Abstract
Bacteriophage φ6 is a complex enveloped double-stranded RNA virus with a segmented genome and replication strategy quite similar to that of the Reoviridae. An in vitro packaging and replication system using purified components is available. The positive-polarity genomic segments are translocated into a preformed polymerase complex (procapsid) particle. This particle is composed of four proteins: the shell-forming protein P1, the RNA polymerase P2, and two proteins active in packaging. Protein P7 is involved in stable packaging, and protein P4 is a homomultimeric potent nucleoside triphosphatase that provides the energy for the RNA translocation event. In this investigation, we used mutational analysis to study P4 multimerization and assembly. P4 is assembled onto a preformed particle containing proteins P2 and P7 in addition to P1. Only simultaneous production of P1 and P4 in the same cell leads to P4 assembly on P1 alone, whereas the P1 shell is incompetent for accepting P4 if produced separately. The C-terminal part of P4 is essential for particle assembly but not for multimerization or enzymatic activity. Altering the P4 nucleoside triphosphate binding site destroys the ability to form multimers.
The complex double-stranded RNA (dsRNA) viruses share a number of characteristics. The genomes are segmented, and the innermost particle in the virion comprises RNA-dependent RNA polymerase activity. The outer protein (or lipid) layers are designed to recognize the host cell and to deliver the polymerase particle into it. The double-stranded genome is contained within the polymerase particle throughout the infection; only the positive-sense transcripts of the genome segments are translocated to the cell interior for protein production and particle assembly. Newly formed polymerase particles express plus-strand synthesis, thus multiplying the number of transcribing particles and allowing the production of large amounts of the structural proteins. Complex dsRNA viruses that infect bacterial, plant, and animal hosts have been found.
Bacteriophage φ6 infects Pseudomonas syringae cells. It is surrounded by a lipid envelope that encloses the nucleocapsid (33). The nucleocapsid is composed of a core, the polymerase complex particle, which is surrounded by a shell of protein P8. The icosahedral core is composed of four protein species and three dsRNA genome segments (15, 32). Protein P1 forms the particle skeleton, and the rest of the core proteins are associated with it (12, 21). Protein P2 contains the RNA polymerase active site, P7 is needed for stable genome packaging (9, 10), and protein P4 is a nonspecific nucleoside triphosphatase (NTPase) that plays a role in providing the energy for the RNA translocation reaction (packaging) (7, 20, 27). The proposed numbers of these proteins in the polymerase complex are 120 for P1, 12 for P2, 120 for P4, and 60 for P7 (10, 11, 14). The genes encoding the polymerase complex proteins are all located in genome segment L (17). A cDNA copy of the L segment expressed in Escherichia coli produces empty polymerase complex particles, procapsids (6). These package plus-sense transcripts and synthesize the corresponding minus strands inside the particle in vitro (8). Protein P8 assembly onto these particles leads to the formation of nucleocapsids that are infectious to spheroplasts of the host cell. This infection process produces infectious enveloped virions (23, 24).
Protein P4 is a nonspecific NTPase cleaving ribo-, deoxyribo-, and dideoxyribonucleoside triphosphates to the corresponding diphosphates (27). The protein is 331 amino acids long (ca. 35 kDa) and forms doughnut-shaped homomultimers in the presence of divalent cations and ATP or ADP (11). The enzymatic activity is associated only with the multimeric form of the protein. The activity is enhanced by calcium and zinc ions as well as single-stranded RNA and is down-regulated by magnesium ions (11, 27). The P4 NTPase is the only one detected in the polymerase particle and, since the RNA packaging reaction is dependent on the presence of nucleotides that can be cleaved by P4, it is considered to be the energy source for the RNA translocation reaction into the procapsid (7, 27). P4 is shown to contain about 30% of both α-helix and β-strand, suggesting an α/β fold with significant amounts of loops and turns (11).
It has previously been shown that proteins P2 and P7 associate with a particle containing proteins P1 and P4 (3, 6, 10). However, due to the difficulties based on the insolubility of P4-deficient particles, it has not been possible to determine the assembly requirements of protein P4. In this investigation, we used mutational analysis to investigate protein P4 assembly behavior. Both random and targeted mutations are produced in gene 4. The corresponding mutant proteins are assayed for solubility, enzymatic activity, multimer formation, and assembly on particles lacking P4. We propose an assembly pathway in which the multimer is formed first and then is assembled on the P1 particle without the aid of any other phage proteins.
MATERIALS AND METHODS
Strains and plasmids.
E. coli JM109 (35), DH5α (30), and HMS174(DE3) (31) were used both for plasmid propagation and protein production. The plasmids used in this study are listed in Table 1. All of the expression plasmids are inducible with isopropyl-β-d-thiogalactopyranoside (IPTG). Pseudomonas phaseolicola HB10Y (HB) (33) and bacteriophage φ6 deletion mutant φ1980 (26) lacking genes 14, 7, 2, and 4 were used in the isolation of mutants in gene 4. Wild-type (wt) proteins P1 and P4 were produced from plasmids pAP4 and pAP2, respectively. To make these plasmids, an expression plasmid, pMF2, was first constructed. The SalI-SalI fragment containing the lacIq gene was deleted from pDMI.1, and the lac promoter (SspI-HindIII fragment) was replaced with a PvuII-HindIII fragment from pJJ2 containing the T7 promoter and multiple cloning site. The P4 expression plasmid pAP2 was constructed by ligating an EcoRI-XbaI fragment of pJTJ7 containing the φ6 gene 4 into pMF2. Gene 1 was copied from plasmid pLM393 by PCR, and an EcoRI-XbaI fragment was ligated into pJJ2 to make pJTJ8. The cloned gene 1 was transferred further into pMF2 as an EcoRI-XbaI fragment to obtain pAP4, the P1 expression plasmid. The P4-null control mutation was constructed by creating an additional MunI site on pLM1159 with the QuickChange site-directed mutagenesis kit (Stratagene) and removing gene 4 by cutting with MunI and religating. The remaining gene 4 flanking regions were exchanged into pLM687 as a SphI-SnaBI fragment to create pAP6.
TABLE 1.
Plasmids used in this study
| Plasmid | Relevant phenotypea | Antibiotic resistanceb | Reference or source |
|---|---|---|---|
| pLM254 | Vector | Amp | 16 |
| pLM523 | Vector | Tet | 8 |
| pT7T3 19U | Vector | Amp | Pharmacia |
| pDMI.1 | Vector | Kan | 4 |
| pMF2 | Vector | Kan | This study |
| pJJ2 | Vector | Amp | 19 |
| pJTJ7 | Produces wt P4 | Amp | 19 |
| pJTJ8 | Produces wt P1 | Amp | This study |
| pAP2 | Produces wt P4 | Kan | This study |
| pAP4 | Produces wt P1 | Kan | This study |
| pAP6 | Produces wt P1, P2, and P7 | Amp | This study |
| pLM393 | Contains gene 1 | Amp | 6 |
| pLM450c | Produces wt P1, P2, P4, and P7 | Tet | 8 |
| pLM624 | Gene 4 in pT7T3 19U | Amp | This study |
| pLM662 | Produces P1, P2, P4*, and P7 | Tet | This study |
| pLM687c | Produces wt P1, P2, P4, and P7 | Amp | 18 |
| pLM743 | pLM624 with point mutation in gene 4 Walker motif; produces P4* (K132QN) | Amp | This study |
| pLM745 | pLM624 with ts411 in gene 4; produces P4*(ts411) | Amp | This study |
| pLM1003 | Contains genes 7, 2, and 4 in pLM254 | Amp | 26 |
| pLM1159 | SphI-ClaI fragment of L segment in pT7T3 19U with an extra MunI near the end of gene 4 | Amp | This study |
| pLM1162 | Contains genes 7, 2, 4, and 1; an extra MunI near the end of gene 4 | Amp | This study |
| pLM1174 | Produces P1, P2, P4*, and P7 (P4* is a product of NTG mutagenesis) | Amp | This study |
| pLM1188 | Same as that of pLM1174 | Amp | This study |
| pLM1223 | Same as that of pLM1174 | Amp | This study |
| pLM1224 | Same as that of pLM1174 | Amp | This study |
| pLM1230 | Same as that of pLM1174 | Amp | This study |
| pLM1233 | Same as that of pLM1174 | Amp | This study |
| pLM1242 | Same as that of pLM1174 | Amp | This study |
| pLM1261 | Same as that of pLM1174 | Amp | This study |
| pLM1272 | Same as that of pLM1174 | Amp | This study |
| pJTJ7.3/3 | Produces truncated P4 (319 aa) | Amp | 19 |
| pJTJ7.3/7 | Produces truncated P4 (311 aa) | Amp | 19 |
| pJTJ7.4/6 | Produces truncated P4 (301 aa) | Amp | 19 |
| pJTJ7.5/7 | Produces truncated P4 (289 aa) | Amp | 19 |
| pJTJ7.6/7 | Produces truncated P4 (247 aa) | Amp | 19 |
| pJTJ7.5/11 | Produces truncated P4 (243 aa) | Amp | 19 |
P4*, protein P4 mutant; aa, amino acids.
Amp, ampicillin (150 μg/ml); Tet, tetracycline (10 μg/ml); Kan, kanamycin (25 μg/ml).
pLM450 and pLM687 both produce active procapsids. The only difference obtained is a higher yield of pLM687 due to the higher copy number of the plasmid.
Isolation of noncomplementing mutants of gene 4.
Plasmid pLM1003 contains a cDNA copy of genes 7, 2, and 4 of segment L in vector pLM254. Cells containing this plasmid can complement mutants in the corresponding genes and can thus complement phage φ1980, which carries a deletion in segment L that encompasses genes 7, 2, and 4 (26). A culture of HB containing pLM1003 was mutagenized with nitrosoguanidine (NTG) and grown overnight on Luria-Bertani (30) ampicillin plates. Plasmid DNA was isolated and used to transform HB cells. Ampicillin-resistant colonies were picked and tested for their ability to complement phage φ1980, which lacks genes 7, 2, and 4. Those that could not complement phage φ1980 were then tested for their ability to complement nonsense mutants of genes 7 and 2 and a temperature-sensitive (ts) mutant of gene 4. Fourteen isolates could not complement gene 4, and nine of these were selected for further studies. Plasmid pLM1162 contains a cDNA copy of the entire L segment but has a MunI site engineered at position 3907, near the end of gene 4. The numbering of φ6 L-segment nucleotides was done as described by Mindich et al. (see 17). The cDNA copy of genes 7, 2, and 4 from the mutant plasmids were excised by cutting with SgrAI and EcoRI and ligated into plasmid pLM1162 that had been cut with SgrAI and MunI. The resulting plasmids (pLM1174, -1188, -1223, -1224, -1230, -1233, -1242, -1261, and -1272) were transformed into E. coli JM109, and these cells were used for the production of procapsid proteins.
Construction of mutants in gene 4 by directed mutagenesis.
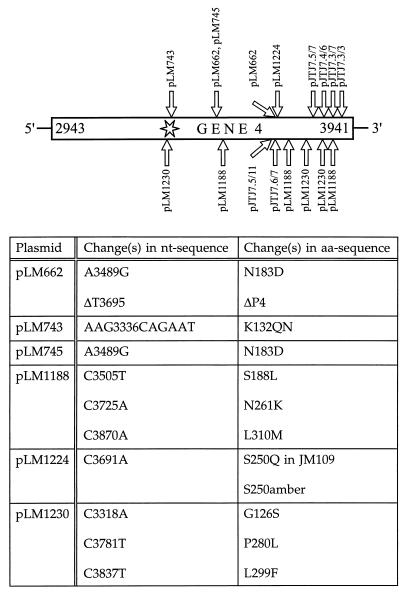
Protein P4 contains the Walker motif A for ATP binding (GSGKS) (34). This was changed by directed mutagenesis (13) to GSGQNS in plasmid pLM624, which has the SphI-ClaI fragment of segment L in plasmid pT7T3 19U. The only intact φ6 gene in this plasmid is gene 4. The mutated plasmid is pLM743. The sequence of a ts mutation in gene 4 (ts411) (28), was determined after reverse transcription-PCR, and the mutation was found to be A3489G, which resulted in amino acid mutation N183D. This mutation was introduced (13) into plasmid pLM624 to form pLM745 and into pLM523 to form pLM662, which also has spontaneously deleted T3695. Due to this deletion, the protein is truncated by 82 amino acids and contains a nonspecific tail of 45 amino acids. The C-terminal truncations of gene 4 have been previously described (19) (Table 1 and Fig. 1).
FIG. 1.
Changes in the nucleotide (nt) and amino acid (aa) sequences of the different gene 4 mutants. The star indicates the location of the Walker A consensus sequence for NTP binding. The length of P4 is 331 amino acids. The exact lengths of the C-terminal truncations (pJTJ7.3/3, -7.3/7, -7.4/6, -7.5/7, -7.6/7, and -7.5/11) are indicated in Table 1. P4 produced by pLM662 is truncated by 82 amino acids and contains a tail of 45 nonspecific amino acids.
Expression of cloned genes.
The recombinant proteins and particles were produced essentially in the same way for several purposes: thin-section electron microscopy, solubility analysis, NTPase activity measurements, P4 multimericity analysis, determination of particle formation, and reconstitution experiments. The expression conditions are summarized in Table 2. Cells were grown overnight in LB medium (30) supplemented with the proper antibiotics. In the morning, the cultures were either diluted into the same medium at a cell density of about 0.6 (A540) and grown to a cell density of about 0.8 (A540) or, for the solubility analysis, diluted 1:20 to 1:15 and grown for 1 h 40 min. In both cases, IPTG was added at this point to a final concentration of 1 mM at the indicated temperatures. The cells were collected after appropriate expression times by centrifugation and lysed by passage twice through a French pressure cell (diameter, 0.375 in.; volume, 3.7 ml; 20,000 lb/in2).
TABLE 2.
Expression conditions
| Assay and plasmids used | Expression temp (°C) | Expression time(s) (h)a | Resuspension buffer |
|---|---|---|---|
| Solubility | |||
| pAP2; pLM743, -1174, -1188, -1223, -1224, -1230, -1233, -1242, -1261, -1272 | 28 | 4–5 | 20 mM Tris (pH 8.0) |
| pLM662, pLM745 | 18 and 28 | 4–5 | 20 mM Tris (pH 8.0) |
| Electron microscopy | |||
| pLM450, -662, -1188, -1224, -1230; pAP4, pAP6 | 28 | ON | 10 mM K phosphate (pH 7.2) |
| NTPase activity | |||
| pLM450, -743, -1174, -1188, -1223, -1224, -1230, -1233, -1242, -1261, -1272; pJTJ7.3/7, -7.4/6, -7.5/7, -7.5/11, -7.6/7; pAP2 | 28 | 5 | 20 mM Tris (pH 7.4) |
| pLM745, pLM662 | 18 and 28 | 5 | 20 mM Tris (pH 7.4) |
| P4 multimericity | |||
| pLM743, pLM1224, pAP2 | 28 | 5 h and ON | 20 mM Tris (pH 8.0) |
| pLM745 | 18 and 28 | 5 h and ON | 20 mM Tris (pH 8.0) |
| Formation of soluble particles | |||
| pLM450, -1174, -1188, -1223, -1224, -1230, -1233, -1242, -1261, -1272 | 28 | 3–6 | 20 mM Tris (pH 8.0) |
| pLM662, pAP6 | 28 | ON | 20 mM Tris (pH 8.0) |
| Reconstitution | |||
| pLM1224, pAP2, pAP4; pJTJ7.3/3, -7.3/7, -7.4/6, -7.5/7 | 28 | ON | 20 mM Tris (pH 8.0) |
| pLM662, pLM745, pAP6 | 18 and 28 | ON | 20 mM Tris (pH 8.0) |
ON, overnight.
P4 solubility analysis.
The solubility of mutant P4 produced in JM109 cells from plasmids pLM743, -745, -1174, -1188, -1223, -1224, -1230, -1233, -1242, -1261, and -1272, in DH5α cells from pLM662, and in HMS174(DE3) cells from pAP2 (Table 1) was analyzed. The collected cells were sonicated twice for 30 s in an ice bath and centrifuged in a microcentrifuge (16,000 × g, 20 min, 5°C). Both the supernatant and the pellet were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (22). The Western blot analysis with nucleocapsid-specific polyclonal antiserum was done essentially as previously described (25). The ratio of P4 in the supernatant to that in the pellet fractions was approximated visually.
Electron microscopy.
Cells producing either protein P1 alone (pAP4) or together with proteins P2 and P7 (pAP6) or with P2, P7, and a P4 mutant (pLM662, -1188, -1224, and -1230) were analyzed by thin-section electron microscopy. Cells producing the procapsids (pLM450) were used as the control. Initially, cell sections were analyzed, but due to the difficulties in analyzing the aggregated material, the aggregates were isolated and partially purified for screening of particle production. Pellets of cell lysates resulting from low-speed centrifugation (16,000 × g, 10 min, 22°C; in a microcentrifuge) were used directly or washed with 430 mM NaCl (5 min, 22°C), followed by washing with 0.9% Triton X-100 (5 min, 22°C) before preparation for sectioning. Thin sections were prepared as previously described (1). The micrographs were taken with a JEOL 1200EX electron microscope operating at 60 kV.
P4 NTPase activity measurement in cell lysates.
The cell lysates were stored in aliquots at −80°C. The NTPase activity was analyzed by thin-layer chromatography (TLC) in principle as previously described (27). The standard reaction mixtures (10 μl) contained 50 mM Tris (pH 7.4), 1 mM CaCl2, 1 mM unlabelled nucleoside triphosphate (NTP), and 0.2 μCi of [α-32P]NTP (Amersham). The reactions were allowed to proceed for an hour at 45°C and were stopped by placement on ice. Aliquots of 2 μl were spotted onto a plastic-backed polyethyleneimine-cellulose F TLC sheet (no. 1.05579; Merck) and developed by ascending chromatography in 0.25 M LiCl and 1 M formic acid. The sheets were dried and exposed to X-ray film. The spots with uncleaved NTP were cut out, the radioactivity was measured by liquid scintillation counting, and the proportions of cleaved NTP were determined by comparison to the control. Alternatively, the radioactivity in the TLC sheets was measured with a phosphorimager (Fuji).
Sedimentation analyses.
The multimeric status of mutant P4 proteins produced from plasmids pLM743, -745, and -1224 in JM109 cells was determined. wt P4 produced from pAP2 in HMS174(DE3) cells was used as the control. The cell lysates were cleared by centrifugation in a microcentrifuge (16,000 × g, 20 min, 5°C). A 100-μl aliquot of the supernatant was sedimented through a sucrose gradient (SW41 Beckman rotor; 28 h 20 min, 158,100 × gav (average), 6°C; 5 to 20% sucrose, 20 mM Tris [pH 8], with or without 4 mM ATP). Six-hundred-microliter fractions were collected and analyzed by SDS-PAGE and by Western blotting using P4-specific polyclonal antiserum. Three-microliter aliquots of every second fraction of selected strains were used for the NTPase activity determination immediately after fraction collection.
The ability to produce soluble particles was analyzed by using plasmids pLM662 in DH5α cells, pAP6 in HMS174(DE3) cells, and pLM1174, -1188, -1223, -1224, -1230, -1233, -1242, -1261, and -1272 in JM109 cells producing the mutated P4 and the other three wt procapsid proteins. Procapsids produced from pLM450 were used as the control. The cell lysates were subsequently extracted with 5 and 3% Triton X-114 (9). The material in the aqueous phase was sedimented through a sucrose gradient (SW41 rotor; 89,000 × gav, 1 h 50 min, 10°C; 5 to 20% sucrose, 20 mM Tris [pH 8.0], 150 mM NaCl). One-milliliter fractions were collected and analyzed by SDS-PAGE and when appropriate by Western blotting using P4-, P7-, and nucleocapsid-specific polyclonal antisera.
For reconstitution of procapsids by P4 assembly, indicated cell lysates were combined 1:1 in the presence or absence of 4 mM ATP, incubated for 45 to 60 min at 18, 22, or 28°C, and either centrifuged in a microcentrifuge (16,000 × g, 10 to 15 min, 5°C) or extracted with 5 and 3% Triton X-114. The material in the supernatant or aqueous phase was sedimented through a sucrose gradient (SW41 rotor, 89,000 × gav, 2 h 10 min, 10°C; 5 to 20% sucrose, 20 mM Tris [pH 8], 150 mM NaCl, with or without 4 mM ATP). Six-hundred-microliter fractions were collected and analyzed by SDS-PAGE and when appropriate by Western blotting using P4-, P7-, and nucleocapsid-specific polyclonal antisera.
RESULTS
Assay for P4-specific NTPase in cell lysates.
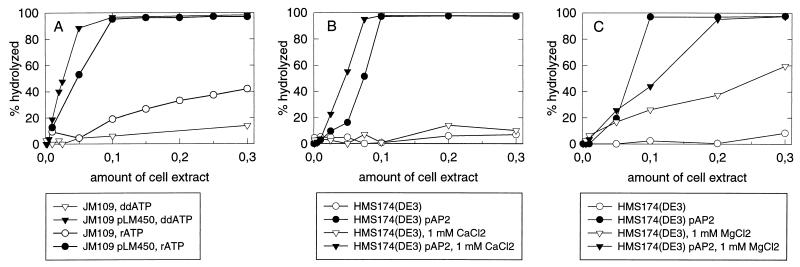
The ability to specifically assay the NTPase activity of protein P4 in cell extracts is a prerequisite for detecting activity even if the protein is strongly aggregative. This assay also makes the activity screening possible without purification of the protein. As a starting point, we used the knowledge (27) that P4 cleaves all tested ribo-, deoxyribo-, and dideoxyribonucleoside triphosphates and is activated by calcium but inhibited by magnesium ions. The high-temperature tolerance of P4 activity was also utilized. By varying the conditions, we measured the cell extract NTPase activity in the presence or absence of protein P4 (Fig. 2). We observed that P4 activity could be assayed both in extracts containing only P4 and in the presence of other procapsid proteins. The usage of ddATP instead of ATP greatly reduced the cellular background. Calcium stimulated the P4-specific but not the cellular activity, whereas magnesium had a stimulatory effect on the cellular NTPases but an inhibitory effect on the P4-specific activity. These results show that it is possible to find conditions in which the cellular NTPase activity is negligible compared to that of protein P4. For further analyses, the following conditions were used to specifically detect the P4 NTPase activity: 50 mM Tris (pH 7.4), 1 mM CaCl2, 1 mM unlabelled ddATP, 0.2 μCi of [α-32P]ddATP (PB10233, Amersham), and incubation for 1 h at 45°C.
FIG. 2.
Analysis of the NTPase activity in the cell extracts. (A) Comparison of ddATP and ATP as the substrate in the presence of 1 mM CaCl2 and in the presence or absence of the polymerase complex; (B and C) effect of 1 mM CaCl2 (B) and 1 mM MgCl2 (C) on the NTPase activity of cell extracts in the presence or absence of protein P4. The substrate is ddATP.
Mutations targeted to gene 4.
The function and assembly of protein P4 were approached by mutational analysis. Both NTG induction and directed mutagenesis were utilized. A previously isolated ts mutation in gene 4 (ts411) (28) and C-terminal truncations (19) were also included in this study. Several NTG-induced mutants of gene 4 as well as the ts mutant were subjected to sequence analysis. Figure 1 illustrates both the change(s) in the nucleotide sequence and the corresponding alteration(s) in the amino acid sequence. The nonfunctional mutations are found scattered over the carboxy-terminal two-thirds of gene 4.
Activity, oligomerization, and assembly analysis of mutant P4.
All of the P4-producing constructs (expressing P4 either alone or together with the other procapsid proteins) were subjected to NTPase analysis (Table 3). The solubility of P4 was determined, and when soluble product was detected, the multimeric status of the free protein P4 was assayed by sedimentation analysis using a wt P4-producing strain [HMS174(DE3)(pAP2)] as a control. Complete and incomplete procapsids were isolated by rate-zonal centrifugation. Their relative sedimentation rates were recorded, and the particle zones were collected and subjected to SDS-PAGE analysis. The complete procapsids were used as a control. The results are summarized in Tables 3 and 4.
TABLE 3.
Analysis of P4 properties
| Plasmid(s) and temp (°C) | Protein(s) produceda | ddATPase activityb | Aggrega-tivity of P4c | Multi-mericity of free P4 |
|---|---|---|---|---|
| pLM662 | ||||
| 18 | P1, P2, P4*, P7 | 0 | 0 | NDd |
| 28 | 0 | +, ++ | ND | |
| pLM1174, -1188, -1223, -1230, -1233, -1242, -1261, -1272 | P1, P2, P4*, P7 | 0 | ++ | ND |
| pLM1224 | P1, P2, P4*, P7 | + | 0 | + |
| pAP6 | P1, P2, P7 | 0 | ND | ND |
| pLM450 | P1, P2, P4, P7 | + | ND | ND |
| pLM743 | P4* | 0 | ND | 0 |
| pLM745 | ||||
| 18 | P4* | + | 0 | + |
| 28 | + | + | + | |
| pJTJ7.3/3, -7.3/7, -7.4/6, -7.5/7 | ΔP4 | + | 0e | +e |
| pJTJ7.5/11, pJTJ7.6/7 | ΔP4 | 0 | +e | 0e |
| pAP2 | wt P4 | + | 0 | + |
P4*, mutated P4; ΔP4, truncated P4.
All the mutant and wt P4 cell extracts were analyzed as described in the legend to Fig. 2. Symbols: 0, activity at the same level as that in the control cell extract; +, activity at the same level as that in the wt P4-containing cell extract. No intermediate activities were found.
The relative amount of aggregative P4 was approximated visually for the centrifugation supernatant and pellet by using Western blots. Symbols: ++, ≥90% in aggregate; +, 50% in aggregate; 0, ≤10% in aggregate.
ND, not determined.
These results have been published previously (19).
TABLE 4.
Production of soluble particles and assembly in reconstitution analysis
| Plasmid used for particle productiona | Soluble particlesb | Reconstitutionc with:
|
|
|---|---|---|---|
| P4 wt (pAP2) | P4 ts411 (pLM745) | ||
| pLM450 (P1, P2, P4, P7) | ++ | NDd | ND |
| pAP6 (P1, P2, P7) | + | + | +e |
| pLM662 (P1, P2, P4*, P7) | + | + | +e |
| pLM1224 (P1, P2, P4*, P7) | ++ | + | ND |
| pLM1174, -1188, -1223, -1230, -1233, -1242, -1261, -1272 (P1, P2, P4*, P7) | 0 | ND | ND |
Procapsid proteins produced are shown in parentheses. P4*, mutated P4 (S250Q; produced from pLM1224).
Symbols: 0, no soluble particles detected; +, small amount of soluble particles detected, with most of the proteins in aggregate; ++, amount of soluble particles detected comparable to that of the procapsid (polymerase complex).
+, P4 binds to the particle.
ND, not determined.
Positive results achieved at both 18 and 28°C.
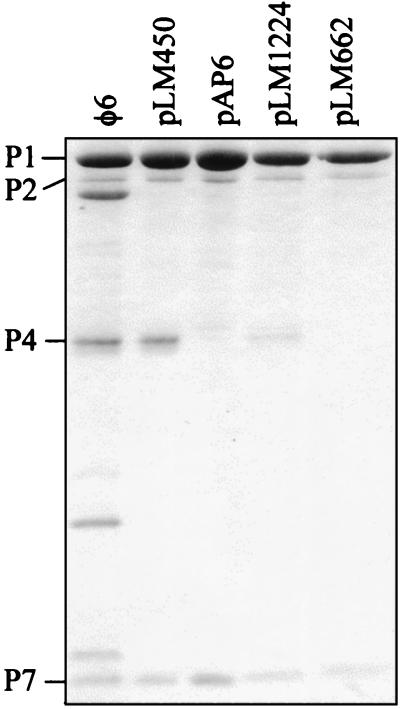
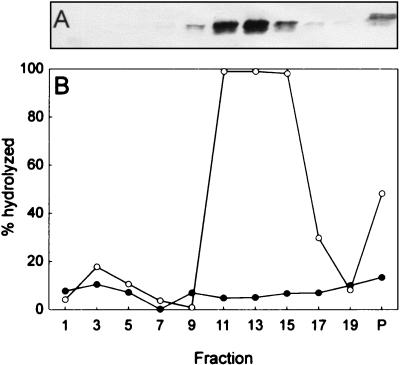
All P4 proteins truncated more than 42 amino acids from the C terminus were enzymatically inactive, whereas less truncated forms were active (Table 3). It has been previously shown that proteins with truncations of more than 42 amino acids were found in aggregates while less truncated proteins were soluble and multimeric (19). A change in the P4 NTP binding site (K132QN; produced from pLM743) also resulted in aggregation of the protein and loss of enzymatic activity (Table 3). The ts411 mutant protein when produced alone (pLM745) was enzymatically active, multimeric, and soluble at a low temperature but increasingly aggregative at a high temperature (Table 3). The truncated ts mutant protein (produced from pLM662) in the presence of the other procapsid proteins did not show enzymatic activity (Table 3), and only a small amount of soluble particles was produced with no or only trace amounts of truncated P4 (Fig. 3). The observed length of the truncation corresponds to that calculated from the sequence. Purification of these particles in sucrose gradients gave similar results to those obtained with the P4-null mutant (produced from pAP6) (Fig. 3 and see Fig. 5A). All NTG-induced P4 mutants obtained in this study (see Materials and Methods) except those produced from pLM1224 were aggregative and enzymatically inactive when expressed with the rest of the procapsid proteins. P4 produced from plasmid pLM1224 was active and multimeric, but only about 10 to 20% of the normal amount of P4 was detected on the particle (Fig. 3) as quantitated by image analysis (BioImage; Whole Band analysis software). Figure 4 shows the sedimentation and enzymatic activity of the free multimeric P4 produced from pLM1224. This is compared to the sedimentation of the monomeric, inactive P4 with a mutated NTP binding site (K132QN; produced from pLM743). The amount of soluble particles produced from pLM1224 was comparable to that obtained in the case of procapsids. Both particles with a reduced amount of P4 (produced from pLM1224) and those without P4 (pLM662 and pAP6) sedimented more slowly than the normal procapsid particles.
FIG. 3.
SDS-PAGE analysis (Coomassie blue stain) of procapsid (pLM450) and particles produced from plasmids pAP6, pLM1224, and pLM662. The φ6 procapsid (polymerase complex) proteins are indicated to the left of the figure.
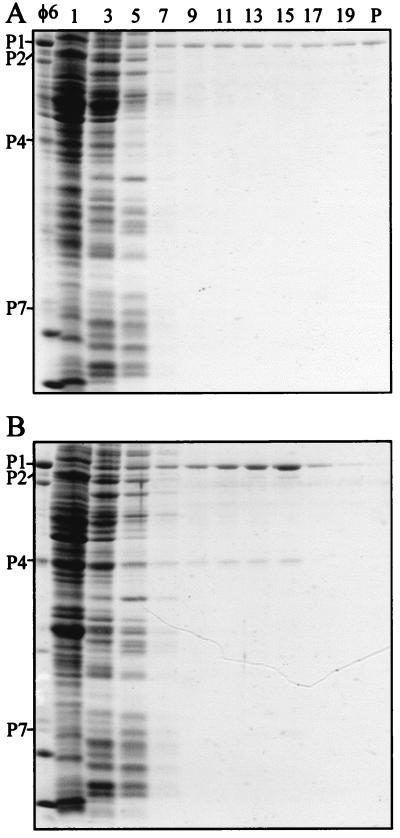
FIG. 5.
SDS-PAGE analysis (Coomassie blue stain) of the reconstitution experiment with cell extracts containing P1,P2,P7 particles (pAP6) and wt P4 (pAP2). (A) Sedimentation of the material produced by pAP6 alone; (B) sedimentation of the particles obtained by combining extracts of P1,P2,P7 (pAP6) and P4 (pAP2) prior to the sedimentation analysis. The φ6 procapsid (polymerase complex) proteins are indicated to the left of the panels. Sedimentation is from left to right.
FIG. 4.
Protein P4 multimericity and NTPase activity analyses of cells containing plasmid pLM1224 or pLM743. (A) Western blot showing the sedimentation of pLM1224 mutant P4 protein; (B) NTPase activity in the fractions shown in panel A for pLM1224 (○) and pLM743 (•). Sedimentation is from left to right.
Reconstitution.
We coexpressed proteins P1 and P4 from separate plasmids (pJTJ8 and pAP2, respectively) in the same cell. The examination of particle production by rate-zonal centrifugation revealed soluble particles that had approximately 50 to 80% of P4 compared to that found in the virion as determined by SDS-PAGE. We also used this assay to test the C-terminally least truncated form of P4 lacking only 13 amino acids. It did not form detectable soluble particles containing P1 (data not shown). This is contrasted with our finding that the protein formed enzymatically active multimers and that even truncations as great as 42 amino acids resulted in enzymatically active multimers.
A system for reconstituting particles by combining cell lysates was developed. It allows us to test the reconstitution of procapsids when cell extracts containing P4 or its derivatives are mixed with extracts containing the rest of the procapsid proteins (produced from pAP6) or those that in addition contain a P4 mutant (produced from pLM662 and pLM1224). When wt P4 and the rest of the polymerase complex proteins were mixed, particles containing P4 were obtained (Fig. 5). It appears that about 50 to 80% of P4 found in the virion was associated with particles, as measured by comparing the P1/P4 ratio by image analysis and using the purified virus as a standard. Under these conditions, wild-type P4, when sedimented alone, was not detected in the sucrose gradient fractions where the particles sediment. The amount of soluble particles increased by a factor of three to five as compared to the amount obtained without the addition of P4 (Fig. 5 and Table 4). In the presence of either 50 mM ATP or ADP, equal amounts of reconstituted particles were obtained. On the contrary, when the reconstitution was done by combining cell extracts containing only wt P1 and P4, no soluble particles could be detected.
The proportion of soluble particles produced from pLM662 (truncated ts P4 and wt P1, P2, and P7) was estimated to increase from 5 to 20% when rescued by wt P4. Also, the ts411 mutant P4 that was produced alone both at low and high temperatures assembled on the P4-deficient particles produced by pAP6. Particles produced from plasmid pLM1224 could bind additional wt P4 to produce particles with close to the normal amount of P4. P4 with the mutated NTP binding site (K132QN; pLM743) could not be bound to particles in reconstitution experiments. In addition, none of the truncated P4 proteins could associate with the particles.
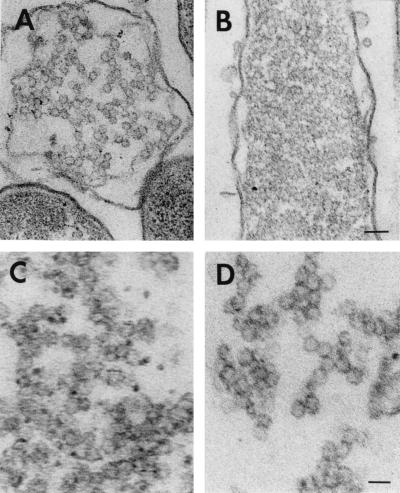
To reveal whether P4 is assembled on preformed particles or whether it induces the assembly of the particle from the protein constituents, we investigated the presence of intracellular particles by thin-section electron microscopy. Both cells and the insoluble aggregate obtained from the cells were studied (Fig. 6 and Table 4). It appeared that in all cases (P1 alone or together with P2 and P7 or in the presence of an aggregating P4 mutant), the aggregated material had assembled into spherical particles approximately the size of the procapsid. It was also observed by SDS-PAGE and Western blot analysis that the salt wash removed protein P7 from the aggregate into a soluble form, also indicating that the P7 is not denatured in the aggregate. The aggregated material had cellular components associated with it, making the particles difficult to be distinguished. The salt-detergent wash applied, however, greatly reduced the particle-associated material, allowing a clear detection of polyhedral particles (Fig. 6D).
FIG. 6.
Thin-section electron microscopy. (A) Cells containing procapsids (pLM450); (B) cells containing P1,P2,P7 particles (pAP6); (C) insoluble aggregative material collected from disrupted pAP6-containing cells; (D) same as panel C but after washing with NaCl and Triton X-100. Bars, 100 (A and B) and 50 (C and D) nm.
DISCUSSION
Recombinant protein P4 oligomerizes in the cell to form enzymatically active multimers. Its NTPase activity can be assayed in cell extracts without the interference of cellular NTPases. The mutational analysis revealed that most changes in the protein led to an inactive aggregated form of the protein. However, a previously isolated ts mutant (ts411; N183D) formed enzymatically active multimers that showed an aggregation tendency only at the high temperature (28°C). A S250Q mutant (produced from pLM1224) obtained by NTG mutagenesis formed soluble, enzymatically active, and multimeric P4. C-terminally truncated proteins longer than 289 amino acids were also enzymatically active. This is in accordance with previous results showing that these truncations were soluble and multimeric (19). We have previously shown that ADP or ATP and divalent cation binding to P4 are driving the multimerization (11). This explains the observation that mutations directed to the nucleotide binding site prevent multimer formation. The rest of the mutations leading to inactive aggregates interfere with either the protein folding or the nucleotide and/or divalent cation binding.
Protein P1 forms the structural framework for the polymerase complex (procapsid) particle. The other three proteins (P2, P4, and P7) are independently associated with this structure (3, 6, 8, 10). The role of protein P4 and its NTPase activity in the procapsid assembly have been unclear. Here we showed that all the constructs with protein P1 alone or with other procapsid proteins produced polyhedral particles that could be detected in thin sections of partially purified cellular aggregates. This indicates that P1 alone can nucleate and accomplish the P1 shell assembly and that P4 or its NTPase activity are not crucial in this process. The P1 particles are, however, very aggregative, as previously shown for virion-derived P1 particles (2).
Soluble particles containing proteins P1 and P4 have previously been produced from an L-genome segment cDNA clone containing genes 1 and 4 (8). Here similar particles were obtained when these proteins were produced in the same cell but from two different plasmids. However, our attempts to assemble P1,P4 particles by combining cell extracts containing recombinant P1 and P4 proteins failed. This is contrary to a situation where a cell extract containing proteins P1, P2, and P7 was incubated with a P4-containing cell extract and particles containing all four proteins were obtained. It may be that the tendency of P1 shells to aggregate when alone in the cell leads to association of cellular material and other P1 particles and thus prevents the association of other procapsid proteins added later. However, if other procapsid proteins are present during P1 shell assembly, their presence in the particle increases the solubility to allow the assembly of other procapsid proteins added later. This is supported by an earlier observation (6) that proteins P2 and P7 can be assembled on P1,P4 particles by mixing cell extracts containing these proteins. Another option is that P1 shell without other procapsid proteins achieves a conformation that is incompetent for correct maturation.
The reconstitution system allowed a test of the ability of mutant P4 proteins to assemble onto the P1,P2,P7 particle. These observations identify two sites in P4 that are involved in forming the interactions or causing changes to the interactive sites that are important in making the contacts with the P1 particle. Mutant S250Q produced particles with a greatly reduced (about 80 to 90%) amount of P4. In this case, the amount of soluble particles was about equal to that obtained with procapsids. In the reconstitution experiments, the mutant P4 did not hinder binding of wt P4 up to almost the amount found in the virion. None of the C-terminal truncations tested associated with the P1,P2,P7 particle, although they were producing enzymatically active multimeric P4. The C-terminal end of P4 has also been shown to be the most antigenic (and probably exposed on the protein surface) (19), which might be consistent with it playing a role in the interaction with P1. The ts mutant (ts411; N183D) protein produced both at the low (18°C) and high (28°C) temperatures assembled normally. The basis of the ts phenotype is not known.
Our observations show that a multimeric P4 is capable of assembling on a preformed particle, indicating an assembly pathway in which the synthesized P4 monomers first form a multimer and then the multimer recognizes and binds to the P1 shell. In vivo, however, these two proteins are cosynthesized, and although a sequential assembly process can be demonstrated, coassembly could take place. The multimerization of P4 has been shown to be independent of P4 NTPase activity (11). Here we showed that addition of competing amounts of ADP did not prevent P4 reconstitution, suggesting that the assembly process is also not associated with the energy production by P4 NTP cleavage. The assembly of P4 on preformed P1,P2,P7 particles indicates a surface location for P4. Our preliminary results with P4-deficient particles confirm the necessity of the presence of P4 for the RNA packaging.
Incomplete orbivirus (reviewed in reference 29) and rotavirus (5) core particles have been produced by coexpression of the cloned genes. These particles can contain the core proteins in several different combinations. So far there has been no report of sequential addition of the proteins to form stable particles in these cases.
ACKNOWLEDGMENTS
Marja-Leena Parälä is acknowledged for skillful technical assistance. M. Frilander and J. Juuti are thanked for providing the pMF2 and pJTJ8 plasmids.
Administrative services were obtained from Institute of Biotechnology, University of Helsinki. Helsinki Graduate School in Biotechnology and Molecular Biology is acknowledged for a fellowship to A.O.P. This work was supported by the Finnish Academy of Sciences grants 37725 and 30562 (D.H.B.), by EU grant BIO4-CT97-2364 (D.H.B.), and by NIH grant GM34352 (L.M.).
REFERENCES
- 1.Bamford D H, Mindich L. Electron microscopy of cells infected with nonsense mutants of bacteriophage φ6. Virology. 1980;107:222–228. doi: 10.1016/0042-6822(80)90287-1. [DOI] [PubMed] [Google Scholar]
- 2.Butcher S J, Dokland T, Ojala P M, Bamford D H, Fuller S D. Intermediates in the assembly pathway of the double-stranded RNA virus φ6. EMBO J. 1997;16:4477–4487. doi: 10.1093/emboj/16.14.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casini G, Qiao X, Mindich L. Reconstitution of active replicase in procapsids of the segmented dsRNA bacteriophage φ6. Virology. 1994;204:251–253. doi: 10.1006/viro.1994.1529. [DOI] [PubMed] [Google Scholar]
- 4.Certa U, Bannwarth W, Stüber D, Gentz R, Lanzer M, Le Grice S, Guillot F, Wendler I, Hunsmann G, Bujard H, Mous J. Subregions of a conserved part of the HIV gp41 transmembrane protein are differentially recognized by antibodies of infected individuals. EMBO J. 1986;5:3051–3056. doi: 10.1002/j.1460-2075.1986.tb04605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crawford S E, Labbé M, Cohen J, Burroughs M H, Zhou Y, Estes M K. Characterization of virus-like particles produced by the expression of rotavirus capsid proteins in insect cells. J Virol. 1994;68:5945–5952. doi: 10.1128/jvi.68.9.5945-5952.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gottlieb P, Strassman J, Bamford D H, Mindich L. Production of polyhedral particle in Escherichia coli from a cDNA copy of the large genomic segment of bacteriophage φ6. J Virol. 1988;62:181–187. doi: 10.1128/jvi.62.1.181-187.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gottlieb P, Strassman J, Mindich L. Protein P4 of the bacteriophage φ6 procapsid has a nucleoside triphosphate-binding site with associated nucleoside triphosphate phosphohydrolase activity. J Virol. 1992;66:6220–6222. doi: 10.1128/jvi.66.10.6220-6222.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gottlieb P, Strassman J, Qiao X, Frucht A, Mindich L. In vitro replication, packaging, and transcription of the segmented double-stranded RNA genome of bacteriophage φ6: studies with procapsids assembled from plasmid-encoded proteins. J Bacteriol. 1990;172:5774–5782. doi: 10.1128/jb.172.10.5774-5782.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Juuti J T, Bamford D H. RNA binding, packaging and polymerase activities of the different incomplete polymerase complex particles of dsRNA bacteriophage φ6. J Mol Biol. 1995;249:545–554. doi: 10.1006/jmbi.1995.0317. [DOI] [PubMed] [Google Scholar]
- 10.Juuti J T, Bamford D H. Protein P7 of phage φ6 RNA polymerase complex, acquiring of RNA packaging activity by in vitro assembly of the purified protein onto deficient particles. J Mol Biol. 1997;266:891–900. doi: 10.1006/jmbi.1996.0817. [DOI] [PubMed] [Google Scholar]
- 11.Juuti J T, Bamford D H, Tuma R, Thomas G J., Jr Structure and NTPase activity of the RNA-translocating protein (P4) of bacteriophage φ6. J Mol Biol. 1998;279:347–359. doi: 10.1006/jmbi.1998.1772. [DOI] [PubMed] [Google Scholar]
- 12.Ktistakis N T, Lang D. The dodecahedral framework of the bacteriophage φ6 is composed of protein P1. J Virol. 1987;61:2621–2623. doi: 10.1128/jvi.61.8.2621-2623.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 14.Mindich L, Bamford D H. Lipid-containing bacteriophages. In: Calendar R, editor. The bacteriophages. Vol. 2. New York, N.Y: Plenum Press; 1988. pp. 475–520. [Google Scholar]
- 15.Mindich L, Davidoff-Abelson R. The characterization of a 120S particle formed during φ6 infection. Virology. 1980;103:386–391. doi: 10.1016/0042-6822(80)90197-x. [DOI] [PubMed] [Google Scholar]
- 16.Mindich L, MacKenzie G, Strassman J, McGraw T, Metzger S, Romantschuk M, Bamford D. cDNA cloning of portions of the bacteriophage φ6 genome. J Bacteriol. 1985;162:992–999. doi: 10.1128/jb.162.3.992-999.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mindich L, Nemhauser I, Gottlieb P, Romantschuk M, Carton J, Frucht S, Strassman J, Bamford D H, Kalkkinen N. Nucleotide sequence of the large double-stranded RNA segment of bacteriophage φ6: genes specifying the viral replicase and transcriptase. J Virol. 1988;62:1180–1185. doi: 10.1128/jvi.62.4.1180-1185.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mindich L, Qiao X, Onodera S, Gottlieb P, Frilander M. RNA structural requirements for stability and minus-strand synthesis in the dsRNA bacteriophage φ6. Virology. 1994;202:258–263. doi: 10.1006/viro.1994.1341. [DOI] [PubMed] [Google Scholar]
- 19.Ojala P M, Juuti J T, Bamford D H. Protein P4 of the double-stranded RNA bacteriophage φ6 is accessible on the nucleocapsid surface: epitope mapping and orientation of the protein. J Virol. 1993;67:2879–2886. doi: 10.1128/jvi.67.5.2879-2886.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ojala P M, Paatero A O, Bamford D H. NTP-binding induces conformational changes in the double-stranded RNA bacteriophage φ6 subviral particles. Virology. 1994;205:170–178. doi: 10.1006/viro.1994.1632. [DOI] [PubMed] [Google Scholar]
- 21.Olkkonen V M, Bamford D H. The nucleocapsid of the lipid-containing double-stranded RNA bacteriophage φ6 contains a protein skeleton consisting of a single polypeptide species. J Virol. 1987;61:2362–2367. doi: 10.1128/jvi.61.8.2362-2367.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olkkonen V M, Bamford D H. Quantitation of the adsorption and penetration stages of bacteriophage φ6 infection. Virology. 1989;171:229–238. doi: 10.1016/0042-6822(89)90530-8. [DOI] [PubMed] [Google Scholar]
- 23.Olkkonen V M, Gottlieb P, Strassman J, Qiao X, Bamford D H, Mindich L. In vitro assembly of infectious nucleocapsids of bacteriophage φ6: formation of a recombinant double-stranded RNA virus. Proc Natl Acad Sci USA. 1990;87:9173–9177. doi: 10.1073/pnas.87.23.9173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olkkonen V M, Ojala P M, Bamford D H. Generation of infectious nucleocapsids by in vitro assembly of the shell protein on to the polymerase complex of the dsRNA bacteriophage φ6. J Mol Biol. 1991;218:569–581. doi: 10.1016/0022-2836(91)90702-8. [DOI] [PubMed] [Google Scholar]
- 25.Olkkonen V M, Pekkala P M, Bamford D H. Monoclonal antibodies to the major structural proteins of bacteriophage φ6. Virology. 1988;165:317–320. doi: 10.1016/0042-6822(88)90693-9. [DOI] [PubMed] [Google Scholar]
- 26.Onodera S, Qiao X, Qiao J, Mindich L. Acquisition of a fourth genomic segment in bacteriophage φ6, a bacteriophage with a genome of three segments of dsRNA. Virology. 1995;212:204–212. doi: 10.1006/viro.1995.1469. [DOI] [PubMed] [Google Scholar]
- 27.Paatero A O, Syväoja J E, Bamford D H. Double-stranded bacteriophage φ6 protein P4 is an unspecific nucleoside triphosphatase activated by calcium ions. J Virol. 1995;69:6729–6734. doi: 10.1128/jvi.69.11.6729-6734.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Revel H R, Ewen M E, Brusslan J, Pagratis N. Generation of cDNA clones of the bacteriophage φ6 segmented dsRNA genome: characterization and expression of L segment clones. Virology. 1986;155:402–417. doi: 10.1016/0042-6822(86)90203-5. [DOI] [PubMed] [Google Scholar]
- 29.Roy P. Orbivirus structure and assembly. Virology. 1996;216:1–11. doi: 10.1006/viro.1996.0028. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 31.Studier F W, Moffat B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 32.Van Etten J, Lane L, Gonzalez C, Partridge J, Vidaver A. Comparative properties of bacteriophage φ6 and φ6 nucleocapsid. J Virol. 1976;18:652–658. doi: 10.1128/jvi.18.2.652-658.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vidaver A K, Koski R K, Van Etten J L. Bacteriophage φ6: a lipid-containing virus of Pseudomonas phaseolicola. J Virol. 1973;11:799–805. doi: 10.1128/jvi.11.5.799-805.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the α and β subunits of ATP synthetase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]