Abstract
Several types of non-coding RNAs such as circRNAs, lncRNAs, and miRNAs have been identified to regulate mRNAs through the mechanism known as the competitive endogenous RNA (ceRNA) network. To explore the role of the ceRNA regulatory network in the immune microenvironment of bladder cancer, whole-transcriptome sequencing of bladder tumor and its peritumoral tissues from 38 bladder cancer patients, with a total of 63 samples, was performed to screen differentially expressed circ-, lnc-, mi-, and mRNAs to construct a circ/lnc-mi-mRNA regulatory network with pruning algorithms. We excavated a key immune-related gene BDNF to build the final ceRNA network as hsa-miR-107 sponged by hsa-circ-000211, AC108488.1, and LINC00163. Finally, a meta-analysis of 7 public datasets demonstrated that low expression of BDNF and high expression of hsa-miR-107 were associated with longer survival. Our study identified a ceRNA regulatory network as a potentially new prognostic marker and molecular therapeutic target of bladder cancer.
Keywords: Bladder cancer, Whole transcriptome sequencing, Competitive endogenous RNA, Immune microenvironment, BDNF
Highlights
-
•
We dissected the immune landscapes of both the tumor and peritumoral tissues.
-
•
Whole-transcriptome sequencing defined abundant circ-, lnc-, mi-, and mRNAs for the ceRNA pipeline construction.
-
•
Low-expressed hsa_circ_0000211, AC108488.1, and LINC00163 might sponge hsa-miR-107 to down-regulate the expression of BDNF.
-
•
Low expression of BDNF and high expression of hsa-miR-107 were associated with a good prognosis of bladder cancer.
1. Introduction
Bladder cancer is known to be immunogenic and highly responsive to immunotherapy including intravesical Bacillus-Calmette–Guerin (BCG) and immune checkpoint inhibitors [[1], [2], [3]]. Currently, the intravesical BCG for the treatment of non-muscle-invasive bladder cancer is the most common and one of the most successful immunotherapies for patients with bladder cancer [4]. Additionally, several lines of evidence have shown that bladder cancer is one of the malignant tumors with high tumor mutation burdens (TMBs) [5], which is a reliable effectiveness predictor of checkpoint inhibitors [6]. However, the heterogeneity of the immunosuppressive and tolerogenic tumor microenvironment (TME) of bladder cancer can not be ignored and the mechanisms of immune evasion need further exploration. As a result, an improved understanding of the immune landscape and immune microenvironment of bladder cancer may lead to the finding of new targets for immunotherapy.
Nowadays, progressing omic techniques have revolutionized cancer biology and immunology. The transcriptome landscape and molecular regulations of bladder cancer have been increasingly described [7,8]. The high-throughput whole transcriptome sequencing has revealed several types of non-coding RNAs (ncRNAs) such as circRNAs, lncRNAs, and miRNAs, which regulate mRNAs through different mechanisms. miRNAs are known as major players in posttranscriptional gene regulation, while circRNAs or lncRNAs function as miRNA sponges and competitively bind to miRNAs to regulate the expression of target mRNAs, which is called the competitive endogenous RNA (ceRNA) hypothesis [9]. For example, circ_ITCH sponges miR-224 upregulating the expression of PTEN[10], circ_NR3C1 sponges miR-27a-3p downregulating cyclin D1 expression [11], and LINC00612 sponges miR-509 elevating the expression of PHF14[12] in bladder cancer. Previous studies have revealed that fifty-five circRNAs are associated with bladder cancer specifically via the ceRNA mechanism [13]. However, the role of the ceRNA regulatory network in the immune microenvironment of bladder cancer remains elusive. Therefore, identifying the immune-related genes that may act as diagnostic or prognostic markers for bladder cancer and disclosing the related circ-, lnc-, and miRNAs via a ceRNA-regulated network will be of great importance.
In this study, we performed whole-transcriptome sequencing of bladder tumor specimens and corresponding peritumoral tissues to identify differentially expressed circ-, lnc-, mi-, and mRNAs and constructed the ceRNA regulatory network with pruning algorithms. The differentially expressed mRNAs were filtered with known immune-related genes and subjected to WGCNA analysis to find out the key immune-related gene. We determined BDNF, known as Brain-Derived Neurotrophic Factor, which was the downregulated gene in bladder tumor tissues, as the key immune-related gene and constructed its ceRNA regulatory network as hsa-miR-107 sponged by one circRNA and two lncRNAs (has-circ-000211, AC108488.1, and LINC00163). Furthermore, the low expression of BDNF and high expression of hsa-miR-107 were found to be associated with better prognosis of bladder cancer patients in the meta-analysis of 7 public bladder cancer datasets. Our study thus gains novel insights into the immune regulatory network, which provides the rationale for exploring new immunotherapy targets.
2. Materials and methods
2.1. Sample collection and preparation
63 samples from 38 patients with bladder cancer who underwent neoadjuvant-free-treatment cystectomy were previously collected in Hubei Cancer Hospital from 2019 to 2022. The samples were prepared and sequenced individually in two batches (26 samples from 15 patients in batch one, 37 samples from 23 patients in batch 2), in which 25 pairs of cancer and peritumoral tissues, and the other 13 cancer tissues alone were pathologically confirmed. All samples were being frozen in liquid nitrogen and preserved at −80 °C before RNA extraction. The clinical and pathological characteristics of the samples were shown in Supplementary Table 1. The experiment had been approved by the Ethics Committee of Huazhong Agricultural University (HZAUHU-2018-012) and all patients had signed preoperative informed consent.
2.2. RNA quantification and qualification
Total RNA degradation and contamination of the frozen samples were monitored on 1 % agarose gels. RNA purity was checked using the NanoPhotometer® spectrophotometer (IMPLEN, CA, USA). RNA concentration was measured using Qubit® RNA Assay Kit in Qubit® 2.0 Fluorometer (Life Technologies, CA, USA). RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA).
2.3. Library construction and sequencing
Sequencing libraries were generated using NEBNext® Multiplex Small RNA Library Prep Set for Illumina® (NEB, USA) following the manufacturer's recommendations. The first-strand cDNA was synthesized using M-MuLV Reverse Transcriptase (RNase H–). PCR amplification was performed using LongAmp Taq 2X Master Mix, SR Primer for Illumina, and index (X) primer. PCR products were purified and harvested. Then, library quality was assessed on the Agilent Bioanalyzer 2100 system using DNA High Sensitivity Chips. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq SR Cluster Kit v3-cBot-HS (Illumia) according to the manufacturer's instructions. After cluster generation, the library preparations were sequenced on an Illumina Hiseq 2500/2000 platform and 50bp single-end reads were generated.
2.4. Differential expression analysis
The upstream sequencing analysis followed the RNA-Seq standard workflow. After quality control of the sequencing data, the analysis of differentially expressed genes (DEGs) was done by comparing tumor and peritumoral samples in each sequencing batch using the R package DESeq2[14]. The thresholds DEGs identification were |log2FC| > 1 and P-value <0.05. The function removeBatchEffect from limma [3] package was applied to correct systemic batch effect in separate batches and the batch effects were visualized with principal components analysis (PCA). The two batches of sequencing data were subsequently merged. Chromosome distributions of DEGs were illustrated with chromPlot [15] package. Chromosome distributions of DE-miRNAs and DE-mRNAs in the TCGA-BLCA dataset were also displayed as a validation. Heatmaps were drawn for the circ-, lnc-, mi-, and mRNA DEGs with R package ComplexHeatmap [16,17] and circlize [18]. Radargrams were generated using ggradar (https://github.com/ricardo-bion/ggradar). Volcano plots were plotted with the above four types of DEGs in R.
2.5. CeRNA network construction
The initial ceRNA network was constructed from several public databases based on co-expression regulation relationships of the lnc-miRNA, circ-miRNA, and mi-mRNA pairs, with a |Pearson's correlation coefficient| of more than 0.4 thresholds for co-expression edges linkage. In which we integrated the circ-miRNA regulatory relationships with the intersection of ENCORI (http://starbase.sysu.edu.cn/) and circbank (http://www.circbank.cn/); and lnc-miRNA regulatory relationships with the intersection of ENCORI, NPInter (http://bigdata.ibp.ac.cn/npinter4), miRcode (http://www.mircode.org/); and mi-mRNA regulatory relationships that intersected in TargetScan (https://www.targetscan.org/), ENCORI; mir2disease (http://www.mir2disease.org/), miRactDB (https://ccsm.uth.edu/miRactDB/), miRcode (http://www.mircode.org/), miRDB (http://mirdb.org/), miRWalk 2.0 (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/custom.html), and NPInter.
To remove the potential redundancy and consideration of the statistic bias as well as biological meaning, a biological function mathematical modeling algorithm inherited from initial ceRNA performance evaluation pruning was established [19]. The first step was to compute the overall regulation similarity of all shared miRNAs on the lnc-mRNA and circ-mRNA pairs. The RegSim (regulation similarity) ! = 0 was defined as regulatory functions.
M is the total number of shared miRNAs, is the number k shared miRNA (, and , and represent the Pearson's correlation coefficients of the number k miRNA and lncRNA, the number k miRNA and circRNA, and the number k miRNA and mRNA, respectively.
The second step was to calculate the circRNA (or lncRNA) competitive mechanisms according to correlation sensitivity (the difference between Pearson's correlation and partial correlation coefficients) for each lnc-mi-mRNA and circ-mi-mRNA triplet. The CorPvalue (sensitivity correlation) threshold was <0.01.
and refer to Pearson's correlation coefficient of lncRNA and mRNA, circRNA and mRNA, respectively.
Based on the commonly shared miRNA and mRNA in these two triplets, the circ/lnc-mi-mRNA filtered quadruple network was constructed and Cytoscape (version, 3.8.2) was used for visualization. Moreover, the target associations within the complicated circ/lnc-mi-mRNA network were visualized as a genome coverage plot created by the R package Circos [20]. In which the outer ring of chromosomes was loaded from UCSC.HG38.Human.CytoBandIdeogram with the default setting of Ideograms and highlights axis. Gene-to-chromosome location mappings were obtained using the DEGs in UCSC genes. The inside tracks were imported from the results of 4 differential types of gene analyses with corresponding ceRNA linkage.
2.6. Single sample gene set enrichment analysis (ssGSEA)
ssGSEA was performed to quantify activity profiles of the 28 immune cell types infiltrating each cancer and peritumoral sample from the DE-mRNAs’ matrix using the R GSVA package [21] with default settings. The scores calculated by ssGSEA of each signature were presented with a heatmap and the 28 cell types were selected to be displayed separately. Gene signature for each immune cell type was obtained from previously published data [22,23]. The TME score of each sample was also calculated by the estimate algorithm (R package estimate [24]), the stomal, immune, and estimate scores were outputted by performing ssGSEA. The ssGSEA enrichment scores and the TME scores were then compared between tumor and peritumoral samples to depict and assess the immune landscape between them by Wilcox t-test. Gene set enrichment analysis (GSEA) of the whole DE-mRNAs was adopted using the R package clusterprofiler [25,26], and the immune-related Biological Process Gene Ontology (GO-BP) terms and the Kyoto Encyclopedia of Genes and Genomes (KEGG) [27] pathways were enriched.
2.7. Weighted gene co-expression network analysis (WGCNA)
Immune-related genes downloaded from ImmPort [28] (https://www.immport.org/home) and InnateDB[29] (https://www.innatedb.com/) were filtered with our DE-mRNAs. Next, the immune-related DE-mRNAs’ matrix was supplied to WGCNA analysis, which was conducted with the R package WGCNA[30]. The biocorrelation algorithm was used to generate the gene correlation matrix and the pickSoftThreshold function was applied to determine the proper soft-threshold power, which was 14. The TOM transformation and Dynamic Tree Cut module finding (minModuleSize = 100) on each network were also carried out. The most tissue-specific modules were identified by comparing the differences between the tumor and the peritumoral tissues. The exportNetworkToCytoscape function, threshold = 0.1, was applied as exporting edges as well as nodes of the top four modules with the most significant co-expression relationships (on a threshold of p-value ≤ 0.01), and those top modules’ genes were sent to Cytoscape. The MCODE plugin of Cytoscape was employed to choose the top two highest MCODE score clusters and the network was then visualized separately.
Thus, based on the intersection of genes from the circ/lnc-mi-mRNA filtered quadruple network built in step 5 and hub genes from MCODE, the only gene BDNF was excavated by Venn diagram using ggVennDiagram [31] package. And following the regulatory relationship between circRNA-miRNA, lncRNA-miRNA, and miRNA-mRNA, the final ceRNA network was constructed. The R package corrplot (https://cran.r-project.org/web/packages/corrplot/vignettes/corrplot-intro.html) was used to generate a correlation heatmap.
2.8. RNA fluorescence in situ hybridization (FISH)
The specific probes for target RNA were designed by spatial FISH Ltd. Fresh samples from a bladder cancer patient after radical cystectomy were fixed with 4 % paraformaldehyde and covered with a reaction chamber. After dehydration and denaturation of samples with methanol, the hybridization buffer with specific targeting probes was added to the chamber for incubation at 37 °C overnight. Afterward, samples were washed three times with PBST, followed by ligation of target probes in ligation mix at 25 °C for 3 h. Next, samples were washed three times again with PBST and underwent rolling circle amplification with Phi29 DNA polymerase at 30 °C overnight. After designing padlock probes with corresponding barcodes for different target genes, the probes target and capture target molecules through in situ hybridization, the signals are amplified in situ in the tissue through rolling circle amplification. Subsequently, the fluorescent detection probes in the hybridization buffer were applied to the samples. Finally, samples were dehydrated with an ethanol series and mounted with a mounting medium. After capturing images, signal dots were decoded to interpret RNA spatial position information.
2.9. Luciferase reporter assay
293T cells were cultured in a DMEM solution containing 10 % FBS, Penicillin-Streptomycin, and Glutamax. 5 % CO2 was added to a humidified incubator at 37 °C for the cells growing. The wild-type or mutant BDNF (WT/Mut-BDNF) were separately amplified and cloned into pmirGLO vectors. In order to co-transfect the luciferase reporter with WT/Mut-BDNF-pmirGLO, hsa-miR-107 mimics, and mimics-NC into 293T cells, Lipofectamine 3000 (Thermofisher) was used according to the manufacturer's guidelines. After 48 h, cells were harvested and luciferase activity was assayed using the Luciferase Assay Reagent II kit from Dual-Luciferase® Reporter Assay System (DLR™, Biyuntian, China). Results were expressed as relative luciferase activity using Renilla luciferase activity as the internal control.
2.10. Meta-analysis of public datasets
To explore how the down-regulated BDNF or the up-regulated hsa-miR-107 influence the survival of patients with bladder cancer, several public databases from The Cancer Genome Atlas (TCGA, https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga), Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/gds), which comprises 7 bladder cancer RNA-Seq or chip datasets (TCGA-BLCA, GSE13507, GSE19423, GSE19915, GSE32894, GSE84525, GSE160693) with follow-up information were integrated meta-analyzed. Among these the chip sequencing data were normalized using the normalizeBetweenArrays function in R package limma. While the TCGA-BLCA dataset was converted from FPKM to TPM. The overall survival of patients with bladder cancer was estimated by Kaplan-Meier methods with the log-rank test.
All statistical analysis was performed using R (version 4.1.3) and R Studio for Windows (version April 1, 1103). The P values ≥ 0.05 = ns, <0.05 = *, <0.01 = **, P values < 0.001 = ***.
3. Results
3.1. Whole-transcriptome sequencing identified key differentially expressed circRNAs, lncRNAs, miRNAs, and mRNAs associated with bladder cancer
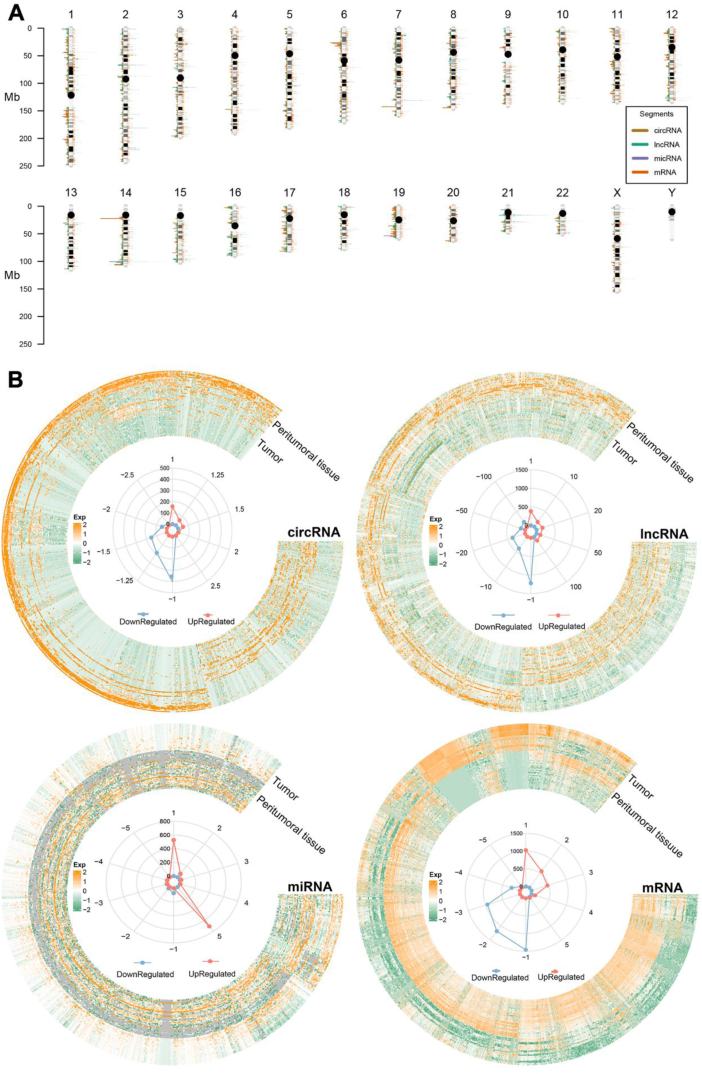
Whole transcriptome RNA-sequencing analysis identified a total of 343 differentially expressed circRNAs (DE-circRNAs), 3198 differentially expressed lncRNAs (DE-lncRNAs), 709 differentially expressed miRNAs (DE-miRNAs), and 6102 differentially expressed mRNAs (DE-mRNAs) (see Methods). Among these DEGs, 111 circ-, 950 lnc-, 606 mi-, and 2199 mRNAs were detected as up-regulated while 406 circ-, 2249 lnc-, 104 mi-, and 3903 mRNAs were down-regulated. Upon PCA, the results showed no obvious bias or batch effect (Supplementary Fig. 1). Chromosome distributions and heatmaps with hierarchical clustering of DE circ-, lnc-, mi-, and mRNAs were shown in Fig. 1. Volcano plots of DEGs were displayed in Supplementary Fig. 2. Overall, the DEGs were distributed evenly along the chromosome without preference, which was consistent with the TCGA-BLCA dataset albeit with DE mi- and mRNAs alone (Supplementary Fig. 3). Similarly, heatmaps showed good differentiation between the up- and down-regulated DEGs: a general trend was elucidated that the up-regulated DE circ-, lnc-, and mRNAs were much more than the down-regulated ones, while DE miRNAs showed the opposite trend. The radargrams also indicated good agreements.
Fig. 1.
Heatmap of Chromosome distributions (A), and heatmaps and radargrams (B) of differentially expressed circ-, lnc-, mi-, and mRNAs.
3.2. Construction of the DE-circ, DE-lnc, DE-mi, and DE-mRNAs regulation network
We analyzed the circRNA-mRNA and lncRNA-mRNA co-expression relationships, as well as the circRNA-miRNA, lncRNA-miRNA, and miRNA-mRNA regulation relationships (see methods). We identified 263 circRNA-miRNA-mRNA interactions, including 5 up-regulated and 16 down-regulated DE-cirRNAs, 7 up-regulated and 6 down-regulated DE-miRNAs, 6 up-regulated and 22 down-regulated DE-mRNAs (Fig. 2 A). Furthermore, 249 lncRNA-miRNA-mRNA interactions were obtained, containing 7 up-regulated and 20 down-regulated DE-lncRNAs, 7 up-regulated and 6 down-regulated DE-miRNAs, 6 up-regulated, and 22 down-regulated DE-mRNAs (Fig. 2 B). We successfully constructed the integrated ceRNA regulatory network composed of the above circ/lnc-mi-mRNAs interactions (Fig. 2C). By employing pruning algorithms, we finally harvested a quadruple network within 35 circRNA-miRNA-mRNA and 41 lncRNA-miRNA-mRNA interactions, containing 2 up-regulated and 10 down-regulated DE-cirRNAs, 3 up-regulated and 12 down-regulated DE-lncRNAs, 2 up-regulated and 6 down-regulated DE-miRNAs, 3 up-regulated and 6 down-regulated DEG-mRNAs (Fig. 2 D). A circos plot in Supplementary Fig. 4 displayed the structural coordinates within the whole genome as well as ncRNA regulatory distributions of the quadruple network. The chromosomes had a homogenous distribution without preference.
Fig. 2.
Regulation networks between DE circ-, lnc-, mi-, and mRNAs. (A) The regulation network between DE circ-, mi-, and mRNAs; (B) The regulation network between DE lnc-, mi-, and mRNAs; (C) The integrated regulation network between DE circ-, lnc-, mi-, and mRNAs; (D) The circ/lnc-mi-mRNA filtered quadruple network after pruning algorithms.
3.3. The landscape of immune microenvironment in tumor and peritumoral tissues
To investigate the difference in the immune microenvironment between the tumor and peritumoral tissues, ssGSEA analysis was conducted on all 6102 DE-mRNAs. The results showed the ssGSEA scores were clearly distinguished (Fig. 3 A). Furthermore, a complex immune landscape that the score of most immune cells such as activated B cell, central memory CD4 T cell, central memory CD8 T cell, effector memory CD8 T cell, immature B cell, regulatory T cell, T follicular helper cell, type 1 T helper cell, eosinophil, macrophage, mast cell, MDSC (Myeloid-derived suppressor cells), natural killer cell, natural killer T cell, and plasmacytoid dendritic cell of the tumor was lower than that of peritumoral tissues (Fig. 3 B). The same effect was observed for the TME scores (Fig. 2C). These results indicate that the peritumoral tissues present a higher degree of immune cell infiltration but lower tumor purity, which was compatible with reality.
Fig. 3.
Analysis of the immune microenvironment in the tumor and peritumoral tissues, enrichment analysis of DE mRNAs, WGCNA analysis of immune-related DE mRNAs and identification of key modules and hub genes. (A) Heatmap of ssGSEA scores of the bladder tumor and peritumoral tissues. (B) Violin plot of the immune microenvironment scores between the bladder tumor and peritumoral tissues. (C) Violin plot of the ssGSEA scores of 28 immune cell types between the bladder tumor and peritumoral tissues. GSEA-GO-BP (D) and GSEA-KEGG (E) analysis of DE mRNAs identification immune-related GO-terms or KEGG-pathways. (F) Unsupervised co-expression module-based clustering classified immune-related DE mRNAs into six modules and (G) the module-trait relationships between all six gene modules of the tumor and peritumoral tissues. (H) (I) The Cytoscape plugin MCODE extracts the top two clusters of hub genes based on the top 4 modules (magenta, green, pink and brown) from WGCNA. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
We next performed GSEA analysis on all 6102 DE-mRNAs and identified numerous GO-BP terms and KEGG pathways that related to immune regulation, immune cell types, immune response, or immune cell states. GO-BP results showed that 10 immune-related terms were enriched such as regulation of T cell activation, regulation of immune system process, immune response, and B cell receptor signaling pathway (Fig. 3 D). KEGG pathway results showed that 10 immune-related pathways were enriched, e.g., the JAK-STAT signaling pathway, cytokine-cytokine receptor interaction, chemokine signaling pathway, and PI3K-Akt signaling pathway (Fig. 3 E).
3.4. WGCNA analysis of DE-mRNAs identified the key modules and hub immune-related genes
A total of 2519 immune-related genes were downloaded from ImmPort and InnateDB, and 624 genes were filtered within our DE-mRNAs. Then the 624 immune-related DE-mRNAs’ matrix was recruited for WGCNA analysis. In total, six distinct coexpression modules (magenta, green, pink, black, brown, and grey) were harvested (Fig. 3 F, G). Among the six modules, the module pink, magenta, green, and brown were the top 4 most significant co-expressional tissue-specific modules, with the |co-expression coefficient| of 0.66 (p-value 4e-9), 0.53 (p-value 1e-5), 0.45 (p-value 2e-4), and 0.39 (p-value 0.002), respectively (Fig. 3 G). The genes in the grey module were considered to be non-moduled.
The Cytoscape plugin MCODE was employed to extract the hub immune-related genes among the top 4 modules and the top 2 clusters with the highest MCODE scores (Fig. 3H and I), and genes contained in each cluster were listed in Supplementary Table 2.
The intersection between ceRNA network genes and hub immune-related genes disclosed 1 overlapped gene BDNF that could be the key immune-related mRNA (Fig. 4 A). According to the ceRNA hypothesis, the expressions of circRNAs or lncRNAs should be positively correlated with mRNAs, while the correlations between miRNAs and mRNAs expressions should be negative. As a result, based on the correlation coefficient of more than 0.4 (Supplementary Fig. 5), the ceRNA regulatory network was constructed (Fig. 4 B).
Fig. 4.
The CeRNA network of BDNF. (A) Venn diagram of the circ/lnc-mi-mRNA filtered quadruple network after pruning algorithms (CeRNA network) and hub genes from MCODE. (B) The CeRNA network of circ-, lnc-, mi-RNAs and BDNF mRNA. (C) FISH experiments of BDNF with its regulatory miRNAs. (D) Circos plot of the CeRNA network in (B). (E) Luciferase reporter assay confirmed the regulatory effects of miR-107 on BDNF.
3.5. Coexpression and coregulation between BDNF and hsa-miR-107
In the FISH experiment, BDNF was detected to be coexpressed with both hsa-miR-17-5p and hsa-miR-107 in tumor, peritumoral, and normal bladder samples, while the expression of BDNF in tumor was lower than that in peritumor or normal tissues but the distribution pattern were different in different tissues (Fig. 4C). Based on the coexistence relationships disclosed by FISH, hsa-miR-107 was selected for the following luciferase reporter assay. According to Maragkakis et al. [32], the energy threshold is inspected by the formula: threshold = region * ratio. With corresponding G: U wobble pairs in the above two miRNAs, the energy ratio was 0.74. However, the absolute energy values interacted with BDNF for hsa-miR-107 and hsa-miR-17-5p were 18.9 m(>17.02 = 23 * 0.74) and 24.9 (|-24.9| < 28.12 = 38 * 0.74), respectively. Thus, hsa-miR-17-5p was below the threshold and was discarded. Luciferase report assay demonstrated that the relative luciferace activities of wild type BDNF (WT-BDNF) was markedly decreased after adding the hsa-miR-107 mimics, confirming that BDNF was a direct target of hsa-miR-107 (Fig. 4 E). A circos plot also displayed the regulatory distributions of the circ/lncRNA, hsa-miR-107 and BDNF network. The chromosome map exhibits a uniform and unbiased distribution.
3.6. Low expression of the key immune-related BDNF and high expression of hsa-miR-107 were associated with good prognosis in bladder cancer patients
To estimate the correlation between BDNF and hsa-miR-107 expressions and bladder cancer prognosis, a multi-database meta-analysis was conducted and the survival analysis was performed with the log-rank test. the Kaplan-Meier survival curve of Overall survival (OS) of bladder cancer patients demonstrated that low expression of BDNF and high expression of hsa-miR-107 were associated with low risk and good prognosis (Fig. 5).
Fig. 5.
Kaplan-Meier survival curve of overall survival (OS) of bladder cancer patients, stratified by the best cut-off of BDNF or has-miR-107 expressions in TCGA-BLCA or GEO datasets (A, TCGA-BLCA; B, GSE13507; C, GSE19423; D, GSE19915_GPL4723; E, GSE32894; F, GSE160693; G, TCGA-BLCA; H, GSE84525).
4. Discussion
Through whole transcriptome sequencing of tumor and peritumoral tissues, our study revealed the molecular landscape of bladder cancer and defined hundreds of differentially expressed ncRNAs and thousands of differentially expressed coding RNAs. Furthermore, we distinguished the up- and down-regulated groups of DEGs as well as tumor and peritumoral samples.
The ceRNA hypothesis has attracted considerable attention recently, with research largely focusing on how the regulation of ceRNA expression affects cancer progression and pathogenicity [33,34]. MiRNAs usually act as negative regulators which affect their target mRNAs' stability and inhibit transcription. circ/lncRNAs may compete the shared miRNAs and thus alleviate mRNA destabilization or transcriptome repression. Therefore, the expression of mRNAs would be sympatric with circ/lncRNAs, whereas opposite to miRNAs. Our whole transcriptome sequencing has confirmed this tendency as shown in heatmaps in Fig. 1 B. What's more, we verified the target regulation relationships between hsa-miR-107 and its target gene, BDNF, via luciferase report assay, and the survival analysis has provided corroborative evidence as they contribute to a converse prognosis of bladder cancers. Besides, whole transcriptome sequencing has maximized efficiency of harvesting both circRNA and lncRNA, which has superior and comprehensive expression information. When adding parallel algorithmic models for competitive mechanisms effectiveness accounting, we believe that our ceRNA construction pipeline comprises the most comprehensive RNA varieties with competitive performance indexes in cancer ceRNA research.
The immune microenvironment plays an essential role in tumor development and immunotherapy [[35], [36], [37]]. Previous studies confirmed that the TME often comprises many different immune cell subpopulations endowed with anti-tumor or pro-tumor activity [38,39]. Therefore, we performed ssGSEA analysis on all DE-mRNAs to define the TME with 28 types of immune cells between bladder tumors and peritumoral tissues. Obviously, the ssGSEA scores of most of the cell types in the tumor samples were significantly lower than those in peritumoral samples, as shown by the violin plots that an apparent discrepancy in the stromal score, immune score, and ESTIMATE score of the two groups: the TME scores in the peritumoral samples were considerably higher than in the tumors. Our results indicated that the peritumoral tissues have more immune infiltration but low tumor purity than tumors, which adequately fits the actual. These inspiring outcomes as well as the variance of immune cell subtypes between tumors and peritumoral tissues prompted the necessity of exploring the immune microenvironment of peritumoral tissues which has rarely been studied.
In the immune microenvironment, the dynamic and reciprocal interactions among diverse immune cell types, together with the stromal cells, determine tumorigenesis and tumor progression. However, the key immune-related genes and the corresponding regulation network remain elusive. In this study, the gene BDNF was demonstrated as the key gene to influence the immunity of bladder cancer and the regulatory ceRNA network was constructed. We hypothesized that the low-expressed has_circ_0000211, AC108488.1, and LINC00163 might sponge hsa-miR-107 to down-regulate the expression of BDNF. Previous studies on the relationship between the ceRNA network and cancer immunity were mostly based on the existing public databases such as TCGA, for instance, two pyroptosis-related lncRNAs that strongly correlated with the tumor immune microenvironment of bladder cancer had been used to construct two ceRNA networks of lncRNA OCIAD1-AS1/miR-141-3p/GPM6B and lncRNA OCIAD1-AS1/miR-200a-3p/AKAP11 regulatory axes, which might had a prognostic value [40]. Also, lncRNA AC107027.3 had been suggested to upregulate fatty acid synthase(FASN) by competitively binding miR-27A-3p to regulate the immunotherapy response in bladder cancer [41]. For which whole transcriptome data are usually unavailable and circRNA information is missing, readers may feel tiresome and tedious. To the best of our knowledge, this was the first study to apply whole transcriptome sequencing to evaluate circ-, lnc-, mi-, and mRNA regulation networks in the immune microenvironment of bladder cancer.
In support of our conclusion, we have validated the identified ceRNA network in independent cohorts. We collected three different categories of data from public databases. These datasets represent different RNA types and contain at least five samples each. The first dataset, GSE190079, is a circRNA sequencing dataset with five bladder cancer tumor tissues and five matched adjacent non-tumor tissues. The second dataset, GSE154261, is a comprehensive collection of data for both lncRNA and mRNA, consisting of 73 discovery cohorts and 26 validation cohorts. The last miRNA dataset is extracted from the TCGA-BLCA dataset and contains 434 samples. To address the issue of data imbalance, random sampling with replacement was employed. The correlation matrix was constructed using the average Pearson correlation coefficients after 100 iterations. The final results are displayed in Supplementary Fig. 6. The graph shows that the verified cohorts' correlation results are consistent with our conclusion. BDNF and two lncRNAs revealed significant negative correlations with miRNA, while circRNA and two lncRNAs displayed significant positive correlations with BDNF.
To consolidate the target relationships between hsa-miRNA-107 and BDNF, extensive literature exploration has been conducted. We found that has-miR-107 and BDNF had been addressed as a direct target pair in many cancers using multiple cell lines. For instance, in non-small-cell lung cancer, BDNF has been demonstrated as a downstream regulator of has-miR-107 and the overexpression of BDNF reversed the inhibition of miR-107 mimics on the proliferation of H1299 and A549 cells [42]. Also, another research using A549 cells has revealed that has-miR-107 exerts its suppressive role in non-small-cell lung cancer cells by repressing BDNF and indirectly regulating the PI3K/AKT signaling pathway [43]. In breast cancer, both in tissues and in MCF-7 and MDA-MB-231 cells, has-miR-107 has been found to have a tumor-suppressive effect through the regulation of BDNF [44]. In gastric cancer, has-miR-107 also has been demonstrated to act as a tumor inhibitor by targeting BDNF expression in SGC-7901 cells [45]. These experiments have all confirmed the robust target regulations between has-miR-107 and BDNF.
Gene BDNF, which encodes a member of the nerve growth factor family of proteins and is one of the most widely distributed and extensively studied neurotrophins in the mammalian brain [46]. Previous research indicated it may promote neuroprotection and neuroregeneration [47], participate in the regulation of stress response and the biology of mood disorders, and has been recognized as a therapeutic factor in depression [48]. However, BDNF overexpression was seen in several human cancers like prostate, lung and breast, associated with poorer prognosis. It potentially enhanced carcinogenesis through tyrosine kinase receptors TrkB and the p75NTR death receptor [49]. Besides, BDNF was identified to own a significant positive correlation with M2 macrophages (CD206+), and an inverse correlation with M1 macrophages (CD86+), which indicates that BDNF might drive an immunosuppressive TME by modulating M2 macrophage polarization in lung adenocarcinoma with brain metastasis [50]. In our study, the role of BDNF has been validated to be down-regulated and lead to a relatively favorable prognosis in bladder cancer, which has been verified by multiple independent datasets in TCGA and GEO. This integrated meta-analysis roughly addressed nearly all the publicly available data with follow-up information for overall survival (OS) estimation.
Our study dissected the immune landscapes of both the tumor and peritumoral tissues and provided a useful resource for immunological studies of bladder cancer. We identified the key immune-related gene BDNF and constructed its ceRNA regulatory network, which provides rationale for exploring new prognostic markers and molecular therapeutic targets of bladder cancer.
5. Conclusion
Our integrated bioinformatic analysis revealed that low-expressed has_circ_0000211, AC108488.1, and LINC00163 might sponge hsa-miR-107 to down-regulate the expression of BDNF, a key immune-related gene that was associated with a good prognosis of bladder cancer.
Data availability statement
Data are available from Gene Expression Omnibus (GEO) database under the accession code GSE236932 and GSE236933. And the main code to generate this analysis and figures are available at http://wang_dafu.gitee.io/crun/index.html.
Patient consent for publication
Consent obtained directly from patient(s).
Ethics approval
Institutional approval was obtained from the local ethics committee.
Funding This work was supported by Hubei Province's Outstanding Medical Academic Leader program (鄂卫通【2019】47号) and the Cancer Research Program of the National Cancer Center (NCC201817B054).
Contributors: SL and YW contributed equally to this work as co-first authors; ZD and SW are co-corresponding authors. SL, LD, and KD collected samples and data. SL and YW performed bioinformatics analysis. ZD, DC, and SW designed the study and wrote the protocol. SL wrote the manuscript. ZD reviewed and revised the manuscript. All authors discussed, read, and approved the final manuscript.
CRediT authorship contribution statement
Sanhe Liu: Writing – original draft, Formal analysis, Data curation. Yiqi Wang: Visualization, Formal analysis. Liqun Duan: Data curation. Diansheng Cui: Project administration, Funding acquisition. Kangli Deng: Project administration. Zhiqiang Dong: Writing – review & editing, Supervision, Project administration. Shaozhong Wei: Supervision, Project administration.
Declaration of competing interest
None declared.
Acknowledgments
The authors thank The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) for the public datasets, and thank the Department of Biobank of Hubei Cancer Hospital for their help in collecting and saving samples, and the Department of Pathology of Hubei Cancer Hospital for their help in hematoxylin-eosin staining.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e29344.
Contributor Information
Zhiqiang Dong, Email: dongz@mail.hzau.edu.cn.
Shaozhong Wei, Email: weishaozhong@163.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.van Wilpe S., Gerretsen E.C.F., van der Heijden A.G., de Vries I.J.M., Gerritsen W.R., Mehra N. Prognostic and predictive value of tumor-infiltrating immune cells in urothelial cancer of the bladder. Cancers. 2020;12(9):2692. doi: 10.3390/cancers12092692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asano T., Ohnishi K., Shiota T., Motoshima T., Sugiyama Y., Yatsuda J., et al. CD169-positive sinus macrophages in the lymph nodes determine bladder cancer prognosis. Cancer Sci. 2018;109(5):1723–1730. doi: 10.1111/cas.13565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7) doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang C., Ding Y., Liu Y., Zhang Q., Xu S., Xia L., et al. Identification of mutated peptides in bladder cancer from exomic sequencing data reveals negative correlation between mutation-specific immunoreactivity and inflammation. Front. Immunol. 2020;11:576603. doi: 10.3389/fimmu.2020.576603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma W., Xue R., Zhu Z., Farrukh H., Song W., Li T., et al. Increasing cure rates of solid tumors by immune checkpoint inhibitors. Exp. Hematol. Oncol. 2023;12(1):10. doi: 10.1186/s40164-023-00372-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu H.-X., Wang Z.-X., Zhao Q., Wang F., Xu R.-H. Designing gene panels for tumor mutational burden estimation: the need to shift from 'correlation' to 'accuracy'. J Immunother Cancer. 2019;7(1):206. doi: 10.1186/s40425-019-0681-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Al-Ahmadie H., Netto G.J. Updates on the genomics of bladder cancer and novel molecular taxonomy. Adv. Anat. Pathol. 2020;27(1):36–43. doi: 10.1097/PAP.0000000000000252. [DOI] [PubMed] [Google Scholar]
- 8.Lindskrog S.V., Prip F., Lamy P., Taber A., Groeneveld C.S., Birkenkamp-Demtröder K., et al. An integrated multi-omics analysis identifies prognostic molecular subtypes of non-muscle-invasive bladder cancer. Nat. Commun. 2021;12(1):2301. doi: 10.1038/s41467-021-22465-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang C., Yuan W., Yang X., Li P., Wang J., Han J., et al. Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression. Mol. Cancer. 2018;17(1):19. doi: 10.1186/s12943-018-0771-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng F., Wang M., Li Y., Huang C., Tao D., Xie F., et al. CircNR3C1 inhibits proliferation of bladder cancer cells by sponging miR-27a-3p and downregulating cyclin D1 expression. Cancer Lett. 2019;460:139–151. doi: 10.1016/j.canlet.2019.06.018. [DOI] [PubMed] [Google Scholar]
- 12.Miao L., Liu H.Y., Zhou C., He X. LINC00612 enhances the proliferation and invasion ability of bladder cancer cells as ceRNA by sponging miR-590 to elevate expression of PHF14. J. Exp. Clin. Cancer Res. 2019;38(1):143. doi: 10.1186/s13046-019-1149-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang X., Ye T., Liu H., Lv P., Duan C., Wu X., et al. Expression profiles, biological functions and clinical significance of circRNAs in bladder cancer. Mol. Cancer. 2021;20(1):4. doi: 10.1186/s12943-020-01300-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verdugo R.A., Orostica K.Y. chromPlot: global visualization tool of genomic data. 2023. https://bioconductor.org/packages/chromPlot R package version 1.30.0. [DOI] [PubMed]
- 16.Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32(18):2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 17.Gu Z. Complex heatmap visualization. iMeta. 2022;1(3) doi: 10.1002/imt2.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu Z., Gu L., Eils R., Schlesner M., Brors B. Circlize Implements and enhances circular visualization in R. Bioinformatics. 2014;30(19):2811–2812. doi: 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- 19.Li R., Qu H., Wang S., Wei J., Zhang L., Ma R., et al. GDCRNATools: an R/Bioconductor package for integrative analysis of lncRNA, miRNA and mRNA data in GDC. Bioinformatics. 2018;34(14):2515–2517. doi: 10.1093/bioinformatics/bty124. [DOI] [PubMed] [Google Scholar]
- 20.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinf. 2013;14(1):7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Charoentong P., Finotello F., Angelova M., Mayer C., Efremova M., Rieder D., et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18(1):248–262. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 23.Lin W., Qiu X., Sun P., Ye Y., Huang Q., Kong L., et al. Association of IDH mutation and 1p19q co-deletion with tumor immune microenvironment in lower-grade glioma. Mol Ther Oncolytics. 2021;21:288–302. doi: 10.1016/j.omto.2021.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshihara K., Shahmoradgoli M., Martínez E., Vegesna R., Kim H., Torres-Garcia W., et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013;4(1):2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation. 2021;2(3) doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu G., Wang L.-G., Han Y., He Q.-Y. clusterProfiler: an R Package for comparing biological themes among gene clusters. OMICS A J. Integr. Biol. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanehisa M., Goto S. KEGG: Kyoto Encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhattacharya S., Dunn P., Thomas C.G., Smith B., Schaefer H., Chen J., et al. ImmPort, toward repurposing of open access immunological assay data for translational and clinical research. Sci. Data. 2018;5(1) doi: 10.1038/sdata.2018.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Breuer K., Foroushani A.K., Laird M.R., Chen C., Sribnaia A., Lo R., et al. InnateDB: systems biology of innate immunity and beyond--recent updates and continuing curation. Nucleic Acids Res. 2013;41(Database issue):D1228–D1233. doi: 10.1093/nar/gks1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9(1):559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao C.-H., Yu G., Cai P. ggVennDiagram: an intuitive, easy-to-use, and highly customizable R package to generate Venn diagram. Front. Genet. 2021;12 doi: 10.3389/fgene.2021.706907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maragkakis M., Alexiou P., Papadopoulos G.L., Reczko M., Dalamagas T., Giannopoulos G., et al. Accurate microRNA target prediction correlates with protein repression levels. BMC Bioinf. 2009;10:295. doi: 10.1186/1471-2105-10-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poliseno L., Pandolfi P.P. PTEN ceRNA networks in human cancer. Methods. 2015;77–78:41–50. doi: 10.1016/j.ymeth.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 34.Florian Reschke M., Ruocco A., Ng C., Chapuy B., Léopold V., et al. The BRAF pseudogene functions as a competitive endogenous RNA and induces lymphoma in vivo. Cell. 2015;161(2):319–332. doi: 10.1016/j.cell.2015.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei S.C., Duffy C.R., Allison J.P. Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov. 2018;8(9):1069–1086. doi: 10.1158/2159-8290.CD-18-0367. [DOI] [PubMed] [Google Scholar]
- 36.Finn O.J. A believer's overview of cancer immunosurveillance and immunotherapy. J. Immunol. 2018;200(2):385–391. doi: 10.4049/jimmunol.1701302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wein L., Luen S.J., Savas P., Salgado R., Loi S. Checkpoint blockade in the treatment of breast cancer: current status and future directions. Br. J. Cancer. 2018;119(1) doi: 10.1038/s41416-018-0126-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang T., Lu R., Kapur P., Jaiswal B.S., Hannan R., Zhang Z., et al. An empirical approach leveraging tumorgrafts to dissect the tumor microenvironment in renal cell carcinoma identifies missing link to prognostic inflammatory factors. Cancer Discov. 2018;8(9):1142–1155. doi: 10.1158/2159-8290.CD-17-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Q., Pan Y., Cao Z., Zhao S. Comprehensive analysis of prognostic value and immune infiltration of chromobox family members in colorectal cancer. Front. Oncol. 2020;10 doi: 10.3389/fonc.2020.582667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mo X., Hu D., Li Y., Nai A., Ma F., Bashir S., et al. A novel pyroptosis-related prognostic lncRNAs signature, tumor immune microenvironment and the associated regulation axes in bladder cancer. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.936305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiong Q., Feng D., Wang Z., Ying Y., Xu C., Wei Q., et al. Fatty acid synthase is the key regulator of fatty acid metabolism and is related to immunotherapy in bladder cancer. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.836939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hong W., Zhang Y., Ding J., Yang Q., Xie H., Gao X. circHIPK3 acts as competing endogenous RNA and promotes non-small-cell lung cancer progression through the miR-107/BDNF signaling pathway. BioMed Res. Int. 2020;2020:1–9. doi: 10.1155/2020/6075902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xia H., Li Y., Lv X. MicroRNA-107 inhibits tumor growth and metastasis by targeting the BDNF-mediated PI3K/AKT pathway in human non-small lung cancer. Int. J. Oncol. 2016;49(4):1325–1333. doi: 10.3892/ijo.2016.3628. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Gao B., Hao S., Tian W., Jiang Y., Zhang S., Guo L., et al. MicroRNA-107 is downregulated and having tumor suppressive effect in breast cancer by negatively regulating brain-derived neurotrophic factor. J. Gene Med. 2017;19(12):e2932. doi: 10.1002/jgm.2932. [DOI] [PubMed] [Google Scholar]
- 45.Cheng F., Yang Z., Huang F., Yin L., Yan G., Gong G. microRNA-107 inhibits gastric cancer cell proliferation and metastasis by targeting PI3K/AKT pathway. Microb. Pathog. 2018;121:110–114. doi: 10.1016/j.micpath.2018.04.060. [DOI] [PubMed] [Google Scholar]
- 46.Colucci-D’Amato L., Speranza L., Volpicelli F. Neurotrophic factor BDNF, physiological functions and therapeutic potential in depression, neurodegeneration and brain cancer. Int. J. Mol. Sci. 2020;21(20):7777. doi: 10.3390/ijms21207777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Palasz E., Wysocka A., Gasiorowska A., Chalimoniuk M., Niewiadomski W., Niewiadomska G. BDNF as a promising therapeutic agent in Parkinson's disease. Int. J. Mol. Sci. 2020;21(3):1170. doi: 10.3390/ijms21031170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murawska-Ciałowicz E., Wiatr M., Ciałowicz M., Gomes De Assis G., Borowicz W., Rocha-Rodrigues S., et al. BDNF impact on biological markers of depression—role of physical exercise and training. Int. J. Environ. Res. Publ. Health. 2021;18(14):7553. doi: 10.3390/ijerph18147553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Malekan M., Nezamabadi S.S., Samami E., Mohebalizadeh M., Saghazadeh A., Rezaei N. BDNF and its signaling in cancer. J. Cancer Res. Clin. Oncol. 2023;149(6):2621–2636. doi: 10.1007/s00432-022-04365-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu Q., Wang Y., Yi G., Yang W., Chen K., Tan X., et al. BDNF is a prognostic biomarker involved in immune infiltration of lung adenocarcinoma and is associated with brain metastasis. Immunology. 2022;168(2):320–330. doi: 10.1111/imm.13581. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available from Gene Expression Omnibus (GEO) database under the accession code GSE236932 and GSE236933. And the main code to generate this analysis and figures are available at http://wang_dafu.gitee.io/crun/index.html.