Abstract
The vaccinia virus 14-kDa protein (encoded by the A27L gene) plays an important role in the biology of the virus, acting in virus-to-cell and cell-to-cell fusions. The protein is located on the surface of the intracellular mature virus form and is essential for both the release of extracellular enveloped virus from the cells and virus spread. Sequence analysis predicts the existence of four regions in this protein: a structureless region from amino acids 1 to 28, a helical region from residues 29 to 37, a triple coiled-coil helical region from residues 44 to 72, and a Leu zipper motif at the C terminus. Circular dichroism spectroscopy, analytical ultracentrifugation, and chemical cross-linking studies of the purified wild-type protein and several mutant forms, lacking one or more of the above regions or with point mutations, support the above-described structural division of the 14-kDa protein. The two contiguous cysteine residues at positions 71 and 72 are not responsible for the formation of 14-kDa protein trimers. The location of hydrophobic residues at the a and d positions on a helical wheel and of charged amino acids in adjacent positions, e and g, suggests that the hydrophobic and ionic interactions in the triple coiled-coil helical region are involved in oligomer formation. This conjecture was supported by the construction of a three-helix bundle model and molecular dynamics. Binding assays with purified proteins expressed in Escherichia coli and cytoplasmic extracts from cells infected with a virus that does not produce the 14-kDa protein during infection (VVindA27L) show that the 21-kDa protein (encoded by the A17L gene) is the specific viral binding partner and identify the putative Leu zipper, the predicted third α-helix on the C terminus of the 14-kDa protein, as the region involved in protein binding. These findings were confirmed in vivo, following transfection of animal cells with plasmid vectors expressing mutant forms of the 14-kDa protein and infected with VVindA27L. We find the structural organization of 14kDa to be similar to that of other fusion proteins, such as hemagglutinin of influenza virus and gp41 of human immunodeficiency virus, except for the presence of a protein-anchoring domain instead of a transmembrane domain. Based on our observations, we have established a structural model of the 14-kDa protein.
Vaccinia virus (VV), a member of the Poxviridae family, is one of the largest and most complex animal viruses. The double-stranded DNA genome of about 187 kb codes for about 200 proteins (21), of which approximately 100 are implicated in virus assembly (37). The mechanisms of entry and release of this virus are not yet completely understood. Understanding the entry process of VV into the cell is complicated due to the existence of two infectious forms which are morphologically different and which apparently bind to different cellular receptors (57). The two VV infectious forms are referred as the intracellular mature virus (IMV), with two tightly apposed membranes derived from a specialized domain between the endoplasmic reticulum and the Golgi complex (47, 54), and the extracellular enveloped virus (EEV), with an additional membrane with respect to IMV (24, 29, 36). The passage from IMV to EEV involves an intermediate form, the intracellular enveloped virus (IEV), which acquires two additional membranes derived from the trans Golgi network cisternae (51), one of which fuses with the plasma membrane, releasing the EEV into the extracellular medium surrounded by three membranes. A proportion of EEV, which varies depending on the virus strain, remains associated with the cell surface and probably mediates direct cell-to-cell spread (4). Recent observations by confocal microscopy have shown that IMV enters by direct fusion with the plasma membrane, while EEV enters by endocytosis (58). The envelopment of IMV to generate IEV and then release the EEV involves at least three proteins: the acylated 37-kDa protein (encoded by gene F13L) (3, 52), gp42 (encoded by gene B5R) (17, 64), and the 14-kDa envelope protein (encoded by gene A27L) (46). While the 37-kDa and gp42 proteins are specific for EEV, the 14-kDa protein is a component of IMV and is localized on its surface (55). In spite of the localization of the 14-kDa protein in the membrane of IMV, the existence of a transmembrane domain needed for anchoring cannot be predicted from its sequence. For this reason, it was suggested that another protein, of 21 kDa, may serve to anchor the 14-kDa protein to the envelope of IMV (42). We have identified this protein as the processed product encoded by the A17L gene, and it contains two large internal hydrophobic domains characteristic of membrane proteins (42, 43).
The 14-kDa protein plays key roles in the biology of VV. The protein is needed for EEV formation, an infectious form required for virus dissemination in cells in culture and in tissues of infected animals (3, 13, 14, 41, 46). The protein is also involved in the entry process, acting in virus-to-cell and cell-to-cell fusions (16, 22). With regard to VV entry, it has been suggested that the 14-kDa protein might act at the level of virus attachment to the cell surface heparan sulfate (11). Another important property of the 14-kDa protein is the ability to confer protection in animals immunized with the purified protein following challenge with lethal doses of VV (15, 32). This protective effect is probably mediated by the induction of neutralizing antibodies (31, 32, 45). The 14-kDa protein is well conserved in members of the poxvirus family (31).
In view of the importance of the VV 14-kDa protein in virus-host cell interactions, the aim of this study was to define the structural organization of this membrane protein by genetic, biochemical, and biophysical approaches. We found that the structural organization of the 14-kDa protein is similar to that of other fusion proteins, such as hemagglutinin (HA) of influenza virus and gp41 of human immunodeficiency virus (HIV), except that the 14-kDa protein does not have a transmembrane domain but contains instead a domain involved in binding to a 21-kDa viral protein for membrane anchoring. Based on our findings, a structural model of the 14-kDa protein is presented.
(This work is in partial fulfillment of the requirement for the Ph.D. degree by the School of Pharmacy, University of Santiago de Compostela, Spain.)
MATERIALS AND METHODS
Peptide synthesis.
The peptide corresponding to amino acids 75 to 106 was synthesized by the DKFZ peptide synthesis service (Heidelberg, Germany) by using Fmoc chemistry and PyBOP (Calbiochem) activation at a 0.025-mmol scale. Peptide homogeneity and identity were analyzed by analytical high-performance liquid chromatography, amino acid analysis, and matrix-assisted laser desorption time-of-flight mass spectrometry. The concentrations of the peptide samples were determined by UV absorbance (20). The peptide synthesized is acetylated at the N terminus and amidated at the C terminus (NDEVLFRLENHAETLRAAMISLAKKIDVQTGR).
Far-UV CD spectroscopy.
Circular dichroism (CD) spectra were recorded on a Jasco-710 dichrograph calibrated with (1S)-(+)-10-camphorsulfonic acid. CD spectra were obtained in the continuous mode by taking point measurements every 0.2 nm with a 100-nm/min scan rate, a response of 1 s, and a 1-nm band width. A total of 30 consecutive scans were averaged. Cells with path lengths of 0.01 and 0.5 cm were used for the analysis of samples with peptide concentrations of around 500 and 10 μM, respectively, and a path length of 0.2 cm was used for the analysis with a purified protein concentration of 13 μM. In some cases a pH analysis was done in the 2.5 to 11.5 range by using the following buffers: pH 2.5 to 3.5, 10 mM glycine-HCl; pH 4.5 to 5.5, 10 mM sodium acetate; pH 6.5 to 7.5, 10 mM sodium phosphate; pH 8.5, 10 mM Tris-HCl; and pH 10.5 to 11.5, NaOH.
Sedimentation equilibrium.
Sedimentation equilibrium experiments were performed with a Beckman XL-A analytical ultracentrifuge equipped with UV-visible absorbance optics and with an An60Ti rotor and standard 12-mm-diameter double sector centerpieces of Epon-charcoal. Protein samples (loading concentration ranged from 20 to 150 μM) in buffer (20 mM Tris-HCl, pH 7.4) were centrifuged at 17,500 and 25,000 × g until sedimentation equilibrium was reached at each speed. Then, absorbance scans at the appropriate wavelengths were taken. The temperature was 20°C. Baselines were determined afterwards by high-speed sedimentation.
Whole-cell apparent weight-average molecular weights (Mw) were obtained by fitting the equation which describes the behavior of a single sedimenting solute at sedimentation equilibrium (60) to the experimental data using the programs XLAEQ and EQASSOC (supplied by Beckman; see reference 35). The partial specific volume of the protein was 0.738 ml/g, calculated from the amino acid composition (33). Self-association models (9) were fitted to the Mw versus concentration data by using a nonlinear least-squares method (40).
Construction of the three-helix bundle models and MD simulations.
The initial backbone coordinates for the construction of the three-helix bundle models were extracted from the X-ray structure of the influenza virus HA (Protein Data Bank entry code, 3hmg). The region of the HA structure from amino acids 80 to 96 of chains B, D, and F (corresponding to three helices of 17 residues each) was used as a starting point to build the triple coiled-coil model. We chose this region of the HA coiled coil because it shows a quite regular diameter with nearly constant interhelical distances. This bundle was then elongated up to 29 residues per helix with the Insight II molecular graphics program (Biosym Technologies). Side chains were built on this template by using the Sparse Matrix Driven program (56), which uses a rotamer database to search for the energetically optimal rotamer combination. Evaluation of model quality was computed with the program PROSA II (53) and the three-dimensional (3-D) profiles of Bowie et al. (6).
Energy minimizations and molecular dynamic (MD) calculations were performed by using a Silicon graphics Octane/R10000 workstation with the forcefield implemented in AMBER 4.1 (38). A distance-dependent dielectric constant and an 8-Å residue-based cutoff were used. The nonbonded interactions pair list was updated every 25 steps. The SHAKE algorithm (59) was used to constrain bond lengths so that it was possible to set the time step to 0.002 ps. The following strategy was used to prepare the model for the MD simulations in vacuo. First, side chains of both models were minimized for 1,000 steps while the backbone was maintained rigid, and then the whole system was minimized for 3,000 steps. A 1-ns MD trajectory was calculated at constant temperature (300 K) including a gradual heating of the system from 10 to 300 K during the first 30 ps. Coordinates were saved on disk every 0.4 ps during the course of the simulation.
Cells and virus.
African green monkey kidney cells (BSC40) and HeLa cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% heat-inactivated newborn calf serum. Recombinant viruses, VVindA17L (43) and VVindA27L (46), were grown and titrated by plaque assay in BSC40 cells in the continuous presence of 5 mM isopropyl-β-d-thiogalactopyranoside (IPTG). These recombinant viruses contain an inducible copy of the A17L and A27L genes, respectively, under the lacI operator-repressor system of Escherichia coli, and in the absence of IPTG there is repression of the corresponding viral proteins (43, 46). VV strain WR (Western Reserve) was obtained by infecting HeLa cells and was then purified by banding after sucrose gradient centrifugation (18, 30).
Generation of mutations in the A27L gene and plasmid vectors.
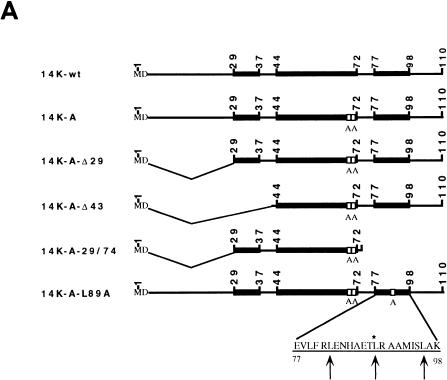
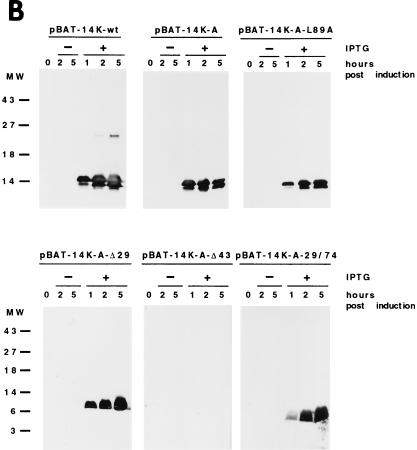
The complete DNA sequence of the 14-kDa protein (encoded by the A27L gene) was obtained by PCR with pT7Nd14K (31) as the template and with the oligonucleotide primers 5′-AC TTT CCA TGG ATG GAA CTC TTT TCC C and 5′-CCC AAG CTT GGG TTA CTC ATA TGG GCG CCG TCC. The specific sites for the restriction endonucleases NcoI and HindIII, respectively, are underlined. To generate point mutations in the A27L gene, we used PCR as previously described by Higuchi et al. (23). The mutagenic primers used were 5′-TA GAA AAG gcT gcT AAA CGC AACG and 5′-GCT GAA ACT gcg AGA GCG. The lowercase letters in the first primer indicate the nucleotides that were changed in order to alter the two contiguous cysteine residues at positions 71 and 72 to alanines, and in the second primer, the lowercase letters indicate the change of the leucine residue at position 89 to alanine. Δ29 and Δ43 deletions at the N-terminal regions of the 14-kDa protein were generated with the primers 5′-CAT GCC ATG GAG GCT AAA CGC GAA GC and 5′-CAT GCC ATG GAC AAT GAG GAA ACT CTC AAAC, respectively, flanked by NcoI restriction sites (underlined). The N-terminal deletions were constructed by using as the template mutant forms of the 14-kDa protein with the two cysteine residues replaced by alanines. The fragment corresponding to amino acids 29 to 74 was obtained with the following oligonucleotides flanked by NcoI and HindIII restriction sites (underlined) (5′ and 3′ ends, respectively): 5′-CAT GCC ATG GAG GCT AAA CGC GAA GC and 5′-CCC AAG CTT TTA GCG TTT agC agC CTT TTC, where small letters are mutations for alanine residues. PCR products flanked by NcoI and HindIII restriction sites at the 5′ and 3′ ends, respectively, were cloned in the E. coli expression vector pBAT-4 (39), which was digested with the same restriction endonucleases. The corrected sequences of the mutant forms of the 14-kDa protein were confirmed by automated DNA sequencing analysis using the sequencing primer T7. All DNA vectors were transformed in BL21(DE3) E. coli cells for expression via IPTG induction. High-level expression was observed at different times postinduction by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analyses after Coomassie blue staining (not shown) of all mutant forms. Schematic drawings of the mutant forms are presented in Fig. 1A with the nomenclature of each one on the left. No detectable levels of the VV 14-kDa protein or its mutant forms were observed in the absence of IPTG, while when the inducer was present, proteins were produced and accumulated during a 5-h period. The protein lacking 43 amino acids at the N terminus did not react with monoclonal antibody C3 (MAbC3), although it was produced to a high level (Fig. 1B).
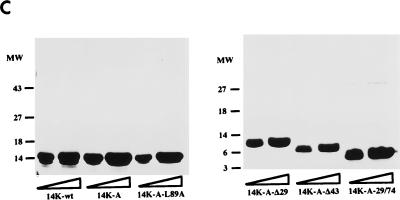
FIG. 1.
(A) Diagram representing mutant forms of the VV 14-kDa protein. Black bars indicate regions predicted as being α-helices in the protein secondary structure. Open boxes correspond to the point mutations introduced in the sequences, and the amino acids changed are indicated below the boxes. The three hepta repeat leucine residues present in the putative third α-helix are indicated by arrows, and the amino acid that was changed is indicated by the asterisk. (B) Expression in E. coli of mutant forms of the VV 14-kDa protein. E. coli BL21(DE3) cells, transformed with different plasmids, were grown to an optical density of 0.4 at 560 nm in the absence or presence of inducer IPTG (1 mM). At various times postinduction (0, 1, 2, and 5 h), aliquots were removed and centrifuged, cells were lysed, and proteins were fractionated by SDS-PAGE and visualized by immunoperoxidase staining after reaction with the 14-kDa-specific MAbC3. Wild-type and mutant forms of the 14-kDa protein were fractionated on SDS–13% PAGE gels, while deletion mutants were fractionated on SDS–15% PAGE gels. The mutant with a 43-amino-acid deletion at the N terminus does not show reactivity with the antibody in spite of high-level expression as revealed by Coomassie staining (not shown). Molecular weight (MW) markers are shown to the left. Plasmid origins are shown on top (see panel A). (C) Coomassie blue staining of purified 14-kDa proteins. The proteins were purified as described in Materials and Methods. Results for two different concentrations (10 and 20 μg/lane) of each protein are shown. The full-length proteins (wild type and point mutation mutants) were fractionated on SDS–13% PAGE gels, and mutant forms with deletions were fractionated on SDS–15% PAGE gels. Molecular weight (MW) markers are shown to the left.
Purification of the wild-type 14-kDa protein and mutant forms.
BL21(DE3) E. coli cells were grown in 1 liter of Luria broth until the optical density reached 0.4 at 560 nm. Thereafter, IPTG was added to a final concentration of 1 mM, and the protein was induced for 5 h. Cells were collected by centrifugation at 4,000 × g for 20 min in a GSA rotor (Beckman). Cell pellets were resuspended in 20 mM Tris-HCl, pH 7.4, with protease inhibitors (1 mM phenylmethylsulfonyl fluoride [PMSF] and 10 μg of leupeptin/ml), and cells were disrupted by sonication. The soluble fraction was separated by centrifugation at 25,000 × g in an SS34 rotor (Beckman), and DNase was added at a final concentration of 1 μg/ml. The samples were filtered through 0.22-μm-pore-size membranes and purified by fast-performance liquid chromatography (Pharmacia) by gel filtration (HiLoad Superdex prep grade 26/60) and ion exchange (Mono Q HR5/5). Briefly, the sample was loaded in 20 mM Tris-HCl (pH 7.4)-equilibrated HiLoad Superdex and eluted at 2 ml/min in the same equilibration buffer, and fractions of 4 ml were collected. Protein peaks were monitored at 280 nm. Fractions that contained the overexpressed protein were loaded on a Mono Q ion exchange column preequilibrated with 20 mM Tris-HCl, pH 7.4. The sample was loaded at 1 ml/min and eluted at the same flow rate with an NaCl gradient (0 to 500 mM). Fractions were dialyzed against 20 mM Tris-HCl, pH 7.4, and were concentrated in a Centriprep device (Amicon) following the manufacturer’s recommendation. Mutant forms of the 14-kDa proteins were obtained at 95% purity. Protein concentrations were determined by the bicinchoninic acid method with bovine serum albumin (BSA) as the standard. Mass spectrometry confirmed the expected molecular mass of each protein and showed that the band migrating at about 12 kDa (Fig. 1B) differed from the 14-kDa protein in only the first two amino acids, methionine and aspartic acid (data not shown), in agreement with the observations of Lai et al. (31).
Purification of the 21-kDa protein of VV and preparation of polyclonal antibodies.
The 21-kDa protein (encoded by the A17L gene) was purified from VV strain WR. About 5 × 1011 virus particles, purified by sucrose gradient centrifugation (18, 30), were treated for 30 min at 37°C with a lysis buffer (50 mM Tris-HCl [pH 8.5], 10 mM MgCl2, 1% Nonidet P-40 [NP-40], and protease inhibitors [1 mM PMSF and 10-μg/ml leupeptin]) in the presence of 50 mM dithiothreitol. After centrifugation at 10,000 × g for 15 min, the pellet was reextracted with lysis buffer plus 0.5% deoxycholic acid–0.1% SDS and was centrifuged again, and the supernatant containing the 21-kDa protein was fractionated by SDS-PAGE. Coomassie blue staining showed a band at 21 kDa, which was then excised from the gel and electroeluted with a buffer containing 9.8 g of NH4HCO3 (pH 9.0)/liter and 0.1% SDS in dialysis tubing at a constant voltage of 50 V for 24 h. The sample was concentrated in a Speed-Vac and precipitated with methanol-acetone (50% [vol/vol]). The pellet was resuspended in 500 μl of phosphate-buffered saline (PBS) containing about 100 μg of protein. Rabbit serum was prepared by repeated immunizations with purified 21-kDa protein in Freund’s complete and incomplete adjuvants. The collected serum was used for immunoprecipitation of 35S-labeled extracts from cells infected with the wild type or with a virus that contains an inducible copy of the A17L gene (VVindA17L), in the absence or presence of IPTG (43). After autoradiography of SDS-PAGE, we found that this polyclonal antibody recognizes specifically the 21-kDa protein (data not shown).
Enzyme-linked immunosorbent assay (ELISA) with specific antibodies against the 14-kDa protein.
To determine the reactivities of monoclonal and polyclonal antibodies for the wild-type 14-kDa protein and its mutant forms, duplicate wells of a microtiter plate were coated with 100 ng of purified proteins in PBS. The coated plate was blocked with borate-buffered saline (0.17 M H3BO4, 0.12 M NaCl [pH 8.5]) plus 0.05% Tween 20–1 mM EDTA–0.25% BSA, washed with PBS and 0.05% Tween 20, and incubated with specific antibody at different dilutions. After the plates were washed with PBS-Tween 20, a second antibody, goat anti-rabbit or goat anti-mouse labeled with peroxidase (Organon Teknica), was added. Antibody reactivity was developed with 50 μl of 0.05 M phosphate citrate buffer (pH 5.0) containing 0.4 mg of o-phenylenediamine dihydrochloride (Sigma)/ml and 30% hydrogen peroxide. The reaction was stopped with 3 M sulfuric acid, and optical densities were measured at 492 nm.
Infection and 35S-labeling of cells.
BSC40 cells grown in 60-mm-diameter plates were infected with the different viruses at 10 PFU/cell. At 6 h postinfection (p.i.) the medium was removed and [35S]methionine-cysteine (Amersham) was added at 10 μCi/ml in methionine-cysteine-free DMEM supplemented 1:10 with regular DMEM. After 16 h, cells were collected, washed with PBS, and resuspended in lysis buffer (20 mM Tris-HCl [pH 8.0], 80 mM NaCl, 20 mM EDTA, 1% NP-40) with protease inhibitors (1 mM PMSF and 10-μg/ml leupeptin). Cells were incubated 30 min on ice and centrifuged at 10,000 × g for 5 min. Supernatants were collected and used for the different binding assays. Uninfected cells were labeled and prepared under the same conditions.
Binding assays with monoclonal and polyclonal antibodies to 14-kDa protein.
Binding assays were performed with purified wild-type protein and mutant forms. Each protein was used in excess to saturate the binding sites of antibodies (100 μg) and was incubated with 150 μl of 10% protein A-Sepharose (PAS) beads coupled to MAbC3 (44) or polyclonal antibodies reactive against the 14-kDa protein (15) for 18 h at 4°C with rotation. Thereafter, the beads were washed three times with PBS and were incubated with 35S-labeled extracts (2 × 106 cells/150 μl of beads) prepared from uninfected BSC40 cells or from cells infected (10 PFU/cell) with VVindA27L in the absence of IPTG. To evaluate the presence of the 14-kDa protein produced by leakiness of the inducible virus, a control was carried out with extracts of 35S-labeled infected cells incubated with the beads lacking the purified 14-kDa protein. The samples were incubated for 18 h at 4°C with rotation and were washed three times with lysis buffer (20 mM Tris-HCl [pH 8.0], 80 mM NaCl, 20 mM EDTA, 1% NP-40) and with PBS, and the beads were resuspended in 25 μl of 2× sample buffer (1.25 M Tris-HCl [pH 6.8], 0.2% SDS, 1% bromophenol blue, 10% 2-β-mercaptoethanol). The beads were boiled for 3 min, and bound proteins were analyzed by SDS-PAGE. The gel was dried, and either the protein profile was revealed by autoradiography or the proteins were transferred to nitrocellulose paper and reacted with anti-21-kDa polyclonal antibody.
Transient expression of mutant forms of the VV 14-kDa protein: immunoprecipitation assay.
Transfection experiments using BSC40 cells were carried out with DNA vectors encoding mutant forms of the 14-kDa protein. The VV early-late promoter and lacZ gene under the control of a VV P7.5 promoter were excised from pSC65 by SmaI and BamHI digestion and cloned into the same restriction sites present in the polylinker of pHGN 3.1 (5). The plasmid obtained, pHLZ, contains a nonfunctional HA gene as well as multiple cloning sites for protein expression under the control of a VV synthetic early-late promoter and produces a functional β-galactosidase in VV-infected cells when X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) is added. The wild-type 14-kDa protein (14K-wt) and mutants formed by point mutation (14K-A and 14K-A-L89A) or deletion of the N-terminal region (14K-A-Δ29) (see above) were excised by NcoI and HindIII digestion from pBAT-4 clones and blunt ended with the Klenow fragment of DNA polymerase. DNA-purified fragments were cloned into the SmaI site of pHLZ. The resulting plasmids, pHLZ-14K-wt, pHLZ-14K-A, pHLZ-14K-A-L89A, and pHLZ-14K-A-Δ29, containing 14K-wt, 14K-A, 14K-L89A, and 14K-A-Δ29, respectively, in the appropriate orientations, were transfected, by using Lipofectamine reagent (Gibco-BRL), into cells infected (10 PFU/cell) with VVindA27L in the absence of IPTG. For each transfection the amount of plasmid DNA was 10 μg/well. Infected cells were transfected and labeled with [35S]methionine-cysteine at 6 h p.i. in methionine-cysteine-free medium and were collected at 24 h p.i. Immunoprecipitation analysis was performed as previously described by Rodriguez et al. (42).
RESULTS
Structure prediction and design of mutations in the VV 14-kDa protein.
The PHD program for secondary structure prediction, which uses multiple-sequence alignment (48–50), predicts that the sequence of the 14-kDa protein (110 amino acids) contains three α-helical regions, from residues 29 to 37, 44 to 72, and 77 to 98, while the first 28 residues are essentially unstructured. Interestingly, the protein contains two contiguous Cys residues at positions 71 and 72 that have been proposed to be responsible for the oligomerization of the 14-kDa protein (trimer formation) through intermolecular disulfide bonds (45). The amino acid sequence of the 14-kDa protein presents a hydrophobic pattern in the central region which is typical of proteins that form coiled-coil structures (hydrophobic-polar-polar-hydrophobic-polar-polar-polar). In concordance with this hypothesis, the algorithm of Wolf et al. (63) predicts for this region a 60% probability of forming a triple coiled coil, while the N terminus portion (amino acids 1 to 42) is predicted to have zero probability of being a coiled coil and the C terminus (amino acids 70 to 110) has a 15% probability of forming a coiled coil (data not shown). The small coiled-coil tendency at the C-terminal region could be due to the distribution of Leu residues spaced every seven positions, as is typical of Leu zippers.
To determine if the structure predictions were correct, as well as to find out if Cys 71 and 72 are critical for protein oligomerization, we designed several mutations in the 14-kDa protein (Fig. 1A). We generated a mutant form with the same amino acid sequence as that of 14K-wt except for the replacement of the two contiguous Cys residues by Ala (14K-A). As a result of this mutation, we obtained a soluble protein which could be concentrated to more than 1 mg/ml, while the 14K-wt protein produces aggregates after purification from E. coli (data not shown). In view of the aggregation of 14k-wt, we decided to do all our studies with the Ala mutant since, as we will explain below, it behaves functionally in a manner which is indistinguishable from that of the wild-type protein. Deletions at the N and C termini of the 14-kDa protein were constructed based on secondary structure prediction methods by using the plasmid with the 14K-A sequence (pBAT-14K-A) as template for PCR, but in these cases it was necessary to add two amino acids, methionine and aspartic acid, at the N terminus to provide a NcoI restriction site and a start codon in the sequence. The resultant plasmids were called pBAT-14K-A-Δ29, pBAT-14K-A-Δ43, and pBAT-14K-A-29/74 for single deletions of 28 or 42 amino acids at the N terminus or double deletions at the N and C termini, respectively (see Fig. 1A). Finally, we tried to construct point mutations in the region predicted to form a coiled coil in the protein with two Cys replaced by two Ala. The idea behind this was that if this region of the protein is involved in oligomerization, the mutation should allow us to obtain monomers. However, mutations in this region abolished expression of the mutant form. In addition, we generated a mutant protein in which Leu 89, located at the second position of the three hepta repeats in the C-terminal region (Leu zipper-like domain), was replaced by Ala (pBAT-14K-A-L89A). This mutant form was expressed at a high level, although it was less stable in solution than 14K-wt. The protein was soluble when made in E. coli but it was prone to degradation with an increasing time of incubation at room temperature.
CD analysis of the purified VV 14-kDa protein.
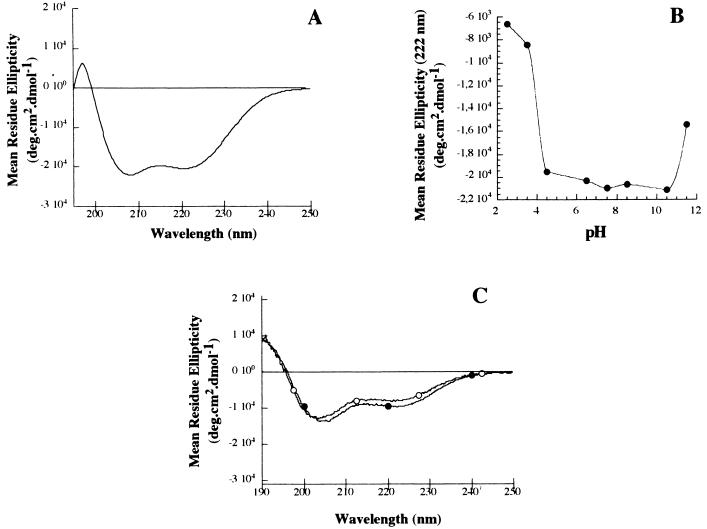
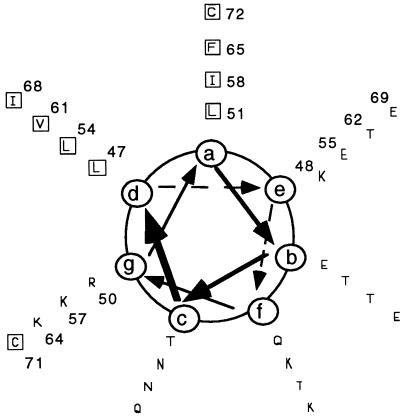
To determine which type of secondary structure is adopted by the 14-kDa protein, we analyzed the 14K-wt protein and some of the mutant forms by CD. The 14K-wt protein gave a typical α-helix CD spectrum with two minima at 222 and 208 nm and a maximum at 193 nm (data not shown). But the inability to obtain a homogeneous preparation without aggregation prevented us from any further characterization of the protein. The mutant 14K-A shows a typical helix spectrum similar to that found for the wild-type protein but without the aggregates (Fig. 2A). Calculation of the helix population from the CD spectra (10) indicates an average helical population of around 30%, which roughly correlates with the percentage expected from the triple coiled-coil prediction (∼28%). Another characteristic in triple coiled-coil structures is that ionic pair interactions between charged residues in adjacent helices normally stabilize the folded conformation. To find out if this was the case here, we pH titrated the 14K-A protein (Fig. 2B). Essentially, we found that the protein is stable in a pH range of 4.5 to 10.5 and denatures below or above these values, respectively. These results support the idea that there are Glu and Lys residues involved in forming ionic stabilizing pairs, in good agreement with the distribution of the amino acids into a helical wheel (Fig. 3) and the 3-D model of the protein (see below). In addition, we analyzed the CD spectrum of a peptide corresponding to the predicted Leu zipper region (see Materials and Methods) to show the concentration dependence of its ellipticity (Fig. 2C).
FIG. 2.
CD spectra of different mutant forms of the 14-kDa protein. (A) Far-UV CD spectrum of the 14K-A mutant form of the 14-kDa protein. (B) pH dependence of the ellipticity at 222 nm for the 14K-A mutant. (C) Far-UV CD spectrum of a peptide corresponding to the predicted Leu zipper region at the C terminus of the 14-kDa protein, at two different concentrations (20 and 500 μM), to show the concentration dependence of its ellipticity. Open circles, 20 μM; solid circles, 500 μM.
FIG. 3.
Helical wheel analysis of the predicted coiled-coil region (residues 44 to 72) by the algorithm of Wolf et al. (63). At the end of the potential coiled-coil region are localized the two cysteine residues. Hydrophobic amino acids (squares), represented by one-letter code, are predominantly aligned at the a and d positions, surrounded by hydrophilic amino acids at other positions. At the e and g positions we found glutamic and lysine residues predominantly. Position numbers of the amino acids in the complete sequence of the 14-kDa protein are indicated.
The region between residues 44 and 72 is involved in the formation of a triple coiled coil in the VV 14-kDa protein.
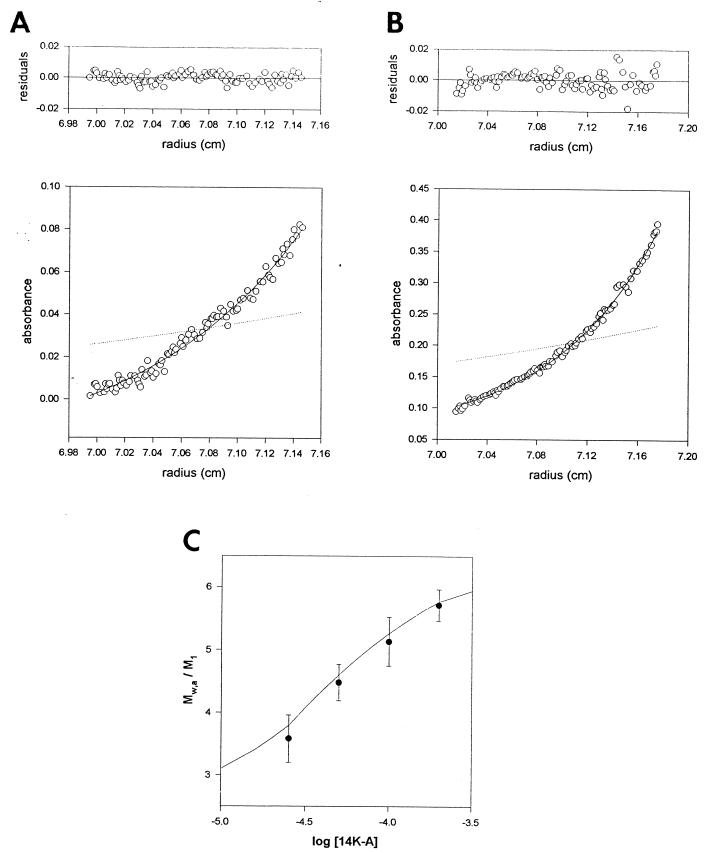
To determine the extent of oligomerization of the 14-kDa protein, we carried out a sedimentation equilibrium analysis of the purified 14K-A protein in the concentration range of 20 to 150 μM. We found that at 20 μM the protein sediments with a Mw of 43,100 ± 5,200 (Fig. 4A), which corresponds to a trimer (monomer molecular weight [M1] = 12,500) (45), while at 150 μM the protein sediments as a hexamer (Mw, 68,000 ± 3,000) (Fig. 4B). This behavior is typical of a protein self-association equilibrium (1). In Fig. 4C the degree of association (Mw/M1) of the 14K-A protein is plotted as a function of protein concentration (see also Table 1). The simplest model compatible with the experimental data is a trimer-hexamer association with an equilibrium constant (K) of 2 × 104 M−1.
FIG. 4.
Sedimentation equilibrium analysis of the 14-kDa protein. (A) Sedimentation equilibrium profile of the 14K-A protein at a concentration of 20 μM in 20 mM Tris-HCl (pH 7.4) buffer at 25,000 × g, 20°C, as described in Materials and Methods. The symbols represent the experimental data. The solid line shows the best-fit function corresponding to a single species at sedimentation equilibrium with an MW of 43,500. The dotted line represents the concentration gradient of the monomer protein (M1 = 12,500). (B) The same as described for panel A with 150 μM 14K-A protein. In this case, the Mw was 69,100 ± 3,000. (C) Dependence of the degree of association (Mw/M1) of the 14-kDa protein on protein concentration. The solid line shows the best-fit function for a trimer-hexamer association (2 × 104 M−1) at sedimentation equilibrium.
TABLE 1.
Effect of protein concentration on Mw at the sedimentation equilibrium
| Protein concn (μM) | Mw |
|---|---|
| 20 | 43,100 ± 5,200 |
| 40 | 56,000 ± 3,600 |
| 80 | 64,300 ± 4,900 |
| 150 | 69,100 ± 3,000 |
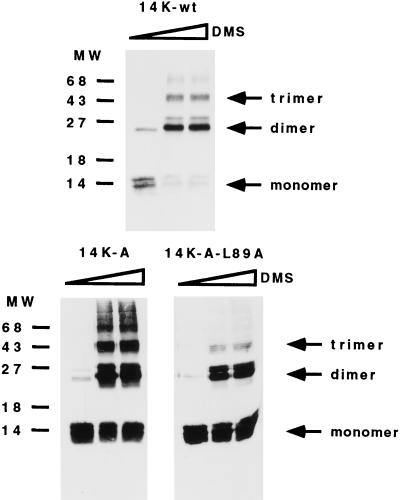
Since the amino acid sequence for the C-terminal region of the protein contains characteristic Leu zippers, it could be that the hexamer is made of two trimers by interaction of the Leu zippers at a high concentration. To find out if this was the case, we performed the same experiments as before with the mutant 14K-A-29/74. In this case, we found trimer formation independent of the concentration used (data not shown), indicating that the region between residues 75 and 110 could be involved in Leu zipper formation, either with another molecule or with itself. In fact, the peptide corresponding to the Leu zipper region analyzed by CD (Fig. 2C) and analytical centrifugation confirm this hypothesis, indicating that this peptide is not a monomer at 80 μM (Mw = 17,000; Mw/M1 = 4.5) (data not shown). This fact explains the formation of the hexamer at high concentrations in the 14K-A protein because of the potential of the region between amino acids 75 and 110 of the 14-kDa protein to form a Leu zipper-type structure. In this case, we expected that the mutant form 14K-A-L89A would have less affinity for this interaction, being a trimer independently of protein concentration. Since the purified mutant protein 14K-A-L89A was broken down during the long run of analytical ultracentrifugation by its C-terminal region, as confirmed by SDS-PAGE and Western blotting (data not shown), we carried out cross-linking experiments to evaluate the effect of the mutation on oligomer formation. As shown in Fig. 5, trimers and higher oligomers were observed after cross-linking with 14K-wt and 14K-A proteins but not with the mutant form 14K-A-L89A. In addition, we observed changes in the CD spectra of the purified protein compared to that of 14K-A, due to the loss in hexamer formation that would be expected from the association constant at the experimental concentration used (data not shown).
FIG. 5.
Oligomerization of the 14-kDa protein occurs independently of the cysteine residues. Cross-linking experiments were carried out with full-length E. coli-expressed 14-kDa wild-type protein and mutant forms 14K-A and 14K-A-L89A. Bacterial lysates obtained from 10-ml cultures of an E. coli expression system at 5 h postinduction were concentrated and resuspended in 100 μl of PBS. Different amounts of dimethyl suberimidate (DMS) (0, 2, and 5 μl) in dimethyl sulfoxide at a stock concentration of 50 mg/ml (freshly prepared) were added to the protein sample, and the mixtures were incubated on ice for 90 min. The reactions were quenched by addition of Tris-HCl (pH 8.0) to a final concentration of 20 mM, and the mixtures were then incubated at 4°C for 15 min. After the addition of sample buffer, proteins were analyzed on SDS–13% PAGE gels and profiles were revealed by Western blotting after reaction with MAbC3. Monomer, dimer, and trimer forms are indicated, and no differences were observed with increasing amounts of DMS. Molecular weight (MW) markers are on the left.
Thus, our results provided evidence for oligomerization of the VV 14-kDa protein independent of disulfide bonds, identified the region involved in the formation of a triple coiled coil, and characterized the homo-intermolecular affinity of the Leu zipper motif at the C terminus.
Molecular modeling of the triple coiled-coil region of the VV 14-kDa protein.
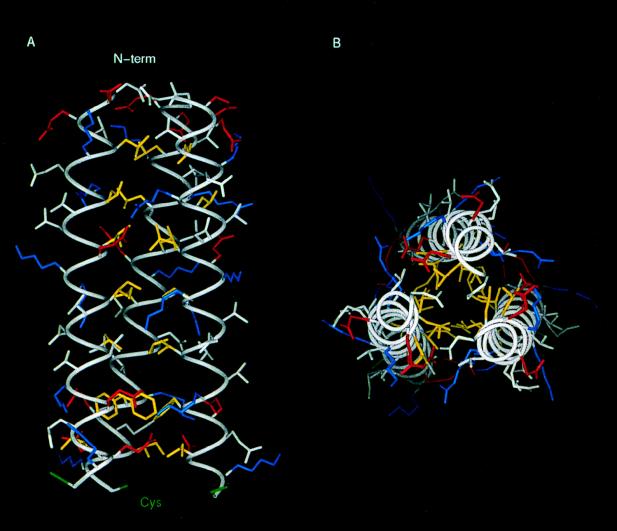
A hypothetical atomic model of the predicted triple coiled coil was built with the central triple coiled-coil region of the 3-D structure of HA as a template (see Materials and Methods). An assessment of model quality was made with the program PROSA II (53) and the 3-D profiles of Bowie et al. (6), which analyze in different ways to what extent a sequence is compatible with a given fold. This analysis did not highlight any significant misfolded region along the sequence of the model, indicating that this triple coiled-coil model is plausible. In order to check the stability of this molecular model, it was submitted to a 1-ns MD simulation in vacuo at 300 K. Results show that, after the initial equilibration period, the triple coiled-coil motif remains very stable except for the last helical turn, where Cys 71 and 72 are located, which tends to open (Fig. 6). During the course of the simulation the model keeps a tight packing of residues in the hydrophobic core. The core is formed by strips of hydrophobic amino acids (Leu, Ile, Val, and Phe) lying on one face of the helices. This arrangement is partly disrupted close to the C-terminal end due to the presence of three bulky Phe residues. This results in a partial opening of the helix C terminus ends. Interestingly enough, Cys 71 and 72 are pointing immediately after these and dispose outwards in the structure. Therefore, their function could be to close the structure stabilizing the triple coiled coil. The model also displays numerous inter- and intrahelical salt bridges, stabilizing the structure, in good agreement with the pH dependence of its stability and a parallel disposition of the three α-helices.
FIG. 6.
3-D model of the triple coiled-coiled region of the 14-kDa protein. Amino acid side chains are shown color coded according to their physical properties. Yellow, hydrophobics (Leu, Ile, Val, and Phe); Blue, positively charged residues (Arg and Lys); Red, negatively charged residues (Asp and Glu). Cys residues are in green and the rest of the residues are in white. (A) Longitudinal view. (B) Transversal view. The hydrophobic core can be seen in panel B, while in panel A the formation of ionic pairs between helices is shown.
Mapping the interaction of 14-kDa protein with 21-kDa protein.
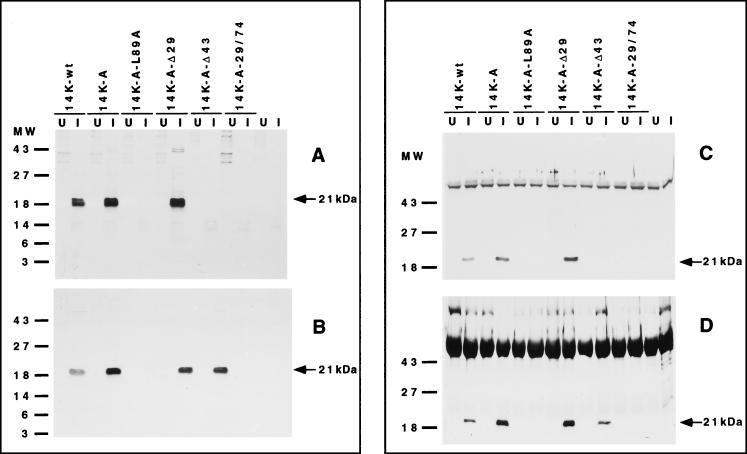
Since the VV 14-kDa protein has to be inserted in the membrane of the virus, our next approach was to define how the 14-kDa protein interacts with the previously described 21-kDa anchoring protein. To map protein-protein interactions, we developed an in vitro binding assay. This assay is based on the ability of a preformed complex, PAS beads–antibodies to 14-kDa protein–purified 14-kDa protein, to bind radioactively labeled proteins from cytoplasmic lysates of uninfected cells or from cells infected with a virus that does not produce the 14-kDa protein during infection (VVindA27L) (46). By this method we expected to find bound 21-kDa protein in the column only when the domains of the 14-kDa protein involved in binding were present. Thus, purified 14-kDa protein, prepared as described in Materials and Methods, was bound to PAS beads coupled to monoclonal (MAbC3) or polyclonal anti-14kDa antibodies. ELISA experiments with these purified proteins showed that the soluble wild-type 14-kDa protein and its mutant forms bind similarly to these antibodies and that the epitope recognized by MAbC3 maps between amino acids 29 and 43 (data not shown). This is also shown in Fig. 1B, where the protein lacking the first 42 amino acids at the N terminus did not react with this monoclonal antibody. Thus, the amount of protein added to form the complex PAS-antibodies-protein was the same (100 μg) and was in excess to ensure saturation of the binding sites in the antibodies. Following incubation with labeled extracts from uninfected or infected cells, the beads were washed and bound proteins were analyzed by SDS-PAGE and autoradiography. As shown in Fig. 7A for beads with bound MAbC3, a protein of about 21-kDa appeared with three of the six different mutant forms of the 14-kDa protein and only in lysates from infected cells. Because MAbC3 binds to the 14-kDa protein in the region between amino acids 29 and 43, the results obtained with the N-terminal mutant form 14K-A-Δ43 protein were expected. However, failure of the 21-kDa protein to bind the mutant 14K-A-L89A and 14K-A-29/74 proteins suggested that the lack of binding was the result of a modification at the C-terminal region. This was further confirmed following analysis of the complex formed using, instead of MAbC3, polyclonal anti-14-kDa antibodies that recognize all different forms of the 14-kDa protein (Fig. 7B). A labeled 21-kDa protein was observed bound with a 43-amino-acid N-terminal deletion of the 14-kDa protein, corroborating the results with mutants at the C-terminal region. Western blotting with MAbC3 revealed that the wild type and mutant forms of the 14-kDa protein had the expected molecular weights, indicating that under the conditions used the proteins were not degraded (data not shown). To demonstrate that the bound protein corresponded to the processed product of the A17L gene, as previously identified by immunoprecipitation analyses (42), the same samples used for Fig. 7A and B were run on SDS-PAGE gels, proteins were transferred to nitrocellulose paper, and filters were reacted with a specific polyclonal antiserum raised against the A17L gene (see Materials and Methods). Western blotting (Fig. 7C and D) identified the 21-kDa protein shown in the autoradiograms of Fig. 7A and B as the processed product of the A17L gene. The proteins appearing above the 21-kDa protein correspond to immunoglobulins. In all cases, a control without the binding to the beads of purified 14-kDa protein was run in parallel, to discard the occurrence of A17L gene product by binding with the small amount of 14-kDa protein produced by leakiness of the inducible virus (shown to the rightmost part in each panel of Fig. 7).
FIG. 7.
In vitro binding assay for selective binding of the 21-kDa protein to the 14-kDa protein. PAS beads, coupled to MAbC3 (A and C) or polyclonal antibodies to the 14-kDa protein (B and D), were incubated first with purified 14-kDa wild type and mutant forms and then with 35S-labeled extracts from uninfected cells (U) or from cells infected with VVindA27L (I) and were processed as described in Materials and Methods. The autoradiograms (A and B) of 15% polyacrylamide gels show the presence of a 21-kDa protein in lanes with infected cells for 14K-wt, 14K-A, and mutants with deletions in the amino terminus (except for Δ43 in panel A due to the lack of reactivity of MAbC3). There was no reactivity when the leucine residue at position 89 was changed to alanine and when the third α-helix was deleted. Uninfected and infected cells with no E. coli purified protein added to the beads were processed in parallel to eliminate the presence of the 21-kDa protein that might be immunoprecipitated due to the 14-kDa protein produced by the inducible system. Panels C and D show Western blots of the same samples used in panels A and B but after reaction with a polyclonal antibody to the 21-kDa protein. Molecular weight (MW) markers are on the left.
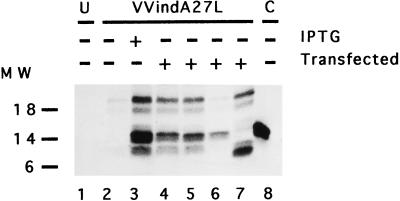
While the above experiments were performed in vitro, it was important to demonstrate that similar protein-protein interaction (14-kDa–21-kDa) also occurs in vivo. With this aim, we generated VV vectors expressing different forms of the 14-kDa protein and tested complex formation in cultured cells. For this, we chose only the mutants that we considered to be the most interesting based on the results of the in vitro experiments. Thus, the genes for the 14K-wt, 14K-A, 14K-A-L89A, and 14K-A-Δ29 proteins were cloned in pHLZ under the control of the VV early-late promoter (see Materials and Methods). These plasmid vectors were transfected into cells infected with VVindA27L (10 PFU/cell) in the absence or presence of IPTG. In the absence of IPTG, the A27L gene is repressed but there is expression of A27L mutants from the transfected plasmids because the promoter is under the control of the VV transcriptional machinery. In the presence of IPTG there is expression of both endogenous and exogenous A27L genes. After [35S]methionine-cysteine labeling, cell extracts were immunoprecipitated with MAbC3. The results shown in Fig. 8 (lane 3) revealed that in cells infected with VVindA27L in the presence of IPTG, both the 21-kDa and 14-kDa proteins are coprecipitated. No bands appeared in uninfected cells (lane 1) or in cells infected in the absence of IPTG (lane 2). As expected, the 14-kDa–21-kDa proteins were observed in cells infected with VVindA27L and transfected with pHLZ-14K-wt (lane 4), pHLZ-14K-A (lane 5), or pHLZ-14K-A-Δ29 (lane 7). Significantly, no such protein complex was observed in cells transfected with pHLZ-14K-A-L89A (lane 6). We included a control based on cells infected with a virus that does not produce the 21-kDa protein (VVindA17L) to show that formation of the complex is dependent on the expression of the 21-kDa protein (lane 8). Clearly, when the 14-kDa protein is modified at the C terminus by a point mutation, leucine to alanine at position 89, the protein is unable to bind the 21-kDa protein under the conditions used in this assay. Since the in vitro (Fig. 7) and in vivo (Fig. 8) findings are in agreement, we conclude that formation of the 14-kDa–21-kDa protein complex is a specific event and that a leucine at position 89 plays a critical role in this interaction.
FIG. 8.
In vivo assay to measure 14-kDa–21-kDa protein-protein interactions. BSC40 cells grown in 12-well plates were infected (10 PFU/cell) with VVindA27L and transfected with plasmid vectors (10 μg/well) expressing different mutant forms of the 14-kDa protein. The cells were labeled with [35S]methionine-cysteine from 6 to 24 h p.i., and cell extracts immunoprecipitated with MAbC3 were analyzed by SDS-PAGE and autoradiography, as described in Materials and Methods. Lane 1, uninfected cells (U); lanes 2 to 7, cells infected with VVindA27L; lane 8, cells infected with VVindA17L (control [C]). Cells in lanes 4 to 7 were transfected with pHLZ-14K-wt, pHLZ-14K-A, pHLZ-14K-A-L89A, and pHLZ-14K-A-Δ29, respectively. Molecular weight (MW) markers are on the left. Densitometric analyses revealed the following ratios for the 14-kDa/21-kDa proteins normalized after subtracting the value in lane 2 (negative control): 1.49 (lane 3), 1.36 (lane 4), 1.3 (lane 5), 162.9 (lane 6), and 1.2 (lane 7).
DISCUSSION
VV is one of the most complex animal viruses, with about 200 polypeptides encoded by the virus genome, of which about 100 proteins are part of the virion. In the last few years, numerous studies of the function of viral proteins and of viral morphogenesis have been carried out. In spite of this, there is little information available concerning the structure of viral proteins. Only one protein of VV, which participates in the modification of both mRNA ends, has been defined structurally by X-ray crystallography (25). Due to the important role of the 14-kDa protein (encoded by the A27L gene) in the biology of poxvirus, in this study we have carried out a structural characterization of this IMV surface protein.
Using different secondary structure prediction algorithms (48–50, 63), we defined four regions in this protein of 110 amino acids: from residues 1 to 28, from 29 to 37, from 44 to 72, and from 77 to 98. The first region is predicted to be unstructured, the second is predicted to form an isolated α-helix, the third is supposed to form a triple coiled-coil structure, and the last one has a Leu zipper sequence fingerprint. CD analysis corroborates the existence of a significant helix population, as expected from the predictions. The helix content changes with pH (Fig. 2B), in concordance with the fact that ionic interactions between oppositely charged amino acids are stabilizing the helix structure. Thus, pHs below 4.5 or above 10.5, which lead to modifications in the structure, correspond with changes in the ionization states of Glu and Lys, amino acids that are located at the e and g positions on a helical wheel (Fig. 3). Adjacent to them, at positions a and d, there are hydrophobic residues which correspond to a 4-3 heptad repeat pattern, characteristic of coiled-coil structures (2, 12, 26, 27). This type of structure is responsible for oligomerization of proteins that normally form dimers, trimers, or tetramers, although evidence has been obtained for a coiled coil of five helices (for a review, see reference 34).
The CD results are in agreement with those obtained by analytical ultracentrifugation, cross-linking experiments, and modeling of the three-dimensional structure, supporting the hypothesis of a parallel three-stranded coiled coil between amino acids 44 and 72. By the use of mutant forms lacking the two contiguous cysteine residues in the sequence we found that cysteines are not required for oligomer formation, in contrast with the hypothesis of disulfide-linked trimers (45). The mutant form 14K-A is a trimer, at least, independent of Cys residues. This 14K-A mutant behaves functionally like the 14K-wt protein, as revealed by the generation of a recombinant VV (VVInd14K-A) which expresses 14K-A constitutively and wild-type 14-kDa protein under regulation (unpublished data). We observed that the degree of association changes with the concentration, and thus at low concentrations (20 μM) the purified 14K-A protein sediments as a trimer while at high concentrations (150 μM) it tends to form hexamers. This feature would be the result of the tendency described for the Leu zipper domain to associate with itself into nonnatural forms at high concentrations. This hypothesis is supported by experiments with several mutant forms and with one synthetic peptide. Thus, C-terminal deletions only form trimers in a manner which is independent of concentration (14K-A-29/74), and the same fact was observed when the leucine residue at the second position in the putative third α-helix was mutated. On the other hand, a synthetic peptide with the same amino acid composition as the C-terminal region tends to aggregate by itself, as shown by CD spectra and analytical ultracentrifugation.
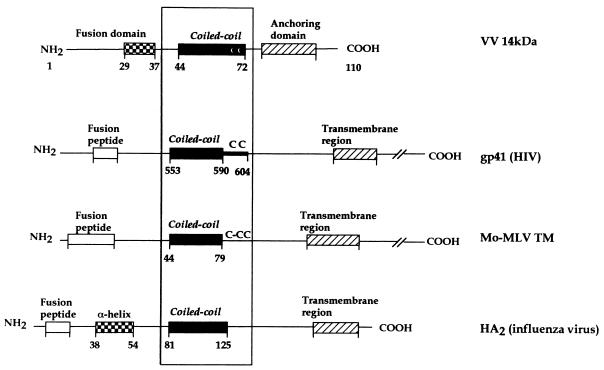
Taking together data from this and previous studies, we defined a central region involved in oligomerization by hydrophobic interactions between amino acids in positions a and d (Fig. 3) on a helical wheel and ionic interactions between residues in adjacent positions (e and g) and the N terminus of the protein involved in fusion (22). If we compare the structure of the 14-kDa protein with that described for other fusion proteins, we can find several homologies (Fig. 9). Data derived from different studies have shown a coiled-coil structure as a common feature of active conformation in several viral fusion proteins, such as the HA2 fragment of influenza virus (7), the transmembrane subunit of Moloney murine leukemia virus (Mo-MLV TM) (19), and the gp41 envelope glycoprotein of HIV (8, 61). If we analyze in more detail the functional regions in these proteins, we find similarities in organization: (i) a hydrophobic fusion peptide that will be immediately amino terminal to a central three-stranded coiled coil, (ii) a three-stranded coiled-coil region with a role in oligomerization and stabilization of the structure, and (iii) a transmembrane domain at the C terminus of the protein needed for anchoring to the surface of the virus. Besides, Mo-MLV TM and gp41 have cysteine residues at the end of the 4-3 hydrophobic repeat region.
FIG. 9.
Schematic comparison of the VV 14-kDa protein, HIV gp41, Mo-MLV TM, and influenza HA2 structures. The four proteins form three-stranded coiled-coil structures involving a central α-helix. For all of them, the hydrophobic fusion peptide would be immediately amino terminal to the oligomerization domain, although for the VV 14-kDa protein the peptide implicated in this process has not been defined yet (22). For the 14-kDa protein an anchoring domain is indicated instead of a transmembrane region of the C terminus. Except for the influenza HA2, these fusion proteins have cysteine residues at the end of the coiled-coil region.
Although in the sequence of the 14-kDa protein we can identify a fusion peptide domain, a coiled-coil domain, and cysteine residues immediately behind it, the protein lacks a transmembrane domain. Thus, how is the 14-kDa protein anchored in the membrane of IMV? In previous reports we suggested, by immunoprecipitation studies (42) and by the use of a conditional lethal mutant virus (43), that another protein of about 21 kDa (A17L gene) was responsible for anchoring the 14-kDa protein on the surface of IMV. To directly show 14-kDa–21-kDa protein interactions and the region of the 14-kDa protein involved in the formation of a stable complex, in this investigation we have carried out binding assays with different mutant forms. The in vitro and in vivo experiments demonstrate that only a C-terminally modified 14-kDa protein (a point mutation in leucine at position 89 or lacking the C terminus) was unable to interact with the 21-kDa protein under different experimental conditions. Deletions at the N terminus do not affect complex formation (Fig. 7 and 8). These findings mean that the 14-kDa hexamers formed in solution at high concentrations of protein are artifacts resulting from a tendency of the protein to bury the hydrophobic residues, which will be used in the virus for interaction with the 21-kDa protein.
While the roles of the three different domains in the protein have been discussed, the role of the conserved cysteine residues in the 14-kDa protein is unclear. The experiments showed that Cys residues are not necessary for oligomerization or for binding with the 21-kDa protein. We suggest a role for Cys in stabilization of the protein structure, as has been described for other proteins. Thus, Zhou et al. (65) demonstrated that insertion of a disulfide bond at a flexible region of the protein (N-terminal or C-terminal end of the coiled coil) could significantly increase protein stability. However, the structure is maintained without the presence of the disulfide bonds, which could be explained by the studies of Hodges et al. (28) showing that exposed disulfide bonds can be reduced without destroying the structure of the protein. In some cases the Cys residues have been described as providing protection from proteolytic degradation and/or as preventing transmission of protein flexibility upstream of the coiled coil in order to facilitate protein function in vivo (62).
In summary, in this study we have defined the structural domains of the VV 14-kDa protein, which has key roles during virus-host cell interactions, and presented a model of protein trimers. We show that this protein shares structural similarities with other fusion proteins, i.e., an N-terminal fusion peptide, a coiled-coil domain, and a C-terminal region for anchoring to the surface of the IMV by formation of a complex with the 21-kDa membrane protein. Among virus membrane proteins, the VV 14-kDa is, as yet, the only protein that lacks a transmembrane domain but uses instead the anchoring domain of another virus protein for membrane insertion.
ACKNOWLEDGMENTS
We thank J. F. Rodriguez for the gift of WR32-7/Ind14K (VVindA27L), B. Moss for pSC65 plasmid, R. W. Moyer for pHGN 3.1, M. Hyvönen for pBAT-4, and Victoria Jiménez and Eva López for expert technical assistance.
M.-I. V. is a recipient of a fellowship from Comunidad Autónoma de Madrid. This investigation was supported by grants BIO95-0022 and SAF95-0072 from CICYT of Spain.
REFERENCES
- 1.Abril A, Salas M, Andreu J M, Hermoso J M, Rivas G. Phage Φ29 protein p6 is in a monomer-dimer equilibrium that shifts to higher association states at the millimolar concentrations found in vivo. Biochemistry. 1997;36:11901–11908. doi: 10.1021/bi970994e. [DOI] [PubMed] [Google Scholar]
- 2.Adamson J G, Zhou N E, Hodges R S. Structure, function and application of the coiled-coil protein folding motif. Curr Opin Biotechnol. 1993;4:428–437. doi: 10.1016/0958-1669(93)90008-k. [DOI] [PubMed] [Google Scholar]
- 3.Blasco R, Moss B. Extracellular vaccinia virus formation and cell-to-cell virus transmission are prevented by deletion of the gene encoding the 37,000-dalton outer envelope protein. J Virol. 1991;65:5910–5920. doi: 10.1128/jvi.65.11.5910-5920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blasco R, Moss B. Role of cell-associated enveloped vaccinia virus in cell-to-cell spread. J Virol. 1992;66:4170–4179. doi: 10.1128/jvi.66.7.4170-4179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bloom D C, Edwards K M, Hager C, Moyer R W. Identification and characterization of two nonessential regions of the rabbitpox virus genome involved in virulence. J Virol. 1991;65:1530–1542. doi: 10.1128/jvi.65.3.1530-1542.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowie J U, Lüthy R, Eisenberg D. A method to identify protein sequences that fold into a known three-dimensional structure. Science. 1991;253:164–170. doi: 10.1126/science.1853201. [DOI] [PubMed] [Google Scholar]
- 7.Bullough P A, Hughson F M, Skehel J J, Wiley D C. Structure of influenza haemagglutinin at the pH of membrane fusion protein. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 8.Chan D C, Fass D, Berger J M, Kim P S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 9.Chatelier R C, Minton A P. Sedimentation equilibrium in macromolecular solutions of arbitrary concentration. I. Self-associating proteins. Biopolymers. 1987;26:507–524. doi: 10.1002/bip.360260405. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y H, Yang J T, Chow K H. Determination of the helix and β-form of proteins in aqueous solution by circular dichroism. Biochemistry. 1974;13:3350–3359. doi: 10.1021/bi00713a027. [DOI] [PubMed] [Google Scholar]
- 11.Chung C-S, Hsiao J-C, Chang Y-S, Chang W. A27L protein mediates vaccinia virus interaction with cell surface heparan sulfate. J Virol. 1998;72:1577–1585. doi: 10.1128/jvi.72.2.1577-1585.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen C, Parry D A D. α-Helical coiled coils and bundles: how to design an α-helical protein. Proteins Struct Funct Genet. 1990;7:1–15. doi: 10.1002/prot.340070102. [DOI] [PubMed] [Google Scholar]
- 13.Dallo S, Esteban M. Isolation and characterization of attenuated mutants of vaccinia virus. Virology. 1987;159:408–422. doi: 10.1016/0042-6822(87)90480-6. [DOI] [PubMed] [Google Scholar]
- 14.Dallo S, Rodriguez J F, Esteban M. A 14K envelope protein of vaccinia virus with an important role in virus-host cell interactions is altered during virus persistence and determines the plaque size phenotype of the virus. Virology. 1987;159:423–432. doi: 10.1016/0042-6822(87)90481-8. [DOI] [PubMed] [Google Scholar]
- 15.Demkowicz W A, Maa J-S, Esteban M. Identification and characterization of vaccinia virus genes encoding proteins that are highly antigenic in animals and are immunodominant in vaccinated humans. J Virol. 1992;66:386–398. doi: 10.1128/jvi.66.1.386-398.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doms R W, Blumenthal R, Moss B. Fusion of intra- and extracellular forms of vaccinia virus with the cell membrane. J Virol. 1990;64:4884–4892. doi: 10.1128/jvi.64.10.4884-4892.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engelstad M, Smith G L. The vaccinia virus 42-kDa envelope protein is required for the envelopment and egress of extracellular virus and for virus virulence. Virology. 1993;194:627–637. doi: 10.1006/viro.1993.1302. [DOI] [PubMed] [Google Scholar]
- 18.Esteban M. Defective vacinia virus particles in interferon-treated, infected cells. Virology. 1984;133:220–227. doi: 10.1016/0042-6822(84)90443-4. [DOI] [PubMed] [Google Scholar]
- 19.Fass D, Harrison S C, Kim P S. Structure of Moloney murine virus envelope domain at 1.7Å resolution. Nat Struct Biol. 1996;3:465–469. doi: 10.1038/nsb0596-465. [DOI] [PubMed] [Google Scholar]
- 20.Gill S C, von Hippel P H. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 21.Goebel S J, Johnson G P, Perkus M E, Davis S W, Winslow J P, Paoletti E. The complete DNA sequence of vaccinia virus. Virology. 1990;179:247–266. doi: 10.1016/0042-6822(90)90294-2. [DOI] [PubMed] [Google Scholar]
- 22.Gong S C, Lai C F, Esteban M. Vaccinia virus induces cell fusion at acid pH and this activity is mediated by the N-terminus of the 14-kDa virus envelope protein. Virology. 1990;178:81–91. doi: 10.1016/0042-6822(90)90381-z. [DOI] [PubMed] [Google Scholar]
- 23.Higuchi R, Krummel B, Saiki R K. A general method of specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hiller G, Weber K. Golgi-derived membranes that contain an acylated viral polypeptide are used for vaccinia virus envelopment. J Virol. 1985;55:651–659. doi: 10.1128/jvi.55.3.651-659.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hodel A E, Gershon P D, Shi X, Quiocho F A. The 1.85 Å structure of vaccinia protein VP39: a bifunctional enzyme that participates in the modification of both mRNA ends. Cell. 1996;85:247–256. doi: 10.1016/s0092-8674(00)81101-0. [DOI] [PubMed] [Google Scholar]
- 26.Hodges R S. Unzipping the secrets of coiled-coils. Curr Biol. 1992;2:122–124. doi: 10.1016/0960-9822(92)90241-2. [DOI] [PubMed] [Google Scholar]
- 27.Hodges R S, Sodek J, Smillie L B, Jurazek L. Tropomyosin: amino acid sequence and coiled-coil structure. Cold Spring Harbor Symp Quant Biol. 1972;37:299–310. [Google Scholar]
- 28.Hodges R S, Zhou N E, Kay C M, Semchuk P D. Synthetic model proteins: contribution of hydrophobic residues and disulfide bonds to protein stability. Peptide Res. 1990;3:123–137. [PubMed] [Google Scholar]
- 29.Ichihashi Y, Matsumoto S, Dales S. Biogenesis of poxviruses: role of A-type inclusions and host cell membranes in virus disemination. Virology. 1971;46:507–532. doi: 10.1016/0042-6822(71)90056-0. [DOI] [PubMed] [Google Scholar]
- 30.Joklik W K. Purification of four strains of poxvirus. Virology. 1962;18:9–18. doi: 10.1016/0042-6822(62)90172-1. [DOI] [PubMed] [Google Scholar]
- 31.Lai C, Gong S C, Esteban M. Structural and functional properties of the 14kDa envelope protein of vaccinia virus synthesized in Escherichia coli. J Biol Chem. 1990;265:22174–22180. [PubMed] [Google Scholar]
- 32.Lai C, Gong S C, Esteban M. The purified 14-kilodalton envelope protein of vaccinia virus produced in Escherichia coli induces virus immunity in animals. J Virol. 1991;65:5631–5635. doi: 10.1128/jvi.65.10.5631-5635.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laue T M, Shah B D, Ridgeway T M, Pelletier S L. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding S E, Rowe A J, Horton J C, editors. Analytical ultracentrifugation in biochemistry and polymer science. London, United Kingdom: Royal Society of Chemistry; 1992. pp. 90–125. [Google Scholar]
- 34.Lupas A. Coiled coils: new structures and new functions. Trends Biochem Sci. 1996;21:375–382. [PubMed] [Google Scholar]
- 35.Minton A P. Conservation of signal: a new algorithm for the elimination of the reference concentration as an independently variable parameter in the analysis of sedimentation equilibrium. In: Schusterand T M, Laue T M, editors. Modern analytical ultracentrifugation. Boston, Mass: Birkhauser; 1994. pp. 81–93. [Google Scholar]
- 36.Morgan C. Vaccinia virus reexamined: development and release. Virology. 1976;73:43–58. doi: 10.1016/0042-6822(76)90059-3. [DOI] [PubMed] [Google Scholar]
- 37.Moss B. Poxviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2637–2671. [Google Scholar]
- 38.Pearlman D A, Case D A, Caldwell J C, Ross W S, Cheatham T E, Fergusson D M, Seibel G L, Chandra-Singh U, Weiner P, Kollman P A. AMBER 4.1. University of California, San Francisco; 1995. [Google Scholar]
- 39.Peräen J, Rikkonen M, Hyvönen M, Kääriäinen L. T7 vectors with modified T7lac promoter for expression of proteins in Escherichia coli. Anal Biochem. 1996;236:371–373. doi: 10.1006/abio.1996.0187. [DOI] [PubMed] [Google Scholar]
- 40.Press W H, Flannery B P, Teukolsky S A, Vetterling W T. Numerical recipes in Pascal: the art of scientific computing. Cambridge, United Kingdom: Cambridge University Press; 1989. [Google Scholar]
- 41.Rodriguez D, Rodriguez J-R, Rodriguez J F, Trauber D, Esteban M. Highly attenuated vaccinia virus mutants for the generation of safe recombinant viruses. Proc Natl Acad Sci USA. 1989;86:1287–1291. doi: 10.1073/pnas.86.4.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodriguez D, Rodriguez J-R, Esteban M. The vaccinia virus 14-kilodalton fusion protein forms a stable complex with the processed protein encoded by the vaccinia virus A17L gene. J Virol. 1993;67:3435–3440. doi: 10.1128/jvi.67.6.3435-3440.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodriguez D, Esteban M, Rodriguez J-R. Vaccinia virus A17L gene product is essential for an early step in virion morphogenesis. J Virol. 1995;69:4640–4648. doi: 10.1128/jvi.69.8.4640-4648.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez J F, Janeczko R, Esteban M. Isolation and characterization of neutralizing monoclonal antibodies to vaccinia virus. J Virol. 1985;56:482–488. doi: 10.1128/jvi.56.2.482-488.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodriguez J F, Paez E, Esteban M. A 14,000-Mr envelope protein of vaccinia virus is involved in cell fusion and forms covalently linked trimers. J Virol. 1987;61:395–404. doi: 10.1128/jvi.61.2.395-404.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodriguez J F, Smith G L. IPTG-dependent vaccinia virus: identification of a virus protein enabling virion envelopment by Golgi membrane and egress. Nucleic Acids Res. 1990;18:5347–5351. doi: 10.1093/nar/18.18.5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roos N, Cyrrklaff M, Cudmore S, Blasco R, Krijnse-Locker J, Griffiths G. A novel immunogold cryoelectron microscopic approach to investigate the structure of the intracellular and extracellular forms of vaccinia virus. EMBO J. 1996;15:2343–2355. [PMC free article] [PubMed] [Google Scholar]
- 48.Rost B, Sander C. Prediction of protein structure at better than 70% accuracy. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 49.Rost B, Sander C. Improved prediction of protein secondary structure by use of sequence profiles and neural networks. Proc Natl Acad Sci USA. 1993;90:7558–7562. doi: 10.1073/pnas.90.16.7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rost B, Sander C. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins. 1994;19:55–72. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- 51.Schmelz M, Sodeik B, Ericsson M, Wolffe E J, Shida H, Hiller G, Griffiths G. Assembly of vaccinia virus: the second wrapping cisterna is derived from the trans Golgi network. J Virol. 1994;68:130–147. doi: 10.1128/jvi.68.1.130-147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmutz C, Payne L G, Gubser J, Wittek R. A mutation in the gene encoding the vaccinia virus 37,000-Mr protein confers resistance to an inhibitor of virus envelopment and release. J Virol. 1991;65:3435–3442. doi: 10.1128/jvi.65.7.3435-3442.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sippl M J. Recognition of errors in three-dimensional structures of proteins. Proteins. 1993;17:355–362. doi: 10.1002/prot.340170404. [DOI] [PubMed] [Google Scholar]
- 54.Sodeik B, Doms R W, Ericsson M, Hiller G, Machamer C E, van’t Hof W, van Meer G, Moss B, Griffiths G. Assembly of vaccinia virus: role of the intermediate compartment between the endoplasmic reticulum and the Golgi stacks. J Cell Biol. 1993;121:521–541. doi: 10.1083/jcb.121.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sodeik B, Cudmore S, Ericsson M, Esteban M, Niles E G, Griffiths G. Assembly of vaccinia virus: incorporation of p14 and p32 into the membrane of the intracellular mature virus. J Virol. 1995;69:3560–3574. doi: 10.1128/jvi.69.6.3560-3574.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tuffery P, Etchebest C, Hazout S, Lavery R. A new approach to the rapid determination of protein side chain conformations. J Biomol Struct Dyn. 1991;8:1267–1289. doi: 10.1080/07391102.1991.10507882. [DOI] [PubMed] [Google Scholar]
- 57.Vanderplasschen A, Smith G L. A novel virus binding assay using confocal microscopy: demonstration that the intracellular and extracellular vaccinia virions bind to different cellular receptors. J Virol. 1997;71:4032–4041. doi: 10.1128/jvi.71.5.4032-4041.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vanderplasschen A, Hollinshead M, Smith G L. Intracellular and extracellular vaccinia virions enter cells by different mechanisms. J Gen Virol. 1998;79:877–887. doi: 10.1099/0022-1317-79-4-877. [DOI] [PubMed] [Google Scholar]
- 59.van Gunsteren W F, Berendsen H J C. Algorithms for macromolecular dynamics and constraint dynamics. Mol Phys. 1977;34:1311–1327. [Google Scholar]
- 60.van Holde K E. Physical biochemistry. 2nd ed. Englewood Cliffs, N.J: Prentice-Hall Inc.; 1985. [Google Scholar]
- 61.Weissenhorn W, Dessen A, Harrison S C, Skehel J J, Wiley D C. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 62.Wiltscheck R, Kammerer R A, Dames S A, Schulthess T, Blommers M J J, Engel J, Alexandrescu A T. Heteronuclear NMR assignments and secondary structure of the coiled coil trimerization domain from cartilage matrix protein in oxidized and reduced forms. Protein Sci. 1997;6:1734–1735. doi: 10.1002/pro.5560060814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wolf E, Kim P S, Berger B. MultiCoil: a program for predicting two- and three-stranded coiled coils. Protein Sci. 1997;6:1179–1189. doi: 10.1002/pro.5560060606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wolffe E J, Isaacs S N, Moss B. Deletion of the vaccinia virus B5R gene encoding a 42-kilodalton membrane glycoprotein inhibits extracellular virus envelope formation and dissemination. J Virol. 1993;67:4732–4741. doi: 10.1128/jvi.67.8.4732-4741.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou N E, Kay C M, Hodges R S. Disulfide bond contribution to protein stability: positional effects of substitution in the hydrophobic core of the two-stranded α-helical coiled-coil. Biochemistry. 1993;32:3178–3187. doi: 10.1021/bi00063a033. [DOI] [PubMed] [Google Scholar]