Abstract
Photoautotrophic cyanobacteria assimilate the greenhouse gas carbon dioxide as their sole carbon source for producing useful bioproducts. However, harvesting the cells from their liquid media is a major bottleneck in the process. Thus, an easy-to-harvest method, such as auto-flocculation, is desirable. Here, we found that cyanobacterium Synechocystis sp. PCC 6803 co-flocculated with a natural fungal contamination in the presence of the antibiotic erythromycin (EM) but not without EM. The fungi in the co-flocculated biomass were isolated and found to consist of five species with the filamentous Purpureocillium lilacinum and Aspergillus protuberus making up 71% of the overall fungal population. The optimal co-cultivation for flocculation was an initial 5 mg (fresh weight) of fungi, an initial cell density of Synechocystis of 0.2 OD730, 10 µM EM, and 14 days of cultivation in 100 mL of BG11 medium with no organic compound. This yielded 248 ± 28 mg/L of the Synechocystis-fungi flocculated biomass from 560 ± 35 mg/L of total biomass, a 44 ± 2% biomass flocculation efficiency. Furthermore, the EM treated Synechocystis cells in the Synechocystis-fungi flocculate had a normal cell color and morphology, while those in the axenic suspension exhibited strong chlorosis. Thus, the occurrence of the Synechocystis-fungi flocculation was mediated by EM, and the co-flocculation with the fungi protected Synechocystis against the development of chlorosis. Transcriptomic analysis suggested that the EM-mediated co-flocculation was a result of down-regulation of the minor pilin genes and up-regulation of several genes including the chaperone gene for pilin regulation, the S-layer protein genes, the exopolysaccharide-polymerization gene, and the genes for signaling proteins involved in cell attachment and abiotic-stress responses. The CuSO4 stress can also mediate Synechocystis-fungi flocculation but at a lower flocculation efficiency than that caused by EM. The EM treatment may be applied in the co-culture between other cyanobacteria and fungi to mediate cell bio-flocculation.
Keywords: Antibiotic, Cyanobacteria, Flocculation, Gene expression, Cell harvest, Stress
Subject terms: Biotechnology, Microbiology, Applied microbiology
Introduction
Synechocystis sp. PCC 6803 (hereafter, Synechocystis) is one of best-studied cyanobacteria that produces useful bioproducts, such as polysaccharides, bioplastic polymers, lipids, hydrogen gas, and pigments1–4. The well-characterized metabolism and the available gene manipulation in Synechocystis enable rational metabolic engineering to produce industrial bioenergy and biochemical compounds5–7.
However, harvesting of the cells from the liquid growth medium is one of the main barriers for producing such products by Synechocystis and other cyanobacteria and algae8,9. To harvest cyanobacterial cells from liquid culture, physical processes (sedimentation, filtration, and centrifugation), and chemical flocculation have been used9–12, but these techniques have a high operation cost and excessive energy consumption9,11,13. Biological flocculation, which requires cell–cell attachment between cyanobacteria-cyanobacteria14–17, cyanobacteria-bacteria15,18, and cyanobacteria-fungi12,19, is a promising advantage because this approach does not require a high cost or extra energy12,19. The co-flocculation between cyanobacteria and fungi is interesting because of its high efficiency of biomass flocculation in a liquid culture20–22.
The co-flocculation of cyanobacteria-fungi has been previously described20–22. For Synechocystis, co-flocculated Synechocystis-Aspergillus fumigatus and Synechocystis-A. oryzae were formed by adding a substantial amount of pre-grown fungal pellets to the Synechocystis culture19,21. The biological benefit for the co-flocculation between cyanobacteria-fungi and between the microalgae-fungi in lichens has been known for nutrient exchange20,23. However, knowledge on the cellular mechanisms of flocculation between cyanobacteria and fungi is still limited24. Pilin, exopolysaccharides (EPS), and S-layer protein on the cell surface of cyanobacteria have all been reported to play a role in cell–cell attachment of cyanobacterial auto-flocculation25–27. In cyanobacteria-fungi co-flocculation, previous studies described that cell surface attachment is mediated by the electrostatic interaction between the negative charges of EPS on the cyanobacterial cell surface and the positive charges of saccharides on the cell surface of fungal hypha21,28. The S-layer protein is also required for cell–cell attachment, since the Synechocystis mutant lacking S-layer protein has been shown to have a significantly reduced biofilm formation25. In addition, pilins are known to be involved in the motility and cell adhesion of bacteria29 and cyanobacteria30–32.
In nature, various microorganisms produce antibiotics that inhibit bacterial growth or kill bacteria33,34. Bacteria can alleviate growth inhibition by antibiotic by forming a biofilm to protect the cells from antibiotic exposure and absorption35,36. The consortium cultivation between bacteria and eukaryotic green algae helps to reduce exposure to antibiotics in liquid culture37,38. However, the consortium between cyanobacteria and other organisms for alleviating antibiotic activity has yet to be studied. In cyanobacterium Synechocystis, among various antibiotics that inhibit cell growth39, erythromycin (EM) is of interest because it exhibits bactericidal activity to Synechocystis39,40 via interfering with bacterial protein synthesis by blocking a nascent peptide exit tunnel of the large ribosomal subunit41.
In this study, Synechocystis-fungi flocculation was observed in a Synechocystis culture with a natural fungal contamination in the presence of EM. This study hypothesized that Synechocystis co-flocculates with the fungi under the presence of EM. To prove this, the fungi were then isolated on agar medium, and the isolated fungi and Synechocystis were co-cultivated in a liquid medium in the presence or absence of EM. This study also aims to identify: (i) the fungal population present in the co-flocculated biomass; (ii) the optimal inoculation amounts of Synechocystis and fungi and the optimal initial EM concentration for the maximal biomass flocculation; (iii) potential Synechocystis genes responsible for Synechocystis-fungi flocculation. This study also confirmed this bioflocculation formation by illustrating the cell morphology of the fungi and Synechocystis in the flocculated biomass using bright-field and fluorescent microscopy.
Methods
Strain and culture medium
The cyanobacterium Synechocystis sp. PCC 6803 was obtained from Institute Pasteur, France. Synechocystis was cultured in the standard BG11 medium (hereafter, BG11)42 supplemented with 20 mM [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid]–NaOH (pH 7.5) but without sodium citrate and with the ferric ammonium citrate being replaced with ferric chloride. Therefore, this BG11 medium does not contain any organic compounds43 in order to limit the growth of the fungi. The fungi were first obtained from a natural contamination in a Synechocystis culture. Subsequently, the fungi were isolated by streaking onto BG11 medium agar plates containing 1.5% (w/v) bacto agar (Himedia, India), which is the sole carbon source of the fungi. For fungal growth, the BG11 agar plates were incubated at 37 °C for 2–3 days in the dark. The contaminated fungi were re-streaked four times.
Synechocystis axenic cultivation, co-cultivation of Synechocystis and fungi, and cell harvesting
All cultivations were operated in 100 mL BG11 liquid medium under continuous cool white fluorescent light (50 μmol m−2 s−1) at 28 °C with constant shaking at 160 rpm, and an atmospheric carbon dioxide (CO2) concentration [0.04% (v/v)] was supplied. The EM was added to the BG11 medium when required. For Synechocystis axenic cultivation, only Synechocystis was cultivated. For the co-cultivation of Synechocystis and fungi, Synechocystis and fresh fungal biomass (harvested from the agar plate) were mixed and cultivated for 7–14 days. Flocculated biomass was harvested from the liquid culture using gravitational filtration through an aluminum filter with a pore size of 0.05 mm. The remaining biomass in the liquid medium after filtration was collected using centrifugation. The flocculated and suspended biomass were dried at 60 °C until at a constant weight was achieved.
Calculation of % flocculated biomass and % suspended biomass with respect to total biomass
Microscopic examination of the flocculated cells
Each Synechocystis-fungi flocculated biomass was visualized under light microscopy (BX51; Olympus Corporation, Tokyo, Japan). For observation of the overall cell morphology, cells were observed under bright-field mode using a standard white light. For examination of chlorophyll in Synechocystis cells, the autofluorescence of chlorophyll (seen as red fluorescent light) was detected using an excitation filter (530–535 nm) and an emission filter (> 580 nm).
Transcriptomics analysis
The transcriptomes of Synechocystis from three different groups were analyzed (Table 2). Each group was performed using biomass from three independent cultures. For RNA extraction, fresh cells were quickly frozen with liquid nitrogen, milled with glass beads (150–212 μm, Cat. No. G9018, Sigma-Aldrich, Missouri, USA), and then TRIzol reagent (Life Technologies, Carlsbad, CA, USA) was used to extract RNA as described previously44. A RNase-free DNase kit (Qiagen, Hilden, Germany) was used to remove DNA contamination. The cDNA library was synthesized, and cDNA sequencing was performed at the OMICS SCIENCE center (Faculty of Science, Chulalongkorn University, Bangkok, Thailand) as previously described44. In brief, RNA strand-specific RNA-Seq libraries were created using the QIAseq FastSelect RNA removal and QIAseq stranded total RNA library preparation kits (QIAGEN). QIAseq FastSelect–rRNA HMR QIAseq FastSelect–5S/16S/23S-rRNA removal solution (QIAGEN, USA) was used to remove ribosomal RNAs. AMPure XP beads (Beckman Coulter Genomic) were used to enrich the RNA sample following cDNA synthesis.
Table 2.
Synechocystis responsive genes potentially involved in cell flocculation that were found in both this study and in previous reports.
| Gene ID | Gene product | Log2-fold change of expression in this studya | Gene expression status in the flocculated Synechocystis reported in previous studiesb | ||
|---|---|---|---|---|---|
| [C + F + E] compared with [C] | [C + F + E] compared with [C + E] | [C + E] compared with [C] | |||
| slr1674 | Hypothetical protein | 7.3 | 1.0 | 6.3 | Up-regulation in flocculated wild type, compared to suspended wild type60 |
| slr2076 (groEL1) | 60 kD Chaperonin 1 | 7.3 | 1.4 | 5.9 | Normal expression in flocculated wild type, compared to down-regulation in suspended in kpsM-deleted strain67 |
| slr0967 | Hypothetical protein | 6.9 | NS | 6.7 | Up-regulation in flocculated wild type, compared to suspended wild type60 |
| sll0416 (groEL2) | 60 kD Chaperonin 2 | 6.5 | 1.6 | 4.9 | Normal expression in flocculated wild type, compared to down-regulation in suspended in kpsM-deleted strain67 |
| slr1704 | Hypothetical S-layer protein | 6.3 | NS | 5.4 | Normal expression in flocculated wild type, compared to down-regulation in suspended sigF-deleted strain68 |
| sll1867 (psbA3) | Photosystem II D1 protein | 6.3 | NS | 6.0 | Normal expression in flocculated wild type, compared to down-regulation in suspended in kpsM-deleted strain67 |
| slr1738 | PerR transcriptional regulator of Fur family | 6.1 | NS | 5.8 | Normal expression in flocculated wild type, compared to down-regulation in suspended in kpsM-deleted strain67 |
| slr1915 | Hypothetical protein | 5.8 | NS | 5.4 | Up-regulation in flocculated wild type, compared to suspended wild type60 |
| sll1823 (purA) | Adenylosuccinate synthetase | 5.5 | 2.0 | 3.5 | Normal expression in flocculated wild type, compared to down-regulation in suspended in kpsM-deleted strain67 |
| ssr0692 | Key regulator of arginine synthesis | 5.2 | − 1.5 | 6.6 | Up-regulation in flocculated wild type, compared to suspended wild type60 |
| slr0923 (wzc) | EPS polymerization, assembly, and export | 4.6 | NS | 4.0 | Normal expression in flocculated wild type, compared to no expression in suspended slr0923-deleted strain25 |
| ssr3341 (hfq) | RNA chaperone regulating pilin formation | 2.6 | NS | 3.4 | Normal expression in flocculated wild type, compared to no expression in suspended ssr3341-deleted strain27 |
| slr0322 (hik43) | CheA-like protein regulating pilin biogenesis | 2.0 | NS | 1.5 | Up-regulation in flocculated wild type under salt stress, compared to suspended wild type under normal culture condition61 |
| sll1951 (slyr) | S-layer protein on cell surface | 1.9 | NS | 2.7 | Normal expression in flocculated wild type, compared to no expression in suspended sll1951-deleted strain25 |
| sll1965 | Hypothetical protein in pilin regulon | 1.7 | NS | 2.0 | Up-regulation in flocculated hik43-overexpressed strain, compared to suspended wild type61 |
| slr0073 (hik36) | Type IV pili sensor histidine kinase regulating pilin biogenesis | 1.7 | NS | 1.3 | Normal expression was detected in flocculated wild type under salt stress. This gene was involved in signal transduction of cell flocculation61 |
| sll0821 (cph2) | Protein involved in inhibition of phototactic motility under blue light | 1.5 | NS | 1.1 | Up-regulation in flocculated wild type under blue light, compared to suspended wild type under normal light69 |
| slr2019 | Hypothetical protein in minor pilin regulon | − 1.2 | NS | − 2.1 | No expression in flocculated slr2019-deleted strain, compared to normal expression in suspended wild type21,69 |
| slr2017 | Minor pilin protein PilA11 | − 1.6 | NS | − 1.3 | No expression in flocculated slr2017-deleted strain, compared to normal expression in suspended wild type21,69 |
| slr2018 | Minor pilin protein PilA12 | − 3.6 | − 1.8 | − 1.8 | No expression in flocculated slr2018-deleted strain, compared to normal expression in suspended wild type21,69 |
| slr2016 | Minor pilin protein PilA10 | − 5.1 | − 3.7 | − 1.4 | No expression in flocculated slr2016-deleted strain, compared to normal expression in suspended wild type21,69 |
aSynechocystis gene expression levels were obtained from the transcriptomic analysis by comparing between the two different experimental groups: [C + F + E], Synechocystis in the Synechocystis-fungi flocculated biomass cultured with EM and with the fungi; [C + E], axenic Synechocystis suspended cells with EM and without the fungi; [C] axenic Synechocystis suspended cells without EM and without Fungi. NS, not significantly different level.
bGene expression status was compared between those from the Synechocystis flocculated cells and the Synechocystis suspended cells, as described in the indicated previous reports.
All cDNA libraries were analyzed for quality using an Agilent 2100 Bioanalyzer (Agilent) after indexing adapters were ligated to the cDNAs. The sequencing libraries were pooled in an equimolar amount after the cDNA libraries were quantified using a DeNovix fluorometer (DeNovix). Pair-end 125 nucleotides read sequencing on an Illumina HiSeq sequencer was done with cluster creation. The quality of the raw read data was examined using the FASTQC software. Fastp software was used to delete adapters and low-quality reads. Salmon software was used to compare high-quality reads to the reference genome sequence (Synechocystis sp. PCC 6803, accession number: ASM972v1). DESeq2 was used for the differential gene expression analysis (FDR 0.05, log2 fold change > 1.0). The detected transcripts from fungi were excluded and were not used for transcriptomic analysis. The transcriptome's raw read data were submitted to the Gene Expression Omnibus (GEO) repository, accession numbers: GSE244152, GSE256451, and GSE256452.
Fungal population determination
Total DNA from Synechocystis-fungi flocculated biomass was purified using DNeasy PowerSoil Pro DNA Kit (Qiagen, USA). The PCR amplification of the fungal internal transcribed spacer (ITS) rDNA was conducted using the ITS1 and ITS2 primers (Illumina, USA), using 2X sparQ HiFi PCR master Mix (QuantaBio, USA). The PCR thermal cycling was 2 min at 98 °C, followed by 30 cycles of 98 °C for 20 s, 60 °C for 30 s, and 72 °C for 1 min, and then followed by a final 72 °C for 1 min. Following enrichment using sparQ Puremag Beads (QuantaBio, USA) and indexing with 5 µL of each Nextera XT index primer in a 50-µL PCR reaction, the ITS amplicons were processed with 8–10 cycles of the above PCR procedure. Cleansing, pooling, and diluting the final PCR products to a final loading concentration of 4 pM were performed. The 250-bp paired-end read sequencing and cluster formation were performed on an Illumina MiSeq at the OMICS SCIENCE center (Faculty of Science, Chulalongkorn University, Bangkok, Thailand).
Results
Synechocystis-fungi flocculation is mediated by EM
A Synechocystis-fungi flocculated biomass was observed in a Synechocystis cultivation that was naturally contaminated with fungi upon the addition of EM. The fungi were subsequently isolated by re-streaking four times on an agar plate (Fig. 1). The isolated fungi exist in septate hyphae filaments (Fig. 2a,b).
Figure 1.
Outlines of the experimental procedures. (a) Flocculated biomass derived from the co-existence of Synechocystis and fungi was found in a Synechocystis culture under 10 μM EM treatment with a natural fungal contamination. (b) The flocculated biomass from (a) was streaked onto agar medium to isolate the fungi. (c) Co-cultivation between Synechocystis and the isolated fungi obtained from (b). This cultivation was optimized for the maximal level of flocculated biomass with respect to the three indicated factors. (d) The optimal co-cultivation yielding the maximal flocculated biomass was subjected to fungal classification by fungal ITS rDNA sequencing, Synechocystis transcriptomic analysis, and microscopic examination.
Figure 2.
Culture and microscopic examination of Synechocystis-fungi flocculation. (a) Isolated fungi cultured on agar medium, as obtained from the procedure described in Fig. 1b. (b) Isolated fungi seen under bright-field (left) and fluorescent (right) microscopy. (c) Axenic Synechocystis cultured without EM. (d) Axenic Synechocystis from (c) as seen by bright-field microscopy (left) and fluorescent microscopy (right) showing the red light resulting from the auto-fluorescence of chlorophyll. (e) Co-cultivation between Synechocystis and the fungi with 10 µM EM showing Synechocystis-fungi flocculation. (f) Synechocystis-fungi flocculated biomass visualized by bright-field microscopy. (g, h) Synechocystis-fungi flocculated biomass as seen by bright-field microscopy (left) and fluorescent microscopy (right) showing the red light resulting from auto-fluorescence of Synechocystis chlorophyll.
To validate that the isolated fungi and Synechocystis co-flocculate in the presence of EM, the co-cultivation of Synechocystis and the isolated fungi was performed using an initial Synechocystis density of an optical density at 730 nm (OD730) of 0.2, a fungi inoculum of 5 mg, and 10 µM EM in 100 mL of the BG11 medium without any organic compounds. Under this condition, a flocculated biomass was clearly visible from day 7 onwards (Fig. 2e). Microscopic examination using the bright-field mode to detect the overall cell morphology and the fluorescent mode to detect autofluorescence of Synechocystis’s chlorophyll showed that the flocculated biomass obtained from the co-culture consisted of both Synechocystis and the fungi, with Synechocystis attached on the surface of the fungal hyphae filament (Fig. 2f–h).
Additionally, the co-cultivation of Synechocystis and the fungi without EM (Figs. 3b and 5a), and the cultivation of Synechocystis with EM but without the fungi (Figs. 3a and 4a), showed no cell flocculation. Thus, the cell flocculation required the presence of Synechocystis, fungi, and EM. It is noted that under 10–20 µM EM, the co-cultivation of both Synechocystis and fungi yielded higher levels of total harvested biomass than that obtained from the cultivation of only Synechocystis without fungi (Fig. 3a,b and Fig. 4a,b).
Figure 3.
Effect of the EM concentration on the levels of Synechocystis-fungi flocculation. Total biomass is the sum of flocculated and suspended biomasses. Values are shown as the mean ± 1SD (n = 4). ND = not detected. (a) Axenic cultivation of Synechocystis at an initial cell density of OD730 = 0.2 in the presence of various EM concentrations. Representative images of the culture flasks are shown. Significantly different levels (*P < 0.05: two-tailed student’s t-test) between the EM-treated cells and the untreated cells at the same cultivation time and the same type of biomass. Representative images of the respective cultivation (culture flasks) are shown. (b) Co-cultivation between Synechocystis (fixed initial cell density of OD730 = 0.2) and the fungi (fixed initial fungi inoculation of 5 mg fresh weight/100 mL) in the presence of various EM concentrations. Significantly different levels (*P < 0.05: two-tailed student’s t-test) between the EM-treated cells and the untreated cells at the same cultivation time and the same type of biomass. Representative images showing cell flocculation from the respective cultivation are shown.
Figure 5.
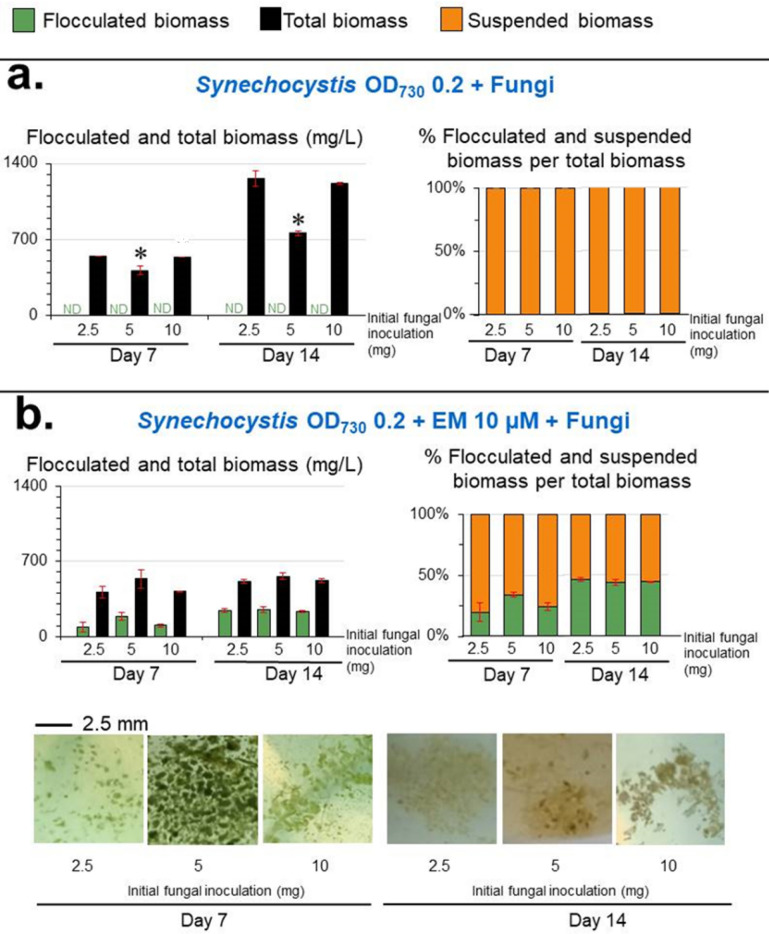
Effect of initial fungi amount on the levels of Synechocystis-fungi flocculation. Total biomass is the sum of flocculated and suspended biomasses. Values are shown as the mean ± 1SD (n = 4), and ND = not detected. (a) Co-cultivation of Synechocystis at an initial cell density of OD730 = 0.2 without EM and with the initial fungal inoculation at 2.5, 5, and 10 mg fresh weight/100 mL. Significantly different levels (*P < 0.05: two-tailed student’s t-test), compared to that of the initial fungal inoculation at 2.5 mg/100 mL of the same cultivation time and the same type of biomass. (b) Co-cultivation of Synechocystis (fixed initial density of OD730 = 0.2) and initial fungal inoculation at 2.5, 5, and 10 mg fresh weight/100 mL with 10 µM EM. Significantly different levels (*P < 0.05: two-tailed student’s t-test) compared to that of the initial fungal inoculation at 2.5 mg/100 mL at the same cultivation time and the same type of biomass. Representative images of the cell flocculation from the respective culture are shown.
Figure 4.
Effect of initial Synechocystis density on the levels of Synechocystis-fungi flocculation. Total biomass is the sum of flocculated and suspended biomasses. Values are shown as the mean ± 1SD (n = 4), and ND = not detected. (a) Axenic cultivation of various initial Synechocystis densities in the presence of 10 µM EM. Significantly different levels (*P < 0.05: two-tailed student’s t-test), compared to that of the initial Synechocystis inoculation at OD730nm = 0.1 of the same cultivation time and the same type of biomass. (b) Co-cultivation between various Synechocystis densities and the fungi (fixed initial fungi inoculation of 5 mg fresh weight/100 mL) in the presence of 10 µM EM. Significantly different levels (*P < 0.05: two-tailed student’s t-test) compared to that of the initial Synechocystis inoculation at OD730nm = 0.1 at the same cultivation time and the same type of biomass. Representative images of the cell flocculation from the respective cultivation are shown.
Fungal species present in the Synechocystis-fungi flocculated biomass were identified following the PCR amplification of fungal ITS rDNA, followed by next-generation sequencing. The fungal population was found to consist of five species (Table 1), with the filamentous Purpureocillium lilacinum and Aspergillus protuberus making up 71% of the overall fungal population (Table 1).
Table 1.
Percent composition of the fungi present in the Synechocystis-fungi flocculated biomass.
| Species of fungi | Detected frequency of ITS rDNA | % Composition in fungal population |
|---|---|---|
| Purpureocillium lilacinum | 94,141 | 40.8 |
| Aspergillus protuberus | 70,920 | 30.7 |
| Sarocladium implicatum | 44,794 | 19.4 |
| Candida tropicalis | 20,459 | 8.9 |
| Dipodascaceae sp. | 465 | 0.2 |
The Synechocystis-fungi flocculated biomass was obtained from the co-cultivation between Synechocystis (initial density of an OD730 of 0.2) and the isolated fungi (initial inoculation of 5 mg fresh weight) in the presence of 10 µM EM for seven days. Fungal identification and frequency determination were obtained from the analysis of total fungal ITS rDNA amplified from total DNA isolated from the flocculated biomass.
Optimal initial inoculum amount of EM, Synechocystis, and fungi for maximal Synechocystis-fungi flocculation.
Increasing the initial EM concentration from 10 to 20 µM significantly reduced the Synechocystis-fungi flocculated biomass and caused a stronger chlorotic effect on Synechocystis (Fig. 3b). With respect to the initial inoculum size of Synechocystis, a Synechocystis inoculum of an OD730 of 0.2 yielded the highest Synechocystis-fungi flocculated biomass (248 ± 28 mg/L) corresponding to a 44 ± 2% flocculation efficiency at day 14 (Fig. 4), compared to the other initial inoculum sizes of Synechocystis. Increasing or decreasing the Synechocystis inoculum from OD730 = 0.2 slightly reduced the Synechocystis-fungi flocculated biomass level and % flocculation efficiency (Fig. 4). For the initial fungi inoculum, an initial inoculum of 2.5, 5, and 10 mg in 100 mL cultivation yielded comparable levels of both Synechocystis-fungi flocculated biomass and % flocculation efficiency at day 14 (Fig. 5).
Assessment of altered gene-expression levels of Synechocystis in the co-flocculated biomass compared to the suspended Synechocystis cells.
Total cellular transcripts were compared between the Synechocystis in the Synechocystis-fungi flocculated biomass and the axenic Synechocystis suspended cells. The detected transcripts from fungi were excluded and were not used for transcriptomic analysis. Decreased transcript levels (down-regulated genes) of Synechocystis in the co-flocculated biomass with respect to Synechocystis suspended cells were found mainly in the genes responsible for metabolic pathways (234 genes), biosynthesis of secondary metabolites (104 genes) and cofactors (56 genes), ABC transporter (53 genes), and carbon metabolism (28 genes) (Fig. 6a). Thus, these cellular processes are likely to be affected by EM exposure.
Figure 6.
Synechocystis responsive genes in the flocculated cells. (a) Synechocystis responsive genes under flocculation. Synechocystis transcriptomics were compared between Synechocystis in [the Synechocystis-fungi flocculated biomass with EM] and [axenic suspended Synechocystis without EM]. The Synechocystis-fungi flocculated biomass was obtained from the co-cultivation between Synechocystis (initial density of OD730 = 0.2) and the fungi (initial inoculation of 5 mg fresh weight in 100 mL cultivation) in the presence of 10 µM EM. Axenic Synechocystis biomass was derived from the axenic culture using an initial density at OD730 = 0.2 without EM. The cultivation time is seven days. Data were obtained from three independent cultures. The transcriptomic data were analyzed, and the responsive genes were categorized by their cellular functions and metabolism using the Kyoto Encyclopedia of Genes and Genomes (KEGG; www.genome.jp/kegg/pathway.html)70. Names of the genes in each category of cellular processes are given in Supplementary Data S1. (b) Synechocystis-responsive genes under flocculation found in this study (shown in a) have also been reported to respond to other abiotic stresses: hydrogen peroxide71, copper72, cadmium73, nitrogen deprivation44, salt74, and heat75. Names of responsive genes in each stress are given in Supplementary Data S2.
Many of the responsive genes in Synechocystis found in the flocculated biomass in this study have also been previously reported to exhibit altered expression levels under various abiotic stresses (Fig. 6b). Of particular significance, more than two hundred responsive genes that might be involved in the flocculation in this study were also previously reported to show altered expression levels under the stress induced by hydrogen peroxide or copper treatment (Fig. 6b). Thus, these data suggested that the flocculated Synechocystis may encounter abiotic stress mediated by EM treatment.
Synechocystis responsive genes that are potentially involved in Synechocystis-fungi flocculation.
Total transcripts were compared between the Synechocystis in [the Synechocystis-fungi flocculated biomass with EM exposure] and [the axenic Synechocystis suspended cells without EM]. The twenty-one Synechocystis genes that showed altered transcript levels in response to the co-flocculation in this study have been previously reported to be involved in the auto-flocculation or biofilm formation of Synechocystis (Table 2).
Interestingly, the altered expression levels of all these 21 genes were mainly caused by the EM exposure as evidenced by the transcriptomic comparison between the Synechocystis in [the axenic Synechocystis suspended cells with EM exposure] and [the axenic Synechocystis suspended cells without EM] (Table 2). The presence of the fungi slightly altered expression levels of only the 7 out of those 21 genes as shown by the transcriptomic comparison between the Synechocystis in [the Synechocystis-fungi flocculated biomass with EM] and [the axenic Synechocystis suspended cells with EM] (Table 2).
Overall, the results showed that altering expression of these 21 genes that are potentially involved in the flocculation were mainly induced by the EM exposure. Of particular importance, the up-regulated genes include the genes involved in pilin formation (slr0322 and slr0073), the gene for EPS polymerization and export (slr0923), the genes for S-layer protein (slr1704, and sll1951), and the genes for the regulatory/signal transduction proteins (slr1738, sll1823, ssr062, and sll0821) (Table 2). Additionally, the down-regulated genes are the three genes for the minor-pilin proteins (slr2016, slr2017, and slr2018).
Synechocystis-fungi flocculation is also mediated by CuSO4 stress
The presence of NaCl at 1–2 M45 and the presence of CuSO4 at 4–8 µM46 that have been shown to effectively induce abiotic stress in Synechocystis, were examined for their ability in mediating the co-flocculation between the fungi and Synechocystis. Interestingly, the presence of 4 and 8 µM CuSO4 can induce the Synechocystis-fungi flocculation, but with a low flocculation efficiency at 4.6–5.0% of the total biomass (Fig. 7b). The presence of 1 and 2 M NaCl results in only a trace amount of the flocculated biomass (< 0.5% of the flocculation efficiency) (Fig. 7a).
Figure 7.
Effect of the NaCl stress and CuSO4 stress on the Synechocystis-fungi flocculation. Total biomass is the sum of flocculated and suspended biomasses. Values are shown as the mean ± 1SD (n = 4). LD: low detection level at < 2 mg/L of the flocculated biomass or < 0.5% of percent flocculated biomass per total biomass. (a) Effect of the NaCl stress. Co-cultivation between Synechocystis (fixed initial cell density of OD730 = 0.2) and the fungi (fixed initial fungi inoculation of 5 mg fresh weight/100 mL) in the presence of NaCl at 1 or 2 M for 7 days. Culture: representative images of the culture flasks. (b) Effect of the CuSO4 stress. Co-cultivation between Synechocystis (fixed initial cell density of OD730 = 0.2) and the fungi (fixed initial fungi inoculation of 5 mg fresh weight/100 mL) in the presence of CuSO4 at 4 or 8 µM for 7 days. Culture: representative images of the culture flasks. Microscope: Synechocystis-fungi flocculated biomass as seen by bright-field microscopy (BF, left) and fluorescent microscopy (FL, right) showing the red light resulting from auto-fluorescence of Synechocystis chlorophyll. (c) The cultivation of the fungi (using the initial inoculation of 5 mg fresh weight/100 mL) in the BG11 medium without Synechocystis and without EM. The fresh weights of the fungi were dried to determine the dry weight.
Discussion
Cyanobacterium-fungi flocculation in liquid culture has been previously reported as follows:
Synechocystis-A. oryzae19, Nostoc sp.-Aspergillus sp.20, Synechocystis-A. fumigatus21, a consortium of four cyanobacterial species and A. niger22, Microcystis aeruginosa-A. oryzae47, and Chroococcus sp.-A. lentulus48. However, all these reported algal-fungi flocculation required an organic carbon supply in the culture medium for fungal growth or the inoculation of a substantial amount of fungal pellet without the presence of a toxic chemical. In this study, we found that the fungi can co-flocculate with Synechocystis in the medium without an organic carbon source, but with the presence of 10–20 µM EM (Figs. 2, 3, 4, 5) exhibiting bactericidal activity against Synechocystis40. Thus, Synechocystis may provide organic nutrients for fungal growth and EM is the primary mediator for Synechocystis-fungi flocculation.
Previous studies reported that co-flocculation between the eukaryotic algae Chlorella vulgaris and A. oryzae occurred when they encountered toxic arsenic. This co-flocculation helped to remove 27% of the 1.3 µM arsenic in the culture medium49. Additionally, co-flocculation between Chlorella vulgaris and A. flavus reduced up to 98% of 1.6 mM Cu(II)50. Thus, these data suggested that the formation of an algae-fungi flocculate decreased the exposure to the toxic compounds in their environment. This study revealed that in the presence of 10 µM EM, the Synechocystis in the Synechocystis-fungi flocculation survived, as evidenced by their normal blue-green color (Figs. 2e–h, 3b). However, the axenic-culture Synechocystis cells under the same EM concentrations showed obvious chlorotic phenotypes (Fig. 3a) and have been shown previously to have a significantly reduced cell viability40. Thus, the formation of Synechocystis-fungi flocculation protected Synechocystis against chlorosis. We speculated that this might be due to the reduced EM level in the culture medium caused by the co-flocculation or the co-flocculation formation helps protect Synechocystis against EM. Note that the actual EM concentration in the liquid media was not determined in this study (such as from the inhibition zone assay of Escherichia coli toward EM using the liquid-culture samples or by HPLC analysis) due to the subtle EM concentrations used in this study (0–10 µM equivalent to 0–7.3 µg/mL), which are below the minimum inhibitory concentration of EM for E. coli at 20 µg/mL51 and the lowest HPLC sensitivity at 1000–7000 µg/mL52,53. A more sensitive method, such as ELISA, might be used to determine the EM level.
It is noted that the abiotic stress by the CuSO4 exposure can also induce the Synechocystis-fungi flocculation but with the lower flocculation efficiently than that of EM (Fig. 7b). The cultivation of only the fungi in the BG11 medium showed that the fungi did not grow after 7 days of cultivation (Fig. 7c).
It has been reported in the co-flocculation between Chlorella pyrenoidosa and A. oryzae in liquid cultivation containing starch that these two species exchange nutrients. The A. oryzae secreted CO2 and enzymes to digest starch and protein into simple sugars and amino acids, which can be subsequently utilized by C. pyrenoidosa, while C. pyrenoidosa generated O2 for A. oryzae respiration23. In symbiotic lichens, algae, and fungi also exchange various nutrients for co-benefits of their metabolism54,55. Thus, one major known cause for these symbioses and co-flocculation between algae and fungi is the advantages of nutrient exchanges. In this study, Synechocystis and the fungi were cultivated in BG11 medium without organic compounds and the co-flocculation was formed in the presence of EM but not in the absence of EM (Fig. 3b). Thus, these results showed a novel finding that the primary mediating factor in the Synechocystis-fungi co-flocculation is the presence of the antibiotic EM, where the Synechocystis that flocculate with the fungi survive better (Fig. 2e–h). To minimize the bactericidal effect of EM, lower concentrations or short-term treatment of EM are needed to be tested for their efficiency in promoting the bioflocculation.
The transcriptomic analysis revealed the altered expression levels of 21 genes of Synechocystis that are potentially involved in the Synechocystis-fungi flocculation (Table 2). The altered expression of these 21 genes was mainly induced by the EM exposure (Table 2). In addition, the Synechocystis axenic culture treated with EM did not form Synechocystis flocculation (Fig. 3A). Thus, the altered expressions of the 21 genes did not cause Synechocystis-self flocculation, but rather contribute to the attachment between Synechocystis cell surface and the fungi filaments as seen under the microscope (Fig. 2g,h).
The transcriptomic comparison also showed that the presence of the fungi slightly affected the expression levels of the 7 out of the 21 genes (Table 2), indicating that the presence of the fungi was not a major cause for altering these gene expressions. These data are consistent with the results that the co-culture between Synechocystis and the fungi without EM did not lead to the Synechocystis-fungi flocculation (Fig. 5A).
For the known mechanism of cell flocculation in bacteria and cyanobacteria, cell surface molecules, such as EPS, S-layer protein, and pilin, play crucial roles in cell–cell attachment25–27,56–58. For algal-fungi flocculation, previous studies described that cell attachment is mediated by the different electrostatic charges on the cell surface between microalgae and fungi21,28, where highly negative charges on the cyanobacterial EPS26 attach to positive charges of saccharides on the cell surface of fungal hyphae21,28. The outer membrane porin responsible for EPS polymerization and transport (encoded by wzc) was reported to be involved in Synechocystis biofilm formation25. This study also found that wzc expression was highly increased up to 4.6 log2-fold (24-fold) in Synechocystis flocculated with the fungi (Table 2), suggesting EPS transport might be involved in the co-flocculation. However, all the other known EPS biosynthetic genes were not up-regulated in this work [Gene Expression Omnibus (GEO) repository, accession number GSE244152].
Inactivating the main S-layer protein gene sll1951 significantly reduced Synechocystis biofilm formation, indicating that the S-layer protein is required in cell–cell attachment25. In this study, sll1951 and the other two hypothetical S-layer protein genes slr1704 and slr1272 were upregulated at 1.9, 6.3, and 1.4 log2-fold, respectively, in the Synechocystis flocculated with fungi [Table 2 and Gene Expression Omnibus (GEO) repository: accession number GSE244152]. The S-layer protein Sll1951 has a negative charge in BG11 medium59, and so increased sll1951 expression would increase the negative charge on the Synechocystis cell surface and would enhance the electrostatic attachment to the positive charges on the fungal surface. In addition, the S-layer has been demonstrated to help protect Synechocystis from EM exposure59. This data is correlated to the up-regulation of the sll1951, slr1704, and slr1272 S-layer protein genes of the flocculated Synechocystis (Table 2) in the presence of EM in this study.
Bacterial type-IV pilins have been reported to play a role in cell mobility and cell adhesion29,30. Reduced expression of the genes for the three minor pilin proteins (pilA10, pilA11, and pilA12; which exist in the same operon58,60) resulted in shorter pilins, and subsequently increased the cell surface contact and promoted Synechocystis auto-flocculation58. In accord, the expression levels of pil10, pil11, and pil12 were decreased by 1.6 to 5.1 log2-fold (Table 2), and sycrp1, which encodes for a positive transcriptional regulator for the pilA9-pil10-pil11-pil12-slr2019 operon58, was also down-regulated in the Synechocystis flocculated with the fungi in this study [Gene Expression Omnibus (GEO) repository, accession number GSE244152]. In contrast to the minor pilin proteins, the presence of the chaperone protein Hfq to regulate pilin development is required for Synechocystis auto-flocculation27. This study also found that hfq was up-regulated by 2.6 log2-fold (Table 2) in the flocculated Synechocystis.
Hik36 and Hik43 regulatory proteins involved in pilin formation were previously reported to promote Synechocystis auto-flocculation under salt stress61. In this study, hik36 and hik43 were also up-regulated in Synechocystis flocculated with the fungi (Table 2). In addition, the proteins Cph2 (regulates the inhibition of cell motility and is involved in auto-flocculation under blue light exposure62), PurA (produces signaling AMP63), PII (regulates various metabolisms64), PerR (responds to peroxide stress65), and GroEL2 (a temperature-stress response protein66) were also significantly up-regulated in Synechocystis flocculated with fungi (Table 2). It is worth further examining whether these stress-responsive proteins are responsible for activating Synechocystis-fungi flocculation.
Conclusion
This study found that EM mediates the co-flocculation between Synechocystis and the five species of fungi and that the co-flocculation helps Synechocystis to survive EM exposure. Transcriptomic results suggested that the flocculation is mediated by the alteration of Synechocystis pilin and cell surface composition, and this process might be regulated by a number of stress-responsive proteins. Thus, further identifying signal transduction pathways as well as characterizing cell surface alteration in both Synechocystis and the fungi are required to understand the mechanism of the co-flocculation. A methodology for determining biomass proportion between the cyanobacteria and the fungi in the flocculated biomass has to be established. Approaches to minimizing the fungal population and maximizing the cyanobacterial population in the flocculated biomass are also required. These approaches might be screening for optimal medium composition, as well as searching for other flocculation-stimulating compounds (especially those not affecting cyanobacterial growth) and other cyanobacterial/fungal species to achieve the maximal bioflocculation. Alternatively, an antifungal compound at a suitable concentration might be used to limit growth of the fungi. The bioflocculation with no requirement of organic compound reported in this study might be applicable for other photosynthetic microalgae and other fungal species as a novel approach for cell harvest and bioscaffold formation for CO2 capture.
Supplementary Information
Acknowledgements
The authors thank Dr. Jittra Piapukiew for the technical instruction on ITS rDNA analysis, Miss Kobkul Boorachokwiwatand for algal cultivation, and Dr. Robert Butcher for critical proofreading.
Author contributions
P.P., J.L., and T.M. developed the concept and designed the experiments with input from P.I. and A.I. J.L., P.P., T.M., and N.S. conducted the experiments and analyzed the data. P.P., J.L., P.I., and T.M. wrote the manuscript. All authors reviewed the manuscript. T.M. is responsible for funding acquisition.
Funding
This research was funded by the Thailand Science Research and Innovation Fund Chulalongkorn University (to T.M).
Data availability
The transcriptomic datasets generated and analysed during the current study are available in the Gene Expression Omnibus (GEO) repository, accession numbers: GSE244152, GSE256451, and GSE256452.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Panutchaya Pichaiyotinkul and Jidapa Leksingto.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-60016-7.
References
- 1.McCormick AJ, et al. Hydrogen production through oxygenic photosynthesis using the cyanobacterium Synechocystis sp. PCC 6803 in a bio-photoelectrolysis cell (BPE) system. Energy Environ. Sci. 2013;6:2682–2690. doi: 10.1039/c3ee40491a. [DOI] [Google Scholar]
- 2.Monshupanee T, Incharoensakdi A. Enhanced accumulation of glycogen, lipids and polyhydroxybutyrate under optimal nutrients and light intensities in the cyanobacterium Synechocystis sp. PCC 6803. J. Appl. Microbiol. 2014;116:830–838. doi: 10.1111/jam.12409. [DOI] [PubMed] [Google Scholar]
- 3.Baebprasert W, Jantaro S, Khetkorn W, Lindblad P, Incharoensakdi A. Increased H2 production in the cyanobacterium Synechocystis sp. strain PCC 6803 by redirecting the electron supply via genetic engineering of the nitrate assimilation pathway. Metab. Eng. 2011;13:610–616. doi: 10.1016/j.ymben.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 4.Klotz A, et al. Awakening of a dormant cyanobacterium from nitrogen chlorosis reveals a genetically determined program. Curr. Biol. 2016;26:2862–2872. doi: 10.1016/j.cub.2016.08.054. [DOI] [PubMed] [Google Scholar]
- 5.Fu P. Genome-scale modeling of Synechocystis sp. PCC 6803 and prediction of pathway insertion. J. Chem. Technol. Biotechnol. Int. Res. Process Environ. Clean Technol. 2009;84:473–483. [Google Scholar]
- 6.Joshi CJ, Peebles CA, Prasad A. Modeling and analysis of flux distribution and bioproduct formation in Synechocystis sp. PCC 6803 using a new genome-scale metabolic reconstruction. Algal Res. 2017;27:295–310. doi: 10.1016/j.algal.2017.09.013. [DOI] [Google Scholar]
- 7.Qi F, Yao L, Tan X, Lu X. Construction, characterization and application of molecular tools for metabolic engineering of Synechocystis sp. Biotech. Lett. 2013;35:1655–1661. doi: 10.1007/s10529-013-1252-0. [DOI] [PubMed] [Google Scholar]
- 8.Grima EM, Belarbi E-H, Fernández FA, Medina AR, Chisti Y. Recovery of microalgal biomass and metabolites: Process options and economics. Biotechnol. Adv. 2003;20:491–515. doi: 10.1016/S0734-9750(02)00050-2. [DOI] [PubMed] [Google Scholar]
- 9.Uduman N, Qi Y, Danquah MK, Forde GM, Hoadley A. Dewatering of microalgal cultures: A major bottleneck to algae-based fuels. J. Renew. Sustain. Energy. 2010;2:012701. doi: 10.1063/1.3294480. [DOI] [Google Scholar]
- 10.Golueke CG, Oswald WJ. Harvesting and processing sewage-grown planktonic algae. J. Water Pollut. Control Feder. 1965;37:471–498. [Google Scholar]
- 11.Wijffels RH, Barbosa MJ. An outlook on microalgal biofuels. Science. 2010;329:796–799. doi: 10.1126/science.1189003. [DOI] [PubMed] [Google Scholar]
- 12.Huang K-X, et al. Integrated culture and harvest systems for improved microalgal biomass production and wastewater treatment. Bioresour. Technol. 2023;376:128941. doi: 10.1016/j.biortech.2023.128941. [DOI] [PubMed] [Google Scholar]
- 13.Pragya N, Pandey KK, Sahoo P. A review on harvesting, oil extraction and biofuels production technologies from microalgae. Renew. Sustain. Energy Rev. 2013;24:159–171. doi: 10.1016/j.rser.2013.03.034. [DOI] [Google Scholar]
- 14.Gerardo ML, Van Den Hende S, Vervaeren H, Coward T, Skill SC. Harvesting of microalgae within a biorefinery approach: A review of the developments and case studies from pilot-plants. Algal Res. 2015;11:248–262. doi: 10.1016/j.algal.2015.06.019. [DOI] [Google Scholar]
- 15.Iasimone F, et al. Insights into bioflocculation of filamentous cyanobacteria, microalgae and their mixture for a low-cost biomass harvesting system. Environ. Res. 2021;199:111359. doi: 10.1016/j.envres.2021.111359. [DOI] [PubMed] [Google Scholar]
- 16.Fisher ML, Allen R, Luo Y, Curtiss R., III Export of extracellular polysaccharides modulates adherence of the cyanobacterium Synechocystis. PLoS ONE. 2013;8:e74514. doi: 10.1371/journal.pone.0074514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iasimone F, et al. Bioflocculation and settling studies of native wastewater filamentous cyanobacteria using different cultivation systems for a low-cost and easy to control harvesting process. J. Environ. Manag. 2020;256:109957. doi: 10.1016/j.jenvman.2019.109957. [DOI] [PubMed] [Google Scholar]
- 18.Magdouli S, Brar SK, Blais J-F. Co-culture for lipid production: Advances and challenges. Biomass Bioenergy. 2016;92:20–30. doi: 10.1016/j.biombioe.2016.06.003. [DOI] [Google Scholar]
- 19.Choi Y-N, et al. Efficient harvesting of Synechocystis sp. PCC 6803 with filamentous fungal pellets. J. Appl. Phycol. 2016;28:2225–2231. doi: 10.1007/s10811-015-0787-y. [DOI] [Google Scholar]
- 20.Jiang L, et al. Evidence for a mutualistic relationship between the cyanobacteria Nostoc and fungi Aspergilli in different environments. Appl. Microbiol. Biotechnol. 2020;104:6413–6426. doi: 10.1007/s00253-020-10663-3. [DOI] [PubMed] [Google Scholar]
- 21.Miranda AF, et al. Lipid production in association of filamentous fungi with genetically modified cyanobacterial cells. Biotechnol. Biofuels. 2015;8:1–18. doi: 10.1186/s13068-015-0364-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oliveira HR, Bassin ID, Cammarota MC. Bioflocculation of cyanobacteria with pellets of Aspergillus niger: Effects of carbon supplementation, pellet diameter, and other factors in biomass densification. Bioresour. Technol. 2019;294:122167. doi: 10.1016/j.biortech.2019.122167. [DOI] [PubMed] [Google Scholar]
- 23.Wang S-K, et al. One-step co-cultivation and flocculation of microalgae with filamentous fungi to valorize starch wastewater into high-value biomass. Bioresour. Technol. 2022;361:127625. doi: 10.1016/j.biortech.2022.127625. [DOI] [PubMed] [Google Scholar]
- 24.Bhattacharya A, Mathur M, Kumar P, Prajapati SK, Malik A. A rapid method for fungal assisted algal flocculation: Critical parameters & mechanism insights. Algal Res. 2017;21:42–51. doi: 10.1016/j.algal.2016.10.022. [DOI] [Google Scholar]
- 25.Allen R, Rittmann BE, Curtiss R., III Axenic biofilm formation and aggregation by Synechocystis sp. strain PCC 6803 are induced by changes in nutrient concentration and require cell surface structures. Appl. Environ. Microbiol. 2019;85:e02192–02118. doi: 10.1128/AEM.02192-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Babiak W, Krzemińska I. Extracellular polymeric substances (EPS) as microalgal bioproducts: A review of factors affecting EPS synthesis and application in flocculation processes. Energies. 2021;14:4007. doi: 10.3390/en14134007. [DOI] [Google Scholar]
- 27.Conradi FD, et al. Factors controlling floc formation and structure in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 2019;201:e00344–00319. doi: 10.1128/JB.00344-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J, Hu B. A novel method to harvest microalgae via co-culture of filamentous fungi to form cell pellets. Bioresour. Technol. 2012;114:529–535. doi: 10.1016/j.biortech.2012.03.054. [DOI] [PubMed] [Google Scholar]
- 29.Strom MS, Lory S. Structure-function and biogenesis of the type IV pili. Annu. Rev. Microbiol. 1993;47:565–596. doi: 10.1146/annurev.mi.47.100193.003025. [DOI] [PubMed] [Google Scholar]
- 30.Nakane D, Nishizaka T. Asymmetric distribution of type IV pili triggered by directional light in unicellular cyanobacteria. Proc. Natl. Acad. Sci. USA. 2017;114:6593–6598. doi: 10.1073/pnas.1702395114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhaya D, Bianco NR, Bryant D, Grossman A. Type IV pilus biogenesis and motility in the cyanobacterium Synechocystis sp. PCC6803. Mol. Microbiol. 2000;37:941–951. doi: 10.1046/j.1365-2958.2000.02068.x. [DOI] [PubMed] [Google Scholar]
- 32.Chen Z, Li X, Tan X, Zhang Y, Wang B. Recent advances in biological functions of thick pili in the cyanobacterium Synechocystis sp. PCC 6803. Front. Plant Sci. 2020;11:241. doi: 10.3389/fpls.2020.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rokem JS, Lantz AE, Nielsen J. Systems biology of antibiotic production by microorganisms. Nat. Product Rep. 2007;24:1262–1287. doi: 10.1039/b617765b. [DOI] [PubMed] [Google Scholar]
- 34.Netzker T, et al. Microbial interactions trigger the production of antibiotics. Curr. Opin. Microbiol. 2018;45:117–123. doi: 10.1016/j.mib.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 35.Høiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents. 2010;35:322–332. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 36.Abebe GM. The role of bacterial biofilm in antibiotic resistance and food contamination. Int. J. Microbiol. 2020;2020:1705814. doi: 10.1155/2020/1705814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Z, et al. Advanced insights on removal of antibiotics by microalgae-bacteria consortia: A state-of-the-art review and emerging prospects. Chemosphere. 2022;307:136117. doi: 10.1016/j.chemosphere.2022.136117. [DOI] [PubMed] [Google Scholar]
- 38.Leng L, et al. Use of microalgae based technology for the removal of antibiotics from wastewater: A review. Chemosphere. 2020;238:124680. doi: 10.1016/j.chemosphere.2019.124680. [DOI] [PubMed] [Google Scholar]
- 39.Pomati F, Netting AG, Calamari D, Neilan BA. Effects of erythromycin, tetracycline and ibuprofen on the growth of Synechocystis sp. and Lemna minor. Aquat. Toxicol. 2004;67:387–396. doi: 10.1016/j.aquatox.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 40.Sukkasam N, et al. Erythromycin treatment under a specific nitrogen supply affects carbon metabolism and increases poly(3-hydroxybutyrate) and glycogen accumulation in cyanobacterium Synechocystis sp. PCC 6803. Algal Res. 2023;72:103142. doi: 10.1016/j.algal.2023.103142. [DOI] [Google Scholar]
- 41.Svetlov MS, et al. High-resolution crystal structures of ribosome-bound chloramphenicol and erythromycin provide the ultimate basis for their competition. RNA. 2019;25:600–606. doi: 10.1261/rna.069260.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. Microbiology. 1979;111:1–61. doi: 10.1099/00221287-111-1-1. [DOI] [Google Scholar]
- 43.Monshupanee T, Nimdach P, Incharoensakdi A. Two-stage (photoautotrophy and heterotrophy) cultivation enables efficient production of bioplastic poly-3-hydroxybutyrate in auto-sedimenting cyanobacterium. Sci. Rep. 2016;6:37121. doi: 10.1038/srep37121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pichaiyotinkul P, Ruankaew N, Incharoensakdi A, Monshupanee T. Enhanced polyglucan contents in divergent cyanobacteria under nutrient-deprived photoautotrophy: Transcriptional and metabolic changes in response to increased glycogen accumulation in nitrogen-deprived Synechocystis sp. PCC 6803. World J. Microbiol. Biotechnol. 2023;39:27. doi: 10.1007/s11274-022-03476-1. [DOI] [PubMed] [Google Scholar]
- 45.Schubert H, Fulda S, Hagemann M. Effects of adaptation to different salt concentrations on photosynthesis and pigmentation of the cyanobacterium Synechocystis sp. PCC 6803. J. Plant Physiol. 1993;142:291–295. doi: 10.1016/S0176-1617(11)80425-6. [DOI] [Google Scholar]
- 46.Giner-Lamia J, López-Maury L, Florencio FJ. Global transcriptional profiles of the copper responses in the cyanobacterium Synechocystis sp. PCC 6803. PLoS ONE. 2014;9:e108912. doi: 10.1371/journal.pone.0108912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nie Y, et al. Bio-flocculation of Microcystis aeruginosa by using fungal pellets of Aspergillus oryzae: Performance and mechanism. J. Hazard. Mater. 2022;439:129606. doi: 10.1016/j.jhazmat.2022.129606. [DOI] [PubMed] [Google Scholar]
- 48.Prajapati SK, Kumar P, Malik A, Choudhary P. Exploring pellet forming filamentous fungi as tool for harvesting non-flocculating unicellular microalgae. Bioenergy Res. 2014;7:1430–1440. doi: 10.1007/s12155-014-9481-1. [DOI] [Google Scholar]
- 49.Li B, Zhang T, Yang Z. Immobilizing unicellular microalga on pellet-forming filamentous fungus: Can this provide new insights into the remediation of arsenic from contaminated water? Bioresource Technology. 2019;284:231–239. doi: 10.1016/j.biortech.2019.03.128. [DOI] [PubMed] [Google Scholar]
- 50.Zhang C, et al. Capturing effects of filamentous fungi Aspergillus flavus ZJ-1 on microalgae Chlorella vulgaris WZ-1 and the application of their co-integrated fungi-algae pellets for Cu(II) adsorption. J. Hazard. Mater. 2023;442:130105. doi: 10.1016/j.jhazmat.2022.130105. [DOI] [PubMed] [Google Scholar]
- 51.Sun Y, et al. Erythromycin loaded by tetrahedral framework nucleic acids are more antimicrobial sensitive against Escherichia coli (E. coli) Bioactive Mater. 2021;6:2281–2290. doi: 10.1016/j.bioactmat.2020.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Griessmann K, Kaunzinger A, Schubert-Zsilavecz M, Abdel-Tawab M. A rapid HPLC-UV method for the quantification of erythromycin in dermatological preparations. Die Pharmazie Int. J. Pharm. Sci. 2007;62:668–671. [PubMed] [Google Scholar]
- 53.Hassib ST, Farag AE, Elkady EF. Liquid chromatographic and spectrophotometric methods for the determination of erythromycin stearate and trimethoprim in tablets. Bull. Facul. Pharm. 2011;49:81–89. [Google Scholar]
- 54.Leng L, et al. Co-culture of fungi-microalgae consortium for wastewater treatment: A review. Bioresour. Technol. 2021;330:125008. doi: 10.1016/j.biortech.2021.125008. [DOI] [PubMed] [Google Scholar]
- 55.Kono M, Kon Y, Ohmura Y, Satta Y, Terai Y. In vitro resynthesis of lichenization reveals the genetic background of symbiosis-specific fungal-algal interaction in Usnea hakonensis. BMC Genomics. 2020;21:671. doi: 10.1186/s12864-020-07086-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Limoli DH, Jones CJ, Wozniak DJ. Bacterial extracellular polysaccharides in biofilm formation and function. Microb. Biofilms. 2015;3:223–247. doi: 10.1128/9781555817466.ch11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karatan E, Watnick P. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol. Mol. Biol. Rev. 2009;73:310–347. doi: 10.1128/MMBR.00041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chandra A, Joubert L-M, Bhaya D. Modulation of type IV pili phenotypic plasticity through a novel chaperone-usher system in Synechocystis sp. Bio Arch. 2017;66:130–278. [Google Scholar]
- 59.Agarwal R, Whitelegge JP, Saini S, Shrivastav AP. The S-layer biogenesis system of Synechocystis 6803: Role of Sll1180 and Sll1181 (E. coli HlyB and HlyD analogs) as type-I secretion components for Sll 1951 export. Biochimica et Biophysica Acta BBA Biomembr. 2018;1860:1436–1446. doi: 10.1016/j.bbamem.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 60.Oeser S, et al. Minor pilins are involved in motility and natural competence in the cyanobacterium Synechocystis sp PCC 6803. Mol. Microbiol. 2021;116:743–765. doi: 10.1111/mmi.14768. [DOI] [PubMed] [Google Scholar]
- 61.Kera K, et al. Hik36–Hik43 and Rre6 act as a two-component regulatory system to control cell aggregation in Synechocystis sp. PCC 6803. Sci. Rep. 2020;10:1–9. doi: 10.1038/s41598-020-76264-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Enomoto G, Wallner T, Wilde A. Control of light-dependent behaviour in cyanobacteria by the second messenger cyclic di-GMP. Microlife. 2023;4:uqad019. doi: 10.1093/femsml/uqad019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vázquez-Ciros OJ, Alvarez AF, Georgellis D. Identification of Z nucleotides as an ancient signal for two-component system activation in bacteria. Proc. Natl. Acad. Sci. USA. 2020;117:33530–33539. doi: 10.1073/pnas.2006209117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Forchhammer K, Selim KA, Huergo LF. New views on PII signaling: From nitrogen sensing to global metabolic control. Trends Microbiol. 2022;30:722–735. doi: 10.1016/j.tim.2021.12.014. [DOI] [PubMed] [Google Scholar]
- 65.Grifantini R, Toukoki C, Colaprico A, Gryllos I. Peroxide stimulon and role of PerR in group A Streptococcus. J. Bacteriol. 2011;193:6539–6551. doi: 10.1128/JB.05924-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sato S, Ikeuchi M, Nakamoto H. Expression and function of a groEL paralog in the thermophilic cyanobacterium Thermosynechococcus elongatus under heat and cold stress. FEBS Lett. 2008;582:3389–3395. doi: 10.1016/j.febslet.2008.08.034. [DOI] [PubMed] [Google Scholar]
- 67.Santos, M. P. F. Unveiling Biosynthetic Pathways for the Production of Extracellular Polymeric Substances (Eps) in Cyanobacteria and Possible Aplications of the Polymers. Ph.D. Thesis, Universidade do Porto (2021).
- 68.de Bento Flores, C. E. Extracellular Polymeric Substances (EPS) from the Cyanobacterium Synechocystis sp. Pcc 6803: From Genes to Polymer Application as Antitumor Agent. Ph.D. Thesis, Universidade do Porto (2019).
- 69.Wallner T, Pedroza L, Voigt K, Kaever V, Wilde A. The cyanobacterial phytochrome 2 regulates the expression of motility-related genes through the second messenger cyclic di-GMP. Photochem. Photobiol. Sci. 2020;19:631–643. doi: 10.1039/c9pp00489k. [DOI] [PubMed] [Google Scholar]
- 70.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li H, Singh AK, McIntyre LM, Sherman LA. Differential gene expression in response to hydrogen peroxide and the putative PerR regulon of Synechocystis sp. strain PCC 6803. J. Bacteriol. 2004;186:3331–3345. doi: 10.1128/JB.186.11.3331-3345.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Giner-Lamia J, López-Maury L, Florencio FJ. Global transcriptional profiles of the copper responses in the cyanobacterium Synechocystis sp. PCC 6803. PLoS ONE. 2014;9:e108912. doi: 10.1371/journal.pone.0108912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ruan G, Mi W, Yin X, Song G, Bi Y. Molecular responses mechanism of Synechocystis sp. PCC 6803 to cadmium stress. Water. 2022;14:4032. doi: 10.3390/w14244032. [DOI] [Google Scholar]
- 74.Klähn S, et al. Integrative analysis of the salt stress response in cyanobacteria. Biol. Direct. 2021;16:26. doi: 10.1186/s13062-021-00316-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Suzuki I, et al. The histidine kinase hik34 is involved in thermotolerance by regulating the expression of heat shock genes in Synechocystis. Plant Physiol. 2005;138:1409–1421. doi: 10.1104/pp.104.059097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The transcriptomic datasets generated and analysed during the current study are available in the Gene Expression Omnibus (GEO) repository, accession numbers: GSE244152, GSE256451, and GSE256452.