Abstract
Two proteins, VP19C (50,260 Da) and VP23 (34,268 Da), make up the triplexes which connect adjacent hexons and pentons in the herpes simplex virus type 1 capsid. VP23 was expressed in Escherichia coli and purified to homogeneity by Ni-agarose affinity chromatography. In vitro capsid assembly experiments demonstrated that the purified protein was functionally active. Its physical status was examined by differential scanning calorimetry, ultracentrifugation, size exclusion chromatography, circular dichroism, fluorescence spectroscopy, and 8-anilino-1-naphthalene sulfonate binding studies. These studies established that the bacterially expressed VP23 exhibits properties consistent with its being in a partially folded, molten globule state. We propose that the molten globule represents a functionally relevant intermediate which is necessary to allow VP23 to undergo interaction with VP19C in the process of capsid assembly.
Herpes simplex virus type 1 (HSV-1) is the prototypical herpesvirus and is among the largest animal viruses known. Its complex virions have a characteristic multilayered structure comprising a nucleocapsid surrounded by a thick, amorphous protein layer (tegument) which is, in turn, enclosed by a glycoprotein-containing lipid envelope (26, 31). The capsid is a regular icosahedron with a diameter of 125 nm and is made up of 150 hexons with 12 pentons at the icosahedral vertices and 320 connecting triplexes (2, 28, 42). The outer shell of the capsid comprises four major proteins: VP5 (encoded by gene UL19 [16]), VP19C (UL38), VP23 (UL18), and VP26 (UL35). The products of a further two genes, UL26 (encoding a protease) and UL26.5 (encoding the main scaffolding protein, preVP22a), form the internal scaffold which is required for capsid assembly but is not present in mature, DNA-containing capsids (11, 26). The major capsid protein, VP5 (149 kDa), is the principal component of both hexons and pentons (20, 42), while VP26 (12 kDa) is located at the distal ends of hexon subunits (35, 41). One copy of VP19C (50 kDa) and two copies of VP23 (34 kDa) form the triplexes which lie between, and link, adjacent capsomers (20, 30).
In vitro assembly experiments have identified a possible precursor to the capsid which has been designated the procapsid (18, 19). The procapsid is a very open structure with only limited contacts between the various subunits. For instance, in typical capsids, horizontal extensions from the bases of the pentons and hexons meet to form the floor of the capsid shell. In procapsids, this floor is not present and the major contact between neighboring capsomers is through their adjacent triplexes (34). The procapsid is also a very unstable structure which dissociates spontaneously when cooled to 0°C. If incubated at room temperature, procapsids undergo spontaneous structural rearrangement into a more angular, polyhedral shape which closely resembles wild-type capsids in structure and stability. The contrast between the fragility of the procapsid and the durability of the wild-type capsid implies that rearrangement strengthens the initial tenuous interactions between the capsid subcomponents, resulting in much stronger interactions between its constituent subunits.
Assembly of virus capsids is a complex, multistep condensation process which requires the participating proteins to establish numerous and often extensive interactions. Little is known about the pathways involved in these condensations or about the manner in which individual protein molecules adopt their final positions and conformations within capsids. In part, this is because of difficulties inherent in analyzing the often unstable or transient intermediate stages. Thus, although the three-dimensional structures of capsids from several diverse virus families have been determined to atomic resolution, relatively few capsid proteins’ structures are known outside the context of the capsid or of subcapsid assemblies.
Here we report the application of a combination of biophysical techniques to examine the structure of the purified HSV-1 triplex protein, VP23. The data obtained suggest that VP23 exists predominantly in the form of a molten globule—a compact, folded, but highly flexible structure which has been implicated in protein folding pathways (23). These findings are discussed in relation to the role of this protein in capsid assembly.
MATERIALS AND METHODS
Expression plasmid construction.
To construct recombinant plasmid pETul18, which expresses a 6×His-tagged form of VP23, the UL18 open reading frame was isolated as a PCR product by using primers 1 and 2. Primer 1 is complementary to nucleotides 36,021 to 36,048 in the HSV-1 genome (16) and contains an EcoRI site at its 5′ end. Primer 2 is complementary to nucleotides 35,083 to 35,105 and contains an XbaI site at its 5′ end. The sequences are as follows: primer 1, 5′GACAGAATTCTGGCGGACGGCTTTGAAACTGACATCG (the EcoRI site is underlined); primer 2, 5′GACATCTAGATCTAGCCGGGCCTTAGGGATAGC (the XbaI site is underlined). The purified PCR fragment was digested with EcoRI and XbaI and ligated into EcoRI/SpeI-digested pET28mod. pET28mod is a derivative of pET28a (Novagen Ltd.) in which sequences between the NdeI and EcoRI sites have been removed and replaced with oligonucleotide A, which contains several restriction enzyme sites, including novel EcoRI and SpeI sites. The sequence of oligonucleotide A is 5′ TATGGGAAT TCCGGATCCACTAGTACACCC T TAAGGCC TAGGTGATCATG T TAA5′ (the EcoRI and SpeI sites, respectively, are underlined). Insertion of the PCR product into pET28mod generated pETul18, which has the 6×His and thrombin sequences from pET28 fused in frame to amino acids 2 to 318 of VP23. The resulting fusion protein is designated VP23His.
Purification of VP23His.
Escherichia coli BL21 DE3 cells were electroporated in the presence of pETul18 and incubated at 37°C, in 750 ml of Luria-Bertani medium containing 50 μg of kanamycin per ml, to an optical density of 0.6 at 630 nm. Expression of VP23His was induced by the addition of 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), followed by a further 16 h of incubation at 18°C. Cells were harvested by centrifugation at 3,000 rpm for 15 min in a Sorvall SLA-3000 rotor (Dupont). The pellet was resuspended in 20 ml of sonication buffer (20 mM Tris, 10% glycerol, 0.1% Nonidet P-40, pH 7.5), sonicated for 10 × 15 s by using a probe sonicator (Soniprep 150), and centrifuged at 10,000 rpm for 15 min in a Sorvall SS34 rotor. The supernatant was mixed for 1 h at room temperature with 1 ml of nickel agarose resin (Qiagen) which had been equilibrated in sonication buffer. The resin was placed in a Bio-Rad Poly-Prep chromatography column (0.8 by 4 cm) and washed sequentially with 4 × 20 ml of sonication buffer containing 0, 50, 100, and 200 mM imidazole, respectively. The 200 mM imidazole fractions contained VP23His that had been purified to homogeneity, as assessed by Coomassie brilliant blue staining of sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels. For most biophysical experiments, the purified protein was dialyzed against 20 mM sodium phosphate buffer (pH 7.5) containing 0.004% Nonidet P-40 (buffer P).
In vitro capsid assembly.
Lysates of Sf21 cells, infected singly at 5 PFU/cell with recombinant baculoviruses expressing the six HSV-1 capsid protein genes (32), were prepared in phosphate-buffered saline as described by Newcomb et al. (19). In vitro capsid assembly was performed by mixing the extracts, which were then incubated overnight at 28°C. In experiments done to test the function of bacterially expressed VP23His, the baculovirus VP23 extract was omitted from the mixture and replaced with an equal volume of purified VP23His in sonication buffer. Following incubation, capsids were pelleted at 30,000 × g in a Beckman TLS-55 rotor.
Ultracentrifugation.
Analytical centrifugation experiments were performed by using Beckman Optima XL-A and XL-I analytical centrifuges with absorption and Rayleigh interference optics. Both centrifuges have full on-line computer data capture and analysis facilities (Beckman, Palo Alto, Calif.). For sedimentation equilibrium experiments, double-sector cells with a 12-mm optical path length were loaded with 100 μl of buffer P and an 80-μl sample, in the solvent and sample channels, respectively, and run at 5°C. Two independent methods of average molecular mass analysis were employed: MSTARA (absorption optics) and MSTARI (interference optics), which use the computerized M* method (5, 6).
Size exclusion chromatography.
Chromatography was carried out on a 25-ml (1 by 30 cm) Superose 12 gel filtration column (Pharmacia) in 20 mM Tris (pH 8.0)–150 mM NaCl–250 mM EDTA–0.1% Tween 80. The column was run at 0.5 ml/min. Protein size markers, β-amylase (Mr, 200,000), alcohol dehydrogenase (Mr, 150,000), bovine serum albumin (Mr, 66,000), ovalbumin (Mr, 45,000), and carbonic anhydrase (Mr, 29,000) from Sigma, were analyzed in the same buffer.
CD.
Circular-dichroism (CD) spectra were obtained on a Jasco-600 spectropolarimeter (Japan Spectroscopic Co., Tokyo, Japan). Near-UV CD spectra (320 to 260 nm) were collected by using a cylindrical quartz cell with a path length of 0.5 cm. The protein concentration was 1.5 mg/ml. Far-UV CD spectra (260 to 190 nm) were obtained by using cylindrical quartz cells with a path length of 0.05 cm. Secondary structure was estimated by using the CONTIN procedure (22). The protein was at a concentration of 0.2 mg/ml in buffer P, and all measurements were recorded at 25°C.
Calorimetry.
Calorimetric measurements were carried out with an MC-2 precision differential scanning microcalorimeter (Microcalc) with a cell volume of 1.5 ml. The rate of heating was 1°C/min, and the excess pressure was kept at 8 × 106 Pa. Protein was used at concentrations in the range of 0.5 to 2.0 mg/ml in buffer P. The molar heat capacity of the protein was estimated by comparison with duplicate samples containing identical buffer from which the protein had been omitted.
Fluorescence and ANS binding.
Fluorescence measurements were recorded on a Perkin-Elmer LS-50B spectrofluorimeter in a 1-ml semimicrocuvette with a 1-cm path length at 25°C. For protein fluorescence, the excitation wavelength was 295 nm and the emission spectra were recorded between 310 and 380 nm. For 8-anilino-1-naphthalenesulfonate (ANS) fluorescence, the excitation wavelength was 370 nm and the emission spectra were recorded between 440 and 540 nm. ANS was added to protein samples to a final concentration of 20 μM. The protein concentration in all experiments was 0.2 mg/ml in buffer P.
RESULTS
Purified VP23His functions in in vitro capsid assembly.
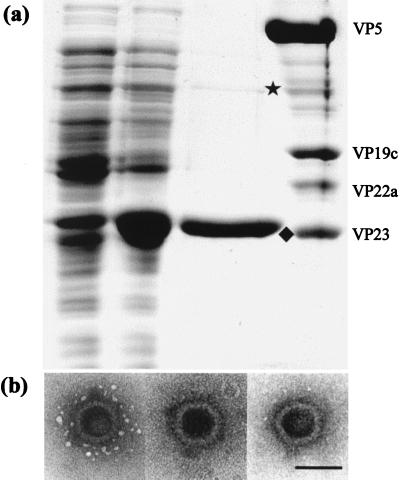
Purified VP23His was readily obtained following Ni-agarose affinity chromatography. The protein was over 95% pure as determined by Coomassie brilliant blue staining (Fig. 1A). The minor band, of approximately 70 kDa, has been shown by Western blotting to be dimeric VP23His, which is resistant to the denaturation conditions used (data not shown). It was important for interpretation of the characterizations described below to establish that the purified, bacterially expressed VP23His was functional. In vitro capsid assembly was therefore carried out. Extracts of infected Sf21 cells containing HSV capsid proteins VP5, VP19C, and pre-VP22a and the UL26 protease were mixed and incubated together, either in the absence of any other proteins, following addition of the purified VP23His, or following addition of a further baculovirus extract containing wild-type VP23. No HSV capsids were detected in the samples lacking any VP23, but characteristic HSV capsids were formed in both of the other samples following incubation at 28°C. Consistently higher numbers of capsids were seen with purified VP23His than with baculovirus extracts containing approximately equivalent amounts of VP23 (as determined by Coomassie brilliant blue staining), and the capsids appeared to have the typical HSV B-capsid structure (Fig. 1B). This confirmed that the bacterially expressed VP23His is capable of interacting normally and efficiently with the other capsid proteins and that the presence of the 6×His tag does not affect its function.
FIG. 1.
Purification and functional analysis of bacterially expressed VP23His. (A) A 10% polyacrylamide gel showing Coomassie brilliant blue-stained profiles of uninduced BL21 cells transformed with pETul18 (lane 1), cells after induction for 16 h with 0.1 mM IPTG (lane 2), the fraction eluted from Ni-agarose by 200 mM imidazole (lane 3), and purified B capsids (lane 4). The positions of monomeric (⧫) and dimeric (★) VP23His are indicated. The B capsid proteins are marked to the right of the gel. (B) In vitro capsid assembly. Extracts of Sf21 cells infected with recombinant baculoviruses expressing genes UL19, UL26, UL26.5, and UL38 were mixed with purified VP23His and incubated at 28°C for 18 h as described in the text. The capsids were pelleted at 30,000 × g, resuspended in phosphate-buffered saline, stained with 1% phosphotungstic acid, and examined by electron microscopy. Bar, 100 nm.
Hydrodynamic properties of VP23His.
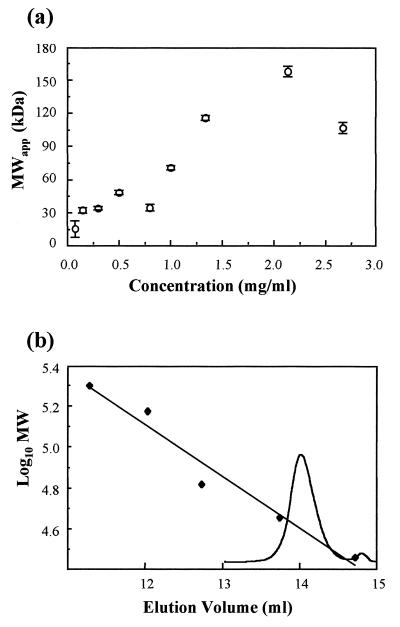
Equilibrium sedimentation was used to determine the apparent molecular mass of purified VP23His over a range of different concentrations (Fig. 2A). The results show that VP23His sedimented with a molecular mass of approximately 34 to 35 kDa at protein concentrations of up to 0.8 mg/ml. The low value of 15 kDa observed at 0.05 mg/ml is probably due to high noise levels found at low protein concentrations. The value of 34 to 35 kDa is in good agreement with that of 36.6 kDa calculated from the amino acid composition of VP23His, suggesting that the protein was predominantly monomeric under these conditions. At VP23His concentrations above 0.8 mg/ml, there was a progressive increase in apparent molecular mass from around 70 kDa at 1.0 mg/ml to between 100 and 160 kDa at higher concentrations. The increase in apparent molecular mass indicates a tendency for VP23His to associate into oligomers at increasing concentrations. Since each of the concentrations analyzed was prepared by dilution from a more concentrated stock sample, it is clear that the oligomerization is a reversible process.
FIG. 2.
Hydrodynamics. (a) Concentration dependence of apparent average molecular mass (MWapp) of VP23His. Purified VP23His in pH 7.5, 20 mM sodium phosphate–0.004% Nonidet P-40 was centrifuged for 16 h at 5°C in a Beckman analytical ultracentrifuge with a rotor speed of 15,000 rpm. (b) Size exclusion chromatography of VP23His. VP23His (0.5 mg/ml) was analyzed on a 25-ml (1 by 30 cm) Superose 12 gel filtration column (Pharmacia) in 20 mM Tris (pH 8.0)–150 mM NaCl–250 mM EDTA–0.1% Tween 80. The column was run at 0.5 ml/min. Protein size markers, β-amylase (Mr, 200,000), alcohol dehydrogenase (Mr, 150,000), bovine serum albumin (Mr, 66,000), ovalbumin (Mr, 45,000), and carbonic anhydrase (Mr, 29,000) from Sigma, were analyzed in the same buffer. The elution profile of VP23His is shown superimposed on the standard curve.
Purified VP23His was also analyzed by size exclusion chromatography (Fig. 2B). A single prominent peak with a calculated size of 36 kDa was detected. Again, this corresponds closely to the calculated size of VP23His, suggesting that the protein is monomeric under these conditions. This peak was confirmed as containing VP23His by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis (data not shown).
Isolated VP23His is a folded protein.
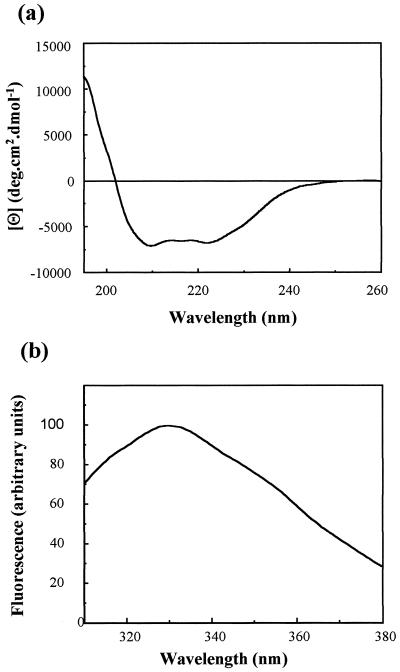
Far-UV CD can be used to examine the secondary structure content of proteins, since the different types of regular structure (α-helix, β-sheet, etc.) give rise to characteristically different spectra in this region (13). The far-UV CD spectrum of VP23His (Fig. 3A) shows that the protein has a significant amount of secondary structure, estimated as 24% α-helix and 30% β-sheet by applying the CONTIN procedure (22) to the data over the range of 240 to 195 nm. These estimates can be compared with the predicted values of 34% α-helix and 20% β-sheet obtained by applying the secondary structure prediction program PHD (27) to the amino acid sequence of the protein. The agreement between the experimental and predicted values can be regarded as satisfactory. It should be noted that the experimental values can be affected by (i) small errors in the determination of the protein concentration and (ii) the inability to use data below 195 nm because of the high level of noise in this region. In addition, the reliability of the predicted values will be affected by the degree of similarity between the VP23 protein and other proteins in the structural database.
FIG. 3.
Far-UV CD (a) and fluorescence (b) spectra of purified VP23His. Purified VP23His at 0.2 mg/ml in 20 mM sodium phosphate buffer (pH 7.5)–0.004% Nonidet P-40 was examined by far-UV CD and fluorescence spectroscopy as described in the text. Both traces were corrected for the effect of the buffer by subtraction of a buffer control spectrum. deg, degrees.
Fluorescence spectroscopy can be used to assess the extent to which tryptophan side chains are buried within a protein interior (9). The emission spectrum of VP23His showed an emission maximum (λmax) at 332.5 nm, significantly blue shifted from the value (356 nm) for tryptophan in small model compounds. This clearly demonstrates that the single Trp side chain in VP23His is at least partially buried within the interior of the folded protein (Fig. 3B).
Conformational stability of VP23His.
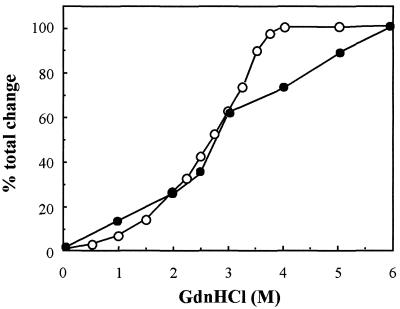
Exposure to high concentrations of a strong denaturant such as urea or guanidinium chloride (GdnHCl) usually causes the protein to adopt a random coil conformation. Therefore, GdnHCl-induced unfolding was used to assess the stability of VP23His. As the concentration of GdnHCl was raised from 0 to 6 M, the unfolding of the protein was monitored by fluorescence spectroscopy and far-UV CD. Fluorescence measurements showed that as the GdnHCl concentration was increased, the buried side chain of the Trp residue of VP23His became exposed to the solvent. The unfolding transition detected by fluorescence intensity measurement takes place largely between 1.5 and 3.5 M GdnHCl (Fig. 4, ○). The end point seen at 4 M GdnHCl probably corresponds to complete exposure of the previously buried Trp side chain (as monitored by fluorescence determination). Far-UV CD measurements showed that this exposure was accompanied by the loss of a large proportion (over 70%) of the secondary structure (Fig. 4, •). The continuing change in the far-UV CD signal up to 6 M GdnHCl probably represents further unfolding of local secondary structural elements. The pattern of the changes in secondary structure and in the degree of exposure of the Trp residue is in marked contrast to the highly cooperative transitions seen with most proteins (13), which arise from the concerted loss of the interactions stabilizing the native tertiary structure. The absence of such transitions is consistent with the proposal that VP23His, although folded and possessing secondary structure, lacks defined tertiary structure.
FIG. 4.
Structural stability of VP23. VP23 (0.2 mg/ml) in 20 mM sodium phosphate buffer (pH 7.5)–0.004% Nonidet P-40 was incubated in increasing GdnHCl concentrations of 0 to 6 M for 15 min at room temperature. Fluorescence and CD spectra were recorded as described in the text. The observed change from the native protein (0 M GdnHCl) in intensity at 325 nm (fluorescence) or ellipticity at 225 nm (CD) at each concentration was expressed as a percentage of the total change in intensity (○) or ellipticity (•) induced by 6 M GdnHCl and plotted against the concentration of GdnHCl.
VP23His has a flexible tertiary structure.
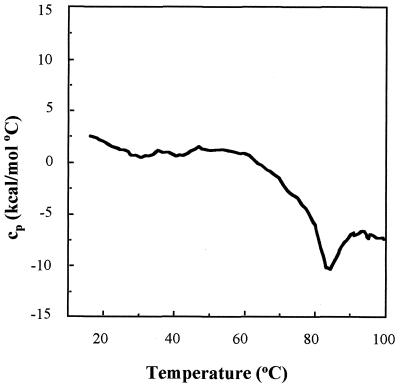
Temperature-induced denaturation of a protein molecule can also be used to assess its structure. The presence of an endothermic transition on a plot of heat capacity versus temperature, is usually considered to result from the cooperative melting of tertiary structure (21). Differential scanning calorimetry (DSC) can therefore be used to analyze the conformational stability of a folded protein. Figure 5 shows the DSC profile obtained with VP23His. The absence of a heat absorption peak indicates that no endothermic transition occurred over the temperature range employed (20 to 100°C). The decline in heat capacity above 60°C is due to the irreversible exothermic aggregation of the protein. Similar results were obtained over a protein concentration range of 0.5 to 2 mg/ml. Thus, no excess energy was being absorbed by the purified VP23His, indicating that endothermic unfolding was not occurring. This result again suggested that purified VP23His does not possess a rigid tertiary structure.
FIG. 5.
Differential scanning calorimetry of VP23His. The heat absorption profile was obtained by heating a solution of purified VP23His at a concentration of 0.7 mg/ml in 20 mM sodium phosphate buffer (pH 7.5)–0.004% Nonidet P-40 through a range of 20 to 100°C. The scan rate was 1°C/min. cp, heat capacity.
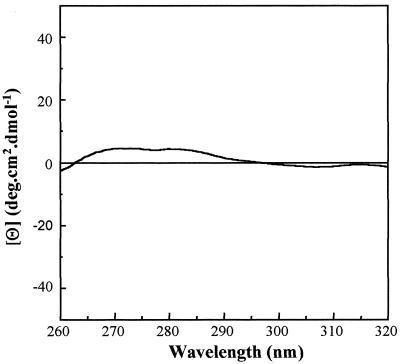
The near-UV CD spectrum of proteins is generated by aromatic amino acid residues (particularly Trp) and is strongly influenced by their local environment (13). The intensity of a CD band reflects the mobility of the aromatic ring and is determined by its surroundings, in particular, whether it is interacting with neighboring aromatic residues. The near-UV CD spectrum therefore reflects the degree to which stable interactions occur between amino acids and provides a measure of the extent of the defined tertiary structure of a protein. For VP23His, the near-UV CD spectrum was a flat trace with no evidence of distinct peaks (Fig. 6). This is consistent with the proposals that tertiary interactions in VP23His are very weak and that the aromatic side chains in the protein do not occupy fixed positions.
FIG. 6.
Near-UV CD analysis of VP23His. Purified VP23His at 1.5 mg/ml in 20 mM sodium phosphate buffer (pH 7.5)–0.004% Nonidet P-40 was examined by near-UV CD (320 to 260 nm) spectroscopy as described in the text. The spectrum was corrected for the effect of the buffer by subtraction of a buffer control spectrum. deg, degrees.
VP23His is a molten globule.
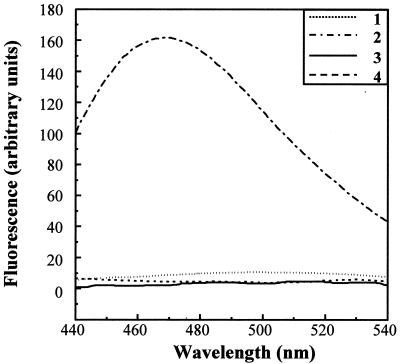
The DSC and near-UV CD data suggest that VP23His does not have a rigid tertiary structure. However, the results of far-UV CD show that the protein has substantial secondary structure, and fluorescence measurements indicate that the Trp side chain is hidden from the solvent. The most likely explanation is that the purified VP23His molecule is in a partially folded state, in which the main structural elements (α-helix and β-sheet) have formed but their geometrical relationship has not become fixed. This condition is characteristic of the molten globule state which has been proposed as an intermediate in protein folding (14, 23, 24). Binding of ANS to a protein (as shown by a characteristic increase in fluorescence at 470 nm following excitation at 370 nm) is generally considered to be a sensitive test for partial folding, since in this state, ANS has access to the hydrophobic interior of the protein (25). Although in some cases, ANS has been shown to bind to proteins in a native, non-molten globule state via solvent-accessible clusters of nonpolar groups, binding to the partially folded state is much stronger than that to either the native or fully unfolded state (29). For example, addition of ANS to a sample of purified major capsid protein VP5 (data not shown) or to purified VP23His which had been unfolded by 6 M GdnHCl did not lead to any increase in ANS fluorescence (Fig. 7). In contrast, ANS bound tightly to purified, undenatured VP23His, as shown by a strong increase in the intensity of fluorescence at 470 nm (Fig. 7). The ANS results therefore complement those from the other analyses described above and strongly support the molten globule model for VP23.
FIG. 7.
ANS binding. ANS was added to purified VP23His in the presence (curve 1) and absence (curve 2) of 6 M GdnHCl. The excitation wavelength was 370 nm, and fluorescence was measured between 440 and 540 nm. The protein concentration was 0.2 mg/ml, and ANS was added to a final concentration of 20 μM. Control fluorescence spectra were measured for ANS in buffer (curve 3) and for VP23His in the absence of ANS (curve 4).
DISCUSSION
Although the structures of the HSV capsid and its component subunits are becoming known in increasing detail, virtually nothing is known about the nature of the constituent proteins prior to their incorporation into the capsid. In this study, we have carried out biophysical analyses of highly purified VP23, an essential capsid component that forms part of the triplex.
According to the ultracentrifugation data shown in Fig. 2A, purified VP23His is monomeric at concentrations below 1 mg/ml but shows a tendency to form oligomers at higher concentrations. Size exclusion chromatography also demonstrated that VP23His is monomeric. Support for this conclusion comes from the observation that VP23 does not interact with itself in the yeast two-hybrid system (8). By contrast, Spencer et al. (30) have recently reported that unpurified VP23 in extracts from baculovirus-infected cells is dimeric. Interestingly, the presence of a dimer band on denaturing protein gels (Fig. 1A) also suggests a propensity of the protein to self-associate, even under unfavorable conditions. The reasons for the apparent variation in oligomeric status in these different assays are not clear. Differences in conditions, such as protein purity and concentration and the presence or absence of detergents, may affect the behavior of the protein. It is possible that the presence of the 6×His tag affects the oligomeric status and folding of VP23His. However, since VP23, both as a monomeric purified protein (Fig. 1B) and as a dimer in cell extracts (30), can assemble into capsids, any such differences do not appear to affect its function. Although the triplex contains two copies of VP23, it is not possible, by examining capsid structures at the present level of resolution, to determine whether the two copies are in direct contact with each other (40). The significance of any ability on the part of VP23 to dimerize therefore remains unclear.
The results of near- and far-UV CD, fluorescence, DSC, and ANS binding studies indicate that the bacterially expressed VP23His contains secondary structure but lacks a defined tertiary structure. This is one of the main characteristics of the molten globule state. A protein molecule in the molten globule state is almost as compact as in the fully folded state and has pronounced secondary structure. It differs from the fully folded molecule mainly in the absence of tight packing of the amino acid side chains in the protein core. As a result, the structure of the protein is much more mobile, allowing considerable movement of secondary structural elements with respect to each other. Many proteins adopt the molten globule state as an equilibrium intermediate under mild denaturing conditions, and it is believed to form as a kinetic intermediate during protein folding (14, 23). Absence of thermal transition was shown, for example, for molten globule forms of human α-fetoprotein (37) and apo-α-lactalbumin (39). The physiological importance of the molten globule state has been demonstrated in a number of different circumstances (3, 7, 12, 15, 38). Molten globule-like states have not been widely described among viral structural proteins. A kinetic intermediate in the refolding of the wild-type coat protein of phage P22 has been described (33) which differed from the native state in its intrinsic fluorescence and binding of ANS, and a molten globule model has recently been proposed for the native scaffolding subunit that functions in P22 procapsid assembly (36).
At different stages in their life cycles, most viruses encounter changing and sometimes harsh environments during transmission from one host to another. During this transmission stage, the virus particle needs to retain its biological integrity and protect the nucleic acid genome. Consequently, many icosahedral virus capsids are robust entities which are capable of resisting considerable physical and chemical assaults. To achieve the necessary structural strength, capsid proteins frequently form extensive and intimate interactions, the creation of which may require the type of flexibility inherent in molten-globule-like intermediate states. For example, in the adenovirus hexon, the three copies of the major capsid protein interpenetrate to form a very compact structure (1). The degree of interaction seen in the adenovirus hexon must be achieved by extensive movements of large fractions of each protein, amounting to the mutually induced refolding of the individual protein subcomponents. Indeed, the nascent hexon protein must form a transient complex with another adenovirus protein which acts as its chaperone and maintains it in a configuration suitable for trimer assembly (4). Similarly, in the bluetongue virus capsid, a trimer is formed from three copies of VP7 which are tightly integrated to form a rigid monolithic structure (10). In this case, each VP7 molecule consists of an upper and a lower domain which are twisted with respect to each other so that the upper domain of one protein sits on top of the lower domain of the neighboring VP7. Here again, the ability of the newly synthesized VP7 proteins to undergo such comprehensive interaction suggests that their conformation must differ considerably from that seen in the trimer. These two examples suggest that the newly synthesized proteins must possess considerable inherent flexibility to allow the formation of such tightly integrated complexes, and it is perhaps significant that neither the bluetongue VP7 nor the adenovirus hexon protein has yet been crystallized as a monomer.
HSV capsids are also rather robust structures which are resistant to physical and chemical disruption. For example, treatment of B capsids with 2 M GdnHCl results in the loss of VP26 and the scaffolding proteins and also removes the pentons and peripentonal triplexes but leaves the hexagonal network of hexons and triplexes largely unaffected (20). However, exposure to 3 M GdnHCl (which causes a substantial loss of structure in VP23His [Fig. 4]) results in the disintegration of the capsid shell (17). Hexons and pentons are formed by six and five copies of VP5, respectively, and the triplexes are formed by two copies of VP23 and one copy of VP19C. Although the atomic structure is not known, examination of the HSV capsid at increasingly high resolutions down to 13Å (40) shows the triplex as a largely uniform, globular mass with little evidence of separate domains which could be attributed to the two copies of VP23 and one of VP19C present. This contrasts markedly with the clear separation between VP5 subunits that is evident in pentons and hexons and suggests that the three proteins which make up the triplex are more closely integrated than are the VP5 subunits. Interactions involving VP5 subunits affect relatively small regions of the protein and may therefore occur between molecules which have already adopted a predominantly folded form, while those involving VP23 affect large portions of the molecule and presumably require large-scale conformational shifts. It is interesting, therefore, that ANS binding suggests that VP23 is in molten globule form whereas VP5 is not. VP23 and VP19C can form complexes in the absence of any other capsid proteins, and the complexes formed are functionally active in in vitro capsid assembly experiments (unpublished data and reference 30). Folding of VP23 molecules into their final form is probably triggered by the presence of VP19C, which induces structural rearrangements that result in the extensive intermingling suggested by the apparent uniformity of the triplex. It is likely that triplex formation represents a very early step in the capsid assembly pathway and that the existence of free VP23 as a molten globule may represent a very short-lived stage immediately following its synthesis.
Clearly, assembly of a structure as complicated as a virus capsid is likely to require many types of protein interaction. However, the description of molten globule forms in the capsids of phage P22, and now in HSV, together with the high degree of molecular entanglement seen in other virus particle substructures, suggests that synthesis of certain proteins as flexible, partially folded intermediates represents a general mechanism through which they are able to form the complex and specific interactions necessary for capsid assembly.
ACKNOWLEDGMENTS
M. D. Kirkitadze and D. A. McClelland were supported by Human Frontier Science Program grant RG-537/96. CD analysis was performed at the Scottish Circular Dichroism Facility, Stirling University, Stirling, United Kingdom; DSC was performed at the Department of Chemistry, Glasgow University, Glasgow, United Kingdom; and ultracentrifugation was performed at the National Centre for Macromolecular Hydrodynamics, Nottingham University, Sutton Bonington Campus, Loughborough, United Kingdom. All of these facilities are supported by the BBSRC.
We thank Alan Cooper and Margaret Nutley for kind assistance with the calorimetry experiments and Kornelia Jumel and Stephan Harding for kind help with the ultracentrifugation experiments. We also thank David McNab and Jim Aitken for excellent technical assistance.
REFERENCES
- 1.Athappilly F K, Murali R, Rux J J, Cai Z, Burnett R M. The refined crystal structure of hexon, the major coat protein of adenovirus type 2, at 2.9 Å resolution. J Mol Biol. 1994;242:430–455. doi: 10.1006/jmbi.1994.1593. [DOI] [PubMed] [Google Scholar]
- 2.Booy F P, Newcomb W W, Trus B L, Brown J C, Baker T S, Steven A C. Liquid-crystalline, phage-like packing of encapsidated DNA in herpes simplex virus. Cell. 1991;64:1007–1015. doi: 10.1016/0092-8674(91)90324-r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bychkova V E, Ptitsyn O B. Folding intermediates are involved in genetic-diseases. FEBS Lett. 1995;359:6–8. doi: 10.1016/0014-5793(95)00004-s. [DOI] [PubMed] [Google Scholar]
- 4.Cepko C L, Sharp P A. Assembly of adenovirus major capsid protein is mediated by a nonvirion protein. Cell. 1982;31:407–415. doi: 10.1016/0092-8674(82)90134-9. [DOI] [PubMed] [Google Scholar]
- 5.Cölfen H, Harding S E. MSTARA and MSTARI: interactive PC algorithms for simple model independent evaluation of sedimentation equilibrium data. Eur Biophys J Biophys Lett. 1997;25:333–346. [Google Scholar]
- 6.Creeth J M, Harding S E. Some observations on a new type of point average molecular-weight. J Biochem Biophys Methods. 1982;7:25–34. doi: 10.1016/0165-022x(82)90033-1. [DOI] [PubMed] [Google Scholar]
- 7.de Jongh H H J, Killian J A, de Kruijff B. A water lipid interface induces a highly dynamic folded state in apocytochrome-c and cytochrome-c, which may represent a common folding intermediate. Biochemistry. 1992;31:1636–1643. doi: 10.1021/bi00121a008. [DOI] [PubMed] [Google Scholar]
- 8.Desai P, Person S. Molecular interactions between the HSV-1 capsid proteins as measured by the yeast two-hybrid system. Virology. 1996;220:516–521. doi: 10.1006/viro.1996.0341. [DOI] [PubMed] [Google Scholar]
- 9.Freifelder D. Physical biochemistry. 2nd ed. New York, N.Y: W. H. Freeman & Co.; 1982. [Google Scholar]
- 10.Grimes J, Basak A, Roy P, Stuart D. The crystal structure of bluetongue virus VP7. Nature. 1995;373:167–170. doi: 10.1038/373167a0. [DOI] [PubMed] [Google Scholar]
- 11.Homa F L, Brown J C. Capsid assembly and DNA packaging in herpes simplex virus. Med Virol. 1997;7:107–122. doi: 10.1002/(sici)1099-1654(199707)7:2<107::aid-rmv191>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 12.Hua Q X, Ladbury J E, Weiss M A. Dynamics of a monomeric insulin analog: testing the molten-globule hypothesis. Biochemistry. 1993;32:1433–1442. doi: 10.1021/bi00057a006. [DOI] [PubMed] [Google Scholar]
- 13.Kelly S M, Price N C. The application of circular dichroism to studies of protein folding and unfolding. Biochim Biophys Acta-Protein Struct Mol Enzymol. 1997;1338:161–185. doi: 10.1016/s0167-4838(96)00190-2. [DOI] [PubMed] [Google Scholar]
- 14.Kuwajima K. The molten globule state as a clue for understanding the folding and cooperativity of globular-protein structure. Proteins Struct Funct Genet. 1989;6:87–103. doi: 10.1002/prot.340060202. [DOI] [PubMed] [Google Scholar]
- 15.Martin J, Langer T, Boteva R, Schamel A, Horwich A L, Hartl F U. Chaperonin-mediated protein folding at the surface of GroEL through a molten globule-like intermediate. Nature. 1991;352:36–42. doi: 10.1038/352036a0. [DOI] [PubMed] [Google Scholar]
- 16.McGeoch D J, Dalrymple M A, Davison A J, Dolan A, Frame M C, McNab D, Perry L J, Scott J E, Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988;69:1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 17.Newcomb W W, Brown J C. Structure of the herpes simplex virus capsid: effects of extraction with guanidine hydrochloride and partial reconstitution of extracted capsids. J Virol. 1991;65:613–620. doi: 10.1128/jvi.65.2.613-620.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newcomb W W, Homa F L, Thomsen D R, Booy F P, Trus B L, Steven A C, Spencer J V, Brown J C. Assembly of the herpes simplex virus capsid: characterization of intermediates observed during cell-free capsid formation. J Mol Biol. 1996;263:432–446. doi: 10.1006/jmbi.1996.0587. [DOI] [PubMed] [Google Scholar]
- 19.Newcomb W W, Homa F L, Thomsen D R, Ye Z, Brown J C. Cell-free assembly of the herpes simplex virus capsid. J Virol. 1994;68:6059–6063. doi: 10.1128/jvi.68.9.6059-6063.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newcomb W W, Trus B L, Booy F P, Steven A C, Wall J S, Brown J C. Structure of the herpes simplex virus capsid: molecular composition of the pentons and the triplexes. J Mol Biol. 1993;232:499–511. doi: 10.1006/jmbi.1993.1406. [DOI] [PubMed] [Google Scholar]
- 21.Privalov P L, Potekhin S A. Scanning microcalorimetry in studying temperature-induced changes in proteins. Methods Enzymol. 1986;131:4–51. doi: 10.1016/0076-6879(86)31033-4. [DOI] [PubMed] [Google Scholar]
- 22.Provencher S W, Glöckner J. Estimation of globular protein secondary structure from circular dichroism. Biochemistry. 1981;20:33–37. doi: 10.1021/bi00504a006. [DOI] [PubMed] [Google Scholar]
- 23.Ptitsyn O B. Molten globule and protein folding. Adv Protein Chem. 1995;47:83–229. doi: 10.1016/s0065-3233(08)60546-x. [DOI] [PubMed] [Google Scholar]
- 24.Ptitsyn O B. Protein folding: hypotheses and experiments. J Protein Chem. 1987;6:273–293. [Google Scholar]
- 25.Ptitsyn O B, Pain R H, Semisotnov G V, Zerovnik E, Razgulyaev O I. Evidence for a molten globule state as a general intermediate in protein folding. FEBS Lett. 1990;262:20–24. doi: 10.1016/0014-5793(90)80143-7. [DOI] [PubMed] [Google Scholar]
- 26.Rixon F J. Structure and assembly of herpesviruses. Semin Virol. 1993;4:135–144. [Google Scholar]
- 27.Rost B, Sander C. Improved prediction of protein secondary structure by use of sequence profiles and neural networks. Proc Natl Acad Sci USA. 1993;90:7558–7562. doi: 10.1073/pnas.90.16.7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schrag J D, Prasad B V V, Rixon F J, Chiu W. Three-dimensional structure of the HSV-1 nucleocapsid. Cell. 1989;56:651–660. doi: 10.1016/0092-8674(89)90587-4. [DOI] [PubMed] [Google Scholar]
- 29.Semisotnov G V, Rodionova N A, Razgulyaev O I, Uversky V N, Gripas A F, Gilmanshin R I. Study of the molten globule intermediate state in protein folding by a hydrophobic fluorescent-probe. Biopolymers. 1991;31:119–128. doi: 10.1002/bip.360310111. [DOI] [PubMed] [Google Scholar]
- 30.Spencer J V, Newcomb W W, Thomsen D R, Homa F L, Brown J C. Assembly of the herpes simplex virus capsid: preformed triplexes bind to the nascent capsid. J Virol. 1998;72:3944–3951. doi: 10.1128/jvi.72.5.3944-3951.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steven A C, Spear P G. Herpesvirus capsid assembly and envelopment. In: Chiu W, Burnett R M, Garcea R, editors. Structural biology of viruses. New York, N.Y: Oxford University Press; 1997. pp. 312–351. [Google Scholar]
- 32.Tatman J D, Preston V G, Nicholson P, Elliott R M, Rixon F J. Assembly of herpes simplex virus type 1 capsids using a panel of recombinant baculoviruses. J Gen Virol. 1994;75:1101–1113. doi: 10.1099/0022-1317-75-5-1101. [DOI] [PubMed] [Google Scholar]
- 33.Teschke C M, King J. Folding of the phage P22 coat protein in vitro. Biochemistry. 1993;32:10839–10847. doi: 10.1021/bi00091a040. [DOI] [PubMed] [Google Scholar]
- 34.Trus B L, Booy F P, Newcomb W W, Brown J C, Homa F L, Thomsen D R, Steven A C. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J Mol Biol. 1996;263:447–462. doi: 10.1016/s0022-2836(96)80018-0. [DOI] [PubMed] [Google Scholar]
- 35.Trus B L, Homa F L, Booy F P, Newcomb W W, Thomsen D R, Cheng N, Brown J C, Steven A C. Herpes simplex virus capsids assembled in insect cells infected with recombinant baculoviruses: structural authenticity and localization of VP26. J Virol. 1995;69:7362–7366. doi: 10.1128/jvi.69.11.7362-7366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tuma R, Thomas G J. Mechanisms of virus assembly probed by Raman spectroscopy: the icosahedral bacteriophage P22. Biophys Chem. 1997;68:17–31. doi: 10.1016/s0301-4622(97)00001-x. [DOI] [PubMed] [Google Scholar]
- 37.Uversky V N, Kirkitadze M D, Narizhneva N V, Potekhin S A, Tomashevski A Y. Structural-properties of α-fetoprotein from human cord serum: the protein molecule at low pH possesses all the properties of the molten globule. FEBS Lett. 1995;364:165–167. doi: 10.1016/0014-5793(95)00379-n. [DOI] [PubMed] [Google Scholar]
- 38.van der Goot F G, Gonzalez-Manas J M, Lakey J H, Pattus F. A ‘molten-globule’ membrane-insertion intermediate of the pore-forming domain of colicin-A. Nature. 1991;354:408–410. doi: 10.1038/354408a0. [DOI] [PubMed] [Google Scholar]
- 39.Yutani K, Ogasahara K, Kuwajima K. Absence of the thermal transition in apo-α-lactalbumin in the molten globule state: a study by differential scanning microcalorimetry. J Mol Biol. 1992;228:347–350. doi: 10.1016/0022-2836(92)90824-4. [DOI] [PubMed] [Google Scholar]
- 40.Zhou Z H, Chiu W, Haskell K, Spears H J, Jakana J, Rixon F J, Scott L R. Refinement of herpesvirus B-capsid structure on parallel supercomputers. Biophys J. 1998;74:576–588. doi: 10.1016/S0006-3495(98)77816-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Z H, He J, Jakana J, Tatman J D, Rixon F J, Chiu W. Assembly of VP26 in herpes simplex virus-1 inferred from structures of wild-type and recombinant capsids. Nat Struct Biol. 1995;2:1026–1030. doi: 10.1038/nsb1195-1026. [DOI] [PubMed] [Google Scholar]
- 42.Zhou Z H, Prasad B V V, Jakana J, Rixon F J, Chiu W. Protein subunit structures in the herpes simplex virus A-capsid determined from 400 kV spot-scan electron cryomicroscopy. J Mol Biol. 1994;242:456–469. doi: 10.1006/jmbi.1994.1594. [DOI] [PubMed] [Google Scholar]