Abstract
Interactions and nutrient exchanges among members of microbial communities are important for understanding functional relationships in environmental microbiology. We can begin to elucidate the nature of these complex systems by taking a bottom-up approach utilizing simplified, but representative, community members. Here, we assess the effects of a toxic stress event, the addition of arsenite (As(III)), on a syntrophic co-culture containing lactate-fermenting Desulfovibrio vulgaris Hildenborough and solvent-dechlorinating Dehalococcoides mccartyi strain 195. Arsenic and trichloroethene (TCE) are two highly prevalent groundwater contaminants in the United States, and the presence of bioavailable arsenic is of particular concern at remediation sites in which reductive dechlorination has been employed. While we previously showed that low concentrations of arsenite (As(III)) inhibit the keystone TCE-reducing microorganism, D. mccartyi, this study reports the utilization of physiological analysis, transcriptomics, and metabolomics to assess the effects of arsenic on the metabolisms, gene expression, and nutrient exchanges in the described co-culture. It was found that the presence of D. vulgaris ameliorated arsenic stress on D. mccartyi, improving TCE dechlorination under arsenic-contaminated conditions. Nutrient and amino acid export by D. vulgaris may be a stress-ameliorating exchange in this syntrophic co-culture under arsenic stress, based on upregulation of transporters and increased extracellular nutrients like sarcosine and ornithine. These results broaden our knowledge of microbial community interactions and will support the further development and implementation of robust bioremediation strategies at multi-contaminant sites.
Keywords: Microbial interactions, TCE, arsenic, dechlorination, co-culture, Dehalococcoides, Desulfovibrio
Graphical Abstract
Introduction
Microorganisms exist in the environment in complex, dynamic communities participating in exchanges and feedbacks among themselves and with the environment. Microbial interactions and syntrophies have been demonstrated and discussed for decades 1,2. However, the advent of more sophisticated genetic and meta-omics tools and analyses has enabled the determination that there are more nutrient, signal, and metabolite interactions occurring than are suggested by simple electron donor/acceptor exchanges 3–9. Rather, there are many interactions occurring within microbial communities in any given environment. For example, Harcombe demonstrated a syntrophic co-culture in which one organism exported the energetically expensive amino acid methionine to fulfill an auxotrophy in its partner because the partner’s metabolic waste provided a substrate for the organism’s catabolism 4. Hom and Murray reported obligate mutualism between two usually free-living organisms resulting from environmental perturbation 5. The depth and complexities of microbial mutualism and community cooperation are still largely unknown, and further investigation will help us understand and more effectively utilize microbial community functions.
Genomic analyses of contaminated sites and enrichments with TCE-reducing capabilities often identify Desulfovibrio as important community members10. One example of a syntrophic partnership in environmental microbiology that has been previously identified in groundwater remediation is that of trichloroethene (TCE)-dechlorinating Dehalococcoides mccartyi strain 195 (Dhc195) and lactate-fermenting Desulfovibrio vulgaris Hildenborough (DvH)11. The physiology of co-cultures containing Dhc195 and DvH and basic, essential metabolic exchanges between them under TCE-dechlorinating conditions have been well characterized 12–14. Past research in bioremediation of chlorinated solvents such as trichloroethene (TCE) using these and other microorganisms has focused largely on the remediation of a single contaminant and its intermediate products, but rarely does a contaminated site contain only TCE. Only recently have co-contaminations come to the forefront of TCE bioremediation research15–20. For example, arsenic, one of the most frequent co-contaminants, has been reported in 283 of 454 sites on the current National Priorities List (NPL) that are reported to contain TCE, according to the Environmental Protection Agency 21, and the ATSDR has identified TCE/arsenic as a contaminant mixture of concern 22. TCE/arsenic contaminated sites on the NPL have reported alternate concentration limits for arsenic remediation as high as 90 μM arsenic23. Although the presence of arsenic as a co-contaminant is common, the effects of arsenic on TCE bioremediation are not well characterized.
We previously reported the effects of arsenic on Dhc195 in axenic culture 24. It was found that trivalent arsenic (As(III)) inhibits TCE dechlorination at concentrations environmentally relevant to contaminated sites, impacts the formation of certain aminoacyl-tRNA molecules, and instigates changes in the import and export of certain metabolites to and from the cell 24. We also found that amendment of an amino acid mixture improved Dhc195 growth under As(III)-stressed conditions24. Although these findings are critical in understanding the fundamental impacts of arsenic on TCE dechlorination, axenic culture studies do not always reflect environmentally relevant conditions. Previous studies indicate that the behavior of Dhc195 can change due to the presence of other microorganisms in a consortium 13,14,25. For example, co-cultures of different D. mccartyi strains with lactate-fermenting DvH were shown to promote robust growth and activity of D. mccartyi, including more complete co-metabolic dechlorination of vinyl chloride (VC) by Dhc195 13. Because DvH has been shown to improve Dhc195 VC reduction and cell growth in co-culture, we hypothesize that growth of Dhc195 with DvH in co-culture will also ameliorate As(III) inhibition of TCE dechlorination due to metabolite exchange.
This study reports the changes in cellular function in a TCE-dechlorinating, syntrophic co-culture containing Dhc195 and DvH (Dhc195/DvH) resulting from an arsenic toxicity event. The effects of arsenic on metabolic function, genetic expression, and extracellular metabolome of the syntrophic co-culture were observed, quantified, and compared to previously published effects of arsenic on Dhc195 in axenic culture.
Methods and Materials
Reagents
TCE, cis-dichloroethene, vinyl chloride, and ethene (99.6%, ACS reagents) were obtained from Acros Organics (Geel, Belgium) or Sigma Aldrich (St. Louis, Missouri, USA). The sodium arsenite solution was synthesized from solid sodium (meta) arsenite obtained from Sigma Aldrich (St. Louis, Missouri, USA) by dissolving solid sodium (meta) arsenite in anoxic water under nitrogen headspace followed by filter sterilization. All chemicals used were of reagent grade quality or higher. Gases (air, nitrogen, nitrogen-CO2 mixture, argon, and hydrogen) were obtained from Praxair, Inc (San Ramon, California, USA).
Culture and growth conditions
Dhc195 was grown in a defined mineral salts growth medium with an H2-CO2 (80:20 vol/vol) headspace in 160 mL serum bottles with 100 mL liquid volume as previously described 26,27. DvH was grown in the same defined mineral salts medium with an N2-CO2 (80:20 vol/vol) headspace. The co-culture Dhc195/DvH was created by amending equal volumes Dhc195 and DvH (5mL and 5mL into 95mL, 4.8% inoculation each by volume) and subculturing five times until the ratio of Dhc195:DvH was approximately 5:113. Dhc195/DvH was grown in the same defined mineral salts medium with N2-CO2 (80:20 vol/vol) headspace, 5mM lactate as electron donor, 70.0 – 203.7 μmol TCE as electron acceptor (in doses of approximately 70.0 μmol), and 0.5 mL vitamin solution 28 containing 20 mg L−1 vitamin B12 (final concentration 100 μg L−1) as additional nutrients. All cultures were incubated without light or agitation at 34°C for the duration of the experiment.
To study the impacts of arsenic on TCE-dechlorination, cellular function, and materials exchange by Dhc195/DvH, the co-culture was grown as described above with concentrations of As(III) varying from 0 μM to 30 μM. In the 5-day experiment, As(III) was amended at time 0 immediately before inoculation. In the 28-day experiment, As(III) was amended on day 3 when approximately 50% of amended TCE had been consumed, 5 hours before the FID-GC timepoint was collected. Subsequent amendments of TCE were added to the cultures once TCE and cis-dichloroethene (cDCE) were depleted. Positive controls containing no arsenic were compared against the experimental conditions. All experiments were performed in triplicate. A list of all conditions tested can be found in Supplemental Table S1.
Minimum inhibitory concentration quantification of As(III) on Dhc195 in Dhc195/DvH
To calculate the minimum inhibitory concentration (MIC) of As(III), Dhc195/DvH was grown with concentrations of As(III) ranging from 5 μM to 30 μM As(III) at 5 μM intervals. As(III) was amended 24 hours after inoculation of the culture to a fresh bottle (5% vol/vol). Cell samples were harvested every 24 hours for 5 days. The MIC of each organism was defined as the concentration that caused a 50% decrease in growth rate as compared to the control condition containing no arsenic 29.
DNA extraction and cell enumeration
Cells from 1.5mL of culture sampled from each replicate were collected via centrifugation (14000g x 10 minutes at 4°C), and DNA was extracted using the DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA, USA) according to the manufacturer instructions for Gram-positive bacteria. DNA quality was evaluated via a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). Quantitative PCR (qPCR) using SYBR-green based detection agents was used to enumerate cell growth, using strain-specific primers (forward, 5′-ATCCAGATTATGACCCTGGTGAA; probe, 5′-TGGGCTATGGCGACCGCAGG; and reverse, 5′-GCGGCATATATTAGGGCATCTT) which targeted the reductive dehalogenase tceA gene in Dhc195, which is present as a single copy gene in Dhc195 and not present in the DvH genome, as previously described 30. TceA standards were obtained as gBlocks from Integrated DNA Technologies (Coralville, IA, USA) (sequence: GGTGCCGCGACTTCAGTTATGCCGAATTTTCACGACTTGGATGAAGTAATTTCTGCTGCTAGTGCCGAGCTCTAGACGCGGATCCATCCAGATTATGACCCTGGTGAACTGGGCTATGGCGACCGCAGGGAAGATGCCCTAATATATGCCGC). Average growth rates over the duration of the experiment were calculated by quantifying Dhc195 at the beginning and end of the experiment.
Transcriptomic analysis
To evaluate the effects of As(III) on Dhc195 and DvH metabolism and cellular function in co-culture, transcriptomic analysis was performed on extracted RNA from Dhc195/DvH and sequenced on a HiSeq 4000 Illumina platform (Illumina, San Diego, CA, USA). Results were analyzed and compared to our previous study evaluating the effects of arsenic on Dhc195 in pure culture 24.
RNA extraction and sequencing
Samples for transcriptomic analysis were taken five hours after addition of As(III) from three conditions (10 μM As(III), 20 μM As(III), positive control containing no arsenic) and at an additional timepoint for cultures containing 20 μM As(III) when TCE dechlorination had resumed and approximately 80% of initial TCE was consumed (22 days after As(III) amendment). Cells for RNA extraction were collected from liquid cultures via vacuum filtration, flash-frozen in liquid nitrogen, and stored at −80°C as previously described31.
RNA extraction and purification from the coculture was performed as previously described 32. The integrity of purified RNA was quantified on an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) and the quantity determined by Qubit 3 Fluorometer (Invitrogen, Carlsbad, CA, USA). RNA samples with an RNA integrity number larger than 8 were subsequently treated by Ribo-Zero rRNA Removal Bacteria Kit (Illumina, San Diego, CA, USA) according to the manufacturer instructions. Processed samples were stored at −80°C prior to sequencing. Library construction and RNA-Seq were performed at QB3 Functional Genomics Laboratory at UC Berkeley (Berkeley, CA, USA) using 100 base pair paired-end reads on the HiSeq 4000 Illumina platform (Illumina, San Diego, CA, USA).
RNA-Seq data analysis
The RNA-Seq data analysis pipeline from UC Davis Bioinformatics Core (Davis, CA, USA) was adopted and modified to perform the alignment and statistical analysis of the raw sequencing data for the purpose of this study. Briefly, the sequence reads were first preprocessed using adapter trimming with R package “scythe” (https://github.com/vsbuffalo/scythe) and quality trimming with R package “sickle” 33. The trimmed sequence reads were then aligned to the genomes of Dhc195 and DvH, respectively, using the aligner “STAR” in R 34. Normalization of the read counts was subsequently carried out with “edgeR” package in R 35. Limma-voom analysis was then performed to estimate the mean-variance relationship and generate a 1-Factor model, allowing the downstream differential expression analysis of the dataset 36. Afterwards, the linear model fit was processed with eBayes and the log base-2 fold-change (logFC) was determined in each comparison with toptable. Genes with an adjusted p-value smaller than 0.05 and logFC < 0.5 or > 2 were considered significantly differentially expressed genes.
Extracellular metabolomic analysis
Samples for extracellular metabolite analysis were taken from cultures amended with 20 μM As(III) and the positive control at 3 timepoints during the experiment (5 hours after As(III) amendment, 5 days after As(III) amendment, and the last timepoint at the time when 80% of originally amended TCE was consumed, correlating to 22 days after As(III) amendment), at which point 1 mL liquid medium was withdrawn from the culture and cells were removed via centrifugation (21,000g x 10 minutes at room temperature ≅ 23°C) as previously described 24. The supernatant was decanted and stored at −80°C until analysis.
Targeted and untargeted metabolomic data was acquired using gas chromatography time-of-flight mass spectrometry (GC-TOF-MS), processed, and normalized at the NIH West Coast Metabolomics Center (Davis, USA) as previously described 37.
Statistical significance for this analysis was defined as changes exhibiting fold-change >2, p-value <0.05, and signal intensity >500. For unknown metabolites with significant fold-changes, metabolite numbers were assigned as identification placeholders and their retention indices and spectra are shown in Supplemental Information, Table S2a.
Interaction network analysis
Interaction networks were developed using two different methods for robustness. First, a Spearman’s rank-order correlation analysis was performed as previously described 38. Second, correlation networks were inferred using Random Matrix Theory (RMT) via the Molecular Ecological Network Approach Pipeline (MENA), a procedure that approximates Pearson correlation through an iterative process 39. In the Spearman’s rank-order correlation analysis, all pair-wise correlations were computed among Dhc195 transcripts, DvH transcripts, and metabolites (both targeted and untargeted), and the approximately 600 correlations (edges) with the lowest p-values (cutoff p-value < 0.000607) were visualized using Cytoscape. The Spearman cutoff p-value was an arbitrary selection to keep the number of visualized edges at a reasonable value. In MENA, the cutoff threshold for Pearson coefficient was 0.95, corresponding to a χ2-test value of 74.4 and p-value < 0.05. Using these cutoff values, 5,138 nodes were mapped among 306 nodes. A table with the full description of MENA parameters can be found in Supplemental Table S3 These interaction networks were visualized with Cytoscape version 3.9.1 40. Correlations among differential regulation in Dhc195 and DvH along with the extracellular metabolite profile were evaluated and used to identify potential methods of biostimulation for more robust TCE dechlorination under arsenic stress.
Untargeted metabolite identification
Untargeted metabolites exhibiting significant changes in relative abundance were investigated further as reported previously.24 Briefly, we subtracted one or two trimethyl silyl moieties from the unknown molecular ion [M-73], [M-146], and matched resulting spectra with published underivatized mass spectral databases. Once candidate compounds were determined from this method and compared to the correlation networks including both metabolite and transcript data, we performed a follow-up experiment in which supernatant from Dhc195/DvH cultures with and without As(III) amendment (0 versus 20 μM As(III)) was analyzed for presence of these metabolite candidates and compared against commercial standards using LC-MS/MS.
Analytical Methods
Chlorinated ethenes and ethene were measured by loading 100 μL headspace gas on an Agilent 7890A gas chromatograph equipped with a flame ionization detector and a 30-m J&W capillary column with an inside diameter of 0.32-mm (Agilent Technologies, Santa Clara, CA, USA). A gradient temperature program method was used as previously described 41.
Measurement of soluble As(III) and As(V) was performed using an Agilent 1260 Infinity high-performance liquid chromatograph (HPLC) (Agilent Technologies, Santa Clara, CA, USA) equipped with orthogonal chromatographic separation consisting of an Aminex HPX-87H Ion Exclusion column (300mm x 7.8mm, BIO-RAD, Hercules, CA, USA) coupled to a Hamilton PRP-X300 Reversed-Phase column (250mm x 4.1mm, Hamilton Co., Reno, NV, USA). The HPLC was coupled to an Agilent 7700 series inductively coupled plasma mass spectrometer (ICP-MS) (Agilent Technologies, Santa Clara, CA, USA) as previously described 42. For each measurement, 0.5 mL liquid medium samples were extracted from the batch incubations for cultures amended with As(III). Liquid medium samples were filtered using 0.2 μM HPLC-grade syringe filters (Pall Life Sciences, England) to remove cells. Filtered samples were diluted 10x or 100x to obtain analytical concentrations between 10ppb and 500ppb.
Results and Discussion
Inhibition of arsenic on metabolic function and cell growth in Dhc195/DvH co-culture
The effects of As(III) in concentrations ranging from 5 μM to 30 μM on cell growth, TCE-dechlorination, and hydrogen production of the Dhc195/DvH co-culture were evaluated. The minimum inhibitory concentration (MIC) of As(III) on cell growth was quantified for Dhc195 in co-culture Dhc195/DvH and compared to that previously measured for the axenic Dhc195 24. The MIC for Dhc195 in Dhc195/DvH was estimated to be 14 ± 0.53 μM As(III) while the MIC for Dhc195 in axenic culture was 9.1 ± 1.0 μM (Figure 1A), indicating more robust growth of Dhc195 under arsenic stress in the Dhc195/DvH co-culture. Additionally, the average TCE dechlorination rate was estimated to decrease by 50% with the amendment of 17.2 μM As(III) in Dhc195/DvH versus 8.9 μM As(III) in Dhc195 axenic culture (Figure 1B). Figures 2A and 2B show TCE consumption and H2 production, respectively, for Dhc195/DvH over 5 days with increasing As(III). At all times, H2 concentrations remained far above the 0.6 nM hydrogen threshold requirement for TCE reductive dechlorination by Dhc195 43. This result indicates that H2 production by DvH was not the limiting factor for TCE dechlorination by in Dhc195/DvH under arsenic stress. Cultures containing no arsenic and 5 μM As(III) consumed 99% of TCE (less than 1 μmol TCE remaining) by the end of day 5, while cultures containing 25 μM and 30 μM As(III) were observed to consume approximately 23% of TCE (approximately 54 μmol TCE remaining) at the end of day 5. Production of cDCE, VC, and ethene were likewise diminished with increasing As(III) (Figure S1), suggesting the initial dechlorination of TCE was inhibited. Molar masses of TCE, cDCE, VC, and ethene were balanced within one standard deviation of experimental triplicates on day 5 as compared to day 0.
Figure 1.
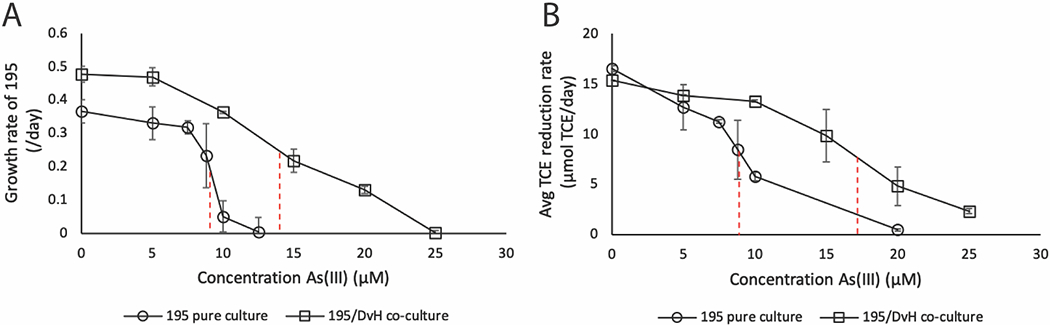
A) Average growth rate (day−1) of Dhc195 with increasing As(III) concentrations in the 195 axenic culture 24 and in co-culture with DvH. B) Average TCE reduction rate (μmol TCE/day) with increasing As(III) concentrations in the 195 axenic culture and in co-culture with DvH. Error bars represent one standard deviation of experimental triplicates. Vertical dashed lines represent the As(III) concentration causing a 50% decrease in growth and TCE reduction rates.
Figure 2.
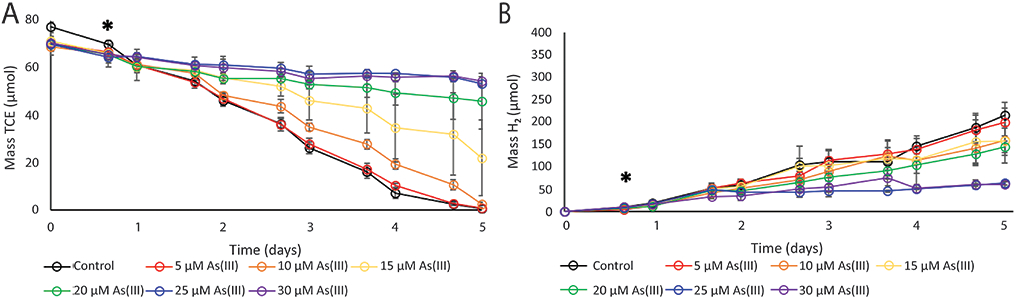
A) TCE consumption reported as mass TCE (μmol) and B) H2 production reported as mass H2 (μmol) over 5 days in Dhc195/DvH with concentrations of As(III) from 5 μM to 30 μM and a no-arsenic control. Mass reported is the sum of aqueous and gaseous masses. Asterisks denote the time that As(III) was added. Error bars represent one standard deviation of experimental triplicates.
TCE dechlorination by Dhc195/DvH was also evaluated over a period of 28 days in co-cultures amended with 10 μM, 20 μM As(III), and a no-arsenic positive control (Figure 3). As(III) was amended on day 3 after approximately 60 percent of the first TCE dose was consumed. In the positive control and the 10 μM As(III)-amended co-culture, 202 ± 8.9 and 204 ± 6.6 μmol TCE, respectively, was consumed per microcosm from 3 doses over 11 days. At the end of 11 days, 68 ± 6.4 μmol VC and 111 ± 9.5 μmol ethene was produced in the no-arsenic control and 98 ± 7.5 μmol VC and 89 ± 5.1 μmol ethene was produced in the 10 μM As(III)-amended co-culture, respectively. In the 20 μM As(III) co-culture, TCE dechlorination stalled after addition of As(III) but resumed at a lower rate after approximately 20 days, consuming the initial dose of TCE (70.0 ± 2.0 μmol) and producing 64 ± 2.4 μmol VC and 2.5 ± 0.21 μmol ethene by day 28 (Figure S2). No changes in arsenic speciation were observed in culture amended with either 10 μM or 20 μM As(III) (Figure S3).
Figure 3.
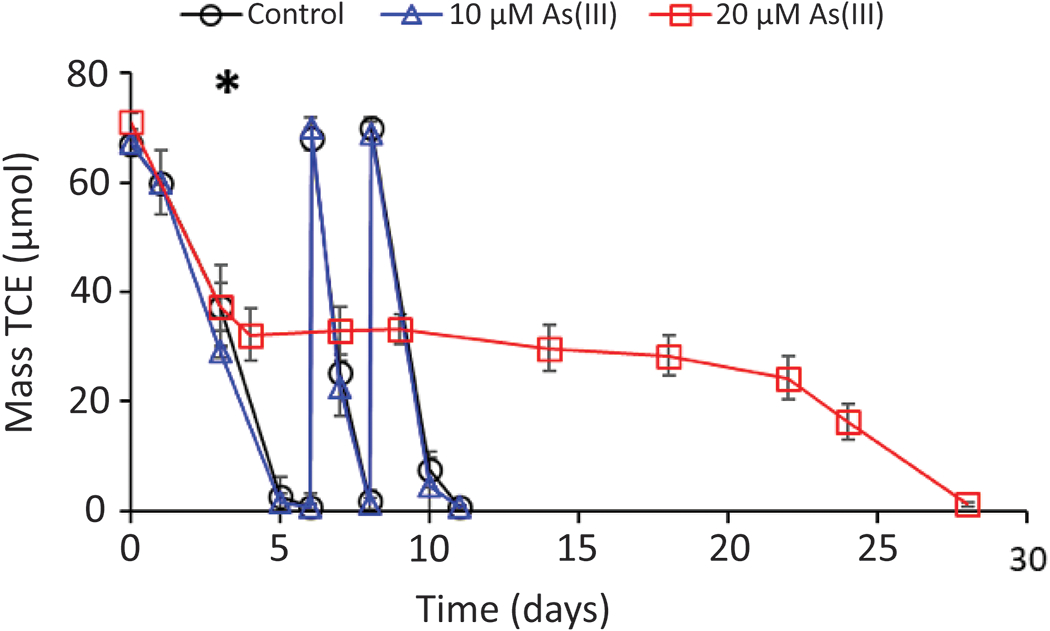
TCE consumption by Dhc195/DvH reported as mass TCE (μmol) versus time (days) over 28 days with 10 μM As(III), 20 μM As(III), and a positive control. Mass reported is the sum of aqueous and gaseous masses. The asterisk denotes the time that As(III) was added to the cultures. Error bars represent one standard deviation of experimental triplicates. No change in the valence state of arsenic was observed in any cultures (Figure S3).
Transcriptomic analysis of arsenic inhibition on Dhc195/DvH co-culture
Analysis of RNA-Seq data from Dhc195/DvH containing 0 μM (control), 10 μM, and 20 μM As(III) was performed. In all cultures, RNA samples were taken 5 hours after amendment of As(III). RNA samples were also taken at 22 days for cultures containing 20 μM As(III). The 22 day timepoint represents the time at which metabolic function was regained and 80% of initially amended TCE was consumed (the same amount of TCE consumed in the control and cultures containing 10 μM As(III) at 5 hours. Gene expression was compared under the experimental conditions as described in Figure 4.
Figure 4.
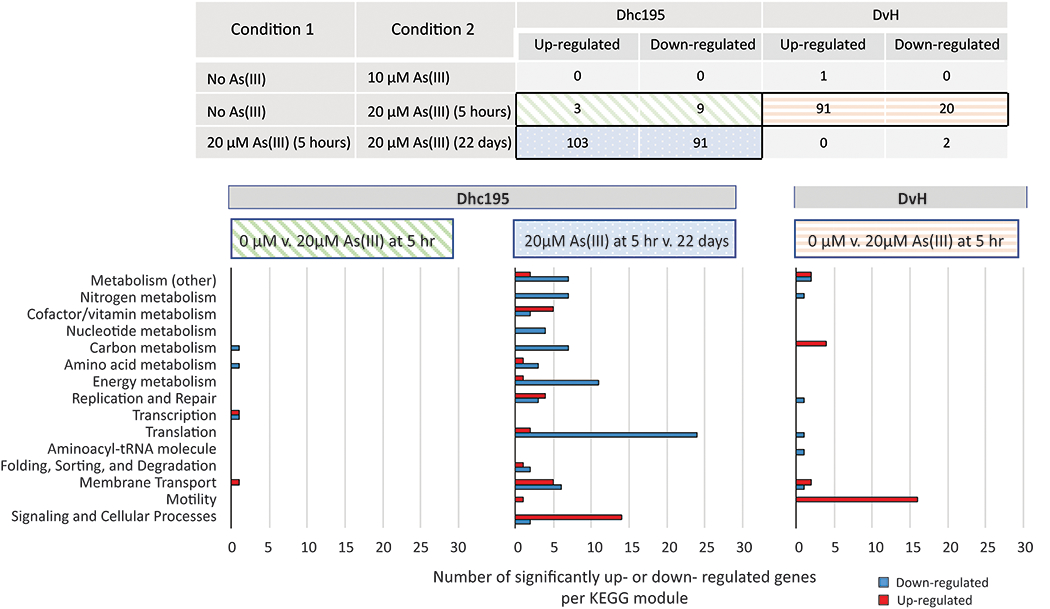
RNA-Seq analyses and corresponding numbers of significantly up- and down-regulated genes in both Dhc195 and DvH in response to As(III). Three analyses yielded notable changes in gene expression: 20 μM As(III) at 5 hr as compared to no As(III) at 5 hr in Dhc195 (green diagonal stripes), 20 μM As(III) at 22 hr as compared to the same culture at 5 hr in Dhc195 (blue with white dots), and 20 μM As(III) at 5 hr as compared to no As(III) at 5 hr in DvH (orange horizontal stripes). The results of these analyses are further described in bar plots of up-regulated (red) and down-regulated (blue) genes as organized by KEGG module. Any genes annotated as coding for hypothetical proteins or not categorized by KEGG module are not included. Changes in gene expression were calculated as (Condition 2/Condition 1).
In cultures containing 10 μM As(III), a single DvH gene (DVU1644) encoding an unidentified permease exhibited significant differential regulation (upregulation with fold-change >2, p-value < 0.05). No Dhc195 genes exhibited significant differential regulation as compared to the control, aligning with the physiological observation that there was not a significant change in TCE dechlorination from the control to cultures containing 10 μM As(III) (Figure 3).
In cultures amended with 20 μM As(III), 5 hours after amendment 91 DvH gene exhibited significant upregulation and 20 genes were significantly downregulated, while 3 Dhc195 genes exhibited significant upregulation and 9 were significantly downregulated as compared to the control. At 22 days after 20 μM As(III) amendment, no DvH genes exhibited significant upregulation compared to the 5 hour timepoint and two genes were significantly downregulated. In the same condition, 103 Dhc195 genes exhibited significant upregulation and 91 were significantly downregulated as compared to the 5 hour timepoint (Figure 4).
At the time when 20 μM As(III) was amended to Dhc195/DvH and TCE dechlorination halted (5 hours), Dhc195 presented upregulation of tceA (DET0078-0079) and arsR (DET1005), but not significantly (Supplemental Table S5). Only three and five genes were significantly up- and down-regulated at 5 hours, respectively. This is dissimilar to previously reported results of Dhc195 responses to arsenic stress in pure culture, in which many amino acid biosynthesis, ribosome biogenesis, and replication and repair proteins were upregulated in response to As(III) 24. The discrepancy in As(III) stress responses by Dhc195 in pure culture and co-culture with DvH indicate one or more interactions between Dhc195 and DvH are beneficial to Dhc195 under stress. In DvH, downregulated genes included genes related to electrochemical potential-driven transport (DVU3026), DNA replication and repair (DVU1649), and tRNA biogenesis (DVU0953) (Figure 4C). Genes upregulated in DvH included genes related to cell motility (DVU0307, 0313, 0316, 0512, 0514, 0516, 0523, 0863, 1441-1445, 2444, 2948, 3231), permease proteins (DVU1644, 2298), and phage genes (DVU0189, 0197-0198, 0200, 0210-0211, 0214-0221, 2859, 2869, 2871-2872). Johnson et al. previously showed upregulation of motility-related genes in DvH in response to oxygen stress 44. However, other studies evaluating the effects of stressors such as nitrate, nitrite, and alkalinity did not share similar responses 45–47.
At day 22 in 20 μM As(III)-amended cultures as compared to the 5 hour timepoint, Dhc195 presented upregulation of 10 putative reductive dehalogenase genes (DET0173, 0175, 0180-0181, 1171, 1535, 1544-1545, 1558-1559). Some of these genes are generally not expressed in Dhc195 11,30. However, DET0180 has also been reported to be upregulated under sulfide stress 11, suggesting that Dhc195 may upregulate generally dormant or minimally expressed reductive dehalogenases to maximize the amount of energy capture under recovery from toxicity stress. Dhc195 also exhibited upregulation of a molybdopterin-containing oxidoreductase (DET0101-0103) (Figure 4B). Arsenic-related proteins contain molybdopterin active sites 48,49. However, molybdopterin-containing oxidoreductases are highly diverse in their substrates, so it is therefore difficult to determine if these proteins act on arsenic substrates in Dhc195. As expected, the arsenical pump (DET0908) was significantly upregulated, suggesting the low background expression of the As(III) efflux pump in Dhc195 is sufficient for the first 5 hours and is upregulated later on. This delay in efflux pump upregulation may also be due to the genomic structure of the arsenic detoxification genes in Dhc195. While many organisms possess arsenic-related genes in operons (e.g., arsRDABC)50, arsenic-related genes in Dhc195 appear to be located farther apart from one another in its genome (i.e., DET0143, 0908, 1005). The heat shock proteins Hsp20 (DET0954) and HtpX (DET1587) were also significantly upregulated, aligning with upregulation of heat shock related proteins in previous prokaryotic studies of arsenic toxicity 51–53, as were ABC transporters (DET0814, 1619-1620). Several aminoacylated-tRNA molecules were also found to be present in significantly higher concentrations at 22 days as compared to 5 hours, including arginyl-tRNA, cystinyl-tRNA, and tryptophanyl-tRNA. We previously reported differential expression of aminoacylated-tRNA molecules in Dhc195 pure culture under arsenic stress 24, however, different aminoacylated-tRNA molecules were present in higher abundance in pure culture than the ones observed in this co-culture study. In the pure culture study, arginyl-tRNA was present in significantly lower abundances, and many more aminoacyl-tRNA molecules had lower abundances24. It is widely known that the primary mode of As(III) toxicity results from binding to sulfhydryl groups54,55, and aminoacyl-tRNA synthetase enzymes commonly contain sulfhydryl groups 56–58. The concomitant return to non-significant differences in aminoacyl-tRNA molecule abundances with Dhc195 recovery of TCE dechlorination function compared to pre-arsenic amended conditions, in combination with these previous results, reinforces the importance of As(III) interaction with aminoacyl-tRNA synthetases as a mode of toxicity.
Dhc195 also exhibited downregulation of several genes related to general stress response and energy conservation, including the H+-ATP synthase operon (DET0559-0565). Previous studies have identified heat shock proteins as stress-inducible regulators of the H+-ATPase in Saccharomyces cerevisiae 59 and reported downregulation of ATPase under toxicity stress in Saccharomyces cerevisiae, Pseudomonas putida, and Salmonella enterica 59–61. Piper et al. identified this ATPase regulatory mechanism as a method of energy conservation under stress 59. Dhc195 also exhibited downregulation of nitrogen fixation (DET1124, 1148, 1153-1155, 1158). Since the growth medium used in these experiments was supplemented with both ammonium and N2, downregulation of the nitrogen fixation operon is also probably for the purpose of energy conservation under stress. Downregulation of several degV-family proteins (DET0105, 0421, 1265-1267), and a few genes related to the central carbon metabolism (DET0449-0451, 0453) was also observed in Dhc195. Previous studies have reported that degV family proteins are related to fatty-acid transport and metabolism 62,63. Downregulation of fatty-acid and other carbon metabolism-related genes may also be part of the effort by Dhc195 to conserve energy under stress. Of the 103 genes significantly up-regulated in Dhc195 under this condition, 31 encoded hypothetical proteins compared to 10 of the 91 down-regulated genes encoding hypothetical proteins.
As compared to the differential gene expression results reported previously 24, Dhc195 only exhibited significant up-regulation of 2 of the same genes and significant down-regulation of 2 of the same genes in co-culture as compared to pure culture in response to As(III). The shared upregulated genes included an ABC transporter (DET0814) and a bacterioferritin domain protein (DET0955). The shared downregulated genes were glutamine synthetase (DET1123) and an ammonium transporter (DET1125). Down-regulation of these genes may also be due to energy conservation. Notably, amino acid biosynthesis pathways were significantly upregulated in Dhc195 in pure culture under arsenic stress 24, but were not found to be significantly differentially expressed under As(III) stress in Dhc195/DvH co-culture. This result may suggest an amino acid exchange between Dhc195 and DvH. Exchange of energetically expensive amino acids has been observed before in which the organism sharing the amino acid required the accepting organism to provide substrate for its metabolism 4. This relationship is not unfounded between Dhc195 and DvH, as DvH requires Dhc195 to consume the H2 in solution to make lactate fermentation energetically favorable and may be supplying amino acids to Dhc195 when under stress. It is important to note these two studies utilized different transcriptomic data processing methods (GeneChip microarray versus RNA-Seq), so the relative expression values cannot be directly compared. However, a comparison of trends in the types of genes and pathways exhibiting differential expression can be discussed.
At 22 days in 20 μM As(III)-amended cultures as compared to 5 hours, DvH exhibited significant downregulation of only 2 genes encoding hypothetical proteins (DVU0132-0133). A comprehensive list of all significantly differentially expressed genes in Dhc195 and DvH under each condition can be found in Supplemental Tables S4–S9.
Extracellular metabolite profile of Dhc195/DvH in the presence of arsenic
Due to the results of the transcriptomic analysis, changes in the extracellular metabolite profile of Dhc195/DvH in response to 20 μM As(III) amendment were evaluated using samples taken at 3 timepoints. Extracellular metabolite samples were taken 5 hours after amendment of As(III), 5 days after amendment of As(III), and 22 days after amendment of As(III) when TCE dechlorination function was regained and 80% of initial TCE had been consumed. Inclusion of data from day 5 provides another point of reference between 5 hours and 22 days to gain a more comprehensive understanding of the response to arsenic stress over time. Figure 5 illustrates fold-changes and p-values of both targeted and untargeted metabolites in As(III)-amended cultures as compared to the positive control at the 3 timepoints.
Figure 5.
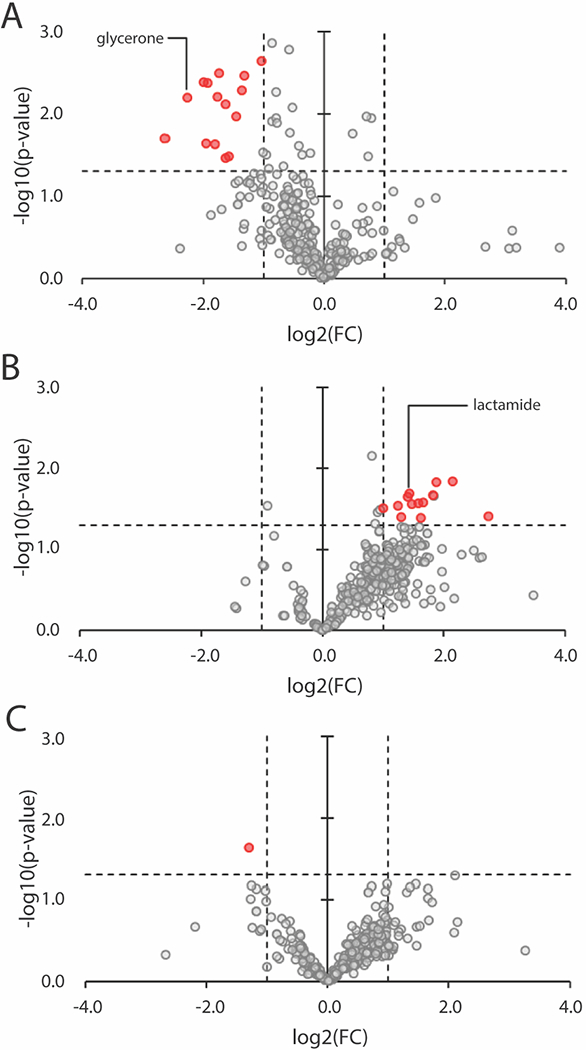
Fold-changes in extracellular metabolite abundances in Dhc195/DvH cultures amended with 20 μM As(III) as compared to the positive control at A) 5 hours after As(III) addition, B) 5 days after As(III) addition, and C) 22 days after As(III) addition and when TCE dechlorination function was regained and 80% of initial TCE had been consumed. Fold-change (FC) is expressed as (As(III)-amended/Control). Vertical dashed lines indicate a 2-fold change and the horizontal dashed line indicates p-value = 0.05. Metabolites falling outside the vertical lines and above the horizontal line are considered statistically significant and are colored red.
Shortly after addition of As(III) to Dhc195/DvH, 15 metabolites were observed to be present in significantly lower abundance as compared to the positive control (Figure 5A). Of the targeted metabolites, glycerone was present in lower abundance. Glycerone (also known as dihydroxyacetone) is a carbohydrate intermediate in carbon metabolism and precursor to glyceraldehyde-3-phosphate and glyceraldehyde-6-phosphate, which are essential in multiple metabolic pathways such as biosynthesis of tryptophan and glycolysis 64–67. The tryptophan biosynthesis pathway was previously reported to be significantly upregulated in Dhc195 under arsenic stress 24, and the uptake (or retention) of glycerone following As(III) amendment may conceivably be related to tryptophan synthesis. There were other metabolites that showed significant fold-changes but whose identities were unknown. 5 days after As(III) amendment, 14 metabolites were observed to be present in significantly higher abundance as compared to the positive control (Figure 5B). Of these, 13 were unknown and the single metabolite identified in targeted metabolomic analysis was lactamide. Lactamide is an amide derived from lactic acid and an isomer of alanine 68. It is possible that the increase in abundance of lactamide and other extracellular metabolites when TCE dechlorination stalled was due to the metabolism of DvH being minimally affected by addition of 20 μM As(III) and Dhc195 conserving energy for toxicity stress-response. When TCE dechlorination had regained function, only one unknown metabolite was observed to be present in significantly different abundance in the As(III)-amended cultures as compared to the control, indicating that the stress response was no longer being expressed. It is possible that this unidentified metabolite is a false positive, but further discussion is difficult without metabolite identification. Comparisons of normalized signal intensities for all targeted metabolites can be found in Supplemental Tables S10–S12.
Interactions Network Results
After transcriptomic and metabolomic analyses were processed, we developed interaction networks among the significant differential gene expressions of Dhc195 and DvH, as well as the significant extracellular metabolite changes using two different network development methods: Spearman’s rank-order correlation analysis and RMT using MENA (Figures S4 and 6, respectively). From the Spearman’s rank-order correlation analysis (Figure S4), molecular weights and mass spectra of the correlated unknown metabolites were compared against intermediates and products of pathways in which citramalate, glycerone (a.k.a. dihydroxyacetone), 3-phenyllactic acid, tyrosine, and putrescine are intermediates. They were also compared against intermediates and precursors of DvH flagellum synthesis. Based on these correlations and the unknown metabolite molecular weights and mass spectra, methyl-2-oxobutanoate, ornithine, and citrulline were identified as potential candidates.
From RMT analysis using MENA, the metabolites lactate, lactamide, allothreonine, valine, and metabolite 366131 clustered with the flagellar synthesis genes in DvH (Figure 6). As flagellar synthesis is dependent on proteins, and flagella are constructed of proteins, it is unsurprising that amino acids clustered with these genes. Based on these correlations and the molecular weight and mass spectra of 366131, we were unable to propose a metabolite. However, it is probable that 366131 is related to amino acid biosynthesis. Additionally, RMT yielded a particularly interesting cluster of amino acids (threonine, alanine, isoleucine), organic acids (fumaric acid, citramalic acid, and glyceric acid), and unknown metabolites (6067, 18173, 31359, 62327, 125472, 210682, 351252) which connected the Dhc195 and DvH clusters. From the molecular weights and mass spectra of the untargeted metabolites and their proximity to certain amino and organic acids, phthalate, glyoxylate, formamide, sarcosine, maleic acid, l-ornithine, and m-2-oxobutanoate were proposed as potential metabolite candidates.
Figure 6.
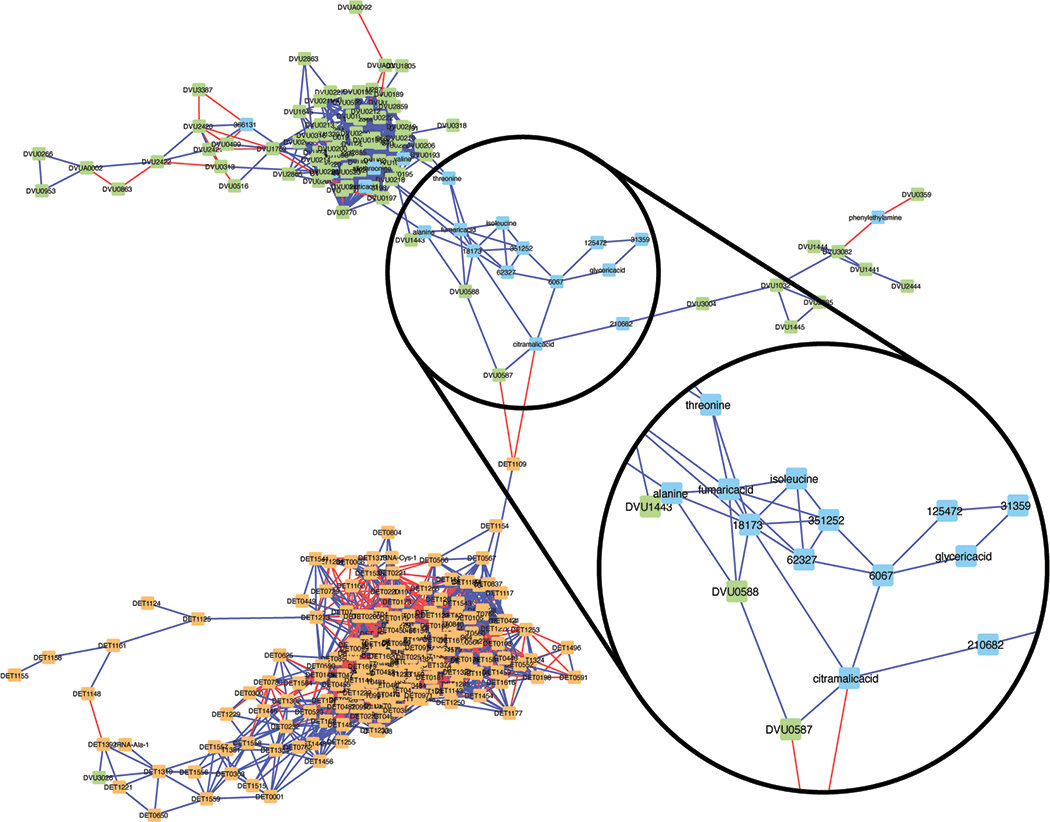
Visualized correlation network of significantly differentially abundant transcripts and metabolites developed using Random Matrix Theory via MENA. Orange nodes represent Dhc195 transcripts, green nodes represent DvH transcripts, and blue nodes represent metabolites. Blue edges represent a positive correlation and red edges represent a negative correlation. An interesting cluster of known and unknown metabolites that bridge the Dhc195 and DvH transcript networks is highlighted which includes known amino acids and organic acids as well as unknown metabolites from untargeted metabolomics. These interactions were used to identify key metabolic pathways and inform potential candidates for untargeted metabolite identification and subsequent verification.
Targeted metabolomics for validation of significantly differing untargeted metabolite spectra
To determine if any of the potential metabolite candidates above were present in significantly different extracellular abundances due to the addition of 20 μM As(III), the experiment was repeated and supernatant analyzed using LC/MS/MS with analytical standards of the unknown metabolite candidates for positive identification. Of the candidates, both sarcosine and l-ornithine were identified to be present in the supernatant of arsenic-free conditions in higher abundances as compared to cultures containing 20 μM As(III) (Figure S5), indicating cellular uptake or retention of these amino acids in Dhc195/DvH under arsenic stress. Sarcosine and l-ornithine are central intermediates in broader amino acid metabolism, supporting the hypothesis that amino acids and central carbon pathway intermediates are key metabolites in robust TCE dechlorination under arsenic stress. We previously reported that amendment of amino acids to Dhc195 in pure culture improved cell growth in As(III)-amended cultures24, helping it to overcome As(III) toxicity, also in line with the results of this study.
This study demonstrates that growth and robustness of Dhc195 in the presence of As(III) in concentrations ranging from 5 to 30 μM is augmented by the presence of supporting organism DvH. In the company of syntrophic partner DvH, Dhc195 is able to regain TCE dechlorination function under As(III) stress. It is possible that DvH exports nutrients and amino acids such as sarcosine, l-ornithine, lactamide, and glycerone based on the metabolomics results and the upregulation of permease proteins under As(III) stress. Dhc195 may, in turn, import these nutrients based on the upregulation of ABC transporters. The upregulation of motility genes in DvH may further augment this beneficial interaction by bringing the nutrients spatially closer to non-motile Dhc195. These results suggest the interactions between Dhc195 and DvH extend beyond hydrogen transfer and other essential exchanges, especially under toxicity stress.
Implications
Engineering solutions for TCE and arsenic co-contaminated sites may include bioaugmentation with Dehalococcoides consortia containing DvH, as the results of this study indicate DvH may export nutrients and amino acids (e.g., sarcosine, l-ornithine, lactamide, glycerone) under arsenic stress when cultured with a syntrophic partner. Since addition of amino acids is not a feasible biostimulation strategy in the field, other solutions and future research directions may include pretreatment of co-contaminated sites to prevent arsenic from entering the aqueous phase. Future studies should also continue to investigate microbial community interactions utilizing tools such as stable-isotope probing and biorthogonal noncanonical amino acid tagging to further elucidate these complex interactions among members of microbial communities.
Supplementary Material
Synopsis:
Co-culture interactions ameliorate arsenic toxicity stress, as suggested by phenotypic and multi-omics analysis.
Acknowledgements
This work was funded by NIEHS grant P42ES004705. This work used the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, supported by NIH S10 OD018174 Instrumentation Grant. Thank you to the West Coast Metabolomics Center for metabolomics services and Ray Keren, Scott Miller, and Aidan Cecchetti for manuscript edits.
Footnotes
Supporting Information
Transcriptomic and metabolomics signal intensity tables, supporting plots, and detailed methods and materials as referenced herein can be found in the Supporting Information.
Conflict of interest
The authors declare no competing interests.
References
- 1.Fildes P Production of tryptophan by Salmonella typhi and Escherichia coli. Microbiology 15, 636–642 (1956). [DOI] [PubMed] [Google Scholar]
- 2.Selwyn SC & Postgate JR A search for the Rubentschikii group of Desulphovibrio. Antonie Van Leeuwenhoek 25, 465–472 (1959). [DOI] [PubMed] [Google Scholar]
- 3.Shou W, Ram S & Vilar JMG Synthetic cooperation in engineered yeast populations. Proc. Natl. Acad. Sci 104, 1877–1882 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harcombe W Novel cooperation experimentally evolved between species. Evol. Int. J. Org. Evol 64, 2166–2172 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Hom EFY & Murray AW Niche engineering demonstrates a latent capacity for fungal-algal mutualism. Science (80). 345, 94–98 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ponomarova O & Patil KR Metabolic interactions in microbial communities: untangling the Gordian knot. Curr. Opin. Microbiol 27, 37–44 (2015). [DOI] [PubMed] [Google Scholar]
- 7.McNally L, Viana M & Brown SP Cooperative secretions facilitate host range expansion in bacteria. Nat. Commun 5, 1–8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kruse S, Türkowsky D, Birkigt J, Matturro B, Franke S, Jehmlich N, von Bergen M, Westermann M, Rossetti S, Nijenhuis I, Adrian L, Diekert G & Goris T Interspecies metabolite transfer and aggregate formation in a co-culture of Dehalococcoides and Sulfurospirillum dehalogenating tetrachloroethene to ethene. ISME J. 15, 1794–1809 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chau ATT, Lee M, Adrian L & Manefield MJ Syntrophic Partners Enhance Growth and Respiratory Dehalogenation of Hexachlorobenzene by Dehalococcoides mccartyi Strain CBDB1. Frontiers in Microbiology vol. 9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maphosa F, Lieten S, Dinkla I, Stams A, Smidt H & Fennell D Ecogenomics of microbial communities in bioremediation of chlorinated contaminated sites. Frontiers in Microbiology vol. 3 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mao X, Polasko A & Alvarez-Cohen L Effects of Sulfate Reduction on Trichloroethene Dechlorination by Dehalococcoides-Containing Microbial Communities. Appl. Environ. Microbiol 83, e03384–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He J, Holmes VF, Lee PKH & Alvarez-Cohen L Influence of Vitamin B12 and Cocultures on the Growth of Dehalococcoides Isolates in Defined Medium. Appl. Environ. Microbiol 73, 2847–2853 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Men Y, Feil H, Verberkmoes NC, Shah MB, Johnson DR, Lee PKH, West KA, Zinder SH, Andersen GL, & Alvarez-Cohen L Sustainable syntrophic growth of Dehalococcoides ethenogenes strain 195 with Desulfovibrio vulgaris Hildenborough and Methanobacterium congolense: global transcriptomic and proteomic analyses. ISME J. 6, 410–421 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhuang W-Q, Yi S, Bill M, Brisson VL, Feng X, Men Y, Conrad ME, Tang YJ & Alvarez-Cohen L Incomplete Wood–Ljungdahl pathway facilitates one-carbon metabolism in organohalide-respiring Dehalococcoides mccartyi. Proc. Natl. Acad. Sci 111, 6419–6424 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao L, Lu X, Polasko A, Johnson NW, Miao Y, Yang Z, Mahendra S & Gu B Co-contaminant effects on 1, 4-dioxane biodegradation in packed soil column flow-through systems. Environ. Pollut 243, 573–581 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Harding-Marjanovic KC ,Yi S, Weathers TS, Sharp JO, Sedlak DL, Alvarez-Cohen L Effects of Aqueous Film-Forming Foams (AFFFs) on Trichloroethene (TCE) Dechlorination by a Dehalococcoides mccartyi-Containing Microbial Community. Environ. Sci. Technol 50, 3352–3361 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Jasmann JR, Gedalanga PB, Borch T, Mahendra S & Blotevogel J Synergistic Treatment of Mixed 1,4-Dioxane and Chlorinated Solvent Contaminations by Coupling Electrochemical Oxidation with Aerobic Biodegradation. Environ. Sci. Technol 51, 12619–12629 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Gushgari-Doyle S, Oremland RS, Keren R, Baesman SM, Akob DM, Banfield JF & Alvarez-Cohen L Acetylene-Fueled Trichloroethene Reductive Dechlorination in a Groundwater Enrichment Culture. MBio 12, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mao X, Oremland RS, Liu T, Gushgari S, Landers AA, Baesman SM & Alvarez-Cohen L Acetylene Fuels TCE Reductive Dechlorination by Defined Dehalococcoides/Pelobacter Consortia. Environ. Sci. Technol 51, 2366–2372 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding C, Rogers MJ & He J Dehalococcoides mccartyi Strain GEO12 Has a Natural Tolerance to Chloroform Inhibition. Environ. Sci. Technol 54, 8750–8759 (2020). [DOI] [PubMed] [Google Scholar]
- 21.U.S.E.P.A. Superfund: National Priorities List (NPL). (2017).
- 22.Risher J, Odin M & Osier M Interaction profile for: Arsenic, Hydrazines, Jet Fuels, Strontium-90, and Trichloroethylene. (2004). [PubMed] [Google Scholar]
- 23.U.S. Environmental Protection Agency, R. 8. Final Close-out Report Midvale Slag Superfund Site Midvale, Utah. https://semspub.epa.gov/work/08/1558757.pdf (2014). [Google Scholar]
- 24.Gushgari-Doyle S & Alvarez-Cohen L Effects of Arsenic on Trichloroethene–Dechlorination Activities of Dehalococcoides mccartyi 195. Environ. Sci. Technol 54, 1276–1285 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He J, Holmes VF, Lee PKH & Alvarez-Cohen L Influence of vitamin B12 and cocultures on the growth of Dehalococcoides isolates in defined medium. Appl. Environ. Microbiol 73, 2847–2853 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He J, Ritalahti KM, Aiello MR & Löffler FE Complete Detoxification of Vinyl Chloride by an Anaerobic Enrichment Culture and Identification of the Reductively Dechlorinating Population as a Dehalococcoides Species. Appl. Environ. Microbiol 69, 996 LP–1003 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He J, Ritalahti KM, Yang K-L, Koenigsberg SS & Löffler FE Detoxification of vinyl chloride to ethene coupled to growth of an anaerobic bacterium. Nature 424, 62–65 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Wolin EA, Wolin MJ & Wolfe RS FORMATION OF METHANE BY BACTERIAL EXTRACTS. J. Biol. Chem 238, 2882–2886 (1963). [PubMed] [Google Scholar]
- 29.Mukhopadhyay A, He Z, Alm EJ, Arkin AP, Baidoo EE, Borglin SC, Chen W, Hazen TC, He Q & Holman H -Y. Salt stress in Desulfovibrio vulgaris Hildenborough: an integrated genomics approach. J. Bacteriol 188, 4068–4078 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson DR, Lee PKH, Holmes VF & Alvarez-Cohen L An internal reference technique for accurately quantifying specific mRNAs by real-time PCR with application to the tceA reductive dehalogenase gene. Appl. Environ. Microbiol 71, 3866–3871 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mao X, Polasko A & Alvarez-Cohen L Effects of sulfate reduction on trichloroethene dechlorination by Dehalococcoides-containing microbial communities. Appl. Environ. Microbiol 83, e03384–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhuang W-Q, Yi S, Feng X, Zinder SH, Tang YJ & Alvarez-Cohen L Selective utilization of exogenous amino acids by Dehalococcoides ethenogenes strain 195 and its effects on growth and dechlorination activity. Appl. Environ. Microbiol 77, 7797–7803 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joshi NA & Fass JN Sickle: A sliding-window, adaptive, quality-based trimming tool for FastQ files (Version 1.33). [Software] Available at https://github.com/najoshi/sickle. [Google Scholar]
- 34.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M & Gingeras TR STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson MD, McCarthy DJ & Smyth GK edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ritchie ME, Phipson B, Wu DI, Hu Y, Law CW, Shi W & Smyth GK limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47–e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fiehn O, Wohlgemuth G, Scholz M, Kind T, Lee DY, Lu Y, Moon S & Nikolau B Quality control for plant metabolomics: reporting MSI-compliant studies. Plant J. 53, 691–704 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Spearman C The proof and measurement of association between two things. (1961). [DOI] [PubMed] [Google Scholar]
- 39.Deng Y, Jiang Y-H, Yang Y, He Z, Luo F & Zhou J Molecular ecological network analyses. BMC Bioinformatics 13, 1–20 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B & Ideker T Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Men Y, Lee PKH, Harding KC & Alvarez-Cohen L Characterization of four TCE-dechlorinating microbial enrichments grown with different cobalamin stress and methanogenic conditions. Appl. Microbiol. Biotechnol 97, 6439–6450 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reuter W, Davidowski L, Neubaur K & Di Bussolo J Speciation of five arsenic compounds in urine by HPLC/ICP-MS, Application Note. (2003). [Google Scholar]
- 43.Mao X, Stenuit B, Polasko A & Alvarez-Cohen L Efficient Metabolic Exchange and Electron Transfer within a Syntrophic Trichloroethene-Degrading Coculture of Dehalococcoides mccartyi 195 and Syntrophomonas wolfei. Appl. Environ. Microbiol 81, 2015–2024 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson MS, Zhulin IB, Gapuzan ME & Taylor BL Oxygen-dependent growth of the obligate anaerobe Desulfovibrio vulgaris Hildenborough. J. Bacteriol 179, 5598–5601 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He Q, Huang KH, He Z, Alm EJ, Fields MW, Hazen TC, Arkin AP, Wall JD & Zhou J Energetic consequences of nitrite stress in Desulfovibrio vulgaris Hildenborough, inferred from global transcriptional analysis. Appl. Environ. Microbiol 72, 4370–4381 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Redding AM, Mukhopadhyay A, Joyner DC, Hazen TC & Keasling JD Study of nitrate stress in Desulfovibrio vulgaris Hildenborough using iTRAQ proteomics. Brief. Funct. Genomics 5, 133–143 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Stolyar S, He Q, Joachimiak MP, He Z, Yang ZK, Borglin SE, Joyner DC, Huang K, Alm EJ & Hazen TC Response of Desulfovibrio vulgaris to alkaline stress. J. Bacteriol 189, 8944–8952 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Silver S & Phung LT Genes and enzymes involved in bacterial oxidation and reduction of inorganic arsenic. Appl. Environ. Microbiol 71, 599–608 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lloyd JR & Oremland RS Microbial transformations of arsenic in the environment: from soda lakes to aquifers. Elements 2, 85–90 (2006). [Google Scholar]
- 50.Lin Y-F, Walmsley AR & Rosen BP An arsenic metallochaperone for an arsenic detoxification pump. Proc. Natl. Acad. Sci 103, 15617–15622 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salotra P, Singh DK, Seal KP, Krishna N, Jaffe H & Bhatnagar R Expression of DnaK and GroEL homologs in Leuconostoc esenteroides in response to heat shock, cold shock or chemical stress. FEMS Microbiol. Lett 131, 57–62 (1995). [DOI] [PubMed] [Google Scholar]
- 52.Parvatiyar K, Alsabbagh EM, Ochsner UA, Stegemeyer MA, Smulian AG, Hwang SH, Jackson CR, McDermott TR & Hassett DJ Global analysis of cellular factors and responses involved in Pseudomonas aeruginosa resistance to arsenite. J. Bacteriol 187, 4853–4864 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Daware V, Kesavan S, Patil R, Natu A, Kumar A, Kulkarni M & Gade W Effects of arsenite stress on growth and proteome of Klebsiella pneumoniae. J. Biotechnol 158, 8–16 (2012). [DOI] [PubMed] [Google Scholar]
- 54.Knowles FC & Benson AA The biochemistry of arsenic. Trends Biochem. Sci 8, 178–180 (1983). [Google Scholar]
- 55.Klemperer NS & Pickart CM Arsenite inhibits two steps in the ubiquitin-dependent proteolytic pathway. J. Biol. Chem 264, 19245–19252 (1989). [PubMed] [Google Scholar]
- 56.Bruton CJ & Hartley BS Sub-unit structure and specificity of methionyl-transfer-ribonucleic acid synthetase from Escherichia coli. Biochem. J 108, 281–288 (1968). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Iaccarino M & Berg P Requirement of sulfhydryl groups for the catalytic and tRNA recognition functions of isoleucyl-tRNA synthetase. J. Mol. Biol 42, 151–169 (1969). [DOI] [PubMed] [Google Scholar]
- 58.Lawrence FJ Studies on Methionyl Transfer RNA Synthetase from Escherichia coli K12 Amino Acid Composition and Relation of Sulfhydryl Groups to Enzyme Activities: Amino Acid Composition and Relation of Sulfhydryl Groups to Enzyme Activities. Eur. J. Biochem 15, 436–441 (1970). [DOI] [PubMed] [Google Scholar]
- 59.Piper PW, Ortiz-Calderon C, Holyoak C, Coote P & Cole M Hsp30, the integral plasma membrane heat shock protein of Saccharmyces cerevisiae, is a stress-inducible regulator of plasma membrane H+-ATPase. Cell Stress Chaperones 2, 12 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang S, Phillippy AM, Deng K, Rui X, Li Z, Tortorello ML & Zhang W Transcriptomic responses of Salmonella enterica serovars Enteritidis and Typhimurium to chlorine-based oxidative stress. Appl. Environ. Microbiol 76, 5013–5024 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Volkers RJM, De Jong AL, Hulst AG, Van Baar BLM, De Bont JAM & Wery J Chemostat-based proteomic analysis of toluene-affected Pseudomonas putida S12. Environ. Microbiol 8, 1674–1679 (2006). [DOI] [PubMed] [Google Scholar]
- 62.Schulze-Gahmen U, Pelaschier J, Yokota H, Kim R & Kim S Crystal structure of a hypothetical protein, TM841 of Thermotoga maritima, reveals its function as a fatty acid–binding protein. Proteins Struct. Funct. Bioinforma 50, 526–530 (2003). [DOI] [PubMed] [Google Scholar]
- 63.Broussard TC, Miller DJ, Jackson P, Nourse A, White SW & Rock CO Biochemical roles for conserved residues in the bacterial fatty acid-binding protein family. J. Biol. Chem 291, 6292–6303 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kanehisa M & Goto S KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hemme CL & Wall JD Genomic insights into gene regulation of Desulfovibrio vulgaris Hildenborough. Omi. a J. Integr. Biol 8, 43–55 (2004). [DOI] [PubMed] [Google Scholar]
- 66.Tang Y, Francesco P, Mukhopadhyay A, Phan R, Hazen TC & Keasling JD Pathway Confirmation and Flux Analysis of Central Metabolic Pathways in Desulfovibrio vulgaris Hildenborough using Gas Chromatography-Mass Spectrometry and Fourier Transform-Ion Cyclotron Resonance Mass Spectrometry. J. Bacteriol 189, 940–949 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ouellette M, Makkay AM & Papke RT Dihydroxyacetone metabolism in Haloferax volcanii. Front. Microbiol 4, 376 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wendt R & Gosting L The diffusion coefficient of lactamide in dilute aqueous solutions at 25 as measured by the Gouy diffusiometer. J. Phys. Chem 63, 1287–1291 (1959). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


