Abstract
A new subgroup of avian leukosis virus (ALV), designated subgroup J, was identified recently. Viruses of this subgroup do not cross-interfere with viruses of the avian A, B, C, D, and E subgroups, are not neutralized by antisera raised against the other virus subgroups, and have a broader host range than the A to E subgroups. Sequence comparisons reveal that while the subgroup J envelope gene includes some regions that are related to those found in env genes of the A to E subgroups, the majority of the subgroup J gene is composed of sequences either that are more similar to those of a member (E51) of the ancient endogenous avian virus (EAV) family of proviruses or that appear unique to subgroup J viruses. These data led to the suggestion that the ALV-J env gene might have arisen by multiple recombination events between one or more endogenous and exogenous viruses. We initiated studies to investigate the origin of the subgroup J envelope gene and in particular to determine the identity of endogenous sequences that may have contributed to its generation. Here we report the identification of a novel family of avian endogenous viruses that include env coding sequences that are over 95% identical to both the gp85 and gp37 coding regions of subgroup J viruses. We call these viruses the ev/J family. We also report the isolation of ev/J-encoded cDNAs, indicating that at least some members of this family are expressed. These data support the hypothesis that the subgroup J envelope gene was acquired by recombination with expressed endogenous sequences and are consistent with acquisition of this gene by only one recombination event.
In 1991, Payne and colleagues reported the isolation of new nonacute transforming avian retroviruses that exhibited a novel subgroup specificity designated subgroup J (1, 4, 5, 32–35). Biological assays revealed that subgroup J viruses differed from the previously characterized avian A to E virus subgroups in terms of patterns of viral interference, cross-neutralization, and host range (4, 34). The subgroup specificity of avian retroviruses is determined by the surface (SU) envelope protein gp85 (4, 9, 10, 17, 34). The second protein encoded by env is the transmembrane (TM) protein, or gp37, which serves to anchor gp85 to the membrane (reviewed in reference 28). Sequence comparisons of env genes of the prototype avian leukosis virus subgroup J (ALV-J) strain HPRS-103 as well as additional ALV-J isolates revealed that the subgroup J gp85 protein, which is the primary determinant of subgroup specificity, showed only a 40% overall level of identity to the gp85 genes of the subgroup A to E viruses (4, 5, 7, 42). This is in contrast to the A to E subgroups, which are over 85% identical to each other, differing primarily in hypervariable and variable regions that determine subgroup specificity and neutralization patterns (9, 10, 17, 41). As reported several years ago, however, the subgroup J env gene does include sequences that are highly related to a member of the ancient endogenous avian virus (EAV) family called E51 (11, 12, 18), as well as other regions that appear unique to subgroup J viruses. These data led to the suggestion that the subgroup J envelope gene might have been generated by multiple recombination events between one or more exogenous and endogenous viruses. Studies in this report were therefore initiated to investigate the origin of the subgroup J envelope gene and particularly to define the potential endogenous virus parent(s) that might have contributed to this gene.
Three major families of endogenous viruses have been identified in chickens. The best characterized are the evs (endogenous viruses). Twenty-one of these proviruses have been identified and characterized in White Leghorn chickens (3, 14, 27, 38). Some of the evs, such as ev-3 and ev-6, are expressed and produce functional envelope glycoproteins, while others, such as ev-1, are classified as transcriptionally silent (6, 23, 24). In addition, several nondefective evs such as ev-2 have been identified that can give rise to infectious virus. All evs that include an env gene exhibit a common subgroup specificity designated E. Sequence comparisons reveal that the E subgroup is as highly related to the A to D subgroups of exogenous viruses as these exogenous virus subgroups are to each other, showing an overall level of identity of 85 to 90%. While most lines of chickens contain several evs, animals have been bred to lack all copies of this family of endogenous viruses (2). These animals are referred to as ev-0 lines.
The second class of endogenous viruses are the ancient EAVs (11, 12, 18). These proviruses were originally identified by low-stringency hybridization of ev-0 cell DNA with avian leukosis-sarcoma virus (ALSV) probes which revealed the presence of approximately 50 copies of EAV per genome in many avians, including the domestic White Leghorn chicken as well as the progenitor of the domestic chicken, red jungle fowl. The EAV family has not been fully characterized, but appears to be diverse. Sequence comparisons of different members of this family (which include EAV-0 proviruses and E51-related proviruses) reveal variability in long terminal repeat (LTR) length and sequence composition. Although diverse, all of the EAV proviruses that have been characterized are defective. In fact, only one member, called E51, contains what appears to be an intact env gene; all others include deletions of part or all of SU. Although grossly intact, sequence analysis of E51 demonstrated that it contains deletions and point mutations throughout env. Thus, even though some EAVs are expressed, there is no evidence that they give rise to functional viral envelope proteins, and their subgroup specificity is therefore unknown.
The third class of endogenous avian viruses are the avian retrotransposons (ART-CH) (22, 30). These elements, also present at approximately 50 copies per genome, are relatively small elements (about 3 kbp in length) that include short regions that show similarity to the ALSV genome. The ART-CH elements are transcriptionally active and include a potentially translated portion of the gag gene, although no protein product encoded by these elements has been described.
To investigate the potential endogenous virus origin of the subgroup J envelope gene, we generated probes specific for the gp85 and gp37 regions of env by using an infectious molecular clone of a U.S. field isolate of a subgroup J virus (ADOL-R5-4) (7, 19, 20). These probes allowed us to isolate and characterize members of a novel family of avian endogenous viruses that we call the ev/J viruses that show over 95% identity to the subgroup J envelope gene in infectious ALV-J isolates. We also report the isolation of ev/J-encoded cDNAs. Based on these data, we propose that the subgroup J envelope gene was acquired entirely from these novel endogenous sequences.
MATERIALS AND METHODS
Oligonucleotide primers, PCRs, and cloning PCR products.
One set of primers was generated to amplify sequences within the transmembrane portion of the ALV-J gp37 gene that is unique to the subgroup J env gene. This region is defined by residues 6796 and 7015 in the published HPRS-103 ALV-J sequence (4). The sequences of the upstream and downstream oligonucleotides are 5′-ccctcgagTTTACGCGCACGTTTG-3′ and 5′-cgctcgagCCCGTCACATCGCGTTC, respectively. The sequences in lowercase represent additional nucleotides added to the 5′ ends of each oligonucleotide that included an XhoI site to facilitate subcloning final PCR products. The SU-specific primers (which also included the terminal XhoI site) were bounded by sequences 6013 and 6182 in the HPRS-103 sequence and were 5′-ccctcgagTTCACCAGTAACGAG-3′ and 5′-cgctcgaGTAAACCCATATG-3′. Another set of primers were designed to amplify intact proviruses that included ALV-J-related envelope genes. These oligonucleotides were synthesized based on partial sequence analysis of the LTRs of cDNA clones described below and included NotI restriction sites on their 5′ ends. The upstream and downstream LTR primer sequences were 5′-atgcggccgcTTCGTGATTGGAGGAAACACTTG-3′ and 5′-atgcggccgcGTTACACTTGGCACACAAAGGTGGCATAAC-3′, respectively.
The TM and SU primers described above were used in PCRs with either the cloned ALV-J isolate (ADOL-R5-4) (7, 19, 20) as a template (to generate probes for Southern blot analysis) or with genomic DNA from ev-0 chicken cells (to clone regions of the ev/J proviruses). Reactions were conducted under similar conditions, included 20 pmol of each primer and 100 ng of purified template DNA, and were conducted according to the manufacturers’ recommendations. The cycle conditions were 95°C for 5 min, followed by 20 to 40 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min, after which time samples were incubated at 72°C for 5 min before holding at 4°C. In all cases, PCR products were gel purified before further manipulations. In the case of the TM and SU probes for Southern blot analysis, PCR products were digested with XhoI, self-ligated, and labeled by nick translation as described previously (16). For subcloning, PCR products were digested with XhoI and cloned into the XhoI site of pBluescript. Subsequent propagation and sequencing were conducted essentially as described previously.
The LTR primers were used in PCRs with ev-0 genomic DNA to obtain products that included intact ev/J proviruses. The reaction conditions for this assay were 95°C for 5 min, followed by 40 cycles of 95°C for 1 min, 60°C for 1 min, and 72°C for 12 min, after which time samples were incubated at 72°C for 10 min before being held at 4°C. PCR products were then digested with NotI, gel purified, ligated into the NotI site of pBluescript (Stratagene), and sequenced. The env gene sequence of one clone (4-1) is included in Fig. 4.
FIG. 4.
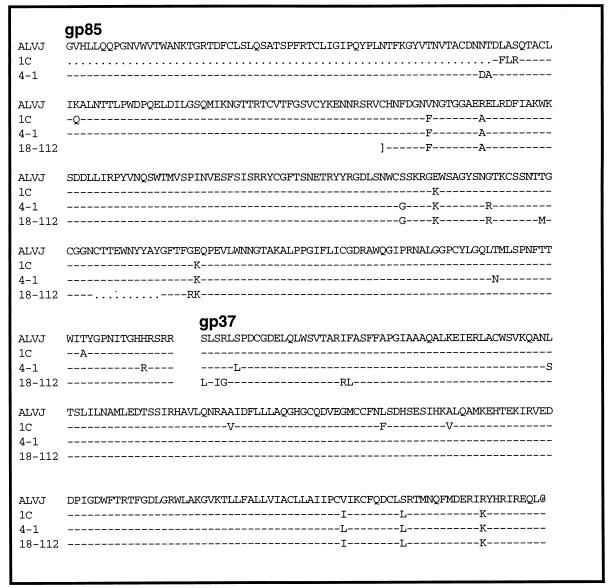
Predicted amino acid sequences of the ALV and ev/J env gene products. Endogenous env genes were sequenced from a phage clone isolated from a genomic library (1-C), a PCR product generated with ev/J-specific LTR primers (clone 4-1), and a c-DNA clone (18-112). The methods used to obtain each type of clone are described in Materials and Methods. The gap in the sequence denotes the boundary between gp85 and gp37. Dashes indicate identity with the ALV-J sequence, brackets define the boundary of sequenced regions, and dots indicate deletions in clones relative to ALV-J.
Southern blot analysis of ev-0 DNA.
Three samples of ev-0 DNA were obtained for genomic Southern blot analysis. One of these DNAs was purified from a continuous line of ev-0 fibroblasts (DF-1) cells, kindly provided by Doug Foster, University of Minnesota. The other two ev-0 DNAs were obtained from erythrocyte DNA of two White Leghorn chickens maintained at the Poultry Research Facility at the University of Minnesota. Isolation, restriction digestion, and Southern blot analysis of genomic DNA were performed essentially as described previously (15).
Identification and isolation of ev/J genomic and cDNA clones.
To identify genomic sequences that included potential ev/J proviruses, a chicken genomic library (15) was screened with an ALV-J SU-specific probe generated with the SU primers described above. Of 4 × 105 plaques screened, 19 positive clones were detected and plaque purified. Preliminary characterization of DNA purified from these clones indicates they include at least two categories of proviruses that differ in length. The env gene of one of these clones (1C) was completely sequenced and is shown in Fig. 4. As indicated, it includes a deletion of the amino-terminal end of SU.
To isolate ev/J-encoded cDNAs, we obtained a cDNA library from Sharon Soodeen-Karamath and Ann Gibbins at the University of Guelph that had been constructed from RNA isolated from 48-h-old White Leghorn chicken embryos. Of approximately 106 plaques screened with the 220 ALV-J-specific TM probe, 3 positive clones were obtained, purified, and sequenced. None of the clones analyzed contained a full-length env coding region, and although virtually identical in sequence, each clone ended at different locations within env. The sequence of the longest of these clones (18-112) is shown in Fig. 4.
The conditions for generation and cloning of the ev/J proviruses from PCR products are described above. The sequence of the most complete product, which includes an intact SU and TM coding region (4-1) is included in Fig. 4.
Nucleotide sequence accession number.
The GenBank accession no. for the sequences presented here are AF082078 (ev/J clone 1C), AF082079 (ev/J clone 4-1), and AF082080 (ev/J cDNA clone 18-112).
RESULTS
Comparison of the subgroup J envelope gene with those of other virus subgroups.
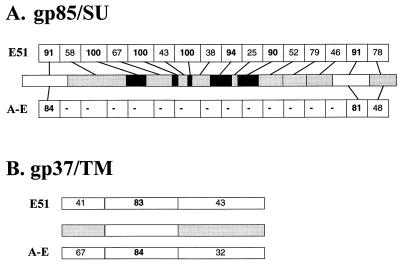
The line drawing in Fig. 1 depicts the ALV-J gp85 (Fig. 1A) and gp37 (Fig. 1B) proteins and the percent identity of discrete regions of these proteins relative to those of previously identified ALSV Env proteins. As shown, the ALV-J gp85 protein is similar to the gp85 proteins of the subgroup A to E viruses only in the amino-terminal 43 amino acids where it shows 84% identity and in two regions near the carboxy terminus which show 81 and 48% identity; other regions of the subgroup J gp85 protein are unrelated to subgroup A to E env proteins as judged by both nucleotide and amino acid-based search programs (4, 5, 7, 42). This is in contrast to the A to E gp85 proteins, which show approximately 85% similarity to each other. The ALV-J gp37 (TM) protein shows an overall identity of approximately 65% to those of the A to E subgroups. However, sequence alignments reveal that this identity is not evenly distributed throughout the protein. Instead, a high degree of identity (84%) between the ALV-J and subgroup A to E TM proteins is evident in the central portion of the protein, while the amino-terminal end and the carboxy-terminal end (which encodes the transmembrane portion of the protein) exhibit 67 and 32% identity, respectively. Thus, while discrete regions of the subgroup J gp85 and gp37 proteins are clearly related to those of the common A to E virus subgroups, the subgroup J proteins are composed largely of sequences unrelated to these previously characterized env genes.
FIG. 1.
The SU and TM proteins of ALV-J include regions that are unique, regions that are similar to those of an ancient endogenous virus (E51), and regions related to those of other ALVs. The diagrams depict the predicted amino acid sequence of the gp85/SU (A) and gp37/TM (B) proteins of ALV-J compared with those of the ancient endogenous virus E51 and of the ALV A to E subgroups. White boxes depict regions that are common (over 80% identity) between all three env proteins, black boxes depict regions that are at least 90% identical between E51 and ALV-J Env, and shaded boxes depict regions that are less than 80% identical between any of the virus types.
These data are in contrast to results obtained when the gp85 protein of ALV-J is compared with the predicted amino acid sequence of gp85 encoded by the ancient EAV E51 (11, 12, 18). The E51 provirus is the only member of the ancient family of EAVs that includes a full-length env gene (11). The subgroup specificity of the E51 env gene is unknown, however, since it contains small deletions and stop codons and is therefore unable to encode a functional envelope glycoprotein. Our comparisons demonstrate that there are seven discrete regions within gp85 of between 6 and 28 amino acids that show 90% or more identity between ALV-J and E51 (this alignment requires correction of the several stop codons and small deletions in the E51 coding region). These regions of high homology, however, are interspersed with regions that show significantly less similarity to E51 (between 25 and 79% identity [Fig. 1]). Similarly, while the central portion of the subgroup J gp37 protein shows an 83% level of identity to E51, the amino and transmembrane portions of the protein are only approximately 40% identical to the ancient EAV sequence (Fig. 1B). Together, these data demonstrate that the subgroup J env gene includes sequences related to those of other subgroups of ALSVs, regions that are highly related to E51, and still other regions that appear unique to the ALV-J env gene. These data suggest either that the ALV-J env gene might have been generated by multiple recombination events between one or more endogenous and exogenous viruses, at least one of which has yet to be identified, or that it was acquired from one currently unidentified source.
Detection of endogenous ALV-J env-specific sequences by Southern blot hybridization.
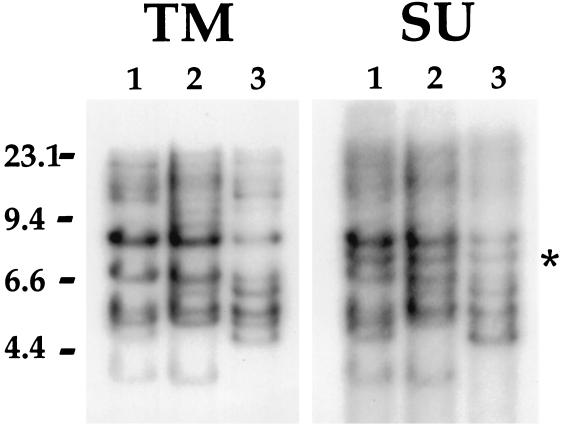
A previous report demonstrated the presence of ALV-J-related sequences in the chicken genome by Southern blot hybridization with a relatively large probe from the SU portion of the ALV-J env gene (5). To further explore the origin of the ALV-J env gene and to identify the potential endogenous virus component that might have contributed to its generation, we probed the chicken genome for ALV-J env gene-related sequences, concentrating initially on sequences within the transmembrane portion of the ALV-J gp37 TM coding region. As reported previously (5), this portion of the ALV-J TM coding protein (nucleotides 6796 to 7015 of the published HPRS-103 sequence) is composed of sequences that are unique to ALV-J; they show no homology to any sequence in the database at the nucleotide level and only an approximate 30 to 40% level of identity to other retroviral TM proteins at the amino acid level. We therefore reasoned that this sequence would be a useful reagent to identify J-related endogenous sequences. Subgroup J TM-specific primers were generated and used to amplify a DNA fragment corresponding to the transmembrane domain coding region by using the cloned ADOL-R5-4 isolate of ALV-J (7, 19, 20) as a template as described in Materials and Methods. The resultant 220-bp fragment was then purified, labeled, and used to probe Southern blots of EcoRI-digested genomic DNA. Results obtained by using DNA from three different ev-0 animals are shown in Fig. 2. As shown, between six and eight EcoRI bands were detected in each sample; additional, higher-molecular-weight bands were also variably present. Some bands were present in all three samples, while others were not. This variability is frequently seen with endogenous virus sequences in the genome and indicates the sequences are segregating in the population. Since the sequences used to probe this blot show no significant homology at the nucleotide level to any previously identified sequence in the database, these data suggest the presence of a novel family of avian endogenous viruses that might have been the source of at least a portion of the subgroup J env gene.
FIG. 2.
ALV-J-specific SU and TM probes detect endogenous sequences in Southern blots of ev-0 chicken genomic DNAs. Total genomic DNA isolated from three different ev-0 samples was purified, digested with EcoRI, and subjected to electrophoresis in 1% agarose gels. After Southern blotting, the filter was hybridized with probes generated by PCR by using the cloned isolate of ALV-J (ADOL-R5-4) as a template as described in Materials and Methods. (A) Results obtained with the 220-bp probe generated from the TM portion of the env gene. (B) Results obtained with the SU-specific probe. The filter was stripped between hybridizations and monitored to verify that the signal seen with the TM probe was removed before rehybridization with the SU probe. Lane 1, DF-1 DNA; lanes 2 and 3, erythrocyte DNA from two White Leghorn ev-0 chickens. As shown in panel A, the TM probe detects six clearly distinct EcoRI bands below 9.4 kbp in each sample; higher-molecular-weight fragments are variably present. Some bands are common to all samples, while others are unique to one sample. As shown in panel B, the SU probe detects the same fragments as were seen with the TM probe in addition to one unique fragment, marked by an asterisk. Numbers on the left are size markers in kilobase pairs.
To determine whether portions of the SU coding region might also detect this or another class of endogenous sequences, primers were used to generate a PCR product (spanning the region between positions 6013 and 6182 of the published HPRS-103 sequence [primers depicted as underlined sequences in Fig. 3B]) from the SU region of the ADOL-R5-4 cloned isolate of ALV-J (7, 19, 20) as described in Materials and Methods. This probe would not be expected to detect the E51 provirus, since it has only approximately 70% homology to E51, and this level of identity would not allow efficient hybridization under the stringent conditions used in these studies. Figure 2B shows the results obtained when the ALV-J SU probe was used as a hybridization probe against the same blot as shown in panel A. As shown, a virtually identical pattern of hybridizing bands was obtained; the only exception was the appearance of one additional band with the SU probe. This finding indicates that the ALV-J-related env sequences within the chicken genome include at least portions of both the TM and SU coding region. Together, these data support the hypothesis that the restriction fragments detected by the SU and TM probes identify a novel class of endogenous viruses in the chicken genome that are related to the ALV-J env gene.
FIG. 3.
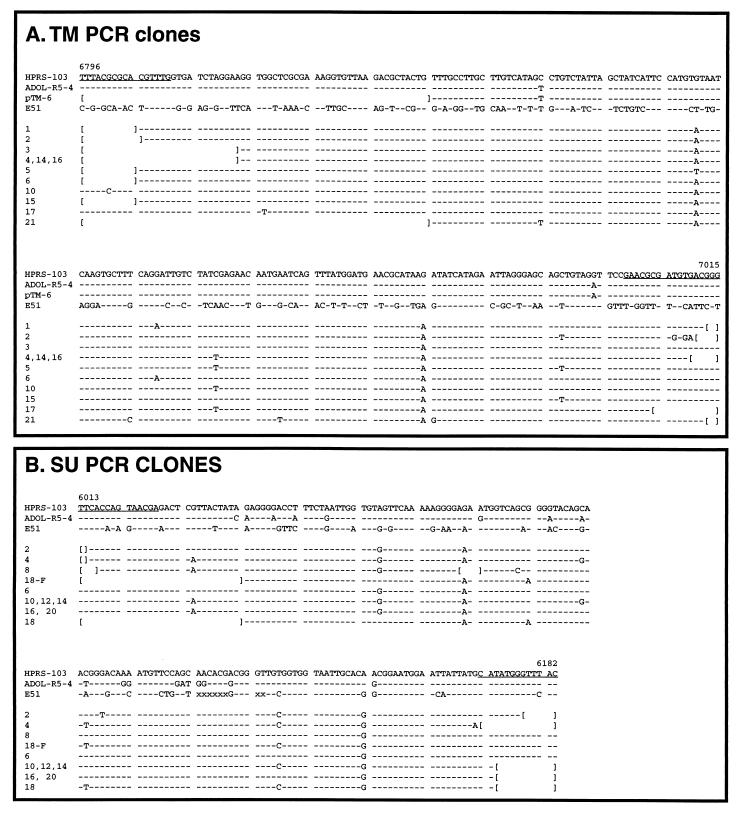
Sequence of endogenous ALV-J SU- and TM-related products amplified from ev-0 DNA. TM (A) and SU (B) sequences were amplified from DF-1 DNA by using the ALV-J-specific primers, cloned, and sequenced as described in Materials and Methods. The pTM6 clone is a subclone of the ADOL-R5-4 TM portion of the env gene that was amplified along with genomic DNA; the sequence obtained was identical to that of the original plasmid. Brackets indicate regions that were not sequenced, and dashes indicate sequence identity with the HPRS-103 sequence. Numbers above the HPRS-103 sequence are from reference 5. The underlined sequences define the oligonucleotides used in amplification reactions.
Analysis of subgroup J SU- and TM-related endogenous sequences.
The data shown above indicate that there are at least six different copies of the ALV-J env-related endogenous viruses in the chicken genome. However, Southern blot hybridization does not provide a detailed picture of the relationship between these endogenous sequences and the ALV-J env gene. In addition, the Southern blotting approach provides only a lower limit of the copy number of ALV-J env-related sequences, since if these sequences include conserved EcoRI sites, then multiple copies could give rise to a common-size restriction fragment. Therefore, to verify that these sequences represent a novel class of ALV-J env-related sequences and to utilize a different approach to estimate their copy number, the same TM- and SU-specific primers described above were used to directly amplify sequences from ev-0 chicken genomic DNA as described in Materials and Methods. The results obtained are shown in Fig. 3. As shown, 12 clones obtained after amplification with the TM primers were sequenced, 10 of which were unique (Fig. 3A). We believe that variations in these clones reflect variations in the genomic templates and are not due to PCR-induced mutations, since sequencing the product of a control amplification with the cloned ADOL-R5-4 isolate of ALV-J as a template yielded a product identical to the original cloned isolate (pTM-6 [Fig. 3A]). Comparison of these endogenous clones to the ALV-J env gene sequences shows that they are at least 97% identical to the TM domain of ALV-J env gene. This is in contrast to the E51 provirus, which shows only a 51% match to ALV-J within this region. Figure 3B shows that similar results were obtained with clones obtained after amplification with SU-specific primers. In this case, eight distinct sequences were obtained from sequencing a total of 11 clones. These clones showed at least 95% identity to the ALV-J env gene, while the E51 provirus shows only a 74% identity over the same region. Together, these data strongly support the hypothesis that at least the portions of the ALV-J env gene examined in these analyses are derived from a novel class of endogenous viruses that are over 95% identical to the ALV-J env genes of infectious viruses and that these endogenous sequences are present in approximately 10 copies in the avian virus genome.
Isolation and sequence analysis of intact endogenous subgroup J-related env genes.
Sequence analyses of the ancient EAVs revealed that most members contain large deletions within env; E51 was the only provirus identified that included the entire SU and TM coding regions (11). However, the E51 env gene is unable to encode a functional protein, since it contains stop codons and small deletions. To determine the content of the ALV-J env-related sequences, we cloned and sequenced endogenous ALV-J env-related sequences by three approaches as described in Materials and Methods. The first two approaches involved isolating phage clones that included ALV-J env-related sequences from a chicken genomic library and from a chicken cDNA library. Through characterization of the cDNA clones, we identified sequences specific for the LTRs of these ALV-J-related proviruses and used this information to generate primers to amplify intact proviral sequences from the genome by using PCR (a full description of these proviruses and the LTR sequences is in preparation). The results of sequencing the env genes obtained from each of these sources are shown in Fig. 4. Clone 1C represents a genomic clone isolated from a phage library; this clone contains a large deletion in the amino-terminal portion of SU, a feature shared by other clones obtained by this approach (data not shown). Clone 4-1 was obtained by PCR amplification of ev-0 DNA by using LTR primers; this clone includes intact SU and TM coding regions. Finally, the 18-112 clone represents a cDNA clone that includes an intact TM portion of the env gene and a partial copy of the SU region; the SU coding region includes a 10-amino-acid in-frame deletion. Although each of the clones analyzed has an overall distinct content, sequence analysis demonstrates that they all represent sequences with extremely high conservation to the ALV-J env gene. For example, the clones show only between a 2.5% (1C and 4-1) and a 4.1% (18-112) variation from the ALV-J env gene within the TM sequence, while all clones showed under a 4% variation within the SU sequence. These data clearly demonstrate that the ALV-J env gene present in infectious virus isolates likely arose from these endogenous sequences and could have in fact been acquired in one round of recombination.
DISCUSSION
The data presented in this report document the presence of a novel family of avian endogenous viruses that exhibit a high degree of sequence identity to the subgroup J envelope gene of infectious ALV-J virus isolates. Based on this similarity, we have designated these proviruses as the ev/J family. Our preliminary characterization of this family indicates that there are between 6 and 10 copies of ev/J proviruses per genome and that at least some of these elements are segregating in the population. The isolation of an ev/J-encoded cDNA indicates that at least some members of this family are expressed. We are currently conducting additional analyses to quantitate the number of these elements and to determine their diversity in terms of structure and sequence variability.
Analyses of the EAV family of endogenous viruses have demonstrated that the majority of proviruses contained large deletions in env that included most of the SU portion of the env gene (11). While the env gene of one EAV provirus (E51) appeared to be intact, sequence analysis demonstrated that it was also defective, since it contained multiple stop codons and small deletions. These results are in contrast to our data with the ev/J family. Southern blot hybridization analysis indicated that all EcoRI fragments that were detected with the ALV-J-specific TM probe were also detected by the ALV-J-specific SU probe, demonstrating that all env genes detected by this technique included at least a portion of SU sequence. In addition, of the 19 genomic phage clones that were originally detected with the SU probe, 17 also scored positive for the TM region, supporting the hypothesis that most of the ev/J env genes include both SU and TM regions (data not shown). Finally, sequence analysis of the 4-1 PCR clone demonstrated that this provirus contained an intact env gene, extending through both the SU and TM coding sequences. The predicted amino acid sequence of this gene differs in only 15 amino acids from that of the ALV-J env gene (an approximate 3% variation); 5 of these changes are conservative changes and 4 more are changes that are also seen in either the genomic phage env gene isolate or in the cDNA clone. While we have not yet isolated an intact cDNA clone, the finding that at least one of the ev/J family members appears to be expressed, together with the finding that at least one member (4-1) contains an intact env gene, supports the hypothesis that the subgroup J envelope gene could have been acquired by an exogenous virus in one recombination event.
The discovery of the ev/J family of proviruses is of interest for several reasons. The first is based on reports that subgroup J viruses, during the course of infection, give rise to antigenically distinct variants that exhibit differences in neutralization properties (35, 42). One mechanism that could give rise to this variation is continued recombination with different members of the ev/J family. While the degree of identity between env genes of infectious isolates and the different ev/J proviruses analyzed to date is high, it is possible that minor differences in env gene sequences between proviruses could affect the biological properties of the protein. Alternatively, it is possible that subgroup J env genes could be efficient substrates for recombination with expressed members of the EAV family of viruses, since they are highly homologous in specific portions of env. This mechanism has been described with feline leukemia virus (FeLV), where env gene variants are generated by recombination of exogenous virus with endogenous FeLV-related sequences (8, 13, 31, 36). Similarly, experiments with the murine system have demonstrated that recombination between ecotropic virus and endogenous sequences can generate virus with an extended host range that is often found in leukemic tissues (reviewed in reference 21). ALV-J provides an interesting system with which to investigate generation of genetic diversity, since it is undergoing high rates of change as it spreads through commercial chicken stocks. In addition, the availability of relatively homogeneous chicken lines that include only a limited number of endogenous ALV-J-related sequences provides an experimental system amenable to precise analyses.
The hypotheses that the ev/J family of proviruses provided the original subgroup J envelope gene by recombination and that these proviruses might be continuing to provide genetic variation to infectious virus by recombination requires that the ev/J proviruses be expressed and that their transcript(s) include packaging signals that mediate efficient incorporation into virions. These features are required, since recombination between retroviruses necessitates that the genomes of the different viruses be copackaged in one virion to generate a heterozygous particle (25, 26, 40). The finding of ev/J-encoded cDNAs indicates that these elements are expressed, at least in early embryo tissues, from which the cDNA library used in our analyses was constructed. However, we have not yet been able to convincingly demonstrate the presence of ev/J-encoded transcripts in RNA isolated from a chicken fibroblast cell line (DF-1 cells) by Northern analysis. This finding suggests that these proviruses are expressed at low levels in the DF-1 cells and/or that these transcripts are not efficiently polyadenylated; we are currently pursuing these possibilities. The failure to detect ev/J-encoded transcripts by Northern blotting analysis does not alter their potential importance as recombination substrates, however, since the ev-1 provirus, which is expressed at under one copy per cell in fibroblasts (6, 23, 24) has been shown to give rise to easily detectable recombinants with exogenous viruses (14).
In addition to providing a potential source of genetic diversity to infecting ALV-J, the presence of ALV-J-related endogenous envelope sequences could impact ALV-J-induced pathogenicity in additional ways. Most pathogenesis studies with ALV-J have been conducted with the prototype English strain HPRS-103 (32, 35, 37) and have demonstrated that this virus induces a high incidence of myeloid tumors in some lines of chickens. The finding that ALV-J can replicate in cultured cells from animals that are resistant to ALV-J-induced disease (4, 34) demonstrates that resistance to ALV-J in vivo is not explained by the absence of a viral receptor in resistant animals. Similar results have been obtained with lines of animals resistant to ALV-induced bursal tumors. Further analyses of lines of animals that are susceptible and resistant to ALV-J lymphomagenesis has demonstrated that resistant lines are able to mount an effective immune response to the virus; they have high titers of circulating neutralizing antibody and are able to clear the virus (32, 35, 37). In contrast, susceptible animals lack significant levels of circulating antibody and develop a persistent viremia and late-onset myeloid tumors (32, 35, 37). It is therefore possible that the expression of endogenous viruses in different lines of animals might affect disease progression, similar to results obtained in other systems. For example, endogenous expression of a functional envelope glycoprotein can be at least partially protective to superinfection by a virus of the same subgroup due to receptor blockage (16, 29). Alternatively, it has been suggested that the endogenous expression of even a truncated envelope protein could induce tolerance and thereby result in increased susceptibility to lymphomagenesis by infecting exogenous virus (39). Ongoing studies are being conducted to investigate ALV-J-induced pathogenesis and a potential role for ev/J sequences in this process.
ACKNOWLEDGMENTS
S.J.B. and B.L.R. contributed equally to this work.
We thank Sharon Soodeen-Karamath and Ann Gibbins at the University of Guelph for providing the cDNA library from which the ev/J-encoded cDNAs were isolated and Doug Foster at the University of Minnesota for his kind gift of DF-1 cells.
This work was supported by Public Health Service grant GM 41571 from the Institute of General Medical Sciences and by a grant from the Leukemia Research Fund. S.J.B. was supported by NIH training grant CA09138.
REFERENCES
- 1.Arshad S S, Bland A P, Hacker S M, Payne L N. A low incidence of histiocytic sarcomatosis associated with infection of chickens with the HPRS-103 strain of subgroup J avian leukosis virus. Avian Dis. 1997;41:947–956. [PubMed] [Google Scholar]
- 2.Astrin S M, Buss E G, Haywards W S. Endogenous viral genes are nonessential in the chicken. Nature. 1979;282:339–341. doi: 10.1038/282339a0. [DOI] [PubMed] [Google Scholar]
- 3.Astrin S M, Robinson H L, Crittenden L B, Buss E G, Wyban J, Hayward W S. Ten genetic loci in the chicken that contain structural genes for endogenous avian leukosis viruses. Cold Spring Harbor Symp Quant Biol. 1980;44:1105–1109. doi: 10.1101/sqb.1980.044.01.119. [DOI] [PubMed] [Google Scholar]
- 4.Bai J, Howes K, Payne L N, Skinner M A. Sequence of host-range determinants in the env gene of a full-length, infectious proviral clone of exogenous Avian Leukosis Virus HPRS-103 confirms that it represents a new subgroup (designated J) J Gen Virol. 1995;76:181–187. doi: 10.1099/0022-1317-76-1-181. [DOI] [PubMed] [Google Scholar]
- 5.Bai J, Payne L N, Skinner M A. HPRS-103 (exogenous avian leukosis virus, subgroup J) has an env gene related to those of endogenous elements EAV-0 and E51 and an E element found previously only in sarcoma viruses. J Virol. 1995;69:779–784. doi: 10.1128/jvi.69.2.779-784.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker B, Robison H, Varmus H E, Bishop J M. Analysis of endogenous avian retrovirus DNA and RNA: viral and cellular determinants of retrovirus gene expression. Virology. 1981;114:8–22. doi: 10.1016/0042-6822(81)90248-8. [DOI] [PubMed] [Google Scholar]
- 7.Benson S J, Ruis B L, Garbers A L, Fadly A M, Conklin K F. Independent isolates of the emerging subgroup J avian leukosis virus derive from a common ancestor. J Virol. 1998;72:1121–98. doi: 10.1128/jvi.72.12.10301-10304.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boomer S, Eiden M, Burns C C, Overbaugh J. Three distinct envelope domains, variably present in subgroup B feline leukemia virus recombinants, mediate Pit1 and Pit2 receptor recognition. J Virol. 1997;71:8116–8123. doi: 10.1128/jvi.71.11.8116-8123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bova C A, Manfredi J P, Swanstrom R. env genes of avian retroviruses: nucleotide sequence and molecular recombinants define host range determinants. Virology. 1986;152:343–354. doi: 10.1016/0042-6822(86)90137-6. [DOI] [PubMed] [Google Scholar]
- 10.Bova C A, Olsen J C, Swanstrom R. The avian retrovirus env gene family: molecular analysis of host range and antigenic variants. J Virol. 1988;62:75–83. doi: 10.1128/jvi.62.1.75-83.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boyce-Jacino M T, O’Donoghue K, Faras A J. Multiple complex families of endogenous retroviruses are highly conserved in the genus Gallus. J Virol. 1992;66:4919–4929. doi: 10.1128/jvi.66.8.4919-4929.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boyce-Jacino M T, Resnick R, Faras A J. Structural and functional characterization of the unusually short long terminal repeats and their adjacent regions of a novel endogenous avian retrovirus. Virology. 1989;173:157–166. doi: 10.1016/0042-6822(89)90231-6. [DOI] [PubMed] [Google Scholar]
- 13.Chakrabarti R, Hofman F M, Pandey R, Mathes L E, Roy-Burman P. Recombination between feline exogenous and endogenous retroviral sequences generates tropism for cerebral endothelial cells. Am J Pathol. 1994;144:348–358. [PMC free article] [PubMed] [Google Scholar]
- 14.Coffin J M, Tsichlis P N, Conklin K F, Senior A, Robinson H L. Genomes of endogenous and exogenous avian retroviruses. Virology. 1983;126:51–72. doi: 10.1016/0042-6822(83)90461-0. [DOI] [PubMed] [Google Scholar]
- 15.Conklin K F, Groudine M. Varied interactions between proviruses and adjacent host chromatin. Mol Cell Biol. 1986;6:3999–4007. doi: 10.1128/mcb.6.11.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crittenden L B, Salter D W. A transgene, alv6, that expresses the envelope of subgroup A avian leukosis virus reduces the rate of congenital transmission of a field strain of avian leukosis virus. Poult Sci. 1992;71:799–806. doi: 10.3382/ps.0710799. [DOI] [PubMed] [Google Scholar]
- 17.Dorner A J, Coffin J M. Determinants for receptor interaction and cell killing on the avian retrovirus glycoprotein gp85. Cell. 1986;45:365–374. doi: 10.1016/0092-8674(86)90322-3. [DOI] [PubMed] [Google Scholar]
- 18.Dunwiddie C T, Resnick R, Boyce-Jacino M, Alegre J N, Faras A J. Molecular cloning and characterization of gag-, pol-, and env-related gene sequences in the ev− chicken. J Virol. 1986;59:669–675. doi: 10.1128/jvi.59.3.669-675.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fadly A M, Smith E J. American Association of Avian Pathologists, Proceedings of Avian Tumor Viruses Symposium. Kennett Square, Pa. 1997. An overview of subgroup J-like avian leukosis virus infection in broiler breeder flocks in the United States; pp. 54–57. [Google Scholar]
- 20.Fadly, A. M., and E. J. Smith. Isolation and some characteristics of a subgroup-J-like avian leukosis virus associated with myeloid leukosis in meat-type chickens in the United States. Avian Dis., in press. [PubMed]
- 21.Fan H. Leukemogenesis by Moloney murine leukemia virus: a multistep process. Trends Microbiol. 1997;5:74–82. doi: 10.1016/S0966-842X(96)10076-7. [DOI] [PubMed] [Google Scholar]
- 22.Gudkov A V, Komarova E A, Nikiforov M A, Zaitsevskaya T E. ART-CH, a new chicken retroviruslike element. J Virol. 1992;66:1726–1736. doi: 10.1128/jvi.66.3.1726-1736.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayward W S. Size and genetic content of viral RNAs in avian oncovirus-infected cells. J Virol. 1977;24:47–63. doi: 10.1128/jvi.24.1.47-63.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayward W S, Braverman S B, Astrin S M. Transcriptional products and DNA structure of endogenous avian proviruses. Cold Spring Harbor Symp Quant Biol. 1980;44:1111–1121. doi: 10.1101/sqb.1980.044.01.120. [DOI] [PubMed] [Google Scholar]
- 25.Hu W S, Temin H M. Genetic consequences of packaging two RNA genomes in one retroviral particle: pseudodiploidy and high rate of genetic recombination. Proc Natl Acad Sci USA. 1990;87:1556–1560. doi: 10.1073/pnas.87.4.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu W S, Temin H M. Retroviral recombination and reverse transcription. Science. 1990;250:1227–1233. doi: 10.1126/science.1700865. [DOI] [PubMed] [Google Scholar]
- 27.Hughes S H, Vogt P K, Stubblefield E, Robinson H, Bishop J M, Varmus H E. Organization of endogenous and exogenous viral and linked nonviral sequences. Cold Spring Harbor Symp Quant Biol. 1980;44:1077–1089. doi: 10.1101/sqb.1980.044.01.116. [DOI] [PubMed] [Google Scholar]
- 28.Hunter E. Viral entry and receptors. In: Coffin J M, Hughes S H, Varmus H, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 71–119. [PubMed] [Google Scholar]
- 29.McDougall A S, Terry A, Tzavaras T, Cheney C, Rojko J, Neil J C. Defective endogenous proviruses are expressed in feline lymphoid cells: evidence for a role in natural resistance to subgroup B feline leukemia viruses. J Virol. 1994;68:2151–2160. doi: 10.1128/jvi.68.4.2151-2160.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nikiforov M A, Gudkov A V. ART-CH: a VL30 in chickens? J Virol. 1994;68:846–853. doi: 10.1128/jvi.68.2.846-853.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Overbaugh J, Riedel N, Hoover E A, Mullins J I. Transduction of endogenous envelope genes by feline leukaemia virus in vitro. Nature. 1988;332:731–734. doi: 10.1038/332731a0. [DOI] [PubMed] [Google Scholar]
- 32.Payne L N, Gillespie A M, Howes K. Myeloid leukaemogenicity and transmission of the HPRS-103 strain of avian leukosis virus. Leukemia. 1992;6:1167–1176. [PubMed] [Google Scholar]
- 33.Payne L N, Gillespie A M, Howes K. Recovery of acutely transforming viruses from myeloid leukosis induced by the HPRS-103 strain of avian leukosis virus. Avian Dis. 1993;37:438–450. [PubMed] [Google Scholar]
- 34.Payne L N, Howes K, Gillespie A M, Smith L M. Host range of Rous sarcoma virus pseudotype RSV(HPRS-103) in 12 avian species: support for a new avian retrovirus envelope subgroup, designated J. J Gen Virol. 1992;73:2995–2997. doi: 10.1099/0022-1317-73-11-2995. [DOI] [PubMed] [Google Scholar]
- 35.Payne L N, Howes K, Smith L M, Venugopal K. Avian Tumor Viruses Symposium, Diagnosis and Control of Neoplastic Diseases of Poultry. Reno, Nev. 1997. Current status of diagnosis, epidemiology and control of ALV-J; pp. 58–62. [Google Scholar]
- 36.Roy-Burman P. Endogenous env elements: partners in generation of pathogenic feline leukemia viruses. Virus Genes. 1995;11:147–161. doi: 10.1007/BF01728655. [DOI] [PubMed] [Google Scholar]
- 37.Russell P H, Ahmad K, Howes K, Payne L N. Some chickens which are viraemic with subgroup J avian leukosis virus have antibody-forming cells but no circulating antibody. Res Vet Sci. 1997;63:81–83. doi: 10.1016/s0034-5288(97)90163-6. [DOI] [PubMed] [Google Scholar]
- 38.Skalka A, DeBona P, Hishinuma F, McClements W. Avian endogenous proviral DNA: analysis of integrated ev 1 and a related gs− chf− provirus purified by molecular cloning. Cold Spring Harbor Symp Quant Biol. 1980;44:1097–1104. doi: 10.1101/sqb.1980.044.01.118. [DOI] [PubMed] [Google Scholar]
- 39.Smith E J, Fadly A M, Levin I, Crittenden L B. The influence of ev6 on the immune response to avian leukosis virus infection in rapid-feathering progeny of slow- and rapid-feathering dams. Poult Sci. 1991;70:1673–1678. doi: 10.3382/ps.0701673. [DOI] [PubMed] [Google Scholar]
- 40.Stuhlmann H, Berg P. Homologous recombination of copackaged retrovirus RNAs during reverse transcription. J Virol. 1992;66:2378–2388. doi: 10.1128/jvi.66.4.2378-2388.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taplitz R A, Coffin J M. Selection of an avian retrovirus mutant with extended receptor usage. J Virol. 1997;71:7814–7819. doi: 10.1128/jvi.71.10.7814-7819.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Venugopal K, Smith L M, Howes K, Payne L N. Antigenic variants of J subgroup avian leukosis virus: sequence analysis reveals multiple changes in the env gene. J Gen Virol. 1998;79:757–766. doi: 10.1099/0022-1317-79-4-757. [DOI] [PubMed] [Google Scholar]