Abstract
We isolated a gram-positive, halotolerant psychrophile from a hypersaline pond located on the McMurdo Ice Shelf in Antarctica. A phylogenetic analysis of the 16S rRNA gene sequence of this organism showed that it is a member of the genus Planococcus. This assignment is consistent with the morphology and physiological characteristics of the organism. A gene encoding a β-galactosidase in this isolate was cloned in an Escherichia coli host. Sequence analysis of this gene placed it in glycosidase family 42 most closely related to an enzyme from Bacillus circulans. Even though an increasing number of family 42 glycosidase sequences are appearing in databases, little information about the biochemical features of these enzymes is available. Therefore, we purified and characterized this enzyme. The purified enzyme did not appear to have any metal requirement, had an optimum pH of 6.5 and an optimum temperature of activity at 42°C, and was irreversibly inactivated within 10 min when it was incubated at 55°C. The enzyme had an apparent Km of 4.9 μmol of o-nitrophenyl-β-d-galactopyranoside, and the Vmax was 467 μmol of o-nitrophenol produced/min/mg of protein at 39°C. Of special interest was the finding that the enzyme remained active at high salt concentrations, which makes it a possible reporter enzyme for halotolerant and halophilic organisms.
Glycosidases (EC 3.2.1, EC 3.2.2, and EC 3.2.3) hydrolyze the bond(s) between two or more carbohydrates or the bond between a carbohydrate moiety and a noncarbohydrate moiety. Because there are so many possible combinations of oligosaccharide isomers, there are a variety of enzymes capable of hydrolyzing these compounds. Traditionally, glycosidases were grouped together based on the ability to hydrolyze similar substrates; for example, enzymes that hydrolyze lactose or its related chromogens o-nitrophenyl-β-d-galactopyranoside (ONPG) and 5-bromo-4-chloro-3-indoyl-β-d-galactoside (X-Gal) were classified as β-galactosidases. One of the most-studied and useful β-galactosidases is the lacZ enzyme of Escherichia coli. However, many other important galactosidases are now being discovered in phylogenetically diverse organisms. The advent of computer databases containing more sequence information now makes it possible to compare and group these enzymes in order to obtain additional information about their evolutionary relationships.
A useful classification for glycosidases was developed by Henrissat and coworkers (4, 6, 9, 10), who organized these enzymes into different families based on amino acid sequence similarities and hydrophobic cluster analysis data. This classification identifies possible common structural domains, thereby defining evolutionary connections and suggesting hydrolytic mechanisms for the glycosidases. The classification of Henrissat et al. divides enzymes that exhibit β-galactosidase activity and have the same Enzyme Commission designation (EC 3.2.1.23) into three distinct families. Sequence analyses of these three families have shown that the individual groups are extremely robust. In addition, phylogenetic analyses have shown that each family appears to be derived from a separate gene lineage because the three families are so distantly related to each other (our unpublished results).
One of the three β-galactosidase families in the classification of Henrissat et al. is family 42, which was first defined on the basis of two genes from thermophilic bacteria (10). During the last few years, other family 42 gene sequences have been added to the database (8, 11, 12, 19, 22, 25, 27). Many of these sequences have resulted from genome-sequencing projects, and there is little information about the enzymes or whether the proposed gene sequences are even transcribed in the organisms. In some cases, there are even two phylogenetically distinct family 42 gene sequences present in the same organism. It has not been determined whether the isozymes diverged following gene duplication events or if they resulted from horizontal gene transfer events. The results of powerful sequence comparisons illustrate the need for further work to define the properties of the different family 42 glycosidases potentially encoded by these sequences.
Information about these enzymes could be especially important because there are now several applications for glycosidases, depending on their substrate specificities and biochemical properties. The suggested industrial uses for β-galactosidases include removal of lactose from milk and whey (14), synthesis of oligosaccharides which modify the intestinal microflora (30), and removal of plant saccharides from fruit beverages (2, 29). The transglycosylation activity of a β-galactosidase from Bacillus circulans has been proposed as an enzymatic route for synthesis of para-nitrophenyl galactosyl-glucoside chromogens (18). Other transglycosylation reactions with different donors could be used to produce a variety of chiral sugar derivatives that could be novel pharmaceuticals.
In addition, new reporter enzymes could be useful in situations in which the E. coli lacZ β-galactosidase cannot function. Schrogel and Allmansberger (21) described the use of a β-galactosidase from Bacillus stearothermophilus as a reporter in Bacillus subtilis. This enzyme had the advantage of being stable after heat shock and at incubation temperatures that inactivated the background β-galactosidase activity in B. subtilis. In a search for a reporter enzyme that could be used in halophiles, Holmes et al. (12) characterized a β-galactosidase from Haloferax alicantei that exhibited activity in the presence of 4 M NaCl, a salt concentration at which the E. coli enzyme is inactive. However, the possible disadvantages related to use of this enzyme included its limited activity in low-salt buffer, a requirement for a stabilizing agent (sorbitol), and the fact that it was irreversibly inactivated when it was purified with an NaCl concentration less than 1 M (in the absence of sorbitol). Furthermore, the Haloferax enzyme exhibited greater activity in the presence of NaCl than in the presence of KCl, which is the intracellular salt in many halophilic microorganisms. These properties could interfere with using the Haloferax family 42 β-galactosidase on a shuttle vector when activity in both E. coli and a halophile would be useful.
As part of our comparison of cold-active enzymes from psychrophilic microorganisms, we have cloned several glycosidase genes. In order to obtain pertinent biochemical information that could enhance our understanding of the functions of some of the glycosidases found in the classification of Henrissat et al. we have not only sequenced the new genes, but we have also identified the organisms and purified and characterized the enzymes. Here we describe characterization of an organism (SOS Orange) that was isolated from a hypersaline pond on the McMurdo Ice Shelf in Antarctica and a gene which encodes a β-galactosidase that is active at low salt concentrations and also maintains between 20 and 40% of its activity in the presence of 4 M NaCl or 4 M KCl. These features plus the smaller size and overall stability of the enzyme make the gene and enzyme ideal candidates for a reporter gene and enzyme that can be used for both nonhalophilic and halophilic organisms.
MATERIALS AND METHODS
Isolation, characterization, and identification of isolate SOS Orange.
A cyanobacterial mat sample was collected in January 1993 from a hypersaline pond (Son of Salt Pond) near Bratina Island on the McMurdo Ice Shelf in Antarctica. The sample was frozen at −80°C until an approximately 2-g sample was inoculated into 5 ml of Instant Ocean broth (15), which was incubated at 10°C until the culture became turbid. Organisms were isolated on Instant Ocean agar by using streak plating techniques, and isolated colonies were subcultured at least three times to ensure purity. Isolates were streaked onto Instant Ocean agar containing X-Gal (100 mg/liter; Sigma Chemical Co., St. Louis, Mo.) in order to determine which isolates exhibited β-galactosidase activity. An orange colony that was designated SOS Orange and hydrolyzed X-Gal was chosen for further study.
We examined the ability of isolate SOS Orange to grow at different temperatures and in the presence of different NaCl concentrations by inoculating cells from a turbid culture grown in Trypticase soy broth (TSB) (Difco Laboratories, Detroit, Mich.) into 5 ml of TSB containing 0, 5, 10, 15, 20, or 25% NaCl. The cultures were incubated aerobically at −2, 10, 20, 26, and 37°C.
16S rRNA gene amplification and β-galactosidase gene cloning.
Genomic DNA was obtained from isolate SOS Orange by using a modification of standard methods (7). The 16S rRNA gene was amplified from chromosomal DNA by performing a PCR with Ready-To-Go beads (Amersham Pharmacia, Piscataway, N.J.) and universal primers 8 F and 1492 R (20, 28). The product was sequenced at the Penn State Nucleic Acid Facility with an ABI model 370 sequencer.
A gene encoding β-galactosidase activity was obtained by partially digesting genomic DNA with PstI, ligating the DNA into vector pΔα18 (26), a derivative of pUC18 that lacks the E. coli lacZ alpha fragment, and transforming competent E. coli JM109 cells. Transformants were selected on the basis of resistance to ampicillin (100 mg/liter) and were screened to obtain transformants capable of hydrolyzing X-Gal. Plasmid DNA from one transformant was purified by using a Genomed maxi prep kit (PGC, Gaithersburg, Md.). The gene encoding the β-galactosidase activity was sequenced at the Penn State Nucleic Acid Facility with an ABI model 370 sequencer.
Phylogenetic analyses of the gene sequences.
All of the enzymes in the GenBank database which exhibited β-galactosidase activity were analyzed by using the MegAlign Program to separate them into individual families, and the gene sequence for the β-galactosidase obtained from isolate SOS Orange was added in order to identify its natural group. The program parameters were adjusted to obtain the shortest tree, and the final multiple-alignment parameters were a gap penalty of 30 and a gap length of 30, while the pairwise alignment parameters were a Ktuple of 1 and a gap penalty of 3. The enzymes most closely related to the β-galactosidase from isolate SOS Orange form a naturally occurring robust clade of family 42 enzymes in a tree containing all of the enzymes that exhibit β-galactosidase activity (data not shown). The phylogenetic relationships between the family 42 enzymes and the β-galactosidase of isolate SOS Orange were determined by using the PAUP (24) and PHYLIP (5) programs.
The 16S rRNA gene sequences were aligned with sequences obtained from the Ribosomal Database Project (16) and from a Blast search of the National Center for Biotechnology Information (NCBI) database. Our alignment was based on 1,454 nucleotide positions. The bootstrap values for both maximum-parsimony trees were based on 1,000 replicates. The phylogenetic relationships were determined by using the PAUP and PHYLIP programs; trees generated with the PAUP program were congruent with trees generated with the PHYLIP program.
Enzyme purification and characterization.
The E. coli transformant containing the β-galactosidase gene from isolate SOS Orange was grown at 37°C in 100 ml of Luria-Bertani broth supplemented with ampicillin (100 mg/liter; Sigma) until the turbidity at 600 nm was 0.6. Cells were cooled at 20°C for 20 min, induced with 1 mM isopropyl-β-d-thiogalactoside (IPTG) (Fisher, Pittsburgh, Pa.), and grown at 20°C for 16 h. Cells were harvested by centrifugation (10,000 × g, 10 min) at 4°C and resuspended in 15 ml of Z buffer (17). The cells were disrupted with a French pressure cell (two treatments at 20,000 lb/in2) and centrifuged again. A saturated solution of ammonium sulfate was added stepwise to the resulting lysate (on ice, with stirring) to obtain final ammonium sulfate concentration increases of 20%. The β-galactosidase precipitated at final ammonium sulfate concentrations between 60 and 80%. The precipitate was resuspended in 5 ml Z buffer and dialyzed twice against 1 liter of Z buffer. The dialyzed enzyme was added to 5 ml of Z buffer containing 2 M (NH4)2SO4 and applied to a Phenyl Sepharose 6 Fast Flow column (Amersham Pharmacia). The protein was eluted with a 1 to 0 M (NH4)2SO4 gradient in Z buffer. Fractions (9 ml) were collected and assayed for activity. The most active fraction had a specific activity of 116 μmol of o-nitrophenol produced/min/mg of protein and was determined to be more than 90% pure by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). This preparation was used to determine appropriate assay conditions as described below.
The optimal temperature for enzyme activity was determined by incubating the enzyme (in 1.0 ml [total volume] of 100 mM morpholinepropanesulfonic acid [MOPS] buffer, pH 6.5) with 200 μl of ONPG (4 mg/ml in MOPS buffer; U.S. Biochemicals, Cleveland, Ohio) for 5 min at temperatures ranging from 0°C to 75°C; the reactions were stopped by adding 500 μl of 1 M Na2CO3. Hydrolysis of the o-nitrophenyl group was detected at 420 nm. The thermostability of the enzyme activity was determined by incubating the enzyme at 20, 35, 42, 48, and 55°C, removing aliquots at 10-min intervals from zero time to 120 min, and assaying the preparations at 39°C.
The optimum pH was determined by assaying the enzyme activity at pH values ranging from 3.0 to 10.0 in 0.5-pH unit increments. The buffers used each had a final ionic strength of 0.1 M. The buffers used were citric acid-sodium citrate for pH 3.0 to 6.0, Na2HPO4-NaH2PO4 for pH 6.0 to 8.0, and KCl-H3BO3 for pH 8.0 to 10.0. Activity was measured as described above at 39°C.
The possible requirement for metal ions was examined by first incubating undiluted enzyme with either 20 or 100 mM EDTA in Z buffer for 3 h at 0°C to remove easily bound divalent cations. The enzyme was then applied to a Sephadex G-25 column (Sigma) and eluted with 100 mM MOPS (pH 6.5). MOPS buffer was substituted for Z buffer, which contained phosphate and sodium ions that interfered with the cation analysis. Because the enzyme was equally active in both buffers, the MOPS buffer was used during the enzyme characterization study. Equivalent numbers of units of EDTA-treated and non-EDTA-treated enzyme (used as a control) were added to 100 mM MOPS (pH 6.5). The EDTA-treated enzyme was assayed at 39°C in the presence of different concentrations of CuCl2, ZnCl2, MgCl2, CaCl2, MnCl2, NiCl2, or CoCl2 and also in the presence of different concentrations of NaCl or KCl.
Enzyme purification for kinetic studies.
In order to obtain purer enzyme for substrate specificity and kinetic studies, a culture was grown, lysed, and centrifuged as described above. The resulting lysate was dialyzed against 0.5× Z buffer overnight at 5°C. The dialyzed lysate (14.25 ml) was applied to a DEAE-Sephacel column (Sigma) and eluted with a 0 to 1 M NaCl gradient. Active fractions were combined and assayed. The purified enzyme had a specific activity of 160 μmol of o-nitrophenol produced/min/mg of protein and was determined by SDS-PAGE to have been purified to homogeneity; a single band at approximately 75 kDa was produced (data not shown). The purified enzyme was subjected to electrophoresis on nondenaturing polyacrylamide gels, and activity was detected by incubating each gel in 50 ml of Z buffer containing 1 ml of X-Gal (20 mg/ml). Only one activity band was produced; based on migration of the markers, the molecular mass of this band was estimated to be approximately 150 kDa.
Biochemical characterization.
The highly purified enzyme was used to examine the substrate specificity of the enzyme and to determine its kinetic parameters. Substrate specificity assays were performed at 39°C by incubating the enzyme (in 1.0 ml [total volume] of 100 mM MOPS, pH 6.5) with 200 μl of nitrophenyl substrate (5 mM in 100 mM MOPS, pH 6.5). The substrates used were ONPG, p-nitrophenyl-β-d-galactopyranoside, o-nitrophenyl-β-d-fucopyranoside, p-nitrophenyl-β-d-fucopyranoside, p-nitrophenyl-β-lactose, p-nitrophenyl-β-cellobiose, p-nitrophenyl-α-galactopyranoside, and p-nitrophenyl-β- xylopyranoside (Sigma). In competition assays we incubated purified enzyme for 10 min at 25°C with the appropriate amounts of ONPG and a cellobiose, sucrose, or lactose solution (in 100 mM MOPS, pH 6.5) to give a final volume of 1,200 μl. Kinetic assays were performed at 1.9, 10, 20, 30, and 39°C with different concentrations of ONPG. Kinetic values were calculated by using the analysis program Enzyme Kinetics (23).
Light scattering.
A 5-mg/ml solution of the enzyme was analyzed by using a model dp-801 dynamic light scattering instrument (Protein Solutions Inc., Charlottesville, Va.). A bimodal regression analysis resulted in an estimated molecular weight of 155,000.
N-terminal sequence.
An Applied Biosystems model 477A protein sequencer was used to determine the first eight amino acids (MINDKLPK) of the highly purified β-galactosidase obtained from isolate SOS Orange. N-terminal amino acid sequencing was performed at the Hershey Medical Center of Pennsylvania State University.
Nucleotide sequence accession numbers.
The accession number for the 16S rRNA gene sequence is AF242541, and the accession number for the sequence of the gene encoding β-galactosidase activity is AF242542.
RESULTS
Characterization of isolate SOS Orange.
Several different colonies were purified from the Antarctic cyanobacterial mat sample obtained from a hypersaline pond on the McMurdo Ice Shelf in Antarctica. One isolate that formed bright orange colonies, grew at 0°C but not at 31°C, and could hydrolyze the chromogen X-gal was designated SOS Orange and used for further study. The cells were gram-positive cocci and were nonmotile. Because the isolate was obtained near a hypersaline pond, we studied its growth at different NaCl concentrations. Isolate SOS Orange was inoculated into a series of tubes containing TSB supplemented with different NaCl concentrations (0, 5, 10, 15, 20, and 25%) and incubated at different temperatures (−2, 10, 20, 26, and 37°C). Isolate SOS Orange was able to grow in the presence of all NaCl concentrations at −2, 10, and 20°C and in the presence of 0, 5, and 10% NaCl at 26°C. No growth occurred at 37°C in the presence of any NaCl concentration or at 26°C in the presence of 15, 20, or 25% NaCl. These results show that the isolate is halotolerant rather than halophilic; it does not require NaCl, but it can grow in the presence of NaCl concentrations as high as 25% (4.3 M) at 25°C and lower temperatures. Although the isolate grows more rapidly at 26°C than at lower temperatures, it appears to be more sensitive to NaCl at the higher temperatures.
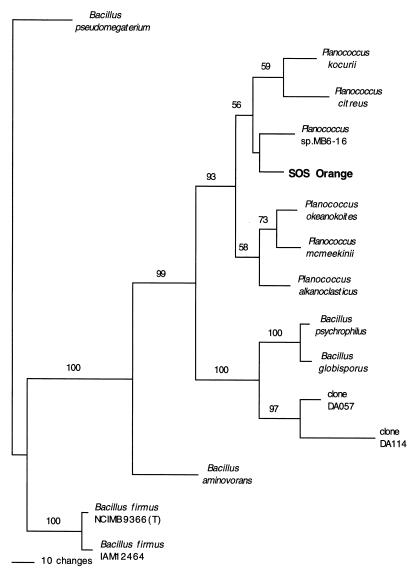
To determine the phylogenetic position of the isolate, the genomic DNA was purified, and the 16S rRNA gene was PCR amplified. The fragment obtained was sequenced, and the results were compared with sequences in the Ribosomal Database Project and NCBI databases. The results suggested that isolate SOS Orange is a member of the genus Planococcus. The phylogenetic relationships of SOS Orange and closely related organisms (Fig. 1) were analyzed by using the PAUP (maximum-parsimony) and PHYLIP (maximum-likelihood) programs (data not shown), and the trees were found to be congruent. The 16S rRNA sequence of isolate SOS Orange differed by about 2% from the 16S rRNA sequences of the two most closely related previously characterized species, Planococcus kocurii and Planococcus citreus (data not shown). Identification of our isolate as a Planococcus isolate is consistent with the physiological properties and habitat of the organism since other Planococcus strains have been isolated from marine environments and Antarctic sea ice brine (1, 13). Based on the phylogenetic and physiological characterization results, we designated our isolate Planococcus sp. isolate SOS Orange.
FIG. 1.
Phylogenetic tree based on maximum-parsimony analysis of the 16S rRNA gene of isolate SOS Orange and closely related species. Gene sequences from the following organisms were used (the numbers in parentheses are GenBank accession numbers): Planococcus sp. strain MB6-16 (U85898), Planococcus okeanokoites (D55729), Planococcus alkanoclasticus (AF029364), Planococcus kocurii (X62173), Planococcus citreus (X62172), Planococcus mcmeekinii (AF041791), Bacillus psychrophilus (D16277), unidentified bacterium 16S rRNA gene clone DA057 (AJ000988), unidentified bacterium 16S rRNA gene clone DA114 (AJ000980), Bacillus globisporus (X68415), Bacillus aminovorans (X62178), Bacillus firmus IAM 12464 (D16268), Bacillus firmus NCIMB 9366 (X60616), and Bacillus pseudomegaterium (X77791).
Characterization of the β-galactosidase gene.
A chromosomal library was prepared from isolate SOS Orange and transformed into E. coli JM109, a transformant capable of hydrolyzing X-Gal was selected, and the fragment insert was characterized. Sequence analysis of the fragment encoding the β-galactosidase activity revealed that a 2,034-nucleotide open reading frame started within 43 nucleotides of the vector. An upstream promoter sequence appeared to be present on this fragment because some β-galactosidase activity was present without induction of the lac promoter on the plasmid and with the gene placed in the orientation opposite that of the lac promoter. With the gene oriented to the lac promoter, however, induction with IPTG increased the β-galactosidase activity more than 100-fold, suggesting that transcription began at the lac promoter and extended into the start of the gene.
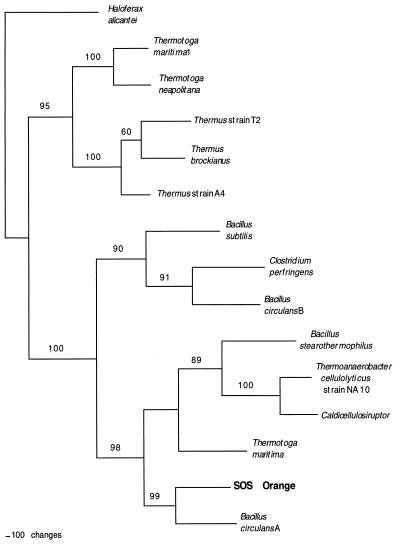
The gene sequence encodes a 677-amino-acid protein that has a calculated Mr of 77,492 and a pI of 5.079. The deduced amino acid sequence was compared with other amino acid sequences in the NCBI database. A phylogenetic analysis of closely related sequences showed that the gene from isolate SOS Orange was most closely related to a β-galactosidase gene from B. circulans (Fig. 2). Other related genes of interest were genes from B. stearothermophilus and the halophile H. alicantei. Alignment with other protein sequences revealed that two glutamic acid residues thought to be involved in the catalytic reaction of the B. circulans β-galactosidase are conserved in the Planococcus isolate (data not shown). The B. circulans gene is a member of the family 42 glycosyl hydrolases and is designated bgaA. Thus, it is likely that the new gene from our isolate is a family 42 glycosyl hydrolase, and this gene is designated the bgaA gene from Planococcus sp. isolate SOS Orange.
FIG. 2.
Phylogenetic tree based on maximum-parsimony analysis of the β-galactosidase gene of isolate SOS Orange and closely related species. Gene sequences from the following organisms were used (the numbers in parentheses are GenBank accession numbers): Bacillus circulans A (AAA22258), Caldicellulosiruptor sp. strain 14B (CAA10365), Bacillus circulans B (AAA22260), Clostridium perfringens (BAA08485), Bacillus stearothermophilus (P19668), Haloferax alicantei (AAB40123), Thermotoga neapolitana (AAC24217), Thermus sp. strain A4 (BAA28362), Thermus sp. strain T2 (CAB07810), Thermus brockianus (AAD33667), Thermotoga maritima 1 (AAD36270), Thermotoga maritima (AAD35398), and Bacillus subtilis (CAB12527).
Enzyme purification and N-terminal sequence determination.
The β-galactosidase was expressed from the cloned gene in E. coli JM109 and was purified as described in Materials and Methods. The enzyme was expressed at high levels, and, in contrast to other glycosidases which we have worked with, it remained in the soluble fraction rather than forming inclusion bodies. The subunit molecular mass determined by SDS-PAGE was about 75 kDa, which was consistent with the predicted Mr of 77,492 based on the deduced amino acid sequence. Light-scattering experiments showed that the active enzyme had an Mr of 155,000, which is consistent with the hypothesis that it is a dimer. The first eight N-terminal amino acids were determined to be MINDKLPK, which matched the amino acid sequence deduced from the cloned gene sequence; this showed that the enzyme was not produced as a fusion product from the plasmid.
Effects of temperature on activity.
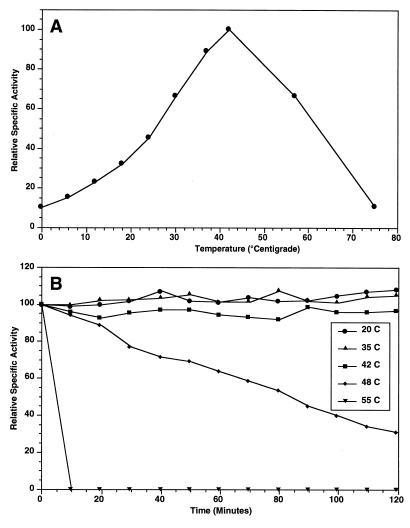
The thermodependency of the enzyme activity was determined by incubating reaction mixtures at different temperatures and determining the activities (Fig. 3A). The optimal temperature for purified enzyme activity is near 42°C. The enzyme is thermostable at temperatures at or below the optimal temperature for activity, but it is rapidly denatured at temperatures above 42°C (Fig. 3B). The enzyme was stable during storage at 5°C and lost no activity during storage for 4 months. Because the enzyme was heat labile at temperatures above 42°C, subsequent assays were performed at 39°C, which is just below the optimum temperature, in order to strike a balance between the temperature at which maximal activity occurs and the inactivation temperature.
FIG. 3.
(A) Plot of relative specific activity of purified SOS Orange β-galactosidase versus temperature. The specific activity that corresponded to 100% was 150 μmol of o-nitrophenol produced/min/mg of protein. (B) Plot of thermostability of purified β-galactosidase from isolate SOS Orange versus time of incubation at different temperatures. The specific activity that corresponded to 100% was 72.5 μmol of o-nitrophenol produced/min/mg of protein.
Effects of pH and salts on activity.
We compared the activities of the enzyme at pH 3 to 10, and the greatest activity was observed at pH 6.5 (data not shown). To examine the possible metal ion requirements of the enzyme, a preparation was first treated with EDTA. No activity was lost during treatment with 20 or 100 mM EDTA in Z buffer for 3 h at 0°C, nor was activity greatly stimulated by the addition of cations (data not shown). Enzyme activity was inhibited by 1 mM zinc and 1 mM copper; the levels of activity decreased to 10% of the untreated control activity. Nickel, cobalt, and manganese at concentrations of 10 mM decreased the enzyme activity to either 40% (for nickel and cobalt) or 60% (for manganese) of the activity in untreated controls. There was no change in enzyme activity in the presence of calcium and magnesium at concentrations up to 50 mM (data not shown).
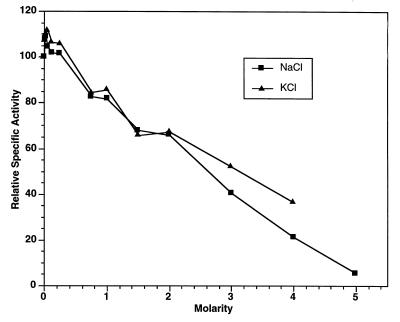
Because isolate SOS Orange was obtained from a hypersaline pond and is halotolerant, the effects of NaCl and KCl on the β-galactosidase activity were examined. Although the EDTA-treated enzyme exhibited a slight increase in relative specific activity when it was assayed in the presence of 50 mM NaCl or 50 mM KCl, it did not exhibit enhanced activity at concentrations greater than 250 mM (Fig. 4). The enzyme was still active when it was assayed in the presence of 4 M NaCl or 4 M KCl, and it was more active in the presence of KCl than in the presence of NaCl.
FIG. 4.
Relative specific activities of purified SOS Orange β-galactosidase determined in the presence of various concentrations of NaCl and KCl. The enzyme activity was assayed at 39°C. The specific activity that corresponded to 100% was 71 μmol of o-nitrophenol produced/min/mg of protein.
Substrate specificity.
Enzyme activity was assayed by using an number of chromogenic substrates (Table 1). The greatest activity was observed with ONPG and p-nitrophenyl-β-d-galactopyranoside, and low levels of activity were observed with o-nitrophenyl-β-d-fucopyranoside and p-nitrophenyl-β-d-fucopyranoside. Various disaccharides were used in competition studies performed with ONPG (Table 2). A slight reduction in ONPG hydrolysis was observed in the presence of lactose; however, a 10-fold-higher concentration of lactose reduced ONPG hydrolysis by only 29%. Adding sucrose and cellobiose had little effect.
TABLE 1.
Activities of the β-galactosidase purified from the E. coli transformant with various nitrophenol-derived chromagenic substrates
| Substratea | % Relative activityb |
|---|---|
| ONPG | 100 |
| PNPG | 92.96 |
| ONPF | 6.11 |
| PNPF | 3.77 |
| PNPL | <0.001 |
| PNPC | <0.001 |
| PNPαG | <0.001 |
| PNPX | <0.001 |
The substrate concentration used in each test was 5.0 mM. Abbreviations: ONPG, o-nitrophenyl-β-d-galactopyranoside; PNPG, p-nitrophenyl-β-d-galactopyranoside; ONPF, o-nitrophenyl-β-d-fucopyranoside; PNPF, p-nitrophenyl-β-d-fucopyranoside; PNPL, p-nitrophenyl-β-lactose; PNPC, p-nitrophenyl-β-cellobiose; PNPαG, p-nitrophenyl-α-galactopyranoside; PNPX, p-nitrophenyl-β-xylopyranoside.
The values are values relative to the activity observed with ONPG, which was 160 μmol of o-nitrophenol produced/min/mg of protein.
TABLE 2.
Substrate competition between disaccharides and ONPG
| ONPG concn (mM) | Competitor | Sp Act | % Relative activity |
|---|---|---|---|
| 2.21 | None | 61.6 | 100 |
| Lactose (2.21 mM) | 59.7 | 96.9 | |
| Sucrose (2.21 mM) | 62.2 | 101 | |
| Cellobiose (2.21 mM) | 61.5 | 99.4 | |
| None | 56.3 | 100 | |
| Lactose (6.63 mM) | 46.2 | 82 | |
| Sucrose (6.63 mM) | 54.8 | 97.3 | |
| Cellobiose (6.63 mM) | 53.1 | 94.3 | |
| 1.105 | None | 36.9 | 100 |
| Lactose (10.0 mM) | 26.3 | 71.3 |
Enzyme kinetics.
The Vmax and apparent Km values for the highly purified enzyme when ONPG was the substrate were determined at five different temperatures (Table 3). As expected from the thermodependency of activity, Vmax was highest at 39°C (467 μmol of o-nitrophenol produced/min/mg of protein) and lowest at 1.9°C (63 μmol of o-nitrophenol produced/min/mg of protein). An energy of activation of 13,516 cal/mol was calculated from an Arrhenius plot by using the linear data in Table 3.
TABLE 3.
Kinetic parameters for purified β-galactosidase determined at different temperaturesa
| Temp (°C) | Vmax (μmol of o-nitrophenol produced/min/mg of protein) | kcat (s−1) | Km (mM ONPG) | Catalytic efficiency (M−1 s−1) |
|---|---|---|---|---|
| 1.9 | 63 | 81.8 | 7.4 | 11,000 |
| 10 | 80 | 104 | 4.5 | 23,000 |
| 20 | 223 | 288 | 5.4 | 54,000 |
| 30 | 392 | 507 | 5.0 | 101,000 |
| 39 | 467 | 603 | 4.9 | 123,000 |
To determine the values, we assumed that there is one active site per subunit in a dimeric enzyme.
DISCUSSION
Antarctic isolate SOS Orange produces an intense orange pigment, is a gram-positive coccus, and grows at −2, 10, 20, and 26°C but not at 37°C. This organism also grows well in media containing up to 10% (1.7 M) NaCl at all of its growth temperatures, and limited growth occurs in the presence of NaCl concentrations up to 25% (4.3 M) at temperatures between −2 and 20°C. Phylogenetic analysis of the PCR-amplified 16S rRNA gene sequence of isolate SOS Orange (Fig. 1) placed this organism in the genus Planococcus, which is consistent with its morphological and growth characteristics. Organisms identified as members of the genus Planococcus have been found in Antarctic sea ice (1), although the ice was collected on the other side of the continent from the Ross Sea. A gram-positive organism isolated from Antarctic sea ice brine (13) was determined to be a new Planococcus species (Planococcus mcmeekinii), and the ice core from which it was isolated was obtained near Dunlop Island in the Ross Sea, which is less than 60 miles from the SOS Orange collection site on the McMurdo Ice Shelf. It is possible that members of the genus Planococcus are more common in Antarctic sea ice and terrestrial lakes and ponds than previously realized. Although the 16S rRNA sequence of isolate SOS Orange is only about 2% different from the sequences of two related species, P. kocurii and P. citreus, we believe that this isolate should not be identified as a member of a novel species of the genus Planococcus until further biochemical, physiological, and ecological characterizations of members of this genus are completed.
Phylogenetic analysis of the SOS Orange β-galactosidase gene sequence (Fig. 2) showed that this sequence is most closely related to the sequence for an enzyme from B. circulans and is also related to the sequences for enzymes from other Bacillus-Clostridium group organisms. The phylogenies based on the 16S rRNA and enzyme gene sequences of isolate SOS Orange are congruent, as these sequences are most closely related to gene sequences of other gram-positive organisms. Therefore, it is not likely that the SOS Orange β-galactosidase gene sequence arose as a gene transfer event. In contrast, the Thermatoga maritima and B. circulans genomes contain two family 42 β-galactosidase sequences. The sequences found in B. circulans are closely related, whereas the sequences found in T. maritima are members of distinct clades; one is related to sequences of gram-positive organisms, and the other is related to sequences of gram-negative thermophiles. The B. circulans sequences probably resulted from a gene duplication event rather than horizontal transfer, as both genes cluster in the gram-positive clade. However, since the two T. maritima isozymes are phylogenetically distinct, it is likely that the sequence in the gram-positive clade resulted from gene transfer rather than from duplication followed by mutation.
There is little biochemical data on the family 42 glycosidases, and it is difficult to assign a physiological function to this family other than that the enzymes hydrolyze β1-4 glycosidic linkages. The β-galactosidase from isolate SOS Orange exhibits a clear preference for substrates containing a galactose moiety (Table 1). The enzyme activity reflects the ability of the organism to grow at low temperatures and to tolerate high-salt conditions. It has an optimal temperature for activity of 42°C (Fig. 3A), retains 10% of this activity at 0°C, and is thermolabile at temperatures above the optimal temperature (Fig. 3B). The enzyme exhibits greater activity under slightly acidic assay conditions, and the optimum pH is 6.5. Two of the interesting features of the enzyme are that it does not appear to have a metal ion requirement and it has a high tolerance for salt, retaining 50% of its activity in the presence of 3 M KCl or 2.5 M NaCl (Fig. 4). Although the enzymes to which it is most closely related are classified as β-galactosidases, their natural substrates and physiological functions have not been determined. It is possible that the enzyme from isolate SOS Orange is used to degrade cyanobacterial cell wall or capsular polysaccharides found in the McMurdo Ice Shelf ponds.
The combination of low-temperature activity and salt tolerance may make the β-galactosidase of isolate SOS Orange useful in a variety of applications. Since other enzymes related to the SOS Orange β-galactosidase are thermophilic, comparisons may lead to insights into the features responsible for the thermal differences of these enzymes. In addition, other workers in our laboratory recently isolated a different glycosidase gene (3) which is closely related to an enzyme from B. stearothermophilus. Because the salt sensitivity of this enzyme contrasts with the salt tolerance of the enzyme from SOS Orange, future comparisons of the two could be useful. We are in the process of determining the X-ray crystal structure of the SOS Orange enzyme in order to initiate these studies.
The β-galactosidase of isolate SOS Orange might also be used in the food industry to digest plant polysaccharides in high-salt processes, or it could be an ideal candidate for a new reporter system on an E. coli-halophile shuttle vector. This enzyme can be expressed in E. coli at high levels, and it exhibits substantial activity in the presence of 3 M KCl, suggesting that it would retain activity in a halophilic host. Because X-Gal is an extremely sensitive indicator, blue colonies would be easily detected even if the SOS Orange β-galactosidase activity were partially inhibited in a halophilic host. Furthermore, unlike the enzyme from H. alicantei, the enzyme from isolate SOS Orange is more active in the presence of high concentrations of KCl than in the presence of high concentrations of NaCl. Because many halophilic Archaea maintain osmotic balance by concentrating KCl in the cytoplasm, the isolate SOS Orange enzyme could be a better reporter gene in halophilic hosts than a related enzyme from an archaeal halophile is.
ACKNOWLEDGMENTS
We thank A. Phillips and M. Tien for helpful discussions and our laboratory coworkers, especially K. R. Gutshall, for suggestions. The light scattering experiment was performed by N. Panasik.
This work was supported by Department of Energy grant DE-FG02-93ER20117. P.S. was also supported by an Alfred P. Sloan Foundation Fellowship in Molecular Evolution from the National Science Foundation and by partial funding from Penn State Astrobiology Center NASA-Ames cooperative agreement NCC2-1057 and grant NSF/IGERT DGE-9972759 from the Biogeochemical Research Initiative for Education (BRIE).
REFERENCES
- 1.Bowman J P, McCammon S A, Brown M V, Nichols D S, McMeekin T A. Diversity and association of psychrophilic bacteria in Antarctic sea ice. Appl Environ Microbiol. 1997;63:3068–3078. doi: 10.1128/aem.63.8.3068-3078.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brenchley J. Psychrophilic microorganisms and their cold-active enzymes. J Ind Microbiol. 1996;17:432–437. [Google Scholar]
- 3.Coombs J M, Brenchley J E. Biochemical and phylogenetic analyses of a cold-active β-galactosidase from the lactic acid bacterium Carnobacterium piscicola BA. Appl Environ Microbiol. 1999;65:5443–5450. doi: 10.1128/aem.65.12.5443-5450.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Durand P, Lehn P, Callebaut I, Fabrega S, Henrissat B, Mornon J-P. Active-site motifs of lysosomal acid hydrolases: invariant features of clan GH-A glycosyl hydrolases deduced from hydrophobic cluster analysis. Glycobiology. 1997;7:277–284. doi: 10.1093/glycob/7.2.277. [DOI] [PubMed] [Google Scholar]
- 5.Felsenstein J. PHYLIP (Phylogeny Inference Package), version 3.5c. Seattle: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 6.Gilkes N R, Henrissat B, Kilburn D G, Miller R C, Jr, Warren R A J. Domains in microbial β-1,4-glycanases: sequence conservation, function, and enzyme families. Microbiol Rev. 1991;55:303–315. doi: 10.1128/mr.55.2.303-315.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giovannoni S J, DeLong E F, Schmidt T M, Pace N R. Tangential flow filtration and preliminary phylogenetic analysis of marine picoplankton. Appl Environ Microbiol. 1990;56:2572–2575. doi: 10.1128/aem.56.8.2572-2575.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gutshall K R, Trimbur D E, Kasmir J J, Brenchley J E. Analysis of a novel gene and β-galactosidase isozyme from a psychrotrophic Arthrobacter isolate. J Bacteriol. 1995;177:1981–1988. doi: 10.1128/jb.177.8.1981-1988.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1991;280:309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henrissat B, Bairoch A. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1993;293:781–788. doi: 10.1042/bj2930781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirata H, Fukazawa T, Negoro S, Okada H. Structure of a β-galactosidase gene of Bacillus stearothermophilus. J Bacteriol. 1986;166:722–727. doi: 10.1128/jb.166.3.722-727.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holmes M L, Scopes R K, Moritz R L, Simpson R J, Englert C, Pfeifer F, Dyall-Smith M L. Purification and analysis of an extremely halophilic β-galactosidase from Haloferax alicantei. Biochim Biophys Acta. 1997;1337:276–286. doi: 10.1016/s0167-4838(96)00174-4. [DOI] [PubMed] [Google Scholar]
- 13.Junge K, Gosink J J, Hoppe H G, Staley J T. Arthrobacter, Brachybacterium and Planococcus isolates identified from Antarctic sea ice brine. Description of Planococcus mcmeekinii, sp. nov. Syst Appl Microbiol. 1998;21:306–314. doi: 10.1016/S0723-2020(98)80038-6. [DOI] [PubMed] [Google Scholar]
- 14.Loveland J, Gutshall K, Kasmir J, Prema P, Brenchley J E. Characterization of psychrotrophic microorganisms producing β-galactosidase activities. Appl Environ Microbiol. 1994;60:12–18. doi: 10.1128/aem.60.1.12-18.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loveland-Curtze J, Sheridan P P, Gutshall K R, Brenchley J E. Biochemical and phylogenetic analyses of psychrophilic isolates belonging to the Arthrobacter subgroup and description of Arthrobacter psychrolactophilus, sp. nov. Arch Microbiol. 1999;177:355–363. doi: 10.1007/s002030050722. [DOI] [PubMed] [Google Scholar]
- 16.Maidak B L, Larsen N, McCaughey M L, Overbeek R, Olsen G J, Fogel K, Blandy J, Woese C R. The Ribosomal Database Project. Nucleic Acids Res. 1994;22:3485–3487. doi: 10.1093/nar/22.17.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 18.Murata T, Akimoto S, Horimoto M, Usui T. Galactosyl transfer onto p-nitrophenyl β-d-glucosidase using β-d-galactosidase from Bacillus circulans. Biosci Biotechnol Biochem. 1997;61:1118–1120. doi: 10.1271/bbb.61.1118. [DOI] [PubMed] [Google Scholar]
- 19.Ohtsu N, Motoshima H, Goto K, Tsukasaki F, Matsuzawa H. Thermostable β-galactosidase from an extreme thermophile, Thermus sp. A4: enzyme purification and characterization, and gene cloning and sequencing. Biosci Biotechnol Biochem. 1998;62:1539–1545. doi: 10.1271/bbb.62.1539. [DOI] [PubMed] [Google Scholar]
- 20.Pace N R, Stahl D A, Lane D J, Olsen G J. The analysis of natural microbial populations by ribosomal RNA sequences. Adv Microb Ecol. 1986;9:1–55. [Google Scholar]
- 21.Schrogel O, Allmansberger R. Optimisation of the BgaB reporter system: determination of transcriptional regulation of stress responsive genes in Bacillus subtilis. FEMS Microbiol Lett. 1997;153:237–243. doi: 10.1111/j.1574-6968.1997.tb10488.x. [DOI] [PubMed] [Google Scholar]
- 22.Shimizu T, Kobayashi T, Ba-Thein W, Ohtani K, Hayashi H. Sequence analysis of flanking regions of the pfoA gene of Clostridium perfringens: β-galactosidase gene (pbg) is located in the 3′-flanking region. Microbiol Immunol. 1995;39:677–686. doi: 10.1111/j.1348-0421.1995.tb03256.x. [DOI] [PubMed] [Google Scholar]
- 23.Stanislawski J. Enzyme Kinetics, version 1.5. Fort Pierce, Fla: Trinity Software; 1991. [Google Scholar]
- 24.Swofford D L. PAUP (Phylogenetic Analysis Using Parsimony.), version 3.1.1. Washington, D.C.: Laboratory of Molecular Systematics, Smithsonian Institution; 1993. [Google Scholar]
- 25.Tran L P, Szabo L, Fulop L, Orosz L, Sik T, Holczinger A. Isolation of a β-galactosidase-encoding gene from Bacillus licheniformis: purification and characterization of the recombinant enzyme expressed in Escherichia coli. Curr Microbiol. 1998;37:39–43. doi: 10.1007/s002849900334. [DOI] [PubMed] [Google Scholar]
- 26.Trimbur D E, Gutshall K R, Prema P, Brenchley J E. Characterization of a psychrotrophic Arthrobacter gene and its cold-active β-galactosidase. Appl Environ Microbiol. 1994;60:4544–4552. doi: 10.1128/aem.60.12.4544-4552.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vian A, Carrascosa A V, Garcia J L, Cortes E. Structure of the β-galactosidase gene from Thermus sp. strain T2: expression in Escherichia coli and purification in a single step of an active fusion protein. Appl Environ Microbiol. 1998;64:2187–2191. doi: 10.1128/aem.64.6.2187-2191.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whitaker J R. New and future uses of enzymes in food processing. Food Biotechnol. 1990;4:669–697. [Google Scholar]
- 30.Yanahira S, Yabe Y, Nakakoshi M, Miura S, Matsubara N, Ishikawa H. Structures of novel acidic galactooligosaccharides synthesized by Bacillus circulans β-galactosidase. Biosci Biotechnol Biochem. 1998;62:1791–1794. doi: 10.1271/bbb.62.1791. [DOI] [PubMed] [Google Scholar]