Abstract
Background & Aims
In the classic form of α1-antitrypsin deficiency (ATD), the misfolded α1-antitrypsin Z (ATZ) variant accumulates in the endoplasmic reticulum (ER) of liver cells. A gain-of-function proteotoxic mechanism is responsible for chronic liver disease in a subgroup of homozygotes. Proteostatic response pathways, including conventional endoplasmic reticulum–associated degradation and autophagy, have been proposed as the mechanisms that allow cellular adaptation and presumably protection from the liver disease phenotype. Recent studies have concluded that a distinct lysosomal pathway called endoplasmic reticulum–to-lysosome completely supplants the role of the conventional macroautophagy pathway in degradation of ATZ. Here, we used several state-of-the-art approaches to characterize the proteostatic responses more fully in cellular systems that model ATD.
Methods
We used clustered regularly interspaced short palindromic repeats (CRISPR)-mediated genome editing coupled to a cell selection step by fluorescence-activated cell sorter to perform screening for proteostasis genes that regulate ATZ accumulation and combined that with selective genome editing in 2 other model systems.
Results
Endoplasmic reticulum–associated degradation genes are key early regulators and multiple autophagy genes, from classic as well as from ER-to-lysosome and other newly described ER-phagy pathways, participate in degradation of ATZ in a manner that is temporally regulated and evolves as ATZ accumulation persists. Time-dependent changes in gene expression are accompanied by specific ultrastructural changes including dilation of the ER, formation of globular inclusions, budding of autophagic vesicles, and alterations in the overall shape and component parts of mitochondria.
Conclusions
Macroautophagy is a critical component of the proteostasis response to cellular ATZ accumulation and it becomes more important over time as ATZ synthesis continues unabated. Multiple subtypes of macroautophagy and nonautophagic lysosomal degradative pathways are needed to respond to the high concentrations of misfolded protein that characterizes ATD and these pathways are attractive candidates for genetic variants that predispose to the hepatic phenotype.
Keywords: Autophagy, Proteasome, Aggregation-Prone Proteins, α1-Antitrypsin Deficiency, Liver Disease
Summary.
This study uses genome engineering in model systems to show how specific genes and pathways facilitate the capacity of cells to adapt to the genetic variant of α1-antitrypsin deficiency, identifying those pathways as candidates for novel therapeutic targets.
Maintenance of protein homeostasis, or what has come to be called proteostasis, is a critical determinant of cellular health. Research over the past several decades has shown that a network of multiple mechanisms, including molecular chaperones and proteolytic machineries, are involved.1 This complex network is designed to ensure that protein synthesis, polypeptide folding, and translocation to the proper destination are performed efficiently and the importance of degradative mechanisms in eliminating misfolded and aggregated proteins has been one of the major advances in the field. One of the first of these degradative mechanisms to be identified is the pathway known as endoplasmic reticulum-associated degradation (ERAD), in which misfolded proteins are translocated from the lumen, or membrane, of the endoplasmic reticulum (ER) into the cytoplasm for degradation by the proteasome.1 It now appears that there are multiple variations in how ERAD works for different cargo, including different chaperones, membrane channels and/or translocators, cytoplasmic adaptors, and components of the ubiquitin system. These various ERAD mechanisms are linked closely to the unfolded protein response (UPR), a transcriptionally activated program designed to restore homeostasis by increasing production of chaperones and degradative machinery while inhibiting protein translation.2
Autophagy has been implicated as the second major pathway that participates in proteostasis. It mediates delivery to the lysosome for degradation of cellular materials including misfolded and aggregated proteins, lipids, as well as parts of subcellular organelles.3 Three distinct types of autophagy have been described: macroautophagy, microautophagy, and chaperone-mediated autophagy. In macroautophagy, cargo is sequestered in newly formed double-membrane vesicles, transported, and delivered to lysosomes by vesicular fusion. In microautophagy, invaginations at the lysosomal membrane internalize cytosolic cargo into small vesicles that then detach into the lumen of the lysosome for degradation. Chaperone-mediated autophagy is a pathway for degradation of soluble cytosolic proteins that are translocated directly across the lysosomal membrane for degradation without the requirement of vesicle formation or fusion. It now appears that macroautophagy and its canonical machinery is actually an umbrella for multiple pathways that can target different subcellular compartments and different types of cargo. Several recent exciting studies have led to the identification of types of autophagy that target cargo in the ER and have been categorized as ER-phagy pathways.4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16 These have included specific cargo that bind to ER surface proteins, such as reticulophagy regulator (FAM134B), reticulon 3 (RTN3L), testis expressed 264, ER-phagy receptor (TEX264), atlastin GTPase 3 (ATL3), vacuolar protein sorting 13 (VPS13), and SEC24 homolog C (SEC24C), which have been referred to as receptors or adaptors and bear microtubule associated protein 1 light chain 3 alpha (LC3)/GABA type A receptor-associated protein (GABARAP)-interacting regions (LIRs/GIMs) to recruit autophagy machinery. Soluble ER proteins also may be subject to 2 recently described distinct ER-phagy pathways, one in which p62 acts in cooperation with the E3-ubiquitin ligase tripartite motif-containing (TRIM13),17 and another involving calcium binding and coiled-coil domain 1 (CALCOCO1) cooperation with vesicle associated membrane protein (VAMP)-associated proteins vamp associated protein A (VAPA) and vamp associated protein B (VAPB).18
There is relatively limited information about how ERAD and autophagy work together or are coordinated. Most of what is known about the 2 overall pathways suggests that ERAD is designed predominantly for unfolded and not for polymerized and/or aggregated proteins because the proteasome could not physically accommodate the larger structures. Autophagy has been thought of as the pathway for polymerized and aggregated proteins. Furthermore, the autophagic mechanisms would appear to be able to adapt to larger total amounts of cargo and therefore is viewed as providing higher overall degradative capacity than ERAD. In terms of coordination, several studies have indicated that autophagy is activated during the UPR, but other studies have shown how autophagy and newer ER-phagy pathways can operate in the absence of the UPR.2,4,19
Proteostasis mechanisms and regulation are very important considerations in understanding the clinical effects of α1-antitrypsin deficiency (ATD) and potential novel therapies. In the classic form of ATD, a point mutation leads to misfolding of one of the most abundant secretory proteins produced by liver cells.20 The α1-antitrypsin Z (ATZ) variant also has a tendency to polymerize and aggregate, all leading to massive globular accumulations of ER that are seen in the liver of affected homozygotes. Together, the abundance of daily production and the tendency to polymerize have led logically to considerations of how the accumulation of the variant affects the cell and how it ultimately is degraded. We know that chronic liver disease is caused by a gain-of-function, proteotoxic mechanism and that a subgroup of homozygotes are severely affected and this has led to the concept that genetic and/or environmental modifiers are predisposing or protective. It would be logical for genetic variants and environmental effects to target components of the proteostasis machinery. This concept has been supported by several studies in which the cellular accumulation and proteotoxicity of ATZ is mitigated by drugs and genetic manipulation, which enhance autophagy in experimental model systems.21,22 Studies in model systems have shown that multiple pathways contribute to cellular proteolysis of ATZ, including ERAD with ubiquitin-dependent and ubiquitin-independent mechanisms, Golgi-to-lysosome, ER-to-lysosome (ERLAD), and several autophagolysosomal pathways.6,16,23, 24, 25, 26, 27
In their article in 2018, Fregno et al28 showed that ATZ may be delivered from the ER to the lysosome by a vesicular pathway that is distinct from macroautophagy. They called this pathway ERLAD, and concluded that it, rather than macroautophagy, is solely responsible for the intracellular degradation of ATZ that is attributable to a lysosomal end point. The concept that a single pathway could account for the degradation of this abundant protein and handle that load in affected patients over long periods of time seemed questionable to us and also seemed contrary to the increasing number of molecules and pathways being implicated in proteostasis of ER proteins. In the current study, we used 2 approaches to address this issue: a screen of proteostasis genes that regulated the cellular accumulation of ATZ using CRISPR-mediated genome editing; deletion of specific autophagy genes in cell line and nematode models of ATZ accumulation. We also used cell line systems with inducible expression of ATZ to evaluate the possibility that temporal factors determine which genes and pathways are involved. The results show that genes of the ERAD system are critically important in an initial time period but autophagy genes became important over time. Knockouts of multiple autophagy genes led to greater cellular accumulation of ATZ, suggesting that multiple subtypes of autophagy and lysosomal pathways are needed to respond to the high concentrations of misfolded protein that characterize this genetic variant and that the ERLAD pathway is not sufficient to account for overall mechanisms needed. Morphologic investigations showed time-dependent evolution of massive globules surrounded by an intensifying autophagosomal reaction with classic autophagosome and autolysosomal structures.
Results
Pooled CRISPR Screening for ATZ Accumulation Identifies Genes From ERAD and Autophagy Pathways as Well as the Chaperonin T-complex 1-Ring Complex as a Novel Modulator
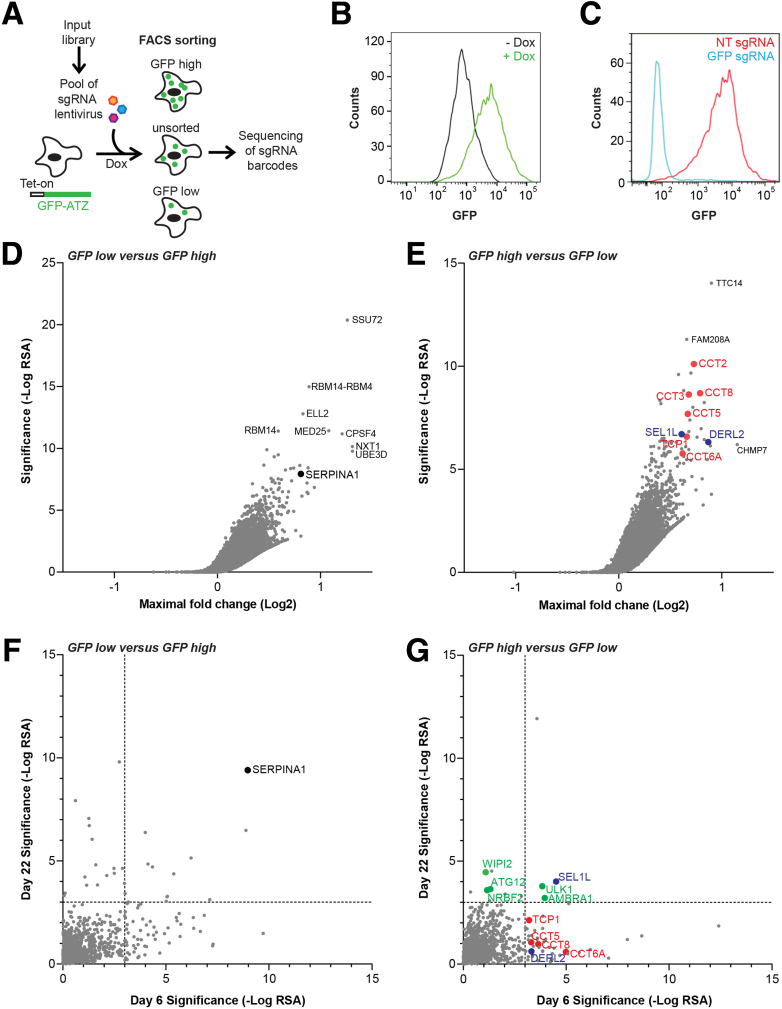
We set out to identify cellular nodes that regulate proteostasis of mutant ATZ by screening for modulators of green fluorescent protein (GFP)-tagged ATZ using a previously validated fluorescence-activated cell sorter (FACS)-based CRISPR screening paradigm29, 30, 31 as depicted in Figure 1A. As a cellular model, we picked human hepatoma cell line HepG232 because of their human origin and because they are characterized by a relatively high degree of hepatocytic differentiation. Although ATZ is expressed in other cell types, its predominant site of synthesis is hepatocytes, and intracellular inclusions containing ATZ are found predominantly in hepatocytes. The HepG2 cell line was engineered to stably express the CRISPR-associated protein Cas9 nuclease and doxycycline-inducible GFP-ATZ (HepG2 Cas9 GFP-ATZ) and then was studied after 7 days in the absence or presence of doxycycline. The HepG2 Cas9 GFP-ATZ cell line contained the regulatory element for expression of GFP-ATZ when doxycycline was added to the medium and FACS analysis confirmed marked expression of GFP-ATZ upon doxycycline treatment (Figure 1B). A GFP-targeting single-guide RNA (sgRNA) robustly reduced GFP fluorescence under doxycycline induction, consistent with efficient CRISPR editing (Figure 1C). We screened a genome-wide pooled sgRNA library, quantifying sgRNA abundance in unsorted, GFP-high and GFP-low cell populations. As a positive control, we found sgRNAs against SERPINA1, the gene encoding α1-antitrypsin (AT), robustly enriched in the GFP-low vs GFP-high cell population (Figure 1D). To identify modulators that stabilize GFP-ATZ, we analyzed sgRNAs enriched in GFP-high vs GFP-low cells (see Supplementary Tables 1 and 2) and observed the ERAD components SEL1L and DERL2 as robust hits (Figure 1E). In addition, several components of the chaperonin TCP1-ring complex components TCP1, chaperonin containing tailless complex polypeptide CCT2, CCT3, CCT5, CCT6A, and CCT8 were enriched significantly in the GFP-high population. The chaperonin TCP1-ring complex has been linked previously to the regulation of protein aggregation33, 34, 35 and is required for the efficient degradation of autophagosomes and autophagy cargo such as mutant Huntingtin.36 However, canonical autophagy components did not score as strong modulators of GFP-ATZ. To confirm the screening results and characterize time-dependency, we rescreened a set of candidates for modulation of GFP-ATZ levels after 6 and 22 days of transgene expression. For this purpose, we used a Cas9 clonal line and screened a mini-pool library consisting of the 300 top-ranking genes from the comparisons of GFP-high vs GFP-low cells, GFP-low vs GFP-high cells, and unsorted cells vs input library. Furthermore, sgRNAs against components of the autophagy machinery were supplemented. The technical validity of the FACS-based mini-pool screen was confirmed by the strong enrichment of SERPINA1-targeting sgRNAs in GFP-low vs GFP-high cells at both time points (Figure 1F). Components of the chaperonin TCP1-ring complex and ERAD components scored as strong modulators of GFP-ATZ after 6 days of transgene expression, consistent with the genome-wide screen, but the effect was less pronounced on day 22 (Figure 1G). By analyzing autophagy components, we observed that unc-51 like autophagy activating kinase 1 (ULK1) and autophagy and beclin 1 regulator 1 (AMBRA1) were enriched in GFP-high vs GFP-low cells at both time points, whereas other autophagy components such as autophagy related 12 (ATG12), WD repeat domain-containing phosphoinositide interacting 2 (WIPI2), and nuclear receptor binding factor 2 (NRBF2) exclusively scored on day 22.
Figure 1.
Genome-wide pooled CRISPR screen for modulators of ATZ proteostasis. (A) Schematic representation of the FACS-based pooled CRISPR screening paradigm for modulators of GFP-ATZ levels. FACS analysis of GFP fluorescence in HepG2 Cas9 Tet-On GFP-ATZ cells cultured in the (B) presence or absence of doxycycline for 7 days and then (C) infected with GFP or nontargeting sgRNAs. Gene-centric view of the genome-wide comparisons of (D) unsorted cells vs the input library, (E) GFP-low vs GFP-high cell populations, and (F and G) of mini-pool comparison of GFP-high vs GFP-low cell populations on day 6 and day 22. SERPINA1 (black), ERAD (blue), autophagy (green), and components of the chaperonin TCP1-ring complex (red) are highlighted. Source data are shown in (D and E) Supplementary Table 1 and (F and G) Supplementary Table 2. Dox, doxycycline; NT, non-targeted control; RSA, redundant siRNA activity.
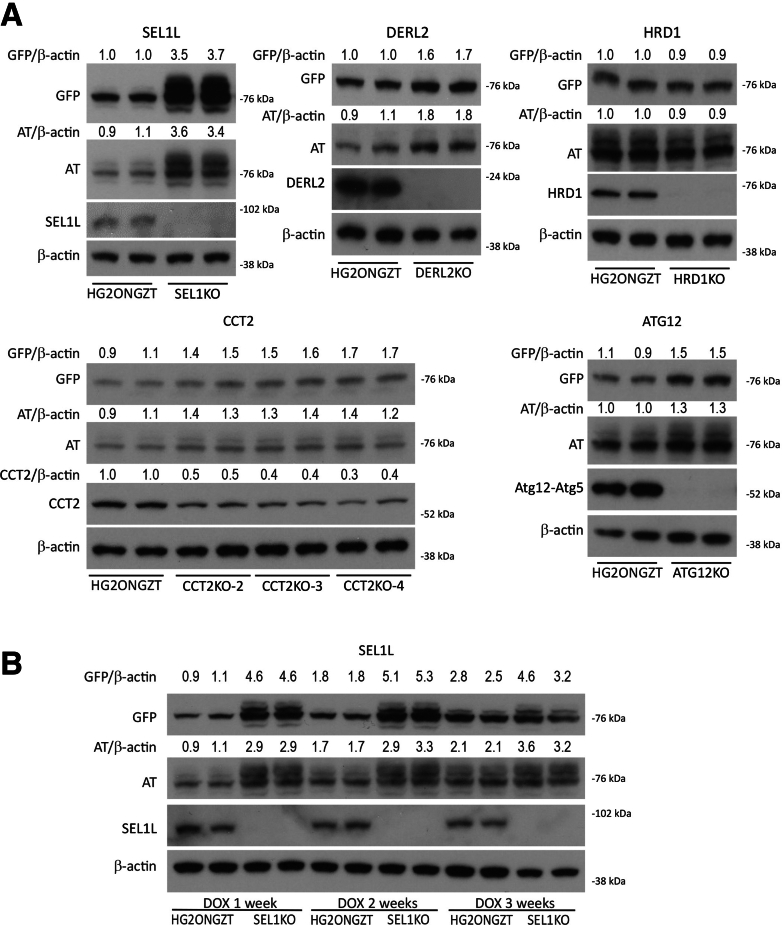
Based on this screening data, we next sought to validate representative genes by selective deletion in the HepG2-GFP-ATZ cell line, including SEL1L1, DERL2, CCT2, and ATG12. Unfortunately, we were only able to recover viable clones with homozygous deletion in the case of SEL1L1, but we were able to investigate the others using pooled heterozygous clones for at least the initial phases of culturing. Figure 2A shows Western blot analysis for steady-state levels in lines deleted for these selected genes 7 days after induction of ATZ expression. The results show increased accumulation when SEL1L1 and DERL2 are deleted but not for hydroxy-3-methylglutaryl reductase degradation 1 (HRD1). There also is increased accumulation of ATZ when CCT2 and ATG12 are targeted for deletion. In each case, the greater accumulation of ATZ was detected by antibodies to GFP and AT. These results provide confirmation of the validity of the screen for identification of proteostasis regulators.
Figure 2.
Steady-state levels of ATZ in the HepG2-GFP-ATZ cell line deleted for specific genes identified in the CRISPR screen. (A) In each case the cells were incubated in the presence of doxycycline for 1 week to induce ATZ expression. Cell homogenates were subjected to immunoblot analysis with antibody to GFP (GFP), polyclonal antibody to all forms of AT (AT), and antibody to the relevant deleted gene. Densitometric quantification is shown at the top of the blots. For CCT2, 3 separate clones of the deleted cell line, CCT2KO-2, -3, and -4, are shown. (B) HepG2-GFP-ATZ cell line with deletion of SEL1L1 was evaluated at 3 different time points after addition of doxycycline. DOX, doxycycline.
To validate the temporal pattern by which ATZ accumulation was regulated by specific genes in the HepG2-GFP-ATZ cell line with specific genomic deletions, we were only able to use the clones with homozygous deletion of SEL1L1 (Figure 2B). The results showed that the effect of SEL1L1 deletion progressively declined from day 7 to day 14, and then day 21, consistent with the concept that ERAD is important in the early phases of cellular ATZ accumulation and suggesting the possibility that other mechanisms of degradation are ramped up over time and can supplant the need for the ERAD mechanism.
Multiple Autophagy Genes Modulate Cellular Accumulation of ATZ, Implicating Multiple Autophagic and Lysosomal Pathways in its Disposal
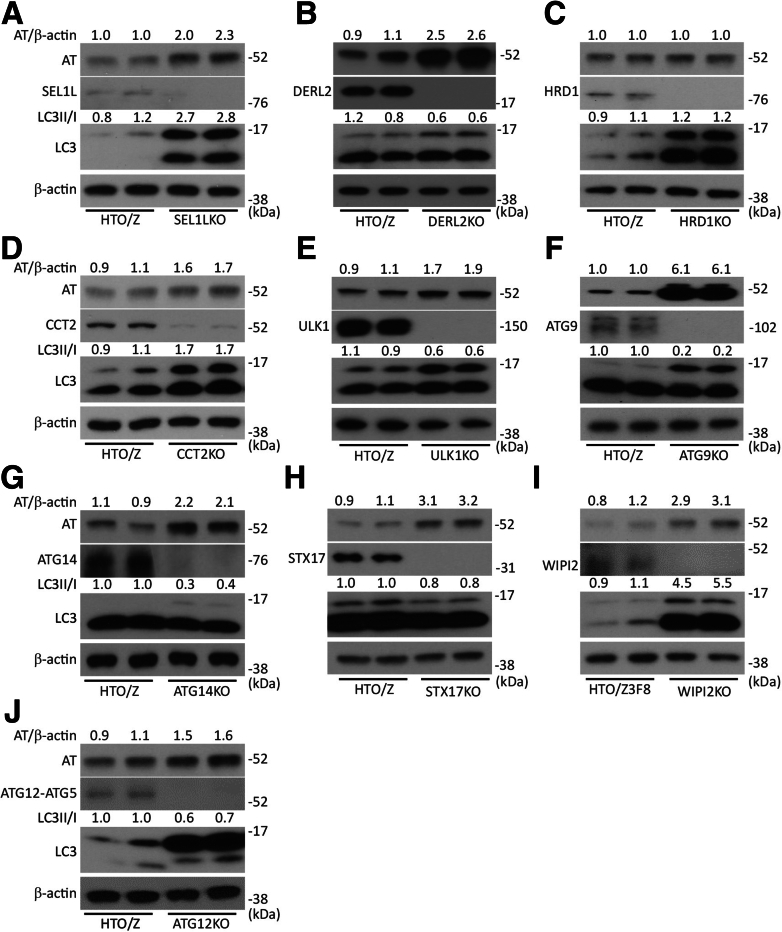
To confirm the importance of the genes identified in the screen and to perform more extensive time course experiments, we investigated the possibility of introducing specific gene deletions in a HeLa-based cell line model with inducible expression of ATZ, the HeLa Tet-off Z (HTO/Z) cell line. The HTO/Z cell line has performed particularly well in reproducibility and time-dependent analyses and we were able to recover viable clones from HTO/Z deleted for almost all of the ERAD and autophagy genes that were identified in the screen. In contrast to the HepG2 Cas9 GFP-ATZ cell line, HTO/Z expresses ATZ without a GFP tag and expression of ATZ in this cell line is induced by withdrawal of doxycycline. First, we investigated the effect of deleting ERAD genes. The results showed increased accumulation when SEL1L1 and DERL2 were deleted but not when HRD1 was deleted (Figure 3A–C), entirely consistent with the results in the HepG2 cell line. Although HRD1 has been considered an essential component of the ERAD mechanism for luminal substrates, our results suggest that it is dispensable for ERAD as it relates to ATZ. Deletion of CCT2, another gene that was identified in the screen, also led to increased accumulation of ATZ in the HTO/Z cell line (Figure 3D), consistent with results in the HepG2 cell line.
Figure 3.
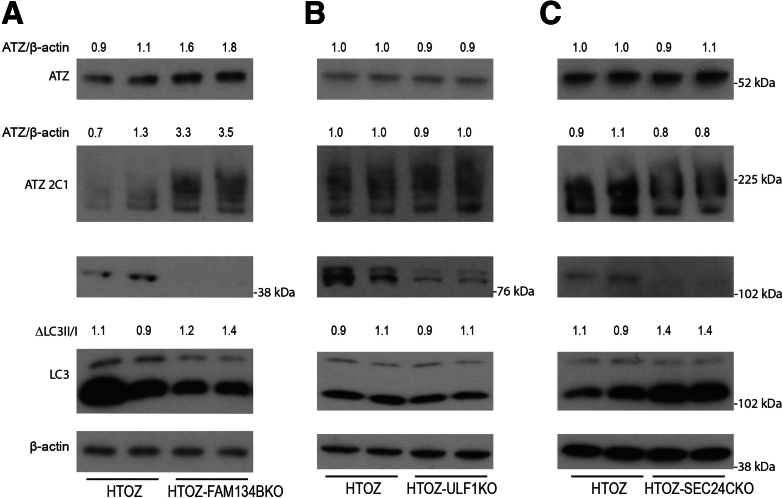
Steady-state levels of ATZ in the HTO/Z cell line deleted for specific autophagy genes. In each case the cells were incubated in the absence of doxycycline for 1–3 weeks to induce ATZ expression. Cell homogenates were subjected to immunoblot analysis with polyclonal antibody to all forms of AT (ATZ), antibody to LC3, antibody to β-actin, and antibody to the relevant autophagy gene. Densitometric quantification is shown at the top of the blots. Deletions are as follows: (A) SEL1L1, (B) DERL2, (C) HRD1, (D) CCT2, (E) ULK1, (F) ATG9, (G) ATG14, (H) STX17, (I) WIPI2, and (J) ATG12.
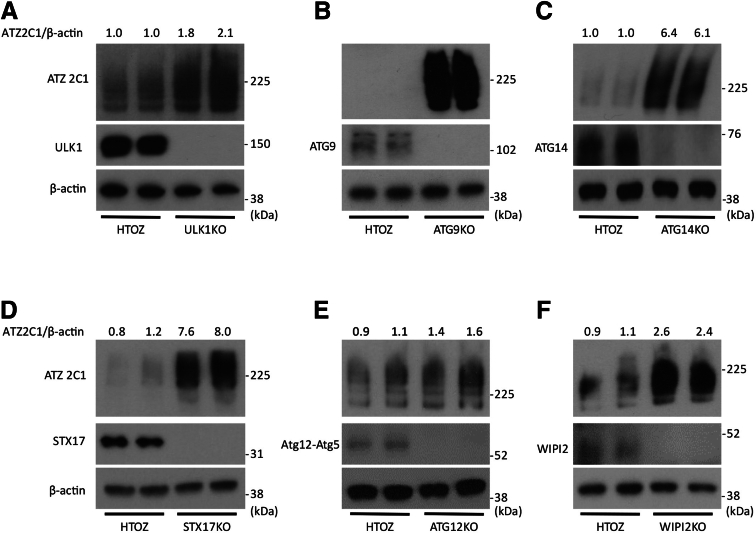
Second, we investigated the effect of CRISPR-mediated selection deletions in autophagy genes on ATZ accumulation in the HTO/Z cell line. We selected several of the autophagy genes that came up in the screen: ULK1, ATG12, and WIP12. In addition, we selected other genes that are critical to the different phases of the canonical macroautophagy pathway, including initiation, nucleation, elongation, and lysosomal fusion. Results in this system showed that deletion of ULK1, ATG9, ATG14, STX17, WIP12, and ATG12 all led to greater accumulation of ATZ (Figure 3E–J). In particular, there was marked accumulation of ATZ in the ATG9 and STX17 knockouts. In each case, the accumulation of ATZ also was greater in the cell lines with deleted autophagy genes when assessed with antibody that only detects the polymeric forms of ATZ (Figure 4). The LC3-II to LC3-I ratio decreased in each of these knockout cell lines except for the cell line deleted for WIPI2, which would be expected to result in increased LC3-II accumulation.37 Taken together, the results of specific deletions of ERAD and autophagy genes on ATZ accumulation in the HTO/Z cell line were entirely consistent with what was observed in the HepG2 system used for the screening.
Figure 4.
Steady-state levels of polymeric ATZ in the HTO/Z cell line deleted for specific autophagy genes. Immunoblot with antibody 2C1 specific for polymeric forms of ATZ with deletions as follows: (A) ULK1, (B) ATG9, (C) ATG14, (D) STX17, (E) ATG12, and (F) WIPI2.
To further understand the role of genes that participate in the autophagy pathway, we investigated several of the genes that recently have been found to be critically important for the ERLAD and other so-called ER-phagy pathways that are distinct from each other (Figure 5). The results showed greater accumulation of monomeric and polymeric ATZ when FAM134B was deleted. We also examined FAM134A, which is related closely to FAM134B but more robustly expressed in liver cells, and its deletion led to greater accumulation of ATZ, including a marked increase in ATZ polymers38. ATZ did not accumulate to a greater extent when UFL1 or SEC24C were deleted. These results could be owing to a lack of complete deletion in the case of UFL1, but it also is important to recognize that adaptations in the cell lines that express proteotoxic molecules are potential explanations for surviving deletions in proteostatic genes.
Figure 5.
Steady-state levels of ATZ in the HTO/Z cell line deleted for specific autophagy receptor genes. Deletions are as follows: (A) FAM134B, (B) UFL1, and (C) SEC24C.
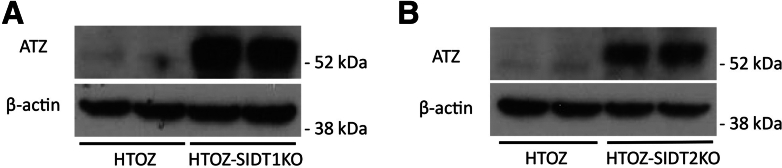
Last, we investigated deletion of SID 1 transmembrane family member 1 (SIDT1) and SID 1 transmembrane family member 2 (SIDT2), mammalian orthologs of nematode sid-1, which have been implicated in the late stages of canonical macroautophagy39 and play a role in ATZ disposal in our nematode model of ATZ accumulation.40 In each case, there was markedly greater accumulation of ATZ (Figure 6), indicating that these 2 genes are particularly important in degradation of ATZ.
Figure 6.
Steady-state levels of ATZ in the HTO/Z cell line deleted for SIDT1 and SIDT2. Immunoblots for ATZ and β-actin levels in cell lines deleted for (A) SIDT1 and (B) SIDT2. Knockout of these genes was validated by polymerase chain reaction.
Further Evidence for Time-Dependent Activation of Autophagy in Mammalian Cell Line Model
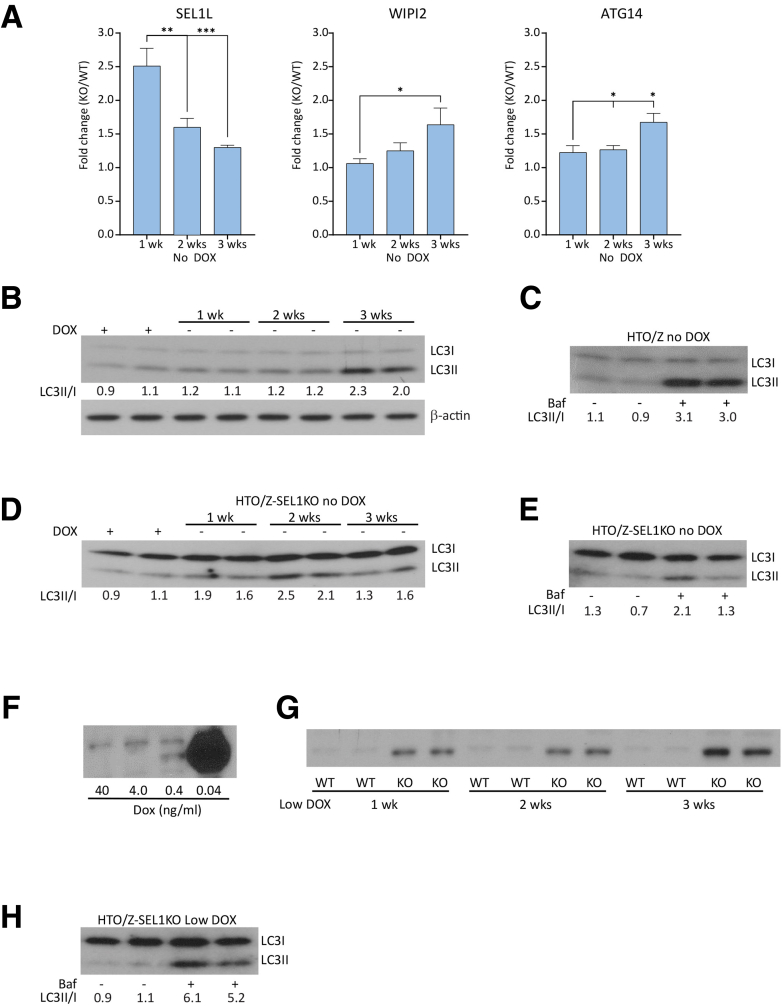
Results of the CRISPR screen suggested that ERAD genes played a role at early time points and autophagy genes became important progressively over time after induction of ATZ expression and accumulation. To further investigate the effect of time on the roles of ERAD and autophagy, we used the HTO/Z cell lines with deletions of select ERAD (SEL1L1) and autophagy genes (WIPI2, ATG14) for effects on ATZ accumulation at 7, 14, and 21 days after withdrawal of doxycycline (Figure 7A). As was the case in the HepG2 system, there was greater ATZ accumulation in HTO/Z cells deleted for SEL1L1, but the impact of SEL1L1 diminished from 1 to 2 weeks and, further, from 2 to 3 weeks. In contrast, the impact of deleting autophagy genes WIPI2 and ATG14 on ATZ accumulation increased progressively from 1 to 3 weeks.
Figure 7.
Time-dependent roles of ERAD and autophagy in the HTO/Z cell line. (A) Steady-state levels of ATZ in the HTO/Z cell line deleted for ERAD or autophagy genes in time course studies. Fold change for deletions of SEL1L1 (left), WIPI2 (center), and ATG14 (right) compared with parent HTO/Z cell line at time points of 1, 2, and 3 weeks after induction of ATZ expression. (B) LC3-II to LC3-I ratio in HTO/Z in the presence of doxycycline or after it was withdrawn for 1, 2, and 3 weeks. (C) LC3-II to LC3-I ratio in HTO/Z that had been out of doxycycline for 3 weeks in the presence or absence of bafilomycin. (D) LC3-II to LC3-I ratio in HTO/Z with SEL1LI deletion in the presence of doxycycline or after it was withdrawn for 1, 2, and 3 weeks. (E) LC3-II to LC3-I ratio in HTO/Z with SEL1L1 deletion that had been out of doxycycline and then incubated in the absence or presence of bafilomycin. (F) ATZ levels in the HTO/Z cell line cultured over 3 weeks in different doses of doxycycline as indicated at the bottom. (G) ATZ levels in HTO/Z with SEL1L1 deletion after 1, 2, and 3 weeks of low-dose doxycycline. WT is parent HTO/Z and KO is HTO/Z with SEL1L1 deletion. (H) LC3-II to LC3-I ratio in HTO/Z with SEL1L1 deletion of 3 weeks of low-dose doxycycline and then incubated in the absence or presence of bafilomycin. Densitometric quantification is shown at the bottom in each case. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001, Student t test. Baf, bafilomycin; DOX, doxycycline; KO, knockout; WT, wild-type.
We also analyzed the LC3-II to LC3-I ratio in the HTO/Z cell line over time after induction of ATZ expression. The results in Figure 7B show a progressive increase from 1 to 3 weeks, with most of the effect from week 2 to week 3. To determine how autophagic flux is impacted we examined the effect of bafilomycin (Figure 7C) and that showed a further increase in the LC3-II to LC3-I ratio, indicating that autophagic flux is increased. These results are entirely consistent with our previous studies, using a transgenic mouse model with liver-specific inducible expression of ATZ on a GFP-LC3 background,27 showing that accumulation of ATZ is sufficient to activate the autophagic response.
Next, we investigated what happens to autophagic flux over time when the ERAD pathway is disrupted. One of the more interesting results of the CRISPR screen was that ERAD genes had a lesser, if any, impact on ATZ accumulation at the later time point after induction of ATZ expression. One possible explanation for this result is that the activation of autophagy reaches a level that makes the ERAD pathway dispensable. Indeed, SEL1L1 deletion in the HepG2-based cell line (Figure 2B) and in the HTO/Z cell line (Figure 7A) showed less impact on ATZ accumulation at later time points. First, we found that the LC3-II to LC3-I ratio also increases in the HTO/Z cell line with specific deletion of SEL1L1 (Figure 7D) and increases further in the presence of bafilomycin (Figure 7E), indicating that autophagic flux has increased.
Last, we sought to determine whether the magnitude or duration of ATZ accumulation played a role in this phenomenon by investigating what happens at the 3-week time point when the magnitude of ATZ accumulation is lowered. First, we determined that a dose of 0.4 ng/mL of doxycycline would lower the accumulation of ATZ in parent HTO/Z by more than 100-fold compared with the same cell line in the absence of doxycycline (Figure 7F). Second, we investigated the effect of this doxycycline dose in the HTO/Z cell line with SEL1L1 deletion (Figure 7G). The results showed a dramatic increase in the level of ATZ accumulation. Furthermore, there was a marked exaggeration in the magnitude of the increase in autophagic flux (Figure 7H), raising the possibility that activation of the autophagic pathway is particularly sensitive to impaired ERAD and/or, alternatively, specific effects of SEL1L1 deletion. Taken together, the results of these experiments provide further evidence that the proteostasis response is influenced by the duration of ATZ accumulation.
Multiple Autophagy Genes Modulate Cellular Accumulation of ATZ in a Nematode Model In Vivo
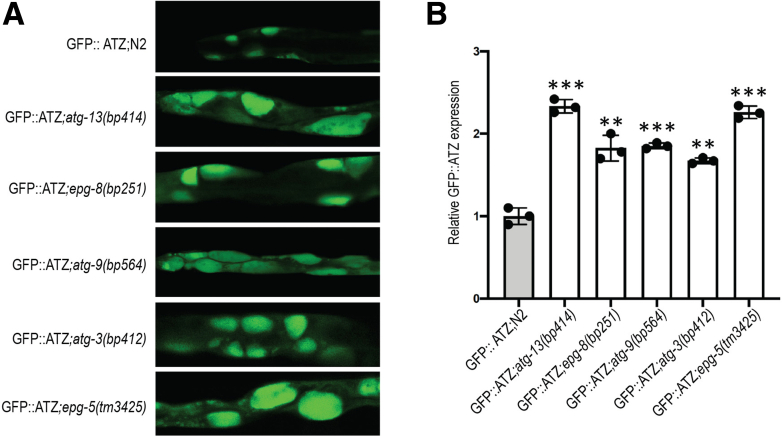
To provide further validation to the importance of multiple autophagy genes in ATZ disposal, we used a Caenorhabditis elegans transgenic model that expresses chimeric GFP-ATZ in intestinal cells.40,41 Our previous studies have shown that this model is ideal for investigating the impact of autophagy on ATZ accumulation at the organism level and with tractable genetic manipulations. The Z worm model was crossed with strains carrying mutations in known autophagy genes. We selected 5 genes that have been purported to have specific key roles during the different stages (initiation, nucleation, phagophore targeting/recycling, vesicle elongation, and fusion with lysosomes) of autophagy. The results showed GFP-positive globules greater in size and number in each of the deleted lines in Figure 8A and quantified in Figure 8B. Taken together, these results indicate that multiple components of the canonical autophagy pathway are important for ATZ clearance in a whole animal model in vivo.
Figure 8.
Effect of autophagy mutants on GFP::ATZ accumulation in ATZ nematode model. (A) Confocal microscopy images of GFP::ATZ–expressing animals crossed with autophagy mutants. (B) Quantification of GFP::ATZ accumulation using CellInsight CX7 high-content image. ∗∗P ≤ .01; ∗∗∗P ≤ .001.
Appearance of Autophagosomes and Marked Mitochondrial Structural Changes Correspond Temporally to Functional Genetic Changes
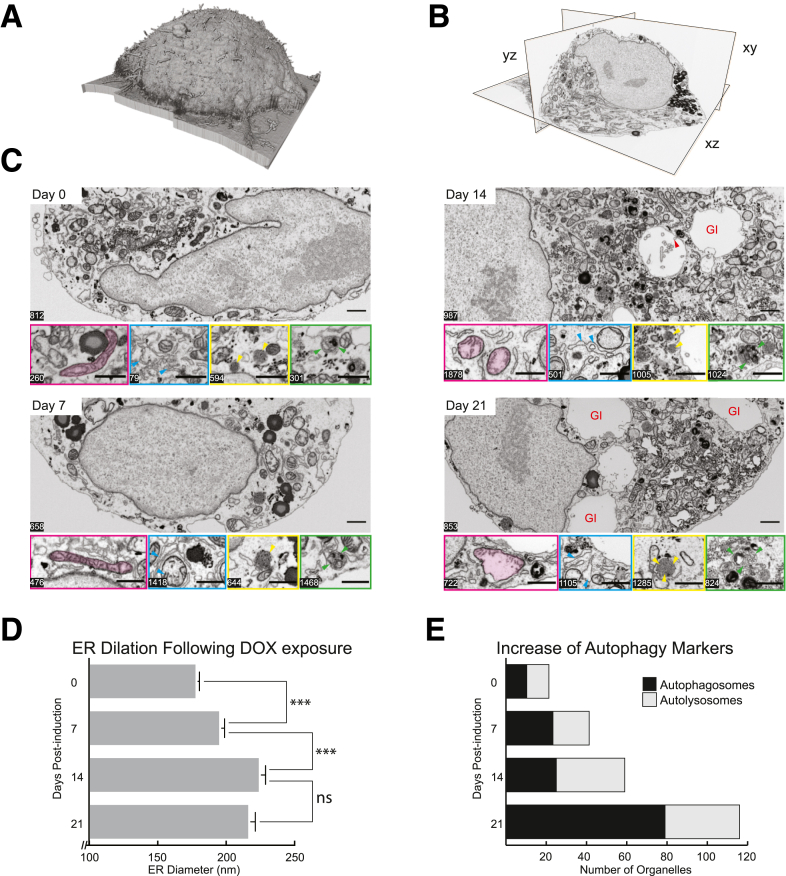
To ascertain both the temporal nature of the ultrastructural changes, as well as their spatial dependence, the HepG2-GFP-ATZ cell line was investigated using focused ion beam–scanning electron microscopy (FIB-SEM) 3-dimensional nanotomography at 0, 7, 14, and 21 days after induction of ATZ expression (Figure 9A and B). We decided to perform these studies in HepG2 rather than the HeLa-based HTO/Z cell line because the morphologic effects of ATZ accumulation have been characterized in hepatocytes. Morphometric analysis of the reconstructed and segmented FIB-SEM data (Figure 9C and D) showed that one of the most pronounced changes in the subcellular architecture was a progressive dilation of the endoplasmic reticulum (Figure 9C, cyan arrowheads). At 0 days, the mean diameter of the ER tubules was 95 ± 55 nm, which increased to 120 ± 75 nm at 14 days, with a reduction to 107 ± 60 nm at 21 days. This change was substantiated further by the increase in the number of ER membranes exceeding 125 nm in diameter after the first week after doxycycline (Figures 9C and 10B). Another striking time-dependent change was the appearance of large globular inclusions at 14 and 21 days after induction (Figure 9C, day 21, red GI notation). These inclusions are surrounded by ER membranes as evidenced by ribosomal studding. In addition, the membranes surrounding the inclusions also are studded by budding vesicles that resemble omegasomes (Figure 9C, day 14, red arrowhead), pinching off from the main body of the globule. In some cases, the membranes of the vesicles appear to be continuous with the ER membranes surrounding the globules, but the imaging resolution of 10-nm voxels prevents definitive determination of this conclusion.
Figure 9.
Nanotomographic analysis of HepG2 cells at different time points postinduction of ATZ expression. (A) Volume rendering and (B) orthogonal view, showing a 21-day postinduction cell in xy, yz, and xz planes. Note the ability to reconstruct the data set in any axis, afforded by the isometric voxels. (C) Representative single-plane images from each of the 4 time points assayed. Inset panels highlight changes in organelle ultrastructure at different z-slice positions, such as increase in ER diameter and the density of autophagic organelle occurence (magenta--mitochondria, cyan--endoplasmic reticulum, yellow--autophagosomes, and green--autolysosomes). Cyan arrowheads illustrate the increased dilation of endoplasmic reticulum over the time course. Yellow arrows indicate the presence of lipid droplets Green arrowheads indicate the presence of autophagosomes. Red arrowhead in day 14 illustrates the presence of omegasomes in the emergent globular inclusions (as denoted by the red GI label). Scale bar: 1 μm. (D) Quantification of the increase in the mean diameter of the endoplasmic reticulum per cell at each of the 4 time points. ∗∗∗ denotes P < .001 (E) Increased density of autophagosomic organelles, as measured by the mean number of autophagosomes and autolysosomes counted per cell at each of the 4 time points. GI, globular inclusions.
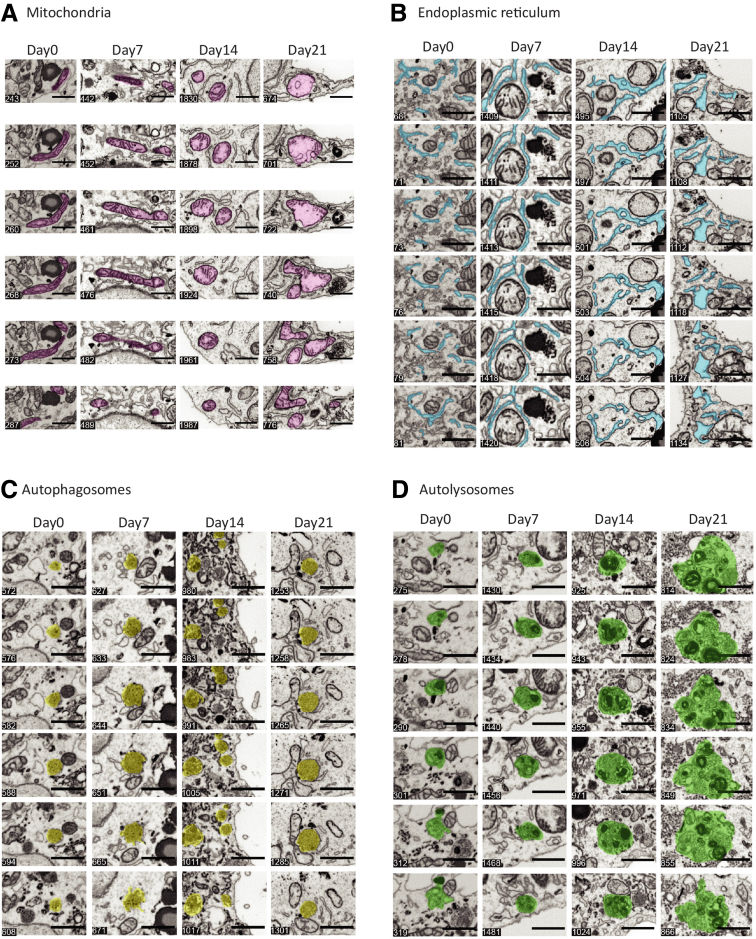
The second major change was the marked increase in the number of autophagosomes and autolysosomes over time (Figures 9C, green and orange arrowheads, respectively; and 10C and D). Quantitative morphometric determinations showed that the number of autophagosomes and autolysosomes dramatically increased by a factor of 4 over the 21-day induction period (Figure 9E). Most strikingly, the change in the number of both organelle types was segregated into 2 distinct time frames. From 0 to 7 days, the increase was approximately a factor of 2, but between 7 and 14 days, the number of autophagosomes remained fairly constant with only an approximate 30% increase in the number of autolysosomes. However, between 14 and 21 days there was a dramatic 3-fold increase in autophagic organelles (Figure 10C and D). Third, a dramatic alteration of mitochondrial structure was observed on days 14 and 21. Overall, the mitochondria became enlarged volumetrically, but, more strikingly, their shape became increasingly dysmorphic with loss of cristae (Figures 9C, green arrowheads, and 10A). On days 14 and 21, there was an increase in autolysosomal structures that contained mitochondria and mitochondrial fragments.
Figure 10.
Time-dependent changes in organelle ultrastructure by 3D FIB-SEM nanotomography. (A) Six z-slices through a single mitochondrion (highlighted in purple) at 0, 7, 14, and 21 days after induction of ATZ expression. Note the canonical tubular appearance on days 0 and 7 transforms to a circular appearance on days 14 and 21, with a significant loss of cristae on day 21. (B) Six z-slices through a region of the endoplasmic reticulum network (highlighted in cyan) at 0, 7, 14, and 21 days after induction of ATZ expression. Note the clear dilation of the tubular ER morphology present on day 14, and the complete loss of tubular morphology on day 21. (C) Six z-slices through a region containing autophagosomes (highlighted in yellow) at 0, 7, 14, and 21 days after induction of ATZ expression. Note the gradual size increase of the autolysosomes at later time points with increased number density present on day 14. Note on day 21 the close association of the autophagosomes with clearly malformed mitochondria. (D) Six z-slices through a region containing autolysosomes (highlighted in green) at 0, 7, 14, and 21 days after induction of ATZ expression. Note both the gradual size increase of the autolysosomes at later time points, and also the complexity of the autolysosomal interior ultrastructure, which show not only autophagosomes, but what also appear to be fragments of mitochondria. Scale bars: 1 μm. Individual z-slice number from each data set is denoted in the bottom left corner of each image.
Discussion
In these studies, we carried out screening for proteostasis genes and, by implication, pathways, that are essential for the cellular response to accumulation of the pathogenic variant ATZ in the ER. This pathogenic variant presents a particularly interesting enigma for the cell to manage because it is produced at very high levels and has a tendency to polymerize/aggregate within the early portion of the secretory pathway. Furthermore, many of the patients that were homozygous for ATZ escaped clinical manifestations of the proteotoxic effects, providing a strong basis for considering the importance of proteostasis mechanisms. The results validate the importance of the 2 major proteostasis mechanisms, ERAD and autophagy, and also provide evidence that different variations of the autophagy mechanism, as well as lysosomal degradation pathways that appear to be distinct from the autophagy as classically defined, are needed to respond to the unabated accumulation of ATZ in model systems. The results also provide evidence for temporal coordination of these proteostasis pathways, with ERAD being essential early and autophagy becoming more important progressively over time as the accumulation of misfolded protein persists. This is consistent with the notion that progressive accumulation, especially with polymers and aggregates, overwhelms the capacity of the ERAD mechanism and invokes the specialized capacity of the autophagy mechanism to handle polymers, aggregates, and larger loads. This specialization and temporal coordination was suggested by early studies of ATZ in yeast,42 but here we were able to use the technological advances of CRISPR screening and cell lines with inducible expression capabilities for more definitive conclusions. Most importantly, the ability to utilize this type of genome-wide analysis at different time points after induction of the proteotoxic condition, and validate the results by varying ATZ levels in the inducible cell line model system, provides 2 separate experimental conditions that reflect what goes on in vivo under disease conditions in which accumulation is a chronic progressive phenomenon and host cells are probably adapting continuously to escape disease consequences.
ERAD genes, particularly SEL1L and DERL2, which have been implicated in ERAD for luminal misfolded proteins, were found to be important in the early, but to a lesser extent in the later, response. Autophagy genes were found to be rate-limiting in the later response, but also may be involved in the early response on the basis of 2 findings. First, autophagy genes ULK1 and AMBRA1 were detected at the early time point in the screening of the mini-pool library. Second, we hypothesized that the TCP1-ring complex genes were essential at this early time point because of the role that they play in autophagy, by promoting the folding of actin required for autophagosome-lysosome fusion.36 Activation of the autophagic response is not surprising because we previously showed that the autophagic response can be activated by accumulation of ATZ in the ER alone.27 It is important to point out, however, that the UPR is not particularly robust in mammalian model systems of ATZ accumulation,20 and this may mean that the activation of ERAD genes is not the result of a classic UPR. Furthermore, activating components of the UPR were not found to be essential in this screen. It is interesting that other autophagy genes become essential at the later time point because this would be consistent with the need for multiple autophagic mechanisms or subpathways to handle the load as the accumulation progresses.
In a recent article, Ordoñez et al43 used a genome-wide CRISPR screen to characterize the response to accumulation of another pathogenic AT variant, the King’s mutant. Although this variant is a much rarer cause of clinical disease, it is prone to misfolding and aggregation and therefore likely has a similar cellular pathobiology to the most common pathogenic variant ATZ. In that study, the AT variant was allowed to accumulate for 1 day and several ER cargo receptors were found to be essential but genes from either proteostasis pathway were not essential. These results provide further understanding of the temporal sequence of events in the cellular response to aggregation-prone AT variants with 3 general phases in the following order: activation of the secretory mechanisms, activation of ERAD pathway, and activation of autophagolysosomal pathways.
Several studies published over the past several years have characterized which ER-phagy pathways that are purported to be distinct from canonical macroautophagy.4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16,28 In several cases these pathways have been likened to microautophagy and, in 1 case, the ERLAD pathway has been described as a nonautophagic, ER-to-lysosome degradation pathway.4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16,28 Indeed, Fregno et al28 posited that the ERLAD pathway was responsible for disposal of ATZ to the exclusion of canonical macroautophagy. However, their studies depended on a read-out of the fate of ATZ in specific types of vesicles and could have missed ATZ molecules that were being disposed through the autophagic and/or lysosomal systems by other vesicles. Furthermore, since their publication, Ji et al18 have shown that ATZ may be degraded in classic autophagosomes through the action of p62 working in concert with the ER E3-ubiquitin ligase TRIM13. The central hypothesis of our study reported here was that multiple pathways, including these ER-phagy and ER-to-lysosome and canonical macroautophagy pathways, are needed to handle the load of ATZ accumulation in the human disease condition, especially because we know that a majority of affected patients escape the proteotoxic consequences of the genotype. To investigate this hypothesis, we tested deletion of specific autophagy genes in our workhorse mammalian cell line model, the HeLa-based HTO/Z cell line, and in our C elegans model of ATZ accumulation. The results are entirely consistent with the hypothesis that multiple pathways, including canonical macroautophagy, are involved. In particular, greater accumulation of ATZ in the presence of deletions of ULK1, ATG9, ATG13, ATG14, and STX17 show that the pathways labeled as ERLAD, Recov-ER phagy, and LC3-associated phagocytosis are not sufficient to handle the ATZ load. The results confirmed the importance of FAM134B, but also show that FAM134A, which is expressed more abundantly in liver than FAM134B, is involved in the response to ATZ accumulation.
The results of this study also provide a framework for how morphology of the cell changes over time as ATZ accumulation progresses, including the enlargement of ER, formation and expansion of proteinaceous globules, and piecemeal digestion of the globules by autophagosomes. This visualization of time–dependent globule formation in a cell line model and the system provides elucidation of that process and whether it is protective or pathogenic. Importantly the appearance of autophagosomes correlates temporally with the functional effects of deleting essential autophagy genes. We previously noted structural alterations of mitochondria in liver cells of the PiZ mouse model as well as biopsy specimens from ATD patients,44 and here we observed very significant damaging changes in the mitochondria over time. Effects on the mitochondria are likely to be very important in the cytotoxic mechanism of the proteinopathy, and recent studies have implicated mitochondrial function in regulating the activation and function of the autophagic response4 as well.
The picture that emerges from these results, and the increasing number of recently described autophagic and lysosomal degradative mechanisms for disposal of misfolded ER proteins, is one wherein cells can call on a repertoire of adaptive responses. Furthermore, the evolution of these adaptive responses is informed by the magnitude and duration of the protein load. This picture is consistent with the variation in prevalence and severity of proteotoxicity that manifests in the population affected by ATD and presumably other misfolded protein disorders. Elucidation of these proteostasis mechanisms, including ERAD and different forms of autophagy and lysosomal pathways, has led to the identification of novel therapeutic approaches for the liver disease of ATD,45,46 as well as other disorders caused by misfolded proteins.47
Materials/Methods
FACS-Based Pooled CRISPR Screening
HepG2 cells with doxycycline-inducible expression of GFP-tagged ATZ (HepG2 Tet-On GFP-ATZ32) were engineered for screening by lentiviral delivery of Cas9 in pNGx-LV-c004 followed by selection with blasticidin as described previously.29,30 The initial genome-wide CRISPR screen was performed by flow cytometry after 7 days of doxycycline-induced ATZ expression. A second mini-pool CRISPR screen was performed using a library of the top-ranked 300 genes enriched in GFP-high and GFP-low cells, the 300 most depleted genes in unsorted cells, and a list of known autophagy genes, and interrogated after 6 or 22 days of doxycycline induction.
The HepG2 Tet-On GFP-ATZ cells were maintained in Dulbecco’s modified Eagle medium with 10% fetal bovine serum, 1% L-glutamine, and 1% penicillin/streptomycin. GFP-ATZ expression was induced by the addition of 1 μg/mL doxycycline (#631311; Clontech). HepG2 Cas9 Tet-On GFP-ATZ cells were generated by lentiviral delivery of Cas9 in pNGx-LV-c004 followed by selection with blasticidin as described previously.29,30 Single clones were isolated by serial dilution. Editing efficiency of the Cas9-expressing cell population and clone #3 was confirmed by infecting cells with pNGx-LV-g003 containing a GFP sgRNA (GGGCGAGGAGCTGTTCACCG) or a nontargeting sgRNA (GTAGCGAACGTGTCCGGCGT) and analyzing GFP fluorescence by FACS after 7 days of doxycycline.
The genome-wide pooled CRISPR screen was performed in the HepG2 Cas9 Tet-On GFP-ATZ stable cell population. The lentiviral genome-wide sgRNA library was infected into 400 million cells at an multiplicity of infection of 0.5 in the presence of 5 μg/mL polybrene (TR-1003-G; Millipore). Twenty-four hours postinfection, 1 μg/mL doxycycline was added and medium was replaced every 2 days. Seven days postinfection, cells were fixed in 1× Mirsky’s fixative (#HS-102; National Diagnostics) for 15 minutes at room temperature, washed twice with phosphate-buffered saline, resuspended in fetal bovine serum Stain Buffer (#554656; BD), and analyzed with a BD FACSCanto II (BD Biosciences US) flow cytometry system. An unsorted sample was collected. Generation of the genome-wide sgRNA library, FACS-based cell sorting, DNA extraction, next-generation sequencing, data analysis, and gene enrichment analysis was performed as described previously.29,30
The mini-pool CRISPR screen was performed in HepG2 Cas9 Tet-On GFP-ATZ clone #3. The mini-pool library was built with the top ranking 300 genes enriched in GFP-high or GFP-low cells, the 300 most depleted genes in unsorted cells, and a list of known autophagy genes. Each gene typically is covered with 5 sgRNA sequences and the construction of the mini-pool was performed as described previously.2,3 The mini-pool was screened in duplicate and cells were analyzed after 6 or 22 days of doxycycline induction.
Ultrastructural Characterization and Quantitative Morphometry
HepG2-GFP-ATZ cells after doxycycline induction for several different time intervals were evaluated by confocal microscopy for fluorescence imaging and then processed further using an OTO protocol48 for 3-dimensional electron nanotomographic analysis. Cellular and subcellular structures of interest were segmented using a customized deep learning approach to facilitate morphometric analysis.
Cultures of the HepG2-GFP-ATZ cells were plated on μ-Dish 35-mm high-grid 50 glass-bottom dishes (ibidi GmbH, Gräfefing, Germany) and fixed with 4% paraformaldehyde in 0.15 mol/L cacodylate buffer containing 2 mmol/L calcium chloride for 1 hour. Fixed cells then were imaged on a Nikon A1RHD25 confocal microscope (Nikon Instruments, Melville, NY) to identify GFP-positive cells that subsequently were targeted for 3-dimensional (3D) nanotomographic analysis. After fixation and fluorescence imaging, samples were processed using an OTO protocol.48 Briefly, cells then were washed 3 times for 10 minutes each in cold 0.15 mol/L cacodylate buffer containing 2 mmol/L calcium chloride, and then were incubated in a solution of 1% osmium tetroxide and 1.5% potassium ferrocyanide in cacodylate buffer for 1 hour in the dark. After incubation, cells were washed 3 times for 10 minutes each in ultrapure water and then incubated for 20 minutes in a 1% thiocarbohydrazide solution, rinsed 3 times for 10 minutes each in ultrapure water, and thereafter incubated in 2% osmium tetroxide for 30 minutes. Cells then were washed 3 times for 10 minutes each in ultrapure water, incubated in a lead aspartate solution at 60°C for 30 minutes, and again washed 3 times for 10 minutes each in ultrapure water. Samples then were dehydrated in sequential steps of 50%, 70%, 90%, 100% × 2 acetone (anhydrous) for 10 minutes in each step, infiltrated with Durcupan ACM resin (Sigma), and polymerized at 60°C for 48 hours. To prepare cells for FIB-SEM analysis, the glass bottom of the ibidi dish was dissolved with hydrofluoric acid to expose the embedded cells and the surface was coated with 6 nm iridium using a Leica EM ACE600 sputter coater (Leica Microsystems, Vienna, Austria).
Three-dimensional electron nanotomography was performed initially on a Zeiss Crossbeam 540 Focused Ion Beam-Scanning Electron Microscope (Carl Zeiss Microscopy, Oberkochen, Germany) for the first cell from each time point and then subsequently on a ThermoFisher Scientific Helios 5 UX Focused Ion Beam-Scanning Electron Microscope (ThermoFisher Scientific, Brno, Czech Republic) for the second and third cells from each time point. Cells were first visualized using secondary electron imaging at 5 keV, with those images correlated with the previously acquired fluorescent images to identify GFP-positive cells. Once targeted, areas containing GFP-positive cells were prepared for 3D nanotomography using ATLAS 5 (FIBICS, Ottawa, Ontario) on the Crossbeam 540 as previously described49 and ASV 4.2 (ThermoFisher Scientific) on the Helios 5 UX. Images on both FIB-SEM instruments were acquired at the same 10-nm isometric voxel resolution using an Inlens backscattered electron detector at an accelerating voltage of 1.9 keV and a probe current of 900 pA. Pixel dwell times were 4 and 5 μs, respectively, and line averaging was 5× and 4×, respectively, on the Crossbeam 540 and the Helios 5 UX FIB-SEM platforms. All FIB-SEM data sets were aligned using Amira 2020.2 (Thermo Fisher, Hillsboro, OR).
Cellular and subcellular structures of interest (cell membrane, nucleus, nucleolus, endoplasmic reticulum, and mitochondria, including cristae, lipid droplets, and globular inclusions) were segmented using a customized deep learning approach to further facilitate morphometric analysis. Briefly, subvolumes (20–50 images) of each of the Crossbeam 540 FIB-SEM 3D data sets were annotated manually to provide the ground truth data for model training. After this manual annotation step, images were cropped randomly into 2-dimensional patches and then passed to a convolutional neural network50 with on-the-fly data augmentation for model training. The Adaptive Moment Estimation method and early stopping were used for optimization and overtraining prevention and dropout regularization was set between 0.5 and 0.8 to prevent overfitting of each model. Predictions were performed initially on unseen subvolumes of a given FIB-SEM data set, which were validated further by manual inspection. Each model underwent additional training until high levels of accuracy (>95%) were achieved for each target structure. Once training was completed, each model was used to predict a given structure of interest on entire 3D data sets. Prediction outputs were segmented masks of each structure stored as binary TIFF files. This binarized output was further postprocessed, proofread, and used for quantitative morphometric analysis. Binary output of the segmented ER was used to quantify ER diameter using the Ridge detection analysis tool described by Schindelin et al,51 which measures the diameter of a given object along a specific vector. Volumetric measurements of segmented structures were calculated using Amira 2020.2 (Thermo Fisher). 3D surface renderings of segmented structures was created using Blender, an oper-source 3D creation software suite distributed under the GNU General Public Library. Finally, all autophagic organelles (both autophagosomes and autolysosomes) were manually identified and counted across all of the acquired FIB-SEM 3D data sets using the Cell Counter plugin in Fiji.
Mammalian Cell Models and Immunoblot Analysis
In addition to HepG2-GFP-ATZ cells, we used a comparable cell line engineered with wild-type AT-GFP chimera (HepG2-GFP-ATM) as an essential control. We also used the HTO/Z cell line20 with inducible expression, however, in contrast to the HepG2-GFP-ATZ, which has Tet-On inducibility, this cell line has Tet-Off inducibility such that ATZ expression is induced by withdrawal of doxycycline. The cell lines with expression of ATZ were engineered by deletion of specific ERAD and autophagy genes using standard CRISPR technology in the Genome Engineering and Stem Cell Center at Washington University in St. Louis. Briefly, depending on the gene structure, 1 or 2 cuts are designed to either introduce out-of-frame indels in an early exon or create deletions of part of the whole coding sequences. Synthetic guide RNAs (gRNAs) were purchased from IDT complexed with recombinant Cas9 protein, and transfected into the HG2 or HTO/Z cells. The transfected cells then were single-cell sorted into 96-well plates, and single-cell clones were analyzed using NGS to identify those only with out-of-frame indels or deletions. Some genes are essential in 1 or both parental lines, judging by the lack of recovered compound homozygous knockout clones and gradually increased wild-type allele portions in the transfected pools over time. In that case, we used either transfected pools at early passages or heterozygous knockout clones.
Steady-state levels of ATZ were analyzed by immunoblot and densitometric quantification as previously described.22 Antibodies used for Western blot were as follows: goat anti-human α-1-antitrypsin (AB707-K; EMD Millipore Corporation), polymeric ATZ (2C1, HM2289; HycultBiotech), ATG9 (ab108338, Abcam), ATG12 (2011; Cell Signaling Technology), ATG14 (5504S; Cell Signaling Technology), β-actin (3700S; Cell Signaling Technology), CCT2 (3561; Cell Signaling Technology), DERL2 (sc-398573; Santa Cruz Biotechnology), HRD1 (14773; Cell Signaling Technology), FAM134A (HPA011170; Sigma), FAM134B (ab151755; Abcam), LC3 (12741; Cell Signaling Technology), SEC24C (8531S; Cell Signaling Technology), STX17 (HPA001204; Sigma), UFL1 (A303-456A; Bethyl Laboratories), ULK1 (8054S; Cell Signaling Technology), p62 (P0067; Sigma), SEL1L1 (ab78298; Abcam), and WIP12 (MABC91; EMD Millipore).
C elegans Strains, Culture Conditions, Imaging, Quantification, and Statistical Evaluation
Transgenic GFP-ATZ-chimera–expressing strains, as previously described,41 were mated to strains deleted for specific autophagy genes. Animals were cultured routinely at 20°C on nematode growth medium plates seeded with E coli strain OP50. C elegans strains, N2, atg-13(bp414), atg-9(bp564), atg-3(bp412), and epg-8(bp251) were obtained from the Caenorhabditis Genetics Center (http://www.cbs.umn.edu/CGC). tm3425(epg-5) was obtained from the National Bioresource Project for the Experimental Animal Nematode C elegans (https://shigen.nig.ac.jp/c.elegans/top.xhtml). Transgenic ATZ-expressing strains, VK2872 and VK3347 (vkIs3347 [Pnhx-2::GFP::ATZ+Pmyo-2::mCherry]) were generated as previously described.41
For microscopic image acquisition, approximately 12 young adult stage animals were transferred to a 35-mm MatTek glass bottom culture dish (MatTek, USA) containing 6 μL of 100 mmol/L sodium azide. Confocal images were collected using a Leica SP8X tandem scanning confocal microscope fitted with a white light laser using a 40× 1.3 NA oil PlanApo objective (Leica Microsystems, Buffalo Grove, IL). Images were displayed as single XY planes. Images were rendered and analyzed using LASX (Leica Microsystems) and Volocity (v6.3; Quorum Technologies) software.
For quantitative analysis of animals expressing fluorescent transgenes, we used the CellInsight CX7 HCS Reader (ThermoFisher) fitted with a 5× objective. Thirty N2 or transgenic young adult stage animals were transferred to 384-well plates containing 40 μL phosphate-buffered saline and anesthetized with 60 μL 2.5 mg/mL tetramizole hydrochloride (levamisole) for 5 minutes before image capture. Images were acquired using a 2-channel [GFP and red fluorescent protein (RFP)] assay and image analysis was performed using the Co-localization BioApplication (ThermoFisher).
Acknowledgments
The authors are grateful for technical assistance in the implementation of the correlative light and electron microscopy (CLEM) imaging pipeline and deep learning data analysis by Matthew Joens, Peter Bayguinov, Michael Shih, and Praveen Krishnamoorthy from the Washington University Center for Cellular Imaging, which is supported by Washington University School of Medicine.
CRediT Authorship Contributions
Jie Li, MD (Conceptualization: Supporting; Data curation: Lead; Methodology: Lead)
Francesca Moretti, PhD (Data curation: Equal; Formal analysis: Equal)
Tunda Hidvegi, PhD (Data curation: Supporting; Formal analysis: Supporting)
Sanja Sviben, PhD (Data curation: Supporting; Formal analysis: Supporting)
James AJ Fitzpatrick, PhD (Conceptualization: Supporting; Data curation: Supporting; Formal analysis: Supporting)
Hemalatha Sundaramoorthi, PhD (Data curation: Supporting; Formal analysis: Supporting)
Stephen C Pak, PhD (Conceptualization: Supporting; Data curation: Supporting; Formal analysis: Supporting)
Gary A Silverman, MD PhD (Conceptualization: Supporting; Data curation: Supporting; Formal analysis: Supporting; Writing – review & editing: Supporting)
Britta Knapp, PhD (Data curation: Supporting; Formal analysis: Supporting)
Ireos Filipuzzi, PhD (Data curation: Supporting; Formal analysis: Supporting)
John Alford, PhD (Data curation: Supporting; Formal analysis: Supporting)
John Reece-Hoyes, PhD (Data curation: Supporting; Formal analysis: Supporting)
Florian Nigsch, PhD (Data curation: Supporting; Formal analysis: Supporting)
Leon Murphy, PhD (Conceptualization: Equal; Formal analysis: Supporting; Investigation: Equal; Methodology: Equal; Writing – review & editing: Supporting)
Beat Nyfeler, PhD (Conceptualization: Equal; Formal analysis: Equal; Investigation: Equal; Methodology: Equal; Writing – review & editing: Equal)
David H. Perlmutter, MD (Conceptualization: Lead; Data curation: Lead; Formal analysis: Lead; Funding acquisition: Lead; Investigation: Lead; Methodology: Equal; Project administration: Lead; Resources: Lead; Supervision: Lead; Validation: Lead; Writing – original draft: Lead; Writing – review & editing: Lead)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding These studies were supported by National Institutes of Health grant P01DK096990 to DHP.
Note: To access the supplementary material accompanying this article, go to the full text version at http://doi.org/10.1016/j.jcmgh.2024.02.006.
Supplementary Material
References
- 1.Klaips C.L., Jayaraj G.G., Hartl F.U. Pathways of cellular proteostasis in aging and disease. J Cell Biol. 2018;217:51–63. doi: 10.1083/jcb.201709072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karagöz G.E., Acosta-Alvear D., Walter P. The unfolded protein response: detecting and responding to fluctuations in the protein-folding capacity of the endoplasmic reticulum. Cold Spring Harb Perspect Biol. 2019;11a doi: 10.1101/cshperspect.a033886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levine B., Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176:11–42. doi: 10.1016/j.cell.2018.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liang J.R., Lingeman E., Louin T., et al. A genome-wide ER-phagy screen highlights key roles of mitochondrial metabolism and ER- resident UFMylation. Cell. 2020;180:1160–1177. doi: 10.1016/j.cell.2020.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khaminets A., Heinrich T., Mari M., et al. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature. 2015;522:354–358. doi: 10.1038/nature14498. [DOI] [PubMed] [Google Scholar]
- 6.Cui Y., Parashar S., Zahoor M., et al. A COPII subunit acts with an autophagy receptor to target endoplasmic reticulum for degradation. Science. 2019;365:53–60. doi: 10.1126/science.aau9263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Omari S., Makareeva E., Roberts-Pilgrim A., et al. Noncanonical autophagy at ER exit sites regulates procollagen turnover. Proc Natl Acad Sci U S A. 2018;115:e10099–e10108. doi: 10.1073/pnas.1814552115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang J.R., Lingemann E., Ahmed S., et al. Atlastins remodel the endoplasmic reticulum for selective autophagy. J Cell Biol. 2018;217:3354–3367. doi: 10.1083/jcb.201804185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loi M., Raimondi A., Morone D., et al. ESCRT-III-driven piecemeal micro-ER-phagy remodels the ER during recovery from ER stress. Nat Commun. 2019;10:5058–5070. doi: 10.1038/s41467-019-12991-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.An H., Ordureau A., Paulo J.A., et al. TEX264 is an endoplasmic reticulum-resident ATG8-interacting protein critical for ER remodeling during nutrient stress. Mol Cell. 2019;74:891–908. doi: 10.1016/j.molcel.2019.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chino H., Hatta T., Natsume T., et al. Intrinsically disordered protein TEX264 mediates ER-phagy. Mol Cell. 2019;74:909–921. doi: 10.1016/j.molcel.2019.03.033. [DOI] [PubMed] [Google Scholar]
- 12.Grumati P., Morozzi G., Hölper S., et al. Full length RTN3 regulates turnover of tubular endoplasmic reticulum via selective autophagy. eLife. 2017;6 doi: 10.7554/eLife.25555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Q., Xiao Y., Chai P., et al. ATL3 is a tubular ER-phagy receptor for GABARAP-mediated selective autophagy. Curr Biol. 2019;29:846–855. doi: 10.1016/j.cub.2019.01.041. [DOI] [PubMed] [Google Scholar]
- 14.Fumagalli F., Noack J., Bermann T.J., et al. Translocon component Sec62 acts in endoplasmic reticulum turnover during stress recovery. Nat Cell Biol. 2016;18:1173–1184. doi: 10.1038/ncb3423. [DOI] [PubMed] [Google Scholar]
- 15.Smith M.D., Harley M.E., Kemp A.J., et al. CCPG1 is a non-canonical autophagy cargo receptor essential for ER-phagy and pancreatic ER proteostasis. Dev Cell. 2018;44:217–232. doi: 10.1016/j.devcel.2017.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen S., Mari M., Parashar S., et al. Vps13 is required for the packaging of the ER into autophagosomes during ER-phagy. Proc Natl Acad Sci U S A. 2020;117:18530–18539. doi: 10.1073/pnas.2008923117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang T.G., Weir N.R., Polyakov N.J., et al. CRISPR screening using an expanded toolkit of autophagy reporters identifies TMEM41B as a novel autophagy factor. PLoS Biol. 2019;17 doi: 10.1371/journal.pbio.2007044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ji C.H., Kim H.Y., Heo A.J., et al. The N-degron pathway mediates ER-phagy. Mol Cell. 2019;75:1058–1072. doi: 10.1016/j.molcel.2019.06.028. [DOI] [PubMed] [Google Scholar]
- 19.Nthiga T.M., Shrestha B.K., Sjøttem E., et al. CALCOCO1 acts with VAMP-associated proteins to mediate ER-phagy. EMBO J. 2020;39 doi: 10.15252/embj.2019103649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hidvegi T., Schmidt B.Z., Hale, et al. Accumulation of mutant α1ATZ in the ER activates caspases-4 and -12, NFκB and BAP 31 but not the unfolded protein response. J Biol Chem. 2005;280:39002–39015. doi: 10.1074/jbc.M508652200. [DOI] [PubMed] [Google Scholar]
- 21.Perlmutter D.H. α1-antitrypsin deficiency: a misfolded protein variant with unique effects on the endoplasmic reticulum. Endoplasmic Reticulum Stress Dis. 2016;3:63–72. doi: 10.1515/ersc-2016-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hidvegi T., Ewing M., Hale P., et al. An autophagy-enhancing drug promotes degradation of mutant α-antitrypsin Ζ and reduces hepatic fibrosis. Science. 2010;329:229–232. doi: 10.1126/science.1190354. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y., Cobanoglu M.C., Li J., et al. An analog of glibenclamide selectively enhances autophagic degradation of misfolded α1-antitrypsin Ζ. PLoS One. 2019;14 doi: 10.1371/journal.pone.0209748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qu D., Teckman J.H., Omura S., et al. Degradation of a mutant secretory protein, α1-antitrypsin Ζ, in the endoplasmic reticulum requires proteasome activity. J Biol Chem. 1996;271:22791–22795. doi: 10.1074/jbc.271.37.22791. [DOI] [PubMed] [Google Scholar]
- 25.Teckman J.H., Gilmore R., Perlmutter D.H. The role of ubiquitin in proteasomal degradation of mutant α1-antitrypsin Ζ in the endoplasmic reticulum. Am J Physiol. 2000;27:G39–G48. doi: 10.1152/ajpgi.2000.278.1.G39. [DOI] [PubMed] [Google Scholar]
- 26.Brodsky J.L., McCracken A.A. ER protein quality control and proteasome-mediated protein degradation. Semin Cell Dev Biol. 1999;10:507–513. doi: 10.1006/scdb.1999.0321. [DOI] [PubMed] [Google Scholar]
- 27.Kamimoto T., Shoji S., Hidvegi T., et al. Intracellular inclusions containing mutant α1-antitrypsin Ζ are propagated in the absence of autophagic activity. J Biol Chem. 2006;281:4467–4476. doi: 10.1074/jbc.M509409200. [DOI] [PubMed] [Google Scholar]
- 28.Fregno I., Fasana E., Bergmann T.J., et al. ER-to-lysosome associated degradation of proteasome-resistant ATZ polymers occurs via receptor-mediated vesicular transport. EMBO J. 2018;37 doi: 10.15252/embj.201899259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeJesus R., Moretti F., McAllister G., et al. Functional CRISPR screening identifies the ufmylation pathway as a regulator of SQSTM1/p62. eLife. 2016;5 doi: 10.7554/eLife.17290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goodwin J.M., Dowdle W.E., DeJesus R., et al. Autophagy-independent lysosomal targeting regulated by ULK1/2-FIP200 and ATG9. Cell Rep. 2017;20:2341–2356. doi: 10.1016/j.celrep.2017.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang T., Birsoy K., Hughes N.W., et al. Identification and characterization of essential genes in the human genome. Science. 2015;350:1096–1101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J., Pak S.C., O’Reilly L.P., et al. Fluphenazine reduces proteotoxicity in C. elegans and mammalian models of alpha-1-antitrypsin deficiency. PLoS One. 2014;9 doi: 10.1371/journal.pone.0087260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voisine C., Søndergaard Pedersen J., Morimoto R.I. Chaperone networks: tipping the balance in protein folding diseases. Neurobiol Dis. 2010;40:12–20. doi: 10.1016/j.nbd.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pouchucq L., Lobos-Ruiz P., Araya G., et al. The chaperonin CCT promotes the formation of fibrillary aggregates of γ-tubulin. Biochim Biophys Acta. 2018;1866:519–526. doi: 10.1016/j.bbapap.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 35.Behrends C., Langer C.A., Boteva R., et al. Chaperonin TriC promotes the assembly of polyQ expansion proteins into nontoxic oligomers. Mol Cell. 2006;23:887–897. doi: 10.1016/j.molcel.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 36.Pavel M., Imarisio S., Menzies F.M., et al. CCT complex restricts neuropathogenic protein aggregation via autophagy. Nat Commun. 2016;7 doi: 10.1038/ncomms13821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ji C., Zhao H., Chen D., et al. β-propeller proteins WD45 and WD45B regulate autophagosome maturation into autolysosomes in neural cells. Current Biol. 2021;31:1666–1677. doi: 10.1016/j.cub.2021.01.081. [DOI] [PubMed] [Google Scholar]
- 38.Tafaleng EN, Li J, Wang Y, et al. Variants in autophagy genes MTMR12 and FAM134A are putative modifiers of the hepatic phenotype in α1-antitrypsin deficiency. Hepatology (in press). [DOI] [PMC free article] [PubMed]
- 39.Chen X., Gu X., Zhang H. Sidt2 regulates hepatocellular lipid metabolism through autophagy. J Lipid Res. 2018;59:404–415. doi: 10.1194/jlr.M073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gosai S.J., Kwak J.H., Luke C.J., et al. Automated high-content live animal drug screening using C. elegans expressing the aggregation prone serpin α1-antitrypsin Z. PLoS One. 2010;5 doi: 10.1371/journal.pone.0015460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Long O.S., Benson J.A., Kwak J.H., et al. A C. elegans model of human α1-antitrypsin deficiency links components of the RNAi pathway to misfolded protein turnover. Hum Mol Genet. 2014;23:5109–5122. doi: 10.1093/hmg/ddu235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kruse K.B., Brodsky J.L., McCracken A.A., et al. Characterization of an ERAD gene as VPS30/ATG6 reveals two alternative and functionally distinct protein quality control pathways: one for soluble Z variant of human alpha-1 proteinase inhibitor (A1PiZ) and another for aggregates of A1PiZ. Mol Biol Cell. 2006;17:203–212. doi: 10.1091/mbc.E04-09-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ordoñez A., Harding H.P., Marciniak S.J., et al. Cargo receptor-assisted endoplasmic reticulum export of pathogenic a1-antitrypsin polymers. Cell Rep. 2021;35 doi: 10.1016/j.celrep.2021.109144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teckman J.H., An J.-K., Blomenkamp K., et al. Mitochondrial autophagy and injury in the liver in alpha 1-antitrypsin deficiency. Am J Physiol Gastrointest Liver. 2014;286:G851–G862. doi: 10.1152/ajpgi.00175.2003. [DOI] [PubMed] [Google Scholar]
- 45.Pastore N., Annunziata F., Colonna R., et al. Increased expression or activation of TRPML1 reduces hepatic storage of toxic alpha-1-antitrypsin. Mol Ther. 2023;31:2651–2661. doi: 10.1016/j.ymthe.2023.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun S., Wang C., Zhao P., et al. Capturing the conversion of pathogenic alpha-1-antitrypsin fold by ATF6 enhanced proteostasis. Cell Chem Biol. 2023;30:22–32. doi: 10.1016/j.chembiol.2022.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oh Y.M., Lee S.W., Kim W.K., et al. Age-related Huntington’s disease progression modeled in directly reprogrammed patient-derived striatal neurons highlights impaired autophagy. Nat Neurosci. 2022;25:1420–1433. doi: 10.1038/s41593-022-01185-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deerinck T.J., Bushong E., Lev-Ram V., et al. Enhancing serial block-face scanning electron microscopy to enable high resolution 3-D nanohistology of cells and tissues. Microscopy and Microanalysis. 2010;16:1138–1139. [Google Scholar]
- 49.Robles H., Park S., Joens M.S., et al. Characterization of the bone marrow adipocyte niche with three-dimensional electron microscopy. Bone. 2019;118:89–98. doi: 10.1016/j.bone.2018.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Falk T., Mai D., Bensch R., et al. U-Net: deep learning for cell counting, detection, and morphometry. Nat Methods. 2019;16:67–70. doi: 10.1038/s41592-018-0261-2. [DOI] [PubMed] [Google Scholar]
- 51.Schindelin J., Arganda-Carreras I., Frise E., et al. FIJI: an open-source platform for biological-image analysis. Nat Methods. 2018;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.