Abstract
Herpes simplex virus type 1 (HSV-1) infection results in the disruption of ND10 (also called nuclear bodies, PODs, or PML-associated bodies), which are nuclear matrix domains of unknown function present in mammalian cells. After ND10 disruption, viral transcription and DNA replication occur in globular nuclear domains called replication compartments. In this report we define four stages of infection by using antibodies to ICP8 (also called SSB and UL29) and the ND10 antigen PML. Immediately after infection, cells contain intact ND10 as detected by staining for PMLs (stage I); within 1 hour, however, ND10 are disrupted and cells begin to exhibit diffuse staining for the major viral DNA binding protein, ICP8 (stage II). After all ND10 have been disrupted, foci which resemble but are not equivalent to ND10 appear, containing both PML and ICP8 (stage III). Cells infected with mutants defective in the helicase-primase or origin binding protein are unable to form stage III foci. Cells infected with a mutant that is null for the polymerase catalytic subunit, however, form stage III-like ICP8 foci which do not contain PML. Thus, stage III foci recruit the cellular PML protein in the presence but not the absence of HSV polymerase. PML was recruited to stage III foci in some but not all cells infected with a mutant defective in the polymerase accessory protein, UL42. Thus, UL42 is not required for the recruitment of PML to viral foci. In wild-type infection, stage III cells are quickly replaced by cells containing replication compartments (stage IV). PML and ICP8 staining are both observed within replication compartments, indicating a potential role for PML in HSV-1 replication. Models for the role of ND10 proteins in the formation of replication compartments are discussed.
Several DNA viruses, including adenovirus, simian virus 40, and herpesviruses, have been shown to affect the partitioning of cellular antigens within ND10 (1, 4, 7, 19, 21, 32, 38, 44). These nuclear matrix-associated domains occur at an average of 10 per nucleus and are also known as PODs, Kr bodies, nuclear bodies, and PML-associated bodies (7, 15, 38). ND10 proteins have been associated with the control of cellular growth, transcription, apoptosis, and the life cycles of several viruses (5, 11, 14, 35). The most extensively studied ND10 protein, PML, is expressed as a fusion with the retinoic acid receptor alpha in individuals with acute promyelocytic leukemia (APL) (18). Expression of the PML-retinoic acid receptor alpha fusion protein in APL cells results in both the disruption of ND10 foci and the loss of growth control. Retinoic acid treatment of APL cell lines results in both the restoration of growth control and the reformation of ND10 (20). The observations that dispersal and reformation of ND10 occur during the cell cycle and in response to stress (34, 43) suggest that the partitioning of ND10 antigens into ND10 foci is linked to cellular growth control. PML has been shown to play a role in cellular growth control (46), and both PML and another ND10 protein, Sp100, have been implicated in transcriptional regulation (14, 49).
Soon after infection with herpes simplex virus type 1 (HSV-1), the viral transactivator ICP0 migrates to ND10 and is believed to initiate the dispersal of PML and other ND10 antigens (8, 10, 12, 31, 47). Everett et al. have recently reported that the disruption of ND10 correlates with the ICP0- and proteosome-dependent loss of several PML isoforms (9). DNA genomes have been observed at the periphery of ND10 and have been reported to preferentially initiate transcription at sites adjacent to ND10 (33). During infection, viral DNA replication proteins are found in large globular nuclear domains called replication compartments (40). Under some experimental conditions replication compartments have been shown to form at sites adjacent to ND10 (15, 26). Taken together, these results suggest that ND10 or sites in the cell close to ND10 play an important role in the establishment of a productive infection (32). One attractive model posits that ND10 antigens mark sites in the nucleus that are necessary for the establishment of a productive viral infection (30); whether these sites represent nuclear matrix attachment sites or some other relatively fixed domains within the nucleus remains to be determined. We and others have also observed that ND10 proteins are recruited to replication compartments in infected cells (24, 39), although the role of these proteins in viral replication is not clear.
In this report, we explore the disruption of ND10 and the formation of replication compartments in cells infected with wild-type and mutant viruses. Based on staining patterns of the ND10 protein PML and the HSV-1 major DNA binding protein (SSB, ICP8, or UL29), we define four distinct stages during early infection leading to replication compartment formation. Stage I cells resemble uninfected cells in that they contain intact ND10 and show no ICP8 staining. Stage II cells show diffuse PML staining and no intact ND10. ICP8 staining is diffuse and nuclear during this stage. After the initial disruption of ND10, cells enter a stage (stage III) in which ICP8 and PML colocalize in punctate foci, which resemble one type of prereplicative site. Prereplicative sites in cells infected with wild-type virus in the presence of polymerase inhibitors and in cells infected with viruses lacking the viral DNA polymerase have previously been described (6, 40). It is now recognized that there are two types of prereplicative sites: (i) “numerous,” thought to represent foci of ICP8 and other viral proteins at cellular replication forks in S-phase cells (24), and (ii) “ND10 associated,” thought to represent actual intermediates in the formation of replication compartments (24, 45). The PML-containing ICP8 foci which appear in stage III cells are likely equivalent to the so-called ND10-associated prereplicative sites. In cells competent to carry out viral DNA synthesis, stage III cells are quickly replaced by cells containing replication compartments (stage IV). In this paper we report the requirements for formation of PML-containing prereplicative sites and replication compartments by studying mutants defective in various HSV replication proteins. The presence of PML in stage III prereplicative sites and in replication compartments implicates ND10 proteins in the establishment of viral infection.
MATERIALS AND METHODS
Cells and viruses.
HEp-2 cells were obtained from the American Type Culture Collection (Manassas, Va.). They were maintained in Dulbecco’s modified Eagle’s medium (ICN Biomedicals Inc., Aurora, Ohio) supplemented with 10% fetal calf serum (Atlanta Biologicals) and 1% penicillin–streptomycin solution (Sigma) at 37°C in a 7% CO2 atmosphere.
The KOS strain of HSV-1 was used as the wild-type strain. ICP6::lacZ insertion mutants in the helicase-primase genes, UL5 (hr99), UL8 (hr80), and UL52 (hr114) and the origin binding protein, UL9 (hr94) were previously described (3, 13, 28, 51). The polymerase null mutant HP66 and ΔB10, in which the polymerase coding sequences were restored, were previously described (29, 50). The UL42 mutant CgalΔ42 was generously provided by P. Johnson and D. Parris (17).
Immunofluorescence and imaging.
Cells were grown on glass coverslips and infected as described above. Cells on coverslips were fixed in 3.7% formaldehyde in phosphate-buffered saline (PBS) for 30 min, washed in PBS, and permeabilized in 1.0% Triton X-100 in PBS for 10 min. The coverslips were again washed in PBS and pretreated with 3% normal goat serum in PBS for several minutes. The polyclonal antibody used to detect the viral ICP8 was anti-ICP8, a kind gift from Bill Ruyechan (State University of New York at Buffalo) (41). The monoclonal antibody used for detection of the ND10 protein, PML, was obtained from Santa Cruz Biotechnology (Santa Cruz, Calif.). All antibodies were used at a dilution of 1:200 in 3% normal goat serum. Cells were stained with the primary antibodies for 30 min. Coverslips were washed extensively with PBS between primary and secondary antibodies. The secondary antibodies, Texas red-conjugated goat anti-rabbit and fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulins, were obtained from Cappel, Organon Teknika Corporation (Durham, N.C.) and were used at a dilution of 1:200 in PBS for the 30-min staining of the coverslips. Coverslips were then washed extensively in PBS and mounted in glycerol gelatin containing 2.5% diazibicyclooctane (DABCO) to retard bleaching. Imaging was performed on a Zeiss Axiovert 135 laser scanning microscope (confocal) equipped with an argon-krypton laser. Texas red was excited at 568 nm; fluorescein isothiocyanate was excited at 488 nm. Emissions were collected separately, and the channels were overlaid by computer for the dual images. Images were collected with a 63X Neofluar lens and arranged and labeled by using Adobe Photoshop 3.0.
RESULTS
Four stages of HSV infection.
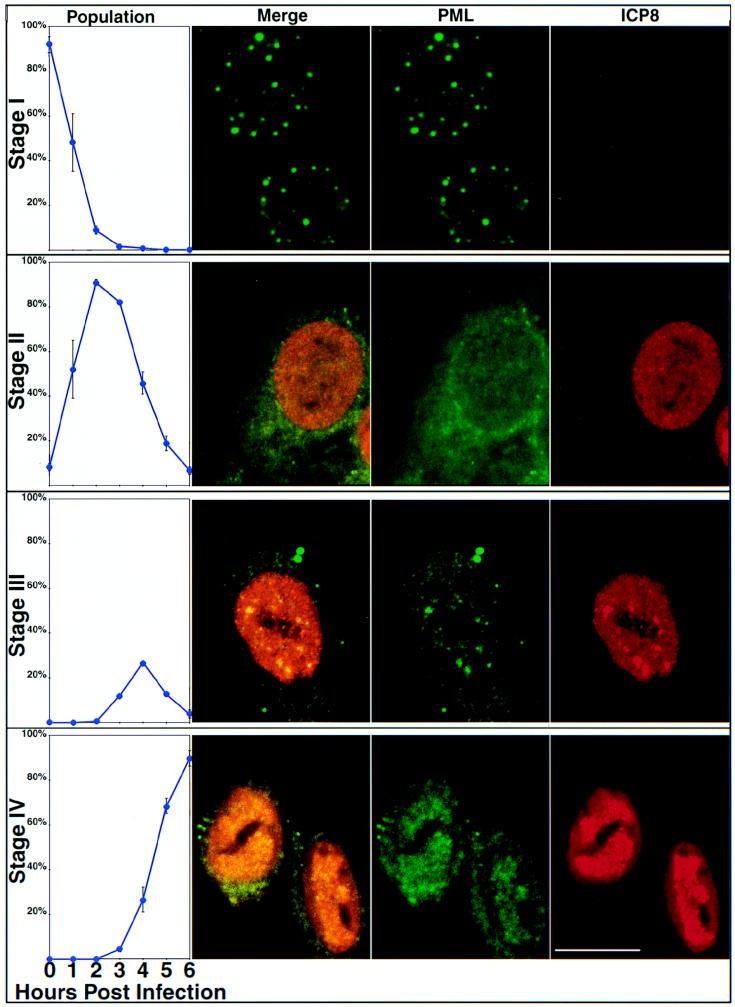
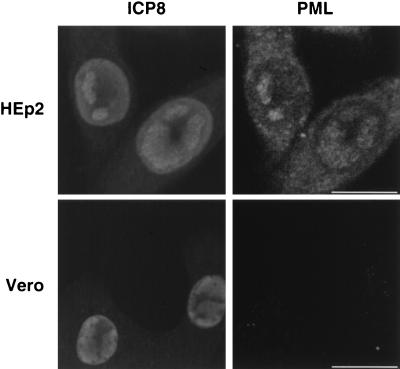
In this report we describe four stages of early HSV-1 infection based on the PML and ICP8 staining patterns observed during the first 6 h of infection. HEp2 cells were infected with KOS at a multiplicity of infection (MOI) of 2 PFU/cell and were harvested at 1-h intervals between 0 and 6 h postinfection. Cells were prepared for immunofluorescence microscopy and stained with a polyclonal antibody specific for ICP8 (41) as a marker for viral replication proteins and a monoclonal antibody specific for PML as a marker for ND10. As a control, cells were stained with one primary antibody and both secondary antibodies: in all cases only the expected staining was observed. This indicates that there was no cross-reactivity of antibodies and no leakage of light from either fluor into the inappropriate channel. A minimum of 100 cells on each coverslip were tallied according to their staining patterns, and the percentages of cells exhibiting each staining pattern are shown in the first column of Fig. 1. At time zero, almost all cells contained PML in intact ND10 and showed no ICP8 staining (Fig. 1, top row); this pattern was designated stage I. Within the first 2 h of infection, PML staining became diffuse and cells exhibited a gradient of ICP8 staining from no ICP8 to strong diffuse nuclear ICP8 staining; we defined this staining pattern stage II (Fig. 1, second row). After the disruption of ND10 and prior to the formation of replication compartments, we observed the unexpected reappearance of PML foci within a background of diffuse nuclear PML staining, peaking at about 3 h postinfection. These nuclear PML foci resembled intact ND10 of uninfected cells and colocalized with ICP8. Although Everett et al. (9) have reported the degradation of several isoforms of PML in cells infected with HSV, some isoforms remain. We suggest that it is these remaining isoforms of PML which appear to be recruited into PML-ICP8 foci. Cells exhibiting these foci were classified as stage III cells (Fig. 1, third row). Stage III appears to be transitory, never exceeding approximately 20% of the cells, and cells in stage III are quickly replaced by cells containing replication compartments (stage IV), which stained for both ICP8 and PML (Fig. 1, bottom row). By approximately 5 h postinfection, almost all cells exhibited replication compartments. In this experiment, some PML staining was seen in the cytoplasm of many cells, although some of this staining may have been perinuclear (Fig. 1). This observation is consistent with the previous report that PML has been shown to shuttle between the cytoplasm and nucleus; however, cells stained with secondary antibodies alone show faint cytoplasmic and/or perinuclear staining but no nuclear staining (data not shown). Therefore, at least some of the cytoplasmic staining is due to background staining with the secondary antibody. The PML staining observed in stage III foci and in the replication compartments in HEp2 cells is not due to cross-reactivity of the PML antibody with viral proteins, because no PML staining was observed in KOS-infected Vero cells (Fig. 2). Vero cells contain a version of PML that does not react with the PML antibody used. These results indicate that the nuclear PML staining observed in infected HEp2 cells is due to PML colocalization with viral replication compartments and not cross-reactivity with viral antigens. In conclusion, we observe and define four different stages of viral infection with respect to PML and ICP8 antigens. Moreover, we confirm that ND10, as observed by PML staining, are disrupted early in viral infection and we suggest that at least some isoforms of the PML protein are present within HSV-1 replication compartments. The most surprising aspect of these experiments was the apparent reformation of PML foci (stage III) after the initial disruption of ND10 during stage II.
FIG. 1.
Early HSV-1 infection occurs in four distinct stages. HEp2 cells were infected with KOS and harvested for immunofluorescence at various time points between 0 and 6 h postinfection. Infected cells were stained with anti-PML monoclonal antibody and anti-ICP8 polyclonal antibody. Over 100 cells for each time point were tallied according to the staining patterns of these two proteins, and four stages of infection were defined. The first column shows the percentage of infected cells in each stage at various time points. The photographs in the second, third, and fourth columns show representative fields of HEp2 cells infected with KOS. The column labeled Merge shows the merged image of staining patterns for PML (green) and ICP8 (red). The columns labeled PML and ICP8 show the single staining patterns of PML (green) and ICP8 (red), respectively. The different rows (graphs and three columns of photographs) represent cells in stages as follows: stage I, cells demonstrating intact ND10 by PML staining and no ICP8; stage II, cells exhibiting disrupted ND10 and diffuse ICP8; stage III, cells exhibiting PML-ICP8 foci against a diffuse nuclear background staining; stage IV, cells exhibiting PML and ICP8 staining in replication compartments. Marker bar = 15 μm.
FIG. 2.
The anti-PML antibody does not cross-react with HSV-1 proteins. HEp2 (top row) and Vero (bottom row) cells were infected with KOS for 5 h, processed for immunofluorescence, and stained with anti-PML and anti-ICP8 as indicated. Marker bar = 15 μm.
Stage III foci may be equivalent to the previously described ND10-associated prereplicative sites.
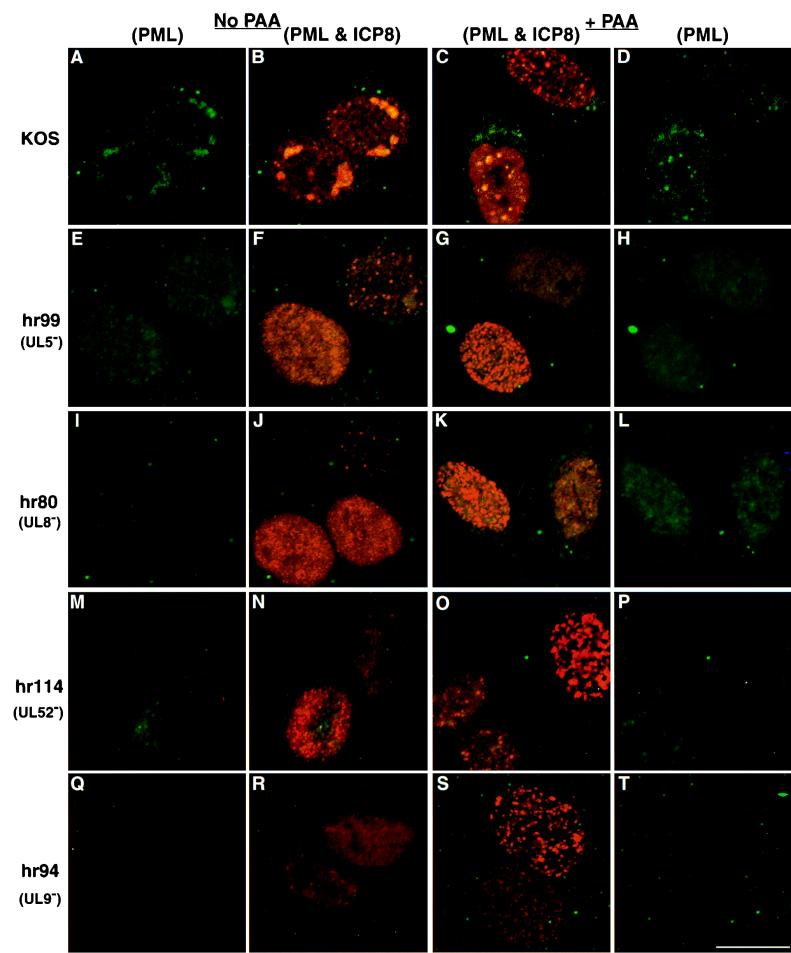
Stage III foci seen during wild-type infection resemble the previously described ND10-associated prereplicative sites observed in cells infected in the presence of polymerase inhibitors such as phosphonoacetic acid (PAA) (24, 45): the ICP8 and PML staining patterns are similar in both cases. The first row of Fig. 3 shows cells infected with wild-type virus. In the absence of PAA (Fig. 3A and B), replication compartments, which stain for both PML and ICP8, were observed. In the presence of PAA (Fig. 3C and D), both types of prereplicative sites were observed (23, 24). The upper cell (Fig. 3C and D) contains numerous prereplicative sites which are thought to represent ICP8 localization to cellular replication forks (24); the majority of these numerous prereplicative sites do not colocalize with PML. The lower cell (Fig. 3C and D) contains ND10-associated prereplicative sites, which are thought to represent intermediates in the formation of replication compartments (23, 24). Note that the cells containing ND10-associated prereplicative sites resemble the stage III cells in Fig. 1, third row. The similarity between these staining patterns prompted us to undertake a more detailed analysis of the kinetics of formation of the ND10-associated prereplicative sites. In order to determine if stage III foci and ND10-associated prereplicative sites form with similar kinetics, we infected HEp2 cells with KOS in the absence and presence of PAA and harvested the infections at 1-h intervals to determine staining patterns of PML and ICP8. In the presence of PAA, cells progressed through stages I (intact ND10), II (disrupted ND10), and III (foci) with the same kinetics as were observed in the absence of PAA (data not shown). In the presence of PAA, however, cells did not progress to stage IV, consistent with the observation that PAA inhibits the formation of replication compartments (40). Since ND10-associated prereplicative sites resemble stage III foci in appearance and formation kinetics, we propose that ND10-associated prereplicative sites represent stage III foci which are not able progress to stage IV because of the presence of PAA. When ND10-associated prereplicative sites were first observed and named, it was assumed these foci appeared prior to the disruption of ND10. The results of the present study with cells infected for shorter periods indicate, however, that these foci formed after, not before, the disruption of ND10. Thus, the observation that ND10 are disrupted before the formation of ICP8-containing foci indicates that the term ND10-associated prereplicative sites may be inappropriate; hereafter in this report we refer to them as PML-containing prereplicative sites. Whether the PML-containing prereplicative sites form at the site of the original ND10 remains to be determined.
FIG. 3.
PML and ICP8 staining in cells infected with various HSV-1 mutants in the presence and absence of PAA. HEp2 cells were infected at an MOI of 10 PFU/cell with HSV-1 strain KOS or mutants lacking helicase-primase subunits or origin binding protein and were processed for immunofluorescence as described in the legend to Fig. 1 at 5 h postinfection. The first two columns represent cells infected with various viruses (as indicated) in the absence of polymerase inhibitors: the first column shows only PML staining, and the second column shows a merged image of the same cells exhibiting both PML and ICP8 staining. The third and fourth columns represent cells infected with various viruses in the presence of the polymerase inhibitor PAA (400 μg/ml): the third column shows the merged image of PML and ICP8 staining, and the fourth column shows PML staining only of the same cells. (A to D) KOS (wild-type)-infected cells; (E to H) cells infected with hr99 lacking the helicase UL5; (I to L) cells infected with hr80 lacking the helicase-primase accessory protein UL8; (M to P) cells infected with hr114 lacking the primase UL52; (Q to T) cells infected with hr94 lacking the origin binding protein, UL9. Marker bar = 15 μm.
HSV-1 mutants defective in DNA synthesis do not form PML-containing prereplicative sites.
To better characterize stage III foci (PML-containing prereplicative sites) and understand the events leading to their formation, we assessed HSV-1 mutants defective in DNA synthesis for their ability to form the PML-containing prereplicative sites. Insertion mutants (hr99, hr80, and hr114) of the HSV-1 helicase-primase (UL5, UL8, and UL52, respectively) and an insertion mutant (hr94) of the origin binding protein (UL9) were previously described (3, 13, 28, 51). ICP8 staining patterns in cells infected with hr99, hr80, hr114, and hr94 were previously shown to be diffuse and nuclear in untreated cells and prereplicative sites in the presence of PAA (23, 25). Since the two types of prereplicative sites had not been distinguished at the time of those observations, we asked whether both types of prereplicative sites are present in HEp2 cells infected with these mutants in the presence and absence of PAA. At 5 h postinfection, cells were fixed for immunofluorescence and stained with antibodies to ICP8 and PML. Cells infected with mutants defective in helicase-primase or origin binding protein in the absence of PAA exhibited similar patterns of ICP8 staining: the untreated cells exhibited diffuse and somewhat granular ICP8 staining which does not colocalize with PML (Fig. 3E, F, I, J, M, N, Q, and R). Cells infected with these mutants in the presence of PAA were able to form the numerous prereplicative sites, but they did not form the PML-containing prereplicative sites, which are fewer in number and colocalize precisely with foci of PML (Fig. 3C, H, K, L, O, P, S, and T). Thus, in the presence of PAA, cells infected with helicase-primase or UL9 mutants exhibited either the diffuse granular pattern of ICP8 staining (as seen in the upper cell in Fig. 3G) or the numerous prereplicative site pattern of ICP8 staining (as seen in the lower cell in Fig. 3G). The fact that PML-containing prereplicative sites (stage III foci) did not form in cells infected with mutants defective in helicase-primase and UL9, proteins necessary for DNA replication, supports the proposal that this subset of prereplicative sites represents a functional intermediate in the formation of replication compartments (24, 45).
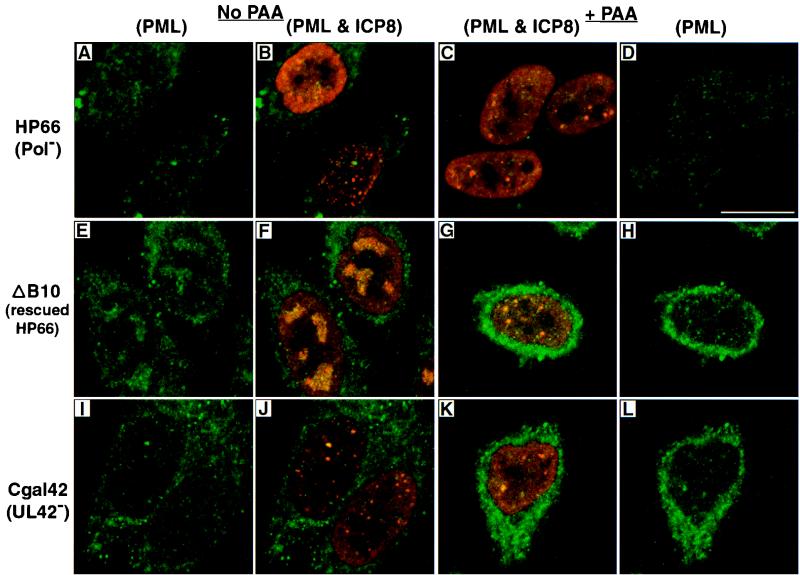
Consistent with previous results (6, 40), cells infected with a mutant defective in the catalytic subunit of the HSV-1 polymerase (HP66) (29) were able to form both types of prereplicative sites in the presence and absence of PAA. Figure 4 shows not only the numerous prereplicative sites (Fig. 4B, bottom cell) but also the less numerous ICP8 foci, which resemble stage III foci (Fig. 4B, top cell; Fig. 4C, all three cells). To our surprise, however, the less numerous ICP8 foci did not stain with PML (Fig. 4A to D). This is in contrast to cells infected with wild-type virus, which exhibited colocalization of ICP8 and PML not only in the absence of PAA (Fig. 1, third row) but also in PML-containing prereplicative sites in the presence of PAA (Fig. 3C and D). The mutant staining pattern is due specifically to the mutation in the polymerase gene, as an HP66-derived virus, ΔB10, in which the polymerase primary coding sequences were restored (29, 50), regains its ability to recruit PML and progress to replication compartment formation (Fig. 4E to H). The observation that PML is not present in the stage III-like foci in cells infected with the polymerase mutant suggests that PML is somehow recruited to these viral structures in the presence of polymerase. This observation raised a question about the apparent interaction of PML with viral foci: is the interaction between PML and (i) the polymerase, (ii) the polymerase accessory protein (UL42), or (iii) a viral complex that is dependent on the polymerase?
FIG. 4.
Viral foci in cells infected with polymerase and polymerase accessory mutant viruses. HEp2 cells were infected at an MOI of 10 PFU/cell with either HP66, a virus lacking the catalytic domain of the HSV-1 polymerase; ΔB10, a virus derived from HP66 whose polymerase coding sequences have been restored by marker rescue; or Cgal 42, a virus lacking the polymerase accessory protein. The first two columns represent cells infected as indicated in the absence of polymerase inhibitors: the first column shows only PML staining, and the second column shows a merged image of the same cells exhibiting both PML and ICP8 staining. The third and fourth columns represent cells infected with various viruses in the presence of the polymerase inhibitor PAA (400 μg/ml): the third column shows the merged image of PML and ICP8 staining, and the fourth column shows PML staining only of the same cells. (A to D) HP66-infected cells; (E to H) ΔB10-infected cells showing a wild-type phenotype; (I to L) Cgal 42-infected cells. The strong cytoplasmic fluorescein staining seen in panels E to L is likely due to the secondary antibody used; control experiments show no secondary antibody staining of the nucleus. Marker bar = 15 μm.
Since polymerase is thought to recruit UL42 to viral replication foci (23), we hypothesized that the recruitment of PML to viral foci might be dependent on the presence of the polymerase accessory protein. We asked whether cells infected with a virus lacking the UL42 gene would show PML recruitment to viral foci. In HEp2 cells infected with the UL42 null virus, CgalΔ42 (17), PML was recruited to viral foci in some, but not all, of the cells showing viral foci (Fig. 4I to L). This intermediate phenotype suggests that the presence of UL42 is not essential for the recruitment of PML to stage III foci.
In sum, viral mutants lacking the helicase-primase subunits or the origin binding protein cannot form stage III foci (PML-associated prereplicative sites). The viral mutant lacking an active polymerase can form ICP8-containing foci resembling stage III foci; however, these foci do not associate with PML as they do during wild-type infection.
DISCUSSION
Several observations regarding the relationship between ND10 and viral infection are made in this report. (i) The early events of HSV-1 replication can be broken down into four stages as identified by confocal microscopy of ICP8 and PML staining patterns. PML appears to be recruited into stage III and stage IV foci after the initial disruption of ND10. (ii) Stage III foci do not form in cells infected with mutants lacking UL5, UL8, UL9, or UL52. (iii) The association of PML with the stage III-like ICP8 foci requires the HSV DNA polymerase catalytic subunit. (iv) UL42, the HSV-1 polymerase accessory protein, enhances but is not absolutely required for the recruitment of PML to viral foci.
In this report we have confirmed that ND10 are disrupted almost immediately upon infection (stage II) (10, 31). An unexpected result, however, was the appearance of PML-containing foci after the initial disruption of ND10 (stage III). Stage III foci, which may represent the earliest replication compartments, resemble the cellular ND10 in their staining with PML antibodies, in their morphology, and in their approximate number per cell, but they form after most cells have undergone complete disruption of ND10 in stage II. The similarity between ND10 and stage III foci raises the question of whether stage III foci form at the sites originally occupied by ND10 or whether they form at unrelated sites in the nucleus. If stage III foci form at the same subnuclear sites which were previously occupied by ND10, it would indicate that ND10 mark sites within the nucleus which are important to the replication of the virus. The concept that ND10 marks important sites on the nuclear matrix was originally proposed by Maul (30). The observations that replication compartments form adjacent to ND10 in transfected cells (26) and in cells infected with an ICP0 mutant (16) support the notion that the PML-containing prereplicative sites form at or near the site previously occupied by ND10. We cannot rule out, however, that PML-containing stage III foci represent the nucleation or aggregation of PML protein at unrelated sites in the nucleus as it is recruited to viral foci after being released from cellular ND10.
Müller et al. (36) and Sternsdorf et al. (42) recently showed that modification of the ND10 protein SUMO, also called PIC1 (2, 27, 37), affects its partitioning within the nucleus (36, 42). When the PML protein is modified by SUMO, it is observed in the nuclear matrix-insoluble cell fraction, or ND10 (36, 42). When, however, the protein is unmodified, the protein is found in the soluble nucleoplasm outside of ND10 (36, 42). The modification and movement appear to be reversible and may represent a nuclear response to influences such as heat shock, interferon treatment, and heavy metal treatment. More recently, Everett et al. have shown that HSV-1 infection has profound effects on the modification state of PML (9). Upon infection, SUMO-modified forms of PML appear to be degraded, leaving only the unmodified, soluble forms of PML in the nucleus (9). Further experiments will be required to determine the modification state of the PML which is recruited to HSV-1-infected cells during stages III and IV. It is possible that the PML observed in stage III and stage IV foci represents unmodified isoforms not targeted for degradation.
In order to better understand the viral requirements for the formation of stage III foci and replication compartments, we studied the viral foci formed in cells infected with viruses defective in DNA replication genes. Since the previously identified ND10-associated prereplicative sites (now designated PML-containing prereplicative sites) resemble stage III foci both in their morphology and in the time of their appearance, we propose that PML-containing prereplicative sites are equivalent to these intermediates but are unable to progress to replication compartments because of polymerase inhibition. In previous studies Liptak et al. (23) and Lukonis and Weller (25) examined the requirements for the formation of prereplicative sites, using viral mutants lacking individual HSV replication proteins such as helicase-primase, origin binding protein, and viral polymerase. Both of these studies, however, were performed before the two types of prereplicative sites were identified. In this study we have examined the involvement of viral DNA replication genes in the formation of the PML-containing prereplicative sites. We show that stage III foci do not form in cells infected with mutants lacking the helicase-primase or the origin binding protein. Furthermore, in the presence of a polymerase inhibitor, the helicase-primase and origin binding protein mutant viruses form the numerous type of prereplicative sites, thought to represent localization of ICP8 to cellular replication forks of cells in S phase, but they do not form the PML-containing prereplicative sites which resemble stage III foci. This result indicates that the formation of PML-containing prereplicative sites (and stage III foci) requires the presence of each of the helicase-primase subunits and the origin binding protein. This supports the model proposed by Liptak et al. in which ICP8, UL5, UL8, UL52, and UL9 form a subassembly of viral replication proteins which later recruits the polymerase and polymerase accessory protein (23).
In addition to containing each of the viral DNA replication proteins, replication compartments have been reported to contain both PML and Sp100 and another ND10 protein recognized by monoclonal antibody 138 (24, 26, 39). A surprising finding made in the present study is that PML is recruited to PML-containing prereplicative sites (stage III foci) and replication compartments in a manner which is dependent on the presence of the viral polymerase. ICP8 foci resembling stage III foci are observed in cells infected with a virus lacking the polymerase, but they do not contain PML. We considered several possible explanations for the inability of a polymerase null mutant to recruit PML into stage III foci. First, it was possible that UL42, the polymerase accessory factor, is responsible for the recruitment of PML to replication foci. However, in cells infected with a UL42 null virus, PML was detected in viral foci in some but not all of the cells. This result indicates that UL42 is not absolutely required for PML recruitment but may enhance PML recruitment in some way. Second, it was possible that the polymerase protein itself is required for PML recruitment. Experiments using small deletions and point mutations of the polymerase catalytic subunit will be required to answer this question. It is also possible that PML is recruited only to areas at which DNA synthetic machinery is intact, although it is interesting that the presence of a polymerase inhibitor, PAA, does not prevent PML recruitment to viral foci. Regardless of how PML is recruited to wild-type viral foci, the recruitment of PML to stage III foci and replication compartments raises questions about its role in these domains. Since PML has never been implicated directly in cellular DNA replication, it seems unlikely that it plays a direct role in DNA synthesis; however, it may play a role in the recruitment of other cellular proteins which are involved either directly or indirectly in DNA replication. The observation that several cellular proteins involved in DNA replication and repair are recruited to viral prereplicative sites and replication compartments (48) is consistent with this hypothesis.
During HSV infection ICP0 is responsible for the selective degradation of some isoforms of PML as well as other cellular proteins including the large subunit of the DNA-dependent protein kinase (9, 22). The loss of the PML isoforms is correlated with the ND10 disruption (9). At least two models which are not mutually exclusive can be envisioned for how ND10 disruption may contribute to the establishment of a productive viral replication. (i) Everett et al. have proposed that the selective degradation of cellular proteins leads to a stimulation of viral gene expression (9). The positive effects on viral gene expression may increase the likelihood that viral infection will progress efficiently. (ii) Viral DNA replication may require association with nuclear sites that are marked by ND10 (30). The disruption of ND10 and the degradation of some isoforms of PML upon infection may expose underlying sites, allowing the viral genome or viral replication proteins to become associated. According to this model, the presence of intact ND10 may inhibit viral DNA replication by masking important nuclear attachment sites.
ACKNOWLEDGMENTS
We thank Bill Ruyechan for providing antisera and P. A. Johnson and D. S. Parris for providing the UL42 mutant virus, CgalΔ42. We also thank the members of our laboratory and Mark Challberg for helpful comments.
This investigation was supported by Public Health Service grants AI21747 to S.K.W. and AI19838 to D.M.C.
REFERENCES
- 1.Ahn J H, Hayward G S. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J Virol. 1997;71:4599–4613. doi: 10.1128/jvi.71.6.4599-4613.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boddy M N, Howe K, Etkin L D, Solomon E, Freemont P S. PIC 1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene. 1996;13:971–982. [PubMed] [Google Scholar]
- 3.Carmichael E P, Kosovsky M J, Weller S K. Isolation and characterization of herpes simplex virus type 1 host range mutants defective in viral DNA synthesis. J Virol. 1988;62:91–99. doi: 10.1128/jvi.62.1.91-99.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carvalho T, Seeler J S, Ohman K, Jordan P, Pettersson U, Akusjarvi G, Carmo-Fonseca M, Dejean A. Targeting of adenovirus E1A and E4-ORF3 proteins to nuclear matrix-associated PML bodies. J Cell Biol. 1995;131:45–56. doi: 10.1083/jcb.131.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen G Q, Shi X G, Tang W, Xiong S M, Zhu J, Cai X, Han Z G, Ni J H, Shi G Y, Jia P M, Liu M M, He K L, Niu C, Ma J, Zhang P, Zhang T D, Paul P, Naoe T, Kitamura K, Miller W, Waxman S, Wang Z Y, de The H, Chen S J, Chen Z. Use of arsenic trioxide (As2O3) in the treatment of acute promyelocytic leukemia (APL): I. As2O3 exerts dose-dependent dual effects on APL cells. Blood. 1997;89:3345–3353. [PubMed] [Google Scholar]
- 6.de Bruyn Kops A, Knipe D M. Formation of DNA replication structures in herpes virus-infected cells requires a viral DNA binding protein. Cell. 1988;55:857–868. doi: 10.1016/0092-8674(88)90141-9. [DOI] [PubMed] [Google Scholar]
- 7.Doucas V, Ishov A M, Romo A, Juguilon H, Weitzman M D, Evans R M, Maul G G. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 1996;10:196–207. doi: 10.1101/gad.10.2.196. [DOI] [PubMed] [Google Scholar]
- 8.Dyck J A, Maul G G, Miller W H, Jr, Chen J D, Kakizuka A, Evans R M. A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell. 1994;76:333–343. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- 9.Everett R D, Freemont P, Saitoh H, Dasso M, Orr A, Kathoria M, Parkinson J. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J Virol. 1998;72:6581–6591. doi: 10.1128/jvi.72.8.6581-6591.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Everett R D, Maul G G. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 1994;13:5062–5069. doi: 10.1002/j.1460-2075.1994.tb06835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fagioli, M., F. Grignani, P. F. Ferrucci, M. Alcalay, A. Mencarelli, I. Nicoletti, F. Grignani, and P. G. Pelicci. 1994. Effect of the acute promyelocytic leukemia PML/RAR alpha protein on differentiation and survival of myeloid precursors. Leukemia 8(Suppl.):7–11. [PubMed]
- 12.Gelman I H, Silverstein S. Co-ordinate regulation of herpes simplex virus gene expression is mediated by the functional interaction of two immediate early gene products. J Mol Biol. 1986;191:395–409. doi: 10.1016/0022-2836(86)90135-x. [DOI] [PubMed] [Google Scholar]
- 13.Goldstein D J, Weller S K. An ICP6::lacZ insertional mutagen is used to demonstrate that the UL52 gene of herpes simplex virus type 1 is required for virus growth and DNA synthesis. J Virol. 1988;62:2970–2977. doi: 10.1128/jvi.62.8.2970-2977.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guiochon-Mantel A, Savouret J F, Quignon F, Delabre K, Milgrom E, de The H. Effect of PML and PML-RAR on the transactivation properties and subcellular distribution of steroid hormone receptors. Mol Endocrinol. 1995;9:1791–1803. doi: 10.1210/mend.9.12.8614415. [DOI] [PubMed] [Google Scholar]
- 15.Ishov A M, Maul G G. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J Cell Biol. 1996;134:815–826. doi: 10.1083/jcb.134.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishov A M, Stenberg R M, Maul G G. Human cytomegalovirus immediate early interaction with host nuclear structures: definition of an immediate transcript environment. J Cell Biol. 1997;138:5–16. doi: 10.1083/jcb.138.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson P A, Best M G, Friedmann T, Parris D S. Isolation of a herpes simplex virus type 1 mutant deleted for the essential UL42 gene and characterization of its null phenotype. J Virol. 1991;65:700–710. doi: 10.1128/jvi.65.2.700-710.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kakizuka A, Miller W H, Jr, Umesono K, Warrell R P, Jr, Frankel S R, Murty V V, Dmitrovsky E, Evans R M. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell. 1991;66:663–674. doi: 10.1016/0092-8674(91)90112-c. [DOI] [PubMed] [Google Scholar]
- 19.Kelly C, Van Driel R, Wilkinson G W. Disruption of PML-associated nuclear bodies during human cytomegalovirus infection. J Gen Virol. 1995;76:2887–2893. doi: 10.1099/0022-1317-76-11-2887. [DOI] [PubMed] [Google Scholar]
- 20.Koken M H, Puvion-Dutilleul F, Guillemin M C, Viron A, Linares-Cruz G, Stuurman N, de Jong L, Szostecki C, Calvo F, Chomienne C, Degos L, Puvion E, de The H. The t(15;17) translocation alters a nuclear body in a retinoic acid-reversible fashion. EMBO J. 1994;13:1073–1083. doi: 10.1002/j.1460-2075.1994.tb06356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korioth F, Maul G G, Plachter B, Stamminger T, Frey J. The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp Cell Res. 1996;229:155–158. doi: 10.1006/excr.1996.0353. [DOI] [PubMed] [Google Scholar]
- 22.Lees-Miller S P, Long M C, Kilvert M A, Lam V, Rice S A, Spencer C A. Attenuation of DNA-dependent protein kinase activity and its catalytic subunit by the herpes simplex virus type 1 transactivator ICP0. J Virol. 1996;70:7471–7477. doi: 10.1128/jvi.70.11.7471-7477.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liptak L M, Uprichard S L, Knipe D M. Functional order of assembly of herpes simplex virus DNA replication proteins into prereplicative site structures. J Virol. 1996;70:1759–1767. doi: 10.1128/jvi.70.3.1759-1767.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lukonis C J, Burkham J, Weller S K. Herpes simplex virus type 1 prereplicative sites are a heterogeneous population: only a subset are likely to be precursors to replication compartments. J Virol. 1997;71:4771–4781. doi: 10.1128/jvi.71.6.4771-4781.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lukonis C J, Weller S K. Characterization of nuclear structures in cells infected with herpes simplex virus type 1 in the absence of viral DNA replication. J Virol. 1996;70:1751–1758. doi: 10.1128/jvi.70.3.1751-1758.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lukonis C J, Weller S K. Formation of herpes simplex virus type 1 replication compartments by transfection: requirements and localization to nuclear domain 10. J Virol. 1997;71:2390–2399. doi: 10.1128/jvi.71.3.2390-2399.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 28.Malik A K, Martinez R, Muncy L, Carmichael E P, Weller S K. Genetic analysis of the herpes simplex virus type 1 UL9 gene: isolation of a LacZ insertion mutant and expression in eukaryotic cells. Virology. 1992;190:702–715. doi: 10.1016/0042-6822(92)90908-8. [DOI] [PubMed] [Google Scholar]
- 29.Marcy A I, Yager D R, Coen D M. Isolation and characterization of herpes simplex virus mutants containing engineered mutations at the DNA polymerase locus. J Virol. 1990;64:2208–2216. doi: 10.1128/jvi.64.5.2208-2216.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maul G. Nuclear domain 10, the site of DNA virus transcription and replication. Bioessays. 1998;20:660–667. doi: 10.1002/(SICI)1521-1878(199808)20:8<660::AID-BIES9>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 31.Maul G G, Everett R D. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J Gen Virol. 1994;75:1223–1233. doi: 10.1099/0022-1317-75-6-1223. [DOI] [PubMed] [Google Scholar]
- 32.Maul G G, Guldner H H, Spivack J G. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0) J Gen Virol. 1993;74:2679–2690. doi: 10.1099/0022-1317-74-12-2679. [DOI] [PubMed] [Google Scholar]
- 33.Maul G G, Ishov A M, Everett R D. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology. 1996;217:67–75. doi: 10.1006/viro.1996.0094. [DOI] [PubMed] [Google Scholar]
- 34.Maul G G, Yu E, Ishov A M, Epstein A L. Nuclear domain 10 (ND10) associated proteins are also present in nuclear bodies and redistribute to hundreds of nuclear sites after stress. J Cell Biochem. 1995;59:498–513. doi: 10.1002/jcb.240590410. [DOI] [PubMed] [Google Scholar]
- 35.Mu Z M, Chin K V, Liu J H, Lozano G, Chang K S. PML, a growth suppressor disrupted in acute promyelocytic leukemia. Mol Cell Biol. 1994;14:6858–6867. doi: 10.1128/mcb.14.10.6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Müller S, Matunis M J, Dejean A. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 1998;17:61–70. doi: 10.1093/emboj/17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okura T, Gong L, Kamitani T, Wada T, Okura I, Wei C F, Chang H M, Yeh E T. Protection against Fas/APO-1- and tumor necrosis factor-mediated cell death by a novel protein, sentrin. J Immunol. 1996;157:4277–4281. [PubMed] [Google Scholar]
- 38.Puvion-Dutilleul F, Chelbi-Alix M K, Koken M, Quignon F, Puvion E, de The H. Adenovirus infection induces rearrangements in the intranuclear distribution of the nuclear body-associated PML protein. Exp Cell Res. 1995;218:9–16. doi: 10.1006/excr.1995.1125. [DOI] [PubMed] [Google Scholar]
- 39.Puvion-Dutilleul F, Venturini L, Guillemin M C, de The H, Puvion E. Sequestration of PML and Sp100 proteins in an intranuclear viral structure during herpes simplex virus type 1 infection. Exp Cell Res. 1995;221:448–461. doi: 10.1006/excr.1995.1396. [DOI] [PubMed] [Google Scholar]
- 40.Quinlan M P, Chen L B, Knipe D M. The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell. 1984;36:857–868. doi: 10.1016/0092-8674(84)90035-7. [DOI] [PubMed] [Google Scholar]
- 41.Shelton L S, Albright A G, Ruyechan W T, Jenkins F J. Retention of the herpes simplex virus type 1 (HSV-1) UL37 protein on single-stranded DNA columns requires the HSV-1 ICP8 protein. J Virol. 1994;68:521–525. doi: 10.1128/jvi.68.1.521-525.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sternsdorf T, Jensen K, Will H. Evidence for covalent modification of the nuclear dot-associated proteins PML and Sp100 by PIC1/SUMO-1. J Cell Biol. 1997;139:1621–1634. doi: 10.1083/jcb.139.7.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stuurman N, de Graaf A, Floore A, Josso A, Humbel B, de Jong L, van Driel R. A monoclonal antibody recognizing nuclear matrix-associated nuclear bodies. J Cell Sci. 1992;101:773–784. doi: 10.1242/jcs.101.4.773. [DOI] [PubMed] [Google Scholar]
- 44.Szekely L, Pokrovskaja K, Jiang W-Q, de The H, Ringertz N, Klein G. The Epstein-Barr virus-encoded nuclear antigen EBNA-5 accumulates in PML-containing bodies. J Virol. 1996;70:2562–2568. doi: 10.1128/jvi.70.4.2562-2568.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uprichard S L, Knipe D M. Assembly of herpes simplex virus replication proteins at two distinct intranuclear sites. Virology. 1997;229:113–125. doi: 10.1006/viro.1996.8430. [DOI] [PubMed] [Google Scholar]
- 46.Wang Z G, Delva L, Gaboli M, Rivi R, Giorgio M, Cordon-Cardo C, Grosveld F, Pandolfi P P. Role of PML in cell growth and the retinoic acid pathway. Science. 1998;279:1547–1551. doi: 10.1126/science.279.5356.1547. [DOI] [PubMed] [Google Scholar]
- 47.Weis K, Rambaud S, Lavau C, Jansen J, Carvalho T, Carmo-Fonseca M, Lamond A, Dejean A. Retinoic acid regulates aberrant nuclear localization of PML-RAR alpha in acute promyelocytic leukemia cells. Cell. 1994;76:345–356. doi: 10.1016/0092-8674(94)90341-7. [DOI] [PubMed] [Google Scholar]
- 48.Wilcock D, Lane D P. Localization of p53, retinoblastoma and host replication proteins at sites of viral replication in herpes-infected cells. Nature. 1991;349:429–431. doi: 10.1038/349429a0. [DOI] [PubMed] [Google Scholar]
- 49.Xie K, Lambie E J, Snyder M. Nuclear dot antigens may specify transcriptional domains in the nucleus. Mol Cell Biol. 1993;13:6170–6179. doi: 10.1128/mcb.13.10.6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yager D R, Marcy A I, Coen D M. Translational regulation of herpes simplex virus DNA polymerase. J Virol. 1990;64:2217–2225. doi: 10.1128/jvi.64.5.2217-2225.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu L, Weller S K. The UL5 gene of herpes simplex virus type 1: isolation of a lacZ insertion mutant and association of the UL5 gene product with other members of the helicase-primase complex. J Virol. 1992;66:458–468. doi: 10.1128/jvi.66.1.458-468.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]