Abstract
We have characterized the nucleotide sequences of the 70-kDa heat shock protein (HSP70) genes of Cryptosporidium baileyi, C. felis, C. meleagridis, C. muris, C. serpentis, C. wrairi, and C. parvum from various animals. Results of the phylogenetic analysis revealed the presence of several genetically distinct species in the genus Cryptosporidium and eight distinct genotypes within the species C. parvum. Some of the latter may represent cryptic species. The phylogenetic tree constructed from these sequences is in agreement with our previous results based on the small-subunit rRNA genes of Cryptosporidium parasites. The Cryptosporidium species formed two major clades: isolates of C. muris and C. serpentis formed the first major group, while isolates of C. felis, C. meleagridis, C. wrairi, and eight genotypes of C. parvum formed the second major group. Sequence variations were also observed between C. muris isolates from ruminants and rodents. The HSP70 gene provides another useful locus for phylogenetic analysis of the genus Cryptosporidium.
Cryptosporidium is an intracellular extracytoplasmic protozoan parasite with a monoxenous life cycle, where all asexual and sexual development occurs within one host. The parasite infects the microvillus border of the gastrointestinal and respiratory epithelium of a wide range of vertebrate hosts, including humans, causing diarrheal diseases. It has been reported to cause waterborne and food-borne outbreaks worldwide (23, 32). Zoonotic infection and person-to-person transmission, however, are also known (2, 6). An understanding of its epidemiology has been hampered by poor knowledge of the species structures and public health importance of various Cryptosporidium species and genotypes.
Tyzzer (35, 36) was the first researcher to recognize the multispecies nature of Cryptosporidium parasites. He described two species in mammals, C. muris and C. parvum, based on the differences in morphology and infection sites. In 1955, a new species, C. meleagridis, was associated with illness and death in turkeys (31). To date, 22 species of Cryptosporidium have been named based on host occurrence, but only 8 are considered valid by some researchers (10). We have recently characterized the small-subunit (SSU) rRNA genes of various Cryptosporidium parasites for phylogenetic analysis. The results show that (i) Cryptosporidium parasites form a multispecies complex having at least four distinct species (C. parvum, C. baileyi, C. muris, and C. serpentis); (ii) there are two distinct genotypes of C. muris and various genotypes (human, bovine, dog, ferret, kangaroo, monkey, mouse, and pig) of C. parvum which are related to C. felis, C. meleagridis, and C. wrairi; and (iii) some of the C. parvum genotypes may be cryptic species (38, 39). These observations are in agreement with other sequence analyses of rRNA genes (24–26).
The heat shock protein (HSP) gene belongs to a multigene family that is highly conserved across the prokaryotes and eukaryotes. Under normal conditions, these proteins function as molecular chaperons for facilitating the folding of proteins in secretion and transport. Their expression, however, is upregulated under environmental stress and is involved in the protection of the cells (9, 13–15, 22). Khramtsov et al. (19) cloned and sequenced the 70-kDa HSP (HSP70) gene of an isolate of the C. parvum bovine genotype. Based on this sequence, several molecular diagnostic techniques have recently been designed for the detection of Cryptosporidium parasites in environmental samples. These techniques have been used for (i) the detection of viable C. parvum oocysts by reverse transcription-PCR (33), (ii) the detection of viable C. parvum oocysts by cell culture reverse transcription-PCR (29), and (iii) the detection of viable C. parvum oocysts by cell culture PCR (7). However, the polymorphic nature of the HSP70 gene sequences used as primers is not clear, which complicates the use of the assay in detecting Cryptosporidium in environmental and clinical samples (5). Therefore, in order to use the HSP70 gene as a diagnostic target for the analysis of clinical and environmental samples, there is a need to characterize the HSP70 genes from different species or genotypes of Cryptosporidium.
In this communication, we present the results of sequence characterization and phylogenetic analysis of various Cryptosporidium isolates from human and animal hosts at the HSP70 gene locus. Our results with the HSP70 gene confirmed our previous observations of the multispecies nature of Cryptosporidium parasites based on the SSU rRNA gene (38, 39). The sequence information generated from this study is also useful in the development of HSP70-based species and genotype diagnostic tools.
MATERIALS AND METHODS
Purification of oocysts and extraction of genomic DNA.
Fecal samples containing oocysts of C. baileyi (chicken and quail), C. felis (cat and human), C. meleagridis (turkey and human), C. muris (cattle, camel, and mouse), C. parvum (human, cattle, cat, dog, ferret, monkey, mouse, kangaroo, koala, and pig), C. serpentis (savanah monitor and snake), C. wrairi (guinea pig), and an unknown Cryptosporidium species (desert monitor) were obtained from infected humans and animals and stored at 4°C in 2.5% potassium dichromate solution until they were used (Table 1). The oocysts were purified by the sucrose and Percoll gradient method (1). DNA was isolated from the purified oocysts as described before (34) and stored at −20°C before use. The concentration of DNA samples was measured by UV absorption at 260 nm. The identities of Cryptosporidium species and genotypes were established based on morphological examinations and sequence analysis of the SSU rRNA gene (38, 39).
TABLE 1.
Cryptosporidium isolates used in this study
| Isolate | Source location | Host | Species and genotype | A+T content (%) |
|---|---|---|---|---|
| 183 | Washington | Human | C. parvum human | 58.8 |
| 497 | Kenya | HIV+ humana | C. parvum human | 58.9 |
| 6 | Ohio | Calf | C. parvum bovine | 58.6 |
| 671 | Australia | Calf | C. parvum bovine | 58.5 |
| 674 | Australia | Calf | C. parvum bovine | 58.6 |
| 244 | Ohio | Dog | C. parvum dog | 48.2 |
| 715a | Maryland | Dog | C. parvum dog | 48.1 |
| 351 | Georgia | Ferret | C. parvum ferret | 58.4 |
| 712a | Washington, D.C. | Black-footed ferret | C. parvum ferret | 58.4 |
| 713a | Washington, D.C. | Black-footed ferret | C. parvum ferret | 58.4 |
| 714a | Washington, D.C. | Black-footed ferret | C. parvum ferret | 58.5 |
| 428 | Australia | Red kangaroo | C. parvum marsupial | 60.1 |
| 587 | Australia | Koala | C. parvum marsupial | 60.1 |
| 518 | Georgia | Rhesus monkey | C. parvum monkey | 58.8 |
| 359 | Maryland | Mouse | C. parvum mouse | 59.1 |
| 411 | Maryland | Mouse | C. parvum mouse | 59.2 |
| 499 | Australia | Pig | C. parvum pig | 59.8 |
| 763 | Australia | Quail | C. baileyi | 65.8 |
| 764 | Maryland | Chicken | C. baileyi | 65.8 |
| 288 | Australia | Cat | C. felis | 51.1 |
| 160 | New Orleans | HIV+ human | C. felis | 51.0 |
| 297 | New Orleans | HIV+ human | C. felis | 51.2 |
| 670 | New Orleans | HIV+ human | C. felis | 51.0 |
| 295 | Maryland | Turkey | C. meleagridis | 58.1 |
| 672 | Kenya | HIV+ human | C. meleagridis | 58.2 |
| 589 | Australia | Cattle | C. muris | 62.2 |
| 697 | Czech Republic | Cattle | C. muris | 62.2 |
| 698 | Czech Republic | Cattle | C. muris | 62.2 |
| 699 | Czech Republic | Cattle | C. muris | 62.2 |
| 700 | Czech Republic | Cattle | C. muris | 62.2 |
| 701 | Czech Republic | Camel | C. muris | 62.6 |
| 703 | Czech Republic | Camel | C. muris | 62.2 |
| 707 | Czech Republic | Mouse | C. muris | 63.5 |
| 708 | Czech Republic | Mouse | C. muris | 63.4 |
| 709 | Spain | Mouse | C. muris | 63.4 |
| 63 | Maryland | Savanna monitor | C. serpentis | 62.2 |
| 64 | Maryland | Amazon tree boa | C. serpentis | 62.2 |
| 517 | Michigan | Guinea pig | C. wrairi | 59.3 |
| 691 | St. Louis | Desert monitor | Cryptosporidium sp. | 61.0 |
| 692 | St. Louis | Desert monitor | Cryptosporidium sp. | 61.0 |
HIV, human immunodeficiency virus.
PCR amplification.
A two-step nested-PCR protocol was used to amplify the HSP70 gene fragments from various Cryptosporidium isolates, using primers complementary to the conserved nucleotide sequences of apicomplexan parasites downloaded from GenBank: the C. parvum bovine genotype (U71181), Eimeria acervulina (Z26134), Plasmodium cynomolgi (M90978), Theileria annulata (J04653), and Toxoplasma gondii (U85648). A PCR product of ∼2,015 bp was amplified using forward (5′-ATG TCT GAA GGT CCA GCT ATT GGT ATT GA-3′) and reverse (5′-TTA GTC GAC CTC TTC AAC AGT TGG-3′) primers. The PCR mixture consisted of 50 ng of DNA, 200 μM (each) deoxynucleoside triphosphate, 1× PCR buffer (Perkin-Elmer, Foster City, Calif.), 3.0 mM MgCl2, 5.0 U of Taq polymerase (GIBCO BRL, Frederick, Md.), and 200 nM (each) primer in a total volume of 100 μl. The reactions were performed for 35 cycles (each cycle was 94°C for 45 s, 55°C for 45 s, and 72°C for 60 s) in a Perkin-Elmer GeneAmp PCR 9700 thermocycler with an initial hot start (94°C for 5 min) and a final extension (72°C for 10 min). For the secondary PCR, a fragment of ∼1,950 bp was amplified using 2.5 μl of primary PCR mixture and a set of nested forward (5′-TA/CT TCA TG/CT GTT GGT GTA TGG AGA AA-3′) and reverse (5′-CAA CAG TTG GAC CAT TAG ATC C-3′) primers. The conditions for the secondary PCR were identical to those for the primary PCR, except for the use of a lower annealing temperature (45°C). The PCR product was analyzed by agarose gel electrophoresis and visualized after ethidium bromide staining.
Sequencing and phylogenetic analysis.
The secondary-PCR products were sequenced on an ABI 377 automated sequencer (Perkin-Elmer) using a Big Dye terminator cycle-sequencing ready-reaction kit (Perkin-Elmer). Sequence accuracy was confirmed by two-directional sequencing and by sequencing of a new PCR product if necessary. Multiple alignments of the DNA sequences were done with the Wisconsin Package version 9.0 (Genetics Computer Group, Madison, Wis.) with manual adjustment.
Two phylogenetic analyses were carried out on the aligned sequences to assess phylogenetic relationships among various species and genotypes. The first analysis was conducted to assess the evolutionary relationship between Cryptosporidium species and other members of the phylum Apicomplexa. In this analysis, the HSP70 gene sequences representing the C. parvum bovine genotype and C. muris were aligned with the published sequences of the C. parvum bovine genotype (U71181 and U69698), E. acervulina (Z26134), Eimeria maxima (Z46964), P. cynomolgi (M90978), Plasmodium falciparum (M19753), T. annulata (J04653), Theileria parva (U40190), Theileria sergenti (D12692), and T. gondii (AF045559 and U85648) obtained from GenBank. A neighbor-joining tree (30) was constructed using the program TreeconW (37), and evolutionary distances were calculated by Kimura two-parameter analysis. The sequence of Babesia microti (GenBank accession no. U53448) was used as an outgroup to assess the relatedness of the genus Cryptosporidium with other members of the phylum Apicomplexa. We chose to use B. microti as an outgroup because construction of unrooted trees suggested that it was the most divergent member of this group.
In the second analysis, a neighbor-joining tree was constructed for all the isolates of Cryptosporidium to assess the relationship among various Cryptosporidium species and within C. parvum genotypes. The tree was rooted using P. cynomolgi (GenBank accession no. M90978) and P. falciparum (GenBank accession no. M19753). The second phylogenetic analysis also included the construction of a neighbor-joining tree based on the deduced amino acid sequences. The reliabilities of these trees were assessed by the bootstrap method (11) with 1,000 pseudoreplicates. We used 95% as the statistically significant value (8); however, values greater than 70% are reported, since the bootstrap method (11) may be a conservative estimate for the reliability of a clade (16).
Nucleotide sequence accession numbers.
The nucleotide sequences of the HSP70 genes of C. baileyi, C. felis, C. meleagridis, C. muris, C. serpentis, C. wrairi, the unknown Cryptosporidium sp., and eight genotypes of C. parvum (human, bovine, dog, ferret, marsupial, monkey, mouse, and pig) have been deposited in the GenBank database under accession no. AF221528 to AF221543.
RESULTS
We sequenced ∼1,950 bp of the HSP70 genes from 17 C. parvum isolates, 2 C. baileyi isolates, 4 C. felis isolates, 2 C. meleagridis isolates, 10 C. muris isolates, 2 C. serpentis isolates, 1 C. wrairi isolate, and 2 isolates from an unknown Cryptosporidium species from a desert monitor. The C. parvum isolates, represented the following genotypes of C. parvum: two isolates of the C. parvum human genotype, three isolates of the C. parvum bovine genotype, two isolates of the C. parvum dog genotype, four isolates of the C. parvum ferret genotype, two isolates of the C. parvum marsupial genotype, one isolate of the C. parvum monkey genotype, two isolates of the C. parvum mouse genotype, and one isolate of the C. parvum pig genotype. HSP70 gene sequences of an unknown Cryptosporidium species from two desert monitors were also obtained. The HSP70 gene of Cryptosporidium parasites was AT rich (58.4 to 65.8%), except for the isolates of the C. parvum dog genotype and C. felis (48.1 to 51.2%). However, within each Cryptosporidium species and C. parvum genotype the A+T contents of different isolates were quite consistent (Table 1).
Multiple alignment of the HSP70 gene sequences revealed distinct sequences for the eight species of Cryptosporidium (C. baileyi, C. parvum, C. meleagridis, C. muris, C. serpentis, C. felis, C. wrairi, and the unknown Cryptosporidium sp.) analyzed in the study. Distinct interspecies variations were also noticed throughout the entire HSP70 gene, including in the regions of PCR primers utilized by Stinear et al. (33), Rochelle et al. (29), and Di Giovanni et al. (7) (Table 2 and Fig. 1). Eight genotypes of C. parvum and two genotypes of C. muris were found by HSP70 gene analyses, in concordance with the results of analysis at the SSU rRNA gene locus (38, 39). The extent of genetic variation in the genus Cryptosporidium was also assessed by comparing the C. parvum bovine genotype nucleotide sequence with other HSP70 gene sequences of different Cryptosporidium species and C. parvum genotypes. The variation between the C. parvum bovine genotype and other C. parvum genotype isolates was low (1.4 to 7.4%), except for the C. parvum dog genotype isolates (12.5%). A significant difference was observed among non-parvum species, such as C. baileyi, C. muris, C. serpentis, C. felis, and the unknown Cryptosporidium sp. (14.7 to 18.7%). The genetic differences between the C. parvum bovine genotype and C. meleagridis and C. wrairi isolates, however, was substantially lower (1.9 to 4.0%) (Table 3). Compared to the C. parvum bovine genotype, the majority of mutations in the HSP70 genes of other Cryptosporidium parasites were synonymous. However, the percentages of nonsilent mutations were higher at the interspecies level and lower at the intergenotype level, except for isolates of C. parvum from humans and a monkey and C. baileyi (Table 3).
TABLE 2.
Evolutionary genetic distances among Cryptosporidium species and eight genotypes of C. parvum
| Species and genotype | No. of nucleotide differences per 100 basesa
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P. falciparum | P. cynomolgi | C. muris bovine | C. muris murine | C. serpentis | C. baileyi | Desert monitor | C. felis | C. Tparvum dog | C. parvum pig | C. parvum marsupial | C. meleagridis | C. parvum ferret | C. wrairi | C. parvum human | C. parvum monkey | C. parvum mouse | C. parvum bovine | |
| P. falciparum | 0.00 | |||||||||||||||||
| P. cynomolgi | 21.03 | 0.00 | ||||||||||||||||
| C. muris bovine | 34.84 | 40.51 | 0.00 | |||||||||||||||
| C. muris murine | 32.73 | 38.79 | 1.76 | 0.00 | ||||||||||||||
| C. serpentis | 34.55 | 40.43 | 5.16 | 4.34 | 0.00 | |||||||||||||
| C. baileyi | 29.27 | 36.74 | 15.50 | 15.60 | 15.74 | 0.00 | ||||||||||||
| Cryptosporidium sp. | 32.04 | 37.25 | 20.44 | 20.40 | 21.11 | 14.80 | 0.00 | |||||||||||
| C. felis | 42.57 | 38.79 | 27.74 | 27.84 | 27.97 | 26.59 | 19.42 | 0.00 | ||||||||||
| C. parvum dog | 43.51 | 38.19 | 29.77 | 30.02 | 29.64 | 28.37 | 20.86 | 15.66 | 0.00 | |||||||||
| C. parvum pig | 33.87 | 37.28 | 20.47 | 20.07 | 20.25 | 16.17 | 11.53 | 18.29 | 18.51 | 0.00 | ||||||||
| C. parvum marsupial | 32.28 | 37.22 | 20.92 | 21.15 | 20.61 | 15.97 | 12.15 | 18.36 | 19.23 | 8.00 | 0.00 | |||||||
| C. meleagridis | 33.08 | 36.97 | 21.23 | 21.17 | 21.22 | 17.35 | 13.67 | 18.58 | 16.54 | 9.61 | 5.79 | 0.00 | ||||||
| C. parvum ferret | 34.38 | 37.82 | 22.08 | 21.77 | 21.90 | 16.73 | 13.75 | 18.65 | 17.17 | 8.86 | 5.31 | 4.02 | 0.00 | |||||
| C. wrairi | 32.60 | 36.80 | 21.21 | 20.85 | 20.62 | 16.21 | 12.46 | 18.24 | 17.87 | 8.12 | 4.03 | 4.04 | 2.57 | 0.00 | ||||
| C. parvum human | 33.77 | 37.07 | 21.71 | 21.31 | 21.72 | 16.37 | 13.55 | 18.45 | 17.49 | 8.55 | 4.90 | 4.47 | 3.68 | 2.29 | 0.00 | |||
| C. parvum monkey | 34.20 | 37.10 | 21.78 | 21.38 | 21.59 | 16.51 | 13.44 | 18.41 | 17.51 | 8.73 | 4.96 | 4.58 | 3.73 | 2.35 | 0.25 | 0.00 | ||
| C. parvum mouse | 33.19 | 36.44 | 21.79 | 21.45 | 21.47 | 16.38 | 13.22 | 18.68 | 17.42 | 8.44 | 4.96 | 4.25 | 2.93 | 2.18 | 2.05 | 2.22 | 0.00 | |
| C. parvum bovine | 34.19 | 37.69 | 21.78 | 21.38 | 21.74 | 16.79 | 14.22 | 18.31 | 16.78 | 8.62 | 4.97 | 4.48 | 3.14 | 1.92 | 2.88 | 2.47 | 1.51 | 0.00 |
The values are nucleotide changes per 100 bp calculated by the Kimura two-parameter method using the Wisconsin Package, version 9.0 (Genetics Computer Group).
FIG. 1.
Variation in the HSP70 gene nucleotide sequences in the primer regions of diagnostic tools by Rochelle et al. (29) (A and B), Di Giovanni et al. (7) (A and B), and Stinear et al. (33) (C).
TABLE 3.
Divergence of Cryptosporidium spp. and other C. parvum genotypes from the C. parvum bovine genotype
| Isolate | Species and genotype | Variation at nucleotide level (%) | No. of amino acids changed | Nonsilent mutations/total mutations (%) |
|---|---|---|---|---|
| 589 | C. muris | 18.7 | 41 | 11.5 |
| 707 | C. muris | 18.4 | 38 | 11.5 |
| 64 | C. serpentis | 17.6 | 43 | 12.1 |
| 764 | C. baileyi | 14.7 | 12 | 4.3 |
| 288 | C. felis | 15.3 | 13 | 4.3 |
| 295 | C. meleagridis | 4.0 | 6 | 7.7 |
| 517 | C. wrairi | 1.9 | 1 | 2.7 |
| 692 | Cryptosporidium sp. | 12.5 | 15 | 6.0 |
| 183 | C. parvum human | 1.5 | 5 | 16.6 |
| 244 | C. parvum dog | 14.7 | 7 | 2.5 |
| 428 | C. parvum marsupial | 4.7 | 3 | 3.2 |
| 351 | C. parvum ferret | 3.0 | 2 | 3.3 |
| 518 | C. parvum monkey | 1.7 | 6 | 18.1 |
| 359 | C. parvum mouse | 1.4 | 0 | 0.0 |
| 499 | C. parvum pig | 7.4 | 7 | 4.8 |
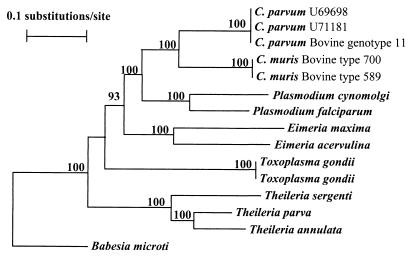
Phylogenetic analysis of C. parvum and C. muris together with published HSP70 sequences of various members of the phylum Apicomplexa, including C. parvum (U71181 and U69698), E. acervulina (Z26134), E. maxima (Z46964), P. cynomolgi (M90978), P. falciparum (M19753), T. annulata (J04653), T. parva (U40190), T. sergenti (D12692), T. gondii (AF045559 and U85648), and B. microti (U534448), revealed a close relationship between the genera Cryptosporidium and Plasmodium (Fig. 2). A neighbor-joining tree showed that the Cryptosporidium clade and the Plasmodium clade clustered together with full statistical reliability. Other intestinal coccidian parasites traditionally associated with Cryptosporidium parasites were placed in a different cluster in this analysis (Fig. 2).
FIG. 2.
Phylogenetic relationship of Cryptosporidium parasites to other apicomplexan parasites inferred from neighbor-joining analysis of HSP70 gene nucleotide sequences.
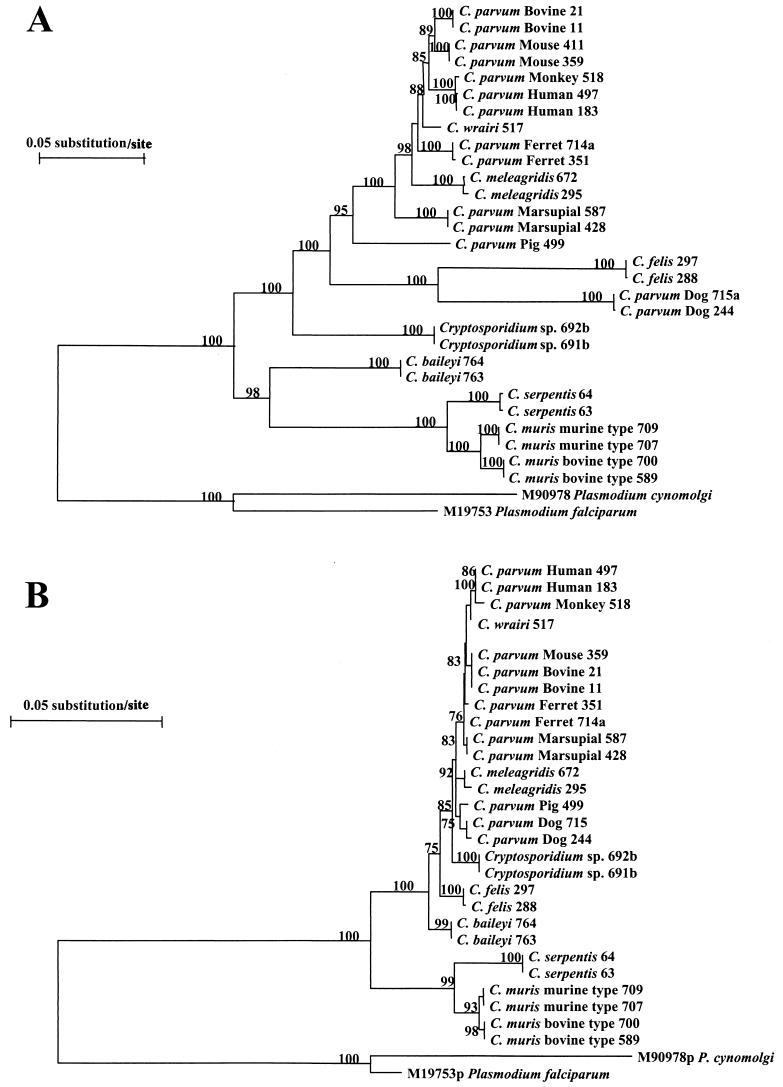
In a second phylogenetic analysis, a neighbor-joining tree was constructed from aligned HSP70 gene sequences of various Cryptosporidium isolates, using nucleotide sequences of P. cynomolgi (M90978) and P. falciparum (M19753) as an outgroup (Fig. 3A). The genus Cryptosporidium formed two distinct clusters in this phylogenetic analysis: the first consisted of two genotypes of C. muris and C. serpentis and C. baileyi isolates, and the second cluster contained isolates of C. felis, C. meleagridis, and C. wrairi, eight different genotypes of C. parvum, and two isolates of the unknown Cryptosporidium species. Within the first cluster, the C. muris bovine type and the C. muris rodent murine isolates formed distinct clades. In the second major cluster, the unknown Cryptosporidium species, C. felis, and the C. parvum dog genotype were separated from the remaining member of the cluster (seven C. parvum genotypes, C. meleagridis, and C. wrairi). Significant intraspecies diversity was seen in C. parvum, as reflected by the presence of eight genotypes. Similar phylogenetic structure was also observed with analysis of deduced amino acid sequences. In the latter, however, C. baileyi clustered together with the broad C. parvum group, and the C. parvum dog genotype and C. felis did not form a clade (Fig. 3B).
FIG. 3.
Phylogenetic relationships among Cryptosporidium parasites inferred from neighbor-joining analysis of nucleotide sequences (A) and deduced amino acid sequences (B).
DISCUSSION
In the present study, nucleotide sequences of the HSP70 gene were obtained from eight Cryptosporidium species and eight different C. parvum genotypes to reassess the species structure of this genus as well as the evolutionary relationship of Cryptosporidium parasites to other members of the phylum Apicomplexa. Results of sequence and phylogenetic analyses are in agreement with our previous observations, based on the SSU rRNA gene, of the presence of multiple species within the genus Cryptosporidium, two genotypes of C. muris, and multiple genotypes of C. parvum (38, 39). The various Cryptosporidium species are placed in different clades and, with the exception of C. wrairi and C. meleagridis, showed interspecies genetic distances comparable to those between different apicomplexan parasites. A close relatedness of Cryptosporidium parasites to Plasmodium parasites was also revealed by phylogenetic analysis of HSP70 sequences, an observation previously suggested by analysis of the structural organization of the rRNA gene (21).
Analysis of the HSP70 gene sequence further suggests that the unknown Cryptosporidium parasite from desert monitors, C. felis, and the C. parvum dog genotype may be valid species. The Cryptosporidium sp. from desert monitors was consistently placed at the bottom of the broad C. parvum cluster. This is also reflected in its genetic distance from other Cryptosporidium parasites, which was >11.53 nucleotide changes per 100 bp. This genetic distance was greater than the distance between C. muris and C. serpentis (4.34 to 5.16) or the C. parvum bovine genotype and C. meleagridis (4.48) and was comparable to the distance between C. baileyi and other Cryptosporidium parasites (14.80 to 17.35). It remains unclear whether this unknown Cryptosporidium sp. is C. saurophilum, a new species identified in desert monitors and other lizards (20). Although, C. felis and the C. parvum dog genotype clustered together with the broad C. parvum clade, their genetic distances from C. muris, C. serpentis, and C. baileyi (26.59 to 30.02 nucleotide changes/100 bp) were far greater than those of other Cryptosporidium spp. (<22 nucleotide changes/100 bp). These isolates also diverged significantly from the rest of the C. parvum genotypes, C. meleagridis, and C. wrairi, with genetic distances (16.54 to 19.23) comparable to those between C. baileyi and other Cryptosporidium species (14.80 to 17.35). This is also reflected in the low G+C contents of HSP70 nucleotide sequences in these two parasites. Previous analysis at the SSU rRNA gene locus also suggested that C. felis, the C. parvum dog genotype, and the unknown Cryptosporidium parasite from desert monitors might be cryptic species (39).
The results of this study suggest that the HSP70 gene offers several advantages over the SSU rRNA gene for phylogenetic studies of Cryptosporidium parasites. Although this gene is under selection pressure (as reflected in the presence of a high percentage of synonymous mutations), the HSP70 gene is apparently more permissive of nucleotide changes. As a result, higher heterogeneity was seen in the HSP70 gene nucleotide sequences than in SSU rRNA gene sequences, which makes it a better target for genotyping. This lower selection pressure in the HSP70 gene is also reflected in the location of nucleotide mutations. Unlike those in the SSU rRNA gene, which restricts nucleotide changes to a certain region of the gene, mutations in the HSP70 gene are spread over the entire sequence. Because deletions and insertions are limited in the HSP70 gene, the alignment of sequences from very different organisms is much easier. Therefore, phylogenetic analysis of Cryptosporidium parasites based on HSP70 gene sequences is much more robust than those based on the SSU rRNA gene, with significantly higher bootstrap values. The minor dissimilarity between the nucleotide and amino acid trees may be explained by the lesser heterogeneity in amino acid sequences due to the predominance of synonymous mutations in the nucleotide sequences. In other eukaryotic systems, the HSP70 gene has also become a useful alternative in the study of molecular evolution (3, 4, 12, 15, 17, 18).
The HSP70 gene sequence generated in this study reveals problems in the current HSP70-gene-based PCR diagnosis tools designed for the use of environmental samples (7, 29, 33). As shown in Fig. 1, the primers used in these protocols matched only sequences from the C. parvum bovine, human, and mouse genotypes. We have found dissimilarities with the C. parvum dog, pig, and marsupial genotypes, C. meleagridis, and C. felis. It is likely that the efficiencies of these primers in amplifying DNA from these organisms may be compromised due to the heterogeneity in the primer regions, especially in the case of environmental samples, which usually have small numbers of organisms. This is a matter of concern, because many of the Cryptosporidium parasites, such as the C. parvum dog genotype, C. felis, and C. meleagridis, have been found in patients with AIDS (27, 28) and in children (L. Xiao, C. Bern, and A. A. Lal, unpublished observation). Indeed, all six C. parvum HSP70 genotypes recently described in water samples (7) belong to the bovine, human, and mouse genotypes in the present study. The minor differences among some of the water genotypes are much smaller than those among other genotypes; thus, they may represent intragenotype diversity or artifacts. The nucleotide sequences of the HSP70 gene generated in this study will be useful in the improvement of these diagnostic tools and in the development of new molecular tools for Cryptosporidium species and genotype differentiation.
ACKNOWLEDGMENTS
This work was supported in part by an interagency agreement (DW75937730-01-0) between the U.S. Environmental Protection Agency and the Centers for Disease Control and Prevention and by Opportunistic Infectious Disease funds from the CDC.
We thank Mike Arrowood, Ron Fayer, and Anne Moore for providing some samples used in the study.
REFERENCES
- 1.Arrowood M J, Sterling C R. Isolation of Cryptosporidium oocysts and sporozoites using discontinuous sucrose and isopycnic Percoll gradients. J Parasitol. 1987;73:314–319. [PubMed] [Google Scholar]
- 2.Awad-El-Kariem F A, Warhurst D C, McDonald V. Detection of Cryptosporidium oocysts using a system based on PCR and endonuclease restriction. Parasitology. 1994;109:19–22. doi: 10.1017/s0031182000077714. [DOI] [PubMed] [Google Scholar]
- 3.Budin K, Philippe H. New insights into the phylogeny of eukaryotes based on ciliate Hsp70 sequences. Mol Biol Evol. 1998;15:943–956. doi: 10.1093/oxfordjournals.molbev.a026010. [DOI] [PubMed] [Google Scholar]
- 4.Bui E T, Bradley P J, Johnson P J. A common evolutionary origin of the mitochondria and hydrogenosomes. Proc Natl Acad Sci USA. 1996;93:9651–9656. doi: 10.1073/pnas.93.18.9651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Champliaud D, Gobet P, Naciri M, Vagner O, Lopez J, Buisson J C, Varga I, Harly G, Mancassola R, Bonnin A. Failure to differentiate Cryptosporidium parvum from C. meleagridis based on PCR amplification of eight DNA sequences. Appl Environ Microbiol. 1998;64:1454–1458. doi: 10.1128/aem.64.4.1454-1458.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cordell R L, Addiss D G. Cryptosporidiosis in child care settings: a review of the literature and recommendations for prevention and control. Pediatr Infect Dis J. 1994;13:311–317. [PubMed] [Google Scholar]
- 7.Di Giovanni G D, Hashemi F H, Shaw N J, Abrams F A, LeChevallier M W, Abbaszadegan M. Detection of infectious Cryptosporidium parvum oocysts in surface and filter backwash water samples by immunomagnetic separation and integrated cell culture-PCR. Appl Environ Microbiol. 1999;65:3427–3432. doi: 10.1128/aem.65.8.3427-3432.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Efron B, Halloran E, Holmes S. Bootstrap confidence levels for phylogenetic trees. Proc Natl Acad Sci USA. 1996;93:13429–13434. doi: 10.1073/pnas.93.23.13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellis R J, van der Vies S M. Molecular chaperones. Annu Rev Biochem. 1991;60:321–347. doi: 10.1146/annurev.bi.60.070191.001541. [DOI] [PubMed] [Google Scholar]
- 10.Fayer R, Spear C A, Dubey J P. The general biology of Cryptosporidium. In: Fayer R, editor. Cryptosporidium and cryptosporidiosis. Boca Raton, Fla: CRC Press; 1997. pp. 1–41. [Google Scholar]
- 11.Felsenstein J. Confidence limits on the phylogenies: an approach using bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 12.Germot A, Philippe H. Critical analysis of eukaryotic phylogeny: a case study based on the HSP70 family. J Eukaryot Microbiol. 1999;46:116–124. doi: 10.1111/j.1550-7408.1999.tb04594.x. [DOI] [PubMed] [Google Scholar]
- 13.Gething M J, Sambrook J. Protein folding in the cell. Nature. 1992;355:33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- 14.Gupta R S, Bustard M, Falah M, Singh D. Sequencing of heat shock protein 70 (DnaK) homologs from Deinococcus proteolyticus and Archaebacteria, Eubacteria, and Thermomicrobium roseum and their integration in a protein-based phylogeny of prokaryotes. J Bacteriol. 1997;179:345–357. doi: 10.1128/jb.179.2.345-357.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta R S, Golding G B. Evolution of the Hsp70 gene and its implications regarding relationships between Archaebacteria, Eubacteria, and Eukaryotes. J Mol Evol. 1993;37:573–582. doi: 10.1007/BF00182743. [DOI] [PubMed] [Google Scholar]
- 16.Hills D M, Bull J J. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst Biol. 1993;8:189–191. [Google Scholar]
- 17.Karasev A V, Kashina A S, Gelfand V I, Dolja V V. Hsp70-related 65 kDa protein of beet yellows closterovirus is a microtubule-binding protein. FEBS Lett. 1992;304:12–14. doi: 10.1016/0014-5793(92)80578-5. [DOI] [PubMed] [Google Scholar]
- 18.Karlin S, Brocchieri L. Heat shock protein 70 family: multiple sequence comparisons, function, and evolution. J Mol Evol. 1998;47:565–577. doi: 10.1007/pl00006413. [DOI] [PubMed] [Google Scholar]
- 19.Khramtsov N V, Tilley M, Blunt D S, Montelone B A, Upton S J. Cloning and analysis of a Cryptosporidium parvum gene encoding a protein with homology to cytoplasmic form HSP70. J Eukaryot Microbiol. 1995;42:416–422. doi: 10.1111/j.1550-7408.1995.tb01605.x. [DOI] [PubMed] [Google Scholar]
- 20.Koudela B, Modry D. New species of Cryptosporidium (Apicomplexa, Cryptosporidiidae) from lizards. Folia Parasitol. 1998;45:93–100. [Google Scholar]
- 21.Le Blancq S V, Khramtsov N V, Zamani F, Upton S J, Wu T W. Ribosomal RNA gene organization in Cryptosporidium parvum. Mol Biochem Parasitol. 1997;90:463–478. doi: 10.1016/s0166-6851(97)00181-3. [DOI] [PubMed] [Google Scholar]
- 22.Lindquist S, Craig E A. The heat shock proteins. Annu Rev Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- 23.Millard P S, Gensheimer K F, Addis D G, Sosin D M, Beckett G A, Houck-Jankoski A, Hudson A. An outbreak of cryptosporidiosis from fresh-pressed apple cider. JAMA. 1994;272:1592–1596. [PubMed] [Google Scholar]
- 24.Morgan U M, Sturdee A P, Singleton G, Gomez M S, Gracenea M, Torres J, Hamilton S G, Woodside D P, Thompson R C A. The Cryptosporidium “Mouse” genotype is conserved across geographic areas. J Clin Microbiol. 1999;37:1302–1305. doi: 10.1128/jcm.37.5.1302-1305.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morgan U M, Buddle J R, Armson A, Elliot A, Thompson R C A. Molecular and biological characterization of Cryptosporidium in pigs. Aust Vet. 1999;77:44–47. doi: 10.1111/j.1751-0813.1999.tb12428.x. [DOI] [PubMed] [Google Scholar]
- 26.Morgan U M, Monis P, Fayer R, Deplazes P, Thompson R C A. Phylogenetic relationships among isolates of Cryptosporidium: evidence for several new species. J Parasitol. 1999;85:1126–1133. [PubMed] [Google Scholar]
- 27.Morgan U M, Xiao L, Sulaiman I, Thompson R C A, Lal A, Deplazes P. Which genotypes/species of Cryptosporidium are humans susceptible to? J Eukaryot Microbiol. 1999;46:42S–43S. [PubMed] [Google Scholar]
- 28.Pieniazek N J, Bornay-Llinares F J, Slemenda S B, da Silva A J, Moura I N, Arrowood M J, Ditrich O, Addiss D G. New cryptosporidium genotypes in HIV-infected persons. Emerg Infect Dis. 1999;5:444–449. doi: 10.3201/eid0503.990318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rochelle P A, Ferguson D M, Handojo T R, Leon R D, Stewart M H, Wolfe R L. An assay combining cell culture with reverse transcriptase PCR to detect and determine the infectivity of waterborne Cryptosporidium parvum. Appl Environ Microbiol. 1997;63:2029–2037. doi: 10.1128/aem.63.5.2029-2037.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saitou, N., and M. Nei. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425. [DOI] [PubMed]
- 31.Slavin D. Cryptosporidium meleagridis (sp. nov.) J Comp Pathol. 1955;65:262–265. doi: 10.1016/s0368-1742(55)80025-2. [DOI] [PubMed] [Google Scholar]
- 32.Smith H V, Rose J B. Water borne cryptosporidiosis: current status. Parasitol Today. 1998;14:14–22. doi: 10.1016/s0169-4758(97)01150-2. [DOI] [PubMed] [Google Scholar]
- 33.Stinear T, Matusan A, Hines K, Sandery M. Detection of a single viable Cryptosporidium parvum oocyst in environmental water concentrates by reverse transcription-PCR. Appl Environ Microbiol. 1996;62:3385–3390. doi: 10.1128/aem.62.9.3385-3390.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sulaiman I M, Xiao L, Yang C, Moore A, Beard C B, Arrowood M J, Lal A A. Differentiating human from animal isolates of Cryptosporidium parvum. Emerg Infect Dis. 1998;4:681–685. doi: 10.3201/eid0404.980424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tyzzer E E. A sporozoon found in the peptic glands of the common mouse. Proc Soc Exp Biol Med. 1907;5:12–13. [Google Scholar]
- 36.Tyzzer E E. An extracellular coccidium, Cryptosporidium muris (gen. & sp. nov.) of the gastric glands of the common mouse. J Med Res. 1910;1:487–590. [PMC free article] [PubMed] [Google Scholar]
- 37.Van de Peer Y, De Wachter R. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci. 1994;10:569–570. doi: 10.1093/bioinformatics/10.5.569. [DOI] [PubMed] [Google Scholar]
- 38.Xiao L, Escalante L E, Yang C, Sulaiman I M, Escalante A A, Montali R, Fayer R, Lal A A. Phylogenetic analysis of Cryptosporidium parasites on the small-subunit ribosomal RNA gene locus. Appl Environ Microbiol. 1999;65:1578–1583. doi: 10.1128/aem.65.4.1578-1583.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao L, Morgan U, Limor J, Escalante A, Arrowood M, Shulaw W, Thompson R C A, Fayer R, Lal A A. Genetic diversity within Cryptosporidium parvum and related species of Cryptosporidium. Appl Environ Microbiol. 1999;65:3386–3391. doi: 10.1128/aem.65.8.3386-3391.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]