Abstract
A functional capsid protein (CP) is essential for host plant infection and insect transmission in monopartite geminiviruses. We studied two defective genomic DNAs of tomato yellow leaf curl virus (TYLCV), Sic and SicRcv. Sic, cloned from a field-infected tomato, was not infectious, whereas SicRcv, which spontaneously originated from Sic, was infectious but not whitefly transmissible. A single amino acid change in the CP was found to be responsible for restoring infectivity. When the amino acid sequences of the CPs of Sic and SicRcv were compared with that of a closely related wild-type virus (TYLCV-Sar), differences were found in the following positions: 129 (P in Sic and SicRcv, Q in Sar), 134 (Q in Sic and Sar, H in SicRcv) and 152 (E in Sic and SicRcv, D in Sar). We constructed TYLCV-Sar variants containing the eight possible amino acid combinations in those three positions and tested them for infectivity and transmissibility. QQD, QQE, QHD, and QHE had a wild-type phenotype, whereas PHD and PHE were infectious but nontransmissible. PQD and PQE mutants were not infectious; however, they replicated and accumulated CP, but not virions, in Nicotiana benthamiana leaf discs. The Q129P replacement is a nonconservative change, which may drastically alter the secondary structure of the CP and affect its ability to form the capsid. The additional Q134H change, however, appeared to compensate for the structural modification. Sequence comparisons among whitefly-transmitted geminiviruses in terms of the CP region studied showed that combinations other than QQD are present in several cases, but never with a P129.
Tomato yellow leaf curl virus (TYLCV) is a geminivirus important in tomato crops of the Mediterranean area, Central America, and Asia. Geminiviruses have characteristic twinned particles, encapsidating a genome of circular single-stranded DNA (ssDNA). Geminiviruses are divided into three genera, according to their genome and insect transmission characteristics (8, 33), i.e., Mastrevirus, Begomovirus, and Curtovirus. Begomoviruses are transmitted by the whitefly Bemisia tabaci, infect dicotyledonous plants, and typically have a genome composed of two DNA molecules (DNA A and B). TYLCV is a member of the genus Begomovirus, but has a single genomic component (22, 27, 28), although in one case, a bipartite genome was isolated (35).
TYLCV DNA (about 2.8 kb) has a molecular organization resembling the DNA A of bipartite geminiviruses and contains six genes bidirectionally organized in two transcription units that are separated by an intergenic region of about 300 bp. The gene encoding the capsid protein (CP) is located on the virion strand DNA, while the genes encoding the replication-associated protein (RepC1), a DNA-binding protein (C2), and the C3 protein are on the complementary strand DNA. Two additional genes (V2 “precoat” and C4 genes) coding for proteins involved in movement and symptom development are also present (21, 41).
In bipartite geminiviruses, proteins involved in movement are encoded by DNA B (19, 20, 29, 36, 39). TYLCV and the monopartites do not have a DNA B, so it is likely that functions analogous to those of the bipartite geminivirus movement proteins BR1 and BL1 are performed by other viral factors.
In geminiviruses, the CP can have different roles. Wartig et al. (41) have shown that the CP of TYLCV is essential for infection of Nicotiana benthamiana and tomato plants. An intact CP is required in the related tomato leaf curl virus (TLCV) from Australia (34) and monopartite geminiviruses of other genera, such as maize streak virus (MSV) (4) and beet curly top virus (BCTV) (6), suggesting that the monopartite viruses move within the plant in the form of viral particles. In contrast, some bipartite geminiviruses such as tomato golden mosaic virus (TGMV) (14), African cassava mosaic virus (ACMV) (13), and bean golden mosaic virus (BGMV) (2) systemically infect plants even when their CPs are absent or truncated.
Geminiviruses are transmitted by leafhoppers or whiteflies. The CP is required for transmission (2) and determines insect specificity (7).
In this study, we have analyzed the abilities to systemically infect plants of several spontaneous and engineered mutants derived from two closely related isolates of TYLCV, TYLCV-Sic (from Sicily) (11) and TYLCV-Sar (from Sardinia) (22). We have also studied the influence of these mutations on virus transmissibility by B. tabaci. We have found that a single amino acid change in the CP of TYLCV completely abolished infectivity in plants and that a compensatory mutation of a nearby amino acid restored infectivity but, concomitantly, prevented transmission by whiteflies. Our results indicate that the region of the CP between amino acids 129 and 134 is essential for both the correct assembly of virions and transmission by the insect vector.
MATERIALS AND METHODS
Plasmid construction.
Table 1 describes the relevant plasmids used in this paper.
TABLE 1.
Relevant TYLCV constructs used in this study
| Construct | Description (reference) |
|---|---|
| Sic | |
| pSic36 | Monomer in pBluescript KS+ (11) |
| pSicSst | Monomer in pUC19 |
| pBinSic1.8mer | 1.8-mer in pBin19 |
| pBinSic2mer | Dimer in pBin19 |
| pBinSic1.2mer | 1.2-mer in pBin19 |
| pSicRcv | Monomer in pUC118 |
| pBinSicRcv | SicRcv 1.2-mer in pBin19 |
| pBinSic708m | 1.2-mer in pBin19 with the CP of SicRcv (PHE combination) |
| pBinSic2025m | 1.2-mer in pBin19 with the RepC1 of SicRcv, but the CP of Sic (PQE combination) |
| Sar | |
| pBinSarQQD | 1.2-mer wild type in pBin19 |
| pBinSarQQE | 1.2-mer with SpeI-SnaBI fragment from pSic36 |
| pBinSarPQD | 1.2-mer with SstII-SpeI fragment from pSic36 |
| pBinSarPQE | 1.2-mer with SstII-SnaBI fragment from pSic36 |
| pBinSarPHD | 1.2-mer with SstII-SpeI fragment from pSicRcv |
| pBinSarPHE | 1.2-mer with SstII-SnaBI fragment from pSicRcv |
| pBinSarQHD | 1.2-mer mutated with primers SARMUT692(+) and SARMUT725(−) |
| pBinSarQHE | pBinSarQHD with SpeI-SnaBI fragment from pSicRcv |
| pSar1.8mer | 1.8-mer wild type in pBin19 (22) |
(i) TYLCV-Sic constructs.
Full-length TYLCV-Sic DNA, originally cloned from an infected tomato plant, was excised as an SstI fragment from pSic36 (11) and cloned into SstI-cut pUC19 (pSicSst). After a SalI-SstI deletion (503 nucleotides [nt]), a full-length TYLCV-Sic DNA was introduced in the single remaining SstI site. By using EcoRI and HindIII, the fragment containing 1.8 copies of the viral genome was moved into the binary vector pBin19, giving rise to pBinSic1.8mer. A TYLCV-Sic head-to-tail dimer (pBinSic2mer) was produced by insertion of two full-length TYLCV-Sic SstI fragments into pUC118 digested with SstI and moved as an EcoRI-HindIII insert into pBin19. The pBinSic1.2mer was obtained by cloning the full-length TYLCV-Sic SstI fragment into SstI-cut pSicVect. pSicVect is a pBin19-based vector in which the 596-nt SstI-BamHI fragment from TYLCV-Sic, encompassing the viral intergenic region, had been inserted.
(ii) TYLCV-SicRcv cloning and sequencing.
To clone SicRcv viral DNA, total DNA from the single infected tomato plant (see below) was extracted and centrifuged to equilibrium in CsCl as described previously (11). The fraction below the plant genomic DNA was collected, phenol extracted, and ethanol precipitated. After BamHI digestion, SicRcv DNA was cloned into BamHI-cut pUC118, giving rise to pSicRcv. The full-length SicRcv insert was moved into the BamHI-cut pSicVect, obtaining pBinSicRcv. For sequencing, the viral DNA was subcloned in both orientations into M13mp18, and the recombinant phage ssDNA was sequenced by the dideoxy method with synthetic primers.
(iii) TYLCV-Sic mutant clones.
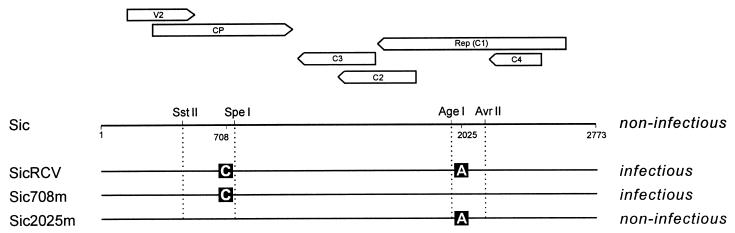
Sic708m and Sic2025m were obtained by substitution into pSicSst of either the 282-nt SstII-SpeI fragment, containing the H134 mutation in the CP, or the 176-nt AgeI-AvrII fragment, including the L198 mutation in RepC1, respectively, derived from pSicRcv (Fig. 1). For agroinoculation, both mutants were moved as SstI fragments into SstI-cut pSicVect, giving rise to pBinSic708m and pBinSic2025m.
FIG. 1.
Scheme of the genomic DNA and genes of TYLCV-Sic and derived mutants. Boxed letters in mutants represent nucleotide changes from TYLCV-Sic. Restriction sites used to obtain Sic708m and Sic2025m by DNA fragment exchanges are indicated. Nucleotide numbering is according to EMBL accession no. Z28390; i.e., nucleotide 1 corresponds to the first T in the conserved TAATATTAC sequence of the stem-loop of geminiviruses.
(iv) TYLCV-Sar clones.
TYLCV-Sar CP mutants were obtained from a TYLCV-Sar SstI clone in pUC118 (pSarSst) by fragment exchange or, in one case, by site-directed mutagenesis.
For fragment exchanges, unique restriction sites present in TYLCV-Sar and TYLCV-Sic were used, i.e., SstII, SpeI, and SnaBI, cutting, respectively, at positions 466, 748, and 1049 (see Fig. 3). SarPQE was constructed by replacing the 583-nt SstII-SnaBI fragment of pSarSst with the corresponding fragment of pSic36. SarPQD was obtained by inserting the 282-nt SstII-SpeI fragment into pSarSst, which had the corresponding SstII-SpeI fragment deleted. The same procedure was adopted for the construction of SarPHE and PHD, except that pSicRcv was used instead of pSic36.
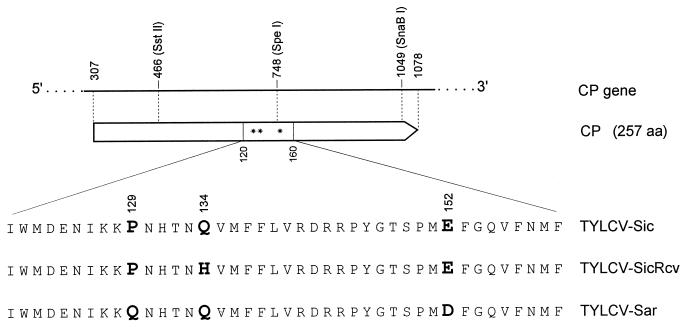
FIG. 3.
Representation of the CP region (amino acids [aa] 120 to 160) containing the three amino acid differences (in boldface) among TYLCV-Sic, SicRcv, and TYLCV-Sar. The upper part shows the location of restriction sites used to construct some of the TYLCV-Sar mutants with amino acid replacements described in Table 1. Asterisks localize positions 129, 134, and 152.
For construction of SarQHD, the Quick Change site-directed mutagenesis kit (Stratagene) was employed, using primers SARMUT692(+) (5′-CAAAATCATACTAACCACGTGATGTTTTTCCTTG) and SARMUT725(−) (5′-CAAGGAAAAACATCACGTGGTTAGTATGATTTTG) and with pSarSst as a template. The mutagenesis induced an A708C substitution (underlined in the first primer sequence), producing a Q134H change in the CP. The mutation was verified by sequencing the surrounding region with primers SIC CP604(+) (5′-GGTATTACTCATAGAGTTGG) and SIC CP873(−) (5′-CCAACAACCGTAGCATGGAACTTCC). To eliminate the effect of possible errors generated in the TYLCV-Sar genome by the Pfu polymerase during the mutagenesis reaction, the SstII-SnaBI fragment of the mutant obtained was substituted in pSarSst, giving rise to the final pSarQHD clone. SarQHE was derived from SarQHD by exchange of the SpeI-SnaBI fragment with that from pSicRcv. Finally, SarQQE was obtained by exchanging the SpeI-SnaBI fragment from pSicRcv in pSarSst.
After fragment exchange and/or mutagenesis, the viral mutants obtained were excised as full-length SstI fragments and inserted into pSarVect, a construct analogous to pSicVect, containing the 596-nt SstI-BamHI fragment encompassing the intergenic region of TYLCV-Sar. Thus all mutants inserted into pSarVect or in pSicVect described above contained 1.2 copies of the TYLCV genome (1.2-mers). Clone pBinSarQQD was made with the full-length wild-type TYLCV-Sar genome inserted in pSarVect. The TYLCV-Sar1.8-mer has already been described (22).
Agroinoculation of plants.
Recombinant binary vectors containing redundant copies of the wild-type and mutant genomes of TYLCV were introduced into Agrobacterium tumefaciens LBA4404 by direct transformation (15). Tomato plants were agroinoculated as described previously (22).
N. benthamiana leaf disc assay.
Leaf discs were agroinoculated as described previously (41). Discs were collected 6 days after inoculation and analyzed for the presence of viral DNA in Southern blots, viral CP and V2 protein in Western blots, and virus particles by electron microscopy (see below).
Analysis of viral DNA.
To assess the presence of viral DNA in plants, tissue blot assays and molecular hybridizations were used (1). Different viral DNA forms were visualized in Southern blots. Total DNA was extracted from tomato plants or N. benthamiana leaf discs with TLES buffer (5% sodium dodecyl sulfate [SDS] 150 mM LiCl, 50 mM Tris-HCl, 5 mM EDTA [pH 9.0]). After electrophoresis in 1% agarose gels containing 0.5 μg of ethidium bromide per ml in 0.5× Tris-borate-EDTA, gels were blotted onto positively charged nylon membranes (Boehringer Mannheim). A digoxigenin-labelled random-primed full-length TYLCV-specific probe was used. Hybridization was performed at 68°C in Standard Hybridization mix, according to the Boehringer protocol, followed by high-stringency washes. CDP-Star (Tropix) was used for final detection.
Western blots.
Total proteins were extracted from plant tissue (tomato plants or N. benthamiana leaf discs) with trichloroacetic acid (TCA) (42). Equal amounts of total proteins were loaded on 18% and 20% SDS-polyacrylamide gels. After blotting, polyvinylidene difluoride (PVDF [PolyScreen; NEN Life Science Products]) membranes were probed with a TYLCV-specific polyclonal antiserum prepared in rabbit against purified virus particles (25) and cross-absorbed with healthy TCA-extracted tomato tissue (final dilution 1/10,000). For V2 detection, a polyclonal antiserum prepared against glutathione S-transferase–V2 (GST-V2) fusion protein, cross-absorbed with healthy tomato and N. benthamiana extracts (40), was used at a 1/10,000 dilution. The antibody reaction was visualized with goat anti-rabbit serum either conjugated with alkaline phosphatase (AP) and detected with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate (NBT-BCIP) or conjugated with horseradish peroxidase (HRP) with a chemiluminescent substrate (Renaissance; NEN Life Science Products).
Detection of virus particles by ISEM.
Young tomato leaves collected 4 to 6 weeks after inoculation or 6-day-old N. benthamiana leaf discs were homogenized in 0.5 M potassium phosphate buffer (pH 7.0). Virions were adsorbed to electron microscope grids previously coated with anti-TYLCV immunoglobulin G (14 μg/ml) (25). After 2 h at room temperature, grids were rinsed, negatively stained with uranyl acetate, and inspected by immunosorbent electron microscopy (ISEM). In order to maximize the amount of infected tissue in the N. benthamiana leaf discs, only margins were used.
Whitefly transmission experiments.
B. tabaci flies were reared and handled as described previously (9). For virus transmission tests, insects were allowed to feed for 48 h on infected tomato plants. Groups of 15 insects were then moved to single tomato test plants (cv. Marmande) for virus inoculation. Two days later, insects were removed and the plants were sprayed with an insecticide and maintained in an insect-free greenhouse. Plants were scored individually for TYLCV infection by tissue blotting 4 and 6 weeks after inoculation. Transmission experiments were generally repeated three times for each mutant. Results were compared by Pearson’s χ2 test. The sequence of the viral DNA progeny in the region of the CP mutations in both source and whitefly-infected test plants was verified. This region was amplified by PCR with primers SIC CP604(+) and SIC CP873(−) and subjected to dye-terminator cycle sequencing with an Applied Biosystems model 373A automated sequencer and one of the primers mentioned above.
RESULTS
Infectivity of spontaneous and engineered TYLCV-Sic DNAs.
TYLCV-Sic DNA, which is able to replicate in tobacco protoplasts (11), was tested for its ability to systemically infect tomato plants by agroinoculation. In an initial attempt, 16 tomato plants were agroinoculated with pBinSic1.8mer, but none became infected (Table 2). Two other clones for agroinoculation were constructed and tested—pBinSic1.2mer and pBinSic2mer. Only one plant, inoculated with the latter, developed typical symptoms, with yellowing and curling of young leaves, and contained viral DNA. From this plant, viral DNA was isolated and cloned (SicRcv [i.e., “recovered”]), and pBinSicRcv was constructed. All plants agroinoculated with pBinSicRcv showed typical TYLCV symptoms. The complete DNA sequence of SicRcv was then determined.
TABLE 2.
Results for agroinoculation and whitefly transmission with the TYLCV-Sic clones
| Clone | Infectivity on tomatoa | Whitefly transmissibilityb |
|---|---|---|
| Sic1.8mer | 0/16 | NTc |
| Sic1.2mer | 0/13 | NT |
| Sic2mer | 1d/36 | NT |
| SicRcv | 13/13 | 0/23 |
| Sic708m | 10/10 | 0/8 |
| Sic2025m | 0/10 | NT |
| Sar1.8mer | 4/4 | 16/16 |
Number of infected plants/number of agroinoculated plants.
Number of infected plants/number of plants inoculated with 15 B. tabaci (each) previously fed on infected plants.
NT, not testable.
Single plant containing SicRcv.
The genome of the infectious SicRcv had the same size as the original noninfectious Sic DNA (2,773 nt) and differed by only 2 nt. One change was at nt 2025 (A instead of T in the plus strand), determining a CAC-to-CUC codon change in the RepC1 mRNA and an H198L replacement in the RepC1 protein. The other mutation was located at nt 708 (C instead of G), determining a CAG-to-CAC codon change in the CP mRNA and a Q134H replacement in the CP.
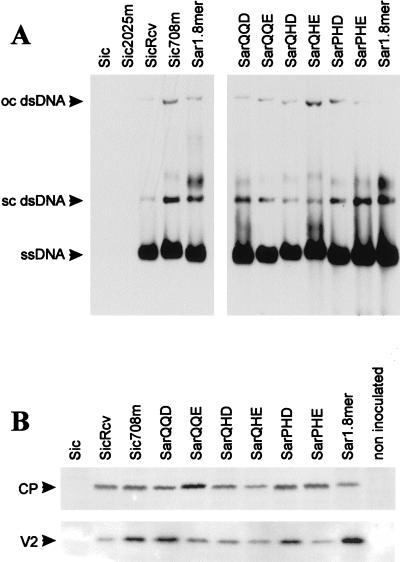
To assess the influence of each change in restoring virus ability to infect plants, the two mutations were separately introduced into the original Sic clone, giving rise to clone Sic2025m, having only the RepC1 mutation, and clone Sic708m, having only the CP mutation (Fig. 1). Clones for agroinoculation of these mutants were prepared in pSicVect. None of the 10 plants inoculated with pBinSic2025m displayed TYLCV symptoms or gave a positive hybridization signal when analyzed by tissue blotting. On the other hand, all plants inoculated with pBinSic708m became infected (Table 2). When total DNA was extracted from these plants and analyzed in Southern blots with a TYLCV-specific probe, both ssDNA and dsDNA were detected, without appreciable differences from SicRcv and TYLCV-Sar (Fig. 2A). These results indicated that the mutation in the CP gene, and not that in the RepC1 gene, was responsible for restoring infectivity.
FIG. 2.
Infectivity of TYLCV-Sic and -Sar clones in tomato plants. (A) Analysis of viral DNA. Total DNA extracted with TLES buffer from plants, 4 to 5 weeks after agroinoculation with the wild-type Sic and Sar and with the mutant clones was separated in 1% agarose gels containing ethidium bromide in 0.5× TBE buffer and blotted onto nylon membrane. Viral DNA was detected in Southern blots with a digoxigenin-labelled random-primed DNA probe synthesized on the full-length TYLCV-Sar wild-type genome. Hybridization was performed at 68°C in Standard hybridization buffer (Boehringer Mannheim). Blots were developed with CDP-Star (Tropix), after incubation with goat antidigoxigenin antibody. ssDNA and the replicative supercoiled (sc) or open-circular (oc) dsDNA forms are indicated. (B) Accumulation of CP and V2 protein. Approximately equal amounts of total proteins extracted with TCA from plants agroinoculated with the TYLCV clones were separated in SDS polyacrylamide gel electrophoresis (18% polyacrylamide for CP and 20% polyacrylamide for V2). Gels were blotted onto PVDF membranes. For Western blots, an anti-TYLCV-Sar serum (for CP) or an anti-GST-V2 serum (for V2) was used (1/10,000 final dilution). Both sera were previously cross-adsorbed with total proteins from healthy plants. Blots were developed either with an anti-rabbit AP-conjugated antibody and NBT-BCIP for CP or with an anti-rabbit HRP-conjugated antibody and a chemiluminescent substrate for V2. The positions of CP (29 kDa) and V2 (14 kDa) are indicated.
Whitefly transmissibility of SicRcv and Sic708m.
Tomato plants infected by SicRcv and Sic708m were used as the virus source in transmission experiments with the insect vector of TYLCV, B. tabaci, together with plants infected by TYLCV-Sar, a whitefly-transmissible isolate (22). Systemic infection was assessed by tissue blotting 4 weeks after inoculation. In spite of repeated attempts, none of the plants inoculated by B. tabaci having fed upon SicRcv- or Sic708m-infected plants became infected (Table 2), whereas all plants inoculated with insects fed on TYLCV-Sar plants became infected. This indicated that the Q134H mutation changed a viral DNA, only capable of replicating in single cells (Sic), into one that was systemically infectious, but not insect transmissible (SicRcv).
Comparison of amino acid sequences of CPs and construction of TYLCV-Sar mutants.
In an attempt to understand more about infectivity and insect transmissibility of TYLCV, the amino acid sequences of the capsid proteins of some TYLCV isolates were compared. In the course of the present work, parts of the original TYLCV-Sar clone were sequenced again, and some differences were found in the CP gene compared to the sequence previously sent to the data bank. These changes were introduced into the EMBL entry. The corrected sequence was used for comparisons.
Sequence alignment of the CP gene of TYLCV-Sic, SicRcv, and the closely related TYLCV-Sar revealed 14 nt differences, but only 3 of them induced changes in the amino acid sequence of the CP (257 amino acids) at positions 129, 134, and 152 (Fig. 3). TYLCV-Sar DNA has high sequence identity (92%) with Sic DNAs, and is infectious and whitefly transmissible (22). In order to confirm the role of the Q134H mutation in restoring infectivity but not transmissibility in SicRcv, and to assess whether only the CP was responsible for the observed phenotypes, eight clones of Sar DNA were constructed that contained all possible combinations of the three amino acids in the CP: Q or P at position 129, Q or H at position 134, and D or E at position 152. For agroinoculation purposes, these mutant DNAs were subcloned into pSarVect, a construct analogous to pSicVect, and named pBinSarQQD, -QQE, -QHD, -QHE, -PHD, -PHE, -PQD, and -PQE, as listed in Table 1.
Infectivity of the TYLCV-Sar mutants in tomato plants.
Tomato plants were inoculated with A. tumefaciens cells carrying each of the eight variants of the Sar genome. Systemic infection was assessed 4 weeks after inoculation by tissue blotting. Results are summarized in Table 3. SarPQE, having a CP identical to the original Sic clone, was not infectious, whereas SarPHE, with the same CP as SicRcv, infected plants systemically. Thus, the proline residue at position 129, combined with glutamine at position 134 and glutamic acid at position 152 (PQE), as originally found in the noninfectious Sic DNA, also abolished infectivity when introduced into the genome of TYLCV-Sar. Concomitantly, the Q134H replacement (PHE) gave rise to an infectious virus, as previously found in SicRcv. Besides SarPQE, only SarPQD was unable to induce a systemic infection. The other six constructs, including the wild type (SarQQD), were infectious. Mutants containing histidine at position 134 and glutamine at position 129, an amino acid combination which is not found in any TYLCV isolate, were infectious.
TABLE 3.
Results for agroinoculation and whitefly transmission with the TYLCV-Sar clonesa
| Clone | Infectivity on tomatob | Whitefly transmissibilityc |
|---|---|---|
| SarQQD | 5/5 | 34/38 |
| SarQQE | 8/9 | 13/16 |
| SarQHD | 7/9 | 26/32 |
| SarQHE | 9/9 | 35/48 |
| SarPHD | 8/9 | 1d/32 |
| SarPHE | 9/9 | 0/39 |
| SarPQD | 0/5 | NTe |
| SarPQE | 0/43 | NT |
Note that all clones showed replication in leaf discs.
Number of infected plants/number of agroinoculated plants.
Number of infected plants/number of plants inoculated with 15 B. tabaci (each) previously fed on infected plants.
Plant containing SarQHDrev.
NT, not testable.
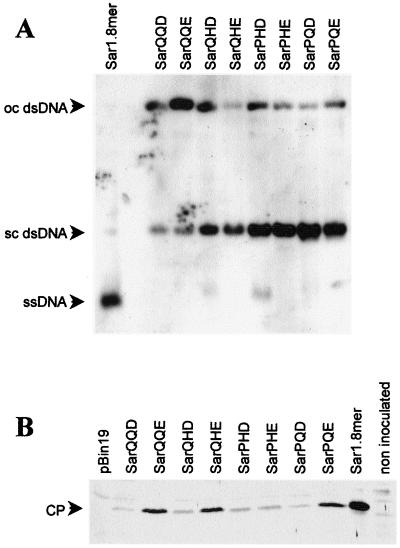
Southern blot analysis of total DNA extracted from plants infected by the six infectious Sar clones (including the wild type) showed that they accumulated similar amounts of ssDNA and dsDNA (Fig. 2A). No viral DNA could be detected in plants inoculated with SarPQE and SarPQD.
When symptom appearance was evaluated, we observed that all plants found infected by the tissue blot assay showed yellow leaf curling. However, plants inoculated with the QHD and QHE mutants displayed less severe symptoms, with milder yellowing and curling of the leaves.
In summary, all clones carrying the amino acid combinations QQ(D/E), QH(D/E), and PH(D/E) established systemic infections in plants, whereas only clones with a PQ(D/E) combination in the CP did not. The presence of D or E, two similar amino acids, at position 152 did not alter the phenotype, which was determined by amino acids 129 and 134.
Whitefly transmissibility of TYLCV-Sar mutants.
To define the role of each amino acid or their combination in insect transmission, tomato plants agroinfected by the TYLCV-Sar mutants were used as source of inoculum. The virus population in the source plants and in some plants infected after inoculation by whiteflies was analyzed by sequencing the region of the CP gene under study. Only in one whitefly-infected plant was a change detected (see below). As shown in Table 3, SarPHE and SarPHD were not transmitted, whereas the other four mutants were transmitted, with similar efficiencies (χ2 = 4.25, df = 11; ρ = 0.962). Thus, SarPHE behaved like SicRcv, the non-whitefly-transmissible mutant. In conclusion, the transmission tests showed that mutants carrying the amino acid combinations QQ(D/E) and QH(D/E) were transmissible, while those carrying PH(D/E) were not. An inability to be transmitted by whiteflies is therefore not determined by the single Q134H replacement, but by the double Q129P and Q134H substitution.
Out of 32 plants inoculated by B. tabaci that had fed on plants infected by mutant SarPHD, one became infected (Table 3). Sequence analysis of the viral DNA from this plant revealed a CCA-to-CAA mutation, causing a P129Q reversion in the CP. This rescued DNA, named SarQHDrev, did not derive from a contamination with whiteflies carrying the SarQHD, because that mutant was constructed by site-directed mutagenesis of SarQQD. Since SarPHD was obtained by inserting the SstII-SpeI fragment from SicRcv DNA into Sar DNA (Table 1), the CP gene of Sar PHD contained some silent nucleotide differences as also were found in the rescued SarQHDrev DNA.
Expression of CP and V2 and particle formation in tomato plants.
In order to see if the CP and the V2 (precoat) protein, which is possibly involved in particle formation (41), were expressed and accumulated, total proteins extracted from infected tomato plants were analyzed by Western blotting. A protein with a size corresponding to that of the CP (29 kDa) was detected by TYLCV-specific antiserum in plants inoculated with the Sic and Sar infectious mutants (Fig. 2B). Similarly, in the same plants, a protein of about 14 kDa was detected with the V2-specific antiserum (Fig. 2C). Both proteins accumulated to similar amounts in all of the infected plants.
When the presence of virions in plants was checked by ISEM, typical geminate particles were observed in tomato plants infected with all the TYLCV-Sic and -Sar mutants (not shown).
Thus, the nontransmissible SarPH(D/E) mutants produced CP, V2 protein, and virions, as did the wild type and the other whitefly-transmissible variants.
Infection of N. benthamiana leaf discs.
Leaf discs are convenient for analyzing both viral replication and gene expression, although they are not suited for studies of viral movement. This system was used to analyze whether mutants that were unable to infect whole plants could accumulate viral DNA, CP and V2 protein, and/or virions. N. benthamiana leaf discs were agroinoculated with all Sar mutants, collected after 6 days, and used for Southern blots of total nucleic acids, Western blots of total proteins, and ISEM assays.
As shown in Fig. 4A, all mutants, including SarPQE and SarPQD, accumulated viral replicative forms (supercoiled and open-circular dsDNA), at similar levels among the different clones. This indicates that inability of the SarPQE and SarPQD mutants to infect plants was not the result of impaired replication. In our test conditions, ssDNA from leaf discs was not easily detected. All mutants produced ssDNA in small amounts that were not reproducible among experiments. This may reflect a differential susceptibility to degradation or extractability of the ssDNA from the callus tissue of the discs, as opposed to differentiated plant tissue.
FIG. 4.
Infectivity of TYLCV-Sar clones in leaf discs of N. benthamiana. (A) Southern blot of total DNA extracted 6 days after inoculation. Discs inoculated with each clone were pooled for extraction. DNAs were extracted and treated as described in Fig. 2A. DNA from a tomato plant infected by TYLCV-Sar1.8mer is shown in the first lane. Genomic ssDNA and the replicative supercoiled (sc) or open circular (oc) dsDNA forms are indicated. (B) Western blot of total proteins extracted with TCA, probed with anti-TYLCV-Sar serum as described for Fig. 2B. Controls included extracts from discs inoculated with A. tumefaciens LBA4404 carrying the pBin19 vector devoid of viral sequences, a tomato plant infected by TYLCV-Sar1.8mer, and a noninoculated plant. The CP (29 kDa) is indicated.
In Western blots, CP (Fig. 4B) and V2 protein (not shown) were produced by all mutants tested, including the noninfectious variants SarPQD and SarPQE. These proteins were not detected when leaf discs were inoculated with A. tumefaciens cells carrying pBin19. Therefore it appears that the amino acid changes present in the mutants did not prevent either CP or V2 expression.
Extracts from leaf discs were also analyzed by ISEM for virus particle formation. In extracts from discs inoculated with all mutants, except SarPQ(D/E), virions were detected, and they did not differ morphologically from the wild type (not shown).
These results indicate that SarPQ(D/E) mutants were able to replicate and produce CP and V2 protein, but failed to form stable particles.
DISCUSSION
Two defective DNAs and a number of engineered TYLCV DNAs allowed us to investigate the role of the viral CP. Specific amino acid changes in the CP, or their combinations, abolished systemic infectivity in plants and transmission by whiteflies.
The original cloned DNA obtained from TYLCV-Sic did not infect plants. Unfortunately, there was no opportunity to isolate and sequence other clones from the same plant sample. It is reasonable to believe that this replication-competent but noninfectious molecule was present as part of the TYLCV population of that plant, where it was encapsidated and moved because of transcomplementation. The presence of variants in natural populations of geminiviruses has been described previously (12, 18), as has the appearance of mutated DNA in plants experimentally inoculated with cloned TYLCV DNA (21, 41). The emergence of SicRcv from Sic and SarQHDrev from SarPHD also indicates that spontaneously arising variants in a replicating population may be selected for.
Comparative analysis of Sic, SicRcv, and the hybrid genomes Sic708m and Sic2025m showed that the mutation in the CP gene, not in the RepC1 gene, was responsible for restoring infectivity in SicRcv; however, it still did not result in a whitefly-transmissible TYLCV. When introduced into TYLCV-Sar, the two capsid protein alterations resulted in the same either noninfectious (SarPQE) or nontransmissible (SarPHE) phenotype.
The CPs of these mutants differed from that of the closely related TYLCV-Sar only in three amino acids. Concerning the role of these replacements, the following conclusions can be drawn. Mutants containing the combinations QQ, QH, and PH at positions 129 and 134 are infectious in plants, whereas those with PQ are not. The PQ mutants can replicate and accumulate CP and V2 protein in leaf discs, but appear unable to produce virus particles. Mutants having the PH combination at positions 129 and 134 infect plants and form apparently normal virions, but are not transmissible by whiteflies. Changing the amino acid at position 152 (D or E) does not influence the phenotype.
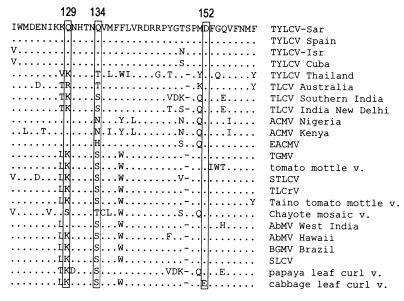
We examined the amino acids around positions 129 and 134 by using the OMIGA sequence analysis software program (Oxford Molecular Group). Such analyses predict a stretch of 12 amino acids (123 to 134) to be mostly external (with a potential target site for N glycosylation at N130), followed by a series of 6 amino acids (135 to 140) that are predicted to be internal, followed by 4 external amino acids. Analyses of several other whitefly-transmitted geminiviruses (WTGs) predict a similar arrangement. By searching the nucleotide and protein databases at the NCBI site (http://www.ncbi.nlm.nih.gov), more than 100 CP sequences of WTGs were retrieved and compared to those of the Sar clones considered in the present study. The QQD combination was found only in the monopartite TYLCVs, while the most common combination appears to be KSD (Fig. 5). At position 129, P was never found. Amino acid 134 is generally Q, S, T, or N, but it is H in a few ACMV isolates, which have a QHD combination. At position 152 there is always a D residue, with a single noteworthy exception: cabbage leaf curl virus, which has an E at this position. P129 is predicted to introduce a bend in the polypeptide chain, potentially modifying the protein structure of the subunits and preventing proper assembly of the virions.
FIG. 5.
Comparison of the CP region between amino acids 120 and 160 of TYLCV-Sar with the corresponding regions of selected WTGs. Dots indicate amino acids identical to those in the reference sequence. Dashes were introduced to maintain alignments. Amino acids corresponding to the three positions concerning this study are boxed. Sequences were extracted from the GenBank, and their accession numbers are given in parentheses: TYLCV-Sar (X61153), TYLCV Spain (Z25751), TYLCV-Isr (X15656), TYLCV Cuba (AJ223505), TLCV Australia (S53251), TLCV Southern India (U38239), TLCV India New Delhi (U15016), ACMV Nigeria (X17095), ACMV Kenya (J02057), East African cassava mosaic virus (EACMV) (Z83256), TGMV (K02029), tomato mottle virus (L14460), Sinaloa tomato leaf curl virus (STLCV) (AF040635), tomato leaf crumple virus (TLCrV) (L34747), Taino tomato mottle virus (AF012300), Chayote mosaic virus (AJ223191), AbMV West India (X15983), AbMV Hawaii (U51137), BGMV Brazil (M88686), SLCV (P27444), papaya leaf curl virus (Y15934), and cabbage leaf curl virus (U65529).
An inability to infect plants can result from defects in virus replication, movement, or assembly. We previously showed that cloned Sic DNA could efficiently replicate in tobacco protoplasts (11) and, in this study, we have shown that the SarPQ(D/E) mutants replicate in leaf discs of N. benthamiana, although they do not systemically infect this host plant (not shown). In spite of the variability in the amounts of ssDNA observed in leaf discs, the fact that no substantial difference in dsDNA accumulation was observed between the PQ mutants and the other mutants confirmed that the PQ mutants were not affected in replication.
Thus, the lack of infectivity of these clones has to be ascribed to causes other than replication. Virus movement in plants is a complex process, and many viruses require the CP for spreading (for a review, see reference 37). As noted above, the CP has different roles in monopartite and bipartite geminiviruses. In bipartite geminiviruses, proteins encoded by DNA B are responsible for viral movement. However, a partial redundancy of functions between CP (called AR1) and the BR1 movement protein of squash leaf curl virus was reported (20), since AR1 could mask the phenotype of some movement-defective BR1 mutants. In addition, Pooma et al. (32) proposed a distinction between CP-dependent systemic movement, occurring in all hosts, and less-efficient CP-independent systemic movement, occurring only in hosts to which a virus is well adapted. The TYLCVs (except the one from Thailand) do not possess a DNA B and must have evolved different strategies for movement within plants. Requirement of the CP for infection has been demonstrated previously (41), and the present study suggests that accurate particle assembly is also necessary. In fact, the PQ mutants, which are unable to assemble virions, accumulate CP in leaf discs, showing that its expression and stability are not altered. Another TYLCV protein, V2, for which a role in virus assembly has recently been suggested (41), was also detected in leaf discs infected with all mutants.
Very little is known about the interaction of geminivirus CP with viral DNA or the mechanism of virion assembly. The importance of a putative zinc finger domain in the CP of tomato leaf curl virus-India has been shown (30), and Liu et al. (23) provided evidence that the CP of the monopartite MSV binds both ssDNA and dsDNA. It is possible that the PQ mutants are not functional because of defects in protein-DNA or protein-protein interaction.
Begomoviruses are reportedly transmitted by whiteflies in a circulative nonpropagative manner. The insect acquires the virions through the stylet during feeding on an infected plant. From the gut, the virus passes into the hemocoel, then to the salivary glands, and is transmitted during subsequent feeding periods.
We have shown that the nontransmissible PH mutants produce and accumulate CP (Fig. 2B) and assemble virions. Furthermore, plants infected by PH mutants and wild-type TYLCV (QQD) appear to contain the same forms of viral DNAs that have accumulated to similar levels (Fig. 2A). Thus, lack of vector transmissibility cannot be attributed to either deficiency of virion assembly or low virus concentration in plants.
TYLCV has been detected only in a very limited number of cells in the phloem (10, 26). The possibility that nontransmissible variants are localized in different tissues remains, although we consider it unlikely, since symptoms were indistinguishable in plants infected by wild-type and nontransmissible TYLCVs. The results of our studies indicate that the reason for nontransmissibility is likely to be at the level of virus-insect interaction.
Geminiviruses that have lost their ability to be transmitted by insects when propagated for long periods in the absence of insect vectors have been reported (3, 24, 43). However, the region of CP including the mutations described in this work has not previously been implicated in transmission. By aligning the CP sequences of two nontransmissible Abutilon mosaic virus (AbMV) isolates with a number of WTGs, only five amino acid differences in the N-terminal portion were suggested to be involved in transmission (43). Hofer et al. (16) compared the CPs of two closely related WTGs, nontransmissible AbMV and the transmissible Sida golden mosaic virus (SiGMV), which show 24 amino acid differences, most of which are located in the N-terminal portion of the protein, and only 1 of which is located in the region studied by us (i.e., Q129 in AbMV, which is K in the transmissible SiGMV). Both of these amino acids are found in other transmissible begomoviruses, so this change is unlikely to be important for transmission.
The specificity of geminivirus transmission may result from an intimate relationship between exposed areas of the CP and certain insect receptors. There is no evidence of the involvement of other virus-coded proteins in insect transmission, as for the aphid transmission factors of caulimoviruses and potyviruses (31) or the read-through domain of luteoviruses (5). As noted previously (16, 44), certain similarities exist between geminiviruses and luteoviruses in terms of virus-vector interactions. Recent results concerning the relationship between luteoviruses and chaperonin-like proteins produced by endosymbiotic bacteria present in the aphid vector (17, 38) raise the possibility that similar mechanisms may also be involved in the interaction between geminiviruses and B. tabaci.
ACKNOWLEDGMENTS
This research was funded in part by the Special Project RAISA, Subproject 2, of the Italian National Research Council (CNR). E.N. at ISV, Gif-sur-Yvette, France, was supported by an EU HCM Project, contract ERBCHBGCT930488.
We thank D. Marian, M. Vecchiati, and F. Veratti for excellent technical assistance; L. Troussard and F. de Kouchkovsky for support in sequencing; S. Crespi and D. Bosco for help with the initial experiments; E. Luisoni and L. Wartig for antibodies, and R. G. Milne and M. Conti for critical reading of the manuscript.
REFERENCES
- 1.Accotto, G. P., A. M. Vaira, E. Noris, and M. Vecchiati. Using non radioactive probes on plants: a few examples. J. Biolumin. Chemilumin., in press. [DOI] [PubMed]
- 2.Azzam O, Frazer J, De La Rosa D, Beaver J S, Ahlquist P, Maxwell D P. Whitefly transmission and efficient ssDNA accumulation of bean golden mosaic geminivirus require functional coat protein. Virology. 1994;204:289–296. doi: 10.1006/viro.1994.1533. [DOI] [PubMed] [Google Scholar]
- 3.Bedford I D, Briddon R W, Brown J K, Rosell R C, Markham P G. Geminivirus transmission and biological characterization of Bemisia tabaci (Gennadius) biotypes from different geographic regions. Ann Appl Biol. 1994;125:311–325. [Google Scholar]
- 4.Boulton M, Steinkellner H, Donson J, Markham P G, King D I, Davies J W. Mutational analysis of the virion-sense genes of maize streak virus. J Gen Virol. 1989;70:2309–2323. doi: 10.1099/0022-1317-70-9-2309. [DOI] [PubMed] [Google Scholar]
- 5.Brault V, van den Heuvel J F J M, Verbeek M, Ziegler-Graff V, Reutenauer A, Herrbach E, Garaud J-C, Guilley H, Richards K, Jonard G. Aphid transmission of beet western yellows luteovirus requires the minor capsid read-through protein P74. EMBO J. 1995;14:650–659. doi: 10.1002/j.1460-2075.1995.tb07043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Briddon R W, Watts J, Markham P G, Stanley J. The coat protein of beet curly top virus is essential for infectivity. Virology. 1989;172:628–633. doi: 10.1016/0042-6822(89)90205-5. [DOI] [PubMed] [Google Scholar]
- 7.Briddon R W, Pinner M S, Stanley J, Markham P G. Geminivirus coat protein gene replacement alters insect specificity. Virology. 1990;177:85–94. doi: 10.1016/0042-6822(90)90462-z. [DOI] [PubMed] [Google Scholar]
- 8.Briddon R W, Markham P G. Geminiviridae. In: Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jaevis A W, Martelli G P, Mayo M A, Summers M D, editors. Virus taxonomy: classification and nomenclature of viruses. Sixth report of the International Committee on Taxonomy of Viruses. New York, N.Y: Springer-Verlag; 1995. pp. 158–165. [Google Scholar]
- 9.Caciagli P, Bosco D, Al-Bitar L. Relationships of the Sardinian isolate of tomato yellow leaf curl geminivirus with its whitefly vector Bemisia tabaci. Gen Eur J Plant Pathol. 1995;101:163–170. [Google Scholar]
- 10.Cherif C, Russo M. Cytological evidence of the association of a geminivirus with the tomato yellow leaf curl disease in Tunisia. Phytopathol Z. 1983;108:221–225. [Google Scholar]
- 11.Crespi S, Noris E, Vaira A M, Accotto G P. Molecular characterization of cloned DNA from tomato yellow leaf curl virus isolate from Sicily. Phytopathol Mediterr. 1995;34:93–99. [Google Scholar]
- 12.Czosnek H, Laterrot H. A worldwide survey of tomato yellow leaf curl viruses. Arch Virol. 1997;142:1391–1406. doi: 10.1007/s007050050168. [DOI] [PubMed] [Google Scholar]
- 13.Etessami P, Watts J, Stanley J. Size reversion of African cassava mosaic virus coat protein gene deletion mutants during infection of Nicotiana benthamiana. J Gen Virol. 1989;70:277–289. doi: 10.1099/0022-1317-70-2-277. [DOI] [PubMed] [Google Scholar]
- 14.Gardiner W, Sunter G, Brand L, Elmer J S, Rogers S G, Bisaro D M. Genetic analysis of tomato golden mosaic virus: the coat protein is not required for systemic spread or symptom development. EMBO J. 1988;7:899–904. doi: 10.1002/j.1460-2075.1988.tb02894.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoefgen R, Willmitzer L. Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res. 1988;16:9877. doi: 10.1093/nar/16.20.9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hofer P, Bedford I D, Markham P G, Jeske H, Frischmuth T. Coat protein gene replacement results in whitefly transmission of an insect nontransmissible geminivirus isolate. Virology. 1997;236:288–295. doi: 10.1006/viro.1997.8751. [DOI] [PubMed] [Google Scholar]
- 17.Hogenhout S A, van der Wilk F, Verbeek M, Goldbach R W, van den Heuvel J F J M. Potato leafroll virus binds to the equatorial domain of the aphid endosymbiotic GroEL homolog. J Virol. 1998;72:358–365. doi: 10.1128/jvi.72.1.358-365.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hong Y G, Harrison B D. Nucleotide sequences from tomato leaf curl viruses from different countries: evidence for three geographically separate branches in evolution of the coat protein of whitefly-transmitted geminiviruses. J Gen Virol. 1995;76:2043–2049. doi: 10.1099/0022-1317-76-8-2043. [DOI] [PubMed] [Google Scholar]
- 19.Ingham D J, Lazarowitz S G. A single missense mutation in the BR1 movement protein alters the host range of the squash leaf curl geminivirus. Virology. 1993;196:694–702. doi: 10.1006/viro.1993.1526. [DOI] [PubMed] [Google Scholar]
- 20.Ingham D J, Pascal E, Lazarowitz S G. Both bipartite geminivirus movement proteins define viral host range, but only BL1 determines viral pathogenicity. Virology. 1995;207:191–204. doi: 10.1006/viro.1995.1066. [DOI] [PubMed] [Google Scholar]
- 21.Jupin I, de Kouchkovsky F, Jouanneau F, Gronenborn B. Movement of tomato yellow leaf curl geminivirus (TYLCV): involvement of the protein encoded by ORF C4. Virology. 1994;204:82–90. doi: 10.1006/viro.1994.1512. [DOI] [PubMed] [Google Scholar]
- 22.Kheyr-Pour A, Bendahmane M, Matzeit V, Accotto G P, Crespi S, Gronenborn B. Tomato yellow leaf curl virus from Sardinia is a whitefly-transmitted monopartite geminivirus. Nucleic Acids Res. 1991;19:6763–6769. doi: 10.1093/nar/19.24.6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu H, Boulton M I, Davies J W. Maize streak virus coat protein binds single- and double-stranded DNA in vitro. J Gen Virol. 1997;78:1265–1270. doi: 10.1099/0022-1317-78-6-1265. [DOI] [PubMed] [Google Scholar]
- 24.Liu S, Bedford I D, Briddon R W, Markham P G. Efficient whitefly transmission of African cassava mosaic geminivirus requires sequences from both genomic components. J Gen Virol. 1997;78:1791–1794. doi: 10.1099/0022-1317-78-7-1791. [DOI] [PubMed] [Google Scholar]
- 25.Luisoni E, Milne R G, Vecchiati M. Purification of tomato yellow leaf curl geminivirus. Microbiologica. 1995;18:253–260. [PubMed] [Google Scholar]
- 26.Masenga, V., and R. G. Milne. Unpublished data.
- 27.Navot N, Pichersky E, Zeidan M, Zamir D, Czosnek H. Tomato yellow leaf curl virus: a whitefly-transmitted geminivirus with a single genomic component. Virology. 1991;185:151–161. doi: 10.1016/0042-6822(91)90763-2. [DOI] [PubMed] [Google Scholar]
- 28.Noris E, Hidalgo E, Accotto G P, Moriones E. High similarity among the tomato yellow leaf curl virus isolates from the West Mediterranean basin: the nucleotide sequence of an infectious clone from Spain. Arch Virol. 1994;135:165–170. doi: 10.1007/BF01309774. [DOI] [PubMed] [Google Scholar]
- 29.Noueiry A O, Lucas W J, Gilbertson R L. Two proteins of a plant DNA virus coordinate nuclear and plasmodesmatal transport. Cell. 1994;76:925–932. doi: 10.1016/0092-8674(94)90366-2. [DOI] [PubMed] [Google Scholar]
- 30.Padidam M, Beachy R N, Fauquet C M. The role of AV2 (“precoat”) and coat protein in viral replication and movement in tomato leaf curl geminivirus. Virology. 1996;224:390–404. doi: 10.1006/viro.1996.0546. [DOI] [PubMed] [Google Scholar]
- 31.Pirone T P, Blanc S. Helper-dependent vector transmission of plant viruses. Annu Rev Phytopathol. 1996;34:227–247. doi: 10.1146/annurev.phyto.34.1.227. [DOI] [PubMed] [Google Scholar]
- 32.Pooma W, Gillette W K, Jeffrey J L, Petty I T D. Host and viral factors determine the dispensability of coat protein for bipartite geminivirus systemic movement. Virology. 1996;218:264–268. doi: 10.1006/viro.1996.0189. [DOI] [PubMed] [Google Scholar]
- 33.Pringle C R. The universal system of virus taxonomy of the International Committee on Virus Taxonomy (ICTV) including new proposals ratified since publication of the Sixth ICTV Report in 1995. Arch Virol. 1998;143:203–210. doi: 10.1007/s007050050280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rigden J E, Dry I B, Mullineaux P M, Rezaian M A. Mutagenesis of the virion-sense open reading frames of tomato leaf curl geminivirus. Virology. 1993;193:1001–1005. doi: 10.1006/viro.1993.1215. [DOI] [PubMed] [Google Scholar]
- 35.Rochester D E, DePaulo J J, Fauquet C M, Beachy R N. Complete nucleotide sequence of the geminivirus tomato yellow leaf curl virus, Thailand isolate. J Gen Virol. 1994;75:477–485. doi: 10.1099/0022-1317-75-3-477. [DOI] [PubMed] [Google Scholar]
- 36.Schaffer R L, Miller C G, Petty I T D. Virus and host specificity adaptation in the BL1 and BR1 genes of bipartite geminiviruses. Virology. 1995;214:330–338. doi: 10.1006/viro.1995.0042. [DOI] [PubMed] [Google Scholar]
- 37.Séron K, Haenni A-L. Vascular movement of plant viruses. Mol Plant-Microbe Interact. 1996;9:435–442. doi: 10.1094/mpmi-9-0435. [DOI] [PubMed] [Google Scholar]
- 38.van den Heuvel J F J M, Bruyére A, Hogenhout S A, Ziegler-Graff V, Brault V, Verbeek M, van der Wilk F, Richards K. The N-terminal region of the luteovirus readthrough domain determines virus binding to Buchnera GroEL and is essential for virus persistence in the aphid. J Virol. 1997;71:7258–7265. doi: 10.1128/jvi.71.10.7258-7265.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.von Arnim A, Stanley J. Determinants of tomato golden mosaic virus symptom development located on DNA B. Virology. 1992;186:286–293. doi: 10.1016/0042-6822(92)90083-2. [DOI] [PubMed] [Google Scholar]
- 40.Wartig L. Ph.D. thesis. Centre d’Orsay, France: Université Paris-Sud; 1994. [Google Scholar]
- 41.Wartig L, Kheyr-Pour A, Noris E, de Kouchkovsky F, Jouanneau F, Gronenborn B, Jupin I. Genetic analysis of the monopartite tomato yellow leaf curl geminivirus: roles of V1, V2, and C2 ORFs in viral pathogenesis. Virology. 1997;228:132–140. doi: 10.1006/viro.1996.8406. [DOI] [PubMed] [Google Scholar]
- 42.Wu F S, Wang M Y. Extraction of proteins for sodium dodecyl sulfate-polyacrylamide gel electrophoresis from protease-rich plant tissue. Anal Biochem. 1984;139:100–103. doi: 10.1016/0003-2697(84)90394-4. [DOI] [PubMed] [Google Scholar]
- 43.Wu Z C, Hu J S, Polston J E, Ullman D E, Hiebert E. Complete nucleotide sequence of a nonvector-transmissible strain of abutilon mosaic geminivirus in Hawaii. Phytopathology. 1996;86:608–613. [Google Scholar]
- 44.Ziegler-Graff V, Brault V, Mutterer J D, Simonis M-T, Herrbach E, Guilley H, Richards K E, Jonard G. The coat protein of beet western yellows luteovirus is essential for systemic infection but the viral gene products P29 and P19 are dispensable for systemic infection and aphid transmission. Mol Plant-Microbe Interact. 1996;9:501–510. [Google Scholar]