Abstract
PANoptosis is a type of programmed cell death (PCD) characterised by apoptosis, necroptosis and pyroptosis. Long non-coding ribonucleic acids (lncRNAs) are participating in the malignant behaviour of tumours regulated by PCD. Nevertheless, the function of PANoptosis-associated lncRNAs in lung adenocarcinoma remains to be investigated. In this work, a PANoptosis-related lncRNA signature (PRLSig) was developed based on the least absolute shrinkage and selection operator algorithm. The stability and fitness of PRLSig were confirmed by systematic evaluation of Kaplan–Meier, Cox analysis algorithm, receiver operating characteristic analysis, stratification analysis. In addition, ESTIMATE, single sample gene set enrichment analysis, immune checkpoints and the cancer immunome database confirmed the predictive value of the PRLSig in immune microenvironment and helped to identify populations for which immunotherapy is advantageous. The present research provides novel insights to facilitate risk stratification and optimise personalised treatment for LUAD.
Keywords: PANoptosis, Lung adenocarcinoma, Long noncoding RNA, Prognosis, Immune microenvironment
1. Introduction
According to the latest worldwide cancer report, among all malignant tumours, lung cancer has the second-highest incidence and the first-highest mortality rate [1]. More than 80 % of lung cancer cases are non-small cell lung cancers, of which lung adenocarcinoma (LUAD) is the most common subtype and accounts for the highest proportion of cases [2]. Despite the advancements of next-generation sequencing, targeted therapies and immunotherapy, which have led to rapid advances in the treatment and prognosis of LUAD, its long-term survival remains dismal [3,4]. Therefore, exploring safe and effective biomarkers of LUAD for predicting prognosis will not only contribute to the clarification of the molecular mechanisms underlying the evolution of LUAD, but also aids in differentiating patients into different subgroups, thus facilitating personalised and precise treatment.
Increasing evidence reveals that different forms of programmed cell death (PCD) play a fundamental and essential part in maintaining the cellular status [[5], [6], [7], [8]]. PANoptosis is a novel type of PCD identified by Malireddi et al. as having the main hallmarks of pyroptosis, necroptosis and apoptosis; however, PANoptosis could not be characterized by only one of these three forms of PCD [9]. It is regulated by molecular signals and upstream receptors that combine to form a multimodal complex, the PANoptosome [10]. The PANoptosome, which also acts as the ‘master switch’ for the initiation of the three PCD pathways, is an attractive intervention target in the treatment of human diseases. Moreover, many investigations have reported that PANoptosis is closely associated with malignant tumour evolution, influencing the regulation of tumour-related molecules and pathways and playing a part in immunotherapeutic response [[11], [12], [13]]. However, there remains a knowledge gap regarding the mechanism of PANoptosis in LUAD. Therefore, it is of interest to explore the potential value of PANoptosis for LUAD.
Long non-coding ribonucleic acids (lncRNAs) regulate gene expression at various transcriptional levels and can be used to not only regulate the biological behaviour of malignant tumours but also act as effective markers for cancer diagnosis and prognosis [14]. Recent evidence suggests that PCD-related lncRNAs not only regulate biological behaviour of cancer, but are also correlated with tumour immune status and treatment response [[15], [16], [17], [18]]. Additionally, studies have confirmed the potential value of apoptosis, necroptosis and pyroptosis-associated lncRNAs as prognostic biomarkers in LUAD [[19], [20], [21]]. Nevertheless, the value of PANoptosis-related lncRNAs as biomarkers for LUAD has not yet been explored.
Here, we establish a PANoptosis-related lncRNA signature (PRLSig) in LUAD based on the least absolute shrinkage and selection operator (LASSO) algorithm, and analysed the relationship between the PRLSig and prognosis, underlying mechanisms, tumour mutation burden (TMB), tumour immune microenvironment (TIME) and individualised therapy in LUAD. In this process, Kaplan-Meier (K-M) curves, univariate (uni-) and multivariate (multi-) Cox analysis, receiver operating characteristic (ROC) analysis, stratified clinicopathological parameters, single sample gene set enrichment analysis (ssGSEA), gene set variation analysis (GSVA), immune checkpoints (ICs), the Cancer Immunome Database (TCIA) and ESTIMATE algorithms were used for the validation and evaluation of PRLSig. Our findings suggest that the PRLSig might be applied as a novel indicator for predicting patient clinical outcomes and characterize the TIME. Thus, this study could provide a basis for personalised therapeutic selections such as immune checkpoint blockades (ICBs) for individuals with LUAD.
2. Materials and methods
2.1. Data collection
RNA-sequencing (RNA-seq) matrix, simple nucleotide variation (SNV) matrix and associated clinicopathological parameters for patients with LUAD were derived from the TCGA repository (https://portal.gdc.cancer.gov/repository). The RNA-seq data was collated using the Strawberry Perl programming language, wherein mRNA and lncRNA matrices were extracted. Data related to the immunophenoscore (IPS) of ICBs therapy in the TCGA-LUAD cohort were derived from TCIA (https://tcia.at/home). The PANoptosis-related genes (PRGs) employed in our research were from previous research [9,10,12,13,[22], [23], [24], [25], [26], [27], [28]] (Supplementary table S1).
2.2. Acquisition of PANoptosis-related lncRNAs (PRLs)
The mRNA expression data of the 24 PRGs were derived from the TCGA-LUAD cohort using ‘limma’ package. PRLs were further obtained via the co-expression algorithm of PRGs mRNAs and lncRNAs (Correlation coefficient >0.4, P < 0.001). The data processing package ‘dplyr’ was used to construct correlation data for PRGs and lncRNAs, and the ‘galluvial’ and ‘ggplot2’ packages were used to visualise the Sankey plots. Additionally, the ‘limma’ was employed to derive differentially expressed PRLs between tumour and normal tissue that met the conditions of fold change >1.5 and FDR <0.05. The ‘pheatmap’ was employed to visualise the differentially expressed PRLs.
2.3. Construction of a PRLSig in LUAD
PRLs significantly associated with prognosis were determined by uni-Cox (P < 0.05). Package ‘survival’ was utilised to map the risk ratio forest for the prognostic lncRNAs, and the ‘pheatmap’ was utilised to visualise the prognosis-related lncRNA expression heat map. The patients were randomly categorised into training and validation cohorts (1:1 ratio). The machine learning algorithm LASSO was utilised to control for overfitting problems such as those occurring at high sample latitudes and obtain the optimal prognosis-related lncRNAs for the PRLSig construction. The above process is implemented by the ‘caret’ and ‘glmnet’.
According to the results of LASSO, we constructed the following risk score formula: , where β represents the screened lncRNA, Coefficient refers to the regression coefficient and Expression represents lncRNA expressions. Risk scores for individuals were derived using the risk equation, and risk stratified all cases utilising the median risk score for the training set.
2.4. Verification of the PRLSig
To verify the prognostic predictive value of the PRLSig in LUAD, we used K-M curves, risk score curves and survivor plots for the assessment. This process was implemented with the ‘survminer’ and ‘survivor’. In addition, uni- and multi-Cox algorithms were implemented to determine if the PRLSig was an independent prognostic factor. Furthermore, the ROC curves were plotted using the packages ‘timeROC’, ‘survminer’ and ‘survival’ to observe the predictive performance of risk scores in relation to different clinicopathological parameters for patients with LUAD.
2.5. Construction of a nomogram in LUAD
Nomograms for 1-, 3- and 5-year overall survival (OS) were developed using the ‘regplot’, ‘survival’ and ‘rms’ packages. In addition, Hosmer–Lemeshow test calibration curves verified the relationship between actual and expectedoutcomes.
2.6. Validation of the PRLSig in different clinical subtypes
The ‘limma’ package compared the differences in risk scores across distinct clinicopathological subgroups and ‘ggpubr’ was used to visualise the results. Finally, K-M curves of the different populations in the various clinicopathological subgroups were plotted using ‘survminer’ and ‘survival’ to determine the stability of the PRLSig.
2.7. Functional analysis of the PRLSig
GSVA is a method that is primarily employed to assess the outcomes of gene set enrichment in the transcriptome [29]. KEGG pathway enrichment was analysed by GSVA in the high-risk and low-risk groups and the correlation between KEGG pathway and lncRNAs expression in PRLSig was analysed. The above process was performed utilising the packages ‘reshape2’, ‘limma’, ‘GSEABase’, ‘pheatmap’, ‘GSVA’ and ‘ggplot2’.
2.8. Correlation between PRLSig and TMB
Perl scripts were utilised to collate the SNV data to generate the TMB values. The ‘limma’ package was utilised to determine the TMB status between the two risk subgroups. Additionally, the ‘survminer’ and ‘survivor’ were used to draw K-M curves of individuals in the different TMB subgroups combined with the different risk subgroups. Furthermore, mutation waterfall for the twenty most frequently mutated genes in each subgroup were plotted using ‘maftools’.
2.9. Predictive value of PRLSig in the TIME of LUAD
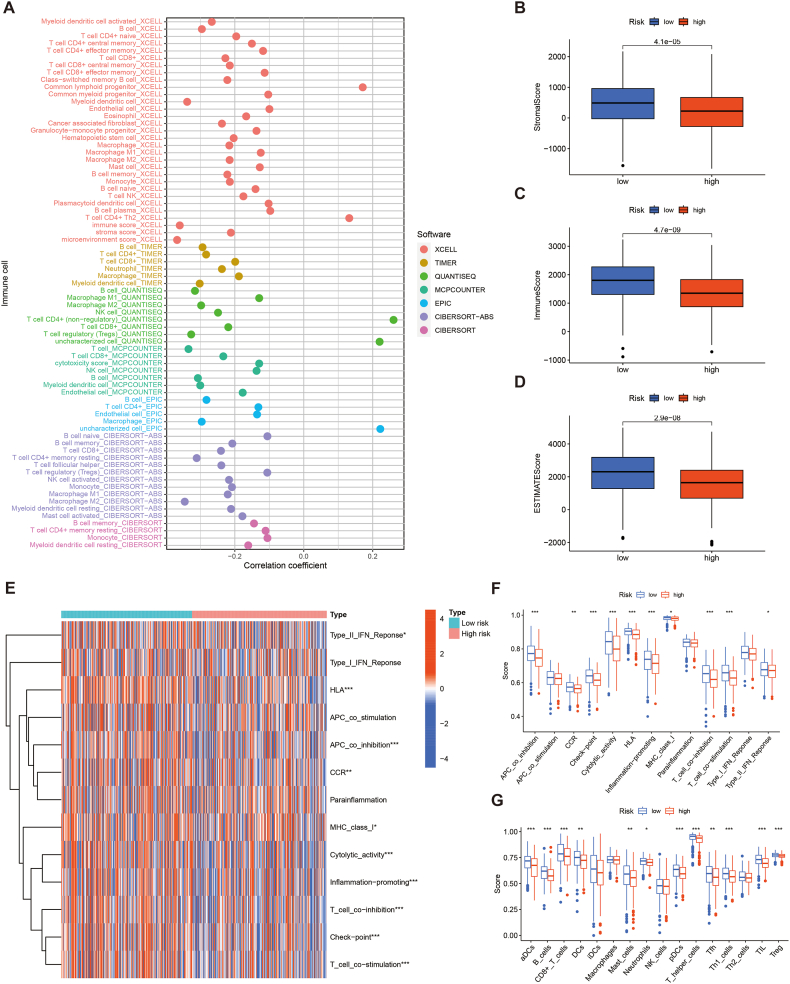
The Tumor IMmune Estimation Resource (TIMER) platform uses different algorithms (CIBERSORT, MCPCOUNTER, XCELL, EPIC, QUANTISEQ and TIMER) to estimate extent of immune cell infiltration in tumours [30,31]. To investigate the correlation between PRLSig and the TIME in LUAD, we downloaded immunological infiltration data of TCGA-LUAD on the TIMER (http://timer.cistrome.org/) and analysed the association between different immuno-infiltrating cells and risk score. Correlation bubble plots were plotted with the packages ‘ggtext’, ‘ggplot2’, ‘scales’, ‘tidyverse’ and ‘ggpubr’.
ESTIMATE method allows inference of the number of tumour and normal cells [32]. We assessed the amount of stromal cells and immuno-infiltrating cells in all LUAD cases using the ‘ESTIMATE’ R package to generate the corresponding stromal cell and immune cell scores, respectively. Box plots of scores between the two risk groups were plotted via the ‘ggpubr’.
Gene set enrichment analysis (GSEA) allows classification of gene sets that share common biological functions [33]. Here, we performed ssGSEA utilising the ‘GSEABase’ and ‘GSVA’ to calculate the proportion of various immune cells among tumour tissues, thereby obtaining immune cell and immune function scores for all individuals. Further differential analysis were performed for the risk groups. The ‘reshape2’, ‘ggpubr’, ‘reshape2’ and ‘pheatmap’ packages were conducted to visualise the results.
ICs regulate the degree of immune activation and act as ‘braking’ signals to suppress the body's immune function. When a tumour develops, ICs are activated, suppressing the immune function of T lymphocytes and inducing an immune escape. This study further explored the variation in ICs between the two risk groups. The PD-1 binds to PD-L1 and promotes tumour cell evasion from immune killing [34]. Moreover, PD-L1 expression in some tumours affects the response to immunotherapy [35,36]. CTLA4 is an important target for other ICBs. Thus, differences in PD-L1, PD1 and CTLA4 expression in the groups were analysed.
The TCIA database provides results of immunogenomic profiling of sequencing data of tumour sequencing data. The immunogenicity of the tumour is scored quantitatively by TCIA to obtain an IPS, which predicts the response to ICBs [37]. We subsequently investigated the potential predictive validity of PRLs by comparing IPS in risk groups to determine the efficacy of immunotherapy.
2.10. Clinical drug predictive value of PRLSig
The package ‘pRRophetic’ was employed to determine IC50 of different chemical drugs in different risk populations, to analyse the potential value of the PRLSig in clinically personalised treatment [38], and to create box plots of agents with significantly different IC50s using the ‘ggpubr’ package.
3. Results
3.1. PRLs in LUAD
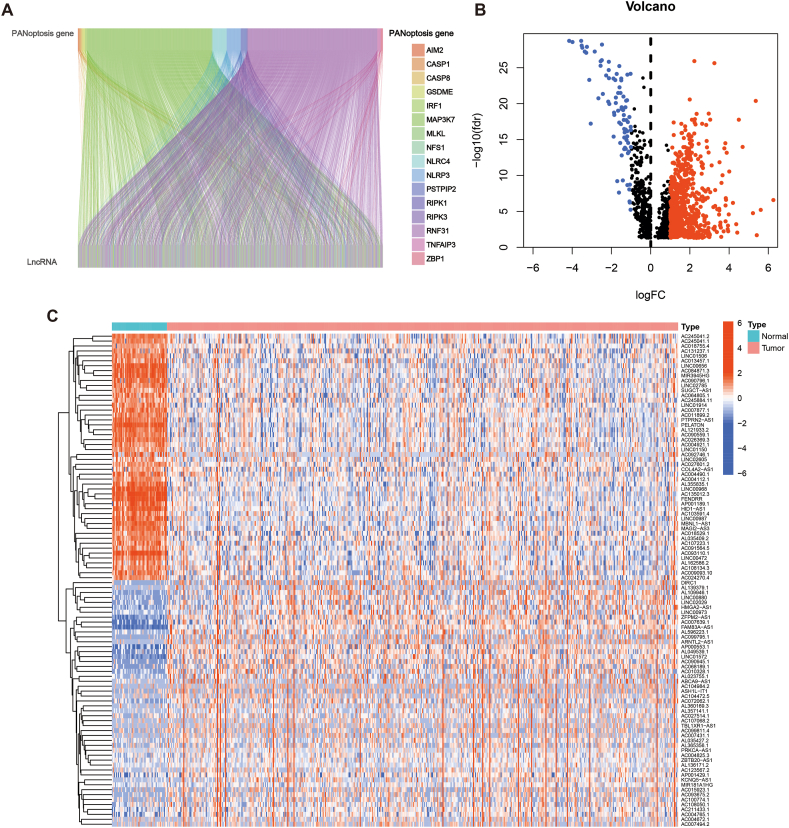
The flow chart of this work is shown in Fig. 1. Co-expression analysis revealed 1883 PRLs (Fig. 2A). Additionally, a total of 797 PRLs were differentially expressed, of which 715 lncRNAs were up-regulated and 82 were down-regulated in tumour samples (Fig. 2B). The heatmap shows the 50 differentially expressed lncRNAs with the highest up- and down-regulation fold (Fig. 2C).
Fig. 1.
Flow chart of the study.
Fig. 2.
PANoptosis-associated lncRNAs in LUAD. (A) Correlation sankey plot of PRGs and lncRNAs. (B) Volcano plots of PRLs expression. (C) Heatmap of the differentially expressed PRLs.
3.2. Construction and validation of PRLSig in LUAD
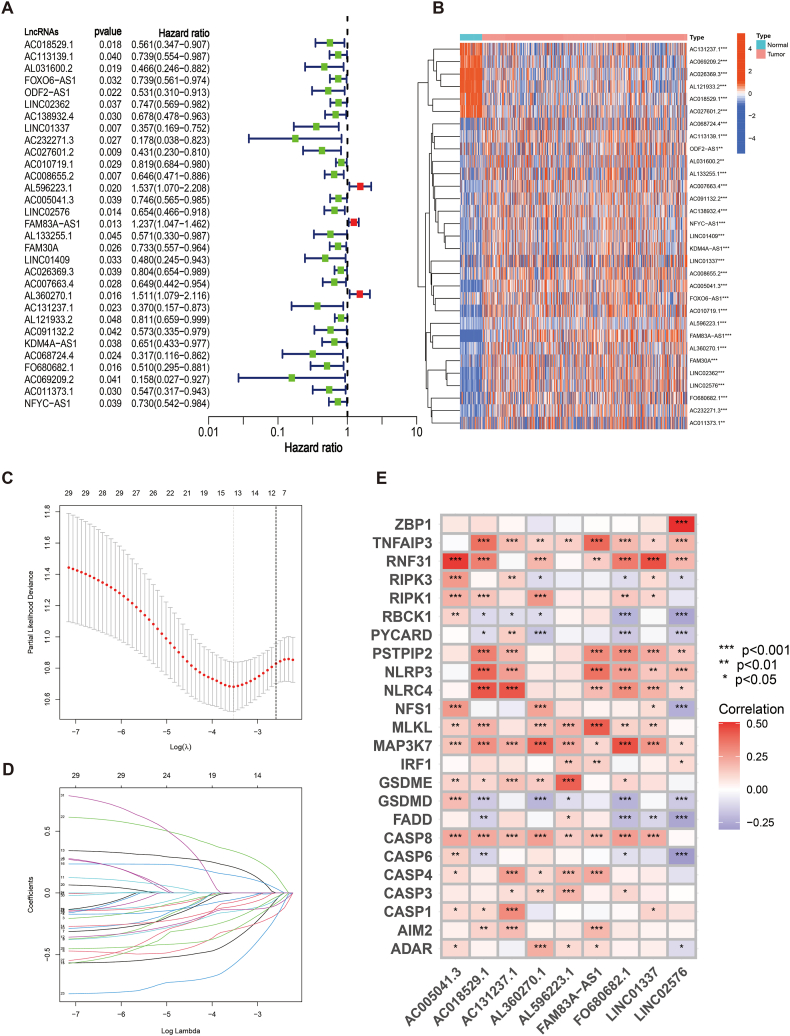
Uni-Cox analysis revealed 31 PANoptosis-related lncRNAs that were correlated with survival (P < 0.05) in the training cohort (Fig. 3A). The expression heat maps of survival-related lncRNAs are shown in Fig. 3B, respectively. To avoid overfitting, LASSO regression analysis was performed (Fig. 3C and D), which identified nine lncRNAs for PRLSig construction (Table 1). The score for every patient was calculated by the risk score equation in PRLSig. Risk score (PRLSig) = AC018529.1 × (−0.688562741) + LINC01337 × (−0.654985608) + AL596223.1 × (0.338025106) + AC005041.3 × (−0.291750665) + LINC02576 × (-0.351975465) + FAM83A-AS1 × (0.237650628) + AL360270.1 × (0.403790433) + AC131237.1 × (−0.817121741) + FO680682.1 × (−0.592519929). The association among the PRLSig-related lncRNAs and PRGs is represented by a heat map in Fig. 3E.
Fig. 3.
Construction of a PRLSig for LUAD. (A) The forest plot identified 31 PRLs that were significantly correlated with prognosis. (B) Heatmap of the 31 PRLs. (C–D) The LASSO regression analysis. (E) Heat map of correlations between the expression of the nine lncRNAs used to construct the signature and the PRGs.
Table 1.
| PANoptosis-related signature in LUAD.
| LncRNA | Coefficient | Hazard ratio | p-value |
|---|---|---|---|
| AC018529.1 | −0.688562741 | 0.561 | 0.018 |
| LINC01337 | −0.654985608 | 0.357 | 0.007 |
| AL596223.1 | 0.338025106 | 1.537 | 0.020 |
| AC005041.3 | −0.291750665 | 0.746 | 0.039 |
| LINC02576 | −0.351975465 | 0.654 | 0.014 |
| FAM83A-AS1 | 0.237650628 | 1.237 | 0.013 |
| AL360270.1 | 0.403790433 | 1.511 | 0.016 |
| AC131237.1 | −0.817121741 | 0.370 | 0.023 |
| FO680682.1 | −0.592519929 | 0.510 | 0.016 |
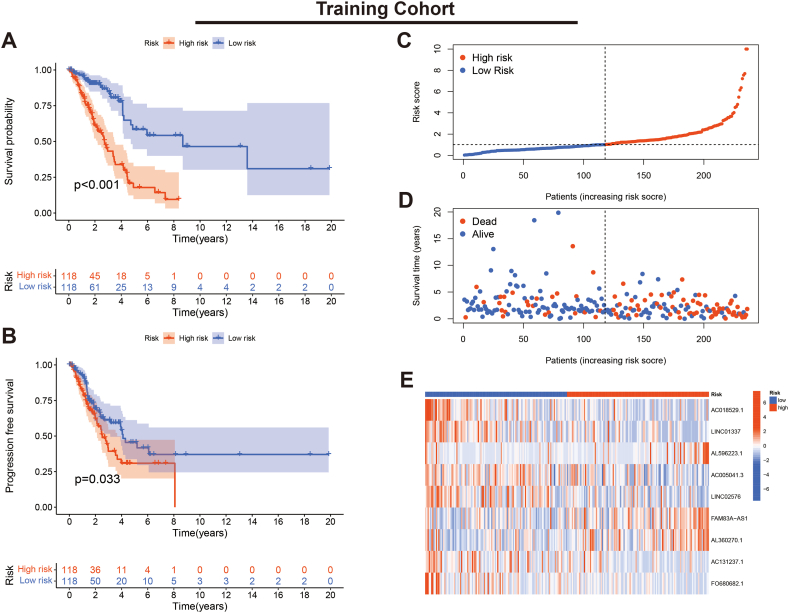
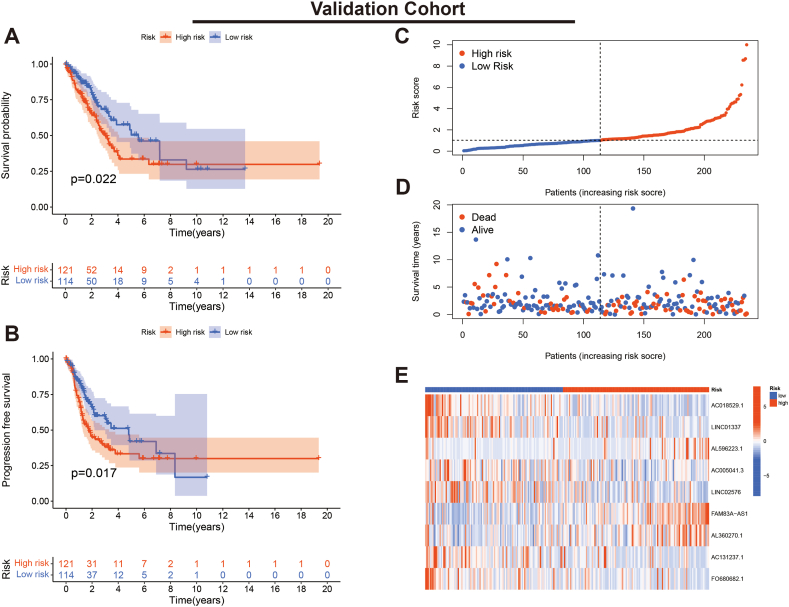
We evaluated the K-M curves, score distribution and survival status of individuals in the training set. The results indicated that individuals in the low-risk subgroup had a significantly better prognosis (Fig. 4A–D). Expression heat maps showed that AL596223.1, FAM83A-AS1 and AL360270.1 were lowly expressed in the low-risk subgroup, while the remaining six lncRNAs showed high expression (Fig. 4E). Furthermore, the consistent results were validated in the validation cohort (Fig. 5A–E).
Fig. 4.
Verification of the PRLSig in training set. (A, B) K-M curves for OS and PFS. (C) Score distribution curves. (D) Survival status map. (E) Heatmap of the expression of the PRLSig-related lncRNAs.
Fig. 5.
Verification of the PRLSig in validation set. (A, B) K-M curves for OS and PFS. (C) Score distribution curves. (D) Survival status map. (E) Heatmap of the expression of the PRLSig-related lncRNAs.
3.3. Assessment of the PRLSig in LUAD
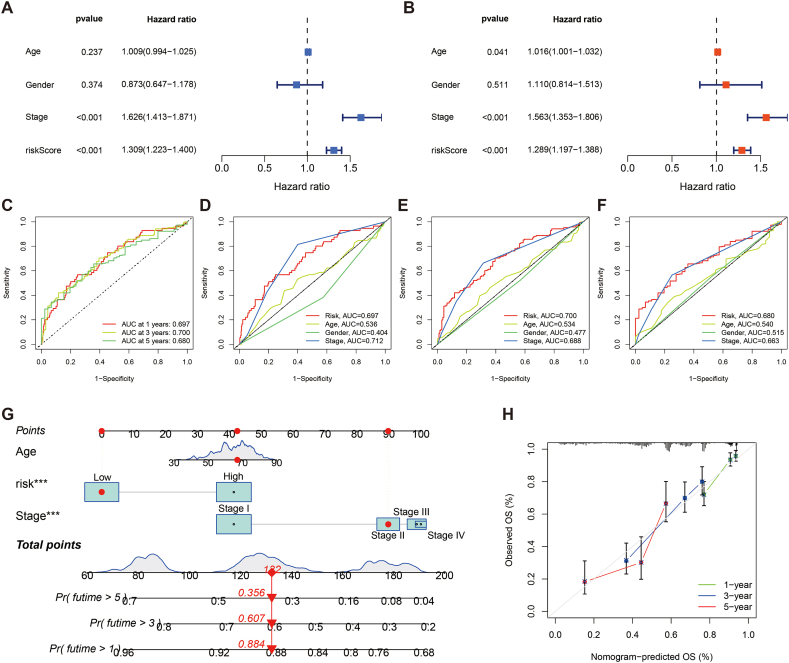
Uni- and multi-Cox regression revealed that the PRLSig is an independent prognostic indicator (P < 0.001) (Fig. 6A and B). Furthermore, ROC curves were used to evaluate the sensitivity of the PRLSig to the prognosis of LUAD. The outcomes revealed that the area under the curve (AUC) for the PRLSig was 0.697, 0.700 and 0.680 at 1, 3 and 5 years (Fig. 6C). Comparing the AUC values of the risk scores and clinical indicators indicated that the PRLSig outperformed other clinicopathological parameters (Fig. 6D–F). Thus, these findings demonstrate the favourable predictive performance of the PRLSig.
Fig. 6.
Evaluation of PRLSig and nomogram. (A–B) The uni- and multi-Cox regression. (C) ROC curves for the PRLSig. (D–F) Comparing PRLSig with age, gender and tumour stage for ROC curves at 1-, 3- and 5-years. (G) Nomogram of risk status, age and tumour stage. (H) Calibration curves elucidating a concordance among the observed and predicted OS rates of individuals.
3.4. Nomogram for LUAD
According to the multi-Cox regression, we developed a nomogram for predicting prognosis in patients with LUAD based on age, tumour stage and risk (Fig. 6G). Moreover, the calibration curves indicate favourable consistency between the actual survival of individuals with LUAD and the predicted outcome (Fig. 6H).
3.5. Correlation of the PRLSig with clinical parameters
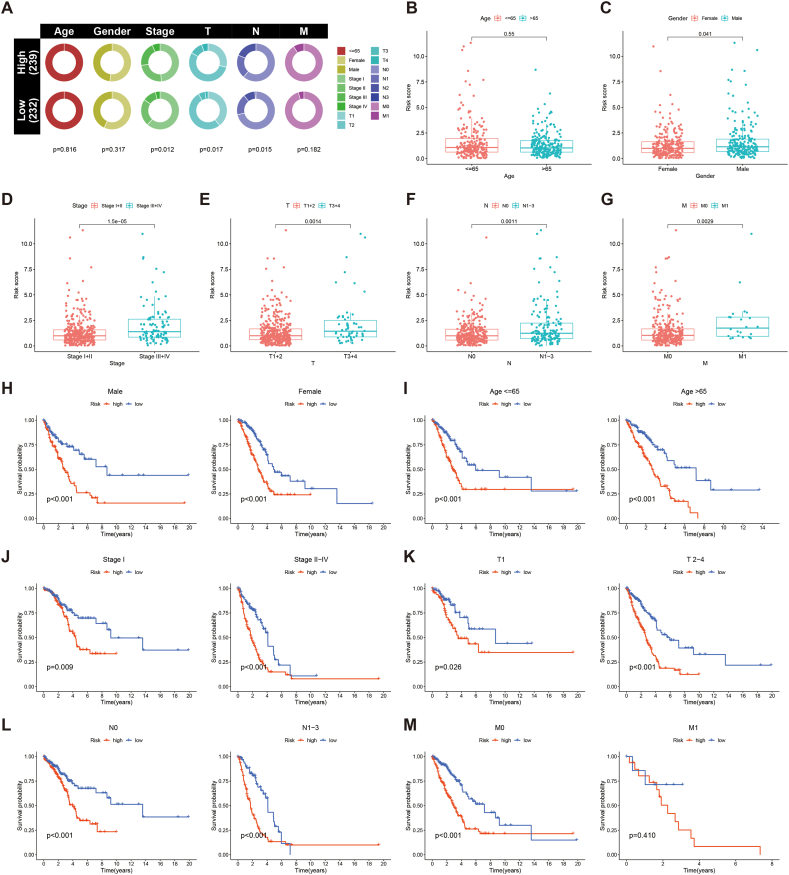
To explore the correlation between the PRLSig and the clinical indicators in LUAD, we first plotted the state proportions of the clinical indicators for the two subgroups (Fig. 7A). The results showed that there were differences in TNM, T and N stages across the two populations. Box plots also showed no difference in risk score between patients with LUAD by age (Fig. 7B), while risk scores for female, patients with low-stage (I-II), low-T-stage (T1-2), no lymph node metastases (N0) and no distant metastases (M0) were lower than those for male, high-stage (III-IV), high-T-stage (T3-4), lymph node metastases (N1-3) and distant metastases (M1) (Fig. 7C–G). The K-M curves showed that patients with LUAD of different age, stage, gender and presence of lymph node or distant metastases had poor survival in the high-risk population (Fig. 7H-M), indicating the applicability of PRLSig to patients with LUAD of different clinicopathological parameters.
Fig. 7.
Correlation of the PRLSig with clinicopathological features. (A) Distribution status of clinicopathological features in different risk cohorts. (B–G) Comparison of risk scores for subgroups of patients by age, gender, tumour stage, T-stage, N-stage and M-stage. (H–M) K–M curves of high- and low-risk populations stratified by different clinicopathological indicators.
3.6. PRLSig-based GSVA
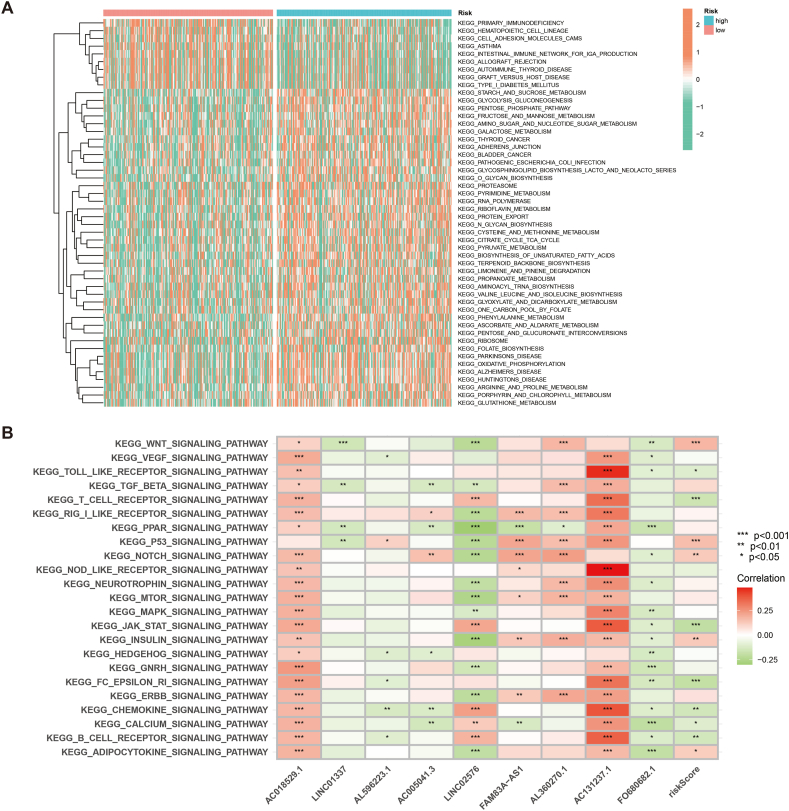
To investigate the relationship between the PRLSig and tumour biological behaviour, the GSVA was utilised to identify differences in KEGG pathway enrichment across risk groups. The heat map showd that the high-risk subgroup was enriched for functions including starch and sucrose metabolism, glycolysis, fructose and mannose, glutathione and fatty acid metabolism. However, functions such as primary immunodeficiency, intestinal immune network and autoimmune thyroid disease were enriched in low-risk population (Fig. 8A). We also analysed the correlation between nine PRLSig lncRNAs and pathways, and the findings demonstrated a close association between these lncRNAs and key signalling pathways (Fig. 8B).
Fig. 8.
PRLSig-based GSVA. (A) GSVA analysis of two risk groups. (B) Association between the PRLSig-related lncRNAs and tumour-associated pathways.
3.7. Correlation of the PRLSig with TMB in LUAD
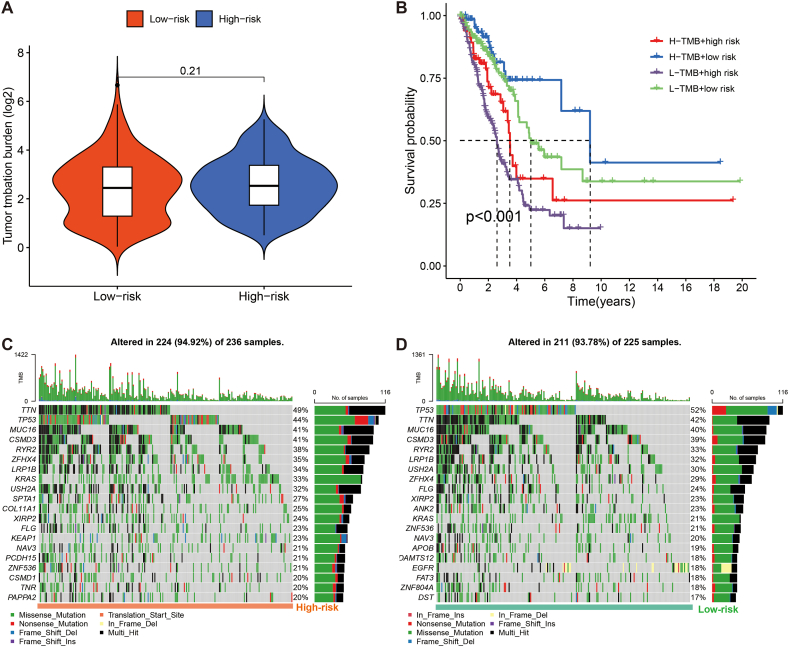
Tumours with high TMB are speculated to have higher levels of neoantigens, which are reported to promote the production of immune cells. TMB is also applied as a quantitative biomarker to predict the treatment response to ICBs in some individuals with tumours [39]. We also used SNV data from the TGCA-LUAD cohort to generate per-sample TMB values. Moreover, box plots revealed no difference in TMB levels across risk groups (Fig. 9A). However, there were significant differences in prognosis between the various combination subgroups (P < 0.001), with the low TMB/high-risk patients having the worst prognosis and the high TMB/low-risk subgroup having the best OS (Fig. 9B). The findings suggest that TMB combined with risk scores could better predict the prognosis in LUAD. Furthermore, the top five genes with the most frequent mutations in the high-risk subgroup were TTN (49 %), TP53 (44 %), MUC16 (41 %), CSMD3 (41 %) and RYR2 (38 %), whereas those in the low-risk population were TP53 (52 %), TTN (42 %), MUC16 (40 %), CSMD3 (39 %) and RYR2 (33 %) (Fig. 9C and D).
Fig. 9.
Association of the PRLSig with TMB in LUAD. (A) TMB status of the different risk groups. (B) K–M curves for TMB combined with risk status. (C, D) Mutant genes' waterfall plots in the two risk groups.
3.8. Role of PRLSig in predicting the TIME in LUAD
Previous studies have shown that PCD performs an essential function in the regulation of TIME [24]. To further explore the correlation between PRLSig and TIME, we first analysed the correlation between immuno-infiltrating cells and PRLSig scores. Bubble plots revealed a negative correlation between risk scores and most immuno-infiltrating cells (Fig. 10A). Scatter plots of correlation coefficients for different immuno-infiltrating cells and risk scores are shown in fig. S1. The ESTIMATE analysis revealed that individuals in the low-risk subgroup had higher stromal scores, immune scores and ESTIMATE scores (Fig. 10B–D). To further validate the results, ssGSEA analysis was performed. The foundings revealed that antigen-presenting cell co-inhibition, human leukocyte antigen, inflammation promotion, major histocompatibility complex I, cytolytic activity, T-cell co-stimulation, T-cell co-inhibition and type II interferon response were significantly stronger in the low-risk group (Fig. 10E and F). In terms of immune cells, CD8+ T cells, neutrophils, dendritic cells, plasma cells, tumour infiltrating lymphocytes and type I helper T lymphocytes were significantly higher in the low-risk cohort (Fig. 10G). Together, the above findings indicate a higher degree of immune cell infiltration status in the low-risk cohort.
Fig. 10.
Association of the PRLSig with the TIME. (A) Bubble diagram illustrates the association coefficient between PRLSig scores and immune cells. (B–D) Stromal cell, immune cell and ESTIMATE scores in two subgroups. (E) Status of immune-related functions in two risk subgroups. (F–G) The ssGSEA analysis between the two groups.
3.9. PRLSig for predicting response to immunotherapy
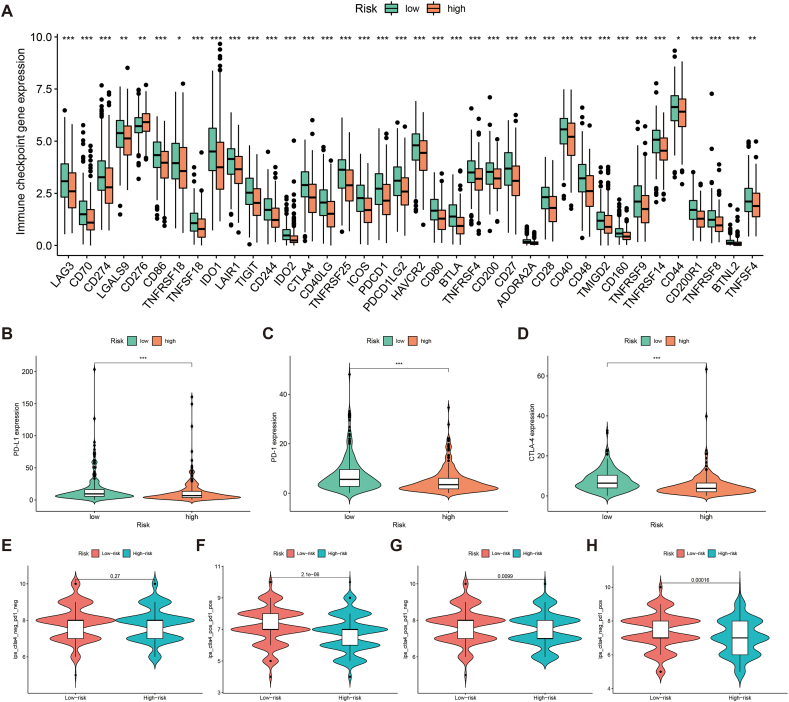
The main targets of the current ICBs include CTLA-4, PD-1 and PD-L1 [40]. Analysis revealed that most immune checkpoint-related genes include CTLA-4, PD-1 and PD-L1 were more highly expressed in the low-risk population (Fig. 11A–D).
Fig. 11.
PRLSig for predicting response to immunotherapy. (A) Box plots illustrates the expression of immune checkpoint-related genes in the two subgroups. (B–D) Differences in the expression of PD-L1, PD-1 and CTLA4 in the two subgroups. (E–H) Violin plots show the different levels of drug response to CTAL-4 and PD-1 ICBs immunotherapy in individuals with high- or low-risk.
To further validate the potential of PRLSig in predicting the efficacy of ICBs, we further explored the correlation between IPS and PRLSig. The findings indicated that there was no difference in IPS across the different risk groups in samples predicted to have a negative response to both immunotherapy regimens (Fig. 11E). In samples with tumours predicted to have a positive response to a single CTLA4 inhibitor regimen, IPS values were significantly higher in the low-risk group (Fig. 11F). Similarly, single anti-PD-1/PD-L1 regimens and anti-PD-1/PD-L1 combined with anti-CTLA4 therapy showed consistent results (Fig. 11G–H). Thus, these results indicate that the low-risk population is the beneficiary population for ICBs treatment.
3.10. The predictive role of the PRLSig on the sensitivity of chemical compounds
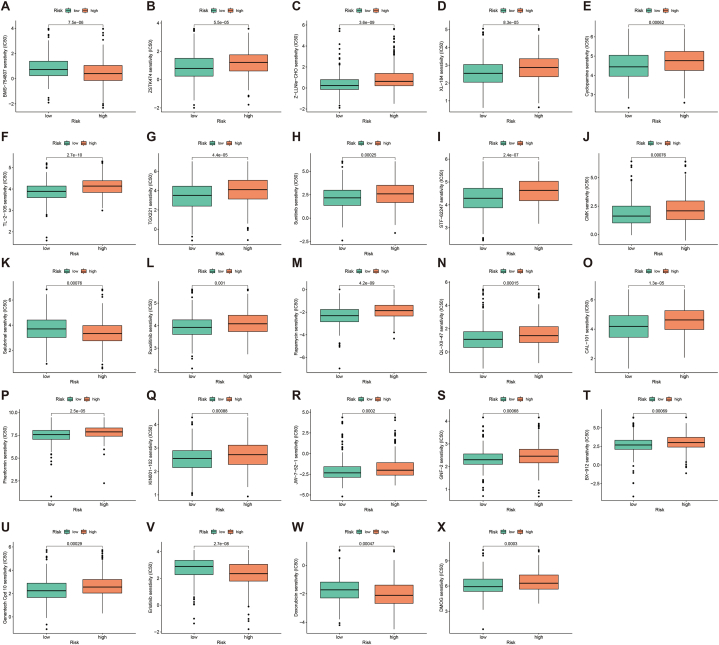
To explore the significance of the PRLSig in guiding personalised treatment regimens, we performed a drug sensitivity analysis. IC50 values for a number of therapeutic medications differ across risk groups (Fig. 12A–X). Among them, the IC50 of the PI3K inhibitor ZSTK474, the Hedgehog pathway antagonist Cyclopamine, Cabozantinib (XL184), TGX221, Sunitinib, Ruxolitinib, Rapamycin and Phenformin were lower in low-risk subgroup. However, for agents such as Salubrinal, Erlotinib and Doxorubicin, the IC50 was higher in low-risk cohort. Scatter plots of the correlation coefficients of the above results are shown are shown in fig. S2.
Fig. 12.
The effect of the PRLSig on the sensitivity of therapeutic agents. (A–X) Differences in IC50 values for different drugs in the two risk subgroups.
4. Discussion
PANoptosis, a novel form of PCD, is regulated by the PANoptosome and has the key characteristics of pyroptosis, apoptosis and necroptosis. Notably, PANoptosis cannot be characterised by any of the death types alone [41]. Compared to the extensive investigations on PANoptosis in the areas of infection [42,43], cancer researches related to PANoptosis still need to be further explored. Nonetheless, considering the similarities between tumour immunity and infection immunity, the research on the interaction between PANoptosis and tumour immune microenvironment is expected to become a new research hotspot [13,44].
LncRNAs typically act as competing endogenous RNAs that regulate cancer development and progression by modulating mRNA translation. Furthermore, increasing evidence supports lncRNAs as critical regulators in lung cancer, including lung tumour cell proliferation, migration, invasion and metastasis [[45], [46], [47]]. Therefore, exploring the potential value of lncRNAs in LUAD helps us to elucidate the development of LUAD and aids us in identifying novel therapeutic strategies.
Although there have been studies on the role of PCD-related lncRNAs as biomarkers in cancer, most are based on a single PCD modality [48,49]. The crosstalk between different PCD modalities allows some molecules to play a regulatory role in multiple PCDs, as evidenced by the introduction of the concept of PANoptosis [9]. However, the role of PANoptosis-associated lncRNAs as predictive biomarkers in cancer prognosis and TIME is unclear. In this work, we generated a novel PRLSig to predict the TIME landscape and clinical outcomes of individuals with LUAD. The PRLSig was revealed to be an independent prognostic indicator for patients. Moreover, its predictive efficacy is superior to other clinicopathological parameters. Among the nine PRLs in the PRLSig, FAM83A-AS1 was shown to promote LUAD cell migration and invasion by targeting miR-150-5p and modifying MMP14 [50] and induce autophagy through the MET-AMPKɑ signalling pathway [51]. Additionally, FAM83A-AS1 promotes tumour proliferation and migration in LUAD through the HIF-1α/glycolytic axis [52]. However, the role of AC018529.1, LINC01337, AL596223.1, AC005041.3, LINC02576, AL360270.1, AC131237.1 and FO680682.1 in lung cancer remains unexplored. Considering that these eight PRLs are key lncRNAs in the construction of the PRLSig, their regulation in LUAD mechanisms deserve further exploration.
Immunotherapy is an epoch-making therapy in the field of tumour treatment, with ICBs providing the promising potential to patients with tumours. Unlike traditional cancer treatments, ICBs can mobilise the body's immune function to fight tumours by restoring the function of T cells and enhancing their recognition of tumour cells. However, the low overall effectiveness of ICBs is a bottleneck in clinical immunotherapy. A major cause of this low efficacy is inadequate infiltration of immune cells in tumours, often referred to as ‘cold tumours’ [53,54]. Contrastingly, ‘hot immune tumours’ are more potentially able to profit from ICBs and are featured by the activation of ICs and high infiltration of effector immune cells [53,55]. Thus, tumour immunophenotype severely influences patient response to treatment with ICBs. In our results, infiltration of most immune cells such as CD8+ T lymphocytes, NK cells, macrophages and neutrophils, were negatively associated with scores in the PRLSig, which was further validated in the ESTIMATE and ssGSEA analyses. These scores suggest a low immune cell infiltration status in high-risk patients, which may partly contribute to the poor clinical outcomes. Furthermore, the fact that most ICs, including PD-L1, were highly expressed in low-risk population, which, combined with the high immune infiltration status of low-risk population, suggests that low-risk individuals are more likely to be characterised with ‘hot tumours’ and could benefit more from treatment with ICBs than high-risk patients. IPS analysis further validated this conclusion. Notably, PD-L1 is currently the only biomarker approved by the US food and drug administration to screen for NSCLC immunotherapy superiority, further suggesting the predictive potential of PRLSig in identifying the therapeutic response of ICBs. TMB is another candidate biomarker for predicting the efficacy of ICBs, with a high TMB theoretically implying an increase in the number of neoantigens and consequently enhanced immunogenicity [[56], [57], [58]]. Our findings suggest that PRLSig combined with TMB may be a better predictor of clinical outcomes in LUAD.
In terms of biological behaviour, LUAD is more aggressive and has a worse prognosis. The discovery of driver-mutated genes has led to breakthroughs in the treatment of LUAD, with the epidermal growth factor receptor (EGFR) being the driver-mutated gene reported in LUAD. Moreover, erlotinib, a small molecule tyrosine kinase inhibitor (TKI) targeting EGFR, significantly prolongs survival and improves the quality of life in individuals with advanced NSCLC [59]. In this study, high-risk individuals were more sensitive to erlotinib, suggesting that the PRLSig combined with the EGFR mutation status of patients with LUAD could better predict the response to EGFR TKIs.
This is the first comprehensive study to report the role of PRLs in the prognosis, immune profile, potential biological behaviour and personalised treatment of patients with LUAD. Therefore, based on current knowledge of the interactions between different PCDs, further studies on the potential mechanisms of PANoptosis in human cancers can be performed.
Despite the different methodological evaluations of the constructed PRLSig in the present study, there are several limitations. First, this study was unable to investigate data bias in our studies. Second, the clinical value of PRLSig for patients with PAAD is yet to be further verified in prospective studies. Furthermore, lncRNA is not yet extensively applied in clinical practice as a predictor of prognosis and efficacy. However, it should be emphasized that studies have shown that circulating lncRNAs are stably available in urine, plasma and serum of tumor patients and could be used as non-invasive biomarkers [60,61]. Therefore, the identification of lncRNA biomarkers and their clinical applications are promising in the future.
5. Conclusions
This study identified a novel prognosis predictive signature, which could provide new insights into the relationship between PANoptosis and LUAD. Furthermore, the developed PRLSig can well predict the clinical outcomes and TIME in LUAD, thereby screening the superior population for ICBs and informing the selection of personalised therapeutic regimens.
Data availability
Data included in article/supp. material/referenced in article.
CRediT authorship contribution statement
Lingling Bao: Conceptualization, Methodology, Writing – original draft. Yingquan Ye: Conceptualization, Methodology, Software. Xuede Zhang: Investigation, Methodology, Data curation, Conceptualization. Xin Xu: Project administration. Wenjuan Wang: Formal analysis, Visualization. Bitao Jiang: Project administration, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Bullet Edits Limited for the linguistic editing and proofreading of the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e29869.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 Countries. CA: a cancer journal for clinicians. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Brody H. Lung cancer. Nature. 2020;587:S7. doi: 10.1038/d41586-020-03152-0. [DOI] [PubMed] [Google Scholar]
- 3.Wang M., Herbst R.S., Boshoff C. Toward personalized treatment approaches for non-small-cell lung cancer. Nature medicine. 2021;27:1345–1356. doi: 10.1038/s41591-021-01450-2. [DOI] [PubMed] [Google Scholar]
- 4.Song Y., Chen D., Zhang X., Luo Y., Li S. Integrating genetic mutations and expression profiles for survival prediction of lung adenocarcinoma. Thoracic cancer. 2019;10:1220–1228. doi: 10.1111/1759-7714.13072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang F., He Y., Zhai Z., Sun E. Programmed cell death pathways in the Pathogenesis of systemic Lupus Erythematosus. Journal of immunology research. 2019;2019 doi: 10.1155/2019/3638562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bedoui S., Herold M.J., Strasser A. Emerging connectivity of programmed cell death pathways and its physiological implications. Nat. Rev. Mol. Cell Biol. 2020;21:678–695. doi: 10.1038/s41580-020-0270-8. [DOI] [PubMed] [Google Scholar]
- 7.Kajarabille N., Latunde-Dada G.O. Programmed cell-death by Ferroptosis: Antioxidants as Mitigators. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20194968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frank D., Vince J.E. Pyroptosis versus necroptosis: similarities, differences, and crosstalk. Cell Death Differ. 2019;26:99–114. doi: 10.1038/s41418-018-0212-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malireddi R.K.S., Kesavardhana S., Kanneganti T.D. ZBP1 and TAK1: master regulators of NLRP3 Inflammasome/pyroptosis, apoptosis, and necroptosis (PAN-optosis) Front. Cell. Infect. Microbiol. 2019;9:406. doi: 10.3389/fcimb.2019.00406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christgen S., Zheng M., Kesavardhana S., Karki R., Malireddi R.K.S., Banoth B., Place D.E., Briard B., Sharma B.R., Tuladhar S., Samir P., Burton A., Kanneganti T.D. Identification of the PANoptosome: a molecular platform Triggering pyroptosis, apoptosis, and necroptosis (PANoptosis) Front. Cell. Infect. Microbiol. 2020;10:237. doi: 10.3389/fcimb.2020.00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan H., Pan J., Li P., Gao J. Characterization of PANoptosis patterns predicts survival and immunotherapy response in gastric cancer. Clin. Immunol. 2022;238 doi: 10.1016/j.clim.2022.109019. [DOI] [PubMed] [Google Scholar]
- 12.Malireddi R.K.S., Karki R., Sundaram B., Kancharana B., Lee S., Samir P., Kanneganti T.D. Inflammatory cell death, PANoptosis, mediated by Cytokines in Diverse cancer Lineages Inhibits tumor growth. ImmunoHorizons. 2021;5:568–580. doi: 10.4049/immunohorizons.2100059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karki R., Sundaram B., Sharma B.R., Lee S., Malireddi R.K.S., Nguyen L.N., Christgen S., Zheng M., Wang Y., Samir P., Neale G., Vogel P., Kanneganti T.D. ADAR1 restricts ZBP1-mediated immune response and PANoptosis to promote tumorigenesis. Cell Rep. 2021;37 doi: 10.1016/j.celrep.2021.109858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huarte M. The emerging role of lncRNAs in cancer. Nature medicine. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 15.Wang L., Cho K.B., Li Y., Tao G., Xie Z., Guo B. Long Noncoding RNA (lncRNA)-Mediated competing endogenous RNA networks provide novel potential biomarkers and therapeutic targets for colorectal cancer. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20225758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhen Q., Gao L.N., Wang R.F., Chu W.W., Zhang Y.X., Zhao X.J., Lv B.L., Liu J.B. LncRNA DANCR promotes lung cancer by Sequestering miR-216a, cancer control. journal of the Moffitt Cancer Center. 2018;25 doi: 10.1177/1073274818769849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y., Zhang Y., Lu J., Liu Z., Zhao S., Zhang M., Lu M., Xu W., Sun F., Wu Q., Zhong Q., Cui Z. Characteristics of prognostic programmed cell death-related long Noncoding RNAs associated with immune infiltration and therapeutic responses to Colon cancer. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.828243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li G., Liu K., Du X. Long non-coding RNA TUG1 promotes proliferation and Inhibits apoptosis of Osteosarcoma cells by Sponging miR-132-3p and Upregulating SOX4 expression. Yonsei Med. J. 2018;59:226–235. doi: 10.3349/ymj.2018.59.2.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo T., Yu S., Ouyang J., Zeng F., Gao L., Huang S., Wang X. Identification of a apoptosis-related LncRNA signature to improve prognosis prediction and immunotherapy response in lung adenocarcinoma patients. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.946939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mao F., Li Z., Li Y., Huang H., Shi Z., Li X., Wu D., Liu H., Chen J. Necroptosis-related lncRNA in lung adenocarcinoma: a comprehensive analysis based on a prognosis model and a competing endogenous RNA network. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.940167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang P, Wang Z, Lin Y, Castellano L, Stebbing J, Zhu L, Peng L. Development of a Novel Pyroptosis-Associated lncRNA Biomarker Signature in Lung Adenocarcinoma. Molecular biotechnology. 2024;66(2):332–353. doi: 10.1007/s12033-023-00757-4. [DOI] [PubMed] [Google Scholar]
- 22.Lee S., Karki R., Wang Y., Nguyen L.N., Kalathur R.C., Kanneganti T.D. AIM2 forms a complex with pyrin and ZBP1 to drive PANoptosis and host defence. Nature. 2021;597:415–419. doi: 10.1038/s41586-021-03875-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng M., Karki R., Vogel P., Kanneganti T.D. Caspase-6 is a key regulator of Innate immunity, inflammasome activation, and host Defense. Cell. 2020;181:674–687 e613. doi: 10.1016/j.cell.2020.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu J., Hong M., Li Y., Chen D., Wu Y., Hu Y. Programmed cell death Tunes tumor immunity. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.847345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karki R., Sharma B.R., Lee E., Banoth B., Malireddi R.K.S., Samir P., Tuladhar S., Mummareddy H., Burton A.R., Vogel P., Kanneganti T.D. Interferon regulatory factor 1 regulates PANoptosis to prevent colorectal cancer. JCI Insight. 2020;5 doi: 10.1172/jci.insight.136720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samir P., Malireddi R.K.S., Kanneganti T.D. The PANoptosome: a Deadly protein complex driving pyroptosis, apoptosis, and necroptosis (PANoptosis) Front. Cell. Infect. Microbiol. 2020;10:238. doi: 10.3389/fcimb.2020.00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin J.F., Hu P.S., Wang Y.Y., Tan Y.T., Yu K., Liao K., Wu Q.N., Li T., Meng Q., Lin J.Z., Liu Z.X., Pu H.Y., Ju H.Q., Xu R.H., Qiu M.Z. Phosphorylated NFS1 weakens oxaliplatin-based chemosensitivity of colorectal cancer by preventing PANoptosis. Signal Transduct Target Ther. 2022;7:54. doi: 10.1038/s41392-022-00889-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sundaram B., Kanneganti T.D. Advances in Understanding activation and function of the NLRC4 inflammasome. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22031048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li T., Fan J., Wang B., Traugh N., Chen Q., Liu J.S., Li B., Liu X.S. TIMER: a Web Server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77:e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li T., Fu J., Zeng Z., Cohen D., Li J., Chen Q., Li B., Liu X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic acids research. 2020;48:W509–w514. doi: 10.1093/nar/gkaa407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshihara K., Shahmoradgoli M., Martínez E., Vegesna R., Kim H., Torres-Garcia W., Treviño V., Shen H., Laird P.W., Levine D.A., Carter S.L., Getz G., Stemke-Hale K., Mills G.B., Verhaak R.G. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yi M., Niu M., Xu L., Luo S., Wu K. Regulation of PD-L1 expression in the tumor microenvironment. J. Hematol. Oncol. 2021;14:10. doi: 10.1186/s13045-020-01027-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu H., Boyle T.A., Zhou C., Rimm D.L., Hirsch F.R. PD-L1 expression in lung cancer. J. Thorac. Oncol. : official publication of the International Association for the Study of Lung Cancer. 2016;11:964–975. doi: 10.1016/j.jtho.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yi M., Jiao D., Xu H., Liu Q., Zhao W., Han X., Wu K. Biomarkers for predicting efficacy of PD-1/PD-L1 inhibitors. Mol. Cancer. 2018;17:129. doi: 10.1186/s12943-018-0864-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Charoentong P., Finotello F., Angelova M., Mayer C., Efremova M., Rieder D., Hackl H., Trajanoski Z. Pan-cancer immunogenomic analyses reveal Genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18:248–262. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 38.Geeleher P., Cox N., Huang R.S. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One. 2014;9 doi: 10.1371/journal.pone.0107468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sha D., Jin Z., Budczies J., Kluck K., Stenzinger A., Sinicrope F.A. Tumor mutational burden as a predictive biomarker in Solid tumors. Cancer Discov. 2020;10:1808–1825. doi: 10.1158/2159-8290.CD-20-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma P., Siddiqui B.A., Anandhan S., Yadav S.S., Subudhi S.K., Gao J., Goswami S., Allison J.P. The next decade of immune checkpoint therapy. Cancer Discov. 2021;11:838–857. doi: 10.1158/2159-8290.CD-20-1680. [DOI] [PubMed] [Google Scholar]
- 41.Malireddi R.K.S., Tweedell R.E., Kanneganti T.D. PANoptosis components, regulation, and implications. Aging. 2020;12:11163–11164. doi: 10.18632/aging.103528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malireddi R.K.S., Kesavardhana S., Karki R., Kancharana B., Burton A.R., Kanneganti T.D. RIPK1 distinctly regulates Yersinia-Induced inflammatory cell death. PANoptosis, ImmunoHorizons. 2020;4:789–796. doi: 10.4049/immunohorizons.2000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Babamale A.O., Chen S.T. Nod-like receptors: critical Intracellular Sensors for host Protection and cell death in Microbial and Parasitic infections. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms222111398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang M., Qi L., Li L., Wu Y., Song D., Li Y. Caspase-8: a key protein of cross-talk signal way in "PANoptosis" in cancer. Int. J. Cancer. 2021;149:1408–1420. doi: 10.1002/ijc.33698. [DOI] [PubMed] [Google Scholar]
- 45.Pan J., Fang S., Tian H., Zhou C., Zhao X., Tian H., He J., Shen W., Meng X., Jin X., Gong Z. lncRNA JPX/miR-33a-5p/Twist 1 axis regulates tumorigenesis and metastasis of lung cancer by activating Wnt/β-catenin signaling. Mol. Cancer. 2020;19:9. doi: 10.1186/s12943-020-1133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang W., Bai M., Liu K., Tan J., Ma J., Zhao J., Hou P. LncRNA surfactant associated 1 activates large tumor suppressor kinase 1/Yes-associated protein pathway via modulating hypoxic exosome-delivered miR-4766-5p to inhibit lung adenocarcinoma metastasis. Int. J. Biochem. Cell Biol. 2022;153 doi: 10.1016/j.biocel.2022.106317. [DOI] [PubMed] [Google Scholar]
- 47.Li Y., Chen C., Liu H.L., Zhang Z.F., Wang C.L. LARRPM restricts lung adenocarcinoma progression and M2 macrophage polarization through epigenetically regulating LINC00240 and CSF1. Cellular & molecular biology letters. 2022;27:91. doi: 10.1186/s11658-022-00376-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang W., Ye Y., Zhang X., Ye X., Liu C., Bao L. Construction of a necroptosis-associated long non-coding RNA signature to predict prognosis and immune response in hepatocellular carcinoma. Front. Mol. Biosci. 2022;9 doi: 10.3389/fmolb.2022.937979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang X., Mei M., Yang J., Guo J., Du F., Liu S. Ferroptosis-related long non-coding RNA signature predicts the prognosis of hepatocellular carcinoma. Aging. 2022;14:4069–4084. doi: 10.18632/aging.204073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao G., Wang P., Zheng X., Liu D., Sun X. FAM83A-AS1 promotes lung adenocarcinoma cell migration and invasion by targeting miR-150-5p and modifying MMP14. Cell Cycle. 2019;18:2972–2985. doi: 10.1080/15384101.2019.1664225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao H., Wang Y., Wu X., Zeng X., Lin B., Hu S., Zhang S., Li Y., Zhou Z., Zhou Y., Du C., Beer D.G., Bai S., Chen G. FAM83A antisense RNA 1 (FAM83A-AS1) silencing impairs cell proliferation and induces autophagy via MET-AMPKɑ signaling in lung adenocarcinoma. Bioengineered. 2022;13:13312–13327. doi: 10.1080/21655979.2022.2081457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Z., Hu Z., Sui Q., Huang Y., Zhao M., Li M., Liang J., Lu T., Zhan C., Lin Z., Sun F., Wang Q., Tan L. LncRNA FAM83A-AS1 facilitates tumor proliferation and the migration via the HIF-1α/glycolysis axis in lung adenocarcinoma. Int. J. Biol. Sci. 2022;18:522–535. doi: 10.7150/ijbs.67556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Galon J., Bruni D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat. Rev. Drug Discov. 2019;18:197–218. doi: 10.1038/s41573-018-0007-y. [DOI] [PubMed] [Google Scholar]
- 54.Bonaventura P., Shekarian T., Alcazer V., Valladeau-Guilemond J., Valsesia-Wittmann S., Amigorena S., Caux C., Depil S. Cold tumors: a therapeutic Challenge for immunotherapy. Front. Immunol. 2019;10:168. doi: 10.3389/fimmu.2019.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu Y.T., Sun Z.J. Turning cold tumors into hot tumors by improving T-cell infiltration. Theranostics. 2021;11:5365–5386. doi: 10.7150/thno.58390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chan T.A., Yarchoan M., Jaffee E., Swanton C., Quezada S.A., Stenzinger A., Peters S. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann. Oncol. : official journal of the European Society for Medical Oncology. 2019;30:44–56. doi: 10.1093/annonc/mdy495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gubin M.M., Artyomov M.N., Mardis E.R., Schreiber R.D. Tumor neoantigens: building a framework for personalized cancer immunotherapy. The Journal of clinical investigation. 2015;125:3413–3421. doi: 10.1172/JCI80008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cristescu R., Mogg R., Ayers M., Albright A., Murphy E., Yearley J., Sher X., Liu X.Q., Lu H., Nebozhyn M., Zhang C., Lunceford J.K., Joe A., Cheng J., Webber A.L., Ibrahim N., Plimack E.R., Ott P.A., Seiwert T.Y., Ribas A., McClanahan T.K., Tomassini J.E., Loboda A., Kaufman D. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science. 2018;362 doi: 10.1126/science.aar3593. New York, N.Y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ricciardi S., Tomao S., de Marinis F. Efficacy and safety of erlotinib in the treatment of metastatic non-small-cell lung cancer. Lung Cancer. 2011;2:1–9. doi: 10.2147/LCTT.S10167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qi P., Zhou X.Y., Du X. Circulating long non-coding RNAs in cancer: current status and future perspectives. Mol. Cancer. 2016;15:39. doi: 10.1186/s12943-016-0524-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin L.Y., Yang L., Zeng Q., Wang L., Chen M.L., Zhao Z.H., Ye G.D., Luo Q.C., Lv P.Y., Guo Q.W., Li B.A., Cai J.C., Cai W.Y. Tumor-originated exosomal lncUEGC1 as a circulating biomarker for early-stage gastric cancer. Mol. Cancer. 2018;17:84. doi: 10.1186/s12943-018-0834-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supp. material/referenced in article.