Abstract
Staphylococcus equorum WS 2733 was found to produce a substance exhibiting a bacteriostatic effect on a variety of gram-positive bacteria. The metabolite was purified to homogeneity by ammonium sulfate precipitation and semipreparative reversed-phase high-performance liquid chromatography. Electrospray mass spectrometry confirmed the high purity of the compound and revealed a molecular mass of 1,143 Da. By two-dimensional nuclear magnetic resonance spectroscopy the substance was identified as micrococcin P1 which is a macrocyclic peptide antibiotic that has not yet been reported for the genus Staphylococcus. A total of 95 out of 95 Listeria strains and 130 out of 135 other gram-positive bacteria were inhibited by this substance, while none of 37 gram-negative bacteria were affected. The antilisterial potential of this food-grade strain as a protective starter culture was evaluated by its in situ application in cheese-ripening experiments under laboratory conditions. A remarkable growth reduction of Listeria monocytogenes could be achieved compared to control cheese ripened with a nonbacteriocinogenic type strain of Staphylococcus equorum. In order to prove that inhibition was due to micrococcin P1, a micrococcin-deficient mutant was constructed which did not inhibit L. monocytogenes in cheese-ripening experiments.
For production of high-quality “red-smear cheese,” complex, undefined consortia composed of many species of bacteria and yeast from different genera are often used (53). Most species of these consortia belong to the “coryneform” bacteria, but many isolates have not yet been classified. Since contamination of red smear cheese with Listeria monocytogenes has caused considerable problems, several studies have been performed on the antilisterial effects of coryneform bacteria (11, 21, 30, 40, 50). Only one bacteriocin produced by Brevibacterium linens has been characterized at the molecular level so far (51, 52). This strain has also been evaluated with respect to its antilisterial potential in situ on model soft cheese under laboratory conditions (15). The bacteriolytic substance linenscin produced by B. linens OC2 was found to inhibit L. monocytogenes, but it was not classified as a bacteriocin (28) and its structure has never been published.
A smaller fraction of the red-smear cheese bacteria belong to genera other than coryneform bacteria, such as Staphylococcus or Micrococcus. By screening the surface ripening flora of German and French smeared cheeses with respect to their antilisterial potential, the highly inhibitory Staphylococcus equorum was isolated from the surface of a traditional French Raclette cheese, where it was found in cell numbers up to 108 per cm2 (the present study).
The genus Staphylococcus is known to produce many antibacterial substances (41, 42). According to Klaenhammer (27), these can be classified into four main groups: Lantibiotics (class I), low-molecular-weight antibiotic-like substances (class II), and high-molecular-weight (100,000 to 500,000 Da) bacteriocins (classes III and IV). Staphylococcins are class III bacteriocins in the classical sense that are sensitive to proteolytic enzymes, often being relatively heat stable (15 min, 120°C). In some cases they were found to be lipoprotein-carbohydrate complexes (class IV, e.g., staphylococcin 1580). Lantibiotics, such as gallidermin, epidermin, Pep5, epicidin, or epilancin, are low-molecular-weight polypeptide antibiotics which are characterized by the presence of unusual amino acids (lanthionine, β-methyllanthionine, forming intramolecular thioether bridges) and a high content of unsaturated amino acids (dehydroalanine and dehydrobutyrine [23, 24, 26, 44]). Class I, III, and IV molecules are ribosomally synthesized as precursor peptides which are sometimes posttranslationally modified and processed (26). In the genus Staphylococcus, some low-molecular-mass (1,200 to 1,400 Da), heat-stable, but non-lanthionine-containing antimicrobial peptides were found (class II), but their structure and their mode of biosynthesis is thus far unknown (7).
In this study S. equorum WS 2733 was shown to produce micrococcin P1, which is an acceptor-site-specific inhibitor of ribosomal protein biosynthesis (12, 35) reported for the first time in the genus Staphylococcus. A potential application of S. equorum as a protective starter culture was evaluated in ripening experiments performed with model soft cheese artificially contaminated with L. monocytogenes.
MATERIALS AND METHODS
Bacterial strains, media, and bacteriocin assay.
The micrococcin P1 producer strain S. equorum WS 2733 was isolated from French Raclette cheese using the HGMF (hydrophobic grid membrane filter) method described previously (11). About 20 cm2 (2 to 3 g) of the cheese surface was scraped off aseptically, homogenized in 100 ml of sterile Peptone-Tween diluent (containing 1.0 g of Peptone and 10.0 g of Tween 80 per liter) and filtered through HGMF membranes filter (QA Life Sciences, Inc., San Diego, Calif.). HGMF membranes were transferred grid-side-up to plate count agar supplemented with 3% NaCl (PC3+). After incubation at 30°C for 3 days, the membranes were transferred to plates of tryptose soft agar (TB; Merck, Darmstadt, Germany) with 8 g of agar/liter inoculated with 100 μl of a 24-h culture of the indicator strain Listeria ivanovii (WSLC 3061) to detect inhibitory effects. Colonies corresponding to inhibition zones were picked from the membranes and purified.
Micrococcin P1-deficient mutants were obtained by insertional mutagenesis. The putative micrococcin synthetase gene has been disrupted by integration of a tetracycline resistance cassette via homologous recombination (M. C. Carnio, K. P. Francis, and S. Scherer, submitted for publication). The wild-type strain, the micrococcin P1-deficient mutants, and the S. equorum type strain DSMZ 20674 were grown on slants of plate count agar (Merck) at 30°C and stored at 4°C. For individual experiments, the cells were subcultured in brain heart infusion broth (BHI; Merck) at 30°C.
Antibacterial activity was determined by the critical dilution method according to the method of Barefoot and Klaenhammer (2) and expressed in arbitrary units (AU) per milliliter. Serial twofold dilutions of sterile-filtered samples in 30 mM sodium phosphate buffer (pH 7.0) were spotted onto tryptose soft agar plates (TB) with 8 g of agar/liter inoculated with 100 μl of an overnight culture of the indicator strain L. ivanovii WSLC 3061 and incubated at 30°C for 24 h (spot-on-the-lawn assay). The titer is defined as the reciprocal of the highest dilution exhibiting complete and clear zones of inhibition on the indicator lawn. L. ivanovii WSLC 3061 and L. monocytogenes WSLC 1364 (Weihenstephan Listeria collection) were grown as described by Valdés-Stauber et al. (50).
For sensitivity screening performed with the sterile-filtered culture supernatant, we used a variety of bacterial strains from the Weihenstephan Culture Collection (WS) housed at the Institute of Microbiology, Forschungszentrum für Milch und Lebensmittel, Weihenstephan, Freising, Germany.
Isolation and purification of micrococcin P1.
Micrococcin P1 was purified from 20-liter cultures of S. equorum WS 2733 grown at 30°C in BHI broth (Merck) for 24 h. The cells were pelleted by centrifugation (12,000 × g, 20 min, 4°C). The antibiotic present in the culture supernatant was concentrated by ammonium sulfate precipitation. The pellet was resuspended in 650 ml of sodium phosphate buffer (50 mM, pH 7.0) and applied to a reversed-phase column (Pharmacia XK 16/70; 700 by 16 mm, silica gel 100 C18, 40 to 63 μm; Fluka) equilibrated with buffer A (0.1% [vol/vol] trifluoroacetic acid [TFA] H2O) at a flow rate of 1 ml/min. After a washing with 240 ml of buffer A the column was eluted at a flow rate of 4 ml/min with increasing amounts of buffer B (80% acetonitrile–20% H2O–0.1% TFA [vol/vol/vol]). The antibiotic-containing fractions were collected, pooled, and redissolved in 50% ethanol (fraction II). Final purification was achieved by reversed-phase high-performance liquid chromatography (RP-HPLC) on an RP column (Eurospher 100-C18, 7 μm, 250 by 8 mm; Knauer, Berlin, Germany) equilibrated with buffer A. Micrococcin P1 was eluted by a linear gradient of 0 to 80% buffer B in 30 min at a flow rate of 3.5 ml/min. All purification steps were performed at room temperature. The chromatographic equipment was obtained from Pharmacia-LKB (Gradi-Frac System; pump, P-50; detectors, Uvicord SII and VWM 2141; detection wavelengths, 280 and 220 nm, respectively) and Perkin-Elmer (Liquid Chromatograph Series 400). In order to monitor purity, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed using a discontinuous 8 to 18% gradient gel (Excel Gel; Pharmacia-LKB). One-half of the gel was silver stained according to the method of Blum et al. (5), while the other half was assayed directly for antimicrobial activity by the modified test described by Bhunia et al. (4).
GC-MS analysis.
The RP-HPLC-purified sample was hydrolyzed under N2 atmosphere in 6 N HCl at 110°C for 24 h. The hydrolyzate was derivatized to n-propyl esters with 3 N HCl in n-propanol (110°C, 10 min). After completion of the reaction the reagents were removed with N2. Reaction with trifluoroacetic anhydride (110°C, 10 min) subsequently gave the trifluoroacetamides. After removal of the reagents with N2, the sample was dissolved in toluene and analyzed on a Carlo-Erba 2900 gas chromatograph (GC). The separation was performed on a fused-silica capillary (20 by 0.25 mm, inner diameter; df ≈ 0.13 μm) coated with Chirasil-γ-Dex (19) as stationary chiral phase. The GC was coupled to a MAT 112S mass spectrometer (MS; Finnigan, Bremen, Germany) with an AMD Intectra Data System. For electron-impact ionization, an energy of 70 eV was used.
MS and NMR spectroscopy.
MS analysis was performed with positive-ion-mode detection on a Apex II-ESI-FT-ICR MS (Bruker Daltonics, Billerica, Mass.) operating at 4.7 T. One- and two-dimensional nuclear magnetic resonance (NMR) experiments were performed on a AMX2-600 spectrometer (Bruker, Karlsruhe, Germany) operating at a proton frequency of 600.13 MHz and equipped with a 5-mm inverse triple resonance probehead with a self-shielded Z-gradient coil. The following experiments were recorded at 298 K using a solution of 1.4 mg of the purified sample dissolved in 0.5 ml CD3OH: 1H NMR, TOCSY (total correlation spectroscopy), NOESY (nuclear overhauser spectroscopy), and HSQC (heteronuclear single quantum coherence). For suppression of the hydroxy resonance of the solvent a Watergate sequence was appended to the homonuclear experiments. An HMBC (heteronuclear multiple bond correlation) spectrum was acquired at 318 K using a solution of 6 mg of the purified sample dissolved in 0.5 ml of CD3OH. HSQC and HMBC experiments were performed with gradient selection. A 13C NMR spectrum of the second sample was recorded on a AC250 spectrometer (Bruker, Karlsruhe, Germany) operating at a carbon frequency of 62.90 MHz and equipped with a 5-mm dual-probe head. Chemical shifts (see Table 2) were referenced to the signals of the solvent at δ(1H) = 3.35 ppm and δ(13C) = 49.0 ppm.
TABLE 2.
1H and 13C chemical shifts observed for micrococcin P1 in CD3OH at 318 K
| No.a | Chemical shift (ppm)
|
No.a | Chemical shift (ppm)
|
|||
|---|---|---|---|---|---|---|
| δ(1H) | δ(13C) | δ(1H) | δ(13C) | |||
| 1 | 1.19 | 20.6 | 28 | 1.55 | 20.8 | |
| 2 | 3.95 | 67.5 | 29 | 170.9 | ||
| 3 | 3.37, 3.25 | 48.3 | 30 | 9.53 | ||
| 4 | 7.91 | 31 | 130.6 | |||
| 5 | 167.4 | 32 | 6.57 | 130.9 | ||
| 6 | 131.2 | 33 | 1.86 | 14.1 | ||
| 7 | 6.73 | 131.8 | 34 | 168.0 | ||
| 8 | 1.85 | 13.7 | 35 | 8.10 | 125.1 | |
| 9 | 9.57 | 36 | 149.7 | |||
| 10 | 162.1 | 37 | 162.5 | |||
| 11 | 151.1 | 38 | 8.48 | |||
| 12 | 8.32 | 126.3 | 39 | 5.24 | 57.4 | |
| 13 | 163.9 | 40 | 2.61 | 34.4 | ||
| 14 | 150.9 | 41 | 1.14 | 19.2 | ||
| 15 | 8.33 | 122.1 | 42 | 1.00 | 20.0 | |
| 16 | 169.9 | 43 | 171.5 | |||
| 17 | 151.6 | 44 | 8.26 | 125.6 | ||
| 18 | 8.32 | 119.9 | 45 | 150.4 | ||
| 19 | 8.28 | 141.1 | 46 | 163.1 | ||
| 20 | 129.9 | 47 | 8.50 | |||
| 21 | 166.1 | 48 | 5.26 | 57.5 | ||
| 22 | 8.20 | 126.2 | 49 | 4.31 | 69.2 | |
| 23 | 151.0 | 50 | 1.18 | 20.5 | ||
| 24 | 162.5 | 51 | 172.2 | |||
| 25 | 7.94 | − | 52 | 7.95 | 121.8 | |
| 26 | 4.79 | 59.3 | 53 | 155.0 | ||
| 27 | 4.60 | 69.6 | 54 | 152.7 | ||
Numbers correspond to those presented in Fig. 3.
Cheese-ripening experiments.
To evaluate the antilisterial potential of the S. equorum strain WS 2733 in situ on soft cheese, model ripening with defined single-strain cultures was performed according to the method of Eppert et al. (15) under laboratory conditions in glass desiccators. Micrococcin-deficient (mic) mutants and the nonbacteriocinogenic (Bac−) S. equorum type strain DSMZ 20674 were used as negative controls. Unripened soft cheese of the type “Weinkäse” were obtained from a local dairy after salting and being hand-smeared in petri dishes using gloves and 50 ml of a brine solution with 5% NaCl inoculated with the test strain and the control strains, respectively. The first day of smearing was designated day 1 of the ripening period. Smearing was applied five times (days 1, 3, 5, 7, and 10) at intervals of 2 or 3 days. The cheese was artificially contaminated with L. monocytogenes WSLC 1364 (from the Vacherin Mont d'Or outbreak) on day 1 of smearing. The contamination levels were 4.5 × 104 CFU/ml of brine (resulting in 1.5 × 102 CFU/cm2 of cheese surface), 1.5 × 103 CFU/ml of brine (10 CFU/cm2), and 2.0 × 102 CFU/ml of brine, respectively. The bacterial counts of the Staphylococcus ripening culture in the smear brine were 1.3 × 108/ml (DSMZ 20674), 1.5 × 108/ml (mic mutant), and 1.8 × 10/ml (WS 2733), respectively. In order to cope with a potentially nonuniform distribution of listeriae on the cheese surface at low contamination levels, a large part of the surface was investigated for cell counts (see below).
For the determination of bacterial cell counts, one cheese for each storage time was examined. First, 20 g of the cheese surface (45 cm2) was homogenized in 180 ml of 1.75% trisodium citrate-dihydrate solution using a stomacher. Serial decimal dilution series of these suspensions were plated on plate count agar supplemented with 3% NaCl (PC3+) for aerobic plate counts (30°C, 48 h) and on Oxford agar (Oxoid, Hampshire, England) for listerial counts (37°C, 48 h). Then, 0.1 ml of each dilution was plated in duplicate except for the early stage of the ripening period, at which 1 ml of the initial suspension (dilution 10−1) was distributed on 10 Oxford agar plates. The cell counts were calculated per square centimeter. The selective enrichment procedure for detection of low levels of L. monocytogenes was performed according to the international IDF standard 143A:1995. A total of 25 g (equivalent to 45 cm2) of the cheese surface was blended with 225 ml of Tryptone soy yeast extract broth using a peristaltic blender. After incubation at 30°C for 48 h, a loopful of the enrichment culture was streaked onto Oxford agar.
RESULTS
Isolation of S. equorum WS 2733 and inhibitory spectrum.
An S. equorum strain, inhibiting a variety of L. monocytogenes strains, was isolated from the surface of a French Raclette cheese. This bacterial consortium was composed of approximately 7.8 × 108 CFU/cm2 (18.5%) orange, 4.4 × 108 CFU/cm2 (10.4%) yellow pigmented, and 2.9 × 109 CFU/cm2 (68.7%) white coryneform bacteria partly identified as B. linens and Corynebacterium fascians. The fraction of the S. equorum strain was 2.5% (1.0 × 108 CFU/cm2) of the total aerobic plate count (4.2 × 109 CFU/cm2). The peptide antibiotic present in the culture supernatant showed a wide spectrum of inhibitory activity that was not restricted to closely related species (Table 1). A total of 95 out of 95 Listeria strains and 130 out of 138 other gram-positive bacteria were inhibited. Nearly all gram-positive bacteria tested were sensitive, whereas no inhibition of 37 gram-negative bacteria was observed. The antimicrobial peptide revealed to be highly inhibitory against all tested L. monocytogenes strains, while only 36% of the tested Bacillus cereus strains were sensitive. The mode of action in the tested concentrations (12,800 AU/ml) was bacteriostatic (data not shown).
TABLE 1.
Inhibitory spectrum of micrococcin P1a
| Species | No. of strains
|
|
|---|---|---|
| Sensitive | Nonsensitive | |
| Gram-positive bacteria | ||
| Staphylococcus spp. | ||
| S. aureus | 15 | 0 |
| S. epidermidis | 2 | 0 |
| S. haemolyticus | 1 | 0 |
| S. caseolyticus | 1 | 0 |
| S. xylosus | 1 | 0 |
| S. saprophyticus | 1 | 0 |
| Enterococcus spp. | ||
| E. faecalis | 1 | 0 |
| E. faecium | 1 | 0 |
| Listeria | ||
| L. monocytogenes | 85 | 0 |
| L. innocua | 5 | 0 |
| L. ivanovii | 2 | 0 |
| L. seeligeri | 2 | 0 |
| L. welshimeri | 1 | 0 |
| Bacillus cereus | 7 | 0 |
| 4 | 7 | |
| Clostridium perfringens | 8 | 0 |
| Lactobacillus sp. | 12 | 0 |
| Coryneform bacteria | 0 | |
| Brevibacterium sp. | 24 | 0 |
| Arthrobacter sp. | 30 | 1 |
| Corynebacterium sp. | 14 | 0 |
| Microbacterium sp. | 4 | 0 |
| Micrococcus sp. | 4 | 0 |
| Gram-negative bacteria | ||
| Salmonella sp. | 0 | 11 |
| Citrobacter freundii | 0 | 5 |
| Enterobacter sp. | 0 | 5 |
| Escherichia coli | 0 | 9 |
| Yersinia enterocolitica | 0 | 3 |
| Pseudomonas sp. | 0 | 4 |
The micrococcin P1-containing sterile-filtered culture supernatant (12,800 AU/ml) was used for sensitivity screening.
Purification of the active substance.
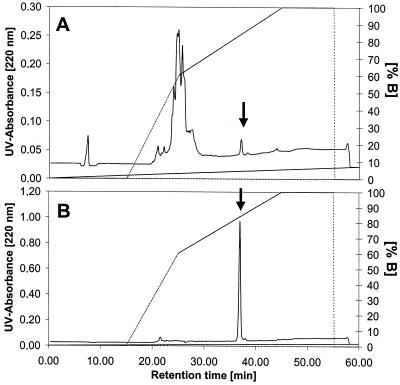
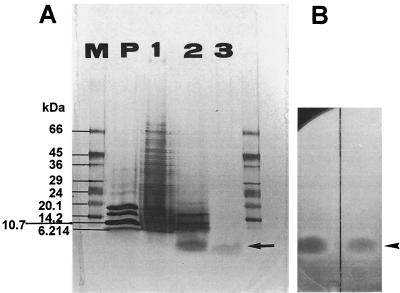
Purification of micrococcin P1 was achieved by RP chromatography. The final polishing step (RP-HPLC) resulted in a 1,600-fold increase in specific activity with 25% recovery. The elution profile from the RP-C18 column is presented in Fig. 1. The antimicrobial substance was eluted at about 84% buffer B (67% acetonitrile). The strong binding of the compound to the C18 column at relatively high acetonitrile concentrations was indicative of a highly hydrophobic molecule. The purification procedure was followed by SDS-PAGE analysis. A single band was detected migrating at an Mr of <2,500 (Fig. 2A, lane 3), which is consistent with a pure preparation and indicative of a relatively small compound. The substance was localized in situ by its antibacterial activity. A clear and distinct zone of inhibition (Fig. 2B), corresponding to an Mr of <2,500 was revealed on the gel overlaid with a L. ivanovii WSLC 3061 indicator lawn.
FIG. 1.
Elution profile of micrococcin P1 (A) RP-HPLC on a Eurosphor 100-C18 column (250 by 8 mm; sample volume, 10 μl; flow rate, 3.5 ml/min; linear acetonitrile gradient, 0 to 80% in 30 min) of crude concentrate (fraction II). All contaminating compounds were separated at the beginning of the linear gradient (0 to 60% B). (B) Analytical separation of pooled fraction IV. The pure antibacterial compound was eluted as a distinct, symmetrical peak at about 84% of solvent B (67% acetonitrile). The arrow indicates the activity.
FIG. 2.
SDS-PAGE of preparations after various stages of purification. (A) Silver-stained Excel Gel (gradient, 8 to 18% T; Pharmacia-LKB). Lane 1, crude concentrate after ammonium sulfate precipitation; lane 2, fraction after preparative RP-LC; lane 3, purified peptide after RP-HPLC; M, molecular weight marker SDS-7L (Sigma); P, peptide length standard (Pharmacia-LKB). (B) Direct detection of antilisterial activity on the gel overlaid with tryptose soft agar seeded with the indicator strain L. ivanovii WSLC 3061. The inhibition zones after incubation overnight at 30°C correspond to the silver-stained bands in Fig. 2A, lanes 2 and 3.
Structure.
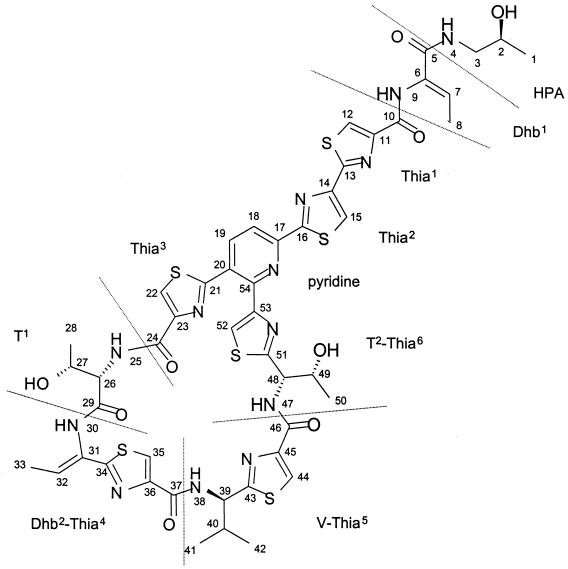
MS analysis confirmed the purity of the sample and revealed a molecular mass of 1,143 Da for the substance. 1H and 13C NMR spectra (data not shown) comprised signals for 46 hydrogens (hydrogens of the three hydroxy groups exchanged fast with the solvent) and 48 carbon atoms. Considering the molecular mass, the moderate number of carbon and hydrogen atoms in the molecule indicated a high content of heteroatoms. The spin systems of one valine (Fig. 3, V), two threonine (T), and two didehydrobutyrine (Dhb) residues, as well as of a 2-hydroxypropylamide (HPA) moiety, were traced in a TOCSY spectrum (data not shown). Sequential connectivities between HPA and Dhb1 and between T1 and Dhb2 (Fig. 3), respectively, were determined by a NOESY experiment by which also the Z-configuration of both Dhb residues was confirmed. The HSQC spectrum contained cross-peaks for eight (hetero)aromatic methine units. Heteronuclear one-bond coupling constants of 1JCH = 196 Hz which were observed in the HMBC spectrum for six of the respective methine units suggested the presence of six thiazole rings in the molecule. Methods for detection of thiazole containing amino acids were described for the microcin B17 structure elucidation (3).
FIG. 3.
Structure of micrococcin P1. Structural units separated by peptide bonds are divided by dashed lines.
The GC-MS analysis of the acid hydrolyzate (data not shown) accounted for one mole each of 1-amino-2-propanol, l-threonine, and 2-(1-amino-2-methylpropyl)-thiazole-4-carboxylic acid and reflected the structural units HPA, T1, and V-Thia5.
These data strongly suggested a close relationship or identity to micrococcin P1 (9). The two-dimensional identity of the substance isolated by us with micrococcin P1 (Fig. 3) was proven by the complete structure elucidation of the molecule relying mainly on HMBC connectivities. Thereby, a detailed assignment of the 1H and 13C chemical shifts was obtained (Table 2), which has never been published for micrococcin P1. Apart from C-26 and C-27 of residue T1, the absolute configurations of the sterocenters were not determined experimentally but were based on biosynthetic considerations and assumed to be as reported for micrococcin P1.
Growth reduction of L. monocytogenes on cheese surface.
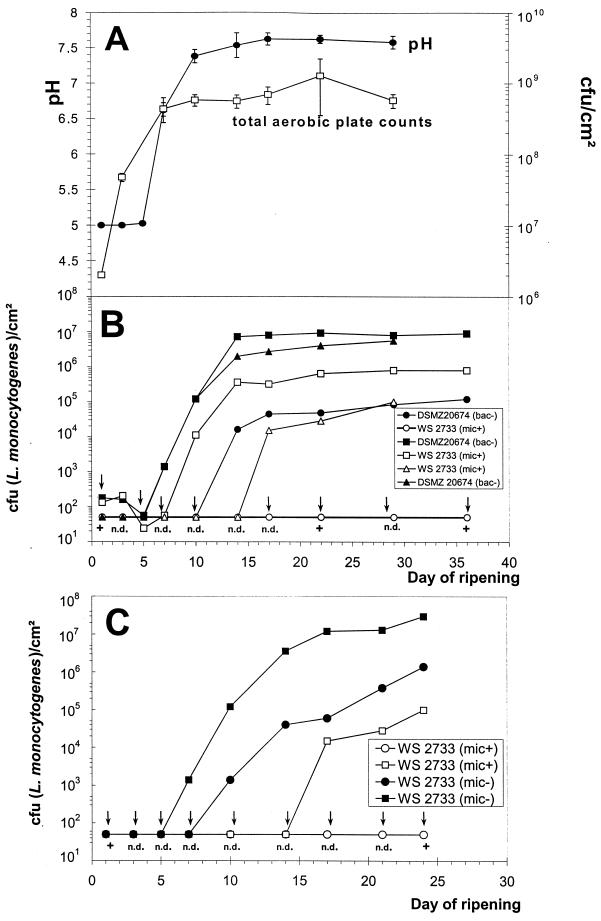
The development of the pH and total aerobic plate counts during the ripening process are shown in Fig. 4A and can be considered typical for the ripening of industrial red-smear cheese using single-strain cultures (15). The total aerobic plates counts of the cheese surface before smearing were about 2.0 × 106 CFU/cm2, and the pH values were in the range of 5.0. At the end of the ripening process all cheeses reached total aerobic cell counts of approximately 6.0 × 108 CFU/cm2. A growth reduction of L. monocytogenes on cheese ripened with the micrococcin P1-producing (Mic+) S. equorum wild-type strain WS 2733 was observed when compared to control cheese ripened with the bacteriocin-negative strain DSMZ 20674 and the micrococcin-deficient mutant (Mic−). The effect was dependent on the contamination level. When challenged with 104 CFU/ml of brine (102 CFU/cm2), a reduction of the viable cell counts by more than 1 log unit could be observed, reaching a final level of 8.0 × 105 CFU/cm2, while listeriae on control cheese grew to 9.0 × 106 CFU/cm2. When challenged with 103 CFU/ml of brine, a growth reduction of 1 to 2 log cycles could be demonstrated. Remarkable effects could be achieved with low contamination levels (102 CFU/ml of brine). The Listeria cells grew to approximately 1.2 × 105 CFU/cm2 on the control cheese ripened with the micrococcin-negative strain DSMZ 20674 (Bac−) and to 1.5 × 106 CFU/cm2 on control cheese ripened with the micrococcin-deficient knockout mutant (Mic−), whereas on cheese inoculated with the wild-type S. equorum WS 2733 the growth of listeriae was completely inhibited.
FIG. 4.
Cheese ripening with defined single-strain cultures of S. equorum WS 2733. (A) Development of pH on the cheese surface and aerobic plate counts (PC3+ agar). (B) Growth reduction of L. monocytogenes WSLC 1364. Listeria cell counts on the cheese surface (Oxford agar) after contamination at day 1 with 104 CFU/ml (102 CFU/cm2) (■ and □), 103 CFU/ml (10 CFU/cm2) (▵ and ▴), and 102 CFU/ml (● and ○), are shown. Control cheeses were ripened with the micrococcin-negative type strain DSMZ 20674 (bac−), closed symbols. (C) Listeria cell counts on the cheese surface (Oxford agar) after contamination at day 1 with 103 CFU/ml (■ and □) and 102 CFU/ml (10 CFU/cm2) (● and ○. Control cheeses were ripened with the micrococcin-deficient mutant (mic−), closed symbols. The micrococcin-producing wild-type strain (mic+) is represented by open symbols. Arrows indicate that even by using enrichment procedures, listeriae could not be detected (n.d.). The measuring points mark the detection limit (50 to 100 CFU/cm2) of the direct plating method. +, Listeriae could be detected only after enrichment procedures.
DISCUSSION
Micrococcin P1 (Fig. 3) is a macrocyclic peptide antibiotic of the thiocillin-thiazolyl class (8, 9, 13, 25, 32, 54). Recently, a complete synthesis was published by Okumura et al. (34). The peptide binds to complexes formed between the L11 protein of the 50S ribosomal subunit and a nucleotide sequence within the 23S rRNA and inhibits the elongation factor EFG-dependent translocation step of the growing peptide chain. Resistant mutants of B. subtilis and B. megaterium either possess a structurally altered 50S subunit or are specifically methylated at a single site (nucleotide A-1067) of the 23S rRNA (39, 45, 46). It has been demonstrated that this macrocyclic thiopeptide antibiotic is synthesized nonribosomally and involves a multifunctional peptide synthetase, which was designated micrococcin synthase (Carnio et al., submitted).
So far, micrococcin P1 has been purified from a bacterium related to Micrococcus varians (20, 48) and from Bacillus pumilus (1, 16), isolated from sewage and soil, respectively. Breiter et al. (7) have reported antibiotically active substances excreted by seven strains belonging to Staphylococcus and Micrococcus species isolated from animal sources (partridge, pig, and dog). Two basic substances were isolated and designated micrococcin M1 and M3. The physicochemical and spectroscopic data, as well as investigations of hydrolysis products, indicated a close relationship to micrococcin P1. However, the structure of these compounds has never been reported. The antibacterial substance secreted by S. equorum WS 2733 was identified as micrococcin P1 by spectroscopic evidence. Based on biosynthetic considerations, the absolute configuration of the sterocenters was assumed to be as reported for micrococcin P1. In this context it is of interest that other thiazole- and oxazole-containing polypeptides such as the bacteriocin microcin B17 originate from ribosomally synthesized precursors which are posttranslationally modified (3).
S. equorum was revealed to be a potent inhibitor of growth of L. monocytogenes on the cheese surface. The phenomenon of complete inhibition of listeriae occurs only when the inhibitory strain is used as sole starter and when contamination with Listeria sp. is performed in the early stage of ripening at a low concentration level (102 CFU/ml of brine), which is a high titer for contamination occurring in the dairy industry. This is in accordance with other studies reporting on the inhibition of Listeria sp. on cheese surfaces (9, 14, 15, 17, 18, 29, 33, 36, 47, 49, 51, 55). The limited effects of antilisterial bacteria clearly show that there is a need for developing hurdle concepts in the dairy industry in which bacteriocinogenic bacteria play a certain role. However, such protective starters cannot replace a good hygienic practice during food processing.
Although micrococcin P1 exhibits a broad activity spectrum, inhibiting nearly all gram-positive bacteria when tested on agar plates (spot-on-the-lawn-assay; Table 1), the antibiotic allows the formation of a stable ecological system on the surface of the French Raclette cheese. However, when combined with other surface-ripening cultures in cheese-ripening experiments under laboratory conditions, S. equorum WS 2733 dominated the ripening flora by suppressing the other gram-positive coryneform bacteria (data not shown). This finding may be explained by the different cheese types: on the surface of a soft cheese the development of the ripening microflora may be completely different from the one on the Raclette surface. It appears, however, to be more likely that the bacterial consortium growing on the surface of the French Raclette cheese constitutes a well-balanced, stable ecosystem which has developed over decades of cheese production. Biocontrol of listeriae on soft cheese by protective ripening bacteria will therefore be more complicated than just adding an antilisterial strain to a well-established smear-cheese flora.
S. equorum was originally isolated from the skin of healthy horses and was first described by Schleifer et al. (43), but it has often been associated with food products. For instance, together with Staphylococcus xylosus, it constitutes the predominant organism present during ripening of an Iberian dry cured ham (37), contributing to the characteristic flavor of the final product. It is also part of the ripening flora from traditional French cheeses (22) and was isolated from goat milk and cheese (31). Moreover, 5 to 15% of the total cell counts of the microflora of the German Tilsit cheese consist of staphylococci classified as S. equorum (6). This species belongs to the group of coagulase-negative staphylococci and has never been reported to be involved in diseases. There would, therefore, be good reason to consider S. equorum a GRAS (generally recognized as safe) organism. In the case of our isolate, however, it should be noted that micrococcin P1 has to be classified as an antibiotic. With respect to its potential pharmaceutical use it may, therefore, be wise to be careful in spreading this strain widely in the human community before its pharmaceutical potential is evaluated.
REFERENCES
- 1.Abraham E P, Heatley N G, Brookes P, Fuller A T, Walker J. Probable identity of an antibiotic produced by a spore-bearing Bacillus of the B. pumilus group with micrococcin. Nature. 1956;178:44–45. doi: 10.1038/178044a0. [DOI] [PubMed] [Google Scholar]
- 2.Barefoot S F, Klaenhammer T R. Detection and activity of lactacin B, a bacteriocin produced by Lactobacillus acidophilus. Appl Environ Microbiol. 1983;45:1808–1815. doi: 10.1128/aem.45.6.1808-1815.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bayer A, Freund S, Jung G. Post-translational heterocyclic backbone modifications in the 43-peptide antibiotic Microcin B17, structure elucidation and NMR study of a 13C, 15N-labelled Gyrase inhibitor. Eur J Biochem. 1995;234:414–426. doi: 10.1111/j.1432-1033.1995.414_b.x. [DOI] [PubMed] [Google Scholar]
- 4.Bhunia A K, Johnson M G. A modified method to directly SDS-PAGE the bacteriocin of Pediococcus acidilactici. Lett Appl Microbiol. 1992;15:5–7. doi: 10.1111/j.1472-765x.1992.tb00721.x. [DOI] [PubMed] [Google Scholar]
- 5.Blum H, Beier H, Gross H J. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis. 1987;8:93–99. [Google Scholar]
- 6.Bockelmann W, Krusch U, Engel G, Klijn N, Smit G, Heller K J. The microflora of Tilsit cheese. Part 1. Variability of the smear flora. Nahrung. 1997;41:208–212. [Google Scholar]
- 7.Breiter J, Metz H, Grigo J. Staphylococcal micrococcins. II. Isolation, purification and identification. Arzneimittelforschung. 1975;25:1244–1248. [PubMed] [Google Scholar]
- 8.Brookes P, Fuller A T, Walker J. Chemistry of micrococcin P. Part I. J Chem Soc. 1957;1957:689–699. [Google Scholar]
- 9.Bycroft B W, Gowland M S. The structures of the highly modified peptide antibiotics micrococcin P1 and P2. J Chem Soc Chem Commun. 1978;1978:256–258. [Google Scholar]
- 10.Carminati D, Neviani E, Ottogalli G, Giraffa G. Use of surface-smear bacteria for inhibition of Listeria monocytogenes on the rind of smear cheese. Food Microbiol. 1999;16:29–36. [Google Scholar]
- 11.Carnio M C, Eppert I, Scherer S. Analysis of the bacterial surface ripening flora of German and French smeared cheeses with respect to their anti-listerial potential. Int J Food Microbiol. 1999;47:89–97. doi: 10.1016/s0168-1605(99)00016-1. [DOI] [PubMed] [Google Scholar]
- 12.Cundliffe E, Thompson J. Concerning the mode of action of micrococcin upon bacterial protein synthesis. Eur J Biochem. 1981;118:47–52. doi: 10.1111/j.1432-1033.1981.tb05484.x. [DOI] [PubMed] [Google Scholar]
- 13.Dean B M, Mijovic M P V, Walker J. Chemistry of micrococcin P. Part IV. Racemisation of 2-(1-amino-2-methylpropyl)thiazole-4-carboxylic acid, and related studies. J Chem Soc. 1961;1961:3394–3400. [Google Scholar]
- 14.Ennahar S, Assobhei O, Hasselmann C. Inhibition of Listeria monocytogenes in a smear-surface soft cheese by Lactobacillus plantarum WHE92, a pediocin AcH producer. J Food Prot. 1998;61:186–191. doi: 10.4315/0362-028x-61.2.186. [DOI] [PubMed] [Google Scholar]
- 15.Eppert I, Valdés-Stauber N, Götz H, Busse M, Scherer S. Growth reduction of Listeria spp. caused by undefined industrial red smear cheese cultures and bacteriocin producing Brevibacterium linens as evaluated in situ on soft cheese. Appl Environ Microbiol. 1997;63:4812–4817. doi: 10.1128/aem.63.12.4812-4817.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuller A T. A new antibiotic of bacterial origin. Nature. 1955;175:722. doi: 10.1038/175722a0. [DOI] [PubMed] [Google Scholar]
- 17.Garcia E, De Paz M, Gaya P, Medina M, Nunez M. Inhibition of Listeria innocua in Manchego cheese by bacteriocin-producing Enterococcus faecalis INIA4. Milchwissenschaft. 1997;52:667–670. [Google Scholar]
- 18.Giraffa G, Picchioni N, Neviani E, Carminati D. Production and stability of an Enterococcus faecium bacteriocin during Taleggio cheese-making and ripening. Food Microbiol. 1995;12:301–307. [Google Scholar]
- 19.Grosenick H, Schurig V. Enantioselective capillary gas chromatography and capillary supercritical fluid chromatography on an immobilized γ-cyclodextrin derivative. J Chromatogr A. 1997;761:181–193. [Google Scholar]
- 20.Heatley N G, Doery H M. The preparation and some properties of purified micrococcin. Biochem J. 1951;50:247–253. doi: 10.1042/bj0500247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hug-Michel Ch, Barben S, Kaufmann U. Hemmumng von Listeria spp. durch Mikroorganismen aus der Rinde von Rotschmiereweichkäsen. Schweiz Milchwirtsch Forsch. 1989;18:46–49. [Google Scholar]
- 22.Irlinger F, Morvan A, El-Solh N, Bergere J L. Taxonomic characterization of coagulase-negative staphylococci in ripening flora from traditional French cheese. Syst Appl Microbiol. 1997;20:319–328. [Google Scholar]
- 23.Jack R, Götz F, Jung G. Lantibiotics. In: Rehm H-J, Reed G, editors. Biotechnology. Vol. 7. Weinheim, Germany: Verlag Chemie; 1997. pp. 323–368. [Google Scholar]
- 24.Jack R W, Bierbaum G, Sahl H-G. Lantibiotics and related peptides. Berlin, Germany: Springer; 1998. [Google Scholar]
- 25.James M N G, Watson K J. Chemistry of micrococcin P. Part IX. The crystal and molecular structure of micrococcinic acid bis-4-bromoanilide. J Chem Soc C. 1966;1966:1361–1371. doi: 10.1039/j39660001361. [DOI] [PubMed] [Google Scholar]
- 26.Jung G, Sahl H-G, editors. Nisin and novel lantibiotics. Leiden, Germany: Escom; 1991. [Google Scholar]
- 27.Klaenhammer T R. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol Rev. 1993;12:39–86. doi: 10.1111/j.1574-6976.1993.tb00012.x. [DOI] [PubMed] [Google Scholar]
- 28.Maisnier-Patin S, Richard J. Activity and purification of Linenscin OC2, an antibacterial substance produced by Brevibacterium linens OC2, an orange cheese coryneform bacterium. Appl Environ Microbiol. 1995;61:1847–1852. doi: 10.1128/aem.61.5.1847-1852.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maisnier-Patin S, Deschamps N, Tatini S R, Richard J. Inhibition of Listeria monocytogenes in Camembert cheese made with a nisin-producing starter. Lait. 1992;72:249–263. [Google Scholar]
- 30.Martin F, Friedrich K, Beyer F, Terplan G. Antagonistische Wirkung von Brevibacterium linens-Stämmen gegen Listerien. Archiv für Lebensmittelhygiene. 1995;46:1–24. [Google Scholar]
- 31.Meugnier H, Bes M, Vernozy-Rozand C, Mazuy C, Brun Y, Freney J, Fleurette J. Identification and ribotyping of Staphylococcus xylosus and Staphylococcus equorum strains isolated from goat milk and cheese. Int J Food Microbiol. 1996;31:325–331. doi: 10.1016/0168-1605(96)00975-0. [DOI] [PubMed] [Google Scholar]
- 32.Mijovic M P V, Walker J. Chemistry of micrococcin P. Part II. J Chem Soc C. 1960;1960:909–916. [Google Scholar]
- 33.Nunez M, Rodriguez J L, Garcia E, Gaya P, Medina M. Inhibition of Listeria monocytogenes by enterocin 4 during the manufacture and ripening of Manchego cheese. J Appl Microbiol. 1997;83:671–677. doi: 10.1046/j.1365-2672.1997.00275.x. [DOI] [PubMed] [Google Scholar]
- 34.Okumura K, Ito A, Yoshioka D, Shin C. Total synthesis of macrocyclic antibiotic micrococcin P1. Heterocycles. 1998;48:1319–1324. [Google Scholar]
- 35.Otaka T, Kaji A. Micrococcin: acceptor-site specific inhibitor of protein synthesis. Eur J Biochem. 1974;50:101–106. doi: 10.1111/j.1432-1033.1974.tb03876.x. [DOI] [PubMed] [Google Scholar]
- 36.Pucci M J, Vedamuthu E R, Kunka B S, Vandenbergh P A. Inhibition of Listeria monocytogenes by using bacteriocin PA-1 produced by Pediococcus acidilactici PAC1.0. Appl Environ Microbiol. 1988;54:2349–2353. doi: 10.1128/aem.54.10.2349-2353.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodríguez M, Núnez F, Córdoba J J, Sanabria C, Bermúdez E, Asensio M A. Characterization of Staphylococcus spp. and Micrococcus spp. isolated from Iberian ham throughout the ripening process. Int J Food Microbiol. 1994;24:329–335. doi: 10.1016/0168-1605(94)90131-7. [DOI] [PubMed] [Google Scholar]
- 38.Rogers M J, Cundliffe E, McCutchan T F. The antibiotic micrococcin is a potent inhibitor of growth and protein synthesis in the malaria parasite. Antimicrob Agents Chemother. 1998;42:715–716. doi: 10.1128/aac.42.3.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosendahl G, Douthwaite S. The antibiotics micrococcin and thiostrepton interact directly with 23S rRNA nucleotides 1067A and 1095A. Nucleic Acids Res. 1994;22:357–363. doi: 10.1093/nar/22.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryser E T, Maisnier-Patin S, Gratadoux J J, Richard J. Isolation and identification of cheese-smear bacteria inhibitory to Listeria spp. Int J Food Microbiol. 1994;21:237–246. doi: 10.1016/0168-1605(94)90030-2. [DOI] [PubMed] [Google Scholar]
- 41.Sahl H G. Staphylococcin 1580 is identical to the lantibiotic epidermin: implications for the nature of bacteriocins from gram-positive bacteria. Appl Environ Microbiol. 1994;60:752–755. doi: 10.1128/aem.60.2.752-755.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sahl H G, Brandis H. Production, purification and chemical properties of an anti-staphylococcal agent produced by Staphylococcus epidermidis. J Gen Microbiol. 1981;127:377–384. doi: 10.1099/00221287-127-2-377. [DOI] [PubMed] [Google Scholar]
- 43.Schleifer K H, Kilpper-Bälz R, Devriese L A. Staphylococcus arlettae sp. nov., S. equorum sp. nov., and S. kloosii sp. nov.: three new coagulase-negative, novobiocin-resistant species from animals. Syst Appl Microbiol. 1984;5:501–509. [Google Scholar]
- 44.Schnell N, Entian K-D, Schneider U, Götz F, Zähner H, Kellner R, Jung G. Prepeptide sequence of epidermin, a ribosomally synthesized polypeptide antibiotic containing four sulphide-rings. Nature. 1988;333:276–278. doi: 10.1038/333276a0. [DOI] [PubMed] [Google Scholar]
- 45.Smith I, Weiss D, Pestka S. A Micrococcin—resistant mutant of Bacillus subtilis: localization of resistance to the 50S subunit. Mol Gen Genet. 1976;144:231–233. doi: 10.1007/BF00341720. [DOI] [PubMed] [Google Scholar]
- 46.Spedding G, Cundliffe E. Identification of the altered ribosomal component responsible for resistance to micrococcin in mutants of Bacillus megaterium. Eur J Biochem. 1984;140:453–459. doi: 10.1111/j.1432-1033.1984.tb08124.x. [DOI] [PubMed] [Google Scholar]
- 47.Stecchini M L, Aquili V, Sarais I. Behaviour of Listeria monocytogenes in Mozarella cheese in presence of Lactococcus lactis. Int J Food Microbiol. 1995;25:301–310. doi: 10.1016/0168-1605(94)00093-l. [DOI] [PubMed] [Google Scholar]
- 48.Su T L. Micrococcin, an antibacterial substance formed by a strain of Micrococcus. Brit J Exp Pathol. 1948;29:473–481. [PMC free article] [PubMed] [Google Scholar]
- 49.Sulzer G, Busse M. Growth inhibition of Listeria spp. on camembert cheese by bacteria producing inhibitory substances. Int J Food Microbiol. 1991;14:287–289. doi: 10.1016/0168-1605(91)90120-e. [DOI] [PubMed] [Google Scholar]
- 50.Valdés-Stauber N, Götz H, Busse M. Antagonistic effects of coryneform bacteria from red smear cheese against Listeria species. Int J Food Microbiol. 1991;13:119–130. doi: 10.1016/0168-1605(91)90054-s. [DOI] [PubMed] [Google Scholar]
- 51.Valdés-Stauber N, Scherer S. Isolation and characterization of Linocin M18, a bacteriocin produced by Brevibacterium linens. Appl Environ Microbiol. 1994;60:3809–3814. doi: 10.1128/aem.60.10.3809-3814.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Valdés-Stauber N, Scherer S. Nucleotide sequence and taxonomical distribution of the bacteriocin gene lin cloned from Brevibacterium linens M18. Appl Environ Microbiol. 1996;62:1283–1286. doi: 10.1128/aem.62.4.1283-1286.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Valdés-Stauber N, Scherer S, Seiler H. Identification of yeasts and coryneform bacteria from the surface microflora of brick cheeses. Int J Food Microbiol. 1997;34:115–129. doi: 10.1016/s0168-1605(96)01171-3. [DOI] [PubMed] [Google Scholar]
- 54.Walker J, Olesker A, Valente L, Rabanal R, Lukacs G. Total structure of the polythiazole-containing antibiotic micrococcin P. A 13C nuclear magnetic resonance study. J Chem Soc Chem Commun. 1977;1977:706–708. [Google Scholar]
- 55.Wan J, Harmark K, Davidson B E, Hillier A J, Gordon J B, Wilcock A, Hickey M W, Coventry M J. Inhibition of Listeria monocytogenes by piscicolin 126 in milk and Camembert cheese manufactured with a thermophilic starter. J Appl Microbiol. 1997;82:273–280. doi: 10.1046/j.1365-2672.1997.00349.x. [DOI] [PubMed] [Google Scholar]