional bias on memory encoding and recall [17].
Over the past three decades, the genetics of AN has been investigated by various approaches including candidate gene studies, linkage analyses, transmission disequilibrium tests (TDT), genome-wide association studies (GWAS), and epigenetic studies, each one bearing distinct advantages and limitations [5, 18, 19]. Candidate gene studies began in the late 1990s exploring the association between specific genes and AN. Among the neurochemical systems investigated, the serotonergic system has received considerable attention due to its role in regulating mood, food intake, and body weight [20], and to alterations in its functioning directly linked to the etiology of AN [21].
In particular, several studies have focused on the rs6311 single nucleotide polymorphism (SNP) of the 5-HT2A receptor gene (5-HTR2A), with G and A alleles, and other receptor gene polymorphisms, including the rs6318 SNP of the 5-HT2C receptor gene (5-HTR2C), with Cys and Ser alleles.
However, many of these studies have shown limitations, leading to frequent inconsistent results and impeding a thorough understanding of the roles of the analyzed polymorphisms [22, 23]. Limiting issues include small sample sizes, low-level replication, geographic variations in genotype/allele frequencies, and population stratifications. Additionally, the intrinsic complexity of AN, lacking a clear biological definition, suggests that its neurobiological etiology is influenced by the activity of multiple genes and significant gene x environment interactions. These factors cast doubt on the reliability of results of these studies.
Previous meta-analyses have focused on studies concerning the 5-HTR2A rs6311 SNP. By analysis of six early studies [24–29], including 556 patients and 1077 controls, Ziegler et al. [29] found no association with AN and high genetic heterogeneity of the samples. In contrast, Gorwood et al. [30] reported a small yet significant association (A vs. G allele, Odds Ratio [OR] = 1.20; 95% Confidence interval [CI] 1.07–1.35; χ2 = 8.14; p = 0.0043), after incorporating three subsequent studies [31–33] with 872 patients and 1656 controls. However, this was coupled to a high heterogeneity (χ2 = 41.7; p = 1.51 × 10–6) that was attributed to a lack of statistical power, variations in allele frequencies among control samples, and methodological differences among the studies. Additionally, a slightly stronger association emerged in the studies involving Caucasian participants (OR = 1.27; 95% CI 1.12–1.44; p = 0.0003), but the small number of reports prevented a deeper examination of geographical differences.
Martásková et al. [34] reported a similar association (A vs. G allele, OR = 1.21; 95% CI 1.09–1.35; p < 0.003), after incorporating two additional studies [34, 35], totaling 1057 patients and 1599 controls. The authors did not provide heterogeneity data but investigated the geographical distribution of European cohorts. Positive association trends were identified in British and Italian cohorts, a Polish cohort and a U.S.A. cohort, but not other European and Asian cohorts, underlining the importance of considering population stratification in further assessments.
In a recent meta-analysis of previous reports and four additional studies [36–39], with 2028 patients and 2725 controls, Yan et al. [40] found a small global association (A vs. G allele, OR = 1.24; 95% CI 1.04–1.48; p = 0.014) and a more significant one in a subgroup of twelve studies on Western samples (A vs. G allele, OR = 1.31; 95% CI 1.11–1.64; p = 0.003), with high heterogeneity in both cases. In contrast, no association and low heterogeneity were observed in a subgroup of three studies on East Asian samples. The authors did not conduct further subgroup analyses.
Due to weaknesses of the existing meta-analyses, including the incorporation of studies that contained virtual controls or samples calculated from TDTs, and the lack of statistical corrections necessary under multiple genetic models, these results cast doubt on the presence of a bona fide association between the 5-HTR2A rs6311 polymorphism and AN.
We have therefore reassessed the findings of serotonin candidate gene studies on AN by an updated and comprehensive systematic review and meta-analysis. Upon evaluation of all reports by the criteria of quality assessment, geographic location, participants’ age and gender, diagnosis, AN subtypes, we performed an analysis whose significant discriminating factor is a detailed geographic evaluation of effect sizes, accompanied by statistical correction for multiple testing.
Methods
Article search and selection
Searches for publications were conducted in PubMed (PubMed, https://pubmed.ncbi.nlm.nih.gov/, last accessed August 16th, 2023), Scopus (Scopus, https://www.scopus.com/search/form.uri#basic, last accessed June 21st, 2023), PsycINFO (PsycINFO, https://web.p.ebscohost.com/ehost/search/ advanced?vid = 2&sid = f36c3704-5378-4d93-8629-ac1da74db433%40redis, last accessed June 12th, 2023) and Web of Science (Web of Science, https://www-webofscience-com.ezproxy.uniroma1.it/wos/woscc/basic-search, last accessed June 20th, 2023) databases following PRISMA guidelines [41], from 1997, when the first candidate gene studies emerged, to December 2022. We initially used the terms “genetic association” AND “anorexia nervosa”, obtaining the following numbers of publications: PubMed: 578, Scopus: 552, PsycINFO: 42, Web of Science: 450. We subsequently used the terms “gene association” AND “anorexia nervosa”, obtaining the following numbers of publications: PubMed: 353, Scopus: 446, PsycINFO: 52, Web of Science: 478.
More specific searches, conducted using the terms “serotonin” AND “genetic association” AND “anorexia nervosa”, and the terms “serotonin” AND “gene association” AND “anorexia nervosa”, yielded the following respective numbers of publications: PubMed: 42, Scopus: 124, PsycINFO: 9, Web of Science: 86; PubMed: 60, Scopus: 109, PsycINFO: 21, Web of Science: 125.
After merging the search results and excluding duplicates, 222 papers were reviewed. Ninety non-genetic papers, papers not concerning serotonin or AN, papers lacking useful genetic information, and 8 papers involving animal models were excluded. The remaining 124 publications were selected by the following criteria:
Study type: we included population-based, case–control genetic association studies. We excluded family- and patient-based studies, TDT studies, mutation analyses, association and/or haplotype analyses, GWAS;
Diagnosis: we included studies in which AN was diagnosed according to the DSM-III-R [42], DSM-IV [43], DSM-IV-TR [44], DSM-5 [45] or ICD-10 [46] and control participants had no diagnosis of any pathology. We excluded studies in which participants were classified by administered or self-administered questionnaires;
Polymorphism type: we included studies that investigated bi-allelic polymorphisms;
Numerosity: we analyzed genetic polymorphisms investigated in at least three independent studies [47].
The search and selection of papers were conducted independently by F. Santini and D. La Porta, reviewed and approved by all authors. Based on the listed criteria, we excluded 52 reviews or methods/perspective papers, 5 out of 8 meta-analyses lacking empirical data, 2 GWAS papers, 7 epigenetic papers; 8 gene mapping, linkage, and mutation analyses, 2 TDTs, 8 papers with no control/patient samples or with virtual control samples, calculated from non-transmitted alleles in the family trios analyzed in TDTs, 2 papers without DSM/ICD diagnoses, 2 case-report papers, 6 papers not meeting the numerosity criterion. The remaining 30 papers on biallelic serotonin gene polymorphisms, including 27 research papers and 3 papers with research and meta-analytical results were further selected. (Fig. 1). After exclusion of 10 papers on the 5-HTTLPR polymorphism of the presynaptic serotonin transporter gene, 20 papers on 5-HTR2A and 5-HTR2C SNPs were meta-analyzed.
Fig. 1.
PRISMA flow of study selection
All papers were in English except for one Polish paper, which however had an English abstract and clear genotype frequencies.
Meta-analysis
For all studies, selection, comparability and exposure qualities were assessed by the Newcastle–Ottawa quality rating scale (NOS) [48]; publication bias was evaluated by the Egger’s test [49]; Hardy–Weinberg (HW) equilibrium was examined by χ2-test.
Data collection from each study was performed independently by F. Santini and D. La Porta, reviewed and approved by all authors. The association of each genetic polymorphism with AN was evaluated by separate analyses for categorical variables. Genotype frequencies of patients and control participants from each study were examined using recessive (AA vs. Aa + aa), dominant (AA + Aa vs. aa), and allele (A vs. a) models, which effectively identify SNP risks in case–control genetic studies, based on mathematical evidence [50]. Additive, co-dominant and over-dominant models were therefore not considered. To control for family-wise error rate, Bonferroni correction was applied to account for multiple hypothesis tests related to our three genetic models (p = 0.05/3) [51]. Thus, P values ≤ 0.0166 were considered statistically significant.
Heterogeneity among the studies was assessed using a χ2-based Q-test [52] and quantified by I2 statistics [53]. Associations were analyzed by calculating the pooled OR and 95% CI, using the Mantel–Haenszel fixed effects (FE) model [54] for studies with low heterogeneity (p > 0.1 or I2 < 50.0%), or the random effects (RE) model for studies with high heterogeneity (p ≤ 0.1 or I2 ≥ 50.0%) [55]. The significance of the pooled OR was determined using a Z-test. The magnitude of the ORs was evaluated by Cohen’s criteria [56] as follows: OR < 1.44: very small effect; 1.44 ≤ OR < 2.48: small effect; 2.48 ≤ OR < 4.27: medium effect; OR ≥ 4.27: large effect.
If I2 exceeded 50.0%, the stability of the relationship was assessed by conducting analyses after removing one study at a time (leave-one-out test). Geographical distribution of the studies was considered to account for population stratifications. Subgrouping criteria based on gender, age, DSM version, NOS, were not applied for the following reasons: most studies included only or predominantly female participants; age distribution, when disclosed, did not significantly differ across the studies, the DSM-IV version was predominantly used, and NOS values were largely consistent. Analyses were performed using Review Manager 5.3 (The Nordic Cochrane Centre, Copenhagen, DM) and MedCalc 20.211 (MedCalc Software Ltd, Ostend, Belgium).
Results
Serotonergic genes investigated in AN in association analyses, TDTs and case–control studies include tryptophan hydroxylase 1 and 2 (TPH1 and TPH2), regulating serotonin biosynthesis, the presynaptic transporter 5-HTT, and 5-HTR1D, 5-HTR2A, 5-HTR2C, 5-HTR3A, 5-HTR3D, 5-HTR7 receptors. Among serotonin receptor gene polymorphisms (Table 1), 5-HTR2A rs6311 and 5-HTR2C rs6318 met our inclusion criteria. Numbers of case–control associations reports are listed in Table 2, their genotyping methods, diagnostic criteria, and NOS values are provided in Table 3. Significance values of publication biases assessed by the Egger’s test are presented in figure legends.
Table 1.
Serotonin receptor gene polymorphisms investigated in the period 1997–2020 and inclusion/exclusion criteria of the present meta-analysis
| Author | Year | Gene | Polymorphism | Included/not included | Criterion for exclusion |
|---|---|---|---|---|---|
| Hinney et al. | 1997 | 5-HTR2A | rs63311 | Yes | |
| Collier et al. | 1997 | 5-HTR2A | rs63311 | Yes | |
| Campbell et al. | 1998 | 5-HTR2A | rs63311 | Yes | |
| Enoch et al. | 1998 | 5-HTR2A | rs63311 | Yes | |
| Sorbi et al. | 1998 | 5-HTR2A | rs63311 | Yes | |
| Nacmias et al. | 1999 | 5-HTR2A | rs63311 | Yes | |
| Ziegler et al. | 1999 | 5-HTR2A | rs63311 | Yes | |
|
Hinney et al. |
1999 | 5-HT1RDbeta | Phe-124-Cys | No | Numerosity |
|
Hinney et al. |
1999 | 5-HTR7 | Pro-279-Leu | No | Numerosity |
| Ando et al. | 2001 | 5-HTR2A | rs63311 | Yes | |
| Karwautz et al. | 2001 | 5-HTR2A | rs63311 | Yes | |
| Karwautz et al. | 2001 | 5-HTR2C | rs6318 | Yes | |
| Nishiguchi et al. | 2001 | 5-HTR2A | rs63311 | Yes | |
| Ricca et al. | 2002 | 5-HTR2A | rs63311 | Yes | |
| Kipman et al. | 2002 | 5-HTR2A | rs63311 | Yes | |
| Westberg et al. | 2002 | 5-HTR2C | rs6318 | Yes | |
|
Gorwood et al. |
2002 | 5-HTR2A | rs63311 | No | TDT |
| Rybakowski et al. | 2003 | 5-HTR2A | rs63311 | Yes | |
| Hu et al. | 2003 | 5-HTR2C | rs6318 | Yes | |
|
Bergen et al. |
2003 | 5-HTR1D |
rs652783 rs604030 rs674386 rs856510 |
No | Linkage analysis |
| Ricca et al. | 2004 | 5-HTR2A | rs63311 | Yes | |
| Rybakowski et al. | 2006 | 5-HTR2A | rs63311 | Yes | |
|
Brown et al. |
2007 | 5-HTR1D |
rs652783 rs604030 rs674386 rs856510 |
No | Association analysis |
| Martásková et al. | 2009 | 5-HTR2A | rs63311 | Yes | |
|
Hammer et al |
2009 | 5-HTR3A | rs1062613 rs1176722 | No | Association analysis |
|
Hammer et al. |
2009 | 5-HTR3B | rs1176744 | No | Association analysis |
|
Slof-Op ‘t Landt et al. |
2011 | 5-HTR1D |
rs605367 rs6300 rs676643 rs674386 |
No | Association analysis |
|
Mas et al. |
2013 | 5-HTR2A |
rs4942577 rs9567733 rs7333412 rs9567736 rs9567737 rs2296972 rs2770298 rs731779 rs1002513 rs927544 rs4942587 rs2296973 rs731245 rs985934 |
No | Association analysis |
|
Mas et al. |
2013 | 5-HTR2C |
rs6318 rs518147 rs1801412 rs3813928 rs3813929 rs6311 rs6313 |
No | Association analysis |
| Kang et al | 2017 | 5-HTR2A | rs63311 | Yes | |
|
Genis Mendoza et al. |
2019 | 5-HTR2A | rs63311 | No | Excessively different patients and control samples |
| Ceccarini et al. | 2020 | 5-HTR2A | rs63311 | Yes |
Where possible, gene polymorphisms are identified by the Reference SNP cluster ID (rs). TDT, transmission disequilibrium test
Table 2.
Biallelic genetic polymorphisms included in the present meta-analysis and numbers of their reports
| Gene | Polymorphism | No. of reports |
|---|---|---|
| 5-HTR2A | rs6311 (− 1438G/A) | 18 |
| 5-HTR2C | rs6318 (Cys23Ser) | 3 |
Only polymorphisms investigated in at least three studies are included
The number of reports, 21, exceeds the number of papers, 20, because Karwautz et al. [50] investigated both polymorphisms
Table 3.
General characteristics of studies included in the present meta-analysis
| Author | Year | Nation | Ethnicity | Polymorphism | Genotyping method | Diagnostic criteria | Individual category | Male | Female | Total | Age | BMI | NOS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hinney et al. | 1997 | Germany | Caucasian | 5-HTR2A rs63311 | PCR–RFLP | DSM-IV | Individuals with AN | – | – | 100 | – | – | 7 |
| Healthy controls | – | – | 355 | – | – | ||||||||
| Collier et al. | 1997 | United Kingdom | Caucasian | 5-HTR2A rs63311 | PCR–RFLP | ICD-10 | Individuals with AN | 0 | 81 | 81 | – | – | 7 |
| Healthy controls | 138 | 88 | 226 | – | – | ||||||||
| Campbell et al. | 1998 | United Kingdom | Caucasian | 5-HTR2A rs63311 | PCR–RFLP | ICD-10 | Individuals with AN | 152 | 152 | – | – | 6 | |
| Healthy controls | 150 | 150 | – | – | |||||||||
| Enoch et al. | 1998 | U.S.A | Caucasian | 5-HTR2A rs63311 | PCR–RFLP | DSM-III-R | Individuals with AN | – | – | 68 | – | – | 6 |
| Healthy controls | – | – | 69 | – | – | ||||||||
| Sorbi et al. | 1998 | Italy | Caucasian | 5-HTR2A rs63311 | PCR–RFLP | DSM-IV | Individuals with AN | 0 | 77 | 77 | 24.6 ± 5.8 |
rAN: 14.9 ± 2.6 pAN: 16.0 ± 1.9 |
7 |
| Healthy controls | 0 | 107 | 107 | 25.3 ± 5.6 | – | ||||||||
| Nacmias et al. | 1999 | Italy | Caucasian | 5-HTR2A rs63311 | PCR–RFLP | DSM-IV | Individuals with AN | 0 | 109 | 109 |
rAN: 19.2 ± 1.8 pAN: 21.8 ± 4.0 |
rAN: 13.8 ± 2.5 pAN: 15.6 ± 2.1 |
6 |
| Healthy controls | 0 | 107 | 107 | 25.3 ± 5.6 | – | ||||||||
| Ziegler et al. | 1999 | Germany | Caucasian | 5-HTR2A rs63311 | PCR–RFLP | DSM-IV | Individuals with AN | – | – | 78 | – | – | 7 |
| Healthy controls | – | – | 170 | – | – | ||||||||
| Ando et al. | 2001 | Japan | Asian | 5-HTR2A rs63311 | PCR–RFLP | DSM-IV | Individuals with AN | 0 | 75 | 75 | 24.4 ± 5.8 | 15.7 ± 2.5 | 6 |
| Healthy controls | 0 | 127 | 127 | – | – | ||||||||
| Karwautz et al. | 2001 | United Kingdom | Caucasian | 5-HTR2A rs63311 | PCR–RFLP | DSM-IV | Individuals with AN | 0 | 44 | 44 | – | – | 8 |
| Healthy controls | 0 | 39 | 39 | – | – | ||||||||
| Nishiguchi et al. | 2001 | Japan | Asian | 5-HTR2A rs63311 | PCR–RFLP | DSM-IV | Individuals with AN | – | – | 62 | 25.3 ± 6.2 | – | 8 |
| Healthy controls | 20 | 354 | 374 | 25.7 ± 10.6 | – | ||||||||
| Ricca et al. | 2002 | Italy | Caucasian | 5-HTR2A rs63311 | PCR–RFLP | DSM-IV | Individuals with AN | – | – | 148 |
rAN: 19.2 ± 1.8 pAN: 21.8 ± 4.0 |
rAN: 13.8 ± 2.5 pAN: 15.6 ± 2.1 |
8 |
| Healthy controls | – | – | 115 | 26.3 ± 6.1 | – | ||||||||
| Kipman et al. | 2002 | France | Caucasian | 5-HTR2A rs63311 | PCR–RFLP | DSM-IV | Individuals with AN | – | – | 145 | – | – | 7 |
| Healthy controls | – | – | 98 | – | – | ||||||||
| Rybakowski et al. | 2003 | Poland | Caucasian | 5-HTR2A rs63311 | PCR–RFLP | DSM-IV | Individuals with AN | – | – | 67 | – | – | 6 |
| Healthy controls | – | – | 114 | – | – | ||||||||
| Ricca et al. | 2004 | Italy | Caucasian | 5-HTR2A rs63311 | PCR–RFLP | DSM-IV | Individuals with AN | – | – | 77 |
rAN: 23.9 ± 8.4 pAN: 23.9 ± 5.4 |
rAN: 14.1 ± 2.7 pAN: 15.9 ± 2.4 |
8 |
| Healthy controls | – | – | 89 | 21.9 ± 2.5 | 24.8 ± 2.7 | ||||||||
| Rybakowski et al. | 2006 | Poland | Caucasian | 5-HTR2A rs63311 | PCR–RFLP | DSM-IV | Individuals with AN | 0 | 131 | 131 | 17.6 ± 2.9 | – | 8 |
| Healthy controls | 0 | 89 | 89 | 20.9 ± 1.6 | – | ||||||||
| Martásková et al. | 2009 | Czechia | Caucasian | 5-HTR2A rs63311 | PCR–RFLP | ICD-10 | Individuals with AN | 75 | 75 | 25.4 ± 6.2 | 14.7 ± 1.4 | 8 | |
| Healthy controls | 65 | 65 | 25.8 ± 5.1 | 20.7 ± 1.9 | |||||||||
| Kang et al. | 2017 | China | Asian | 5-HTR2A rs63311 | SNaP Shot assay | DSM-IV-TR | Individuals with AN | – | – | 249 | 19.1 ± 4.5 | – | 8 |
| Healthy controls | – | – | 228 | 20.4 ± 1.5 | – | ||||||||
| Ceccarini et al. | 2020 | Italy | Caucasian | 5-HTR2A rs63311 | RT-PCR | DSM-5 | Individuals with AN | 5 | 306 | 311 | 22.4 ± 8.9 | 14.3 ± 2.0 | 8 |
| Healthy controls | 38 | 317 | 355 | 27.7 ± 7.9 | 21.5 ± 2.5 | ||||||||
| Karwautz et al. | 2001 | United Kingdom | Caucasian | 5-HTR2C rs6318 | PCR–RFLP | DSM-IV | Individuals with AN | 0 | 44 | 44 | – | – | 8 |
| Healthy controls | 0 | 39 | 39 | – | – | ||||||||
| Westberg et al. | 2002 | Sweden | Caucasian | 5-HTR2C rs6318 | PCR–RFLP | DSM-III-R | Individuals with AN | 0 | 41 | 41 | – | 15.0 ± 2.0 | 8 |
| Healthy controls | 0 | 91 | 91 | – | – | ||||||||
| Hu et al. | 2003 | United Kingdom | Caucasian | 5-HTR2C rs6318 | PCR–RFLP | DSM-IV | Individuals with AN | – | – | 118 | – | 12.9 ± 2.1 | 7 |
| Healthy controls | – | – | 316 | – | – |
Age: years ± S.D.; BMI: kg/m2 ± S.D.
NOS: Newcastle–Ottawa Scale; DSM: Diagnostic and Statistical Manual of Mental Disorders; ICD: International Classification of Diseases; PCR–RFLP: polymerase chain reaction-restriction fragment length polymorphism; RT-PCR: real time polymerase chain reaction; SNaP Shot assay: fluorescence labeled single base extension chain termination reaction-based typing assay. M: male; F: female; rAN: patients with restricting type AN; pAN: patients with purging type AN
Rs6311 polymorphism of 5-HTR2A
Several studies have evaluated the association of the 5-HTR2A rs6311 SNP with AN [24–29, 31–39, 57–61]. Ziegler et al. [29] provided genotype frequencies missing in the Hinney et al. [24] study, and correct the data for those miscalculated in the Collier et al. [25] study. We excluded the Gorwood et al. [37] study, included in previous meta-analyses [34, 40], due to virtual calculation of its control participants from non-transmitted alleles in family trios analyzed in TDTs. The Genis-Mendoza et al. study [61] was also excluded due to a considerably smaller number of AN participants (30) compared to control participants (292), a notable difference in age distribution (15 years vs. 30, respectively) and an undefined gender distribution. Details on excluded studies are reported in Table 1.
The eighteen studies included a total of 4926 participants, comprising 2049 patients and 2877 controls), with a NOS quality rating of 7.17 ± 0.75. Genotype and allele frequencies in patients and control cohorts were in HW equilibrium across all studies.
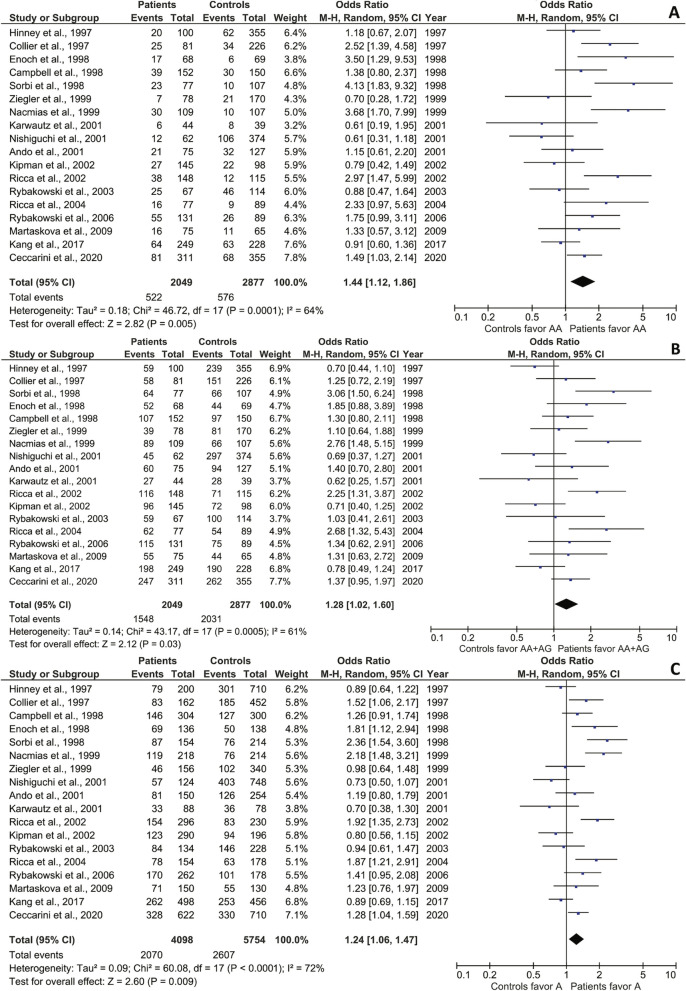
In an initial global assessment, under the recessive model, we found a small but significant association of the AA genotype with AN (RE, OR 1.44 (95% CI 1.12–1.86); Z = 2.82, p = 0.005) as a risk factor (Fig. 2A), thus as a risk factor. Although substantial effect size heterogeneity and variation were observed (χ2 = 46.72; df = 17; p = 0.0001; I2 = 64%), the pooled OR did not change significantly in the leave-one-out test, suggesting good result stability. Conversely, according to the dominant model, the pooled AA and AG genotypes did not show an association with AN (RE, OR = 1.28 (95% CI 1.02–1.60); Z = 2.12, p = 0.03) (Fig. 2B), with high heterogeneity and variation among the studies (χ2 = 43.17; p = 0.0005; I2 = 61%). Under the allele model, the A allele was the predominant one among AN participants, representing a risk factor, with a very small but significant association [RE, OR = 1.24 (95% CI 1.06–1.47); Z = 2.60; p = 0.009]. Again, high heterogeneity and variation were observed (χ2 = 60.08; df = 17; p < 0.00001; I2 = 72%) (Fig. 2C) although coupled to good result stability, as evidenced by non-significant changes of the pooled OR in the leave-one-out test.
Fig. 2.
Odds ratios, 95% confidence intervals, and forest plots of individual studies and relative pooled results between AN and the 5-HT2A -1438G/A SNP in different genetic models: A recessive (AA vs. AG + GG); B dominant (AA + AG vs. GG); and C, allele (A vs. G). Egger’s test for publication bias: A p = 0.4020; B p = 0.4337; C p = 0.5992
Subsequently, we divided the studies into three geographical regions based on the origins of all cohorts: Central Europe, Britain, Southern Europe, and Asia. NOS quality ratings did not exhibit significant variations among these subgroups (data not shown).
The Central European subgroup comprised 591 patients and 891 controls from Germany [24, 29]; France [33], Poland [35, 59], and Czechia [34]. In this subgroup, no association emerged according to all models: (a) recessive model, [FE, OR = 1.10 (95% CI 0.84–1.43); Z = 0.67; p = 0.50], with low heterogeneity and variation (χ2 = 5.31; df = 5; p = 0.38; I2 = 6%); (b) dominant model, [FE, OR = 0.92 (0.71–1.17); Z = 0.70, p = 0.48], with low heterogeneity and no variation (χ2 = 4.53; df = 5; p = 0.48; I2 = 0%); (c) allele model, [FE, OR = 1.00 (95% CI 0.85–1.17); Z = 0.05; p = 0.96, with low heterogeneity and variation (χ2 = 5.74; df = 5; p = 0.33; I2 = 13%).
The British subgroup [25, 26, 57] comprised 277 patients and 415 controls. No association was observed in this subgroup according to all models: (a) recessive model, [RE, OR = 1.47 (95% CI 0.76–2.85); Z = 1.15; p = 0.25], with low heterogeneity but high variation (χ2 = 5.17; df = 2; p = 0.08; I2 = 61%); (b) dominant model, [FE, OR = 1.16 (95% CI 0.83–1.62); Z = 0.86, p = 0.39], with very low heterogeneity and variation (χ2 = 2.01; df = 2; p = 0.37; I2 = 1%); (c) allele model, [RE, OR = 1.19 (95% CI 0.83–1.70); Z = 0.96; p = 0.34], with low heterogeneity but substantial variation (χ2 = 4.46; df = 2; p = 0.11; I2 = 55%).
The Southern European subgroup comprised Italian studies [28, 31, 39, 58, 60] totaling 722 patients and 733 controls. Significant associations were observed in this subgroup according to all models: (a) recessive model, with a medium association [RE, OR = 2.55 (95% CI 1.62–4.02); Z = 4.05, p < 0.0001], low heterogeneity but substantial variation (χ2 = 9.08; df = 4; p = 0.06; I2 = 56%); (b) dominant model, with a small association [FE, OR = 1.98 (95% CI 1.56–2.51); Z = 5.66, p < 0.00001], low heterogeneity and some variation (χ2 = 7.41; df = 4; p = 0.12; I2 = 46%); and (c) allele model, with a small association [RE, OR = 1.82 (95% CI 1.41–2.37); Z = 4.55; p < 0.00001], some heterogeneity but substantial variation (χ2 = 10.93; df = 4; p = 0.03; I2 = 63%). Upon conducting leave-one-out tests, exclusion of the Ceccarini et al. [39] study, conducted in Umbria, resulted in an increase in association values and a decrease in heterogeneity and variation among the four other studies, all conducted in Tuscany, comprising 411 patients and 378 controls, according to all models: (a) recessive model [FE, OR = 3.23 (95% CI 2.18–4.78); Z = 5.86; p < 0.00001, χ2 = 1.04; df = 3; p = 0.79; I2 = 0%]; (b) dominant model [FE, OR = 2.62 (95% CI 1.91–3.59); Z = 5.97; p < 0.00001, χ2 = 0.52; df = 3; p = 0.91; I2 = 0%]; (c) allele model [FE, OR = 2.07 (95% CI 1.69–2.52); Z = 7.18; p < 0.00001, χ2 = 0.80; df = 3; p = 0.85; I2 = 0%]. Exclusion of other studies did not result in substantial variations.
The Asian subgroup comprised Japanese [32, 36] and Chinese Han [38] cohorts, totaling 386 patients and 729 controls. No association emerged in this subgroup according to all models: (a) recessive model [FE, OR = 0.87 (95% CI 0.64–1.18); Z = 0.88; p = 0.38], with very low heterogeneity and no variation (χ2 = 1.89; df = 2; p = 0.39; I2 = 0%); (b) dominant model, [FE, OR = 0.86 (95% CI 0.62–1.19); Z = 0.89, p = 0.37], with small heterogeneity and some variation (χ2 = 2.64; df = 2; p = 0.27; I2 = 24%); (c) allele model, [FE, OR = 0.90 (95% CI 0.75–1.09); Z = 1.06; p = 0.29], with low heterogeneity and some variation (χ2 = 3.05; df = 2; p = 0.22; I2 = 34%).
The Enoch et al. [27] study, performed on North American (U.S.A.) participants, suggested a positive association but was not included in any subgroup.
For all subgroups and across all models, except for the Italian subgroup, the leave-one-out tests did not alter the pooled OR significantly, indicating the stability of the results.
In summary, the geographic subgroup analysis revealed an association of the 5-HTR2A rs6311 polymorphism according to all models only in Italian studies, particularly within the Tuscany cohorts. This suggests a potential regional specificity for the association of this polymorphism and AN.
Rs6318 polymorphism of 5-HTR2C
The rs6318 (Cys23Ser) SNP of 5-HTR2C has been investigated in three studies [57, 62, 63], with 649 total participants, comprising 203 patients and 446 controls. The NOS quality rating of the studies was 7.67 ± 0.58, and genotype and allele frequencies were in HW equilibrium across all cohorts. Under the recessive model, the SerSer genotype was not associated with AN [FE, OR 1.34 (95% CI 0.48–3.72); Z = 0.55, p = 0.58] (Fig. 3A), with negligible heterogeneity and variation (χ2 = 1.40; df = 2; p = 0.50; I2 = 0%). Under the dominant model, presence of the Ser allele displayed a slight tendency towards association with AN as a risk factor [RE, OR = 1.96 (95% CI 0.95 – 4.04); Z = 1.83, p = 0.07] (Fig. 3B), albeit with moderate heterogeneity but high variation (χ2 = 5.82; df = 2; p < 0.05; I2 = 66%). Similar results were observed under the allele model [RE, OR = 1.78 (95% CI 0.96 – 3.31; Z = 1.83; p = 0.07], suggesting a tendency for the association of the Ser allele with AN as a risk factor, but with moderate heterogeneity and high variation (χ2 = 5.56; df = 2; p = 0.06; I2 = 64%) (Fig. 3C). The available evidence does not support the existence of an association between this SNP and AN.
Fig. 3.
Odds ratios, 95% confidence intervals, and forest plots of individual studies and relative pooled results between AN and the 5-HT2C Cys23Ser SNP in different genetic models: A recessive [SerSer (SS) vs. SerCys (SC) + CysCys (CC)]; B, dominant (SS + SC vs. CC); and C allele (S vs. C). Egger’s test for publication bias: A p = 0.1650; B p = 0.9612; C p = 0.8586
Discussion
In this paper, we describe meta-analyses of candidate gene studies on AN, focusing on 5-HTR2A rs6311 and 5-HTR2C rs6318 SNPs, allowing a reevaluation of previously reported information for the former and providing novel results for the latter.
Rs6311 polymorphism of 5-HTR2A
5-HTR2A, mapped on chromosome 13q14-q21, has been implicated in various neuropsychiatric disorders, including depression [64], and central nervous system pathologies, such as epilepsy [65]. The rs6311 (-1438G/A) SNP is located in the distal promoter region of 5-HTR2A. Its functional role remains uncertain: while some studies have linked the A allele with increased transcriptional [66, 67] and translation efficiency [68], as well as higher serotonin binding [69, 70], compared to the G allele, these findings have not been replicated in other studies [71, 72].
Results of numerous reports and previous meta-analyses [29, 30, 34, 40] have suggested a potential association of this SNP with AN, depending on the geographic distribution of the cohorts involved.
Our meta-analysis incorporates three additional studies to those of Yan et al. [40], and specifically addresses this issue. Upon global analysis, results of the existing studies confirm the presence of an association according to the recessive and allele models, but not the dominant model, with the AA genotype and A allele representing risk factors for AN. In this assessment, taking into account participant numbers and ORs, the estimated attributable risk in the development of AN is 7.8% for the AA genotype and 9.7% for the A allele [73].
However, our geographical partition reveals the presence of a clear association only in the Italian subgroup. In its cohorts, the estimated attributable risk for the AA genotype and A allele in the development of AN is higher, at 15.7% and 10.1%, respectively [73]. Furthermore, a more robust association is observed in the Tuscany cohorts, suggesting a dependence of the association on specific geographic locations and differences in ethnic groups.
In summary, our findings indicate that the positive association observed globally in present and previous meta-analyses originates from a limited number of studies, mainly conducted in Italy. This observation emphasizes the need for further investigations, including molecular and cellular studies focused on gene expression and receptor-mediated transduction signaling, respectively, to assess the potential functional role of the rs6311 SNP of 5-HTR2A in the integration of neuronal activity.
Rs6318 polymorphism of 5-HTR2C
The 5-HTR2C gene, mapped on chromosome Xq24, has attracted attention due to pharmacological evidence suggesting its involvement in food intake regulation and AN in both rodents [74, 75] and humans [76, 77].
The rs6318 (Cys23Ser) SNP of 5-HTR2C involves a cysteine (Cys) to serine (Ser) substitution at position 23 in the N-terminal extracellular domain of the receptor [78]. Studies exploring the association of this SNP with AN emerged in 2001, supported by the finding of serotonin-binding differences between the two allele variants [79], but waned after 2003, likely due to observations of no functional differences between the alleles [80]. Although our meta-analysis does not reveal an association with AN, recent research has identified pharmacological and subcellular localization differences between them [81]. Consequently, further investigations on this SNP promise for uncovering significant insights.
Conclusions
This meta-analysis elucidates potential roles of 5-HTR2A rs6311 and 5-HTR2C rs6318 SNPs in the etiology of AN. While evidence points to an association between the 5-HTR2A rs6311 polymorphism and AN in Italian cohorts, no associations are observed from other geographic regions. These findings unify the results of all previous meta-analyses and support the conclusions of recent reviews [82, 83]. They underscore the notion that this polymorphism should be viewed within a multifactorial inheritance framework that accounts for the interplay of multiple genes and potential environmental factors in the development of a psychiatric disorder.
As for the 5-HTR2C rs6318 polymorphism, no evidence for an association with AN was observed.
In conclusion, while the involvement of serotonin in regulating eating behaviors and contributing to the development of AN is acknowledged, attributing this role to individual polymorphisms of serotonin receptor genes remains ambiguous. In the future, multiple genetic factors involved in AN will likely be identified through genome-wide investigations. Additionally, analysis of the interaction of multiple genes, such as those encoding for 5-HTT and the norepinephrine transporter [84] or 5-HTT and monoamine-oxidase [85], would offer further insights. Lastly, it will be imperative to acquire more information on the etiologic roles of psychological, environmental, familial and social factors [86]. In this context, the advancement of epigenetic research holds significance, by allowing the evaluation of gene x environment interactions potentially involved in AN.
In a bio-psycho-social framework, identifying genuine genetic and epigenetic markers would be valuable in an integrated strategy aimed at early risk detection. Under a pharmacogenomic approach, this could also facilitate the development of improved standard or nutritional therapies tailored to individual responses. From a clinical perspective, understanding the genetic factors associated with AN could offer support to patients and their families, potentially alleviating the burden of blame often linked with the disorder [87]. However, no definitive clinical implications can be drawn from individual serotonin gene polymorphisms considered in the present meta-analysis. Consequently, engaging in discussions about the genetics of AN with affected individuals and their families remains challenging, and the possibility of implementing prevention strategies or genomic approaches to the pathology appears to be postponed into the future.
Strength and limits
This meta-analysis evaluates the outcomes of serotonin candidate gene studies on AN, focusing on the 5-HTR2A rs6311 polymorphism and the 5-HTR2C rs6318 polymorphism. It has various strengths: a) it was conducted by an up-to-date search of articles published between 1997 and 2022; b) article selection was rigorous, employing specific inclusion/exclusion criteria; c) all results underwent statistical correction for multiple testing; and d) it includes a comprehensive geographic evaluation of effect sizes for the 5-HTR2A rs6311 polymorphism.
Similar to all meta-analyses, we acknowledge the possibility of missing publications, despite our rigorous search strategy. Other limitations are inherent of published reports, including small sample sizes of many studies, relatively low numbers of total individuals, subgroup analyses limited to geographic regions.
Finally, this meta-analysis does not differentiate between the binge eating/purging subtype and restricting subtype of AN, since this issue is reported only in some studies. Increasing the number of genetic studies focusing on these two diagnoses would be advisable to gain deeper insights into these phenotype variants.
What is already known on this subject?
Existing research of the subject has seen several case–control candidate gene studies examining the association between serotonin receptor genes and AN from 1997 to 2022. However, these studies have yielded inconsistent results. Additionally, various meta-analyses focusing on the 5-HTR2A rs6311 polymorphism have been conducted between 1999 and 2021. Despite these efforts, methodological and/or statistical weaknesses in these studies have led to conflicting conclusions, raising doubts about the presence of a definitive association between this polymorphism and AN. Conducting an updated and rigorous meta-analysis to gather further information on the 5-HTR2A rs6311 polymorphism, and potentially other serotonin receptor gene polymorphisms, could provide valuable insights into their involvement in AN.
What this study adds?
This meta-analysis focuses on the association of the 5-HTR2A rs6311 polymorphism and the 5-HTR2C rs6318 polymorphism with AN. The findings indicate the absence of association between the 5-HTR2A rs6311 polymorphism and AN when analyzed globally or in various geographic subgroups but pinpoint presence of association in an Italian subgroup, primarily represented by cohorts from Tuscany. They also report no evidence of an association between the 5-HTR2C rs6318 polymorphism and AN.
With the exclusion of the results relative to the 5-HTR2A rs6311 polymorphism in the Italian region that would need further examination, these observations underscore the notion that an etiological role in AN cannot be attributed to individual polymorphisms of serotonin receptor genes. This strengthens the importance of exploring multifactorial inheritance paradigms to understand factors involved in the disorder.
Under a clinical perspective, these findings do not allow to conceive, at present, the implementation of prevention strategies or genomic approaches in supporting and treating subjects with AN.
Acknowledgements
We gratefully acknowledge Valentina Rovacchi for her precious help in literature collection.
Author contributions
DLP and AB conceived the study. FS and DLP independently performed the database search for articles and data collection. All authors performed article selection and data check, FS and AB performed statistical analyses. AB and SC wrote the article. All authors reviewed and approved the article.
Funding
Open access funding provided by Università degli Studi di Roma La Sapienza within the CRUI-CARE Agreement. This study was financially supported by Sapienza “Ateneo” Grant No. RM12117A584F7580 to AB.
Availability of data and materials
Data used for the present meta-analysis are available from the corresponding author upon reasonable request.
Declarations
Ethics approval and consent to participate
Not applicable.
Competing interests
The authors report no competing of interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.American Psychiatric Association . Diagnostic and statistical manual of mental disorders (DSM-5-TR) 5. Washington DC: American psychiatric association; 2022. [Google Scholar]
- 2.Bulik CM, Sullivan PF, Kendler KS. Heritability of binge-eating and broadly defined bulimia nervosa. Biol Psychiatry. 1998;44:1210–1218. doi: 10.1016/s0006-3223(98)00280-7. [DOI] [PubMed] [Google Scholar]
- 3.Klump KL, Miller KB, Keel PK, McGue M, Iacono WG. Genetic and environmental influences on anorexia nervosa syndromes in a population-based twin sample. Psychol Med. 2001;31:737–740. doi: 10.1017/s0033291701003725. [DOI] [PubMed] [Google Scholar]
- 4.Ben-Dor DH, Laufer N, Apter A, Frisch A, Weizman A. Heritability, genetics and association findings in anorexia nervosa. Isr J Psychiatry Relat Sci. 2002;39:262–270. [PubMed] [Google Scholar]
- 5.Rask-Andersen M, Olszewski PK, Levin AS, Schiotch HB. Molecular mechanisms underlying anorexia nervosa: focus on human gene association studies and systems controlling food intake. Brain Res Rev. 2010;62:147–164. doi: 10.1016/j.brainresrev.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 6.Trace SE, Baker JH, Peñas-Lledó E, Bulik CM. The genetics of eating disorders. Annu Rev Clin Psychol. 2013;9:589–620. doi: 10.1146/annurev-clinpsy-050212-185546. [DOI] [PubMed] [Google Scholar]
- 7.Hübel C, Leppä V, Breen G, Bulik CM. Rigor and reproducibility in genetic research on eating disorders. Int J Eat Disord. 2018;51(7):593–607. doi: 10.1002/eat.22896. [DOI] [PubMed] [Google Scholar]
- 8.Paolacci S, Kiani AK, Manara E, Beccari T, Ceccarini MR, Stuppia L, Chiurazzi P, Dalla Ragione L, Bertelli M. Genetic contributions to the etiology of anorexia nervosa: new perspectives in molecular diagnosis and treatment. Mol Genet Genomic Med. 2020;8(7):1244. doi: 10.1002/mgg3.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herpertz-Dahlmann B, Seitz J, Konrad K. Aetiology of anorexia nervosa: From a “psychosomatic family model” to a neuropsychiatric disorder? Eur Arch Psychiatry Clin Neurosci. 2011;261:177–181. doi: 10.1007/s00406-011-0246-y. [DOI] [PubMed] [Google Scholar]
- 10.Berge JM, Wall M, Larson N, Eisenberg ME, Loth KA, Neumark-Sztainer D. The unique and additive associations of family functioning and parenting practices with disordered eating behaviors in diverse adolescents. J Behav Med. 2014;37:205–217. doi: 10.1007/s10865-012-9478-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tambelli R, Cimino S, Cerniglia L, Ballarotto G. Early maternal relational traumatic experiences and psychopathological symptoms: a longitudinal study on mother-infant and father-infant interactions. Sci Rep. 2015;5:13984. doi: 10.1038/srep13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Langdon-Daly J, Serpell L. Protective factors against disordered eating in family systems: a systematic review of research. J Eat Disord. 2017;5:12. doi: 10.1186/s40337-017-0141-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Treasure J, Zipfel S, Micali N, Wade T, Stice E, Claudino A, Schmidt U, Frank GK, Bulik CM, Wentz E. Anorexia nervosa. Nat Rev Dis Primers. 2015;1:15074. doi: 10.1038/nrdp.2015.74. [DOI] [PubMed] [Google Scholar]
- 14.Smink FR, van Hoeken D, Hoek HW. Epidemiology of eating disorders: incidence, prevalence and mortality rates. Curr Psychiatry Rep. 2012;14(4):406–414. doi: 10.1007/s11920-012-0282-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mayhew AJ, Pigeyre M, Couturier J, Meyre D. An evolutionary genetic perspective of eating disorders. Neuroendocrinology. 2018;106(3):292–306. doi: 10.1159/000484525. [DOI] [PubMed] [Google Scholar]
- 16.Stedal K, Broomfield C, Hay P, Touyz S, Scherer R. Neuropsychological functioning in adult anorexia nervosa: a meta-analysis. Neurosci Biobehav Rev. 2021;130:214–226. doi: 10.1016/j.neubiorev.2021.08.021. [DOI] [PubMed] [Google Scholar]
- 17.Keeler JL, Konyn CY, Treasure J, Cardi V, Himmerich H, Tchanturia K, Mycroft H. “Your mind doesn’t have room for anything else”: a qualitative study of perceptions of cognitive functioning during and after recovery from anorexia nervosa. J Eat Disord. 2022;10(1):201. doi: 10.1186/s40337-022-00723-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibson G. Rare and common variants: twenty arguments. Nat Rev Genet. 2012;13(2):135–145. doi: 10.1038/nrg3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bevilacqua A. The limits of association studies in behavioral genetics: a revenge of sexual reproduction? Organisms. J Biol Sci. 2019;3(1):21–24. doi: 10.13133/2532-5876_5.6. [DOI] [Google Scholar]
- 20.Wyler SC, Lord CC, Lee S, Elmquist JK, Liu C. Serotonergic control of metabolic homeostasis. Front Cell Neurosci. 2017;20(11):277. doi: 10.3389/fncel.2017.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaye WH, Frank GK, Bailer UF, Henry SE. Neurobiology of anorexia nervosa: clinical implications of alterations of the function of serotonin and other neuronal systems. Int J Eat Disord. 2005;37(Suppl):S15–9. doi: 10.1002/eat.20109. [DOI] [PubMed] [Google Scholar]
- 22.Sher L. Psychiatric diagnoses and inconsistent results of association studies in behavioral genetics. Med Hypotheses. 2000;54(2):207–209. doi: 10.1054/mehy.1999.0022. [DOI] [PubMed] [Google Scholar]
- 23.Monteleone P, Maj M. Genetic susceptibility to eating disorders: associated polymorphisms and pharmacogenetic suggestions. Pharmacogenomics. 2008;9(10):1487–1520. doi: 10.2217/14622416.9.10.1487. [DOI] [PubMed] [Google Scholar]
- 24.Hinney A, Ziegler A, Nöthen MM, Remschmidt H, Hebebrand J. 5-HT2A receptor gene polymorphisms, anorexia nervosa, and obesity. Lancet. 1997;350(9087):1324–1325. doi: 10.1016/S0140-6736(05)62485-3. [DOI] [PubMed] [Google Scholar]
- 25.Collier DA, Arranz MJ, Li T, Mupita D, Brown N, Treasure J. Association between 5-HT2A gene promoter polymorphism and anorexia nervosa. Lancet. 1997;350(9075):412. doi: 10.1016/S0140-6736(05)64135-9. [DOI] [PubMed] [Google Scholar]
- 26.Campbell DA, Sundaramurthy D, Markham AF, Pieri LF. Lack of association between 5-HT2A gene promoter polymorphism and susceptibility to anorexia nervosa. Lancet. 1998;351(9101):499. doi: 10.1016/S0140-6736(05)78688-8. [DOI] [PubMed] [Google Scholar]
- 27.Enoch MA, Kaye WH, Rotondo A, Greenberg BD, Murphy DL, Goldman D. 5-HT2A promoter polymorphism -1438G/A, anorexia nervosa, and obsessive-compulsive disorder. Lancet. 1998;351(9118):1785–1786. doi: 10.1016/S0140-6736(05)78746-8. [DOI] [PubMed] [Google Scholar]
- 28.Sorbi S, Nacmias B, Tedde A, Ricca V, Mezzani B, Rotella CM. 5-HT2A promoter polymorphism in anorexia nervosa. Lancet. 1998;351(9118):1785. doi: 10.1016/S0140-6736(05)78745-6. [DOI] [PubMed] [Google Scholar]
- 29.Ziegler A, Hebebrand J, Görg T, Rosenkranz K, Fichter M, Herpertz-Dahlmann B, Remschmidt H, Hinney A. Further lack of association between the 5-HT2A gene promoter polymorphism and susceptibility to eating disorders and a meta-analysis pertaining to anorexia nervosa. Mol Psychiatry. 1999;4(5):410–412. doi: 10.1038/sj.mp.4000561. [DOI] [PubMed] [Google Scholar]
- 30.Gorwood P, Kipman A, Foulon C. The human genetics of anorexia nervosa. Eur J Pharmacol. 2003;480:163–170. doi: 10.1016/j.ejphar.2003.08.103. [DOI] [PubMed] [Google Scholar]
- 31.Nacmias B, Ricca V, Tedde A, Mezzani B, Rotella CM, Sorbi S. 5-HT2A receptor gene polymorphisms in anorexia nervosa and bulimia nervosa. Neurosci Lett. 1999;277(2):134–136. doi: 10.1016/s0304-3940(99)00859-9. [DOI] [PubMed] [Google Scholar]
- 32.Nishiguchi N, Matsushita S, Suzuki K, Murayama M, Shirakawa O, Higuchi S. Association between 5HT2A receptor gene promoter region polymorphism and eating disorders in Japanese patients. Biol Psychiatry. 2001;50(2):123–128. doi: 10.1016/s0006-3223(00)01107-0. [DOI] [PubMed] [Google Scholar]
- 33.Kipman A, Bruins-Slot L, Boni C, Hanoun N, Adès J, Blot P, Hamon M, Mouren-Siméoni M, Gorwood P. 5-HT(2A) gene promoter polymorphism as a modifying rather than a vulnerability factor in anorexia nervosa. Eur Psychiatry. 2002;17(4):227–229. doi: 10.1016/s0924-9338(02)00657-0. [DOI] [PubMed] [Google Scholar]
- 34.Martásková D, Slachtová L, Kemlink D, Záhoráková D, Papezová H. Polymorphisms in serotonin-related genes in anorexia nervosa. The first study in Czech population and metaanalyses with previously performed studies. Folia Biol (Praha). 2009;55(5):192–197. doi: 10.14712/fb2009055050192. [DOI] [PubMed] [Google Scholar]
- 35.Rybakowski F, Slopien A, Dmitrzak-Weglarz M, Czerski P, Rajewski A, Hauser J. The 5-HT2A -1438 A/G and 5-HTTLPR polymorphisms and personality dimensions in adolescent anorexia nervosa: association study. Neuropsychobiology. 2006;53(1):33–39. doi: 10.1159/000090701. [DOI] [PubMed] [Google Scholar]
- 36.Ando T, Komaki G, Karibe M, Kawamura N, Hara S, Takii M, Naruo T, Kurokawa N, Takei M, Tatsuta N, Ohba M, Nozoe S, Kubo C, Ishikawa T. 5-HT2A promoter polymorphism is not associated with anorexia nervosa in Japanese patients. Psychiatr Genet. 2001;11(3):157–160. doi: 10.1097/00041444-200109000-00008. [DOI] [PubMed] [Google Scholar]
- 37.Gorwood P, Adès J, Bellodi L, Cellini E, Collier DA, Di Bella D, Di Bernardo M, Estivill X, Fernandez-Aranda F, Gratacos M, Hebebrand J, Hinney A, Hu X, Karwautz A, Kipman A, Mouren-Siméoni MC, Nacmias B, Ribasés M, Remschmidt H, Ricca V, Rotella CM, Sorbi S, Treasure J, for EC Framework V '‘Factors in Healthy Eating'’ consortium The 5-HT(2A) -1438G/A polymorphism in anorexia nervosa: a combined analysis of 316 trios from six European centres. Mol Psychiatry. 2002;7(1):90–94. doi: 10.1038/sj.mp.4000938. [DOI] [PubMed] [Google Scholar]
- 38.Kang Q, Chen J, Yu S, Yuan A, Zhang Y, Zhang R, Jiang W, Zhang C, Zhang H, Zhang M, Xiao Z. Association of the 5-HT2A receptor gene promoter polymorphism-1438G/A with anorexia nervosa and psychopathological traits in the Chinese Han population: a preliminary study. Asia Pac Psychiatry. 2017 doi: 10.1111/appy.12284. [DOI] [PubMed] [Google Scholar]
- 39.Ceccarini MR, Tasegian A, Franzago M, Patria FF, Albi E, Codini M, Conte C, Bertelli M, Dalla Ragione L, Stuppia L, Beccari T. 5-HT2AR and BDNF gene variants in eating disorders susceptibility. Am J Med Genet B Neuropsychiatr Genet. 2020;183(3):155–163. doi: 10.1002/ajmg.b.32771. [DOI] [PubMed] [Google Scholar]
- 40.Yan P, Gao B, Wang S, Wang S, Li J, Song M. Association of 5-HTR2A -1438A/G polymorphism with anorexia nervosa and bulimia nervosa: a meta-analysis. Neurosci Lett. 2021;11(755):135918. doi: 10.1016/j.neulet.2021.135918. [DOI] [PubMed] [Google Scholar]
- 41.Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, Shamseer L, Tetzlaff JM, Akl EA, Brennan SE, Chou R, Glanville J, Grimshaw JM, Hróbjartsson A, Lalu MM, Li T, Loder EW, Mayo-Wilson E, McDonald S, McGuinness LA, Stewart LA, Thomas J, Tricco AC, Welch VA, Whiting P, Moher D. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021;29(372):n71. doi: 10.1136/bmj.n71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.American Psychiatric Association (APA) Diagnostic and statistical manual of mental disorders. 3. Washington DC: American Psychiatric Association; 1987. [Google Scholar]
- 43.American Psychiatric Association (APA) Diagnostic and statistical manual of mental disorders. 4. Washington, DC: American Psychiatric Association; 1994. [Google Scholar]
- 44.American Psychiatric Association (APA) Diagnostic and statistical manual of mental disorders. 4-TR. Washington DC: American Psychiatric Association; 2000. [Google Scholar]
- 45.American Psychiatric Association (APA) Diagnostic and statistical manual of mental disorders (DSM-5) 5. Washington DC: American Psychiatric Association; 2013. [Google Scholar]
- 46.World Health Organization (WHO) The ICD-10 classification of mental and behavioural disorders. Geneva: World Health Organization; 1993. [Google Scholar]
- 47.Crocetti E. Systematic reviews with meta-analysis: why, when, and how? Emerg Adulthood. 2016;4(1):3–18. doi: 10.1177/2167696815617076. [DOI] [Google Scholar]
- 48.Stang A. Critical evaluation of the Newcastle–Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25:603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 49.Egger M, Smith GD, Phillips AN. Meta-analysis: principles and procedures. BMJ. 1997;315(7121):1533–1537. doi: 10.1136/bmj.315.7121.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu H-M, Zheng J-P, Yang D, Liu Z-F, Li Z, Hu Z-Z, Li Z-N. Recessive/dominant model: alternative choice in case-control-based genome- wide association studies. PLoS ONE. 2021;16(7):e0254947. doi: 10.1371/journal.Pone.,0254947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pigott TD, Polanin JR. The use of meta-analytic statistical significance testing. Res Synth Methods. 2015;6(1):63–73. doi: 10.1002/jrsm.1124. [DOI] [PubMed] [Google Scholar]
- 52.Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med. 1997;127(9):820–826. doi: 10.7326/0003-4819-127-9-199711010-00008. [DOI] [PubMed] [Google Scholar]
- 53.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 54.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 55.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 56.Cohen J. Statistical power analysis for the behavioral sciences. 2. New York: Routledge; 1988. [Google Scholar]
- 57.Karwautz A, Rabe-Hesketh S, Hu X, Zhao J, Sham P, Collier DA, Treasure JL. Individual-specific risk factors for anorexia nervosa: a pilot study using a discordant sister-pair design. Psychol Med. 2001;31(2):317–329. doi: 10.1017/s0033291701003129. [DOI] [PubMed] [Google Scholar]
- 58.Ricca V, Nacmias B, Cellini E, Di Bernardo M, Rotella CM, Sorbi S. 5-HT2A receptor gene polymorphism and eating disorders. Neurosci Lett. 2002;323(2):105–108. doi: 10.1016/s0304-3940(02)00088-5. [DOI] [PubMed] [Google Scholar]
- 59.Rybakowski F, Słopień A, Dmitrzak-Weglarz M, Czerski P, Hauser J, Rajewski A. Badanie asocjacyjne polimorfizmu genu receptora serotoniny 5-HT2A w jadłowstrecie psychicznym w populacji polskiej (association study of 5-HT2A receptor gene polymorphism in anorexia nervosa in Polish population) Psychiatr Pol. 2003;37(1):47–55. [PubMed] [Google Scholar]
- 60.Ricca V, Nacmias B, Boldrini M, Cellini E, di Bernardo M, Ravaldi C, Tedde A, Bagnoli S, Placidi GF, Rotella CM, Sorbi S. Psychopathological traits and 5-HT2A receptor promoter polymorphism (-1438 G/A) in patients suffering from anorexia nervosa and bulimia nervosa. Neurosci Lett. 2004;365(2):92–96. doi: 10.1016/j.neulet.2004.04.057. [DOI] [PubMed] [Google Scholar]
- 61.Genis-Mendoza AD, Ruiz-Ramos D, López-Narvaez ML, Tovilla-Zárate CA, Rosa García A, Cortes Meda G, Martinez-Magaña JJ, González-Castro TB, Juárez-Rojop IE, Nicolini H. Genetic association analysis of 5-HTR2A gene variants in eating disorders in a Mexican population. Brain Behav. 2019;9(7):e01286. doi: 10.1002/brb3.1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Westberg L, Bah J, Råstam M, Gillberg C, Wentz E, Melke J, Hellstrand M, Eriksson E. Association between a polymorphism of the 5-HT2C receptor and weight loss in teenage girls. Neuropsychopharmacology. 2002;26(6):789–793. doi: 10.1016/S0893-133X(01)00417-1. [DOI] [PubMed] [Google Scholar]
- 63.Hu X, Giotakis O, Li T, Karwautz A, Treasure J, Collier DA. Association of the 5-HT2c gene with susceptibility and minimum body mass index in anorexia nervosa. NeuroReport. 2003;14(6):781–783. doi: 10.1097/00001756-200305060-00001. [DOI] [PubMed] [Google Scholar]
- 64.Zięba A, Stępnicki P, Matosiuk D, Kaczor AA. Overcoming depression with 5-HT2A receptor ligands. Int J Mol Sci. 2021;23(1):10. doi: 10.3390/ijms23010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guiard BP, Di Giovanni G. Central serotonin-2A (5-HT2A) receptor dysfunction in depression and epilepsy: the missing link? Front Pharmacol. 2015;17(6):46. doi: 10.3389/fphar.2015.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Polesskaya OO, Sokolov BP. Differential expression of the “C” and “T” alleles of the 5-HT2A receptor gene in the temporal cortex of normal individuals and schizophrenics. J Neurosci Res. 2002;67(6):812–822. doi: 10.1002/jnr.10173. [DOI] [PubMed] [Google Scholar]
- 67.Parsons MJ, D’Souza UM, Arranz MJ, Kerwin RW, Makoff AJ. The-1438A/G polymorphism in the 5-hydroxytryptamine type 2A receptor gene affects promoter activity. Biol Psychiatry. 2004;56:406–410. doi: 10.1016/j.biopsych.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 68.Smith RM, Papp AC, Webb A, Ruble CL, Munsie LM, Nisenbaum LK, Kleinman JE, Lipska BK, Sadee W. Multiple regulatory variants modulate expression of 5-hydroxytryptamine 2A receptors in human cortex. Biol Psychiatry. 2013;73(6):546–554. doi: 10.1016/j.biopsych.2012.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Turecki G, Briere R, Dewar K, Antonetti T, Lesage AD, Seguin M, Chawky N, Vanier C, Alda M, Joober R, Benkelfat C, Rouleau GA. Prediction of level of serotonin 2A receptor binding by serotonin receptor 2A genetic variation in postmortem brain samples from subjects who did or did not commit suicide. Am J Psychiatry. 1999;156:1456–1458. doi: 10.1176/ajp.156.9.1456. [DOI] [PubMed] [Google Scholar]
- 70.Khait VD, Huang Y, Zalsman G, et al. Association of serotonin 5-HT2A receptor binding and the T102C polymorphism in depressed and healthy Caucasian subjects. Neuropsychopharmacol. 2005;30(1):166–172. doi: 10.1038/sj.npp.1300578. [DOI] [PubMed] [Google Scholar]
- 71.Myers RL, Airey DC, Manier DH, Shelton RC, Sanders-Bush E. Polymorphisms in the regulatory region of the human serotonin 5-HT2A receptor gene (HTR2A) influence gene expression. Biol Psychiatry. 2007;61(2):167–173. doi: 10.1016/j.biopsych.2005.12.018. [DOI] [PubMed] [Google Scholar]
- 72.Marinova Z, Monoranu C-M, Fetz S, Walitza S, Grünblatt E. Region-specific regulation of the serotonin 2A receptor expression in development and ageing in post mortem human brain. Neuropathol Appl Neurobiol. 2015;41(4):520–532. doi: 10.1111/nan.12167. [DOI] [PubMed] [Google Scholar]
- 73.Coughlin SS, Benichou J, Weed DL. Attributable risk estimation in case-control studies. Epidemiol Rev. 1994;16(1):51–64. doi: 10.1093/oxfordjournals.epirev.a036144. [DOI] [PubMed] [Google Scholar]
- 74.Nonogaki K, Ohba Y, Wakameda M, Tamari T. Fluvoxamine exerts anorexic effect in 5-HT2C receptor mutant mice with heterozygous mutation of beta-endorphin gene. Int J Neuropsychopharmacol. 2009;12(4):547–552. doi: 10.1017/S1461145708009619. [DOI] [PubMed] [Google Scholar]
- 75.Tecott LH, Sun LM, Akana SF, Strack AM, Lowenstein DH, Dallman MF, Julius D. Eating disorder and epilepsy in mice lacking 5–HT2c serotonin receptors. Nature. 1995;374:542–546. doi: 10.1038/374542a0. [DOI] [PubMed] [Google Scholar]
- 76.Halmi KA, Eckert E, LaDu TJ, Cohen J. Anorexia nervosa. Treatment efficacy of cyproheptadine and amitriptyline. Arch Gen Psychiatry. 1986;43(2):177–181. doi: 10.1001/archpsyc.1986.01800020087011. [DOI] [PubMed] [Google Scholar]
- 77.La Via MC, Gray N, Kaye WH. Case reports of olanzapine treatment of anorexia nervosa. Int J Eat Disord. 2000;27:363–366. doi: 10.1002/(sici)1098-108x(200004)27:3<363::aid-eat16>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 78.Lappalainen J, Zhang L, Dean M, Oz M, Ozaki N, Yu DH, Virkkunen M, Weight F, Linnoila M, Goldman D. Identification, expression, and pharmacology of a Cys23-Ser23 substitution in the human 5–HT2c receptor gene (HTR2C) Genomics. 1995;27:274–279. doi: 10.1006/geno.1995.1042. [DOI] [PubMed] [Google Scholar]
- 79.Okada M, Northup JK, Ozaki N, Russell JT, Linnoila M, Goldman D. Modification of human 5-HT2C receptor function by Cys23Ser, an abundant, naturally occurring amino-acid substitution. Mol Psychiatry. 2004;9(1):55–64. doi: 10.1038/sj.mp.4001357. [DOI] [PubMed] [Google Scholar]
- 80.Fentress H, Grinde E, Mazurkiewicz JE, Backstrom JR, Herrick-Davis K, Sanders-Bush E. Pharmacological properties of the Cys23Ser single nucleotide polymorphism in human 5-HT2C receptor isoforms. Pharmacogenomics J. 2005;5:244–254. doi: 10.1038/sj.tpj.6500315. [DOI] [PubMed] [Google Scholar]
- 81.Land MA, Chapman HL, Davis-Reyes BD, Felsing DE, Allen JA, Moeller FG, Elferink LA, Cunningham KA, Anastasio NC. Serotonin 5-HT2C receptor Cys23Ser single nucleotide polymorphism associates with receptor function and localization in vitro. Sci Rep. 2019;9(1):16737. doi: 10.1038/s41598-019-53124-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Baker JH, Schaumberg K, Munn-Chernoff MA. Genetics of anorexia nervosa. Curr Psychiatry Rep. 2017;19(11):84. doi: 10.1007/s11920-017-0842-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Abou Al Hassan S, Cutinha D, Mattar L. The impact of COMT, BDNF and 5-HTT brain-genes on the development of anorexia nervosa: a systematic review. Eat Weight Disord. 2021;26(5):1323–1344. doi: 10.1007/s40519-020-00978-5. [DOI] [PubMed] [Google Scholar]
- 84.Urwin RE, Bennetts BH, Wilcken B, Beumont PJ, Russell JD, Nunn KP. Investigation of epistasis between the serotonin transporter and norepinephrine transporter genes in anorexia nervosa. Neuropsychopharmacology. 2003;28(7):1351–1355. doi: 10.1038/sj.npp.1300204. [DOI] [PubMed] [Google Scholar]
- 85.Urwin RE, Nunn KP. Epistatic interaction between the monoamine oxidase A and serotonin transporter genes in anorexia nervosa. Eur J Hum Genet. 2005;13:370–375. doi: 10.1038/sj.ejhg.5201328. [DOI] [PubMed] [Google Scholar]
- 86.Frank GK. The Perfect Storm—a bio-psycho-social risk model for developing and maintaining eating disorders. Front Behav Neurosci. 2016;10(10):44. doi: 10.3389/fnbeh.2016.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bulik CM, Blake L, Austin J. Genetics of eating disorders: what the clinician needs to know. Psychiatr Clin North Am. 2019;42(1):59–73. doi: 10.1016/j.psc.2018.10.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data used for the present meta-analysis are available from the corresponding author upon reasonable request.