Abstract
Differences in methylmercury (CH3Hg) production normalized to the sulfate reduction rate (SRR) in various species of sulfate-reducing bacteria (SRB) were quantified in pure cultures and in marine sediment slurries in order to determine if SRB strains which differ phylogenetically methylate mercury (Hg) at similar rates. Cultures representing five genera of the SRB (Desulfovibrio desulfuricans, Desulfobulbus propionicus, Desulfococcus multivorans, Desulfobacter sp. strain BG-8, and Desulfobacterium sp. strain BG-33) were grown in a strictly anoxic, minimal medium that received a dose of inorganic Hg 120 h after inoculation. The mercury methylation rates (MMR) normalized per cell were up to 3 orders of magnitude higher in pure cultures of members of SRB groups capable of acetate utilization (e.g., the family Desulfobacteriaceae) than in pure cultures of members of groups that are not able to use acetate (e.g., the family Desulfovibrionaceae). Little or no Hg methylation was observed in cultures of Desulfobacterium or Desulfovibrio strains in the absence of sulfate, indicating that Hg methylation was coupled to respiration in these strains. Mercury methylation, sulfate reduction, and the identities of sulfate-reducing bacteria in marine sediment slurries were also studied. Sulfate-reducing consortia were identified by using group-specific oligonucleotide probes that targeted the 16S rRNA molecule. Acetate-amended slurries, which were dominated by members of the Desulfobacterium and Desulfobacter groups, exhibited a pronounced ability to methylate Hg when the MMR were normalized to the SRR, while lactate-amended and control slurries had normalized MMR that were not statistically different. Collectively, the results of pure-culture and amended-sediment experiments suggest that members of the family Desulfobacteriaceae have a greater potential to methylate Hg than members of the family Desulfovibrionaceae have when the MMR are normalized to the SRR. Hg methylation potential may be related to genetic composition and/or carbon metabolism in the SRB. Furthermore, we found that in marine sediments that are rich in organic matter and dissolved sulfide rapid CH3Hg accumulation is coupled to rapid sulfate reduction. The observations described above have broad implications for understanding the control of CH3Hg formation and for developing remediation strategies for Hg-contaminated sediments.
The presence of Hg in freshwater and marine environments continues to generate concerns related to biological exposure. The incident involving Hg poisoning in Minamata Bay, Japan, illustrated the potential hazards associated with chronic exposure to Hg, particularly CH3Hg (24). The lipophilic nature of CH3Hg enhances its ability to be bioaccumulated compared to inorganic Hg, and this results in enhanced biomagnification of CH3Hg in the food chain (15, 23, 30). Faust and Osman (12) have reported that typically 90 to 99% of the total Hg in the environment is associated with the sediment, while <1% of the total Hg accumulates in the biota. However, only 1 to 10% of the CH3Hg is associated with sediment, while 90 to 99% of CH3Hg accumulates in the biota. Thus, CH3Hg is the Hg species that causes the most concern regarding human exposure. The U.S. Environmental Protection Agency has stated that consumption of fish and shellfish contaminated with CH3Hg is the primary route of human exposure to Hg (11).
Previous work has shown that Hg methylation is dominated by the activities of sulfate-reducing bacteria (SRB) in sediments (1, 5, 8, 10, 16–20, 27). Perhaps the best evidence for the link between sulfate reduction and Hg methylation in sediments was provided by studies in which the researchers used molybdate, a metabolic inhibitor of sulfate reduction; the results of these studies indicated that Hg methylation was almost completely inhibited in the presence of molybdate (8, 18, 20, 27). Other studies have shown that in pure cultures SRB grown in the absence of sulfate do not generate CH3Hg from available inorganic Hg (33). Although the preponderance of evidence indicates that sulfate reduction is linked to Hg methylation, the mechanism(s) by which methylation occurs is poorly understood, and the SRB groups which mediate methylation in situ have not been described in detail.
In previous pure-culture studies the researchers have utilized primarily one SRB, Desulfovibrio desulfuricans, to determine the Hg methylation potential of the entire SRB population (7, 8, 17, 32, 33). Microorganisms capable of sulfate reduction are currently thought to be much more physiologically and phylogenetically diverse than originally thought. To date, 19 genera of SRB have been described, and the SRB include gram-negative and gram-positive mesophiles, members of the archaea, and members of other thermophilic groups (37). Each phylogenetically distinct group could have a different potential to methylate Hg on a per-cell basis. This problem becomes more complex if the relationship between the mercury methylation rate (MMR) and the sulfate reduction rate (SRR), both of which are thought to be enzymatic processes that respond to environmental variables, is different for each SRB group.
The diversity of the SRB has recently been explored by using oligonucleotide probes designed to target the 16S rRNA molecules in freshwater and marine sediments (10, 35, 36, 40). There have been several observations that have related community structure to the function of SRB consortia, and it has been suggested that members of the acetate-utilizing family of mesophilic SRB, the family Desulfobacteriaceae, may be more prevalent and active in marine sediments than members of other SRB families are (36). Similar to carbon metabolism during sulfate respiration, the microbiological controls that link Hg methylation to sulfate reduction in sediments are likely to be equally complex and dependent on SRB physiology. In order to elucidate possible CH3Hg production by members of different SRB groups, Hg methylation was studied in a range of pure cultures and in marine sediments, in which SRB consortia were monitored by using molecular probes.
MATERIALS AND METHODS
Pure-culture experiments.
The pure cultures of SRB used in this study included pure cultures of the freshwater strains Desulfovibrio desulfuricans ATCC 13541, Desulfobulbus propionicus ATCC 33891, and Desulfococcus multivorans ATCC 33890, which were obtained from the American Type Culture Collection, as well as pure cultures of the marine strains Desulfobacter sp. strain BG-8 and Desulfobacterium sp. strain BG-33 obtained from Richard Devereux (U.S. Environmental Protection Agency, Gulf Breeze, Fla.). BG-8 was shown to exhibit 96.6% sequence identity with Desulfobacter curvatus, while BG-33 exhibited 96.6% sequence identity with Desulfobacterium niacini (37).
The SRB were maintained by using standard Hungate techniques for growth of anaerobic bacteria under strictly anoxic conditions. The multipurpose minimal culture medium (freshwater version containing 1 ppt of NaCl) was prepared and manipulated as described by Widdel and Bak (41). This medium contained only mineral salts, a small amount of vitamins, and 1 to 2 mM dissolved sulfide, which was added as a reductant. Carbon substrates (acetate and lactate) were each added from a sterile anoxic stock solution to a final concentration of 10 mM. Sterile medium components were combined, and the medium was dispensed into serum bottles which were sealed with butyl rubber stoppers under a 90% N2–10% CO2 gas stream. Inocula were transferred by using gas-tight syringes (fitted with 20-gauge needles) that were flushed with nitrogen. The initial cell density in each pure culture was ∼104 cells ml−1. Cultures were incubated in the dark at 28°C.
In pure-culture experiments, four replicate culture bottles were prepared for each SRB group studied. Mercuric nitrate was added 120 h after inoculation to a final concentration of 100 ng ml−1 as soluble Hg, and radiolabeled 35SO42− (ICN Biomedical, Inc.) was added 12 h before inorganic Hg was added. At specified times, subsamples were removed and used to determine optical densities, cell counts, sulfate concentrations, SRR, total Hg concentrations in filtered samples, and CH3Hg concentrations in unfiltered samples. Cultures were sampled immediately after inorganic mercury was added and for an additional 96 h (samples were collected at 24-h intervals).
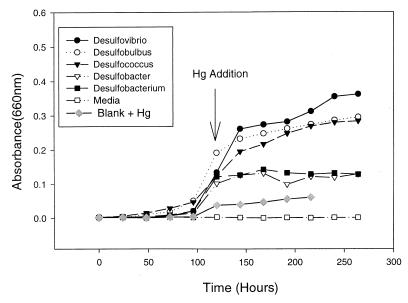
Cell density was monitored by measuring optical density at 660 nm with a Shimadzu spectrophotometer and by determining direct counts by epifluorescence. To determine direct counts, 1-ml culture samples were preserved with 100 μl of 37% formaldehyde (final concentration, 3.4%). Cells were then stored at 4°C in the dark until they were counted. Cells were diluted in buffer, stained with DAPI (4′,6′-diamidino-2-phenylindole), and counted as described by Williams et al. (42). It should be noted that a low level of absorbance was observed when 100 ng of Hg2+ per ml was added (Fig. 1); this was probably due to mineral precipitation, and the absorbance values were adjusted accordingly.
FIG. 1.
Growth curves for five pure cultures of SRB during Hg methylation experiments. Inorganic Hg (100 ng ml−1) was added to cultures 120 h after inoculation.
Direct cell counts were determined at each time point after Hg was added, and the average counts based on the data for five timepoints are shown in Table 1. The SRR and the corresponding standard error were determined based on the results of a linear regression analysis of the amount of sulfate reduced over time after inorganic Hg was added. MMR and standard error calculations were based on the results of a linear regression analysis of CH3Hg concentrations produced over time.
TABLE 1.
Results of pure-culture experiments
| Taxon | No. of cells ml−1 | SRR
|
MMR
|
MMR/SRR | ||
|---|---|---|---|---|---|---|
| nmol ml−1 h−1 | nmol cell−1 h−1 | pg ml−1 h−1 | pg cell−1 h−1 | |||
| Desulfovibrio | 1.08 × 108 ± 6.4 × 108a | 15.23 ± 7.6 | 1.4 × 10−7 ± 7.9 × 10−7 | 4.48 ± 11.8 | 4.2 × 10−8 ± 12.2 × 10−8 | 1.37 × 10−6 ± 1.8 × 10−7 |
| Desulfobulbus | 2.44 × 108 ± 13.0 × 108 | 16.98 ± 12.9 | 7.0 × 10−8 ± 14.6 × 10−8 | 1.05 ± 30.4 | 4.3 × 10−9 ± 32.1 × 10−9 | 2.80 × 10−7 ± 9.0 × 10−8 |
| Desulfococcus | 1.68 × 107 ± 15.9 × 107 | 4.15 ± 20.0 | 2.7 × 10−7 ± 20.4 × 10−7 | 4.62 ± 21.4 | 2.7 × 10−7 ± 23.9 × 10−7 | 4.60 × 10−6 ± 1.30 × 10−6 |
| Desulfobacter | 1.27 × 106 ± 5.0 × 106 | 1.97 ± 12.2 | 1.4 × 10−6 ± 13.7 × 10−6 | 1.55 ± 21.3 | 1.6 × 10−6 ± 21.5 × 10−6 | 4.10 × 10−6 ± 9.5 × 10−7 |
| Desulfobacterium | 1.21 × 106 ± 5.7 × 106 | 1.36 ± 8.3 | 1.1 × 10−6 ± 8.7 × 10−6 | 7.53 ± 9.9 | 6.2 × 10−6 ± 10.4 × 10−6 | 2.58 × 10−5 ± 2.80 × 10−6 |
Mean ± standard error.
Sediment slurry experiments.
Sediment samples were obtained from the bank of the Skidaway River (in a salt marsh dominated by the macrophyte Spartina alterniflora) near the Skidaway Institute of Oceanography in Savannah, Ga. The background concentrations of Hg and CH3Hg in the river water and sediments were below the minimal detectable limit (1 pg ml−1).
Sediment slurries were prepared by using 150-ml beakers with gas-tight caps. Three tubes were placed in each cap and sealed with silicone sealant. One of the tubes was used to continuously add N2-CO2 (9:1) to the slurry at a rate of 40 ml min−1. This removed any oxygen that might have been present in the reactor. The second tube served as a gas release port, while the third tube reached the bottom of the reactor and was used as a sample port.
Approximately 20.0 g (wet weight) of sediment (moisture content, ∼60%) was mixed with 45 ml of Skidaway River water (salinity, 17.39 ppt) to obtain a final sediment content of 12 to 14% (approximately 140 g liter−1 on a dry weight basis). Amended slurries were treated with either 20 mM lactate or 10 mM acetate. It is important to note that the control reactor was constructed 24 h before inorganic Hg was added, while the amended sediment experiments were initiated with the different organic acids 20 days before inorganic Hg was added. Skidaway River water was purged with nitrogen gas for 30 min before sediment was added, and the entire reactor was continually shaken at 100 rpm and maintained at 25°C throughout the incubation period. Three parallel slurries were prepared for each sediment treatment. At specified times, subsamples were withdrawn in order to determine the total soluble Hg, CH3Hg, soluble organic acid, dissolved sulfate content, and the SRR. At the end of the experiment, sediments obtained from the parallel slurries were pooled and used for 16S rRNA determinations. Pore water was separated from the solid phase by centrifugation and was filtered by using 0.2-μm-pore-size cellulose acetate syringe filters that were flushed with nitrogen gas.
To prepare unamended slurries, 8 μl of a 1-μCi ml−1 radioactive 35SO42− solution (2.2 mCi ml−1; catalog no. 64040; ICN Biomedicals, Inc.) was added 12 h before inorganic Hg was added. Mercuric nitrate was added at a concentration of 950 ng g (dry weight) of sediment−1 12 h after 35SO42− was added to avoid any short-term artifacts that may have been associated with the tracer addition. Slurries that were amended with lactate or acetate received a similar aliquot of 35SO42− 19.5 days after substrate was added.
Sample analysis.
Unless otherwise stated, the same method was used for both pure cultures and sediment slurries. Sulfate concentrations were determined by using the turbidometric method of Tabatabi (39). The SRR was determined by using the radiotracer technique developed by Jørgensen (25) and a one-step sulfide distillation procedure performed as described by Fossing and Jørgensen (13). For sediment slurries, the radiotracer technique was modified as described by King et al. (27). For pure cultures, 3-ml aliquots were removed from culture bottles at different times and placed into 30-ml portions of an anoxic 20% zinc acetate solution. The resulting sulfide precipitate was then distilled as described above. CH3Hg and total soluble Hg concentrations were determined by using methods described by King et al. (27). Briefly, CH3Hg was quantified by using cold vapor atomic fluorescence spectrometry (CVAFS) as described by Liang et al. (29) and modified by King et al. (27).
The methods used for extraction and purification of microbial 16S rRNA from sediment slurries were based on the methods described by Moran et al. (31). The oligonucleotide probes used to determine the quantities of SRB 16S rRNA present in sediment slurries were developed by Devereux et al. (9). Oligonucleotide probes were synthesized at the Molecular Genetics Facility of the University of Georgia by using an ABI model 394 DNA-RNA synthesizer. Labeling and probing of oligonucleotides and probing of 16S rRNA were carried out as described by Devereux et al. (9). The appropriate 32P-labeled probe was added to the blots, and this was followed by addition of 15 ml of fresh prehybridization solution that had been warmed to 55°C. Hybridization was allowed to occur for 18 h in a shaking circulating water bath at 55°C. Blots for each probe were washed at 55°C three times (20 min each) in deep dishes that contained 50 ml of wash buffer. After the final washes, the blots were exposed to film (Fuji Medical X-ray film; 20.3 by 25.4 cm; Fuji Medical Systems U.S.A.) in autoradiograph cassettes (type FBXC 810; Fisher Scientific) with an enhancement screen at −80°C. The film was taken out of the cassettes and developed by using methods of Frischer et al. (14). To determine the specificity of probes and to quantify 16S rRNA, a model 420 OE densitometer (PDI Inc.) was used.
RESULTS
Pure cultures. (i) Cell density.
Growth curves showed that the cultures were exposed to 100 ng of Hg2+ ml−1 at 120 h after inoculation during the late log phase of growth. From zero time to 264 h, the cell densities of Desulfovibrio, Desulfobulbus, and Desulfococcus strains increased approximately 180-, 145-, and 140-fold, respectively (Fig. 1). During the same period, the cell numbers in both Desulfobacter sp. strain BG-8 and Desulfobacterium sp. strain BG-33 cultures increased approximately 65-fold (Fig. 1). Table 1 shows the average cell number during measurements of Hg methylation for each of the phylogenetic groups studied.
(ii) SRR.
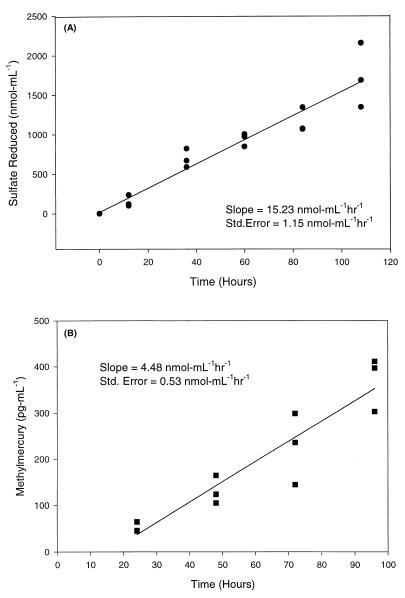
SRR were determined for each of the SRB strains grown in pure culture. Figure 2A shows the amounts of sulfate reduced over time in cultures of Desulfovibrio desulfuricans, and these results are representative of the sulfate depletion results obtained with other pure cultures. A linear regression analysis, based on triplicate assays, revealed that the average SRR was 15.23 nmol ml−1 h−1 for Desulfovibrio desulfuricans (Fig. 2A). Table 1 shows the calculated SRR and standard error for each of the five SRB cultures examined.
FIG. 2.
(A) Sulfate reduction in Desulfovibrio desulfuricans cultures over a 120-h period. The SRR was calculated from the results of a linear regression analysis of the amount of sulfate reduced over time. Inorganic Hg (100 ng ml−1) was added at 12 h. (B) MMR in pure cultures of Desulfovibrio desulfuricans. Inorganic Hg (100 ng ml−1) was added at zero time. A linear regression analysis was performed with CH3Hg concentrations obtained between 24 and 96 h.
(iii) CH3Hg production.
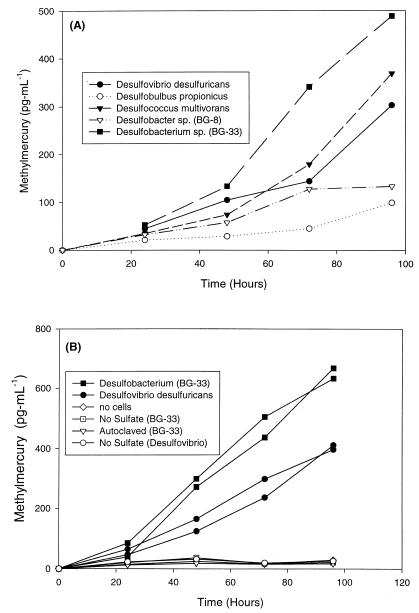
CH3Hg concentrations were determined over time for each of the five SRB pure cultures studied (Fig. 3). All five strains generated substantial concentrations of CH3Hg compared to uninoculated or autoclaved control cultures (Fig. 3). The final concentrations of CH3Hg ranged from 85 to 472 pg ml−1 and the order for the taxa studied was as follows (from lowest concentration to highest concentration): Desulfobulbus < Desulfobacter sp. strain BG-8 < Desulfovibrio < Desulfococcus < Desulfobacterium sp. strain BG-33 (Fig. 3A).
FIG. 3.
(A) CH3Hg concentrations in pure cultures of SRB. (B) CH3Hg production in the presence of Desulfovibrio and Desulfobacterium cells and in control cultures. Inorganic Hg (100 ng ml−1) was added at zero time in both experiments.
To show that CH3Hg production was indeed coupled to sulfate reduction in SRB which differ phylogenetically, a number of treatments were used with Desulfovibrio and Desulfobacterium cultures. The CH3Hg concentrations in Desulfobacterium sp. strain BG-33 cultures growing in a sulfate-containing medium were approximately 52 and 647 pg ml−1 at 24 and 96 h, respectively. Similarly, sulfate-containing Desulfovibrio cultures contained approximately 55 and 404 pg of CH3Hg ml−1 at 24 and 96 h, respectively. In sulfate-depleted media, however, Desulfobacterium and Desulfovibrio cultures produced little or no CH3Hg and the CH3Hg concentrations in these cultures overlapped with the concentrations in uninoculated or killed controls (Fig. 3B).
Based on the fact that all five SRB strains tested exhibited a lag in CH3Hg production in the first 24 h after inorganic Hg was added, the MMR was determined for each strain based on a plot of the CH3Hg concentrations at 24 to 96 h. Figure 2B shows a representative plot of the CH3Hg concentrations in triplicate Desulfovibrio cultures. A linear regression analysis of the data revealed an MMR of 4.48 pg ml−1 h−1 (Fig. 2B). Table 1 shows the observed MMR and the standard error calculated (as shown for the Desulfovibrio culture) for each pure culture studied. For all of the cultures tested, a linear regression analysis resulted in correlation coefficients (r2) greater than 0.85, and the standard errors of the slopes were generally less than 25%. This linear response in CH3Hg production occurred after the initial 24 h and validated the use of linear regression analysis to determine MMR based on CH3Hg concentrations observed at 24 to 96 h.
Sediment slurries. (i) SRR.
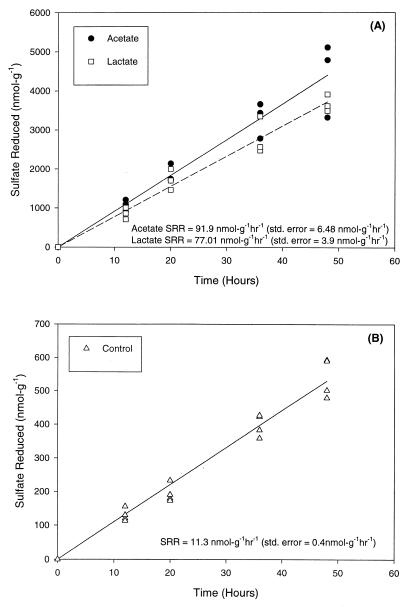
The amounts of sulfate reduced in control and carbon substrate (electron donor)-amended slurries were determined over time (12 h before inorganic Hg was added and for the next 48 h). Figure 4 shows the amount of sulfate reduced over time for each of the sediment slurry treatments. The average SRR for the lactic acid- and acetic acid-amended preparations (91.90 and 77.01 nmol g−1 h−1, respectively) were 7.0- to 8.3-fold greater than the SRR observed in the unamended preparations (11.30 nmol g−1 h−1).
FIG. 4.
(A) Sulfate reduction in sediment slurries amended with lactate and acetate. Lactate or acetate was added to slurries 20 days before inorganic Hg was added. Inorganic Hg (950 ng g−1) was added at zero time. (B) Sulfate reduction in unamended or control sediment slurries. Inorganic Hg (950 ng g−1) was added to slurries at zero time. The data points at each time represent samples acquired from three slurries that received the same treatment, and the lines represent the results of linear regression analysis which was used to generate an average SRR for the duration of the experiment.
(ii) CH3Hg production.
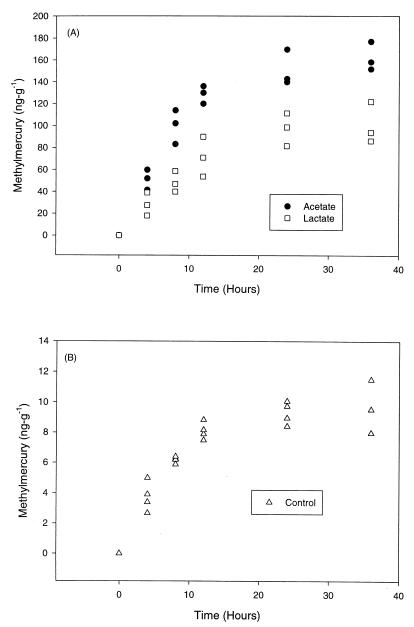
Increases in CH3Hg concentrations over time were observed for all sediment slurry treatments (Fig. 5). The highest maximum concentrations of CH3Hg were observed in the acetate-amended slurries (162 ± 6.01 ng g−1), followed by the lactate-amended slurries (101 ± 6.23 ng g−1). In agreement with the observed SRR, the maximum CH3Hg production values were 10 to 20 times higher in organic acid-amended slurries than in unamended slurries (Fig. 5).
FIG. 5.
(A) CH3Hg production in sediment slurries amended with lactate and acetate. (B) CH3Hg concentrations in control sediment slurries. Inorganic Hg (950 ng g−1) was added to slurries at zero time. The data points each represent triplicate samples which received the same sediment treatment. The slurries represented above were the same slurries as those used in the experiments whose results are shown in Fig. 4.
(iii) Group-specific oligonucleotide probe analysis of sediment SRB consortia.
Data obtained from probe experiments indicated that addition of acetate for 20 days altered the SRB profiles such that Desulfobacterium and Desulfobacter species accounted for 53.0 and 41.6% of the total population, respectively (Table 2), while addition of lactate and incubation for 20 days resulted in bacterial profiles that were similar (in terms of percent composition) to the profiles of unamended (control) sediment slurries (Table 2). In slurries amended with 20 mM lactate, the 16S rRNA concentrations, as determined by the probe signals, were significantly greater with probes that identified the Desulfovibrio (probe 687), Desulfobulbus (probe 660), and Desulfococcus-Desulfosarcina-Desulfobotulus (probe 814) groups than in the controls (P < 0.05). The concentrations of 16S rRNA determined with probes 687, 660, and 814 in lactate-amended slurries were approximately 2.6-, 2.9-, and 3.7-fold greater, respectively, than the concentrations in control unamended sediment slurries (Table 2). The 16S rRNA concentrations in slurries amended with acetate were 5.2- and 12.9-fold lower than the concentrations in control slurries for the Desulfovibrio and Desulfobulbus groups, respectively. However, the 16S rRNA concentrations for the Desulfobacter groups (probe 129) and the Desulfobacterium groups (probe 221) in acetate-amended slurries were 2.6- and 3.5-fold higher than the control concentrations, respectively (Table 2). A comparison of the 16S rRNA profiles obtained for the lactate-amended and acetate-amended slurries revealed that there were significant differences in the concentrations for all of the phylogenetic groups studied (P < 0.05) (Table 2). The Desulfobacter and Desulfobacterium 16S rRNA concentrations in the acetate-amended slurries were approximately 1.7- and 3.1-fold greater, respectively, than the concentrations in the lactate-amended slurries. However, the 16S rRNA concentrations determined by using probes 687, 660, and 814 with lactate-amended slurries were 13.5-, 37.1-, and 5.8-fold higher, respectively, than the concentrations determined for acetate-amended slurries (Table 2). The amount of 16S rRNA identified by the specific probes in each reactor was expressed as a percentage of the additive total 16S rRNA from members of the five SRB groups studied (Table 2).
TABLE 2.
Percentages of SRB in sediment slurries as determined by the summation of labeled probe values
| Treatment | % of SRB
|
||||
|---|---|---|---|---|---|
| Probe 687 (Desulfovibrio sp.) | Probe 660 (Desulfobulbus sp.) | Probe 814 (Desulfococcus sp.) | Probe 129 (Desulfobacter sp.) | Probe 221 (Desulfobacterium sp.) | |
| Control | 13.1 ± 7.1a | 15.9 ± 2.2 | 10.6 ± 3.3 | 30.7 ± 4.5 | 29.6 ± 2.1 |
| Lactate amended | 16.7 ± 2.7 | 22.8 ± 2.7 | 19.2 ± 1.0 | 24.2 ± 2.3 | 16.8 ± 0.6 |
| Acetate amended | 1.2 ± 0.3 | 0.63 ± 0.1 | 3.4 ± 1.5 | 41.6 ± 3.2 | 53.0 ± 2.8 |
Mean ± standard error.
(iv) Total soluble Hg concentrations.
The total soluble Hg concentrations remained within the standard error (<10%) and did not change significantly with time in a given pure culture. Also, there were no significant differences in the mean soluble Hg concentrations in cultures of all of the SRB examined (P > 0.05, as determined by the t test).
The soluble inorganic Hg concentrations over time were similar for all of the sediment slurry treatments, and the amount of inorganic Hg that was present in the slurry water decreased with time (Table 3). The maximum concentrations of Hg were observed at the beginning of the experiment (263 to 198 ng l−1), and the concentrations were sixfold lower at 36 h (55.0 to 42.0 ng l−1) (Table 3).
TABLE 3.
Calculated MMR and average total soluble mercury concentrations in marine sediment incubations
| Prepn | Reactor no. | Time (h) | MMR (ng g−1 h−1)a | Avg total soluble mercury concn (ng liter−1)b |
|---|---|---|---|---|
| Control | 1 | 0–4 | 0.85 | 218.3 |
| 4–8 | 0.71 | 198.4 | ||
| 8–12 | 0.65 | 153.8 | ||
| 12–24 | 0.10 | 92.8 | ||
| 24–36 | 0.11 | 53.3 | ||
| 2 | 0–4 | 0.98 | 169.4 | |
| 4–8 | 0.57 | 113.7 | ||
| 8–12 | 0.43 | 74.7 | ||
| 12–24 | 0.09 | 55.2 | ||
| 24–36 | 0.05 | 49.7 | ||
| 3 | 0–4 | 1.2 | 198.9 | |
| 4–8 | 0.4 | 141.0 | ||
| 8–12 | 0.3 | 88.2 | ||
| 12–24 | 0.08 | 56.0 | ||
| 24–36 | −0.04 | 51.7 | ||
| 4 | 0–4 | 0.7 | 184.0 | |
| 4–8 | 0.8 | 135 | ||
| 8–12 | 0.6 | 83.7 | ||
| 12–24 | 0.1 | 57.4 | ||
| 24–36 | −0.05 | 47.5 | ||
| Lactate amended | 1 | 0–4 | 9.7 | 237.7 |
| 4–8 | 5.0 | 195.6 | ||
| 8–12 | 7.8 | 96.4 | ||
| 12–24 | 0.7 | 62.7 | ||
| 24–36 | −1.0 | 68.4 | ||
| 2 | 0–4 | 6.9 | 207.3 | |
| 4–8 | 4.8 | 147.0 | ||
| 8–12 | 6.0 | 93.7 | ||
| 12–24 | 3.7 | 60.6 | ||
| 24–36 | 0.9 | 49.5 | ||
| 3 | 0–4 | 4.5 | 201.6 | |
| 4–8 | 5.5 | 169.6 | ||
| 8–12 | 3.5 | 131.0 | ||
| 12–24 | 2.3 | 96.1 | ||
| 24–36 | 1.0 | 68.8 | ||
| Acetate amended | 1 | 0–4 | 13.0 | 237.7 |
| 4–8 | 12.5 | 195.6 | ||
| 8–12 | 8.5 | 141.3 | ||
| 12–24 | 2.8 | 89.9 | ||
| 24–36 | 0.6 | 62.3 | ||
| 2 | 0–4 | 15.0 | 226.7 | |
| 4–8 | 13.5 | 176.8 | ||
| 8–12 | 4.0 | 135.9 | ||
| 12–24 | 1.1 | 98.8 | ||
| 24–36 | 0.7 | 58.4 | ||
| 3 | 0–4 | 10.4 | 165.3 | |
| 4–8 | 10.4 | 115.3 | ||
| 8–12 | 9.1 | 104.0 | ||
| 12–24 | 1.7 | 84.6 | ||
| 24–36 | 1.54 | 51.0 |
MMR were calculated by subtracting the methylmercury concentrations measured at the beginning of the time interval from the concentration measured at the end of the time interval and then normalizing by the duration of time that had elapsed between measurements.
Soluble mercury concentrations were calculated by averaging the soluble mercury concentrations determined at the beginning and end of each time interval.
DISCUSSION
Mercury methylation by members of different phylogenetic groups of SRB.
Methylation of Hg has been extensively studied in order to determine its relationship to sulfate reduction activity in sediments, especially in freshwater environments (10, 18, 20, 32) and to a lesser extent in marine environments (8, 27). In addition, using pure-culture experiments, researchers have begun to characterize the mechanisms by which Hg methylation may be coupled to sulfate reduction; in a few studies workers have suggested operative enzyme pathways (7, 17, 33).
However, despite the progress mentioned above, the exact mechanisms used for coupling and the limiting factors for methylation by SRB remain elusive. Most pure-culture experiments (8, 18, 33) have been carried out with members of one group of SRB, the genus Desulfovibrio, which may not be the predominant, active group in marine sediments (36, 37). Furthermore, in no previous work have researchers comprehensively determined or identified the operative SRB populations responsible for methylation activity in sediments. In the past 5 to 10 years, the SRB have been shown to be much more physiologically and phylogenetically broad ranging than was originally thought (37, 41). Using oligonucleotide probes designed to target the 16S rRNA molecule, workers have begun to elucidate the role of relatively new SRB groups with respect to sediment biogeochemistry (10, 35, 36, 40). In light of the results of these new studies, the biological controls that relate Hg methylation to sulfate reduction in natural environments are likely to be complex and to depend on the physiology of the prevailing SRB present.
In this study, we began to characterize how bacterial physiology may limit Hg methylation by constraining methylation activity in a number of phylogenetically and physiologically distinct groups of SRB. The results obtained with Desulfobacterium pure cultures show that methylation does not occur in the absence of sulfate, indicating that Hg methylation is directly coupled to sulfate respiration (Fig. 3B; Table 1). This observation is potentially significant for a number of reasons. In contrast to members of the well-studied genus Desulfovibrio, members of the genus Desulfobacterium are capable of complete acetate oxidation and incapable of fermentation (6, 41). Furthermore, the genus Desulfobacterium is a member of the recently defined family Desulfobacteriaceae, and it has been shown that several members of this group may play a predominant role in sulfate reduction coupled to carbon cycling in marine sediments (37).
Our pure-culture studies also revealed that Hg methylation activities, normalized to cell number, varied more than 3 orders of magnitude for phylogenetically distinct SRB groups under similar culture conditions (Table 1). The rates at which SRB methylated Hg were determined to be in the following order: Desulfobacterium ≫ Desulfobacter ≈ Desulfococcus ≫ Desulfovibrio ≈ Desulfobulbus.
The results described above suggest that members of the family Desulfobacteriaceae methylate Hg at higher rates than members of the family Desulfovibrionaceae methylate Hg. Furthermore, it appears that the SRB that are capable of acetate utilization (all strains except Desulfovibrio and Desulfobulbus strains) methylate Hg more effectively. An explanation for the correlation between Hg methylation and acetate metabolism may be found by consulting previous studies of carbon metabolism in various SRB. It has been shown that methyl transferase enzymes, which are thought to be important contributors to Hg methylation (7, 17), are induced during complete oxidation of acetate by Desulfobacterium strains and other SRB (22, 38).
We suggest that the differential Hg methylation which we observed may be explained by the presence of constitutive and induced methyl transferase pathways. SRB utilize acetate by completely oxidizing the acetyl group of acetyl coenzyme A to CO2 by two entirely different mechanisms. Desulfobacter strains employ the citric acid cycle, while Desulfobacterium strains utilize the carbon monoxide dehydrogenase pathway (38). Since the carbon monoxide dehydrogenase pathway includes several tetrahydrofolate enzymes which have been implicated in Hg methylation (7), it is tempting to suggest that induction of these enzymes was responsible for the rapid Hg methylation rates observed in Desulfobacterium pure cultures.
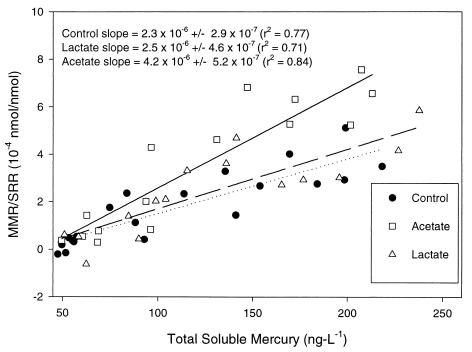
The relationship among the SRB community, carbon metabolism, and Hg methylation activity was further elucidated by studying marine sediments. Adding known electron donors for SRB (26, 34), acetate and lactate, resulted in levels of CH3Hg in marine sediments which were 10 to 20 times higher than the levels in unamended sediments (Fig. 5). Sediment slurries in which acetate was the predominant carbon source methylated Hg at higher rates than lactate-amended or control slurries methylated Hg when the MMR was normalized to the SRR (Fig. 6). In the same sediments, members of the Desulfobacteriaceae dominated other SRB groups in the presence of added acetate, when significantly higher Hg methylation rates were observed (Table 2; Fig. 6). In contrast, members of the Desulfobacteriaceae and Desulfovibrionaceae were more equal contributors to the SRB population in the presence of added lactate or in unamended slurries (Table 2), and lower Hg methylation rates normalized to the SRR were observed (Fig. 6). Here we show that the methylation rates were correlated with the SRB consortia present in abundance, and this relationship appeared to depend on carbon metabolism, among other factors.
FIG. 6.
Plot of MMR normalized to SRR in sediment slurries as a function of total soluble Hg concentrations. The MMR for each sediment slurry was normalized to the observed SRR for the same slurry and was plotted with respect to the average observed total soluble Hg concentration.
Therefore, by using two independent experimental approaches, we obtained evidence which suggests that Hg methylation is related to the SRB group present and to carbon metabolism. These observations have important implications in both marine and freshwater sediments, in which SRB have been shown to be abundant and important in carbon cycling. We clearly demonstrated that acetate stimulates Hg methylation by SRB, and we suggest that the acetate-utilizing SRB methylate Hg at higher rates than other SRB groups methylate Hg. An alternate interpretation of the data is that acetate stimulates methylation via transmethylase induction in a more diverse SRB community. Additional experiments are needed to identify the physiological linkages and enzyme pathways involved in coupling Hg methylation to sulfate reduction in different SRB groups.
Implications for biogeochemical control of CH3Hg formation in sedimentary environments.
Bioavailability (2–4) and the phylogenetic affiliation of SRB appear to act together to control methylation activity in marine sediments. In acetate-amended sediments in which the SRB population was dominated by members of the Desulfobacteriaceae, the MMR was significantly higher than the MMR in other sediments, and total soluble Hg concentrations were highly correlated with normalized MMR in all sediment slurries (Fig. 6). The salt marsh sediments used in this study contain high levels of both organic matter, including humic acids, and sulfides. Therefore, Hg bioavailability in these sediments could be limited by association of inorganic Hg with either dissolved organic carbon, as observed by Barkay et al. (2), or dissolved sulfide, as explained by Benoit et al. (3, 4).
Previous work has suggested that Hg methylation in sediments is inhibited by dissolved sulfide concentrations greater than 10 μM due to precipitation of Hg sulfide minerals (20). However, we have observed methylation activity in the presence of 30 mM sulfate and dissolved sulfide at concentrations in the millimolar range in both pure cultures and marine sediments (27) (Fig. 5), and the level of methylation overlapped with the level found in freshwater sediments (20, 28). We have observed that methyl Hg accounts for up to 1 to 2% of the total Hg in organic compound-rich salt marsh sediments contaminated with Hg (J. E. Kostka, M. Frischer, and K. Maruya, unpublished results), while Gilmour et al. (20) observed that methyl Hg accounts for up to 2% of total Hg in the organic compound-rich freshwater sediments of the northern Everglades. Although Hg methylation may be inhibited by sulfur chemistry to some extent in these sulfidic sediments, we have observed that high SRR may result in significant methyl Hg accumulation in the presence of high sulfur concentrations in marine sediments.
Methyl Hg production by SRB could be a substantial problem source of Hg in the food chains of contaminated estuaries. In support of this, the SRR measured in the Georgia salt marsh (up to >6,000 nmol cm−3 day−1) overlap some of the highest SRR measured in organic compound-rich sediments anywhere on earth (21). In addition, our observations are consistent with those of Winger et al. (43), who suggested that methyl Hg was responsible for sediment toxicity in a contaminated salt marsh and estuary near the place where sediments were collected for our study. Inhibition of Hg methylation by sulfur chemistry and its relationship to the bioavailability of Hg should be studied further, especially in contaminated marine sediments. For example, more sophisticated assays for methylation activity, such as the radiotracer method developed by Gilmour and Riedel (19), should be applied to marine sediments that contain high levels of dissolved sulfide.
Conclusions.
Here we describe novel experimental results which indicate that the physiology of SRB consortia may limit methyl Hg formation. Our results correlate Hg methylation activity with the phylogeny of the SRB for the first time. Finally, we obtained evidence that these relationships occur in marine sediments by performing 16S rRNA oligonucleotide probe analyses of natural SRB consortia. Although we indicate certain enzyme pathways which could be responsible for coupling Hg methylation to acetate metabolism during sulfate reduction (in the Desulfobacteriaceae), we recognize that acetate could also stimulate methylation by transmethylase induction in a more diverse SRB community.
We show that, under sulfidic conditions (in the presence of millimolar amounts of dissolved sulfide) in organic compound-rich marine sediments, rapid CH3Hg accumulation is coupled to rapid sulfate reduction. Our observations expand the small database available for Hg methylation in marine sediments, and they could prove to be vital for directing future studies on the limitations of microbially mediated methyl Hg formation in sediments and could lead to the development or refinement of remediation strategies for Hg-contaminated sediments.
ACKNOWLEDGMENTS
This work was funded by grants from the U.S. Environmental Protection Agency Hazardous Substance South and Southwest Research Center and the Office of Naval Research.
Lori Cowden, Jean Danforth, and Debbie Craven are thanked for their expert technical assistance.
REFERENCES
- 1.Andersson I, Parkman H, Jernelov A. The role of sediments as sink or source for environmental contaminants: a case study of Hg and chlorinated organic compounds. Limnologica. 1990;20:347–359. [Google Scholar]
- 2.Barkay T, Gillman M, Turner R R. Effects of dissolved organic carbon and salinity on the bioavailability of mercury. Appl Environ Microbiol. 1997;63:4267–4271. doi: 10.1128/aem.63.11.4267-4271.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benoit J M, Gilmour C G, Mason R P, Heyes A. Sulfide controls on mercury speciation and bioavailability to methylating bacteria in sediment pore waters. Environ Sci Technol. 1999;33:951–957. [Google Scholar]
- 4.Benoit J M, Mason R P, Gilmour C G. Estimation of mercury-sulfide speciation in sediment pore waters using octanol-water partitioning and implication for the availability to methylating bacteria. Environ Toxicol Chem. 1999;18:2138–2141. doi: 10.1002/etc.5620181004. [DOI] [PubMed] [Google Scholar]
- 5.Blum J E, Bartha R. Effects of salinity on methylation of Hg. Bull Environ Contam Toxicol. 1980;25:404–408. doi: 10.1007/BF01985546. [DOI] [PubMed] [Google Scholar]
- 6.Brock T D, Madigan M T, Martinko J M, Parker J. Biology of microorganisms. Englewood Cliffs, N.J: Prentice-Hall, Inc.; 1992. [Google Scholar]
- 7.Choi S, Chase T, Bartha R. Metabolic pathways leading to mercury methylation in Desulfovibrio desulfuricans LS. Appl Environ Microbiol. 1994;60:4072–4077. doi: 10.1128/aem.60.11.4072-4077.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Compeau G, Bartha R. Sulfate reducing bacteria: principal methylators of Hg in anoxic estuarine sediments. Appl Environ Microbiol. 1985;50:498–502. doi: 10.1128/aem.50.2.498-502.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devereux R, Kane M D, Winfrey J, Stahl D A. Genus- and group-specific hybridization probes for determinative and environmental studies of sulfate-reducing bacteria. Syst Appl Microbiol. 1992;15:601–609. [Google Scholar]
- 10.Devereux R, Winfrey M R, Winfrey J, Stahl D A. Depth profile of sulfate-reducing bacterial ribosomal RNA and mercury methylation in an estuarine sediment. FEMS Microbiol Ecol. 1996;700:1–9. [Google Scholar]
- 11.Environmental Protection Agency. EPA mercury study report to Congress. Publication EPA-452/R-97-009. Washington, D.C.: Office of Air Quality and Standards and Office of Research and Development, U.S. Environmental Protection Agency; 1997. [Google Scholar]
- 12.Faust S D, Osman M A. Chemistry of natural waters. Ann Arbor, Mich: Ann Arbor Science Publishers, Inc.; 1981. Mercury, arsenic, lead, cadmium, selenium, and chromium in aquatic environments; pp. 200–225. [Google Scholar]
- 13.Fossing H, Jørgensen B B. Measurement of bacterial sulfate reduction in sediments: evaluation of a single-step chromium reduction method. Biogeochemistry. 1989;8:205–222. [Google Scholar]
- 14.Frischer M E, Floriani P J, Nierzwicki-Bauer S A. Differential sensitivity of 16S rRNA targeted oligonucleotide probes used for fluorescence in situ hybridization is a result of ribosomal higher order structure. Can J Microbiol. 1996;42:1061–1071. doi: 10.1139/m96-136. [DOI] [PubMed] [Google Scholar]
- 15.Gardiner E E. Differences between ducks, pheasants, and chicken tissue Hg retention, depletion, and tolerance to increasing levels of dietary mercury. Can J Anim Sci. 1972;52:419–427. [Google Scholar]
- 16.Gilmour C C, Capone D G. Relationship between mercury methylation and the sulfur cycle in estuarine sediments. EOS Trans Am Geophys Union. 1987;68:1718–1725. [Google Scholar]
- 17.Gilmour C C, Henry E A. Mercury methylation in aquatic systems affected by acid deposition. Environ Pollut. 1991;71:131–169. doi: 10.1016/0269-7491(91)90031-q. [DOI] [PubMed] [Google Scholar]
- 18.Gilmour C C, Henry E A, Mitchell R. Sulfate stimulation of mercury methylation in freshwater sediments. Environ Sci Technol. 1992;26:2281–2287. [Google Scholar]
- 19.Gilmour C C, Riedel G S. Measurement of mercury methylation in sediments using high specific-activity 203Hg and ambient incubation. Water Air Soil Pollut. 1995;80:747–756. [Google Scholar]
- 20.Gilmour C C, Riedel G S, Ederington M C, Bell J T, Benoit J M, Gill G A, Stordal M C. Methylmercury concentrations and production rates across a trophic gradient in the northern Everglades. Biogeochemistry. 1998;40:327–345. [Google Scholar]
- 21.Habicht K S, Canfield D E. Sulfur isotope fractionation during bacterial sulfate reduction in organic-rich sediments. Geochim Cosmochim Acta. 1997;61:5351–5361. doi: 10.1016/s0016-7037(97)00311-6. [DOI] [PubMed] [Google Scholar]
- 22.Heijthuijsen J H F G, Hansen T A. Anaerobic degradation of betaine by marine Desulfobacterium strains. Arch Microbiol. 1989;152:393–396. [Google Scholar]
- 23.Heinz G. Effects of low dietary levels of methylmercury on mallard reproduction. Bull Environ Contam Toxicol. 1974;13:554–559. doi: 10.1007/BF01684947. [DOI] [PubMed] [Google Scholar]
- 24.Hosokawa J. Remediation work for Hg contaminated bay—experiences of Minamata Bay Project, Japan. Water Sci Technol. 1995;28:338–348. [Google Scholar]
- 25.Jørgensen B B. A comparison of methods for the quantification of bacterial sulfate reduction in coastal marine sediments. Geomicrobiol J. 1978;1:11–27. [Google Scholar]
- 26.Jørgensen B B, Bak F. Pathways and microbiology of thiosulfate transformations and sulfate reductions in marine sediments. Appl Environ Microbiol. 1991;39:847–856. doi: 10.1128/aem.57.3.847-856.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.King J K, Saunders F M, Lee R F, Jahnke R A. Coupling mercury methylation rates to sulfate reduction rates in marine sediments. Environ Toxicol Chem. 1999;18:1362–1369. [Google Scholar]
- 28.Korthals E T, Winfrey M R. Seasonal and spatial variations in mercury methylation and demethylation in an oligotrophic lake. Appl Environ Microbiol. 1987;53:2397–2404. doi: 10.1128/aem.53.10.2397-2404.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang L, Horvat M, Bloom N S. An improved speciation method for mercury by CV/CVAFS after aqueous phase ethylation and room temperature precollection. Talanta. 1994;41:371–379. doi: 10.1016/0039-9140(94)80141-x. [DOI] [PubMed] [Google Scholar]
- 30.Matida Y, Kumada H, Kimura S, Saiga Y, Nose T, Yokote M, Kawatsu H. Toxicity of mercury compounds to aquatic organisms and accumulation of the compounds by the organisms. Bull Freshwater Fish Res Lab (Tokyo) 1971;21:197–227. [Google Scholar]
- 31.Moran M A, Torsvik V L, Torsvick T, Hodson R E. Direct extraction and purification of rRNA for ecological studies. Appl Environ Microbiol. 1993;59:915–918. doi: 10.1128/aem.59.3.915-918.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pak K R, Bartha R. Mercury methylation and demethylation in anoxic lake sediments and by strictly anaerobic bacteria. Appl Environ Microbiol. 1998;64:1013–1017. doi: 10.1128/aem.64.3.1013-1017.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pak K R, Bartha R. Mercury methylation by interspecies hydrogen and acetate transfer between sulfidogens and methanogens. Appl Environ Microbiol. 1998;64:1987–1990. doi: 10.1128/aem.64.6.1987-1990.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parkes R J, Dowling N J E, White D C, Herbert R A, Gibson G R. Characterization of sulphate-reducing bacterial populations within marine and estuarine sediments with different rates of sulphate reduction. FEMS Microbiol Ecol. 1993;102:235–250. [Google Scholar]
- 35.Purdy K J, Nedwell D B, Embley T M, Akii S. Use of oligonucleotide probes to investigate the occurrence and selection of sulfate-reducing bacteria in response to nutrient addition to sediment slurry microcosms from Japanese estuary. FEMS Microbiol Ecol. 1997;24:221–234. [Google Scholar]
- 36.Rooney-Varga J N, Devereux R, Evans R S, Hines M E. Seasonal changes in the relative abundance of uncultivated sulfate-reducing bacteria in salt marsh sediment and in the rhizosphere of Spartina alterniflora. Appl Environ Microbiol. 1997;63:3895–3901. doi: 10.1128/aem.63.10.3895-3901.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rooney-Varga J N, Sharak-Genthner B R, Devereux R, Willis S G, Freidman S D, Hines M E. Phylogenetic and physiological diversity of sulphate-reducing bacteria isolated from salt marsh sediments. Syst Appl Microbiol. 1998;21:557–568. doi: 10.1016/s0723-2020(98)80068-4. [DOI] [PubMed] [Google Scholar]
- 38.Schauder R, Preall A, Jerren M, Fuchs G. Oxidative and reductive acetyl CoA/carbon monoxide dehydrogenase pathway in Desulfobacterium autotrophicum. Demonstration of the enzymes of the pathway and comparison of CO dehydrogenase. Arch Microbiol. 1989;151:84–89. [Google Scholar]
- 39.Tabatabi M A. A rapid method for determination of sulfate in water samples. Environ Lett. 1974;7:237–243. [Google Scholar]
- 40.Trimmer M, Purdy K J, Nedwell D B. Process measurement and phylogenetic analysis of the sulfate reducing bacterial communities of two contrasting benthic sites in the upper estuary of the Great Ouse, Norfolk, UK. FEMS Microbiol Ecol. 1997;24:333–342. [Google Scholar]
- 41.Widdel F, Bak F. Gram negative mesophilic sulfate reducing bacteria. In: Balows A, Truper H G, Dworkin M, Harder W, Schleifer K H, editors. The prokaryotes. A handbook on the biology of bacteria: ecophysiology, isolation, identification, and applications. 2nd ed. Vol. 2. New York, N.Y: Springer-Verlag; 1992. pp. 3352–3378. [Google Scholar]
- 42.Williams S C, Hong Y, Danavall D C A, Howard-Jones M H, Gibson D, Frischer M E, Verity P G. Distinguishing between living and nonliving bacteria: evaluation of vital stain propidium iodide and the combined use with molecular probes in aquatic samples. J Microbiol Methods. 1998;32:225–236. [Google Scholar]
- 43.Winger P V, Lasier P J, Geitner H. Toxicity of sediments and pore water from Brunswick estuary, Georgia. Arch Environ Contam Toxicol. 1993;25:371–376. [Google Scholar]