Abstract
The coronavirus disease 2019 (COVID-19) pandemic is unceasingly spreading across the globe, and recently a highly transmissible Omicron SARS-CoV-2 variant (B.1.1.529) has been discovered in South Africa and Botswana. Rapid identification of this variant is essential for pandemic assessment and containment. However, variant identification is mainly being performed using expensive and time-consuming genomic sequencing. In this study, we propose an alternative RT-qPCR approach for the detection of the Omicron BA.1 variant using a low-cost and rapid SYBR Green method. We have designed specific primers to confirm the deletion mutations in the spike (S Δ143-145) and the nucleocapsid (N Δ31-33) which are characteristics of this variant. For the evaluation, we used 120 clinical samples from patients with PCR-confirmed SARS-CoV-2 infections, and displaying an S-gene target failure (SGTF) when using TaqPath COVID-19 kit (Thermo Fisher Scientific, Waltham, USA) that included the ORF1ab, S, and N gene targets. Our results showed that all the 120 samples harbored S Δ143-145 and N Δ31-33, which was further confirmed by whole-genome sequencing of 10 samples, thereby validating our SYBR Green-based protocol. This protocol can be easily implemented to rapidly confirm the diagnosis of the Omicron BA.1 variant in COVID-19 patients and prevent its spread among populations, especially in countries with high prevalence of SGTF profile.
Keywords: variants of concern, B.1.1.529, SARS-CoV2 RT-PCR, SGTF
Introduction
The World Health Organization has declared the coronavirus disease 2019 (COVID-19) outbreak in China a global pandemic on March 2020. Since then, the causative agent, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has been extensively investigated, with more than 16,533,571 million genomic sequences obtained and made freely available on the GISAID database on 17 February 2024. As the SARS-CoV-2 spreads over the globe, a natural process of random mutations and evolution continues [1, 2]. In some circumstances, a mutation provides an evolutionary advantage for the virus, resulting in the emergence of a novel viral lineage that outnumbers previous forms. Many SARS-CoV-2 variants have emerged in various regions of the world, and five of them, Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), and, most recently, Omicron (BA.1), have been designated as variants of concern (VOC) due to their increased transmissibility, virulence, and/or ability to evade immunity [3–8]. Because of its unusual mutational profile and rapid increase in prevalence, the Omicron variant, first found in South Africa and Botswana in November 2021, acquired its VOC classification within days, displacing pre-existing lineages in that country. Omicron has since surpassed pre-existing variants in Europe, the United States, and a number of other countries, causing a new worldwide outbreak. Omicron (BA.1) displays more than 55 mutations in its genome, with 33 of them occurring only in the spike (S) protein including three deletions (Δ69-70, Δ143-145, Δ211) and one insertion (ins214EPE). Also, the nucleocapsid (N) protein contains four mutations including one deletion Δ31-33, and three substitutions (P13L, R203K, and G204R) [8–10].
The rapid spreading of Omicron across the globe urges the need to establish a rapid and low-cost diagnostic tool that could quickly and efficiently detect and track it in order to initiate response and proper policy for pandemic containment. Until now, sequencing is considered as the gold-standard method for identifying SARS-CoV-2 variants. This method is accurate but time-consuming and expensive. Alternatively, rapid and low-cost RT-qPCR detection methods were proposed recently including the use of a range of primers specific for mutations common to VOCs [11–17], or utilizing commercial kits such as TaqPath COVID-19 diagnostic tests (Thermo Fisher Scientific, Waltham, USA) that included the ORF1ab, S, and N gene targets. Some of the SARS-CoV-2 VOCs, Alpha (B.1.1.7) and Omicron (BA.1), generate dropout of the S-gene result in TaqPath kit, with positive results for the other targets (ORF1ab and N genes). This feature has been used as an indicator or screening method to identify these particular variants. The failure of the S-gene target is caused by a deletion mutation Δ69-70 in the respective gene and is called the S-gene target failure (SGTF). Confirmation of the Omicron variant can be performed by specific RT-PCR assays targeting mutations that are characteristic to this variant (S Δ143-145 and N Δ31-33) with mutation-matched primers or probes, and identification through amplification and melting curve analyses [18]. However, at least a subset of samples should be further characterized by sequencing to increase the confidence and reliability of the obtained results [15].
In 17 February 2024, 9,054,259 Omicron sequences have been shared worldwide through the GISAID, among them 8,087,133 sequences belong to the BA.1 sub-variant (89,31%), and all harboring the S Δ143-145 and N Δ31-33 mutations or at least one of them. Consequently, we report here a low-cost SYBR Green-based qPCR protocol for the rapid and specific detection of these two deletion mutations. We propose primer sets, specific to S Δ143-145, and N Δ31-33, that could provide a comparatively inexpensive, simple, rapid, highly sensitive, and specific protocol alternative to commercial kits or can be applied as a second step to confirm the diagnosis when the number and proportion of SGTF are steadily increasing.
Materials and methods
Clinical specimens
A total of 220 clinical nasopharyngeal samples were collected and selected for this study from 3 to 22 December 2021. All these samples were previously tested for the presence of SARS-CoV-2 at the Laboratory of Molecular Biology and Cancer Immunology of the Lebanese University, using the Applied Biosystems™ TaqPath™ COVID-19 assay which targeted the RdRp, N, and S genes. 120 of these clinical samples were positive for SARS-CoV-2 by TaqPath kit with SGTF profile, and 100 clinical samples were negative for SARS-CoV-2. Fifteen clinical nasopharyngeal samples previously confirmed Delta variant, with Ct (cycle threshold) value between 21.6 and 31, were used as negative control. Written informed consent was provided by all participants.
RNA extraction and SARS-CoV-2 qRT-PCR
RNA was extracted from 200 µL of VTM from the clinical samples on Kingfisher flex purification system (Thermo Fisher) using MagMAX™ Viral/Pathogen Nucleic Acid Isolation Kit (Thermo fisher). Reactions were performed in a 20 µL final volume reaction containing 5 µL of extracted RNA. RT-qPCR was performed using QuantStudio 5 real-time PCR detection system (Thermo fisher) and TaqPath 2019-nCoV real-time PCR kit (Thermo fisher), which targeted the RdRp, N and S genes of SARS-CoV-2.
For the validation of Omicron variant in SARS-CoV-2 positive samples, total RNA was retrotranscribed into cDNA using iScript cDNA synthesis kit (BioRad), following the manufacturer’s recommended procedures. Quantitative RT-PCR was carried out using iTaq universal SYBR Green super mix (Bio Rad). Real-time PCR was performed using Bio Rad CFX96 Real‐Time PCR Machine. The thermal cycling conditions used were as follows: 94°C for 2 min, followed by 40 cycles of amplification at 94°C for 10 seconds, and 60°C for 1 minute. The reaction was completed by determining the dissociation curve of all amplicons.
Primer design
Primers were designed based on the full sequence of the Wuhan SARS-CoV-2 genome sequence from NCBI nucleotide database (NC_045512) and the full sequence of B.1.1.529 (BA.1) variant (EPI_ISL_6640917) from GISAID EpiCoV database collected in November 2021 (Fig 1). Bioinformatics tools were used to design and verify the SARS-CoV-2 specific primers. We validated our PCR primers in-silico with the PCR Primer-Blast tool which allows to investigate the amplification targets of primers to thereby ensuring adequate specificity. The primer sets used in this study were synthesized and delivered by Macrogen (Republic of Korea) (Table 1).
Figure 1.
The Localization and design of the primers used. The nine nucleotides deleted in S gene and in N gene of Omicron variant are marked in bold.
Table 1.
List of primer sets.
| Target gene | Primer name | Primer sequence (5’-3’) |
|---|---|---|
| N | N WT 31-33 forward | GTAACCAGAATGGAGAACGCA |
| N | N del 31-33 forward | ACTGGCAGTAACCAGAATGGTG |
| N | N reverse | CGGTAGTAGCCAATTTGGTCATC |
| S | Spike WT 143-145 forward | ATGATCCATTTTTGGGTGTTTATT |
| S | Spike del 143-145 forward | GTAATGATCCATTTTTGGACCA |
| S | Spike reverse | ACACAAATTCCCTAAGATTTTTGA |
| S | Spike control forward | CAACAAAAGTTGGATGGAAAGTG |
| S | Spike control reverse | GATCACGCACTAAATTAATAGGC |
Deleted nucleotides are in bold.
Genome sequencing and lineage analysis
Ten samples displaying the Omicron variant profile by RT-PCR analysis were selected for genome sequencing. Samples were sequenced using the Nanopore MinION methodology, and consensus sequences were generated using the bioinformatics SOP (https://artic.network/ncov-2019/ncov2019-bioinformatics-sop.html) in Nanopolish mode. Sequences obtained were deposited in GISAID database under the accession numbers EPI_ISL_7651286 to 192 EPI_ISL_7651289, EPI_ISL_11327269, EPI_ISL_11327268, EPI_ISL_11327263, 193 EPI_ISL_11327262, EPI_ISL_11327252, and EPI_ISL_11327251. were collected from 3 to 6 December 2021.
Results
Surveillance data, in the Laboratory of Molecular Biology and Cancer Immunology at the Lebanese University, revealed the re-emergence of SGTF profile with a traveler coming from Abidjan on 3 December 2021. This sample was identified as Omicron BA.1 by whole-genome sequencing. Since then, the number and the proportion of samples with SGTF profile were steadily increasing and have reached, as of 10 January 2022, approximately 95% of positive cases among passengers coming to Lebanon. This dramatic increase in SGTF percentage, suspected as Omicron BA.1 variant, urges the need for a rapid and accurate diagnostic tool to detect and track this variant. In this study, we developed a SYBR Green-based RT-qPCR assay for the detection of Omicron BA.1 variant. Our design was based on the differences in gene sequence from the original SARS-CoV-2 sequence (Fig. 1, Table 1).
The Omicron variants contains several specific mutations that could potentially serve as a good tool for timely detection. These include nine nucleotide deletions in the S gene Δ143-145, and other nine nucleotide deletions in the N gene Δ31-33. Accordingly, two groups of primers were designated. The first group (spike del 143-145, and N del 31-33) was designed to detect these two mutations, whereas the second group (spike WT 143-145 and N WT 31-33) was designed to detect variants not harboring these deletions. A last primer pair (spike control) was designed from within the S gene in a region common to all variants including Omicron and was used as a control for cDNA synthesis and to ensure RNA integrity.
Calibration curve and limit of detection
In order to validate their accuracy and efficacy, the newly designed primer set underwent optimization by testing them on a serial 10-fold dilution of RNA template as follows: 50, 5, 0.5, 0.05, 0.005, 0.0005, and 0.00005 ng/µl. All diluted samples were tested in triplicates by the gold-standard TaqMan RT-qPCR (TaqPath kit) and by our SYBR Green-based RT-qPCR protocol (Table 2).
Table 2.
Determination of the sensitivity by seven serials 10-fold dilutions.
| CT values (SYBR Green) |
Ct values (Taq Path kit, ORF1ab probe) | |||
|---|---|---|---|---|
| Concentration of RNA Dilution Factor 1/10 | Spike del 143-145 | N del 31-33 | S control | |
| 50 ng/μl | 19.51 ± 0.354 | 19.43 ± 0.489 | 19.07 ± 0.518 | 17.73 ± 0.154 |
| 5 ng/μl | 21.89 ± 0.612 | 21.59 ± 0.342 | 21.42 ± 0.485 | 20.79 ± 0.267 |
| 0.5 ng/μl | 24.99 ± 0.3004 | 25 ± 0.189 | 24.65 ± 0.461 | 24.05 ± 0.251 |
| 0.05 ng/µl | 28.58 ± 0.436 | 28.69 ± 0.325 | 28.34 ± 0.297 | 27.89 ± 0.153 |
| 0.005 ng/ul | 30.94 ± 0.640 | 32.1 ± 0.425 | 33.52 ± 0.543 | 31.61 ± 0.075 |
| 0.0005 ng/ul | 35.56 ± 0.338 | 35.4 ± 0.120 | 34.32 ± 0.33 | 34.05 ± 0.250 |
| 0.00005 ng/ul | ND | ND | ND | ND |
Diluted samples were tested by our SYBR Green-based protocol and by TaqMan RT-qPCR (TaqPath kit). ND: not detected.
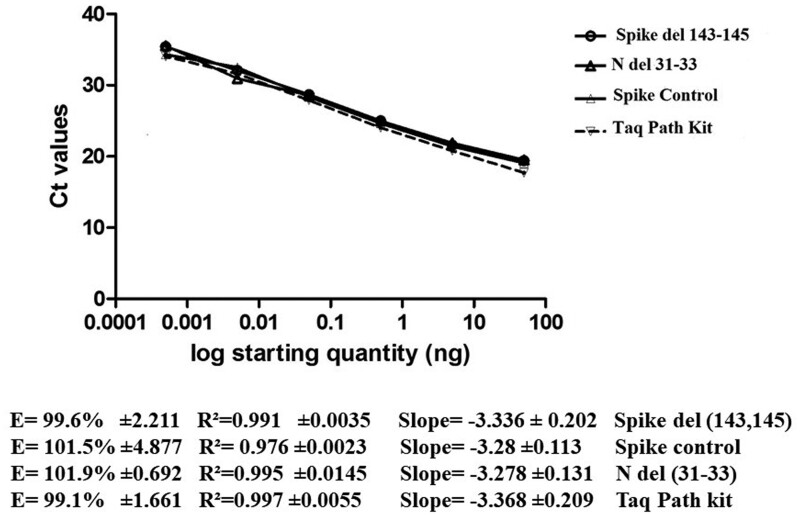
A calibration curve was generated for all the primer sets listed in Table 2, using the serial dilution results. Linear regression performed for these primer sets demonstrated strong correlation (Fig. 2). A limit of detection (LOD) was determined for each primer set and identified as 0.0005 ng/µL for the two sets (Table 2, Fig. 2), or with maximal Ct of 34.05 or 35.5 obtained by TaqMan when targeting ORF1ab and SYBR Green RT-PCR, respectively. According to the manufacturer, the LOD of TaqPath kit is 800 copies/mL when Ct value of ORF1ab is 34 and is 1560 copies/ml when its Ct is 33 (Ion AmpliSeq SARS CoV 2 Research Panel SARS-CoV-2). So, the LOD of our protocol is between 800 and 1560 copies/ml.
Figure 2.
Calibration curves of SYBR Green-based qPCR primers Spike del 143-145, N del 31-33, Spike control and TaqPath kit. Serially 10-fold diluted RNA containing the mutations S Δ143-145 and N Δ31-33 were amplified and analyzed. (a) The threshold cycle (Ct) mean values were plotted against the Log starting quantity (ng/μl). slope, R2 and Efficiency (E) were determined. Each dilution was assayed in triplicate.
Moreover, the specificity of our primers was investigated by four different ways: (i) using 15 clinical positive samples for SARS-CoV-2 confirmed Delta variant; (ii) using 100 clinical negative samples for SARS-CoV-2 by TaqPath kit; they did not manifest a signal when tested with such negative controls; (iii) using in silico prediction analyses as described in the Material and Methods section; and (iv) by WGS of 10 samples.
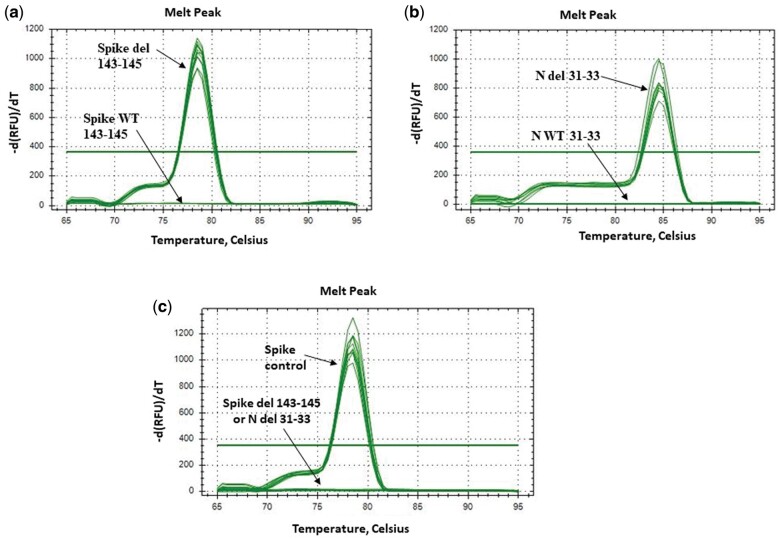
To avoid any false-positive signals resulting from nonspecific products or primer-dimers, a melting curve analysis was included at the end of each PCR assay to determine the specificity and efficiency of each RT-qPCR reaction. Accuracy of our primers to amplify single amplicon is represented as single peak in melting curves (Fig. 3).
Figure 3.
The melting curves for the products amplified with the SYBR Green-based qPCR protocol in S-gene target failure samples using the primers: (a) Spike del 143-145; (b) N del 31-33. The melting curves in Delta samples using the primers: (c) Spike control and Spike del 143-145 or N del 31-33 reveal specific melt peak for each primer set.
Clinical evaluation of spike del 143-145 and N del 31-33 primer sets
After full optimization of the newly designed primer sets and the verification of their sensitivity and specificity in our qPCR assay, they were employed on 120 clinical samples positive for SARS-CoV-2, collected between 3 and 22 December 2021, and they displayed a Ct < 30 with an SGTF profile. Our results showed that the spike control-gene amplicons were detected in all samples, providing evidence of cDNA synthesis and sample integrity preservation (Supplementary Table 1, Fig. 3). Importantly, the S del 143-145-gene amplicons were detected in all the 120 samples, indicating the presence of the S Δ143-145 deletion and obviously the absence of the amino acids 143, 144, and 145 in the spike protein of these samples (Supplementary Table 1, Fig. 3a). Similar results were obtained for the N del 31-33-gene, and amplicons were also detected in all the 120 samples indicating the presence of the N Δ31-33 deletion and the absence of the amino acids 31, 32, and 33 in the nucleocapsid protein of these samples (Supplementary Table 1, Fig. 3b). The use of S WT 143-145 and N WT 31-33 primers did not result in amplified product in any of the samples, thereby indicating that the S Δ143-145 and N Δ31-33 deletions are located within the genomic region targeted by these two primer sets (Fig. 3a and b). Moreover, the S del 143-145 and N del 31-33 primers did not generate any amplification in the 15 Delta samples, providing evidence of the specificity of our protocol, whereas for the spike control primers amplicons were detected in Delta samples demonstrating the synthesis of cDNA (Fig. 3c).
The amplification results obtained with spike del 143-145 and N del 31-33 primers were fully concordant with S WT 143-145 and N WT 31-33 primers, 100% of the SGTF samples harboring both S Δ143-145 and the N Δ 31-33 deletions (Supplementary Table 1).
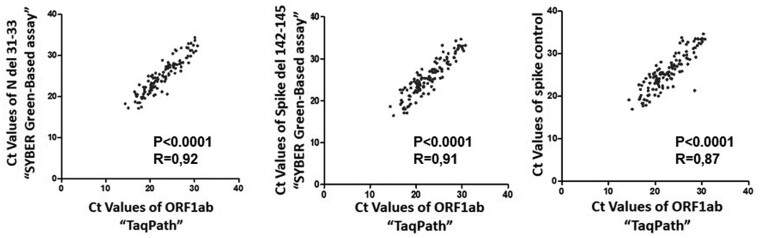
An additional advantage of this protocol is shown in Fig. 4 where we observe a strong correlation between the Ct values generated by TaqPath kit and by our primer sets. This protocol could be used as a comparatively inexpensive, simple, and non-commercial alternative to commercial kits to facilitate monitoring of the spread of the Omicron variants.
Figure 4.
Analyses of the correlation between the Ct values of ORF1ab gene tested by TaqPath™ assay and Ct values of N del 31-33, Spike del 142-145 and Spike control genes tested by our assay. Pearson’s correlation coefficient (R) was used to test the strength of correlation. Black dots denote individual samples (n = 120). P-values <0.05 are considered as significant.
Furthermore, 10 of the S-negative samples were sequenced by WGS at the Microbial Pathogenomics Laboratory of the Lebanese American University and sequences obtained were deposited in GISAID database under the accession number EPI_ISL_7651286 to 192 EPI_ISL_7651289, EPI_ISL_11327269, EPI_ISL_11327268, EPI_ISL_11327263, 193 EPI_ISL_11327262, EPI_ISL_11327252, and EPI_ISL_11327251. Sequence data analysis revealed that the 10 samples belonged to the Omicron variant.
Conclusion
The protocol we described herein is faster, simpler, and more cost-effective than the genome sequencing-based method. The uniqueness of the targeted mutations (S Δ143-145 and N Δ31-33) renders this protocol highly accurate and amply adequate to the early identification of the suspected samples as Omicron BA.1 when genome sequencing is unavailable. In conclusion, this mutation-specific PCR assay would allow any laboratory having the ability to conduct PCR assays to rapidly and reliably screen for Omicron variants in order to enhance surveillance capacity to identify cases and support decision making for interrupting transmission.
Supplementary Material
Contributor Information
Fadi Abdel-Sater, Laboratory of Molecular Biology and Cancer Immunology (COVID-19 Unit), Faculty of science I, Lebanese University, Rafik Hariri Campus, Hadat. Lebanon.
Rawan Makki, Laboratory of Molecular Biology and Cancer Immunology (COVID-19 Unit), Faculty of science I, Lebanese University, Rafik Hariri Campus, Hadat. Lebanon.
Alia Khalil, Laboratory of Molecular Biology and Cancer Immunology (COVID-19 Unit), Faculty of science I, Lebanese University, Rafik Hariri Campus, Hadat. Lebanon.
Nader Hussein, Laboratory of Molecular Biology and Cancer Immunology (COVID-19 Unit), Faculty of science I, Lebanese University, Rafik Hariri Campus, Hadat. Lebanon.
Nada Borghol, Laboratory of Molecular Biology and Cancer Immunology (COVID-19 Unit), Faculty of science I, Lebanese University, Rafik Hariri Campus, Hadat. Lebanon.
Ziad Abi Khattar, Laboratory of Molecular Biology and Cancer Immunology (COVID-19 Unit), Faculty of science I, Lebanese University, Rafik Hariri Campus, Hadat. Lebanon.
Aline Hamade, Laboratory of Molecular Biology and Cancer Immunology (COVID-19 Unit), Faculty of science I, Lebanese University, Rafik Hariri Campus, Hadat. Lebanon.
Nathalie Khreich, Laboratory of Molecular Biology and Cancer Immunology (COVID-19 Unit), Faculty of science I, Lebanese University, Rafik Hariri Campus, Hadat. Lebanon.
Mahoumd El Homsi, Laboratory of Molecular Biology and Cancer Immunology (COVID-19 Unit), Faculty of science I, Lebanese University, Rafik Hariri Campus, Hadat. Lebanon.
Hussein Kanaan, Laboratory of Molecular Biology and Cancer Immunology (COVID-19 Unit), Faculty of science I, Lebanese University, Rafik Hariri Campus, Hadat. Lebanon.
Line Raad, Laboratory of Molecular Biology and Cancer Immunology (COVID-19 Unit), Faculty of science I, Lebanese University, Rafik Hariri Campus, Hadat. Lebanon.
Najwa Skafi, Laboratory of Molecular Biology and Cancer Immunology (COVID-19 Unit), Faculty of science I, Lebanese University, Rafik Hariri Campus, Hadat. Lebanon.
Fatima Al-Nemer, Laboratory of Molecular Biology and Cancer Immunology (COVID-19 Unit), Faculty of science I, Lebanese University, Rafik Hariri Campus, Hadat. Lebanon.
Zeinab Ghandour, Laboratory of Molecular Biology and Cancer Immunology (COVID-19 Unit), Faculty of science I, Lebanese University, Rafik Hariri Campus, Hadat. Lebanon.
Nabil El-Zein, Laboratory of Molecular Biology and Cancer Immunology (COVID-19 Unit), Faculty of science I, Lebanese University, Rafik Hariri Campus, Hadat. Lebanon.
Mhamad Abou-Hamdan, Laboratory of Molecular Biology and Cancer Immunology (COVID-19 Unit), Faculty of science I, Lebanese University, Rafik Hariri Campus, Hadat. Lebanon.
Haidar Akl, Laboratory of Molecular Biology and Cancer Immunology (COVID-19 Unit), Faculty of science I, Lebanese University, Rafik Hariri Campus, Hadat. Lebanon.
Eva Hamade, Laboratory of Molecular Biology and Cancer Immunology (COVID-19 Unit), Faculty of science I, Lebanese University, Rafik Hariri Campus, Hadat. Lebanon.
Bassam Badran, Laboratory of Molecular Biology and Cancer Immunology (COVID-19 Unit), Faculty of science I, Lebanese University, Rafik Hariri Campus, Hadat. Lebanon.
Kassem Hamze, Laboratory of Molecular Biology and Cancer Immunology (COVID-19 Unit), Faculty of science I, Lebanese University, Rafik Hariri Campus, Hadat. Lebanon.
Supplementary data
Supplementary data are available at Biology Methods and Protocols online.
Author contributions
Fadi Abdel-Sater (Conceptualization [equal], Investigation [equal], Methodology [equal], Project administration [equal], Supervision [equal], Visualization [equal], Writing—original draft [equal], Writing—review & editing [equal]), Rawan Makki (Methodology [lead], Writing—original draft [equal], Writing—review & editing [equal]), Alia Khalil (Methodology [equal], Writing—review & editing [equal]), Nader Hussein (Methodology [equal], Writing—review & editing [equal]), Nada Borghol (Methodology [equal], Writing—review & editing [supporting]), Ziad Abi Khattar (Methodology [equal], Writing—review & editing [lead]), Aline Hamade (Methodology [equal], Writing—review & editing [equal]), Nathalie Khreich (Methodology [equal], Writing—review & editing [equal]), Mahmoud Homsi (Methodology [equal], Writing—review & editing [supporting]), Hussein Kanaan (Methodology [equal], Writing—review & editing [supporting]), Line Raad (Methodology [equal], Writing—review & editing [equal]), Najwa Skafi (Methodology [equal], Writing—review & editing [supporting]), Fatima Nemer (Methodology [equal], Writing—review & editing [supporting]), Zeinab Ghandour (Methodology [equal], Writing—review & editing [supporting]), Nabil Zein (Methodology [equal], Writing—review & editing [supporting]), Mohamad Abou Hamdan (Methodology [equal], Writing—review & editing [supporting]), Haidar Akl (Methodology [supporting], Writing—review & editing [supporting]), Eva Hamade (Methodology [equal], Project administration [equal], Writing—original draft [supporting], Writing—review & editing [equal]), Bassam Badran (Funding acquisition [equal], Supervision [equal], Writing—review & editing [supporting]), and Kassem Hamze (Conceptualization [lead], Formal analysis [equal], Investigation [equal], Methodology [equal], Supervision [equal], Validation [equal], Visualization [equal], Writing—original draft [lead], Writing—review & editing [equal])
Competing interests
The authors declare no competing interests.
Funding
This work was supported by Grants from the Lebanese University.
Availability of data and materials
We declared that materials described in the manuscript, including all relevant raw data, will be freely available to any scientist wishing to use them for non-commercial purposes, without breaching participant confidentiality.
Ethics approval and consent to participate
All presented cases or their legal guardian provided consent to data collection and use according to institutional guidelines.
Consent for publication
All presented cases or their legal guardian provided consent to publish according to institutional guidelines.
References
- 1. Habas K, Nganwuchu C, Shahzad F. et al. Resolution of coronavirus disease 2019 (COVID-19). Expert Rev anti Infect Ther 2020;18:1201–11. 10.1080/14787210.2020.1797487 [DOI] [PubMed] [Google Scholar]
- 2. Li X, Wang W, Zhao X. et al. Transmission dynamics and evolutionary history of 2019-nCoV. J Med Virol 2020; 92:501–11. 10.1002/jmv.25701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Davies NG, Jarvis CI, Edmunds WJ, CMMID COVID-19 Working Group et al. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature 2021;593:270–4. 10.1038/s41586-021-03426-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Faria NR, Mellan TA, Whittaker C. et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science 2021;372:815–21. 10.1126/science.abh2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tegally H, Wilkinson E, Giovanetti M. et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature 2021;592:438–43. 10.1038/s41586-021-03402-9 [DOI] [PubMed] [Google Scholar]
- 6. Otto SP, Day T, Arino J. et al. The origins and potential future of SARS-CoV-2 variants of concern in the evolving COVID-19 pandemic. Curr Biol 2021; 31:R918–R929. 10.1016/j.cub.2021.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cele S, Jackson L, Khoury DS, COMMIT-KZN Team et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature 2021;602:654–6. 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Planas D, Saunders N, Maes P. et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature 2021;602:671–5. 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- 9. Callaway E. Heavily mutated Omicron variant puts scientists on alert. Nature 2021;600:21. 10.1038/d41586-021-03552-w. [DOI] [PubMed] [Google Scholar]
- 10. Kannan S, Shaik Syed Ali P, Sheeza A.. Omicron (B.1.1.529) - variant of concern—molecular profile and epidemiology: a mini review. Eur Rev Med Pharmacol Sci 2021; 25:8019–22. 10.26355/eurrev_202112_27653. [DOI] [PubMed] [Google Scholar]
- 11. Abdel Sater F, Younes M, Nassar H. et al. A rapid and low-cost protocol for the detection of B.1.1.7 lineage of SARS-CoV-2 by using SYBR Green-based RT-qPCR. Mol Biol Rep 2021; 48:7243–9. 10.1007/s11033-021-06717-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Corman VM, Landt O, Kaiser M, RECOVER project and collaborating networks et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 2020;25: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Boršová K, Paul ED, Kováčová V. et al. Surveillance of SARS-CoV-2 lineage B.1.1.7 in Slovakia using a novel, multiplexed RT-qPCR assay. Sci Rep 2021; 11:20494. 10.1038/s41598-021-99661-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Granato PA, Kimball SR, Alkins BR. et al. Comparative evaluation of the Thermo fisher TaqPath™ COVID-19 combo kit with the Cepheid Xpert® Xpress SARS-CoV-2 assay for detecting SARS-CoV-2 in nasopharyngeal specimens. BMC Infect Dis 2021; 21:623. 10.1186/s12879-021-06347-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zelyas N, Pabbaraju K, Croxen MA. et al. Precision response to the rise of the SARS-CoV-2 B. 1.1. 7 variant of concern by combining novel PCR assays and genome sequencing for rapid variant detection and surveillance. Microbiol Spectr 2021; 9:e00315-21. 10.1128/Spectrum.00315-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vega-Magaña N, Sánchez-Sánchez R, Hernández-Bello J. et al. RT-qPCR assays for rapid detection of the N501Y, 69-70del, K417N, and E484K SARS-CoV-2 mutations: A screening strategy to identify variants with clinical impact. Front Cell Infect Microbiol 2021; 11:672562. 10.3389/fcimb.2021.672562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bedotto M, Fournier PE, Houhamdi L. et al. Implementation of an in-house real-time reverse transcription-PCR assay for the rapid detection of the SARS-CoV-2 Marseille-4 variant. J Clin Virol 2021; 139:104814. 10.1016/j.jcv.2021.104814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dächert C, , MuenchhoffM, , Graf Aet al. . Rapid and sensitive identification of omicron by variant-specific PCR and nanopore sequencing: paradigm for diagnostics of emerging SARS-CoV-2 variants. Med Microbiol Immunol 2022;211:71–7. 10.1007/s00430-022-00728-7 35061086 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We declared that materials described in the manuscript, including all relevant raw data, will be freely available to any scientist wishing to use them for non-commercial purposes, without breaching participant confidentiality.