Abstract
Borna disease virus (BDV) p24 RNA was detected in the peripheral blood mononuclear cells (PBMCs) of psychiatric patients and blood donors by nested reverse transcriptase PCR (RT-PCR). The prevalences of BDV p24 RNA in patients with mood disorders (4%) and schizophrenia (4%) were not significantly different from that in blood donors (2%). This finding was inconsistent with previous reports that showed either a high prevalence or absence of BDV p24 RNA in patients with psychiatric disorders. The differences in BDV p24 RNA prevalence in these studies may be due to differences in the criteria for positivity, the number of PBMCs used for RNA extraction, or the amount of RNA tested for nested RT-PCR or to laboratory contamination. Sequence analysis of BDV p24 RNA from the PBMCs of patients and blood donors showed a high nucleotide sequence conservation but definite nucleotide mutations compared with horse BDV p24 RNA sequences. In comparison with human BDV p24 RNA sequences previously reported from Japan and Germany, there were several positions with silent nucleotide mutations among these clones.
Epidemiological data suggests that not only genetic factors but also environmental factors may play an important role in the etiology of psychiatric disorders, including schizophrenia and mood disorders (10, 40). The association of viral infections, such as cytomegalovirus, herpes simplex virus, and influenza virus, with psychiatric disorders has been implied (1, 18, 29); however, the role of viral etiology in psychiatric disorders remains unclear.
Borna disease (BD) is an acute meningoencephalitis of horses and sheep caused by BD virus (BDV) (4). BDV is a noncytolytic neurotrophic virus that infects several vertebrate species, including birds and primates (9, 27, 37, 38, 43), and causes central nervous system dysfunction with various manifestations depending on the age, immune status, and species of the host (26, 28, 39). For example, BDV induces a severe cellular immune response to infected cells in the brains of adult rats with movement and behavioral abnormalities, or a persistent infection with only cognitive impairment in neonatal rats (3). The viral nucleic acids and antigens are found in limbic pyramidal and extrapyramidal motor systems (8, 25, 36). BDV is a nonsegmented, negative, single-stranded RNA virus (7, 13, 41). The 8.9-kb RNA genome of BDV contains five major open reading frames (ORFs), three of which encode viral polypeptides of 40, 24, and 14.5 kDa (11, 30). RNA splicing is used for the expression of BDV-specific mRNA (12).
The wide host range of the virus and behavioral disturbances in animals with BD suggest that BDV infection may be associated with human psychiatric disorders (2, 32). Seroepidemiological data and the detection of BDV RNA in the peripheral blood mononuclear cells (PBMCs) of psychiatric patients have also suggested a possible involvement of BDV in human psychiatric disorders (5, 20, 22, 34, 41, 42). However, there is controversy over the prevalence of BDV antibody and BDV RNA in the PBMCs of patients with psychiatric disorders. Furthermore, recently the isolation of infectious BDV from the PBMCs of psychiatric patients and the sequencing of the major part of the viral genome have been reported (6). There is also controversy as to whether the human BDV p24 sequence is highly conserved or variable with respect to the horse BDV sequence.
In the present study, to confirm the prevalence of BDV infection in psychiatric patients, we have investigated the prevalence of BDV p24 RNA in the PBMCs of patients with mood disorders or schizophrenia and in those of blood donors by nested reverse transcriptase PCR (RT-PCR). Furthermore, sequence analysis of the human BDV p24 cDNA showed high nucleotide sequence conservation of p24 but a distinct nucleotide mutation in comparison with horse BDV sequences.
MATERIALS AND METHODS
Subjects.
The number and profiles of patients and blood donors included in this study are summarized in Table 1. The patients consisted of 49 with mood disorders and 77 with schizophrenia. All patients met DSM-IV criteria (American Psychiatric Association, 1994) for the diagnosis of mood disorders and schizophrenia. The blood donors were matched by age and sex to the patients. This study was approved by the Ethical Committee of Fukushima Medical University, and all patients gave their informed consent.
TABLE 1.
Detection of BDV p24 RNA in PBMCs of psychiatric patients and blood donors
| Source | Age (mean yr ± SD) | No. positive/no. tested (% positive)
|
||||
|---|---|---|---|---|---|---|
| Sex
|
Treatment
|
Total | ||||
| Male | Female | In-patient | Out patient | |||
| Psychiatric patients | ||||||
| Mood disorder | 47.5 ± 12.6 | 1/25 (4) | 1/24 (4) | 1/30 (5) | 1/19 (7) | 2/49 (4) |
| Schizophrenia | 45.4 ± 10.3 | 0/45 (0) | 3/32 (9) | 2/69 (5) | 1/8 (8) | 3/77 (4) |
| Blood donors | 45.2 ± 10.2 | 2/49 (2) | 0/35 (0) | 2/84 (2) | ||
Preparation of RNA.
For each subject, PBMCs were isolated from 15 ml of blood by using heparin or EDTA and centrifugation on Ficoll-Conray solution (density, 1.077 g/ml). Total cellular RNA was prepared from 107 PBMCs with an RNA extraction kit (Isogen; Nippon-Gene, Tokyo, Japan) according to the manufacturer’s manual. The approximate concentration of extracted RNA was determined by spectrophotometry. Total RNA was also isolated from Madin-Darby canine kidney (MDCK) cells persistently infected with BDV (BDV-MDCK) originally isolated from a horse and was used as a positive control for nested RT-PCR. The BDV-MDCK was kindly provided by R. Rott (19).
Detection of BDV p24 RNA by nested RT-PCR.
BDV p24 RNA was detected by nested RT-PCR. Approximately 1 μg of cellular RNA was reverse transcribed, and the first PCR was performed with a thermostable rTth RT RNA PCR kit (Perkin-Elmer Co., Foster City, Calif.) with BDV p24-specific primers as described by Kishi et al. (20). Primer pairs used for the reverse transcription and first PCR were 5′-TGACCCAACCAGTAGACCA-3′ (P24-OF; nucleotides [nt] 1387 to 1405) and 5′-GTCCCATTCATCCGTTGTC-3′ (P24-OR; nt 1865 to 1847). The second pair of primers were 5′-TCAGACCCAGACCAGCGAA-3′ (P24-MF; nt 1443 to 1461) and 5′-AGCTGGGGATAAATGCGCG-3′ (P24-MR; nt 1834 to 1816). The numbering of the BDV nucleotide sequence followed that reported for the horse BDV sequence adapted in C6 cells (C6BV) (11). Reverse transcription was performed at 70°C for 15 min followed by 60°C for 15 min in a 10-μl (total volume) mixture; after reverse transcription, the first PCR was performed in a 50-μl mixture according to the manufacturer’s instructions. The conditions of the first and second PCRs were as follows: 92°C for 1 min (1 cycle) and 40 cycles of 92°C for 1 min, 55°C for 1 min, and 75°C for 1 min in a thermal cycler (OmniGene; Hybaid, Teddington, United Kingdom). For the second PCR, 0.5 μl of the first PCR product was amplified in a 25-μl mixture containing 0.5 μM (each) nested primers (P24-MF and P24-MR), 0.2 mM (each) deoxynucleoside triphosphates, 10 mM Tris-HCl (pH 8.9), 1.5 mM MgCl2, 80 mM KCl, 500 μg of bovine serum albumin/ml, 0.1% sodium cholate, 0.1% Triton X-100, and 0.5 U of Tth polymerase (Toyobo, Osaka, Japan). The PCR products were separated by 1.4% agarose gel electrophoresis and stained with ethidium bromide. The specificity of the amplification products was demonstrated by Southern blot hybridization with an alkaline phosphatase (ALP)-labeled oligonucleotide specific for BDV p24 nucleotides (5′-ALP-TCAGCGGTGCGACCACTCCGATAGC-3′) (ALP-P24; nt 1637 to 1661).
To evaluate the quality of the RNA extraction, RT-PCR for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was carried out with 10 ng of total RNA as described previously (16). To prevent contamination of the amplified DNA, PCR was performed according to a strict protocol recommended by Kwok and Higuchi (23). RNA samples and the PCR mixture were prepared in a biosafety hood in an exclusive room where the amplified DNA had never been used. Amplification reactions and analysis of PCR products were performed in a room separate from that used for RNA preparation. Filter tips were used to avoid aerosols that might have contained amplified sample DNA. To confirm the presence of BDV RNA, nested RT-PCR was repeated in the positive cases. We regarded a sample as positive if it showed positive twice. In some positive cases, nested RT-PCR without the RT enzyme in the reverse transcription step were performed to rule out contamination with amplicons of BDV p24 sequences.
Generation of BDV p24 RNA fragments.
The first PCR fragment of BDV p24 (479 bp) amplified from total RNA of BDV-infected MDCK cells was purified and ligated into pGEM-T Easy vector (Promega, Madison, Wis.). The cloned vector was linearized with SalI and in vitro transcribed with T7 RNA polymerase (Life Technologies, Inc., Rockville, Md.) to generate BDV p24 RNA fragments, followed by DNase I (Life Technologies, Inc., Rockville, Md.) digestion of DNA template. The RNA was extracted with phenol and precipitated with ethanol. The concentration of generated RNA was estimated by spectrophotometry and by ethidium bromide staining in comparison with a known amount of tRNA.
Statistical analysis.
Differences in the prevalence of BDV p24 RNA between subject subgroups were analyzed by using Fisher’s exact probability method.
Cloning and sequencing of PCR products.
PCR products were purified and cloned in pGEM-T vectors. Three to five clones from each PCR product were sequenced on both strands by the dideoxy chain termination method with T7 or p24-MF and SP6 primers and a SequiTherm Long-Read Cycle-Sequencing Kit-LC (Epicentre Technologies, Madison, Wis.) and a DNA-sequencing system (model 4000 automated DNA sequencer; LI-COR, Lincoln, Nebr.). Sequence analyses were performed with MacMolly Tetra (Soft Gene GmbH, Berlin, Germany).
Mutations experimentally induced by nested RT-PCR.
To evaluate the sequence heterogeneity of BDV in humans, it is necessary to distinguish between real nucleotide substitutions and those experimentally induced by the nested RT-PCR assay. To determine the experimentally induced mutation frequency, two different nested RT-PCR tests were performed with 100 copies of in vitro-transcribed BDV p24 RNA, which is the limit of sensitivity of our nested RT-PCR. Both second-PCR products were cloned in pGEM-T vectors, and each group of four clones was sequenced. Mutation frequencies among four clones (intragroup) and between two groups (intergroup) were determined.
RESULTS
Sensitivity of nested RT-PCR for BDV p24 RNA.
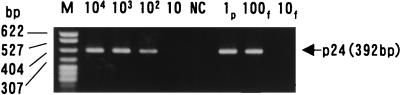
To determine the sensitivity of nested RT-PCR for BDV p24, the in vitro-transcribed BDV p24 RNA fragment was 10-fold serially diluted with tRNA solution (1 μg/μl) and tested by nested RT-PCR. BDV p24 RNA could be consistently detected from 100 molecules of the generated BDV p24 RNA fragment by nested RT-PCR and Southern blot hybridization (Fig. 1). Similarly, 100 fg of total RNA of BDV-infected MDCK cells was constantly detected. The nested RT-PCR specifically yielded a predicted 392-bp DNA fragment. In each RT-PCR experiment, a positive control containing 100 fg of RNA from BDV-MDCK-infected cells was included to evaluate the efficacy of the nested RT-PCR, and it was always found positive.
FIG. 1.
Sensitivity of nested RT-PCR for BDV p24 RNA. Serially diluted in vitro-transcribed BDV p24 RNA fragments (104, 103, 102, and 10 copies) and RNA from BDV-MDCK cells (1 pg [1p], 100 fg [100f], and 10 fg (10f]) were amplified by nested RT-PCR and analyzed by 1.4% agarose gel electrophoresis and ethidium bromide staining. Deionized distilled water was used as a negative control (NC). M, molecular size markers.
Detection of BDV p24 RNA sequences in PBMCs from psychiatric patients and blood donors.
To evaluate the prevalence of BDV RNA in psychiatric patients, RNA samples from the PBMCs were tested by nested RT-PCR. As shown in Table 1, BDV p24-specific DNA fragments were detected by Southern blot analysis in 2 of 49 (4%) patients with mood disorders, 3 of 77 (4%) patients with schizophrenia, and 2 of 84 (2%) blood donors. All positive samples were confirmed to be positive by repeating the nested RT-PCR. Figure 2 illustrates representative results obtained with samples from psychiatric patients. A 598-bp GAPDH fragment was amplified as an internal control in all samples analyzed, suggesting that the quality of the RNA tested was sufficient. There were no significant differences in the prevalence of BDV RNA among the three groups. To rule out possible contamination with amplified BDV p24 sequences in the positive cases, nested RT-PCR without RT enzyme in a reverse transcription step was performed with the two samples of the control group, and the reaction was found to be RT dependent (data not shown). However, in the positive cases from the patient groups, a nested RT-PCR assay without the reverse transcription step was not performed because of a lack of RNA samples from the patients.
FIG. 2.
Detection of BDV p24 RNA in PBMCs of psychiatric patients by nested RT-PCR. Representative results for 10 samples from patients are shown in lanes 1 to 10. PCR products of BDV p24 were analyzed by 1.4% agarose gel electrophoresis and ethidium bromide staining (A) and by Southern blot hybridization with a BDV p24-specific oligonucleotide labeled with ALP (B). One hundred femtograms of RNA extracted from BDV-infected MDCK cells was used as a positive control (100fg), and three independent tubes of distilled water were used as negative controls (NC [three lanes]). As a control for RNA quality, cellular GAPDH mRNA was analyzed by RT-PCR and ethidium bromide staining (C). BDV p24- and GAPDH-specific fragments are indicated at 392 and 598 bp, respectively. M, molecular size markers.
Sequence analysis of the BDV p24 PCR fragments derived from human PBMCs.
To determine the sequences of the human BDV p24 fragments representing 55% of the p24 gene, and to compare them with reported horse and human BDV sequences, PCR products from two patients with mood disorders, three patients with schizophrenia, and two blood donors were cloned and three to five clones of each PCR product were sequenced. The results of the sequence analysis are presented in Fig. 3 along with the sequences of horse BDV strain V (35), C6BV (11), and BDV-MDCK. We sequenced 15 clones from BDV-MDCK, and a predominant consensus sequence is shown in Fig. 3. In total, 30 different p24 clones derived from seven different PBMC samples were analyzed. Cloning and sequencing of the p24 fragments, corresponding to nt 1480 to 1814 of ORF p24, confirmed their BDV specificity and showed a high level of sequence conservation with the sequences of three horse BDV strains. The sequence analysis of human BDV and BDV-MDCK showed the lowest divergency (0 to 1.79%), and the divergency between human BDV and C6BV (3.58 to 5.07%) was the highest. The divergency with strain V was intermediate (1.79 to 3.28%). As shown in Fig. 3, no position with a human-specific mutation was found in all clones from all individuals. However, compared to BDV-MDCK sequences, we identified two positions with mutations, at nt 1535 and 1617. The first silent mutation (C→T), at nt 1535 with no amino acid change, was found in all individuals except one patient with a mood disorder (D1) and blood donor H1-5. The second mutation (G→A), at nt 1617, found in one patient with mood disorders (D1), had a conservative amino acid change (Val→Met). One-point substitutions with no clusters were randomly distributed in human BDV p24 sequences. Interindividual divergency was slightly higher than intraindividual divergency (0 to 2.69 versus 0 to 2.09%).
FIG. 3.
Nucleotide sequences of BDV p24 cDNA clones isolated from PBMCs of two patients with mood disorders (D1 and D2), three patients with schizophrenia (S1, S2, and S3), and two blood donors (H1 and H2) in comparison with those of horse BDVs (BDV-MDCK, strain V, and C6BV). The nucleotide sequence of BDV-MDCK was determined in this study, and the sequences of strain V (35) and C6BV (11) were based on those published previously. The nucleotide numbers above the sequence follow the numbering previously reported for C6BV (11). The dots indicate nucleotide identity with the BDV-MDCK consensus sequence.
Experimentally induced nucleotide mutations during nested RT-PCR for in vitro-transcribed BDV p24 RNA.
To determine the experimentally induced mutation frequency during nested RT-PCR, two different nested RT-PCR tests were performed with 100 copies of in vitro-transcribed BDV p24 RNA, and four clones of each second-PCR product were sequenced. Both the intragroup divergencies among four clones and the intergroup divergencies between two nested RT-PCR tests were 0 to 1.49%.
DISCUSSION
In the present study, we have detected BDV RNA in the PBMCs of psychiatric patients and blood donors by nested RT-PCR. The prevalences of BDV p24 RNA in patients with mood disorders (4%) and schizophrenia (4%) were not significantly different from that in blood donors (2%). We performed nested RT-PCR experiments carefully to avoid BDV DNA contamination, as described in Materials and Methods. Three water samples used as negative controls were always negative in every PCR assay. Furthermore, the nucleotide (T) at nt 1535 of human BDV p24 RNA from patient PBMCs was clearly different from that (C) at the same position in a horse BDV-MDCK sequence which was used as the positive control in nested RT-PCR assays. In addition, RT dependency was confirmed in the two positive cases among blood donors, although in the five positive cases in the patient groups RT dependency could not be tested because of a lack of RNA samples. These findings suggest that we can rule out contamination with horse BDV p24 RNA or amplified BDV DNA sequences in our nested RT-PCR system.
In previous studies, there has been controversy as to the prevalence of BDV RNA in the PBMCs of psychiatric patients. Kishi et al. reported a significantly high prevalence (37%) of BDV RNA in Japanese psychiatric patients compared with that in healthy individuals (6.5%) (20). However, a method of nested RT-PCR almost the same as that they established was used in our study (except for the RT-PCR kit), and the sensitivity of our nested RT-PCR was slightly higher. The reason for the difference in prevalence may be due to differences in the criteria for a positive result. Sauder et al. also reported a high prevalence (38.5%) of BDV RNA in psychiatric patients in Homburg, Germany (34). The sensitivity of the nested RT-PCR, the number of PBMCs used for RNA extraction, and the amount of RNA used for RT-PCR were similar to those in our method. One of the reasons for the difference in prevalence may be geographical. On the other hand, a low prevalence (1.9%) of BDV p24 RNA in psychiatric patients and no viral RNA in control subjects have been reported in Japan (22). The difference between the prevalences in our study and theirs may be due to the lower sensitivity of their single RT-PCR assay. Richt et al. (31) and Lieb et al. (24) reported that there was no evidence for the presence of BDV RNA in PBMCs from either psychiatric patients or normal controls. Since the sensitivity of the nested RT-PCR performed by Lieb et al. was similar to ours (100 molecules), the difference in prevalence may be due to the difference in the number of PBMCs used for RNA extraction and/or the amount of RNA tested for nested RT-PCR. The difference in prevalence in the study of Richt et al. may be due to differences in the number of subjects and/or the sensitivity of nested RT-PCR. Other reasons which may account for the differences in prevalence in previous studies are different criteria for positivity and laboratory contamination of amplified BDV DNA. For a precise comparison of the prevalences of BDV RNA, standardization of the PCR method (e.g., the number of PBMCs used for RNA extraction, the sensitivity of the PCR, and the criteria for positivity) is necessary.
Sequence analysis of BDV p24 RNA from the PBMCs of patients and blood donors showed a high nucleotide sequence conservation with horse BDV p24 sequences (Fig. 3). This high homology is consistent with two previous reports of psychiatric patients from Germany (15, 34) and inconsistent with one previous report from Japan (21). The human BDV sequences in our study were homologous with those of horse BDV in the following decreasing order: BDV-MDCK, strain V, and C6BV. In previous reports on the divergency between human and horse BDV p24 sequences, the BDV p24 sequences of psychiatric patients in Berlin, Germany, exhibited higher conservation with strain V than with C6BV, while the sequences of patients in Homburg, Germany, showed higher homology with C6BV than with strain V (15, 34). The BDV sequence from Kagoshima, Japan (22), was identical to that of the BDV-MDCK sequence and more homologous with strain V. These findings may suggest the existence of heterogeneity in human BDV sequences.
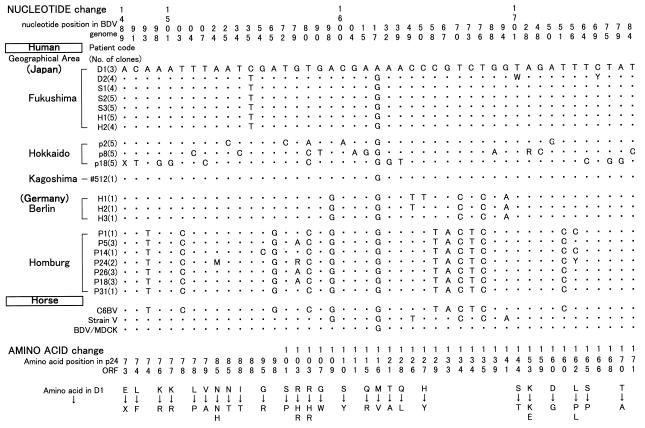
Figure 4 summarizes nucleotide and amino acid mutations among previously reported human BDV p24 sequences (335-bp sequence) in comparison with horse BDV sequences. Nucleotide sequences which are not presented in Fig. 4 are identical among human BDV sequences. Amino acid substitutions which are specific and common to all human BDV sequences were not observed. However, at the nucleotide level, we detected three silent substitutions, at positions 1598, 1670, and 1676, which can distinguish between Japanese and German clones. Furthermore, there are eight positions with silent mutations without amino acid changes (nt 1493, 1503, 1565, 1580, 1658, 1667, 1673, and 1751) which are Homburg specific and identical to those of C6BV. C-to-T silent substitution at nt 1535 without an amino acid change was found in all of the Fukushima clones except those from patient D1. C-to-T and G-to-A silent substitutions at nt 1649 and 1694 were found only in Berlin clones, which were identical to the horse strain V sequence. These findings suggest that human BDV p24 sequences are conserved in general but some sequences differ among clones in different areas.
FIG. 4.
Nucleotide mutations in the partial human BDV p24 sequences derived from psychiatric patients in different geographical areas. The nucleotide numbers above the sequence follow the numbering of the previously reported C6BV sequence (11). The dots indicate nucleotide identity with the sequence of D1. The numbers in parentheses indicate the number of clones sequenced. M, R, W, Y, and X indicate A or C, A or G, A or T, C or T, and deletion, respectively. The sequences obtained in Hokkaido, Kagoshima, Berlin, and Homburg are from references 21, 22, 15, and 34, respectively. Amino acid changes corresponding to nucleotide mutations are shown at the bottom of the figure.
Interindividual and intraindividual divergencies among the patients in our study were 0 to 2.69% and 0 to 2.09%, respectively. To clarify whether these mutations were experimentally induced or were due to sequence heterogeneity already present in the individuals, we also examined the mutation frequency during nested RT-PCR by using in vitro-transcribed BDV p24 RNA. Both intergroup and intragroup divergencies in nested RT-PCR were 0 to 1.49% in 335 bp. Considering these mutation frequencies, most of the one-point substitutions, except those at nt 1535 and 1617, which were randomly distributed in the human BDV sequences, are likely due to experimentally induced mutations. The sequences from psychiatric patients in Hokkaido, Japan, have been reported to be significantly variable (21). It was pointed out that the nested RT-PCR procedure used by the Japanese researchers produced several mutations, probably due to the negative effect of manganese during DNA polymerization (34). In our RT-PCR system, manganese was chelated before the first PCR to reduce the number of artificial mutations.
Although the results of the present study of the detection of BDV RNA in PBMCs did not confirm the association of BDV infection with mood disorders and schizophrenia, the existence of BDV or a BDV-related agent was shown in some humans. In addition, the possible existence of heterogeneity in human BDV sequences was suggested. Recently it was reported that BDV RNA was detected in the brains of both psychiatric patients (14, 33) and healthy individuals (17). To clarify the association of BDV with psychiatric disorders, it is important not only to detect the BDV genome in the brains of psychiatric patients but also to elucidate the effect of BDV infection on brain function. Further studies are necessary to clarify the role of BDV infection in the etiology and pathogenesis of psychiatric disorders.
ACKNOWLEDGMENTS
We thank Yuichi Endo for valuable discussions; Francis A. Ennis for reviewing the manuscript; Hiroki Suzuki, Kiyoshi Ariga, Koichi Osonoe, Minako Sato, Mitsuhiro Ito, Noboru Yokoyama, Shin-ichi Ogata, and Yoshihiko Numata for obtaining samples from patients; and Chiharu Takahashi, Manabu Kikuchi, Yumiko Kanno, Sachiko Sakai, Kazuhiro Mochizuki, and Yasuki Takeuchi for technical assistance.
REFERENCES
- 1.Albrecht P, Torrey E F, Boone E, Hicks J T, Daniel N. Raised cytomegalovirus-antibody level in cerebrospinal fluid of schizophrenic patients. Lancet. 1980;ii:769–772. doi: 10.1016/s0140-6736(80)90386-4. [DOI] [PubMed] [Google Scholar]
- 2.Amsterdam J D, Winokur A, Dyson W, Herzog S, Gonzalez F, Rott R, Koprowski H. Borna disease virus. A possible etiologic factor in human affective disorders? Arch Gen Psychiatry. 1985;42:1093–1096. doi: 10.1001/archpsyc.1985.01790340077011. [DOI] [PubMed] [Google Scholar]
- 3.Bautista J R, Rubin S A, Moran T H, Schwartz G J, Carbone K M. Developmental injury to the cerebellum following perinatal Borna disease virus infection. Dev Brain Res. 1995;90:45–53. doi: 10.1016/0165-3806(96)83485-7. [DOI] [PubMed] [Google Scholar]
- 4.Binz T, Lebelt J, Niemann H, Hagenau K. Sequence analyses of the p24 gene of Borna disease virus in naturally infected horse, donkey and sheep. Virus Res. 1994;34:281–289. doi: 10.1016/0168-1702(94)90128-7. [DOI] [PubMed] [Google Scholar]
- 5.Bode, L., R. Ferszt, and G. Czech. 1993. Borna disease virus infection and affective disorders in man. Arch. Virol. 7(Suppl.):159–167. [DOI] [PubMed]
- 6.Bode L, Durrwald R, Rantam F A, Ferszt R, Ludwig H. First isolates of infectious human Borna disease virus from patients with mood disorders. Mol Psychiatry. 1996;1:200–212. [PubMed] [Google Scholar]
- 7.Briese T, Schneemann A, Lewis A J, Park Y S, Kim S, Ludwig H, Lipkin W I. Genomic organization of Borna disease virus. Proc Natl Acad Sci USA. 1994;91:4362–4366. doi: 10.1073/pnas.91.10.4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carbone K M, Duchala C S, Griffin J W, Kincaid A L, Narayan O. Pathogenesis of Borna disease in rats: evidence that intra-axonal spread is the major route for virus dissemination and the determinant for disease incubation. J Virol. 1987;61:3431–3440. doi: 10.1128/jvi.61.11.3431-3440.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carbone K M, Rubin S A, Sierra-Honigmann A M, Lederman H M. Characterization of a glial cell line persistently infected with Borna disease virus (BDV): influence of neurotrophic factors on BDV protein and RNA expression. J Virol. 1993;67:1453–1460. doi: 10.1128/jvi.67.3.1453-1460.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crow T J. Is schizophrenia an infectious disease? Lancet. 1983;i:173–175. doi: 10.1016/s0140-6736(83)92768-x. [DOI] [PubMed] [Google Scholar]
- 11.Cubitt B, Oldstone C, de la Torre J C. Sequence and genome organization of Borna disease virus. J Virol. 1994;68:1382–1396. doi: 10.1128/jvi.68.3.1382-1396.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cubitt B, Oldstone C, Valcarcel J, de la Torre J C. RNA splicing contributes to the generation of mature mRNAs of Borna disease virus, a non-segmented negative strand RNA virus. Virus Res. 1994;34:69–79. doi: 10.1016/0168-1702(94)90120-1. [DOI] [PubMed] [Google Scholar]
- 13.de la Torre J C, Carbone K M, Lipkin W I. Molecular characterization of the Borna disease agent. Virology. 1990;179:853–856. doi: 10.1016/0042-6822(90)90154-j. [DOI] [PubMed] [Google Scholar]
- 14.de la Torre J C, Gonzalez-Dunia D, Cubitt B, Mallory M, Mueller-Lantzsch N, Grasser F A, Hansen L A, Masliah E. Detection of borna disease virus antigen and RNA in human autopsy brain samples from neuropsychiatric patients. Virology. 1996;223:272–282. doi: 10.1006/viro.1996.0479. [DOI] [PubMed] [Google Scholar]
- 15.de la Torre J C, Bode L, Durrwald R, Cubitt B, Ludwig H. Sequence characterization of human Borna disease virus. Virus Res. 1996;44:33–44. doi: 10.1016/0168-1702(96)01338-x. [DOI] [PubMed] [Google Scholar]
- 16.Gao X, Honn K V, Grignon D, Sakr W, Chen Y Q. Frequent loss of expression and loss of heterozygosity of the putative tumor suppressor gene DCC in prostatic carcinomas. Cancer Res. 1993;53:2723–2727. [PubMed] [Google Scholar]
- 17.Haga S, Yoshimura M, Motoi Y, Arima K, Aizawa T, Ikuta K, Tashiro M, Ikeda K. Detection of Borna disease virus genome in normal human brain tissue. Brain Res. 1997;770:307–309. doi: 10.1016/s0006-8993(97)00903-7. [DOI] [PubMed] [Google Scholar]
- 18.Halonen P E, Rimon R, Arohonka K, Jantti V. Antibody levels to herpes simplex type I, measles and rubella viruses in psychiatric patients. Br J Psychiatry. 1974;125:461–465. doi: 10.1192/bjp.125.5.461. [DOI] [PubMed] [Google Scholar]
- 19.Herzog S, Rott R. Replication of Borna disease virus in cell cultures. Med Microbiol Immunol. 1980;168:153–158. doi: 10.1007/BF02122849. [DOI] [PubMed] [Google Scholar]
- 20.Kishi M, Nakaya T, Nakamura Y, Zhong Q, Ikeda K, Senjo M, Kakinuma M, Kato S, Ikuta K. Demonstration of human Borna disease virus RNA in human peripheral blood mononuclear cells. FEBS Lett. 1995;364:293–297. doi: 10.1016/0014-5793(95)00406-y. [DOI] [PubMed] [Google Scholar]
- 21.Kishi M, Arimura Y, Ikuta K, Shoya Y, Lai P K, Kakinuma M. Sequence variability of Borna disease virus open reading frame II found in human peripheral blood mononuclear cells. J Virol. 1996;70:635–640. doi: 10.1128/jvi.70.1.635-640.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kubo K, Fujiyoshi T, Yokoyama M M, Kamei K, Richt J A, Kitze B, Herzog S, Takigawa M, Sonoda S. Lack of association of Borna disease virus and human T-cell leukemia virus type 1 infections with psychiatric disorders among Japanese patients. Clin Diagn Lab Immunol. 1997;4:189–194. doi: 10.1128/cdli.4.2.189-194.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwok S, Higuchi R. Avoiding false positives with PCR. Nature. 1989;339:237–238. doi: 10.1038/339237a0. [DOI] [PubMed] [Google Scholar]
- 24.Lieb K, Hallensleben W, Czygan M, Stitz L, Staeheli P the Bornavirus Study Group. No Borna disease virus-specific RNA detected in blood from psychiatric patients in different regions of Germany. Lancet. 1997;350:1002. doi: 10.1016/s0140-6736(05)64066-4. [DOI] [PubMed] [Google Scholar]
- 25.Lipkin W I, Travis G H, Carbone K M, Wilson M C. Isolation and characterization of Borna disease agent cDNA clones. Proc Natl Acad Sci USA. 1990;87:4184–4188. doi: 10.1073/pnas.87.11.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lipkin W I, Schneemann A, Solbrig M V. Borna disease virus: implications for human neuropsychiatric illness. Trends Microbiol. 1995;3:64–69. doi: 10.1016/s0966-842x(00)88877-0. [DOI] [PubMed] [Google Scholar]
- 27.Lundgren A L, Zimmermann W, Bode L, Czech G, Gosztonyi G, Lindberg R, Ludwig H. Staggering disease in cats: isolation and characterization of the feline Borna disease virus. J Gen Virol. 1995;76:2215–2222. doi: 10.1099/0022-1317-76-9-2215. [DOI] [PubMed] [Google Scholar]
- 28.Narayan O, Herzog S, Frese K, Scheefers H, Rott R. Behavioral disease in rats caused by immunopathological responses to persistent borna virus in the brain. Science. 1983;220:1401–1403. doi: 10.1126/science.6602380. [DOI] [PubMed] [Google Scholar]
- 29.O’Callaghan E, Sham P, Takei N, Glover G, Murray R M. Schizophrenia after prenatal exposure to 1957 A2 influenza epidemic. Lancet. 1991;337:1248–1250. doi: 10.1016/0140-6736(91)92919-s. [DOI] [PubMed] [Google Scholar]
- 30.Pyper J M, Richt J A, Brown L, Rott R, Narayan O, Clements J E. Genomic organization of the structural proteins of borna disease virus revealed by a cDNA clone encoding the 38-kDa protein. Virology. 1993;195:229–238. doi: 10.1006/viro.1993.1364. [DOI] [PubMed] [Google Scholar]
- 31.Richt J A, Alexander R C, Herzog S, Hooper D C, Kean R, Spitsin S, Bechter K, Schuttler R, Feldmann H, Heiske A, Fu Z F, Dietzschold B, Rott R, Koprowski H. Failure to detect Borna disease virus infection in peripheral blood leukocytes from humans with psychiatric disorders. J Neurovirol. 1997;3:174–178. doi: 10.3109/13550289709015807. [DOI] [PubMed] [Google Scholar]
- 32.Rott R, Herzog S, Fleischer B, Winokur A, Amsterdam J, Dyson W, Koprowski H. Detection of serum antibodies to Borna disease virus in patients with psychiatric disorders. Science. 1985;228:755–756. doi: 10.1126/science.3922055. [DOI] [PubMed] [Google Scholar]
- 33.Salvatore M, Morzunov S, Schwemmle M, Lipkin W I the Bornavirus Study Group. Borna disease virus in brains of North American and European people with schizophrenia and bipolar disorder. Lancet. 1997;349:1813–1814. doi: 10.1016/s0140-6736(05)61693-5. [DOI] [PubMed] [Google Scholar]
- 34.Sauder C, Müller A, Cubitt B, Mayer J, Steinmetz J, Trabert W, Ziegler B, Wanke K, Mueller-Lantzsch N, de la Torre J C, Grässer F A. Detection of Borna disease virus (BDV) antibodies and BDV RNA in psychiatric patients: evidence for high sequence conservation of human blood-derived BDV RNA. J Virol. 1996;70:7713–7724. doi: 10.1128/jvi.70.11.7713-7724.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schneider P A, Briese T, Zimmermann W, Ludwig H, Lipkin W I. Sequence conservation in field and experimental isolates of Borna disease virus. J Virol. 1994;68:63–68. doi: 10.1128/jvi.68.1.63-68.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Solbrig M V, Koob G F, Fallon J H, Lipkin W I. Tardive dyskinetic syndrome in rats infected with Borna disease virus. Neurobiol Dis. 1994;1:111–119. doi: 10.1006/nbdi.1994.0014. [DOI] [PubMed] [Google Scholar]
- 37.Sprankel H, Richarz K, Ludwig H, Rott R. Behavior alterations in tree shrews (Tupaia glis, Diard 1820) induced by Borna disease virus. Med Microbiol Immunol. 1978;165:1–18. doi: 10.1007/BF02121228. [DOI] [PubMed] [Google Scholar]
- 38.Stitz L, Krey H, Ludwig H. Borna disease in rhesus monkeys as a model for uveo-cerebral symptoms. J Med Virol. 1981;6:333–340. doi: 10.1002/jmv.1890060408. [DOI] [PubMed] [Google Scholar]
- 39.Stitz, L., T. Bilzer, J. A. Richt, and R. Rott. 1993. Pathogenesis of Borna disease. Arch. Virol. 7(Suppl.):135–151. [DOI] [PubMed]
- 40.Torrey E F, Miller J, Rawlings R, Yolken R H. Seasonality of births in schizophrenia and bipolar disorder: a review of the literature. Schizophr Res. 1997;28:1–38. doi: 10.1016/s0920-9964(97)00092-3. [DOI] [PubMed] [Google Scholar]
- 41.VandeWoude S, Richt J A, Zink M C, Rott R, Narayan O, Clements J E. A borna virus cDNA encoding a protein recognized by antibodies in humans with behavioral diseases. Science. 1990;250:1278–1281. doi: 10.1126/science.2244211. [DOI] [PubMed] [Google Scholar]
- 42.Waltrip R W, II, Buchanan R W, Carpenter W T, Jr, Kirkpatrick B, Summerfelt A, Breier A, Rubin S A, Carbone K M. Borna disease virus antibodies and the deficit syndrome of schizophrenia. Schizophr Res. 1997;23:253–257. doi: 10.1016/s0920-9964(96)00114-4. [DOI] [PubMed] [Google Scholar]
- 43.Weisman Y, Huminer D, Malkinson M, Meir R, Kliche S, Lipkin W I, Pitlik S. Borna disease virus antibodies among workers exposed to infected ostriches. Lancet. 1994;344:1232–1233. doi: 10.1016/s0140-6736(94)90550-9. [DOI] [PubMed] [Google Scholar]