Abstract
Remdesivir (RDV) is a broad-spectrum nucleotide analog prodrug approved for the treatment of COVID-19 in hospitalized and non-hospitalized patients with clinical benefit demonstrated in multiple Phase 3 trials. Here we present SARS-CoV-2 resistance analyses from the Phase 3 SIMPLE clinical studies evaluating RDV in hospitalized participants with severe or moderate COVID-19 disease. The severe and moderate studies enrolled participants with radiologic evidence of pneumonia and a room-air oxygen saturation of ≤94% or >94%, respectively. Virology sample collection was optional in the study protocols. Sequencing and related viral load data were obtained retrospectively from participants at a subset of study sites with local sequencing capabilities (10 of 183 sites) at timepoints with detectable viral load. Among participants with both baseline and post-baseline sequencing data treated with RDV, emergent Nsp12 substitutions were observed in 4 of 19 (21%) participants in the severe study and none of the 2 participants in the moderate study. The following 5 substitutions emerged: T76I, A526V, A554V, E665K, and C697F. The substitutions T76I, A526V, A554V, and C697F had an EC50 fold change of ≤1.5 relative to the wildtype reference using a SARS-CoV-2 subgenomic replicon system, indicating no significant change in the susceptibility to RDV. The phenotyping of E665K could not be determined due to a lack of replication. These data reveal no evidence of relevant resistance emergence and further confirm the established efficacy profile of RDV with a high resistance barrier in COVID-19 patients.
Keywords: remdesivir, SARS-CoV-2, resistance, genotyping, phenotyping, Nsp12
1. Introduction
Remdesivir (RDV) is a nucleotide analog prodrug that is intracellularly metabolized into an analog of adenosine triphosphate (RDV-TP) that inhibits viral RNA polymerases [1,2]. RDV has broad spectrum activity against coronavirus (SARS-CoV, MERS-CoV, and SARS-CoV-2), filovirus, paramyxovirus, and pneumovirus families [3,4,5,6]. RDV is approved for the treatment of COVID-19 in both hospitalized and non-hospitalized patients, with clinical benefit demonstrated in multiple Phase 3 trials. The efficacy data from the pivotal Phase 3, randomized, double-blind, placebo-controlled study ACTT-1 showed a statistically significant shorter median time to recovery for participants with severe and critical COVID-19 pneumonia treated with RDV compared with those in the placebo arm [7]. In the Phase 3 SIMPLE studies (NCT04292899 and NCT04292730), RDV was evaluated in hospitalized participants with severe or moderate COVID-19 pneumonia, respectively. In participants with severe COVID-19 (pneumonia and room-air oxygen saturation of ≤94%), an analysis of the primary endpoint demonstrated that treatment with RDV for up to 5 days and treatment with RDV for up to 10 days resulted in similar odds of improved clinical status on day 14 [8]. In participants with moderate COVID-19 (pneumonia and room-air oxygen saturation of >94%), the primary endpoint results showed that RDV administered for 5 days resulted in significantly better odds of improvement in clinical status on day 11, as assessed by a 7-point ordinal scale, compared with those who received only standard-of-care treatment [9]. Subsequently, in the Phase 3 randomized, double-blind, placebo-controlled PINETREE study, treatment with RDV for 3 days, administered in an outpatient setting to participants with early-stage COVID-19 who were at risk of disease progression, resulted in a statistically significant reduction of 87% in COVID-19-related hospitalization or all-cause death by day 28 compared with the placebo [10].
The development of resistance to RDV has been investigated in in vitro selection experiments using RDV or its parent nucleoside analog GS-441524. In one study using GS-441524 and SARS-CoV-2, virus pools emerged after 13 passages expressing amino acid substitutions V166A, N198S, S759A, V792I, C799F, and C799R in Nsp12, the SARS-CoV-2 RNA-dependent RNA polymerase (RdRp) and target of RDV. The phenotypic testing of virus from passage 13 resulted in RDV 50% inhibition of virus replication (EC50) fold changes between 2.6 and 10 from the SARS-CoV-2 A lineage WA1 reference strain [11]. In a second in vitro selection experiment, using RDV and a SARS-CoV-2 isolate containing the P323L substitution in Nsp12, a single amino acid substitution at V166L emerged after 17 passages. Phenotypic testing of recombinant SARS-CoV-2 viruses containing P323L alone or P323L + V166L in combination resulted in RDV EC50 fold changes between 1.2 and 1.5 from the WA1 reference strain [12]. In another study, resistance selection with RDV and SARS-CoV-2 identified a single Nsp12 amino acid substitution, E802D, in two virus populations with a minimal loss of susceptibility to RDV (2.5-fold reduction in susceptibility to RDV) [13].
To investigate possible drug resistance development in patients treated with RDV in the Phase 3 SIMPLE studies, SARS-CoV-2 genotypic and phenotypic analyses were conducted retrospectively from stored virology samples.
2. Materials and Methods
2.1. SIMPLE Study Designs
Design details for the SIMPLE studies have been previously published [8 (NCT04292899), 9 (NCT04292730)]. Briefly, the two Phase 3, randomized studies evaluated the safety and antiviral activity of RDV in participants with severe and moderate COVID-19, respectively. The studies were conducted early in the pandemic, and participants were enrolled between March and May 2020 (last patient, last visit June 2020 for both studies). The trial protocol was designed such that all core testing could be performed locally at the site; thus, although the study was conducted at high-quality centers, most did not have local sequencing capabilities. All patients or their legally authorized representative provided written informed consent prospectively, including for future testing of samples collected during the study. The trials were approved by the institutional review board or ethics committee at each site and were conducted in compliance with the Declaration of Helsinki Good Clinical Practice guidelines and local regulatory requirements.
The severe COVID-19 study enrolled hospitalized participants with confirmed SARS-CoV-2 infection with radiographic evidence of pulmonary infiltrates and who had either an oxygen saturation < 94% or were receiving supplemental oxygen.
The moderate COVID-19 study enrolled hospitalized participants with confirmed SARS-CoV-2 infection with moderate COVID-19 pneumonia (defined as any radiographic evidence of pulmonary infiltrates and an oxygen saturation > 94% on room air).
2.2. Virology Data Collection
The collection of virology samples by the study sites for resistance testing was optional in the study protocols. To conduct virology analysis, study sites with virology sample collection and sequencing capabilities were identified through a questionnaire. Sequencing and related viral load data were obtained retrospectively from 10 of 174 study sites for Parts A and B of the severe COVID-19 study and from 6 of 139 study sites for Parts A and B of the moderate COVID-19 study. The sequencing analysis included any participant with a virology sample collected with detectable viral load who received at least one dose of RDV or was assigned to standard of care.
For the severe COVID-19 study, the sample types collected included nasal swabs, nasal/oropharyngeal swabs, naso-oral swabs, nasopharyngeal swabs, throat swabs, swabs (not specified), sputum, endotracheal tube aspirate, and bronchoalveolar lavage. For the moderate COVID-19 study, the sample types collected included nasal swabs, naso-oral swabs, nasopharyngeal swabs, oropharyngeal swabs, and swabs (not specified).
2.3. Sequencing Methodologies
Sequencing was performed retrospectively from stored upper- and lower-respiratory samples. The SARS-CoV-2 whole genome or Nsp12 was amplified by gene-specific primers or enriched by probes and sequenced by locally available assays using either Sanger sequencing, Illumina MiSeq, Illumina Novaseq 6000, or the Ion Torrent platform.
The Nsp12 sequencing was conducted as follows: Nsp12 was amplified using conventional nested PCR methodology using Phusion Hot start Flex Master Mix (New England Biolabs, Ipswich, MA, USA). The outer and inner PCR reaction generated products of 1481 and 1048 base pairs, respectively. Population sequencing (Sanger) was performed, and primer sequences were removed prior to data analysis, generating a sequence with length of 1028 nucleotides.
SARS-CoV-2 whole-genome sequencing was conducted using the following 5 methodologies available at the local study sites: (1) The Twist Bio hybrid capture target enrichment protocol (Twist Bioscience, South San Francisco, CA, USA): Briefly, the nucleic acid was extracted, and cDNA was generated according to the manufacturer’s instructions. Libraries were amplified from the cDNA using a PCR thermal cycler. The SARS-CoV-2 virus sequence was then targeted and enriched during a hybridization reaction with a biotin-attached probe specific to the SARS-CoV-2 virus. The enriched SARS-CoV-2 targeted regions were sequenced by Illumina MiSeq. (2) Illumina Ampliseq SARS-CoV-2 Research panel (Illumina, San Diego, CA, USA) [14]: Briefly, the nucleic acid was extracted, and cDNA and amplicons were generated according to the manufacturer’s instructions. The use of proprietary primer modifications allowed for the removal of the primer sequenced during library preparation. The generated libraries were sequenced using Illumina Novaseq 6000 sequencer. (3) SARS-CoV-2 amplification using ARTIC V3 primer set [15]: Briefly, the nucleic acid was extracted, and cDNA was generated using random hexamers. SARS-CoV-2 was amplified using ARTIC V3 primers with adapter sequences according to the instructions from Illumina COVIDseq Test (Illumina, San Diego, CA, USA), nCoV-2019 sequencing protocol v3 [16], or Qiagen SARS-CoV-2 research panel (Qiagen, Hilden, Germany). The generated libraries were sequenced using Illumina MiSeq (single or paired-end reads). (4) Ion AmpliSeq™ SARS-CoV-2 Research panel (Life Technologies Corporation, Carlsbad, CA, USA): Briefly, the nucleic acid was extracted. cDNA synthesis and the preparation of libraries was performed according to the manufacturer’s instructions using primer pools that target 237 amplicons specific to SARS-CoV-2. The libraries were sequenced using the Ion Torrent platform. (5) SARS-CoV-2 was sequenced using the method described by Gonzalez-Reiche [17]. Briefly, nucleic acid was extracted using QIAamp Viral RNA Minikit (QIAGEN, Cat. No. 52904) per the manufacturer’s instructions. In brief, cDNA synthesis was performed using random hexamers. Sample preparation for sequencing was performed using whole-genome amplification with custom-designed tiling primers generating ~1.5 and ~2 kb amplicons with ~200 bp overlaps between each region and the Artic Consortium protocol v3 (https://artic.network/ncov-2019, accessed on 1 February 2021), with modifications. The libraries were sequenced using Illumina MiSeq.
2.4. Sequencing Data Processing
Sequencing data were received from the sites as FASTQ files (single or paired-end) that were split per sample and amplification pool. Internally developed software was used to process and align deep sequencing data. Briefly, sequence reads from FASTQ files were evaluated to remove adapter sequences and trim low-quality phred quality scores using Trimmomatic [18]. Reads were aligned to the host and SARS-CoV-2 reference sequence and reads matching the host were removed. Reads aligned to the SARS-CoV-2 reference sequence (Wuhan-Hu-1, NC_045512.2) using a SMALT aligner [19] were further evaluated. Read ends that overlapped with the amplification primers were clipped from the aligned reads based on genomic coordinates. Variant calls at nucleotide and amino acid levels were rolled up across amplification pools, if relevant, and reported per sample. To report in-frame insertions and deletions (indels), the amino acid realignment of reads with indels was performed. Reads containing frameshift indels were trimmed to exclude the region with an indel from further amino acid variant analysis. All aligned, trimmed reads were then translated in-frame, and amino acid changes from the reference sequence were tabulated. Nucleotide variants were reported in the consensus sequence, and mixture codes were used when more than one base was present at ≥15% of the viral population. Indels were reported in the consensus sequence when present at ≥50% of the viral population.
2.5. SARS-CoV-2 Lineage Determination
SARS-CoV-2 lineage was determined by Pangolin Software v.2.3.8 (pangoLEARN version 2021-04-21, https://github.com/cov-lineages/pangolin, accessed on 21 April 2021) using SARS-CoV-2 whole-genome consensus sequences. A sequence coverage threshold of least 70% of the SARS-CoV-2 coding region was used for lineage determination.
2.6. SARS-CoV-2 Sequence Analysis
The primary analysis was focused on the identification of amino acid substitutions in the SARS-CoV-2 Nsp12 RdRp gene. Amino acid substitutions identified in SARS-CoV-2 Nsp8, Nsp10, Nsp13, and Nsp14 genes were investigated as secondary analyses. Any pretreatment sequence (day 1 or earlier) was considered as the baseline sequence. Amino acid substitutions in SARS-CoV-2 Nsp12 compared with the reference sequence (Wuhan-Hu-1, NC_045512.2) that occurred in ≥2 participants at baseline were reported. Post-baseline sequences were compared with participant-specific baseline sequences to determine whether any amino acid substitutions had emerged in the Nsp12 gene during or after treatment.
2.7. SARS-CoV-2 Phenotypic Analysis
Phenotypic analysis was attempted on post-baseline clinical isolates with identified treatment-emergent amino acid substitutions compared with their baseline sample in the SARS-CoV-2 Nsp12. Site-directed mutants were generated and tested using a replicon assay. The replicon system and the procedures required to assemble and produce non-infectious SARS-CoV-2 replicon were adapted and modified from Xie [20] and Zhang [21]. Briefly, the four plasmids that encode SARS-CoV-2 genes for the non-structural proteins and the nucleocapsid were prepared. The plasmids were verified by restriction enzyme BsaI digestion and Sanger sequencing to exclude the introduction of any undesired mutations into the plasmids prior to assembly of the full-length (minus structural gene) SARS-CoV-2 replicon DNA (24.2 kb). DNA fragments for ligation by restriction enzyme BsaI digestion of the Maxiprep plasmids were prepared and assembled into full-length SARS-CoV-2 replicon DNA in vitro using a T4 DNA ligase. The full-length ligation product was verified by The LabChip® GX Touch™ (Perkin Elmer, Boston, MA, USA) and purified by AMPure PB magnetic beads. Full-length replicon RNA was generated by in vitro transcription (IVT) and purified with a MegaClear RNA purification Kit. Huh7-1CN cells were resuspended in cold Opti-MEM to 1.125 × 107 cells per mL. Fifteen micrograms of RNA were mixed with cells and immediately electroporated. In a 96-well plate, compounds were dispensed directly into 20 μL of incubation medium as 3-fold serial dilutions. To the diluted compound solutions, 180 μL electroporated cells were added. The plates were then incubated at 37 °C with 5% CO2. At 48 h post-electroporation, 10 μL of supernatant was added to fresh 96-well plates containing 10 μL of lysis solution in each well. The luciferase assay 1× solution was prepared, and 100 μL was added to each well. The relative luciferase signals were calculated by normalizing the luciferase signals of the compound-treated groups to that of the DMSO-treated groups (set as 100%). EC50 values were calculated using a nonlinear four-parameter variable slope regression model as the concentration at which there was a 50% decrease in the luciferase reporter signal relative to the DMSO vehicle alone (0% virus inhibition) and uninfected control culture (100% virus inhibition). Two experiments were performed with technical triplicates. The fold change values were calculated by dividing the variant mean EC50 by the SH01 reference strain mean EC50.
3. Results
3.1. Sequencing Data Collection
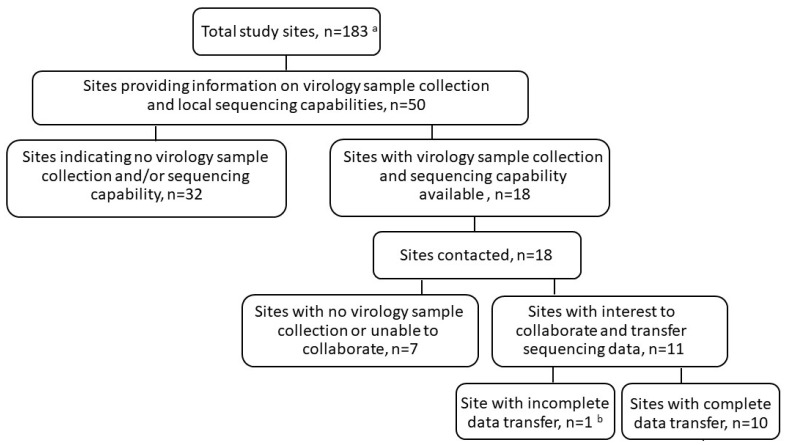
The site collection of virology samples was optional in the study protocols. Sequencing and related viral load data were obtained retrospectively from study sites with sample collection and local sequencing capabilities (10 of 183 sites; Figure 1; Table S1). Sequencing was restricted to samples with positive SARS-CoV-2 PCR with sufficient viral copy number to permit sequencing. Overall, sequencing was obtained from 73 of 4838 (1.5%) participants at baseline and 32 (0.7%) participants post-baseline in the severe COVID-19 study, and 14 of 1087 (1.3%) participants at baseline and 6 (0.6%) participants post-baseline in the moderate COVID-19 study. Among these, 19 (0.4%) participants in the severe COVID-19 study and 4 (0.4%) participants in the moderate COVID-19 study had sequencing data available at both baseline and post-baseline timepoints (Table 1). In the RDV treatment group, a majority of participants received 10 days of RDV treatment.
Figure 1.
Identification of study sites with virology sample collection and sequencing capabilities. a The severe and moderate COVID-19 studies comprised 183 sites in total. b Site unable to provide participant-specific identification numbers to link sequencing data to the corresponding participants in the clinical studies.
Table 1.
Summary of SARS-CoV-2 sequencing data from the SIMPLE studies.
| Study Part | Treatment Group | Participants | Participants with Sequencing Data | |||
|---|---|---|---|---|---|---|
| Baseline | Post-Baseline | Both Baseline and Post-Baseline | ||||
| Severe COVID-19 study a | A | RDV for up to 5 days | 200 | 2 | 1 | 0 |
| RDV for up to 10 days | 197 | 1 | 1 | 1 | ||
| B | Non-randomized expanded access, RDV for up to 10 days | 3597 | 51 | 16 | 9 | |
| Invasive mechanical ventilation, RDV for up to 10 days | 844 | 19 | 14 | 9 | ||
| Total | 4838 | 73 | 32 | 19 | ||
| Moderate COVID-19 study b | A | RDV for up to 5 days | 191 | 3 | 0 | 0 |
| RDV for up to 10 days | 193 | 0 | 1 | 0 | ||
| SOC | 200 | 4 | 2 | 2 | ||
| B | Non-randomized expanded access, RDV for up to 10 days | 503 | 7 | 3 | 2 | |
| Total | 1087 | 14 | 6 | 4 | ||
RDV, remdesivir; SOC, standard of care. a The Severe COVID-19 study (GS-US-540-5773) was divided into two parts. Part A of the study included 397 participants, and Part B of the study included 4441 participants. All participants in both Parts A and B received RDV treatment (randomized for up to 5 days or up to 10 days in Part A, and non-randomized up to 10 days in Part B; access for patients on invasive mechanical ventilation began upon activation of Part B). The primary efficacy endpoint was clinical status assessed using a 7-point ordinal scale on day 14 of Part A as previously described [8]. b The Moderate COVID-19 study (GS-US-540-5774) was divided into two parts. Part A of the study included 584 participants, and Part B of the study included 503 participants. Of the 1087 total participants, 200 participants received SOC, and 887 received RDV treatment (5 days or 10 days). The primary efficacy endpoint was clinical status assessed using a 7-point ordinal scale on day 11 of Part A as previously described [9].
3.2. Lineage and Baseline Nsp12 Substitutions
In both the moderate and severe COVID-19 studies, P323L was the most frequently observed amino acid substitution in SARS-CoV-2 Nsp12 at baseline compared with the reference sequence (Wuhan-Hu-1, NC_045512.2). Among the participants with sequence coverage at position 323, this substitution was observed in 50 of 53 participants (94.3%) in the severe COVID-19 study and 8 of 9 participants (88.9%) in the moderate COVID-19 study (Table 2). The Nsp12 substitution A554V was observed in two participants at baseline in the severe COVID-19 study, and both reverted to wildtype during RDV treatment.
Table 2.
Amino acid substitutions in Nsp12 observed at baseline (compared with Wuhan-Hu-1 reference sequence).
| Number of Participants with Amino Acid Substitutions Occurring in ≥2 Participants at Baseline, n (%) a | ||
|---|---|---|
| Severe COVID-19 Study | Moderate COVID-19 Study | |
| P323L | 50/53 (94%) | 8/9 (88%) |
| A554V | 2/64 (3.1%) | 0/13 (0%) |
a The denominator is based on the number of participants with sequence coverage at the site of the substitution.
The high prevalence of P323L is consistent with its presence in most circulating SARS-CoV-2 variants, including B.1, B.1.1, B.1.119, and B.1.610, which were the most common lineages in these studies. P323L has been phenotyped using the recombinant SARS-CoV-2 system and does not impact susceptibility to RDV in vitro [22]. No participants with variants of concern or interest (VOC/VOI), as defined by the World Health Organization (WHO) or Centers for Disease Control and Prevention (CDC), were identified in these studies conducted in the early portion of the pandemic (enrollment concluded by the end of May 2020).
3.3. Post-Baseline Amino Acid Substitutions in SARS-CoV-2 Nsp12
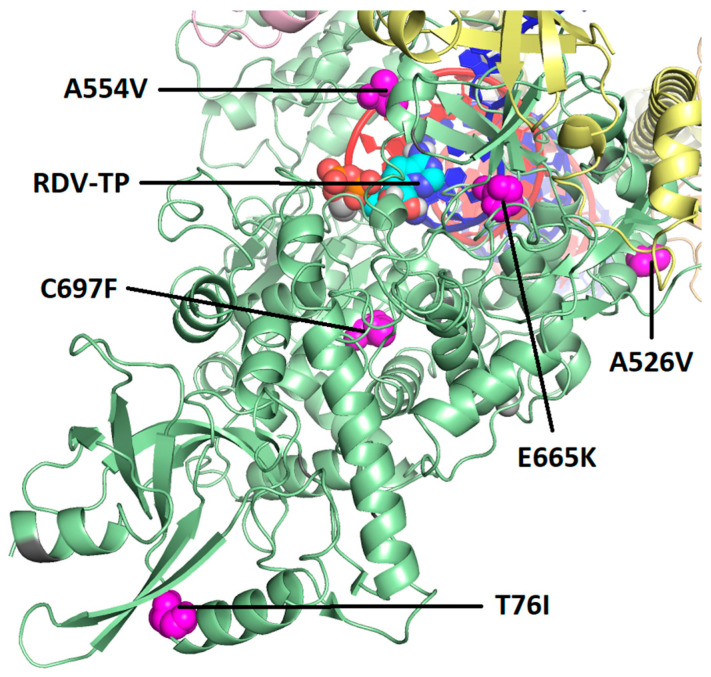
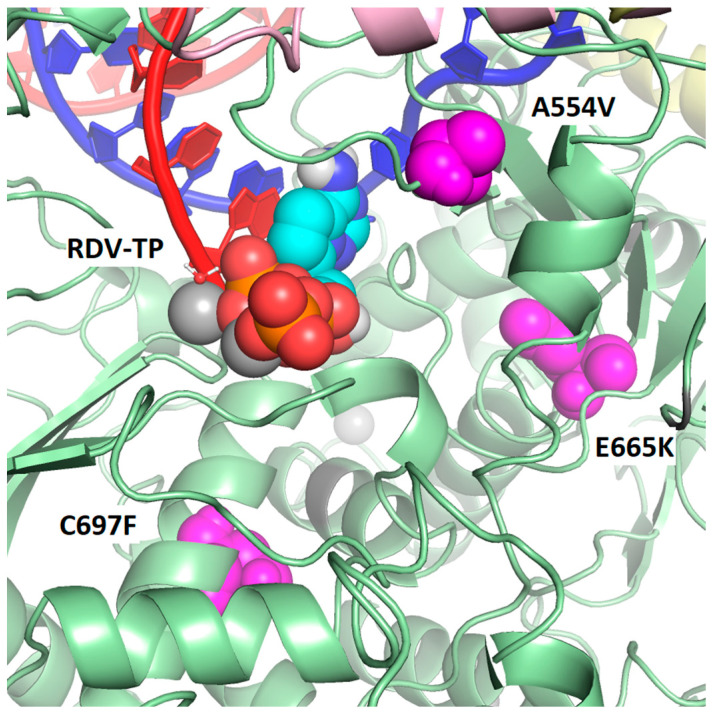
Both baseline and post-baseline sequencing data were obtained from 19 participants in the severe COVID-19 study, all of whom received RDV, and from 4 participants in the moderate COVID-19 study, 2 of whom received RDV and 2 of whom received the standard of care. Post-baseline substitutions in SARS-CoV-2 Nsp12 were observed in 4 of 19 participants (21.1%) in the severe COVID-19 study and none of the 4 participants in the moderate COVID-19 study. The following five substitutions emerged during or after RDV treatment in the severe COVID-19 study: T76T/I, A526V, A554V, E665K, and C697C/F (Table 3). A554V and E665K were observed on day 13 in the same participant, but in endotracheal tube aspirate and bronchoalveolar lavage, respectively. None of the substitutions were observed in >1 participant. Based on the analysis of a cryo-EM structure of the SARS-CoV-2 polymerase complex with a pre-incorporated RDV-TP [23], T76I and A526V are located on the surface of the Nsp12 protein distant from the polymerase active site and viral RNA (Figure 2). In this structure, A554V, E665K, and C697F are located closer to the active site but have no direct interaction with the RNA or the incoming nucleotide (Figure 3). As measured from the C1′ atom of RDV-TP to the amino acid Cα, A554V is 13.9 Å, C697F is 16.9 Å, and E665K is 18.0 Å. Based on this analysis, none of the substitutions were expected to impact susceptibility to RDV.
Table 3.
Amino acid substitutions in Nsp12 detected post-baseline.
| Number of Participants with Emergent Amino Acid Substitutions at Post-Baseline, n (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Severe COVID-19 Study | Moderate COVID-19 Study | |||||||||
| Part A | Part B | Part A | Part B | |||||||
| RDV for up to 5 Days (n = 0) |
RDV for up to 10 Days (n = 1) |
Non-randomized Expanded Access (RDV for up to 10 Days) (n = 9) |
Invasive Mechanical Ventilation (RDV for up to 10 Days) (n = 9) |
Total (n = 19) |
RDV for up to 5 Days (n = 0) |
RDV for up to 10 Days (n = 0) |
SOC (n = 2) |
Non-Randomized Expanded Access (RDV for up to 10 Days) (n = 2) |
Total in RDV Arms (n = 2) |
|
| T76T/I | NA | 0 | 1/9 (11%) | 0 | 1/19 (5%) | NA | NA | 0 | 0 | 0 |
| A526V | NA | 0 | 0 | 1/9 (11%) | 1/19 (5%) | NA | NA | 0 | 0 | 0 |
| A554V a | NA | 0 | 0 | 1/9 (11%) a | 1/19 (5%) | NA | NA | 0 | 0 | 0 |
| E665K a | NA | 0 | 0 | 1/9 (11%) a | 1/19 (5%) | NA | NA | 0 | 0 | 0 |
| C697C/F | NA | 0 | 1/9 (11%) | 0 | 1/19 (5%) | NA | NA | 0 | 0 | 0 |
NA = not applicable; RDV = remdesivir; SOC = standard of care. a A554V and E665K were observed on day 13 in the same participant, but in endotracheal tube aspirate and bronchoalveolar lavage, respectively.
Figure 2.
Map of the observed post-baseline amino acid substitutions on the cryo-EM structure of the SARS-CoV-2 polymerase complex with pre-incorporated RDV-TP. RDV-TP = triphosphate form of remdesivir. Pictured is a modified version of the cryo-EM structure, 7UO4 [23], which captures RDV-TP in its pre-incorporated state. The missing primer 3′OH and catalytic metal were added. The full Nsp12 protein is in green, with the locations of the observed post-baseline amino acid substitutions shown in magenta. Pink is Nsp7, and yellow is Nsp8 (two subunits). The template RNA strand is shown in blue and nascent RNA strand in red.
Figure 3.
Map of observed post-baseline amino acid substitutions closest to the active site of Nsp12 and on cryo-EM structure of the SARS-CoV-2 polymerase complex with pre-incorporated RDV-TP. None of the observed post-baseline substitutions is in direct contact with RDV-TP or the RNA primer or template strands. However, three of the substitutions are located within 20 Å of the pre-incorporated RDV-TP (as measured from Cα to C1′). A554V is 13.9 Å, C697F is 16.9 Å, and E665K is 18.0 Å. The Nsp12 protein is green with the locations of the substitutions shown in magenta. The template RNA strand is shown in blue and nascent RNA strand in red. Pink is Nsp7, and yellow is Nsp8 (two subunits).
3.4. Phenotypic Analysis of Emergent Nsp12 Substitutions
Phenotyping was conducted using SARS-CoV-2 replicons containing site-directed mutants (SDMs) in Huh7-1CN cells. SDMs were generated for the emergent Nsp12 substitutions T76I, A526V, A554V, E665K, and C697F. The substitutions T76I, A526V, A554V, and C697F had EC50 fold changes of ≤1.5 (Table 4), suggesting a similar susceptibility to RDV as the wildtype ancestral reference. For E665K, transfection of the mutant replicons was attempted twice; however, phenotypic results could not be generated due to a lack of replication.
Table 4.
RDV EC50 against site-directed mutants.
| Substitution in Nsp12 | RDV EC50 (nM) | EC50 Fold Change ± SD from WA1 | ||||
|---|---|---|---|---|---|---|
| 1st Replicate | 2nd Replicate | 3rd Replicate | 4th Replicate | Mean ± SD | ||
| Wildtype (SH01) | 8.83 | 8.44 | 9.51 | 13.03 | 9.95 ± 2.10 | 1.00 |
| T76I | - | - | 7.16 | 9.48 | 8.32 ± 1.63 | 0.84 |
| A526V | 10.69 | 13.08 | - | - | 11.89 ± 1.69 | 1.19 |
| A554V | 14.67 | 14.67 | - | - | 14.45 ± 0.31 | 1.45 |
| E665K | No replication | NA | NA | |||
| C697F | 10.33 | 9.43 | - | - | 9.88 ± 0.64 | 0.99 |
EC50 = half-maximal effective concentration; NA = not applicable; RDV = remdesivir; SD = standard deviation; SH01 = wildtype reference SARS-CoV-2 replicon generated from clinical isolate from Shanghai (lineage B).
3.5. Post-Baseline Amino Acid Substitutions Emerging in Other Proteins of the Polymerase Complex
Of the 19 participants with both baseline and post-baseline sequence data, sequencing of Nsp8, Nsp10, Nsp13, and Nsp14 was obtained from 12 (0.2%) of 4838 participants in the severe COVID-19 study and none in the moderate COVID-19 study. Sequencing coverage across Nsp8, Nsp10, Nsp13, and Nsp14 varied per participant and timepoint (Table 5). Of the participants with sequencing coverage, a single amino acid substitution in Nsp8 was observed in 1 of 9 (11%) participants. No amino acid substitutions were observed in Nsp10 in any of the 10 participants with sequence coverage. A single amino acid substitution in Nsp13 was observed in 1 of 10 (10%) participants, and amino acid substitutions in Nsp14 were observed in 2 of 11 (18%) participants. Overall, none of the substitutions occurred in >1 participant or have been associated with resistance to RDV.
Table 5.
Amino acid substitutions emerging in Nsp8, Nsp10, Nsp13, and Nsp14 at post-baseline in the severe COVID-19 study.
| Change from Baseline | Number of Participants with Baseline and Post-Baseline Sequencing Data | ||||
|---|---|---|---|---|---|
| A | B | Total (n = 12) |
|||
| RDV for up to 5 Days (n = 0) |
RDV for up to 10 Days (n = 0) |
Non-Randomized Expanded Access (RDV for up to 10 Days) (n = 6) a |
Invasive Mechanical Ventilation (RDV for up to 10 Days) (n = 6) |
||
| Nsp8 A89T | NA | NA | 0 | 1/6 (17%) | 1/9 (11%) |
| Nsp13 F581L | NA | NA | 1/4 (25%) | 0 | 1/10 (10%) |
| Nsp14 T21R | NA | NA | 0 | 1/6 (17%) | 1/11 (9%) |
| Nsp14 I101V + A287V | NA | NA | 1/5 (20%) | 0 | 1/11 (9%) |
NA = not applicable; RDV = remdesivir. a The denominator is based on sequencing coverage for Nsp8, Nsp10, Nsp13, and Nsp14 at baseline and post-baseline and could vary in the different regions of the polymerase complex.
4. Discussion
The Nsp12 polymerase target of RDV is highly conserved across coronaviruses, with close to 100% identity of the enzyme active site [22]. The low diversity and high genetic stability of the RNA replication complex suggests a minimal global risk of pre-existing SARS-CoV-2 resistance to RDV [24]. With approval globally, RDV has been broadly used in patients with COVID-19. The emergence of drug-resistant strains is of concern during the widespread use of a therapeutic. Here we investigated the possible development of drug resistance in a subset of participants with severe and moderate COVID-19 who were treated with RDV in the SIMPLE clinical studies conducted early in the pandemic.
Across both the SIMPLE severe and moderate studies, P323L was the most frequently observed baseline amino acid substitution in SARS-CoV-2 Nsp12 compared with the reference sequence. This is consistent with the presence of P323L in most circulating SARS-CoV-2 variants, including B.1, B.1.1, B.1.119, and B.1.610, which were the most common lineages observed in the SIMPLE studies. During the COVID-19 pandemic, new SARS-CoV-2 variants have been emerging and circulating around the world [25,26,27]. Changes in the SARS-CoV-2 genome in “variants of concern” are mostly located in the Spike region, which is the primary antigen under selective pressure from the immune system [28]. The Nsp12 target of RDV has been shown to be highly conserved, and only 2 prevalent changes (P323L and G671S) were observed in an analysis of nearly 6 million publicly available variant isolate sequences [22]. RDV retains potency to both P323L and G671S and their combination [22]. In addition, the antiviral activity of RDV to SARS-CoV-2 variants of concern have been tested in vitro, and RDV maintains potent antiviral activity against clinical isolates of SARS-CoV-2 Alpha, Beta, Gamma, Delta, Epsilon, and Omicron subvariants [22,29,30,31,32].
The in vitro selection of RDV-resistant strains of SARS-CoV-2 requires a high number of passages [11,12,13], suggesting a high barrier to the development of RDV resistance. Substitutions identified during the RDV in vitro resistance selection experiments were rare or not detected across >6 million publicly available Nsp12 consensus sequences [11,12,33]. Interestingly, none of the substitutions observed in the in vitro resistance selection experiments were observed in patients with sequencing data who were treated with RDV in the SIMPLE studies. Among the participants with both baseline and post-baseline sequencing data, post-baseline substitutions in SARS-CoV-2 Nsp12 were observed in 4 of 19 participants in the severe COVID-19 study and none of the 2 participants treated with RDV in the moderate COVID-19 study. Five substitutions emerged during or after RDV treatment. Based on the structural analysis, T76I and A526V are located on the surface of the Nsp12 protein distant from the polymerase active site or viral RNA and are therefore unlikely to impact susceptibility to RDV. The other three substitutions, A554V, E665K, and C697F, are located closer to the active site but have no direct interaction with the RNA or the incoming nucleotide. The phenotypic testing of T76I, A526V, A554V, and C697F generated by site-directed mutagenesis in the replicon system resulted in a ≤1.5-fold change in EC50 from the reference wildtype replicon, suggesting a similar susceptibility to RDV as the wildtype reference. The phenotyping of E665K could not be determined due to a lack of replication. As no substitutions with reduced susceptibility to RDV were observed in the SIMPLE studies, it suggests that RDV has a high barrier to resistance development in COVID-19 patients. This observation is concordance with the resistance analysis from the ACTT-1 Phase 3 clinical study, where a similar rate of emergent amino acid substitutions in Nsp12 and other proteins of the replication/transcription complex was observed in participants in the RDV and placebo groups. Importantly in the ACTT-1 study, only 2 participants had an emergence of substitutions associated with low-level reduced remdesivir susceptibility (V792I and C799F; ≤3.4-fold change in EC50 compared with wildtype and associated with reduced fitness). Notably, clinical recovery was similar between participants with and without emergent Nsp12 substitutions in the RDV arm, suggesting no impact on clinical outcome [34]. Rare cases of the prolonged shedding of SARS-CoV-2 have been described in older or immunocompromised patients [35,36,37]. Fortunately, surveillance reports continue to show a high sequence conservation of Nsp12 [24,38,39], and the prevalence of substitutions associated with reduced susceptibility to remdesivir is exceedingly low [24,33,40,41].
This study has important limitations. The severe and moderate COVID-19 SIMPLE studies were conducted early in the pandemic (March to June 2020), and due to supply shortages during this time (including personal protective equipment, swabs, viral transport media, and the availability of viral load and sequencing assays), virology sample collection was optional in the study protocol with only a few study sites collecting and analyzing samples. In addition, sequencing was restricted to samples with positive SARS-CoV-2 PCR and sufficient viral copy number to permit sequencing. Thus, a limited number of virology samples were available for sequencing and only a subset of participants had sequencing data at both baseline and post-baseline. All laboratory testing, including sequencing, was performed by local laboratories at the study sites, rather than through a central laboratory, due to logistical constraints at the time.
Taken together, the phenotypic analysis of emergent Nsp12 substitutions observed in participants with sequencing data available in the SIMPLE studies showed no change in RDV susceptibility, supporting a high barrier to RDV resistance in COVID-19 patients. The fortuitous discordance between in vitro escape mutations and their rare emergence in vivo are consistent with the postulate that the acquisition of mutations in the SARS-CoV-2 Nsp12 with reduced susceptibility to RDV incurs a fitness penalty, preserving RDV susceptibility at the population level. Additional studies are ongoing from RDV clinical trials to further characterize the resistance profile of RDV.
Acknowledgments
Medical writing and editorial support were provided by Becky Norquist, funded by Gilead Sciences, Inc. We thank the patients who participated in this trial, their families, and all participating investigators and support staff from the participating centers: University Hospital rechts der Isar, Technical University of Munich, Germany; Baylor University Medical Center, USA; Academische Ziekenhuis Leiden, The Netherlands; Hospital Universitari Vall d’Hebron, Spain; Klinik für Infektionskrankheiten und Spitalhygiene, Switzerland, UH Clinical Research Center/University Hospital Case Western, USA; Icahn School of Medicine at Mount Sinai, USA.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/v16040546/s1; Table S1: Sequence data obtained per study site.
Author Contributions
Methodology, D.H. (Dong Han) and L.R.; formal analysis, C.H. and D.P.P.; investigation, C.D.S., U.P., D.H. (Dieter Hoffmann), C.K., R.L.G., M.A., M.R., J.J.C.d.V., E.C.C., R.F., A.A., C.A., V.C., H.F.G., M.H., G.A.M., N.S., J.A.A. and H.v.B.; data curation, R.M., J.L. and A.P.; writing—original draft preparation, C.H.; writing—review and editing, C.H., C.D.S., U.P., D.H. (Dieter Hoffmann), C.K., R.L.G., M.A., M.R., J.J.C.d.V., E.C.C., R.M., J.L., D.H. (Dong Han), L.R., A.P., J.K.P., R.F., A.A., C.A., V.C., H.F.G., M.H., G.A.M., N.S., J.A.A., H.v.B. and D.P.P. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The trials were approved by the institutional review board or ethics committee at each site and were conducted in compliance with the Declaration of Helsinki Good Clinical Practice guidelines and local regulatory requirements. The trials were designed and conducted by the sponsor (Gilead Sciences, protocol codes GS-US-540-5773 and GS-US-540-5774) in collaboration with the principal investigators and in accordance with the protocol and amendments. The sponsor collected the data, monitored the conduct of the trial, and performed the statistical analyses.
Informed Consent Statement
All patients or their legally authorized representative provided written informed consent prospectively, including for future testing of samples collected during the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors have the following conflicts of interest: C.D.S., R.L.G., M.R., R.F., H.F.G., G.A.M., and J.A.A. served as investigators for the SIMPLE clinical studies and received funds to their institution from Gilead Sciences. C.S. received grants, personal fees, and non-financial support from Gilead Sciences; R.L.G. reports participation on advisory boards and/or consulting fees from AbbVie, AstraZeneca, Eli Lilly, Gilead Sciences, GSK Pharmaceuticals, and Roche, honoraria for lectures from Gilead Sciences and Pfizer (the latter unrelated to infectious diseases), travel support from Gilead Sciences, de minimis investment in AbCellera, a gift-in-kind to his institution from Gilead Sciences to facilitate an unrelated academic-sponsored clinical trial (NCT03383419) and grants or contracts to his institution from Regeneron and Roivant Sciences (Kinevant Sciences); H.F.G. reports having received honoraria from Gilead Sciences, Merck, ViiV, GSK, Janssen, Johnson and Johnson, and Novartis for serving on DSMB and/or advisory boards and has received a travel grant from Gilead Sciences. In addition, he has received grants from the Swiss National Science Foundation (SNSF), the Swiss HIV Cohort Study, the Yvonne Jacob Foundation, and the NIH, and unrestricted research grants from Gilead Sciences, all paid to the institution. In addition, he was the site investigator for the current study at affiliation; G.A.M. has been a consultant for Gilead Sciences; N.S. received funds from Gilead Sciences to support the sequencing costs for this study; and C.H., R.M., J.L., D.H. (Dong Han), L.R., A.P., J.K.P. and D.P.P. are employees of and own stock in Gilead Sciences. All other authors reported no conflict of interest with respect to this manuscript.
Funding Statement
This research was funded by Gilead Sciences.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Kokic G., Hillen H.S., Tegunov D., Dienemann C., Seitz F., Schmitzova J., Farnung L., Siewert A., Höbartner C., Cramer P. Mechanism of SARS-CoV-2 polymerase stalling by remdesivir. Nat. Commun. 2021;12:279. doi: 10.1038/s41467-020-20542-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bravo J.P.K., Dangerfield T.L., Taylor D.W., Johnson K.A. Remdesivir is a delayed translocation inhibitor of SARS-CoV-2 replication. Mol. Cell. 2021;81:1548–1552. doi: 10.1016/j.molcel.2021.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gordon C.J., Tchesnokov E.P., Woolner E., Perry J.K., Feng J.Y., Porter D.P., Götte M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020;295:6785–6797. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lo M.K., Jordan R., Arvey A., Sudhamsu J., Shrivastava-Ranjan P., Hotard A.L., Flint M., McMullan L.K., Siegel D., Clarke M.O., et al. GS-5734 and its parent nucleoside analog inhibit Filo-, Pneumo-, and Paramyxoviruses. Sci. Rep. 2017;7:43395. doi: 10.1038/srep43395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sheahan T.P., Sims A.C., Graham R.L., Menachery V.D., Gralinski L.E., Case J.B., Leist S.R., Pyrc K., Feng J.Y., Trantcheva I., et al. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 2017;9:eaal3653. doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Warren T.K., Jordan R., Lo M.K., Ray A.S., Mackman R.L., Soloveva V., Siegel D., Perron M., Bannister R., Hui H.C., et al. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., et al. Remdesivir for the treatment of COVID-19–Final report. N. Engl. J. Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldman J.D., Lye D.C.B., Hui D.S., Marks K.M., Bruno R., Montejano R., Spinner C.D., Galli M., Ahn M.Y., Nahass R.G., et al. Remdesivir for 5 or 10 days in patients with severe COVID-19. N. Engl. J. Med. 2020;383:1827–1837. doi: 10.1056/NEJMoa2015301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spinner C.D., Gottlieb R.L., Criner G.J., Arribas López J.R., Cattelan A.M., Soriano Viladomiu A., Ogbuagu O., Malhotra P., Mullane K.M., Castagna A., et al. Effect of remdesivir vs standard care on clinical status at 11 days in patients with moderate COVID-19: A randomized clinical trial. JAMA. 2020;324:1048–1057. doi: 10.1001/jama.2020.16349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gottlieb R.L., Vaca C.E., Paredes R., Mera J., Webb B.J., Perez G., Oguchi G., Ryan P., Nielsen B.U., Brown M., et al. Early remdesivir to prevent progression to severe COVID-19 in outpatients. N. Engl. J. Med. 2022;386:305–315. doi: 10.1056/NEJMoa2116846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stevens L.J., Pruijssers A.J., Lee H.W., Gordon C.J., Tchesnokov E.P., Gribble J., George A.S., Hughes T.M., Lu X., Li J., et al. Mutations in the SARS-CoV-2 RNA dependent RNA polymerase confer resistance to remdesivir by distinct mechanisms. Sci. Transl. Med. 2022:eabo0718. doi: 10.1126/scitranslmed.abo0718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Checkmahomed L., Carbonneau J., Du Pont V., Riola N.C., Perry J.K., Li J., Paré B., Simpson S.M., Smith M.A., Porter D.P., et al. In vitro selection of remdesivir-resistant SARS-CoV-2 demonstrates high barrier to resistance. Antimicrob. Agents Chemother. 2022;66:e0019822. doi: 10.1128/aac.00198-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szemiel A.M., Merits A., Orton R.J., MacLean O.A., Pinto R.M., Wickenhagen A., Lieber G., Turnbull M.L., Wang S., Furnon W., et al. In vitro selection of remdesivir resistance suggests evolutionary predictability of SARS-CoV-2. PLoS Pathog. 2021;17:e1009929. doi: 10.1371/journal.ppat.1009929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carbo E.C., Mourik K., Boers S.A., Munnink B.O., Nieuwenhuijse D., Jonges M., Welkers M.R.A., Matamoros S., van Harinxma Thoe Slooten J., Kraakman M.E.M., et al. A comparison of five Illumina, Ion Torrent, and nanopore sequencing technology-based approaches for whole genome sequencing of SARS-CoV-2. Eur. J. Clin. Microbiol. Infect. Dis. 2023;42:701–713. doi: 10.1007/s10096-023-04590-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quick J. nCoV-2019 Sequencing Protocol v3 (LoCost) V.3. Protocols.io. 2020. [(accessed on 1 May 2022)]. Available online: https://www.protocols.io/view/ncov-2019-sequencing-protocol-v3-locost-bp2l6n26rgqe/v3.
- 16.Tyson J.R., James P., Stoddart D., Sparks N., Wickenhagen A., Hall G., Choi J.H., Lapointe H., Kamelian K., Smith A.D., et al. Improvements to the ARTIC multiplex PCR method for SARS-CoV-2 genome sequencing using nanopore. bioRxiv. 2020 doi: 10.1101/2020.09.04.283077. bioRxiv: 2020.09.04.283077. [DOI] [Google Scholar]
- 17.Gonzalez-Reiche A.S., Hernandez M.M., Sullivan M.J., Ciferri B., Alshammary H., Obla A., Fabre S., Kleiner G., Polanco J., Khan Z., et al. Introductions and early spread of SARS-CoV-2 in the New York City area. Science. 2020;369:297–301. doi: 10.1126/science.abc1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee W.P., Stromberg M.P., Ward A., Stewart C., Garrison E.P., Marth G.T. MOSAIK: A hash-based algorithm for accurate next-generation sequencing short-read mapping. PLoS ONE. 2014;9:e90581. doi: 10.1371/journal.pone.0090581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie X., Muruato A.E., Zhang X., Lokugamage K.G., Fontes-Garfias C.R., Zou J., Liu J., Ren P., Balakrishnan M., Cihlar T., et al. A nanoluciferase SARS-CoV-2 for rapid neutralization testing and screening of anti-infective drugs for COVID-19. Nat. Commun. 2020;11:5214. doi: 10.1038/s41467-020-19055-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y., Song W., Chen S., Yuan Z., Yi Z. A bacterial artificial chromosome (BAC)-vectored noninfectious replicon of SARS-CoV-2. Antiviral Res. 2021;185:104974. doi: 10.1016/j.antiviral.2020.104974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pitts J., Li J., Perry J.K., Du Pont V., Riola N., Rodriquez L., Lu X., Kurhade C., Xie X., Camus G., et al. Remdesivir and GS-441524 retain antiviral activity against Delta, Omicron, and other emergent SARS-CoV-2 variants. Antimicrob. Agents Chemother. 2022;66:e0022222. doi: 10.1128/aac.00222-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malone B., Perry J.K., Olinares P.D.B., Lee H.W., Chen J., Appleby T.C., Feng J.Y., Bilello J.P., Ng H., Sotiris J., et al. Structural basis for substrate selection by the SARS-CoV-2 replicase. Nature. 2023;614:781–787. doi: 10.1038/s41586-022-05664-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin R., Li J., Parvangada A., Perry J., Cihlar T., Mo H., Porter D., Svarovskaia E. Genetic conservation of SARS-CoV-2 RNA replication complex in globally circulating isolates and recently emerged variants from humans and minks suggests minimal pre-existing resistance to remdesivir. Antiviral Res. 2021;188:105033. doi: 10.1016/j.antiviral.2021.105033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fontanet A., Autran B., Lina B., Kieny M.P., Karim S.S.A., Sridhar D. SARS-CoV-2 variants and ending the COVID-19 pandemic. Lancet. 2021;397:952–954. doi: 10.1016/S0140-6736(21)00370-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mahase E. COVID-19: What new variants are emerging and how are they being investigated? BMJ. 2021;372:n158. doi: 10.1136/bmj.n158. [DOI] [PubMed] [Google Scholar]
- 27.van Oosterhout C., Hall N., Ly H., Tyler K.M. COVID-19 evolution during the pandemic-Implications of new SARS-CoV-2 variants on disease control and public health policies. Virulence. 2021;12:507–508. doi: 10.1080/21505594.2021.1877066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., COVID-19 Genomics UK (COG-UK) Consortium et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bojkova D., Widera M., Ciesek S., Wass M.N., Michaelis M., Cinatl J., Jr. Reduced interferon antagonism but similar drug sensitivity in Omicron variant compared to Delta variant SARS-CoV-2 isolates. Cell Res. 2022;32:319–321. doi: 10.1038/s41422-022-00619-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vangeel L., De Jonghe S., Maes P., Slechten B., Raymenants J., André E., Leyssen P., Neyts J., Jochmans D. Remdesivir, molnupiravir and nirmatrelvir remain active against SARS-CoV-2 Omicron and other variants of concern. Antiviral Res. 2022;198:105252. doi: 10.1016/j.antiviral.2022.105252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown S.M., Katz M.J., Ginde A.A., Juneja K., Ramchandani M., Schiffer J.T., Vaca C., Gottlieb R.L., Tian Y., Elboudwarej E., et al. Consistent effects of early remdesivir on symptoms and disease progression across at-risk outpatient subgroups: Treatment effect heterogeneity in PINETREE study. Infect. Dis. Ther. 2023;12:1189–1203. doi: 10.1007/s40121-023-00789-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mackman R.L., Kalla R.V., Babusis D., Pitts J., Barrett K.T., Chun K., Du Pont V., Rodriguez L., Moshiri J., Xu Y., et al. Discovery of GS-5245 (obeldesivir), an oral prodrug of nucleoside GS-441524 that exhibits antiviral efficacy in SARS-CoV-2-infected African green monkeys. J. Med. Chem. 2023;66:11701–11717. doi: 10.1021/acs.jmedchem.3c00750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Focosi D., Maggi F., McConnell S., Casadevall A. Very low levels of remdesivir resistance in SARS-CoV-2 genomes after 18 months of massive usage during the COVID19 pandemic: A GISAID exploratory analysis. Antiviral Res. 2022;198:105247. doi: 10.1016/j.antiviral.2022.105247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hedskog C., Rodriguez L., Roychoudhury P., Huang M.L., Jerome K.R., Hao L., Ireton R.C., Li J., Perry J.K., Han D., et al. Viral resistance analyses from the remdesivir Phase 3 Adaptive COVID-19 Treatment Trial-1 (ACTT-1) J. Infect. Dis. 2023;228:1263–1273. doi: 10.1093/infdis/jiad270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Owusu D., Pomeroy M.A., Lewis N.M., Wadhwa A., Yousaf A.R., Whitaker B., Dietrich E., Hall A.J., Chu V., Thornburg N., et al. Persistent SARS-CoV-2 RNA shedding without evidence of infectiousness: A cohort study of individuals with COVID-19. J. Infect. Dis. 2021;224:1362–1371. doi: 10.1093/infdis/jiab107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou C., Zhang T., Ren H., Sun S., Yu X., Sheng J., Shi Y., Zhao H. Impact of age on duration of viral RNA shedding in patients with COVID-19. Aging. 2020;12:22399–22404. doi: 10.18632/aging.104114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu K., Chen Y., Yuan J., Yi P., Ding C., Wu W., Li Y., Ni Q., Zou R., Li X., et al. Factors associated with prolonged viral RNA shedding in patients with coronavirus disease 2019 (COVID-19) Clin. Infect. Dis. 2020;71:799–806. doi: 10.1093/cid/ciaa351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Showers W.M., Leach S.M., Kechris K., Strong M. Longitudinal analysis of SARS-CoV-2 spike and RNA-dependent RNA polymerase protein sequences reveals the emergence and geographic distribution of diverse mutations. Infect. Genet. Evol. 2022;97:105153. doi: 10.1016/j.meegid.2021.105153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nesterenko P.A., McLaughlin J., Tsai B.L., Burton Sojo G., Cheng D., Zhao D., Mao Z., Bangayan N.J., Obusan M.B., Su Y., et al. HLA-A∗02:01 restricted T cell receptors against the highly conserved SARS-CoV-2 polymerase cross-react with human coronaviruses. Cell Rep. 2021;37:110167. doi: 10.1016/j.celrep.2021.110167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gandhi S., Klein J., Robertson A., Peña-Hernández M.A., Lin M.J., Roychoudhury P., Lu P., Fournier J., Ferguson D., Mohamed Bakhash S.A., et al. De novo emergence of a remdesivir resistance mutation during treatment of persistent SARS-CoV-2 infection in an immunocompromised patient: A case report. Nat. Commun. 2021;13:1547. doi: 10.1038/s41467-022-29104-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hogan J.I., Duerr R., Dimartino D., Marier C., Hochman S., Mehta S., Wang G., Heguy A. Remdesivir resistance in transplant recipients with persistent COVID-19. Clin. Infect. Dis. 2023;76:342–345. doi: 10.1093/cid/ciac769. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available on request from the corresponding author.