Abstract
Strain SF3, a gram-negative, anaerobic, motile, short curved rod that grows by coupling the reductive dechlorination of 2-chlorophenol (2-CP) to the oxidation of acetate, was isolated from San Francisco Bay sediment. Strain SF3 grew at concentrations of NaCl ranging from 0.16 to 2.5%, but concentrations of KCl above 0.32% inhibited growth. The isolate used acetate, fumarate, lactate, propionate, pyruvate, alanine, and ethanol as electron donors for growth coupled to reductive dechlorination. Among the halogenated aromatic compounds tested, only the ortho position of chlorophenols was reductively dechlorinated, and additional chlorines at other positions blocked ortho dechlorination. Sulfate, sulfite, thiosulfate, and nitrate were also used as electron acceptors for growth. The optimal temperature for growth was 30°C, and no growth or dechlorination activity was observed at 37°C. Growth by reductive dechlorination was revealed by a growth yield of about 1 g of protein per mol of 2-CP dechlorinated, and about 2.7 g of protein per mole of 2,6-dichlorophenol dechlorinated. The physiological features and 16S ribosomal DNA sequence suggest that the organism is a novel species of the genus Desulfovibrio and which we have designated Desulfovibrio dechloracetivorans. The unusual physiological feature of this strain is that it uses acetate as an electron donor and carbon source for growth with 2-CP but not with sulfate.
Substantial amounts of halogenated aromatic compounds have been released to the environment and many of them have accumulated in groundwater and sediments (11, 14, 24, 26). Many of these compounds are resistant to aerobic microbial metabolism often because the chlorine substitution blocks oxygenase attack. Fortunately, many halogenated compounds can be dehalogenated by anaerobic microorganisms. Several bacteria capable of dehalogenating halogenated aromatic compounds have been isolated and characterized (2, 3, 4, 5, 18, 25, 27, 28, 29). One previous isolate has been described which reductively dechlorinates 2-chlorophenol (2-CP) to phenol (5). This organism was isolated from freshwater sediment and is unique for dechlorinators in that it could also grow as a microaerophile, although it did not dechlorinate under this condition. Bacteria, such as this one, which use the dehalogenation reaction as their sole electron acceptor for energy generation and growth are termed halorespirers (25) or dehalorespirers (11).
To date, much of the dehalogenation research has been directed toward understanding the fate and behavior of halogenated pollutants in freshwater sediments, soils, and sludges. Comparatively little is known about the fates of these pollutants or dehalogenating organisms from the marine environment despite the fact that marine biota produce a remarkable array of halogenated compounds (12). For example, studies by King (15) indicate that 2,4-dibromophenol occurred at concentrations of up to several hundred micromolar in hemichordate burrow walls and that this chemical was dehalogenated in these sediments under anaerobic conditions. This result suggested that bacterial populations from some marine habitats may have developed enzymatic capabilities to degrade these naturally occurring organohalides. Hence, marine sources may reveal further diversity of dehalogenating microorganisms.
Here, we describe enrichment, isolation, and characterization of a novel marine bacterium capable of growth in a synthetic seawater medium on 2-CP and acetate. This new isolate dechlorinates ortho-chlorophenol, producing phenol as a product. Phenotypic and 16S ribosomal DNA (rDNA) phylogenetic studies indicate that the organism belongs to the Desulfovibrio group of the sulfate-reducing bacteria. To our knowledge, this is the first member of the Desulfovibrio group that is capable of oxidizing acetate.
MATERIALS AND METHODS
Media and growth conditions.
Cultures were grown in 160-ml serum bottles with 50 or 100 ml of anaerobic synthetic seawater medium or in 30-ml anaerobic culture tubes with 20 ml of medium. The medium was modified from standard seawater media to remove sulfate so that dechlorinators rather than sulfate reducers could be enriched, and to achieve an Na+ concentration of 0.46 M, which approximates that of seawater. It contained the following mineral salts (in grams/liter): NaCl, 25; MgCl2, 1.4; KH2PO4, 0.2; NH4Cl, 0.3; KCl, 0.5; and CaCl2, 0.1. A trace element solution was added to give the following final concentrations (in milligrams/liter): MnCl2 · 6H2O, 5; H3BO3, 0.5; ZnCl2, 0.5; CoCl2 · 6H2O, 0.5; NiSO4 · 6H2O, 0.5; CuCl2 · 2H2O, 0.3; and NaMoO4 · 2H2O, 0.1. In addition, the medium contained 0.003 mg of NaSeO3 and 0.008 mg of Na2WO4 per liter and 10 mg of resazurin per liter. The medium was boiled under oxygen-free N2 and cooled to room temperature under N2-CO2 (95:5). Na2S (as a reductant) and NaHCO3 were then added to final concentrations of 1 and 30 mM, respectively, and the pH of the medium was adjusted to 7.3 to 7.5 by varying the CO2 concentration in the headspace. The medium was dispensed into N2-CO2-flushed serum bottles or culture tubes capped with butyl rubber stoppers and sterilized by autoclaving. The sterile medium was amended with an anaerobic sterile Wolin vitamin solution (34) plus thiamine, 1,4-naphthoquinone, nicotinamide, hemin, and lipoic acid at concentrations of 0.05, 0.2, 0.5, 0.05, and 0.05 mg/liter, respectively. MgCl2 and CaCl2 were added from sterile anaerobic stock solutions to avoid precipitation.
Sediment samples were collected from tidal muds near Palo Alto Baylands Preserve on the west shore of San Francisco Bay and transported in headspace-free, air tight jars. About 10 g of sediment was transferred under anaerobic conditions to 160-ml serum bottles with 100 ml of seawater medium amended with 2-CP to the final concentration of 250 μM. The cultures were incubated stationary in the dark at 25°C.
Enrichment and isolation of dechlorinating microorganisms.
Dechlorinating sediment microcosms were fed additional 2-CP several times, and then a 10% transfer was made to a fresh seawater medium. After 2 mM 2-CP was consumed in the second transfer, a series of cultures (5% inoculum, diluted 10−1 to 10−7) was made for further enrichment of 2-CP dechlorinators in the seawater medium amended with 250 μM 2-CP and 2.5 mM acetate as an electron donor. Dechlorinating organisms were isolated from this enrichment using deep agarose shake cultures which consisted of 10 ml of anaerobic seawater medium solidified with 1% low-gelling-temperature agarose and supplemented with 2-CP and acetate to final concentrations of 250 μM and 2.5 mM, respectively.
Gram staining, microscopy, and test of desulfoviridin.
Gram staining was done by standard methods (8). Phase-contrast photomicrographs of cells spread on dry agarose coated slides were taken using a Zeiss microscope. The presence of desulfoviridin was assayed by the method of Postgate (22).
Phenotypic characterization.
To test the salt dependency of the isolate, duplicate 20-ml cultures of seawater medium were amended with 2-CP plus acetate, along with various concentrations of NaCl, KCl, and sucrose. Growth was measured by monitoring the depletion of 2-CP and the appearance of phenol by high-performance liquid chromatography (HPLC).
To test the range of electron donors used, duplicate 20-ml cultures of seawater medium were amended with 2-CP and electron donors at final concentrations of 250 μM and 5 mM, respectively. Acetate at 50 to 100 μM was added as the carbon source when formate and hydrogen were tested as electron donors for dechlorination and sulfate reduction. Growth was determined by measuring the depletion of 2-CP, the production of phenol and the increase of visual culture turbidity over three successive feedings.
To determine the range of electron acceptors used, the indicated halogenated aromatic compounds were added at 250 μM to seawater medium with 2.5 mM acetate in duplicate 20-ml cultures. Fumarate, sulfate, sulfite, thiosulfate, and nitrate were also tested as electron acceptors at 5 mM with each of three different electron donors: acetate, lactate, and pyruvate (5 mM each). The cultures were periodically monitored for the consumption of electron donors, depletion of electron acceptors, appearance of products, and increase in culture turbidity over three successive feedings. A 1% inoculum from an actively dechlorinating culture grown on acetate and 2-CP in seawater medium was used for all of the above experiments. Strict anaerobic conditions were used, and all cultures were incubated at 25°C.
To test if the reductive dechlorination activity was inducible, two sets of duplicate cultures were grown on fumarate plus pyruvate and on 2-CP plus acetate. When 500 μM 2-CP and 5 mM fumarate had been depleted, both sets of cultures were amended with 100 μM 2-CP. Samples were taken at 3-h intervals after 2-CP addition and then analyzed for 2-CP removal.
14C-acetate utilization.
The isolate was grown with 14CH314COOH and 2-CP, sulfate, or no electron acceptor. The cultures were incubated for 3 to 4 weeks and then analyzed as follows: 1 ml of culture fluid was taken from which 0.25 ml was added to the vials containing 100 μl of 2 N HCl and 0.25 ml was added to the vials containing 100 μl of 2 N KOH. The remaining 0.5 ml were filtered through 0.22-μm (pore-size) filters, and then the filters were rinsed with 20 ml of deionized water and placed in scintillation vials to determine the 14C assimilated. KOH (5 ml of a 2 N concentration) was also added to the original cultures, and 0.25 ml was taken for the detection of all the 14C present, including the 14CO2 in the headspace. The vials with HCl were degassed with forced air to drive off CO2. A biodegradable scintillation cocktail (5 ml) was added to each vial, the contents were shaken until clear, and then the radioactivity was determined by scintillation counting. The 14C in bacterial cells was measured in vials with the filter. The 14C in bacterial cells and in the culture solution including dissolved 14CO2 was measured in vials with KOH. At the same time, vials with HCl gave the 14C in bacterial cells and culture solution excluding 14CO2 in the culture solution. The production of total 14CO2 was calculated after subtracting 14C values in vials with HCl from the total 14C recovered.
Growth rate and protein yield.
Because of the low cell density, growth rates were estimated from the rate of phenol production. Samples were taken from duplicate cultures every 12 h and analyzed for phenol production. To measure growth yield, replicate cultures were grown in serum bottles in seawater medium amended with 2.5 mM acetate and with or without 250 μM 2-CP or 2,6-dichlorophenol (2,6-DCP). After about 1 mmol of the substrates was consumed, the cultures were harvested and analyzed for substrate transformation and protein yield.
Chemical analysis.
Phenol and chlorophenols were analyzed by HPLC with a Hibar RP-18 (10-μm) column, a flow rate of 1.5 ml/min, an eluent of H2O-CH3CN-H3PO4 (66:33:0.1), and a UV detector set to 218 nm. The appearance of products and the disappearance of substrates were verified by comparison with authentic standards and with zero time samples. Acetate, fumarate, pyruvate, lactate, formate, and succinate were analyzed by ion-exclusion HPLC. Sulfate, nitrate, and nitrite were analyzed by ion chromatography. Ammonia was analyzed by a Lachat flow injection analyzer. H2 was analyzed by a Hewlett-Packard gas chromatograph with a reduction gas detector. Protein yield was measured by the method of Lowry after alkaline hydrolysis (13).
Genomic fingerprinting.
As a test for culture purity, the isolate was cultured under three different growth conditions: 2-CP plus acetate, sulfate plus lactate, and Luria-Bertani (LB) medium. Samples (4 ml) were collected from these cultures for repetitive extragenic palindromic PCR using the primers and procedure of Rademaker et al. (23).
16S rRNA gene sequencing and analysis.
DNA was extracted by a method for diverse bacteria (30) from a 20-ml culture. The 16S rRNA gene was amplified by using primers FD1 and RD1 (31) and a PCR protocol for this purpose (5). The PCR product was purified using Wizard Purification System (Promega) and sequenced in both directions with an automated fluorescent dye terminator sequencer. The primers used for sequencing corresponded to conserved regions of the 16S rRNA gene sequence (33).
The resulting sequence was analyzed, and the phylogenetic placement was obtained using Ribosomal Database Project (RDP-II, version 7.1) (19). A maximum-likelihood phylogenetic tree was created with the program fastDNAml (21).
Nucleotide sequence accession numbers.
The 16S rDNA sequence described above was deposited under GenBank accession number AF230530. The 16S rDNA sequence of strain TBP-1 was obtained from GenBank as accession number AF090830. The 16S rDNA sequences for the following organisms were obtained in aligned format from RDP-II (19) (RDP-II and GenBank accessions are in parentheses): Desulfovibrio profundus (Dsv.profun, U90726), D. aespoeensis (Dsv.aespoe, X95230), D. halophilus (Dsv.halph2, X99237), “D. oxyclinae” (Dsv.spPIB, U33316), D. salexigens (Dsv.salexi, M34401), D. zosterae (Dsv.zoster, Y18049), D. senezii (Dsv.senezi, AF050100), D. aminophilus (Dsv.amphil, AF067964), D. africanus (Dsv.afric2, X99236), D. alcoholovorans (Dsv.alvora, AF053751), D. fructosovorans (Dsv.frvora, AF050101), D. gabonensis (Dsv.gabonn, U31080), “D. fairfieldensis” (Dsv.fairfl, U42221), D. cuneatus (Dsv.cuneat, X99501), D. litoralis (Dsv.litora, X99504), D. acrylicus (Dsv.acryli, U32578), and Escherichia coli (E. coli, J01695).
RESULTS
Isolation of strain SF3.
Dechlorination activity was obtained in cultures diluted to 10−7 of the third serial transfer. The active 10−7 dilution was fed additional 2-CP and then diluted to inoculate deep agarose shake cultures. Small white colonies became visible after about 1 week. Thirty individual colonies were picked from the 10−6 and 10−7 dilutions and transferred to homologous liquid media and tested for dechlorination activity. After 2 weeks, 2-CP disappeared from 27 cultures with the concomitant appearance of an approximately equal amount of phenol. To further ensure culture purity, one culture was randomly chosen and further purified through a second round of dilution and deep agarose shake cultures. Eight colonies were picked, and all of them showed dechlorination activity within 2 weeks. One of the cultures was selected for further study; it was designated as strain SF3, indicating that it was enriched from San Francisco Bay sediment. Culture purity was demonstrated by the following methods: (i) dechlorination activity was recovered in dechlorination medium when a 1% inoculum was made from cultures grown on LB medium, sulfate plus lactate, or fumarate plus pyruvate; (ii) dechlorination activity was recovered from the colonies grown on sulfate plus lactate and on LB plates; and (iii) repetitive extragenic palindromic PCR patterns were identical from cultures grown on the three media.
Strain characteristics.
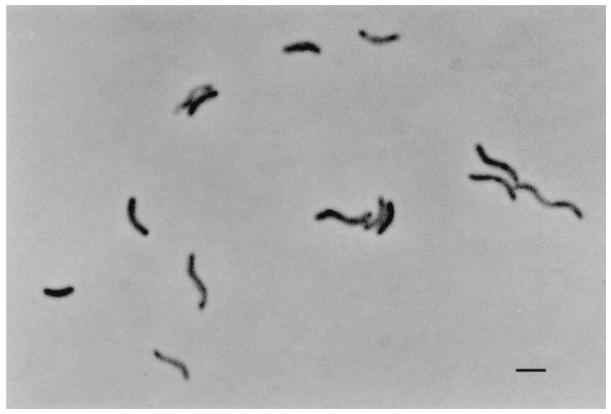
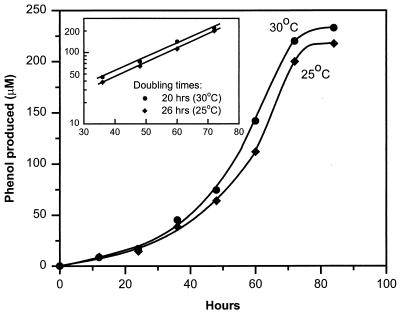
Strain SF3 is a gram-negative, motile, short curved rod 1 to 4 μm long by 0.4 to 0.6 μm wide (Fig. 1). Desulfoviridin was present in the organism. Anaerobic conditions were required for reductive dechlorination and growth, since both ceased when oxygen was introduced into the liquid cultures. The optimal temperature for dechlorination and growth was 30°C, and no dechlorination activity or growth was observed at 37°C. The doubling times on 2-CP plus acetate, inferred from the rate of phenol appearance, at 25 and 30°C were 26 and 20 h, respectively (Fig. 2).
FIG. 1.
Phase-contrast micrograph of strain SF3. Reference bar is 1 μm.
FIG. 2.
Estimation of growth rate based on phenol production. Data are averaged from duplicate cultures on 2-CP plus acetate at 25 and 30°C, respectively.
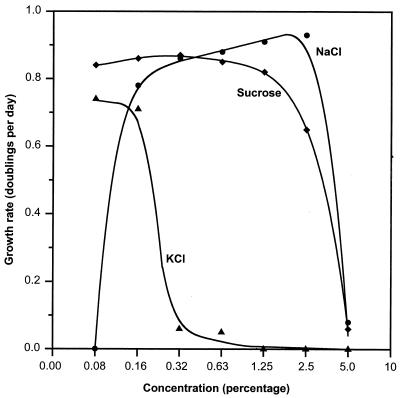
Reductive dechlorination activity occurred at similar rates at concentrations of NaCl ranging from 0.16 to 2.5% (Fig. 3). No dechlorination or growth occurred in the absence of NaCl. Similar dechlorination activity was obtained when strain SF3 was cultured in high concentrations (0.16 to 2.5%) of sucrose but KCl above 0.32% inhibited dechlorination and growth.
FIG. 3.
Growth of strain SF3 in NaCl, sucrose, and KCl amended seawater medium. Growth was measured by the rate of reductive dechlorination of 2-CP in duplicate cultures. NaCl (0.16%) was provided in all media amended with sucrose and KCl.
Electron donors and acceptors.
Besides acetate, strain SF3 also used fumarate, lactate, propionate, pyruvate, alanine, and ethanol as electron donors for reductive dechlorination and growth but did not use hydrogen, formate, phenol, benzoate, or butyrate. Fumarate and propionate produced slower dechlorination rates than the other electron donors. Pyruvate was fermented stoichiometrically to acetate. Among the halogenated electron acceptors tested, only the ortho position of chlorophenols was dechlorinated, and additional chlorines at other positions blocked ortho dechlorination (Table 1). Strain SF3 converted fumarate to succinate and also used sulfate, sulfite, thiosulfate, and nitrate as electron acceptors to oxidize lactate or pyruvate incompletely to acetate. Acetate was not further oxidized when any of these inorganic compounds were the electron acceptors.
TABLE 1.
Test of halogenated aromatic compounds and other chemicals as electron acceptors for strain SF3
| Electron acceptor | Growtha
|
Product(s) | |
|---|---|---|---|
| Acetate | Lactateb | ||
| 2-CP | + | + | Phenol |
| 2-BP | − | − | |
| 3-CP | − | − | |
| 4-CP | − | − | |
| 2,3-DCP | − | − | |
| 2,4-DCP | − | − | |
| 2,5-DCP | − | − | |
| 2,6-DCP | + | + | 2-CP, phenol |
| 2,4,6-Trichlorophenol | − | − | |
| Fumarate | + | + | Succinate |
| Nitrate | − | + | Nitrite, acetate |
| Sulfate | − | + | Acetate |
| Sulfite | − | + | Acetate |
| Thiosulfate | − | + | Acetate |
A plus sign indicates growth assessed by measuring the consumption of the electron donor and acceptor and the appearance of culture turbidity. Data are from duplicate cultures.
The same results were obtained when pyruvate was used as the electron donor.
Induction of dechlorination reaction.
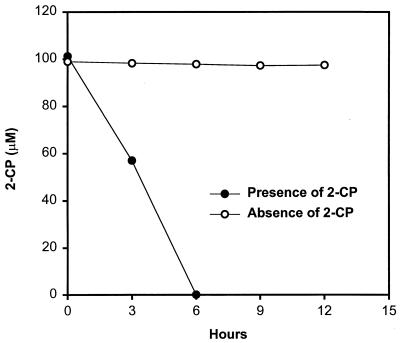
Dechlorination activity was induced by growth on 2-CP, since the culture previously exposed to 2-CP rapidly dechlorinated the additional 2-CP, while the culture previously grown on fumarate plus pyruvate showed no significant decrease in 2-CP concentration within 12 h after addition of 2-CP (Fig. 4).
FIG. 4.
Induction of dechlorination activity was determined by comparison of two sets of cultures previously grown in the presence or absence of 2-CP. Fumarate was substituted as the electron acceptor for growth in the absence of 2-CP.
Assimilation and oxidation of acetate.
Incorporation of 14C-acetate into cells and the production of 14CO2 occurred when 2-CP was present, but not when sulfate was present (Table 2). The ratio of 14CO2 produced per 14C assimilated is consistent with oxidative growth on acetate. These results indicate that strain SF3 is able to couple the oxidation of acetate to the reductive dechlorination of 2-CP.
TABLE 2.
Acetate utilization during anaerobic respiration with 2-CP, sulfate, or no electron acceptor
| Growth conditions (culture no.)a | Fate of 14C from 14C-acetate (dpm/μl)
|
|||
|---|---|---|---|---|
| Total 14C recovered | Filter | Non-acid volatileb | Acid volatile (14CO2)c | |
| 2-CP (1) | 117 | 3.12 | 95.4 | 21.6 |
| 2-CP (2) | 116 | 3.14 | 89.4 | 26.6 |
| Sulfate (1) | 121 | 0.112 | 118 | 3 |
| Sulfate (2) | 118 | 0.113 | 116 | 2 |
| None (1) | 118 | 0.060 | 122 | −4 |
| None (2) | 117 | 0.070 | 122 | −5 |
Data for each replicate culture receiving 14C-acetate are indicated by “1” and “2” in parentheses. Cultures were inoculated with a 1% transfer from an active dechlorinating culture, and reductive dechlorination of 2-CP was measured by HPLC analysis.
14C in acid solution which includes 14C in cells.
Total 14CO2 = total 14C recovered − 14C in acid solution.
Protein yield coupled to reductive dechlorination.
The growth of strain SF3 via reductive dechlorination was demonstrated by measuring the protein yield in the presence or absence of 2-CP and 2,6-DCP with acetate as the electron donor (Table 3). Although both the 2-CP and 2,6-DCP cultures produced approximately 1 mM phenol the amount of cell protein more than doubled with 2,6-DCP as the electron acceptor. This is consistent with the increased available free-energy per mol of electron acceptor with the extra chlorine substituent on 2,6-DCP.
TABLE 3.
Protein yield for strain SF3 grown on acetate and ortho-chlorophenols
| Substrate (culture no.)a | Substrate consumed (μmol) | Phenol produced (μmol) | Acetate consumed (μmol) | Protein (μg) | Substrate/ acetate ratio | Protein/ phenol ratio (g/mol) |
|---|---|---|---|---|---|---|
| 2-CP (1) | 992 | 973 | 370 | 1,995 | 2.63 | 1.07 |
| 2-CP (2) | 997 | 987 | 300 | 1,970 | 3.2 | 1.03 |
| 2,6-DCP (1) | 989 | 957 | 620 | 3,725 | 1.54 | 2.9 |
| 2,6-DCP (2) | 1,004 | 1,001 | 500 | 3,433 | 2.0 | 2.48 |
| None | 950 |
Data for each replicate culture are indicated by “1” and “2” in parentheses. The protein yield was calculated after subtracting protein measured in the controls. None, average of data from duplicate control cultures.
Phylogeny of strain SF3.
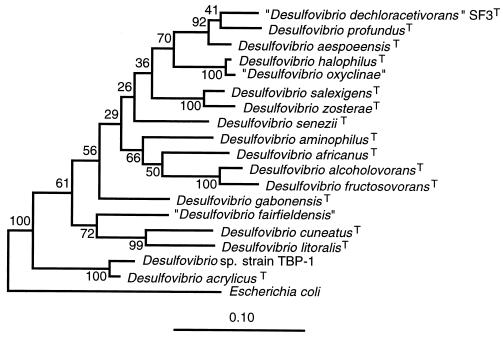
Comparison of the 16S rDNA sequence of strain SF3 with those in RDP-II indicated that strain SF3 is a member of the Desulfovibrio group of the sulfate-reducing bacteria (Fig. 5). Its closest relatives are Desulfovibrio aespoeensis and Desulfovibrio profundus, with similarities of 95.3 and 93.5%, respectively. Phylogenetic analysis of 16S rRNA genes from strain SF3 and selected Desulfovibrio strains indicated that it shares a specific relationship with D. profundus and D. aespoeensis to the exclusion of other Desulfovibrio spp., even though the individual species of Desulfovibrio could not be well resolved (indicated by low bootstrap values). Additional analysis using parsimony and neighbor-joining methods gave similar results (not shown). With all methods, strain SF3 grouped with D. profundus and D. aespoeensis, while the debrominating strain TBP-1 (3) grouped with D. acrylicus.
FIG. 5.
Maximum-likelihood phylogenetic tree based on the 16S rDNA sequences of strain SF3 and representative bacteria. Numbers at internal nodes are the percentage of 100 bootstrap samples in which the group to the right of the node was monophyletic. The scale is the expected number of substitutions per position. T, type strain (as specified in Materials and Methods).
DISCUSSION
The results presented here suggest that the new isolate gains energy for growth from the dechlorination reaction. Acetate supported growth with 2-CP as an electron acceptor, producing CO2, while no growth occurred when only acetate was present. The protein yield was proportional to the amount of chlorine removed and was about twice as much on 2,6-DCP as on 2-CP, implying that, like strains DCB-1 (9, 10, 20) and 2CP-1 (5), strain SF3 gains energy by using the chlorinated aromatic substrate as a respiratory electron acceptor. Also, the stoichiometry of chlorine removed to acetate consumed is in agreement with the theoretical maximal value of four electron pairs produced per acetate oxidized to CO2, with the remaining reducing equivalents going to the biomass. The average fraction of equivalents going to the biomass (fs) was calculated from the protein yield data and 14C-acetate assimilation data to be 0.14. This is similar to values reported by Löffler et al. (17), and the corresponding fraction of electrons used for reductive dechlorination (0.86 = fe) is not far from values reported for halorespiring cultures (17). Based on the data a stoichiometric equation for strain SF3 can be written as follows: CH3COO− + 3.44 2-CP + 0.056 NH4+ + 0.944 H+ + 0.99 H2O→3.44 phenol + 1.72 CO2 + 0.056 C5H7O2N (cells) + 3.44 HCl.
It is very interesting that acetate cannot be oxidized by coupling growth to sulfate, sulfite, thiosulfate, and nitrate but can be used as an electron donor for reductive dechlorination. Growth by coupling acetate oxidation to reductive dechlorination was observed in strain SF3 within 2 weeks with a 1% inoculum. Sulfate reduction, however, was not observed in strain SF3 within 6 months when acetate was provided as the electron donor. That the same organic compound was not used under different electron-accepting conditions was unexpected. Perhaps, strain SF3 employs different electron transport systems for dechlorination and sulfate reduction. To our knowledge, except for strain SF3, no sulfate-reducing bacteria in the genus Desulfovibrio can oxidize acetate under any electron-accepting conditions.
Strain SF3 seems well adapted to the marine and estuarine environments since it grows at concentrations of NaCl ranging from 0.16 to 2.5%. This result is different from a salinity-dependent debrominating bacterium isolated from estuarine sediments for which the highest growth rate and yield were achieved with 3.75% NaCl (3). Since no dechlorination activity was obtained when NaCl was substituted by sucrose or KCl, strain SF3 must have an Na+ requirement. The concentration of Na+ in the river waters is about 5 to 40 mg/liter (32), which is much lower than that required by strain SF3 and that used for enrichment and isolation of some dechlorinating microorganisms from freshwater lake sediment and soil samples (2, 5). It is particularly interesting that under the same enrichment conditions used here, except for the high NaCl concentration, soil and freshwater environments yielded halorespiring myxobacteria (5, 24). These myxobacteria, however, would not grow in the presence of NaCl concentrations equivalent to seawater (J. R. Cole, A. L. Cascarelli, and J. M. Tiedje, unpublished data.).
The evaluation of substrate range for reductive dechlorination indicated that only the ortho position of chlorophenols was dechlorinated and that additional chlorines at other positions blocked ortho dechlorination. Other chlorophenol-dehalogenating microorganisms also only dehalogenated substituents ortho to the phenolic group. Strain 2CP-1, a myxobacterium, dehalogenated several ortho-halogenated aromatics including 2-CP, 2-bromophenol (2-BP), 2,5-DCP, and 2,6-DCP (5). A gram-positive Desulfitobacterium transformed several chlorinated phenols, including 2,3-DCP, 2,3,4-trichlorophenol, and 2,4,6-trichlorophenol (29). It is surprising that 2-BP was not a dehalogenation substrate for strain SF3, since most of the previous 2-CP dechlorinating bacteria also used 2-BP (5, 24). The extremely limited range of dehalogenated substrates used by strain SF3 suggests that chlorophenol dechlorination activity is not a fortuitous or cometabolic reaction. The fact that the dechlorination is inducible is further evidence for a more specific, enzymatically catalyzed dechlorination reaction. The historical reason for such an enzymatic activity is less clear, although some halogenated aromatic compounds occur in natural environments and especially in marine sediments as a consequence of animal and algal activities (1, 6, 12, 15, 16).
Comparison of the 16S rDNA sequence of strain SF3 with a database of 16S rDNA sequences indicated that the organism is a member of the delta subdivision of proteobacteria, as are other 2-CP dechlorinating bacteria such as strains 2CP-1 and 2CP-C (5, 24). The 16S rDNA sequence of strain SF3, however, does not place the organism among the myxobacteria, but instead maps it to the Desulfovibrio group of the sulfate-reducing bacteria which have nutritional versatility and phylogenetic diversity (7). Strain SF3's cell morphology, motility, and sulfate reduction are also characters in common with this genus. However, halorespiration, acetate oxidation, the lack of use of H2, and a 5% difference in 16S rDNA sequence from the nearest relative suggest that strain SF3 represents a new species of the genus Desulfovibrio.
Description of Desulfovibrio dechloracetivorans sp. nov.
D. dechloracetivorans (de.chlor.a.ce.ti.vo′rans. L. pref. de, off, away; Gr. n. chloro, referring to the group VII element chlorine; L. n. acetum, vinegar; L. part. adj. vorans, consuming; M. L. part. adj. dechloracetivorans, removing chlorine and consuming acetate, referring to the characteristic of coupling acetate oxidation to reductive dechlorination for growth). Cells are gram-negative, anaerobic, motile, short curved rods 1 to 4 μm long by 0.4 to 0.6 μm wide. Cells are able to grow by coupling the oxidation of acetate to the reductive dechlorination of 2-CP. ortho-Chlorophenols, sulfate, sulfite, thiosulfate, nitrate, and fumarate are used as electron acceptors. Acetate, fumarate, lactate, propionate, pyruvate, alanine, and ethanol are used as electron donors for reductive dechlorination. Lactate and pyruvate are incompletely oxidized to acetate during growth on sulfate or nitrate. Growth occurs at NaCl concentrations of 0.16 to 2.5%. The optimal growth temperature is 30°C. Colonies grown on LB agar medium, lactate-plus-sulfate medium and 2-CP–plus–acetate medium are white and round with a diameter of 1 to 2 mm after 1 week of growth. The type strain is SF3. The organism was isolated from San Francisco Bay sediment. It has been deposited in the American Type Culture Collection as strain ATCC 700912.
ACKNOWLEDGMENTS
We thank Junko Munakata-Marr for help collecting the sediment sample and Frank Dazzo for microscopy.
This research was supported by ONR grant N00014-95-1-0115 and the Center for Microbial Ecology through NSF grant DEB 9120006.
REFERENCES
- 1.Ashworth R B, Cornier M J. Isolation of 2,6-dibromophenol from the marine hemichordate, Balanoglossus biminiensis. Science. 1967;156:158–159. doi: 10.1126/science.155.3769.1558. [DOI] [PubMed] [Google Scholar]
- 2.Bouchard B, Beaudet R, Villemur R, McSween G, Lépine F, Bisaillon J-G. Isolation and characterization of Desulfitobacterium frappieri sp. nov., an anaerobic bacterium which reductively dechlorinates pentachlorophenol to 3-chlorophenol. Int J Syst Bacteriol. 1996;46:1010–1015. doi: 10.1099/00207713-46-4-1010. [DOI] [PubMed] [Google Scholar]
- 3.Boyle A W, Phelps C D, Young L Y. Isolation from estuarine sediments of a Desulfovibrio strain which can grow on lactate coupled to the reductive dehalogenation of 2,4,6-tribromophenol. Appl Environ Microbiol. 1999;65:1133–1140. doi: 10.1128/aem.65.3.1133-1140.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christiansen N, Ahring B K. Desulfitobacterium hafniense sp. nov., an anaerobic, reductively dechlorinating bacterium. Int J Syst Bacteriol. 1996;46:442–448. [Google Scholar]
- 5.Cole J R, Cascarelli A L, Mohn W W, Tiedje J M. Isolation and characterization of a novel bacterium growing via reductive dechlorination of 2-chlorophenol. Appl Environ Microbiol. 1994;60:3536–3542. doi: 10.1128/aem.60.10.3536-3542.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Craigie J S, Gruening D E. Bromophenols from red algae. Science. 1967;157:1058–1059. doi: 10.1126/science.157.3792.1058. [DOI] [PubMed] [Google Scholar]
- 7.Devereux R, He S-H, Doyle C L, Orkland S, Stahl D A, LeGall J, Whitman W B. Diversity and origin of Desulfovibrio species: phylogenetic definition of a family. J Bacteriol. 1990;172:3609–3619. doi: 10.1128/jb.172.7.3609-3619.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doetsch R N. Determinative methods of light microscopy. In: Gerhardt P, Murray R G E, Costilow R N, Nester E W, Wood W A, Krieg N R, Phillips G B, editors. Manual of methods for general bacteriology. Washington, D.C.: American Society for Microbiology; 1981. pp. 21–33. [Google Scholar]
- 9.Dolfing J. Reductive dechlorination of 3-chlorobenzoate is coupled to ATP production and growth in an anaerobic bacterium, strain DCB-1. Arch Microbiol. 1990;153:264–266. doi: 10.1007/BF00249079. [DOI] [PubMed] [Google Scholar]
- 10.Dolfing J, Harrison B K. Gibbs free energy of formation of halogenated aromatic compounds and their potential roles as electron acceptors in anaerobic environments. Environ Sci Technol. 1992;26:2213–2218. [Google Scholar]
- 11.Fetzner S. Bacterial dehalogenation. Appl Microbiol Biotechnol. 1998;50:633–657. doi: 10.1007/s002530051346. [DOI] [PubMed] [Google Scholar]
- 12.Gribble G W. Naturally occurring organohalogen compounds—a survey. J Nat Prod. 1992;55:1353–1395. doi: 10.1021/acs.jnatprod.3c00803. [DOI] [PubMed] [Google Scholar]
- 13.Hanson R S, Phillips J A. Chemical composition. In: Gerhardt P, Murray R G E, Costilow R N, Nester E W, Wood W A, Krieg N R, Phillips G B, editors. Manual of methods for general bacteriology. Washington, D.C.: American Society for Microbiology; 1981. pp. 328–364. [Google Scholar]
- 14.Holliger C, Wohlfarth G, Diekert G. Reductive dechlorination in the energy metabolism of anaerobic bacteria. FEMS Microbiol Rev. 1999;22:383–398. [Google Scholar]
- 15.King G M. Inhibition of microbial activity in marine sediments by a bromophenol from a hemichordate. Nature. 1986;323:257–259. [Google Scholar]
- 16.King G M. Dehalogenation in marine sediments containing natural sources of halophenols. Appl Environ Microbiol. 1988;54:3079–3085. doi: 10.1128/aem.54.12.3079-3085.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Löffler F E, Tiedje J M, Sanford R A. Fraction of electrons consumed in electron acceptor reduction and hydrogen thresholds as indicators of halorespiratory physiology. Appl Environ Microbiol. 1999;65:4049–4056. doi: 10.1128/aem.65.9.4049-4056.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Madsen T, Licht D. Isolation and characterization of an anaerobic chlorophenol-transforming bacterium. Appl Environ Microbiol. 1992;58:2874–2878. doi: 10.1128/aem.58.9.2874-2878.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maidak B L, Cole J R, Lilburn T G, Parker C T, Jr, Saxman P R, Stredwick J M, Garrity G M, Li B, Olsen G J, Pramanik S, Schmidt T M, Tiedje J M. The RDP (Ribosomal Database Project) continues. Nucleic Acids Res. 2000;28:173–174. doi: 10.1093/nar/28.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohn W W, Tiedje J M. Evidence for chemiosmotic coupling of reductive dechlorination and ATP synthesis in Desulfomonile tiedjei. Arch Microbiol. 1991;157:1–6. [Google Scholar]
- 21.Olsen G J, Matsuda H, Hagstrom R, Overbeek R. fastDNAml: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comput Appl Biosci. 1994;10:41–48. doi: 10.1093/bioinformatics/10.1.41. [DOI] [PubMed] [Google Scholar]
- 22.Postgate J R. A diagnostic reaction for Desulfovibrio desulphuricans. Nature. 1959;183:481–482. doi: 10.1038/183481b0. [DOI] [PubMed] [Google Scholar]
- 23.Rademaker J L W, Louws F J, de Bruijn F J. Characterization of the diversity of ecologically important microbes by rep-PCR genomic fingerprinting. In: Akkermans A D L, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 1–27. [Google Scholar]
- 24.Sanford R A. Characterization of microbial populations in anaerobic food webs that reductively dechlorinate chlorophenols. Ph.D. thesis. East Lansing: Michigan State University; 1996. [Google Scholar]
- 25.Sanford R A, Cole J R, Löffler F E, Tiedje J M. Characterization of Desulfitobacterium chlororespirans sp. nov., which grows by coupling the oxidation of lactate to the reductive dechlorination of 3-chloro-4-hydroxybenzoate. Appl Environ Microbiol. 1996;62:3800–3808. doi: 10.1128/aem.62.10.3800-3808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharak Genther B R, Price II W A, Pritchard P H. Anaerobic degradation of chloroaromatic compounds in aquatic sediments under a variety of enrichment conditions. Appl Environ Microbiol. 1989;55:1466–1471. doi: 10.1128/aem.55.6.1466-1471.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shelton D R, Tiedje J M. Isolation and partial characterization of bacteria in an anaerobic consortium that mineralizes 3-chlorobenzoic acid. Appl Environ Microbiol. 1984;48:840–848. doi: 10.1128/aem.48.4.840-848.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steward C C, Dixon T C, Chen Y P, Lovell C R. Enrichment and isolation of a reductively debrominating bacterium from the burrow of a bromometabolite-producing marine hemichordate. Can J Microbiol. 1995;41:637–642. [Google Scholar]
- 29.Utkin I, Woese C, Wiegel J. Isolation and characterization of Desulfitobacterium dehalogenans gen. nov. sp. nov., an anaerobic bacterium which reductively dechlorinates chlorophenolic compounds. Int J Syst Bacteriol. 1994;44:612–619. doi: 10.1099/00207713-44-4-612. [DOI] [PubMed] [Google Scholar]
- 30.Visuvanathan S, Moss M T, Stanford J L, Hermon-Taylor J, McFadden J. Simple enzymic method for isolation of DNA from diverse bacteria. J Microbiol Methods. 1989;10:59–64. [Google Scholar]
- 31.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wetzel R G. Salinity of inland waters. In: Wetzel R G, editor. Limnology. Philadelphia, Pa: The W. B. Saunders Co.; 1975. pp. 142–165. [Google Scholar]
- 33.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolin E A, Wolin M J, Wolfe R S. Formation of methane by bacterial extracts. J Biol Chem. 1963;238:2882–2886. [PubMed] [Google Scholar]