Abstract
Objective
Metabolomics has been extensively utilized in bladder cancer (BCa) research, employing mass spectrometry and nuclear magnetic resonance spectroscopy to compare various variables (tissues, serum, blood, and urine). This study aimed to identify potential biomarkers for early BCa diagnosis.
Methods
A search strategy was designed to identify clinical trials, descriptive and analytical observational studies from databases such as Medline, Embase, Cochrane Central Register of Controlled Trials, and Latin American and Caribbean Literature in Health Sciences. Inclusion criteria comprised studies involving BCa tissue, serum, blood, or urine profiling using widely adopted metabolomics techniques like mass spectrometry and nuclear magnetic resonance. Primary outcomes included description of metabolites and metabolomics profiling in BCa patients and the association of metabolites and metabolomics profiling with BCa diagnosis compared to control patients. The risk of bias was assessed using the Quality Assessment of Studies of Diagnostic Accuracy.
Results
The search strategy yielded 2832 studies, of which 30 case-control studies were included. Urine was predominantly used as the primary sample for metabolite identification. Risk of bias was often unclear inpatient selection, blinding of the index test, and reference standard assessment, but no applicability concerns were observed. Metabolites and metabolomics profiles associated with BCa diagnosis were identified in glucose, amino acids, nucleotides, lipids, and aldehydes metabolism.
Conclusion
The identified metabolites in urine included citric acid, valine, tryptophan, taurine, aspartic acid, uridine, ribose, phosphocholine, and carnitine. Tissue samples exhibited elevated levels of lactic acid, amino acids, and lipids. Consistent findings across tissue, urine, and serum samples revealed downregulation of citric acid and upregulation of lactic acid, valine, tryptophan, taurine, glutamine, aspartic acid, uridine, ribose, and phosphocholine.
Keywords: Metabolite, Metabolomics, Bladder cancer, Metabolomics profile
1. Introduction
Bladder cancer (BCa) is the sixth most common cancer in the United States, being the third most common cancer in men and the eleventh in women [1,2]. Most cancers derived from transitional epithelium are transitional cell carcinomas (urothelial carcinoma). These can be classified into low-grade and high-grade, distinguished by their propensities to invade the muscle and metastasize to other parts of the body, rather than by the risk of recurrence (as both often recur after treatment) [3].
This type of cancer is also divided into non-muscle invasive BCa (NMIBC), which includes carcinoma in situ, Ta, and T1 tumors; and muscle invasive BCa (MIBC), which refers to tumors with a stage of ≥T2, based on invasion of the muscularis propria (detrusor muscle). Most people with BCa (75%–85%) have NMIBC and frequently recur [4].
Regarding diagnostics, the macroscopic and microscopic hematuria is a clear sign to perform cystoscopy, which is an invasive diagnostic method. The preferred method for diagnosis and treatment of the BCa is transurethral bladder tumor resection, since it allows to have the surgical specimen including part of the detrusor muscle to analyze its compromise. In addition, urinary cytology is a noninvasive and low-cost test, with an especially high specificity and poor sensitivity especially in low-grade BCa [5].
Metabolomics could be useful in BCa research to identify potential non-invasive and highly sensitive biomarkers. This technique has been extensively used in oncologic research, with studies on BCa using mass spectrometry (MS) and nuclear magnetic resonance (NMR) spectroscopy to compare different variables (tissue, serum, blood, and urine) between patients with and without this type of cancer, in order to identify differences [6,7].
Metabolomics is a rapidly growing field that was defined as the quantitative study of metabolites (molecules smaller than 1500 kDa) in a biological system and alterations of their concentrations due to environmental or genetic effects. Research has focused on areas such as toxicology, biomedical sciences, nutrition, genetics, innate errors of metabolism, diabetes, cancers, diagnostic tests, and neuronal diseases. These applications are based on the theory that metabolites are the functional outputs of an organism. This technique might identify a disturbance in the system's homeostasis that could occur before the appearance of symptoms of a particular disease, as a single metabolite may be the substrate for several different enzymes involved in complex metabolic pathways [6,8,9].
One of the main advantages of applying metabolomics is the ability to detect hundreds of metabolites in parallel [10], thereby efficiently monitoring biochemical alterations. Additionally, the metabolic profiles of biological specimens can be affected by factors such as diet, age, ethnicity, lifestyle, medications, or microbiota, and these factors should be controlled to obtain disease-specific information [11].
Two of the essential techniques currently used in metabolomics are NMR and MS. NMR requires little to no preparation, is rapid and non-invasive, does not destroy tissue, and has highly reproducible results (coefficient of variation 1%–2% for technical replicates). Combining NMR with MS might increase the diagnostic yield. However, the data obtained from NMR and/or MS experiments are quite complex. They provide qualitative and quantitative information on several metabolites; however, distinguishing statistically between disease and control markers could be challenging [6,10].
NMR has several advantages and disadvantages. It is known as one of the most relevant diagnostic methods used in metabolomics for a broad range of diseases. Among the advantages are the use of liquid, gassy, solid, and tissue samples, making it a more complete tool. Meanwhile, its disadvantages can be seen in its low sensitivity range, therefore, a challenge for upcoming developments in this matter. On the other hand, MS has higher specificity and sensitivity results, which makes it a more useful technique. It also allows the detection of non-carbon and non-protonic ions, resulting in a great analytical tool for metabolic detection in any sample (Table 1) [[12], [13], [14]].
Table 1.
Advantages and limitations of NMR compared to MS in metabolomics applications.
| Metabolomics technique | Advantage | Disadvantage | Applicable substance |
|---|---|---|---|
| NMR spectroscopy |
|
|
|
| GC-MS |
|
|
|
| LC-MS |
|
|
MS, mass spectrometry; GC, gas chromatography; LC, liquid chromatography; NMR, nuclear magnetic resonance.
Methodologies used in metabolomics may be separated into two big categories, the targeted and untargeted metabolomics. The former studies defined molecules, already known, and biochemically characterized metabolites and their measurement. Meanwhile, the latter analyzes all metabolites that can be measured within a sample including those chemically unknown [[15], [16], [17]].
There are few studies using different samples and analytical platforms with similar results for some metabolites; nonetheless, it is essential to standardize these two fundamental variables to establish a way to diagnose BCa nowadays. We aimed to describe metabolites and metabolomics profiling in patients with BCa that could serve as biomarkers for its early diagnosis.
2. Methods
We performed this review according to the recommendations of the Cochrane Collaboration and following the Preferred Reporting Items for Systematic reviews and Meta-Analyses (PRISMA) statement [18]. The protocol was previously published [19].
2.1. Eligibility criteria
2.1.1. Study designs
We searched for clinical trials, descriptive and analytical observational studies. Also, we looked for molecular studies.
2.1.2. Participants
We included studies with the following characteristics: profiling studies on BCa tissue, serum, blood, and urine; and profiling studies using techniques widely used in metabolomics, such as MS and NMR.
2.1.3. Primary outcomes
-
i.
The description of metabolites and/or metabolomics profiling in patients with BCa that could serve as biomarkers for early diagnosis.
-
ii.
To examine the association between metabolites and/or metabolomics profiling in patients diagnosed with BCa and control participants.
2.1.4. Exclusion criteria
We excluded recurrence BCa, benign bladder disease, and animal-related studies.
2.2. Information sources
We obtained our information through Medline, Embase, Caribbean Literature in Health Sciences, and the Cochrane Central Register of Controlled Trials from inception to nowadays (Appendix 1). To ensure literature saturation, we scanned references from the most relevant articles identified through our search, conferences, thesis databases, Open Grey, Google scholar, and ClinicalTrials.gov, among others. We contacted authors by e-mail in case of any missing information. There were not any language or timing restrictions.
2.3. Data collection and analysis
Two researchers (Dávila-Raigoza AM and García-Perdomo HA) reviewed each reference by the title and abstract, then scanned full texts of relevant studies, applied pre-specified inclusion and exclusion criteria, and extracted the data. Disagreements over the inclusion of papers were discussed and worked out, and whenever they could not be resolved, a third reviewer (Korkes F) mediated the conflict.
Two trained reviewers (Dávila-Raigoza AM and García-Perdomo HA) using a standardized form independently extracted the following information from each article: study design, geographic location, author names, title, objectives, inclusion and exclusion criteria, number of patients included, lost to follow-up, timing, definitions of outcomes, results and association measures, and funding sources.
2.4. Risk of bias
We assessed the risk of bias with the Quality Assessment of Studies of Diagnostic Accuracy tool [20]. We used RevMan 5.4 for the graphical representation.
2.5. Data analysis and synthesis of results
We qualitatively synthesized the results. We could not perform any statistical analysis due to the high heterogeneity of data.
3. Results
3.1. Selection of studies
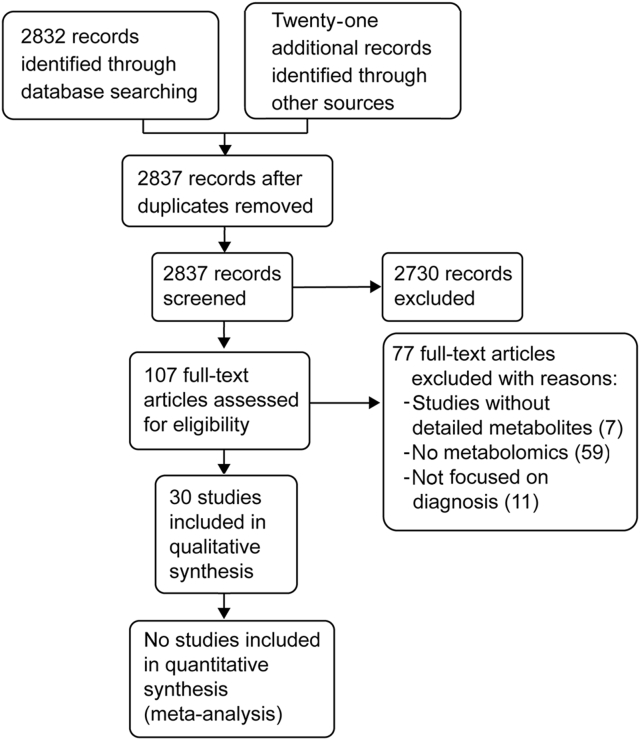
The search strategy in the databases delivered 2832 records. After reviewing the titles, abstracts, and the full texts, we selected 30 studies for qualitative synthesis (Fig. 1) [[21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42], [43], [44], [45], [46], [47], [48], [49], [50]].
Figure 1.
Flowchart of selected studies.
3.1.1. Characteristics of the included studies
All were case-control studies. They accounted for a total of 3269 patients. The biological samples used in metabolomics analyses included blood (n=1), tissue (n=2), serum (n=4), and urine (n=23) (Table 2).
Table 2.
Characteristics of the included studies.
| Study | Country | Participant, n | Gender | Age, year | Analytical platform | Sample type | Cancer Pt, n | Control, n | Metabolite up-regulated in BCa | Metabolite down-regulated in BCa |
|---|---|---|---|---|---|---|---|---|---|---|
| Srivastava et al., 2010 [21] | India | 103 |
|
|
Urine | 33 |
|
|
|
|
| Kim et al., 2010 [23] | Republic of Korea | 19 |
|
|
|
Urine | 11 |
|
|
|
| Pasikanti et al., 2010 [22] | Singapore | 75 |
|
|
Urine | 24 |
|
|
|
|
| Putluri et al., 2011 [24] | USA | 134 |
|
NR |
|
Urine | 83 |
|
|
|
| Gamagedara et al., 2012 [26] | USA | 23 |
|
NR |
|
Urine | 11 |
|
|
|
| Huang et al, 2011 [25] | China | 59 |
|
|
|
Urine | 27 |
|
|
|
| Cao et al., 2012 [27] | China | 110 |
|
|
Serum | 37 |
|
|
|
|
| Huang et al., 2013 [31] | China | 43 |
|
|
|
Urine | 19 |
|
|
|
| Jobu et al., 2012 [32] | Japan | 16 |
|
|
|
Urine | 9 |
|
|
|
| Lin et al., 2012 [28] | China | 68 |
|
|
|
Serum | 20 |
|
|
|
| Tripathi et al., 2013 [35] | USA | 59 |
|
|
|
Tissue | 33 |
|
|
|
| Bansal et al., 2013 [30] | India | 99 |
|
|
|
Serum | 67 |
|
|
|
| Pasikanti et al., 2013 [29] | Singapore | 99 |
|
|
Urine | 38 |
|
|
|
|
| Jin et al., 2014 [33] | Republic of Korea | 259 |
|
|
Urine | 138 |
|
|
|
|
| Peng et al., 2014 [34] | China and Canada | 285 |
|
|
Urine | 135 |
|
|
|
|
| Wittmann et al, 2014 [36] | USA | 440 |
|
|
|
Urine | 95 |
|
|
|
| Shen et al., 2015 [37] | China | 44 |
|
|
Urine | 23 |
|
|
|
|
| Zhou et al., 2016 [38] | China | 140 |
|
|
Plasma | 92 |
|
|
|
|
| Shao et al., 2017 [40] | China | 122 |
|
|
Urine | 87 |
|
|
|
|
| Tan et al., 2017 [39] | China | 172 |
|
|
|
Serum | 120 |
|
|
|
| Cheng et al, 2018 [42] | China | 284 |
|
|
Urine | 167 |
|
|
|
|
| Yumba Mpanga et al., 2018 [41] | Poland | 80 |
|
|
|
Urine | 40 |
|
|
|
| Wei et al., 2019 [44] | China | 30 |
|
|
|
Blood | 15 |
|
|
|
| Jacyna et al, 2019 [43] | Poland | 48 |
|
|
|
Urine | 24 |
|
|
|
| Loras et al., 2019 [45] | Spain | 34 |
|
|
|
Tissue and urine | 21 |
|
|
|
| Lin et al, 2021 [46] | China | 124 |
|
|
Urine | 63 |
|
|
|
|
| Luczykowski et al, 2021 [47] | Poland | 48 |
|
|
Urine | 24 |
|
|
|
|
| Pinto et al., 2021 [48] | Portugal | 109 |
|
|
Urine | 53 |
|
|
|
|
| Li et al, 2022 [49] | China | 95 |
|
|
Urine | 57 |
|
|
|
|
| Jacyna et al., 2022 [50] | Poland | 48 |
|
|
|
Urine | 24 |
|
|
|
Pt, patient; BCa, bladder cancer; UTI, urinary tract infection; HU, hematuria; BS, bladder stone; NMR, nuclear magnetic resonance; H-NMR, proton NMR; MS, mass spectrometry; GC, gas chromatography; TOFMS, time-of-flight MS; NR, not reported; LC, liquid chromatography; HC, healthy controls; CP, calculi patient; TURBT, transurethral resection of bladder tumor; BPH, benign prostatic hyperplasia, ; HR-MAS-NMR, high resolution-magic angle spinning-NMR; GC×GC-TOFMS, two-dimensional GC TOFMS; UHPLC-MS/MS, ultrahigh-performance liquid chromatography/tandem MS; UPLC-HRMS, ultra-performance liquid chromatography and direct infusion HR MS; SIM, selected ion monitoring; UPLCTOF, ultra-performance LC TOF; UHPLC-QTOFMS, UHPLC-quadrupole TOFMS; HPLC, high-performance liquid chromatography; RP-HPLC-QQQ/MS, reverse-phase HPLC coupled with triple quadrupole MS; HS-GC-MS, chromatography-MS coupled with a headspace generator sampler; HS-SPME-GC-MS, headspace solid-phase microextraction GC MS; LBC, low-grade bladder cancer; HBC, high-grade bladder cancer; Asp, aspartic acid; Gly, glycine; Trp, tryptophan; Cys, cysteine; Ala, alanine; Lys, lysine; Thr, threonine; Gln, glutamine; Tyr, tyrosine; Leu, leucine; Ile, isoleucine; Phe, phenylalanine; Val, valine; Pro, proline; Tau, taurine.
Values are presented as mean±standard deviation.
3.2. Risk of bias assessment
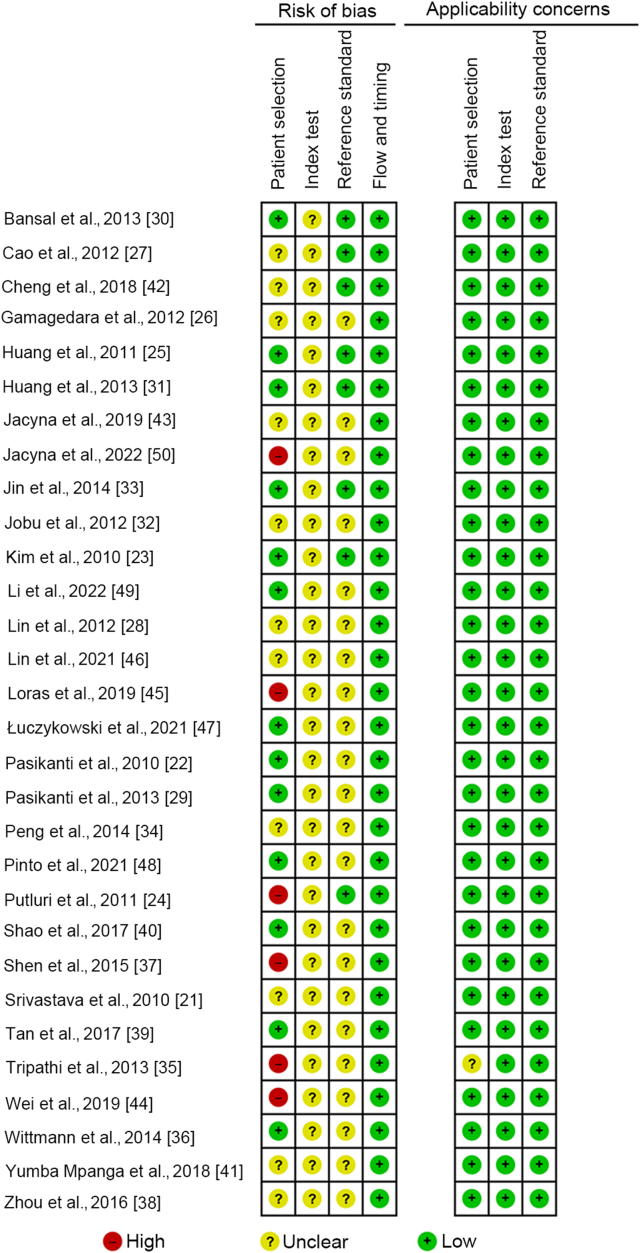
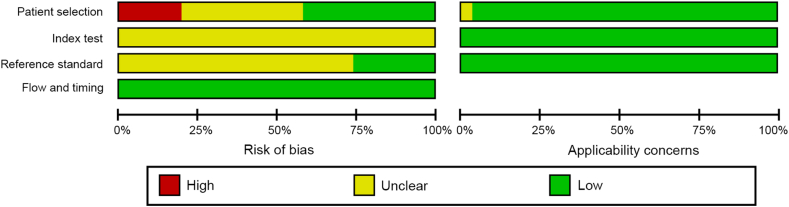
We evaluated a large part of the studies as unclear risk due to the lack of information regarding patient selection, blinding of index test assessment, and blinding of the reference standard assessment. Nevertheless, there were no applicability concerns for this systematic review (Figs. 2 and 3).
Figure 2.
Risk of bias assessment within studies.
Figure 3.
Risk of bias assessment across studies.
3.3. Glucose metabolism
Glucose metabolism implies multiple processes including glycolysis, glycogenolysis, glycogenesis, and gluconeogenesis. Glycolysis is the most crucial process. The end products of which are two molecules of pyruvic acid that turn into acetyl coenzyme A, which then decomposes and releases energy as ATP through Krebs's cycle or the tricarboxylic acid cycle [51,52]. The metabolites intermediate with significant variations in this pathway are shown in Table 3. Glucose levels were found to be upregulated in serum [27]. Conversely, fructose exhibited downregulation in two urine studies [22,36]. In terms of pyruvic acid, its regulation showed divergent patterns. One study reported an upregulation [33], whereas another study observed a downregulation [36]. These contrasting findings highlight the complexity of pyruvic acid's regulatory mechanisms. Moving on to lactose, it was consistently downregulated in plasma [38]. The regulation of lactic acid displayed inconsistent results. One study observed upregulation in serum [30], whereas another study reported downregulation [27]. Interestingly, in tissue, lactic acid was found to be upregulated according to one study [35], emphasizing the need for further investigation into the tissue-specific regulation of lactic acid. Several studies focused on urine regulation, revealing intriguing findings, and three of them reported upregulation [36,43,50]; however, another study noted changes in regulation without specifying the direction of regulation [45], underscoring the complexity of urine metabolite regulation (Table 3).
Table 3.
Glucose metabolism-related metabolites in bladder cancer.
| Study | Sample type | Analytical platform | Anaerobic oxidation (glycolysis) |
Aerobic oxidation (the TCA cycle) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Glucose | Fructose | Pyruvic acid | Lactose | Lactic acid | Citrate or citric acid | Succinate | Malic acid | Fumaric acid | Itaconate | |||
| Srivastava et al., 2010 [21] | Urine | H-NMR | ↓ | |||||||||
| Kim et al., 2010 [23] | Urine | GC-MS | ||||||||||
| Pasikanti et al., 2010 [22] | Urine | GC/TOFMS | ↓ | ↓ | ||||||||
| Cao et al., 2012 [27] | Serum | H-NMR | ↑ | ↓ | ↓ | |||||||
| Bansal et al., 2013 [30] | Serum | H-NMR | ↑ | |||||||||
| Pasikanti et al., 2013 [29] | Urine | GC×GC-TOFMS | ↓ | |||||||||
| Tripathi et al., 2013 [35] | Tissue | HR-MAS-NMR GC-MS | ↑ | |||||||||
| Jin et al., 2014 [33] | Urine | HPLC-QTOFMS | ↑ | ↑ | ||||||||
| Wittmann et al., 2014 [36] | Urine | UHPLC-MS/MS GC-MS | ↓ | ↓ | ↑ | ↓ | ↓ | ↓ | ||||
| Zhou et al., 2016 [38] | Plasma | GC-MS-SIM | ↓ | ↑ | ↑ | |||||||
| Jacyna et al., 2019 [43] | Urine | MS and H-NMR | ↑ | |||||||||
| Loras et al., 2019 [45] | Urine | NMR | ↓↑ | ↓↑ | ↓↑ | |||||||
| Jacyna et al., 2022 [50] | Urine | MS and H-NMR | ↑ | |||||||||
TCA, tricarboxylic acid cycle; NMR, nuclear magnetic resonance; H-NMR, proton NMR; GC, gas chromatography; MS, mass spectrometry; TOFMS, time-of-flight MS; GC×GC-TOFMS, two-dimensional GC-TOFMS; HR-MAS-NMR, high resolution-magic angle spinning NMR; HPLC-QTOFMS, high-performance liquid chromatography-quadrupole TOFMS; UHPLC-MS/MS, ultrahigh-performance liquid chromatography/tandem MS; SIM, selected ion monitoring. ↓, downregulated; ↑, upregulated; ↓↑, undetermined trend.
Citric acid emerged as a consistently regulated metabolite, particularly in urine. Four studies found downregulation [21,22,29,36] and another study reported changes in its regulation without specifying the trend [45]. It was also downregulated in serum [27]. Succinate exhibited inconsistent regulation. One study found upregulation in urine [33], while another study reported downregulation [36]. Another study described changes in succinate regulation without specifying the trend [45]. Regarding plasma metabolites, one study identified upregulation of malic acid and fumarate [38], offering insights into potential alterations in energy metabolism or cellular processes involving these metabolites.
3.4. Amino acid metabolites
Amino acids, the building blocks of protein, based on their physiological and nutritional roles, can be differentiated as essential and non-essential [53]. Table 4 shows the significant variations of amino acid metabolites in BCa samples. In terms of essential amino acids, leucine exhibited upregulation in urine [23,24,36] as well as in tissue [35], while it was downregulated in serum [27]. On the other hand, the regulation of lysine showed inconsistency. It was downregulated in urine in two studies [23,37] and upregulated in one [24]. As for tissue regulation, one study reported upregulation [35], while another [45] described changes without specifying the trend in both urine and tissue.
Table 4.
Metabolites of amino acid metabolism in bladder cancer.
| Study | Sample type | Analytical platform | Essential amino acid |
Non-essential amino acid |
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Leu | Met | Lys | Val | Ile | Phe | Trp | Thr | Ser | Cys | Asp | Pro | Arg | Tyr | Asn | Glu | Gly | Ala | His | Tau | Gln | |||
| Srivastava et al., 2010 [21] | Urine | H-NMR | ↓ | ↑ | |||||||||||||||||||
| Kim et al., 2010 [23] | Urine | GC-MS | ↑ | ↓ | ↑ | ↑ | ↑ | ↓ | ↑ | ↑ | ↑ | ↓ | ↑ | ↓ | |||||||||
| Pasikanti et al., 2010 [22] | Urine | GC/TOFMS | ↑ | ||||||||||||||||||||
| Gamagedara et al., 2012 [26] | Urine | LC-MS | ↑ | ||||||||||||||||||||
| Putluri et al., 2011 [24] | Urine | LC-MS | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ||||||||||||||
| Huang et al., 2011 [25] | Urine | LC-MS | ↑ | ||||||||||||||||||||
| Cao et al., 2012 [27] | Serum | H-NMR | ↓ | ↓ | ↓ | ↓ | ↓ | ||||||||||||||||
| Bansal et al., 2013 [30] | Serum | H-NMR | ↑ | ↑ | ↑ | ||||||||||||||||||
| Tripathi et al., 2013 [35] | Tissue | HR-MAS-NMR GC-MS | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | |||||||||
| Wittmann et al., 2014 [36] | Urine | UHPLC-MS/MS GC-MS | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | |||||||||||||||
| Shen et al., 2015 [37] | Urine | UPLC-HRMS | ↓ | ↑ | ↓ | ↑ | ↑ | ↓ | |||||||||||||||
| Yumba Mpanga et al., 2018 [41] | Urine | RP-HPLC-QQQ/MS | ↑ | ↑ | |||||||||||||||||||
| Jacyna et al., 2019 [43] | Urine | MS and H-NMR | ↑ | ↑ | |||||||||||||||||||
| Loras et al., 2019 [45] | Tissue and urine | H-NMR | ↑↓ | ↑↓ | ↑↓ | ↑↓ | ↑↓ | ↑↓ | ↑↓ | ↑↓ | |||||||||||||
| Łuczykowski et al., 2021 [47] | Urine | LC-MS | ↓ | ↓ | |||||||||||||||||||
| Li et al., 2022 [49] | Urine | LC-MS | ↓ | ↓ | ↑ | ||||||||||||||||||
| Jacyna et al., 2022 [50] | Urine | MS and H-NMR | ↑↓ | ↑↓ | |||||||||||||||||||
Leu, leucine; Met, methionine; Lys, lysine; Ile, isoleucine; Val, valine; Phe, phenylalanine; Trp, tryptophan; Thr, threonine; Ser, serine; Cys, cysteine; Asp, aspartic acid; Pro, proline; Arg, arginine; Tyr, tyrosine; Asn, asparagine; Glu, glutamate; Gly, glycine; Ala, alanine; His, histidine; Tau, taurine; Gln, glutamine; MS, mass spectrometry; NMR, nuclear magnetic resonance; H-NMR, proton NMR; GC, gas chromatography; TOFMS, time-of-flight MS; LC, liquid chromatography; HR-MAS-NMR, high resolution magic angle spinning NMR; UHPLC-MS/MS, ultrahigh-performance liquid chromatography/tandem MS; UPLC-HRMS, ultra-performance liquid chromatography and direct infusion high-resolution MS; RP-HPLC-QQQ/MS, reverse-phase high-performance liquid chromatography coupled with triple quadrupole MS. ↓, downregulated; ↑, upregulated; ↓↑, undetermined trend.
Valine, one of the metabolites, exhibited more consistent regulation. Four urine studies found upregulation [[22], [23], [24],36], along with tissue [35] and serum [30]; however, one study indicated changes in regulation without specifying the trend in both urine and serum [45]. Isoleucine showed upregulation in urine in two studies [24,36], as well as in tissue [35], while it was downregulated in serum [27]. Phenylalanine exhibited downregulation in urine in two studies [21,47], as well as in serum [27], while it was upregulated in tissue [35]. Tryptophan consistently showed upregulation in urine [23,36,37,41]. Lastly, threonine was found to be upregulated in urine in one study [23], while another study indicated an indeterminate regulation [45].
When it comes to non-essential amino acids, serine exhibited inconsistent regulation in urine. One study reported downregulation [23], while another study indicated upregulation [24]. Cysteine showed downregulation in urine [37]. Aspartic acid demonstrated upregulation in urine [23,37] and tissue [35]. Proline showed changes in regulation without a specific trend in both urine and tissue [45]. Arginine was downregulated in urine [49].
Regarding tyrosine, consistent upregulation was observed in urine in three studies [23,24,43] and one study reported changes in regulation without specifying the trend [50]. In tissue, it was upregulated [35], while in serum, it was downregulated [27]. Arginine was downregulated in urine [49], while asparagine exhibited upregulation [23].
Glutamate showed upregulation in tissue [35]. However, another study described changes in regulation without specifying the trend in both urine and tissue [45]. Glycine yielded inconsistent results, with upregulation in urine in one study [37] and downregulation in another [23]. In serum, it was downregulated [27], while in tissue, it was upregulated [35].
Alanine displayed inconsistent regulation in urine, with upregulation in one study [23] and downregulation in another [37]. In tissue, it showed upregulation [35]. However, both urine and tissue studies described changes in regulation without a specific trend [45]. Similarly, histidine showed inconsistent results. In urine, two studies reported upregulation [24,36], while two studies reported downregulation [47,49]. It was upregulated in serum [30].
Taurine exhibited consistent regulation, with upregulation in urine in five studies [21,25,26,41,49], and one study reported upregulation in tissue [35]. However, one study indicated changes in regulation without specifying the trend [45]. Glutamine showed upregulation in urine in two studies [36,43], while one study reported downregulation [23]. It was upregulated in serum [30] and tissue [35], while two studies described changes in regulation without specifying the trend [45,50] (Table 4).
3.5. Nucleotide metabolites
Nucleotides are the building blocks of nucleic acids. They have main roles in many cellular processes. These building blocks are composed by a five-carbon sugar (ribose or deoxyribose), a nitrogen base, and one or more phosphate groups. Nitrogenous bases are classified into purines and pyrimidines. The purines include adenine and guanine, while the pyrimidines are cytosine, thymine, and uracil [54]. Table 5 shows the significant variations of nucleotide metabolites in BCa samples.
Table 5.
Nucleotide metabolites in bladder cancer.
| Study | Sample type | Analytical platform | Purine |
Pyrimidine |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Adenine | Guanine | Cytosine | Thymine | Uridine | Uracil | Inosine | Hypoxanthine | Ribose | |||
| Pasikanti et al., 2010 [22] | Urine | GC/TOFMS | ↑ | ||||||||
| Putluri et al., 2011 [24] | Urine | LC-MS | ↑ | ↑ | ↑ | ||||||
| Tripathi et al., 2013 [35] | Tissue | H-NMR, GC-MS | ↑ | ||||||||
| Pasikanti et al., 2013 [29] | Urine | GC×GC-TOFMS | ↑ | ↑ | |||||||
| Peng et al., 2014 [34] | Urine | LC-MS | ↓ | ||||||||
| Wittmann et al., 2014 [36] | Urine | UHPLC-MS/MS GC-MS | ↓ | ↑ | |||||||
| Zhou et al., 2016 [38] | Plasma | GC-MS-SIM | ↑ | ↓ | ↑ | ||||||
| Tan et al., 2017 [39] | Serum | UHPLC-Q-TOFMS | ↑ | ↑ | |||||||
| Yumba Mpanga et al., 2018 [41] | Urine | RP-HPLC-QQQ/MS | ↑ | ||||||||
| Jacyna et al., 2019 [43] | Urine | MS, H-NMR | ↑ | ||||||||
| Lin et al., 2021 [46] | Urine | GC-MS | ↑ | ||||||||
| Li et al., 2022 [49] | Urine | LC-MS | ↓ | ||||||||
| Jacyna et al., 2022 [50] | Urine | MS, H-NMR | ↑↓ | ||||||||
GC, gas chromatography; MS, mass spectrometry; TOFMS, time-of-flight MS; LC, liquid chromatography; H-NMR, proton nuclear magnetic resonance; UHPLC-MS/MS, ultrahigh-performance liquid chromatography/tandem MS; SIM, selected ion monitoring; UHPLC-QTOFMS, UHPLC-quadrupole TOFMS; RP-HPLC-QQQ/MS, reverse-phase high-performance liquid chromatography coupled with triple quadrupole MS. ↓, downregulated; ↑, upregulated; ↓↑, undetermined trend.
Regarding purines, adenine was downregulated in urine [36,49], while guanine was upregulated [24,36]. In terms of pyrimidines, uridine exhibited the most consistent regulation. In urine, four studies reported upregulation [22,29,41,43]. However, one study observed downregulation [34], and another study reported a change in regulation without specifying the trend [50]. Additionally, uridine was upregulated in tissue [35] and plasma [38]. Continuing with pyrimidines, thymine and uracil were upregulated in urine [24]. Furthermore, inosine showed upregulation in serum [39], while hypoxanthine exhibited upregulation in serum [39] and downregulation in plasma [38]. Ribose also demonstrated consistent regulation, being upregulated in urine [29,46] and in plasma [38].
3.6. Lipid metabolites
The BCa patients showed significant variations in the concentration of lipid metabolites in urine, serum, and tissue, as depicted in Supplementary Table 1. In urine, choline [36,49], phosphocholine [36,47], and carnitine [24,33,36] exhibited upregulation, while ethanolamine [36] and glycerol [22,29] showed downregulation. In tissue, choline, phosphocholine, glycerophosphocholine, inositol, and triglycerides were upregulated [35]. Conversely, in serum, choline was downregulated, while acetoacetate was upregulated [27]. Furthermore, a study was conducted in urine and tissue describing changes in the regulation of choline, phosphocholine, and glycerophosphocholine, without specifying their trends [45].
3.7. Aldehydes metabolites
Aldehydes, the carbonyl compounds derived from both natural and artificial origin, can be found everywhere [55]. Endogen aldehydes are part of lipidic, amino acids, vitamins, and carbohydrates metabolism. Regarding cancer, human tumoral cells produce large amounts of free radicals that increase the aldehyde concentration. Consequently, these molecules can be considered as possible cancer biomarkers [56]. Supplementary Table 2 presents the findings concerning aldehyde-derived metabolites. Formaldehyde showed an upregulation in blood [44]; however, in urine, it exhibited downregulation [48]. As for ethanal, propanal, butanal, pentanal, heptanal, and hexanal, they displayed upregulation in blood [44]. However, hexanal also showed downregulation in urine [48]. On the other hand, nonenal and dodecanal demonstrated upregulation in urine [32].
4. Discussion
4.1. Summary of findings
As a result, we found endogen aldehydes in blood samples. In urine were citric acid, valine, tryptophan, taurine, aspartic acid, uridine, ribose, phosphocholine, and carnitine. In addition, there was an increase in lactic acid, amino acid, and lipidic metabolites in tissue samples. Besides, the most consistent results in all samples (tissue, urine, serum, and blood) were downregulation of citric acid and upregulation of lactic acid, valine, tryptophan, taurine, glutamine, aspartic acid, uridine, ribose, and phosphocholine.
4.2. Clinical applications of metabolomics
Nowadays the clinical application of metabolomics is limited; however, the number of potential applications has been increasing and might change the clinical practice in the future [57]. In oncology, the metabolomics profiling has been associated with the diagnosis of several kinds of tumors, more noticeable in breast cancer, where there have been observed over 30 endogenous metabolites that might be used as biomarkers [57,58].
In addition to its potential in diagnosis, metabolomics has also been used to guide oncologic surgeries. Inglese et al. [59] managed to couple rapid ionization MS with electrosurgical tools to characterize the margins while performing the tumor dissection. This approach enables a real-time metabolomics and surgical process with promising live results, although at high-cost, resulting in an important limitation of its use. Besides the already mentioned applications in oncology [57], Tenori et al. [60] found the relationship between the metastatic breast cancer survival rate and phenylalanine serum levels prior treatment.
In neurology, it allows to obtain the metabolomics profiling to identify patients who have suffered an ischemic event; due to studies carried out on its most common etiologies and the correlation with specific metabolites, metabolomics has the potential to achieve a significant role in the study of cryptogenic ictus [61]. In endocrinology, it has been used to quantify type 2 diabetes mellitus risk. Wang et al. [62], suggested that the 2-aminoadipic acid metabolite is a diabetes risk marker as they found that its concentration in the highest quartile elevated four times diabetes risk, and this could be identified 12 years before the disease becomes clinically evident.
In conclusion, metabolomics is becoming a widely used tool in multiple areas of medical practices and specializations as mentioned before. Its main utility is seeing in novel research with the characterization of metabolic pathways in order to find, as shown in this review, specific biomarkers identified different types of diseases in early stages.
4.3. Contrast with literature
The BCa prognosis might improve if an early diagnosis of the disease is made by the identification of biomarkers and especially if it is possible to be done in a non-invasive way, therefore reducing the morbidity of the tests. Urine is the ideal sample to perform the tests due to the proximity of the bladder to the urine itself; however, blood, tissue, and serum are also useful. It is known that disturbances of several metabolic routes are involved in BCa, which represents an essential tool for its diagnosis [63].
Cheng et al. [64] pooled 11 studies which described metabolites to detect BCa in a systematic review, with different techniques, methodological limitations, and high heterogeneity. They found significant changes in metabolite concentrations of glucose, amino acids, nucleotides, and lipidic pathways [64]. In our study, we identified metabolites involved in a novel metabolic pathway, known as the endogenous aldehyde pathway [32,44,48].
Regarding the altered expression of glucose metabolites, their results were inconclusive about glucose, fructose, or lactic acid, since they were found up in some studies and down in others [64]. Nevertheless, citric acid was found down in different studies. Among BCa patients, the low concentration of citric acid and high concentration of lactic acid in urine were the most representative metabolites which were altered in at least three studies from the same kind of samples in our review, making them potential biomarkers for the diagnose of this kind of cancer.
One of the main characteristics of cancerous cells is the energy metabolism disturbance. Normal cells produce ATP using two mechanisms: glycolysis and the tricarboxylic acid cycle (the Krebs cycle). The representative disturbance of the energy metabolism in cancerous cells is the increase of glucose absorption and its fermentation into lactate. This metabolic change towards anaerobic glycolysis, even in an environment with a normal oxygen concentration, is what can be called an “aerobic glycolysis”, also known as Warburg effect [65]. This process results in the biosynthesis of necessary precursors to trigger growth and cellular division, such as ribose, acetyl-coenzyme A, and glycolytic intermediates to produce nucleotides, fat acids, and non-essential amino acids, respectively. Furthermore, the lactic acid increase reduces the pH of the microenvironment leading to the death of non-cancerous cells because they do not possess the mechanisms to adapt the extracellular acidity (such as p53 mutation), choosing those resistant cells and allowing the tumoral progression. As glycolysis heightens, it produces not only an increase in lactic acid but also a corresponding decrease in the metabolites related to the tricarboxylic acid cycle, therefore reducing the synthesis of citric acid, which is compatible with the findings from a systematic review [66].
Concerning the altered expression of amino acid metabolites, essential amino acids (threonine, phenylalanine, valine, isoleucine, lysine, methionine, and leucine) and non-essential amino acids (glutamate, histidine, arginine, aspartic acid, tyrosine, glutamine, and serine) were upregulated. In our review, the amino acids found upregulated in at least three studies were leucine, taurine, valine, isoleucine, tryptophan, aspartic acid, tyrosine, and glutamine, making them potential biomarkers for the diagnose of BCa. Histidine was upregulated in three studies [24,30,36]; nonetheless, it was downregulated in other two, therefore considered an inconsistent result [47,49].
The findings are consistent with the fact that the glutaminolysis rate increases within cancerous cells as an energy source for its proliferation [67]. In addition, tryptophan metabolism alterations have been broadly associated with BCa. Serotonin, indole, and kynurenine metabolic pathways use tryptophan. If there is an excessive absorption of tryptophan plus a vitamin B6 deficiency, it might result in an accumulation of the mentioned kynurenine pathway metabolites within the bladder which can combine with nitrite becoming an oncogenic promoter as mutagenic nitrosamines and, in this location, can produce BCa. They can also form reactive oxygen species when interacting with transition metals such as copper and iron, and when tryptophan is exposed to ultraviolet light or visible light, it can also form more carcinogenic products [68]. Lee et al. [69] established that kynurenine to tryptophan proportions in both plasma and urine were significantly higher in patients with BCa. Cheng et al. [64] found that metabolites involved in tryptophan metabolism were upregulated in the high-grade NMIBC patients when compared with the metabolites for the low-grade NMIBC ones. In addition, expression of tryptophan 2,3-dioxygenase has been reported as a potential target in immunotherapy of BCa [70].
About nucleotides, the previously mentioned authors found elevated levels of guanine, cytosine, thymine, hypoxanthine, uracil, and ribose in the urine of BCa patients. In our review, the most representative metabolites were uridine and ribose which were upregulated in at least three studies, while in two separate studies, adenine was found to be downregulated [36,49] and guanine was upregulated [24,36]. Both studies utilized urine samples. The levels of hypoxanthine were inconsistent [38,39].
Damaged metabolic pathways develop within the tumor as they grow. Recent discoveries showed that tumor growth and immune system inhibition will occur whenever there is an aberrant nucleotide metabolism. There are a few studies about this matter but they are on a fast-experimenting development. The more discoveries about these pathways and mechanisms, the more strategies can be generated to intervene, regulate, and alter them to prevent or treat the tumors and their development. According to studies from Tan et al. [39], hypoxanthine and inosine serum levels increased in a significant way alongside cancer progression. For instance, in patients with low degree BCa, the increment was higher than that in healthy patients. Additionally, significantly higher levels of hypoxanthine and inosine were encountered in the serum of patients with advanced-stage BCa compared to those with a lower stage. These results suggest an upregulation of purines metabolism [39].
We observed in our study the rise in most of the intermediate lipidic metabolites, which was consistent with findings by Cheng et al. [64]. In our study, the metabolites found to be upregulated in at least three studies were choline, phosphocholine, and carnitine. It has been observed in multiple studies that lipidic metabolism has a vital role in the carcinogenesis and metastasis of cancer. Growth and tumor proliferation demand a critical amount of energy which translates into an increase in lipidic synthesis to ensure the survival of cancerous cells [71]. The importance of the lipidic metabolism is such that in the carcinogenesis simvastatin observed by Wang et al. [72], one of the 3-hydroxy-3-methyl glutaryl coenzyme A reductase inhibitors could reduce BCa cell proliferation and inhibit metastasis producing a stop in the cell cycle.
Endogen aldehydes are intermediate or final metabolic products of a broad spectrum of biochemical and physiological processes, such as oxidative stress and cellular activities. These compounds are generated from free radical induced reactions with cellular lipids. Biochemical processes that contribute to their formation are glycation, aminoacidic, alcohol, and sugar metabolism, with lipidic peroxidation being the main endogen source for them [73]. Due to the increased oxidative stress produced by human tumor cells, the upregulation of endogen aldehydes has been considered as a tumor biomarker.
In addition, Wei et al. [44] identified that average aldehyde concentrations in cancer patients' blood were several times higher than those in healthy controls. This resulted in a remarkable significant difference according to what is described for methanal, ethanal, propanal, pentanal, hexanal, and heptanal (p<0.001) and a significant difference for hexanal (p<0.01). Jobu et al. [32] found five substances in urine with the potential to become biomarkers for BCa; two of them belong to endogen aldehyde metabolism (nonenal and dodecanal).
4.4. Strengths and limitations
Regarding the strengths, we followed the standard recommendation of Cochrane and Preferred Reporting Items for Systematic reviews and Meta-Analyses. We found this study as a very focused, comprehensive systematic review that serves as a starting point to diagnose BCa non-invasively. In our review, the inclusion of endogenous aldehydes as components of novel metabolic pathways implicated in cancer development proved to be of utmost significance. These findings, in particular, garnered significant attention and are regarded as potentially valuable tumor biomarkers.
On the other side, as limitations, the results were heterogeneous due to diverse samples and techniques. The discrepancy on patients' origins within the studies, as well as their economic status and diet may be responsible for resulting in different metabolic alterations. Urine samples, as the most used ones among the studies, vary drastically depending on lifestyle and diet [74,75]. Besides, there is also another factor making this tool even more complicated and is the high variation of a cancer cell metabolism which is subject to different signals produced by the tumor itself. In addition, some researches were carried out listing NMIBC and MIBC in the same category when examining different samples even though their metabolic profilings are most likely different from each other, all these resulting in a quite heterogenic metabolic profiling. Besides, there was not enough information to correctly classify the risk of bias regarding the patient selection, the blinding of the index test, or the reference standard assessment.
Based on the current literature, it is not possible to determine whether the metabolites detected are predisposing factors or a consequence of BCa. Further epidemiologic studies investigating this issue would be of great value.
5. Conclusion
BCa has disturbances in multiple metabolic pathways, mainly those involved in energy synthesis. In our review, the most consistent metabolites found in urine were citric acid, valine, tryptophan, taurine, aspartic acid, uridine, ribose, phosphocholine, and carnitine. In blood samples, endogen aldehydes are highlighted as potential biomarkers. In tissue, there was an increase in lactic acid, amino acid, and lipidic metabolites. The most consistent results in all samples (tissue, urine, and serum) were downregulation of citric acid and upregulation of lactic acid, valine, tryptophan, taurine, glutamine, aspartic acid, uridine, ribose, and phosphocholine, which led to consider them as BCa biomarkers. Nevertheless, the clinical applicability of metabolomics in the diagnostic of BCa requires further studies.
Author contributions
Study concept and design: Herney Andrés García-Perdomo, Angélica María Dávila-Raigoza.
Data acquisition: Herney Andrés García-Perdomo, Angélica María Dávila-Raigoza, Fernando Korkes.
Data analysis: Herney Andrés García-Perdomo, Angélica María Dávila-Raigoza, Fernando Korkes.
Drafting of manuscript: Herney Andrés García-Perdomo, Angélica María Dávila-Raigoza, Fernando Korkes.
Critical revision of the manuscript: Herney Andrés García-Perdomo, Angélica María Dávila-Raigoza, Fernando Korkes.
Conflicts of interest
The authors declare no conflict of interest.
Footnotes
Peer review under responsibility of Tongji University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ajur.2022.11.005.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 3.Burger M., Catto J.W.F., Dalbagni G., Grossman H.B., Herr H., Karakiewicz P., et al. Epidemiology and risk factors of urothelial bladder cancer. Eur Urol. 2013;63:234–241. doi: 10.1016/j.eururo.2012.07.033. [DOI] [PubMed] [Google Scholar]
- 4.Babjuk M., Burger M., Capoun O., Cohen D., Compérat E.M., Dominguez Escrig J.L., et al. European Association of Urology guidelines on non-muscle-invasive bladder cancer (Ta, T1 and carcinoma in situ) Eur Urol 2020. 2022;81:75–94. doi: 10.1016/j.eururo.2021.08.010. [DOI] [PubMed] [Google Scholar]
- 5.Comploj E., Trenti E., Palermo S., Pycha A., Mian C. Urinary cytology in bladder cancer: why is it still relevant? Urologia. 2015;82:203–205. doi: 10.5301/uro.5000129. [DOI] [PubMed] [Google Scholar]
- 6.Garcia-Perdomo H., Vallejo F., Sanchez A. Metabolic profiling based on nuclear magnetic resonance spectroscopy and mass spectrometry as a tool for clinical application. Urol Sci. 2019;30:144–150. [Google Scholar]
- 7.Issaq H.J., Nativ O., Waybright T., Luke B., Veenstra T.D., Issaq E.J., et al. Detection of bladder cancer in human urine by metabolomic profiling using high performance liquid chromatography/mass spectrometry. J Urol. 2008;179:2422–2426. doi: 10.1016/j.juro.2008.01.084. [DOI] [PubMed] [Google Scholar]
- 8.Roux A., Lison D., Junot C., Heilier J.F. Applications of liquid chromatography coupled to mass spectrometry-based metabolomics in clinical chemistry and toxicology: a review. Clin Biochem. 2011;44:119–135. doi: 10.1016/j.clinbiochem.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 9.Smolinska A., Blanchet L., Buydens L.M.C., Wijmenga S.S. NMR and pattern recognition methods in metabolomics: from data acquisition to biomarker discovery: a review. Anal Chim Acta. 2012;750:82–97. doi: 10.1016/j.aca.2012.05.049. [DOI] [PubMed] [Google Scholar]
- 10.Gowda G.A.N., Zhang S., Gu H., Asiago V., Shanaiah N., Raftery D. Metabolomics-based methods for early disease diagnostics. Expert Rev Mol Diagn. 2008;8:617–633. doi: 10.1586/14737159.8.5.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nicholson J., Lindon J., Holmes E. “Metabonomics”: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29:1181–1189. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- 12.Bujak R., Struck-Lewicka W., Markuszewski M.J., Kaliszan R. Metabolomics for laboratory diagnostics. J Pharm Biomed Anal. 2015;113:108–120. doi: 10.1016/j.jpba.2014.12.017. [DOI] [PubMed] [Google Scholar]
- 13.Sethi S., Hayashi M.A., Barbosa B.S., Pontes J.G., Tasic L., Brietzke E. In: Sussulini A., editor. vol. 965. Springer; New York: 2017. Metabolomics: from fundamentals to clinical applications. (Advances in Experimental Medicine and Biology). [DOI] [Google Scholar]
- 14.Emwas A.H. The strengths and weaknesses of NMR spectroscopy and mass spectrometry with particular focus on metabolomics research. Methods Mol Biol. 2015;1277:161–193. doi: 10.1007/978-1-4939-2377-9_13. [DOI] [PubMed] [Google Scholar]
- 15.Ribbenstedt A., Ziarrusta H., Benskin J.P. Development, characterization and comparisons of targeted and non-targeted metabolomics methods. PLoS One. 2018;13:1–18. doi: 10.1371/journal.pone.0207082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roberts L.D., Souza A.L., Gerszten R.E., Clish C.B. Targeted metabolomics. Curr Protoc Mol Biol. 2012;Chapter 30 doi: 10.1002/0471142727.mb3002s98. Unit 30.2.1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fiehn O. Metabolomics by gas chromatography-mass spectrometry: the combination of targeted and untargeted profiling. Curr Protoc Mol Biol. 2016;114:30–32. doi: 10.1002/0471142727.mb3004s114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liberati A., Altman D., Tetzlaff J. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. Ann Intern Med. 2009;151:W65–W94. doi: 10.7326/0003-4819-151-4-200908180-00136. [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Perdomo H.A., Dávila-Raigoza A., Korkes F. 2020. Metabolomics for the diagnosis of bladder cancer. A protocol for a systematic review. p.1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whiting P.F., Rutjes A.W.S., Westwood M.E., Mallet S., Deeks J.J., Reitsma J.B., et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 21.Srivastava S., Roy R., Singh S., Kumar P., Dalela D., Sankhwar S.N., et al. Taurine—a possible fingerprint biomarker in non-muscle invasive bladder cancer: a pilot study by 1H NMR spectroscopy. Cancer Biomark. 2010;6:11–20. doi: 10.3233/CBM-2009-0115. [DOI] [PubMed] [Google Scholar]
- 22.Pasikanti K.K., Esuvaranathan K., Ho P.C., Mahendran R., Kamaraj R., Wu Q.H., et al. Noninvasive urinary metabonomic diagnosis of human bladder cancer. J Proteome Res. 2010;9:2988–2995. doi: 10.1021/pr901173v. [DOI] [PubMed] [Google Scholar]
- 23.Kim J.W., Lee G., Moon S.M., Park M.J., Hong S.K., Ahn Y.H., et al. Metabolomic screening and star pattern recognition by urinary amino acid profile analysis from bladder cancer patients. Metabolomics. 2010;6:202–206. [Google Scholar]
- 24.Putluri N., Shojaie A., Vasu V.T., Vareed S.K., Nalluri S., Putluri V., et al. Metabolomic profiling reveals potential markers and bioprocesses altered in bladder cancer progression. Cancer Res. 2011;71:7376–7386. doi: 10.1158/0008-5472.CAN-11-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Z., Lin L., Gao Y., Chen Y., Yan X., Xing J., et al. Bladder cancer determination via two urinary metabolites: a biomarker pattern approach. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M111.007922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gamagedara S., Shi H., Ma Y. Quantitative determination of taurine and related biomarkers in urine by liquid chromatography-tandem mass spectrometry. Anal Bioanal Chem. 2012;402:763–770. doi: 10.1007/s00216-011-5491-4. [DOI] [PubMed] [Google Scholar]
- 27.Cao M., Zhao L., Chen H., Xue W., Lin D. NMR-based metabolomic analysis of human bladder cancer. Anal Sci. 2012;28:451–456. doi: 10.2116/analsci.28.451. [DOI] [PubMed] [Google Scholar]
- 28.Lin L., Huang Z., Gao Y., Chen Y., Hang W., Xing J., et al. LC-MS-based serum metabolic profiling for genitourinary cancer classification and cancer type-specific biomarker discovery. Proteomics. 2012;12:2238–2246. doi: 10.1002/pmic.201200016. [DOI] [PubMed] [Google Scholar]
- 29.Pasikanti K.K., Esuvaranathan K., Hong Y., Ho P.C., Mahendran R., Raman Nee Mani L., et al. Urinary metabotyping of bladder cancer using two-dimensional gas chromatography time-of-flight mass spectrometry. J Proteome Res. 2013;12:3865–3873. doi: 10.1021/pr4000448. [DOI] [PubMed] [Google Scholar]
- 30.Bansal N., Gupta A., Mitash N., Shakya P.S., Mandhani A., Mahdi A.A., et al. Low- and high-grade bladder cancer determination via human serum-based metabolomics approach. J Proteome Res. 2013;12:5839–5850. doi: 10.1021/pr400859w. [DOI] [PubMed] [Google Scholar]
- 31.Huang Z., Chen Y., Hang W., Gao Y., Lin L., Li D.Y., et al. Holistic metabonomic profiling of urine affords potential early diagnosis for bladder and kidney cancers. Metabolomics. 2013;9:119–129. [Google Scholar]
- 32.Jobu K., Sun C., Yoshioka S., Yokota J., Onogawa M., Kawada C., et al. Metabolomics study on the biochemical profiles of odor elements in urine of human with bladder cancer. Biol Pharm Bull. 2012;35:639–642. doi: 10.1248/bpb.35.639. [DOI] [PubMed] [Google Scholar]
- 33.Jin X., Yun S.J., Jeong P., Kim I.Y., Kim W.J., Park S. Diagnosis of bladder cancer and prediction of survival by urinary metabolomics. Oncotarget. 2014;5:1635–1645. doi: 10.18632/oncotarget.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peng J., Chen Y.T., Chen C.L., Li L. Development of a universal metabolome-standard method for long-term LC-MS metabolome profiling and its application for bladder cancer urine-metabolite-biomarker discovery. Anal Chem. 2014;86:6540–6547. doi: 10.1021/ac5011684. [DOI] [PubMed] [Google Scholar]
- 35.Tripathi P., Somashekar B.S., Ponnusamy M., Gursky A., Dailey S., Kunju P., et al. HR-MAS NMR tissue metabolomic signatures cross-validated by mass spectrometry distinguish bladder cancer from benign disease. J Proteome Res. 2013;12:3519–3528. doi: 10.1021/pr4004135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wittmann B.M., Stirdivant S.M., Mitchell M.W., Wulff J.E., McDunn J.E., Li Z., et al. Bladder cancer biomarker discovery using global metabolomic profiling of urine. PLoS One. 2014;9:1–19. doi: 10.1371/journal.pone.0115870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen C., Sun Z., Chen D., Su X., Jiang J., Li G., et al. Developing urinary metabolomic signatures as early bladder cancer diagnostic markers. Omi A J Integr Biol. 2015;19:1–11. doi: 10.1089/omi.2014.0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou Y., Song R., Zhang Z., Lu X., Zeng Z., Hu C., et al. The development of plasma pseudotargeted GC-MS metabolic profiling and its application in bladder cancer. Anal Bioanal Chem. 2016;408:6741–6749. doi: 10.1007/s00216-016-9797-0. [DOI] [PubMed] [Google Scholar]
- 39.Tan G., Wang H., Yuan J., Qin W., Dong X., Wu H., et al. Three serum metabolite signatures for diagnosing low-grade and high-grade bladder cancer. Sci Rep. 2017;7:1–11. doi: 10.1038/srep46176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shao C.H., Chen C.L., Lin J.Y., Chen C.J., Fu S.H., Chen Y.T., et al. Metabolite marker discovery for the detection of bladder cancer by comparative metabolomics. Oncotarget. 2017;8:38802–38810. doi: 10.18632/oncotarget.16393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yumba Mpanga A., Siluk D., Jacyna J., Szerkus O., Wawrzyniak R., Markuszewski M., et al. Targeted metabolomics in bladder cancer: from analytical methods development and validation towards application to clinical samples. Anal Chim Acta. 2018;1037:188–199. doi: 10.1016/j.aca.2018.01.055. [DOI] [PubMed] [Google Scholar]
- 42.Cheng X., Liu X., Liu X., Guo Z., Sun H., Zhang M., et al. Metabolomics of non-muscle invasive bladder cancer: biomarkers for early detection of bladder cancer. Front Oncol. 2018;8:1–11. doi: 10.3389/fonc.2018.00494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jacyna J., Wawrzyniak R., Balayssac S., Gilard V., Malet-Martino M., Sawicka A., et al. Urinary metabolomic signature of muscle-invasive bladder cancer: a multiplatform approach. Talanta. 2019;202:572–579. doi: 10.1016/j.talanta.2019.05.039. [DOI] [PubMed] [Google Scholar]
- 44.Wei Y., Wang M., Liu H., Niu Y., Wang S., Zhang F., et al. Simultaneous determination of seven endogenous aldehydes in human blood by headspace gas chromatography-mass spectrometry. J Chromatogr B Anal Technol Biomed Life Sci. 2019;1118–1119:85–92. doi: 10.1016/j.jchromb.2019.04.027. [DOI] [PubMed] [Google Scholar]
- 45.Loras A., Suárez-Cabrera C., Martínez-Bisbal M.C., Quintás G., Paramio J.M., Martínez-Máñez R., et al. Integrative metabolomic and transcriptomic analysis for the study of bladder cancer. Cancers (Basel) 2019;11:686. doi: 10.3390/cancers11050686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin J.Y., Juo B.R., Yeh Y.H., Fu S.H., Chen Y.T., Chen C.L., et al. Putative markers for the detection of early-stage bladder cancer selected by urine metabolomics. BMC Bioinformatics. 2021;22:305. doi: 10.1186/s12859-021-04235-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Łuczykowski K., Warmuzińska N., Operacz S., Stryjak I., Bogusiewicz J., Jacyna J., et al. Metabolic evaluation of urine from patients diagnosed with high grade (HG) bladder cancer by SPME-LC-MS method. Molecules. 2021;26:2194. doi: 10.3390/molecules26082194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pinto J., Carapito Â., Amaro F., Lima A.R., Carvalho-Maia C., Martins M.C., et al. Discovery of volatile biomarkers for bladder cancer detection and staging through urine metabolomics. Metabolites. 2021;11:199. doi: 10.3390/metabo11040199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li J., Cheng B., Xie H., Zhan C., Li S., Bai P. Bladder cancer biomarker screening based on non-targeted urine metabolomics. Int Urol Nephrol. 2022;54:23–29. doi: 10.1007/s11255-021-03080-6. [DOI] [PubMed] [Google Scholar]
- 50.Jacyna J., Kordalewska M., Artymowicz M., Markuszewski M., Matuszewski M., Markuszewski M.J. Pre- and post-resection urine metabolic profiles of bladder cancer patients: results of preliminary studies on time series metabolomics analysis. Cancers (Basel) 2022;14:1210. doi: 10.3390/cancers14051210. https://10.3390/cancers14051210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakrani M.N., Wineland R.H., Anjum F. StatPearls [Internet] StatPearls Publishing; Treasure Island: 2023. Physiology, glucose metabolism.https://www.ncbi.nlm.nih.gov/books/NBK560599/ [PubMed] [Google Scholar]
- 52.Zoidis E., Papamikos V. In: Food science. Caballero B., Finglas P.M., Toldrá F., editors. Elsevier Inc.; The Netherlands: 2016. Glucose: metabolism and regulation. p.233–p238. [Google Scholar]
- 53.Wu G. Amino acids: metabolism, functions, and nutrition. Amino Acids. 2009;37:1–17. doi: 10.1007/s00726-009-0269-0. [DOI] [PubMed] [Google Scholar]
- 54.Moffatt B.A., Ashihara H. Purine and pyrimidine nucleotide synthesis and metabolism. Arab B. 2002;1 doi: 10.1199/tab.0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O'Brien P., Siraki A., Shangari N. Aldehyde sources, metabolism, molecular toxicity mechanisms, and possible effects on human health. Crit Rev Toxicol. 2005;35:609–662. doi: 10.1080/10408440591002183. [DOI] [PubMed] [Google Scholar]
- 56.Toyokuni S., Okamoto K., Yodoi J., Hiai H. Persistent oxidative stress in cancer. FEBS Lett. 1995;358:1–3. doi: 10.1016/0014-5793(94)01368-b. [DOI] [PubMed] [Google Scholar]
- 57.Ashrafian H., Sounderajah V., Glen R., Ebbels T., Blaise B.J., Kalra D., et al. Metabolomics: the stethoscope for the twenty-first century. Med Princ Pract. 2021;30:301–310. doi: 10.1159/000513545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spratlin J.L., Serkova N.J., Eckhardt S.G. Clinical applications of metabolomics in oncology: a review. Clin Cancer Res. 2009;15:431–440. doi: 10.1158/1078-0432.CCR-08-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Inglese P., McKenzie J.S., Mroz A., Kinross J., Veselkov K., Holmes E., et al. Deep learning and 3D-DESI imaging reveal the hidden metabolic heterogeneity of cancer. Chem Sci. 2017;8:3500–3511. doi: 10.1039/c6sc03738k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tenori L., Oakman C., Claudino W.M., Bernini P., Cappadona S., Nepi S., et al. Exploration of serum metabolomic profiles and outcomes in women with metastatic breast cancer: a pilot study. Mol Oncol. 2012;6:437–444. doi: 10.1016/j.molonc.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mauri-Capdevila G., Jove M., Suarez-Luis I., Portero-Otin M., Purroy F. [Metabolomics in ischaemic stroke, new diagnostic and prognostic biomarkers] Rev Neurol. 2013;57:29–36. [Article in Spanish] [PubMed] [Google Scholar]
- 62.Wang T.J., Ngo D., Psychogios N., Dejam A., Larson M.G., Vasan R.S., et al. 2-Aminoadipic acid is a biomarker for diabetes risk. J Clin Invest. 2013;123:4309–4317. doi: 10.1172/JCI64801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim W.T., Yun S.J., Yan C., Jeong P., Kim Y.H., Lee I.S., et al. Metabolic pathway signatures associated with urinary metabolite biomarkers differentiate bladder cancer patients from healthy controls. Yonsei Med J. 2016;57:865–871. doi: 10.3349/ymj.2016.57.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cheng Y., Yang X., Deng X., Zhang X., Li P., Tao J., et al. Metabolomics in bladder cancer: a systematic review. Int J Clin Exp Med. 2015;8:11052–11063. [PMC free article] [PubMed] [Google Scholar]
- 65.Liberti M.V., Locasale J.W. The Warburg effect: how does it benefit cancer cells? Trends Biochem Sci. 2016;41:211–218. doi: 10.1016/j.tibs.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Massari F., Ciccarese C., Santoni M., Iacovelli R., Mazzucchelli R., Piva F., et al. Metabolic phenotype of bladder cancer. Cancer Treat Rev. 2016;45:46–57. doi: 10.1016/j.ctrv.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 67.Jin L., Alesi G.N., Kang S. Glutaminolysis as a target for cancer therapy. Oncogene. 2016;35:3619–3625. doi: 10.1038/onc.2015.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chung K., Gadupudi G.S. Possible roles of excess tryptophan metabolites in cancer. Environ Mol Mutagen. 2011;52:81–104. doi: 10.1002/em.20588. [DOI] [PubMed] [Google Scholar]
- 69.Lee S.H., Mahendran R., Tham S.M., Thamboo T.P., Chionh B.J., Lim Y.X., et al. Tryptophan-kynurenine ratio as a biomarker of bladder cancer. BJU Int. 2021;127:445–453. doi: 10.1111/bju.15205. [DOI] [PubMed] [Google Scholar]
- 70.Pilotte L., Larrieu P., Stroobant V., Colau D., Dolusic E., Frédérick R., et al. Reversal of tumoral immune resistance by inhibition of tryptophan 2,3-dioxygenase. Proc Natl Acad Sci U S A. 2012;109:2497–2502. doi: 10.1073/pnas.1113873109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Long J., Zhang C.J., Zhu N., Du K., Yin Y.F., Tan X., et al. Lipid metabolism and carcinogenesis, cancer development. Am J Cancer Res. 2018;8:778–791. [PMC free article] [PubMed] [Google Scholar]
- 72.Wang G., Cao R., Ge Q., Xiao Y., Wang X. The role of lipids metabolism in bladder cancer. Cancer Res. 2017;77:2511. doi: 10.1158/1538-7445.AM2017-2511. [DOI] [Google Scholar]
- 73.Serrano M., Gallego M., Silva M. Analysis of endogenous aldehydes in human urine by static headspace gas chromatography-mass spectrometry. J Chromatogr A. 2016;1437:241–246. doi: 10.1016/j.chroma.2016.01.056. [DOI] [PubMed] [Google Scholar]
- 74.Crowder S.L., Playdon M.C., Gudenkauf L.M., Ose J., Gigic B., Greathouse L., et al. A molecular approach to understanding the role of diet in cancer-related fatigue: challenges and future opportunities. Nutrients. 2022;14:1–14. doi: 10.3390/nu14071496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang Y., Hodge R.A., Stevens V.L., Hartman T.J., McCullough M.L. Identification and reproducibility of urinary metabolomic biomarkers of habitual food intake in a cross-sectional analysis of the cancer prevention study-3 diet assessment sub-study. Metabolites. 2021;11:248. doi: 10.3390/metabo11040248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.