Abstract
CD8+ T cells from human immunodeficiency virus (HIV)-infected individuals can suppress HIV replication in cultured CD4+ cells by a noncytotoxic mechanism. Efficient suppression of HIV replication (>90% reduction) does not require HLA class I or class II histocompatibility between the effector CD8+ T cells and the infected target CD4+ T cells. However, maximal control of HIV production occurs when the CD8+ effector cells and CD4+ target cells are syngeneic. In some cases, more than 20-fold fewer syngeneic CD8+ T cells were required to achieve the same degree of HIV inhibition as HLA-mismatched CD8+ T cells. The increased antiviral activity seen in the syngeneic setting did not map exclusively to either the HLA class I or class II locus. These findings suggest that genetic compatibility (potentially, but not necessarily, at the HLA class I and class II loci) regulates CD8+ T-cell noncytotoxic antiviral activity against infected CD4+ T cells.
CD8+ T cells from healthy human immunodeficiency virus (HIV)-infected individuals can suppress HIV replication in cultured CD4+ T cells without any apparent killing of infected cells (16, 28). This noncytotoxic CD8+-cell antiviral response is detected by a reduction in HIV p24 antigen and/or reverse transcriptase (RT) levels in culture fluids upon mixing CD8+ T cells with the infected target CD4+ T cells. This cellular immune response can suppress replication of HIV type 1 (HIV-1), HIV-2, and simian immunodeficiency virus (references 16 and 29 and unpublished observations) and has been observed in several species of nonhuman primates (2, 6, 8, 13, 23).
Unlike the antigen-specific cytotoxic mechanism of HLA class I-restricted CD8+ cytotoxic T lymphocytes (CTL), the CD8+-T-cell noncytotoxic anti-HIV activity does not appear to require HLA compatibility to effectively control HIV replication (17). Although initial evidence suggested a more efficient control of HIV in autologous settings (27, 28), a number of laboratories have shown that CD8+ cells (polyclonal or clonal) can efficiently inhibit HIV replication (by >90%) in heterologous peripheral blood mononuclear cells (PBMC) or CD4+ T cells (1, 3, 16, 20, 26, 28, 29). Importantly, the antiviral activity in these heterologous systems does not seem to be dependent on alloreactivity between the CD4+ target cells and the CD8+ effector cells (29).
To determine if HLA genetics regulate this type of cellular anti-HIV response, we investigated the HLA compatibility requirements for efficient control of HIV replication by CD8+ T cells. The results suggest that the extent of this antiviral CD8+-T-cell noncytotoxic response is dependent on the genetic relatedness between the effector and target cells, but not necessarily on HLA class I or class II determinants.
MATERIALS AND METHODS
Subjects.
Heparinized peripheral blood samples were obtained by venipuncture from HIV-1-seropositive and -seronegative donors. Among these, two pairs of identical twins, discordant for HIV infection, were studied. The HIV-infected twins were clinically healthy, with CD4+-T-cell counts of around 400/μl (14 to 16%) in one case and just over 300/μl (24 to 26%) in the other. Both individuals have been infected for over 10 years. In addition, cells from four other long-term-asymptomatic HIV-infected subjects were studied. Their CD4+-T-cell counts ranged from 319 to 940 cells/μl (23 to 36%). Blood samples from HIV-seronegative donors were obtained from laboratory volunteers or were provided by Irwin Memorial Blood Centers (San Francisco, Calif.). The study received the approval of the Committee on Human Research, University of California, San Francisco.
Assay for CD8+-cell noncytotoxic antiviral activity.
Purified CD4+ cells isolated from uninfected (or, in some cases, infected) blood donors were stimulated and acutely infected with HIV-1 as described previously (20). In brief, the CD4+ cells, purified with anti-CD4 immunomagnetic beads (Dynal, Lake Success, N.Y.), were cultured for 3 days with 3 μg of phytohemagglutinin (PHA) (Sigma Chemical Co., St. Louis, Mo.)/ml in RPMI 1640 medium containing 2 mM glutamine, 1% antibiotics (100 U of penicillin/ml, 100 μg of streptomycin/ml), 10% heat-inactivated (56°C; 30 min) fetal calf serum, and 100 U of human recombinant interleukin 2 (Collaborative Research, Bedford, Mass.)/ml. In each case, the purity of the CD4+-cell populations was 95% or greater and >95% were CD3+ as assessed by flow cytometric analysis (15). The stimulated cells were then washed and treated with Polybrene (2 μg/ml for 30 min at 37°C) and subsequently acutely infected with 104 50% tissue culture infective doses of HIV-1SF33. This virus is a highly cytopathic strain (25) and is not sensitive to the antiviral effects of β-chemokines (18). After 1 h of incubation with virus at 37°C, the cells were washed and resuspended in the RPMI 1640 growth medium. This method of infection routinely yields 15 to 35% HIV antigen-positive cells as detected by immunofluorescence at the peak of HIV replication (5 to 7 days) (20).
Immediately after the 1-h virus inoculation, the CD4+ cells were cultured with various amounts of CD8+ cells previously purified from an HIV-infected subject’s PHA-stimulated PBMC by using anti-CD8 immunomagnetic beads as described previously (20). The amount of HIV replication in the cultures was measured by a standard particle-associated RT assay (10). The extent of CD8+-cell antiviral activity (indicated by the percent suppression of HIV replication) was determined by comparison to the amount of RT activity in culture fluids of infected CD4+ cells grown alone.
HLA typing for class I and class II antigens.
The subjects’ HLA class I phenotypes were determined by serologic or molecular methods. Class I antigens were detected by the use of the standard microlymphocytotoxicity technique (11), while the genetic alleles were determined by DNA-typing techniques, using sequence-specific primers (5, 21). The HLA class II phenotype was determined by similar molecular methods.
RESULTS
HLA compatibility requirements: HIV-discordant identical twins. (i) Genetic similarity confers maximal suppression of HIV.
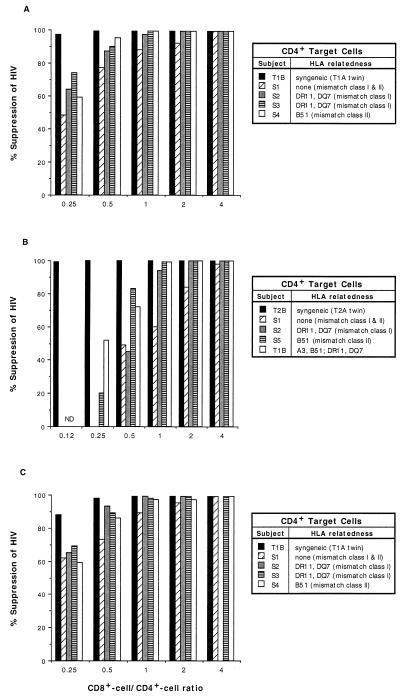
The relative degree of antiviral activity of CD8+ T cells from healthy HIV-infected individuals was assessed by using experimentally infected CD4+ T cells of differing genetic relatedness to the effector cells (Table 1). We first analyzed HLA compatibility requirements with CD4+ cells from seronegative donors as targets and CD8+ cells from two pairs of HIV-discordant monozygotic twins. Thus, in one case, PHA-stimulated CD8+ T cells from subject T1A were cocultured at various input ratios with HIV-1SF33-infected CD4+ T cells that were syngeneic (from subject T1A’s uninfected twin, subject T1B), HLA mismatched in the class I and II loci, or mismatched in only the class I or class II locus. The highest degree of HIV suppression was observed when syngeneic infected target CD4+ T cells were used (Fig. 1A). Almost-complete inhibition of HIV replication (97% reduction) was seen at the lowest CD8+-cell/CD4+-cell ratio tested (0.25). In contrast, in cultures of completely mismatched cells, only half the suppression of HIV replication (48%) occurred at the same CD8+-cell/CD4+-cell ratio (0.25). From a different perspective, even at the CD8+-cell/CD4+-cell ratio of 2.0, HIV suppression in HLA class I- and class II-mismatched CD4+ T cells did not reach the level of that observed when syngeneic target cells were used at the CD8+/CD4+ cell ratio of 0.25 (88 versus 99%, respectively). Thus, greater than eightfold more CD8+ T cells were required to inhibit HIV replication in HLA-unrelated CD4+ T cells than in HLA-identical CD4+ T cells.
TABLE 1.
HLA class I and class II phenotypes of effector and target cells studied
| Cell type | Subject | HIV status | Class I phenotypea | Class II phenotypeb |
|---|---|---|---|---|
| Effectorc | T1A | Infected | A3 A29 B44 B51 | DR7 DR11 DQ2 DQ7 |
| T2A | Infected | A2 A3 B14 B51 | DR2 DR11 DR52 DQ1 DQ7 | |
| Targetd | T1B (T1A twin) | Seronegative | A3 A29 B44 B51 | DR7 DR11 DQ2 DQ7 |
| T2B (T2A twin) | Seronegative | A2 A3 B14 B51 | DR2 DR11 DR52 DQ1 DQ7 | |
| S1 | Seronegative | A24 Ax B27 B61 CW3 CW7 | DR14 DR15 DQ5 | |
| S2 | Seronegative | A1 A24 B35 B37 CW4 | DR10 DR11 DQ5 DQ7 | |
| S3 | Seronegative | A2 A30 B13 B62 CW6 | DR11 DR14 DQ5 DQ7 | |
| S4 | Seronegative | A2 A24 B51 B61 CW3 | DR2 DR4 DQ1 DQ3 | |
| S5 | Seronegative | A11 A31 B51 B60 CW3 | DR4 DR9 DQ3 |
Determined by serological typing as described in Materials and Methods.
Determined by molecular methods as described in Materials and Methods.
Purified CD8+ cells isolated from PHA-stimulated PBMC.
Purified CD4+ cells that had been PHA stimulated for 3 days and then infected with HIV-1SF33.
FIG. 1.
Extent of CD8+-T-cell anti-HIV activity exhibited against HIV replication in CD4+ T cells of different genetic relatedness. The anti-HIV activities of PHA-stimulated CD8+ T cells from two HIV-infected twins, T1A (A) and T2A (B), and unstimulated T1A CD8+ T cells (C) were titrated against HIV-1SF33-infected CD4+ T cells from their respective syngeneic twins, an HLA class I- and II-mismatched subject, HLA class I-mismatched subjects, and HLA class II-mismatched subjects. The amount of antiviral activity exhibited (% suppression of HIV) reflects the extent of reduction in RT activity in the cell culture fluid compared to the activity in the respective infected CD4+ T cells cultured alone. ND, not done. The peak amounts of HIV replication (RT activity on day 6) in the control T1B, S1, S2, S3, and S4 CD4+ cell cultures (A and C) were 120 × 103, 206 × 103, 120 × 103, 212 × 103, and 140 × 103 cpm/ml, respectively. The amounts of HIV production in the T2B, S1, S2, S5, and T1B CD4+ target cells (B) were 400 × 103, 561 × 103, 497 × 103, 567 × 103, and 312 × 103 cpm/ml, respectively. These results are representative of two separate experiments with each twin, including additional examples of class II- and class I/class II partially mismatched cells.
The extent of CD8+-cell inhibition of HIV production by target cells that were mismatched at only the class I or only the class II locus was intermediate to those observed with completely mismatched and syngeneic cultures (Fig. 1A). In these studies, CD8+-cell/CD4+-cell ratios of 1.0 or more were required to achieve >95% suppression of HIV replication. No consistent difference in the extent of suppression was noted whether the effector and target cells were mismatched in the class I or the class II region (Fig. 1A).
CD8+ cells from the seronegative twin T1B did not suppress HIV production from his own experimentally infected CD4+ cells nor from the CD4+ cells of a genetically unrelated subject at any CD8+-cell/CD4+-cell ratio tested (data not shown). These results indicated that HLA histocompatibility by itself is not sufficient to confer HIV-suppressing activity on mitogen-stimulated CD8+ cells and that if allogeneic responses are occurring in these short-term cultures, they do not induce CD8+ cells from uninfected people to block HIV production.
Similar results with anti-HIV responses were observed with CD8+ cells from a different set of twins, T2A and T2B (Fig. 1B). Again, potent antiviral activity was seen in the syngeneic setting. In contrast, 32-, 16-, 8-, and 8-fold more CD8+ cells were needed to achieve the same level of suppression of virus replication in CD4+ cells that were class I and II mismatched, class I mismatched, class II mismatched, or class I and class II partially mismatched, respectively (Fig. 1B).
(ii) Genetic regulatory effect is not dependent on in vitro activation of CD8+ cells.
To further evaluate the extent to which HLA genetics regulate CD8+-cell antiviral responses, analogous experiments were performed with CD8+ cells that were not previously mitogen activated (as opposed to the experiments described above). As with mitogen-stimulated CD8+ cells, the most efficient control by unstimulated CD8+ cells was observed against syngeneic CD4+ cells (Fig. 1C). Unstimulated CD8+ cells showed less antiviral activity against target cells that were mismatched at class I, class II, or both loci.
Results with CD4+ target cells from HIV-infected subjects.
In order to further establish the significance of these findings, CD8+ cells from four additional HIV-infected subjects were evaluated. Because no other twins discordant for HIV infection were available, syngeneic target cells could only be obtained by using autologous CD4+ cells. Thus, CD4+ cells from HIV-infected subjects of different HLA relatedness were experimentally infected with HIV-1 and used as target cells (Table 2). Typically, endogenous virus is not released into culture at appreciable levels until 12 days poststimulation (unpublished observations). It is, therefore, unlikely that endogenous virus contributes much to the total peak HIV replication measured (on day 6 or 9) in these cultures. Reciprocal pairings of effector and target cells were performed to control for possible variations in the replicative capacity of the different CD4+-cell populations used.
TABLE 2.
| Subject | HIV status | Class I phenotypec | Class II phenotyped |
|---|---|---|---|
| T1A | Infected | A3 A29 B44 B51 C15 C16.1 | DR7.1 DR11.4 DQA2 DQA5 DQB2 DQB3.1 |
| S6 | Infected | A2.1 A29 B51 B53 C4 C14 | DR1.2 DR8.1 DQA1.1 DQA4.1 DQB4.2 DQB5.1 |
| S7 | Infected | A3 A26 B27 B57 C2 C6.2 | DR1.1 DR13 DQA1.1 DQA1.3 DQB5.1 DQB6.3 |
| S8 | Infected | A2.1 A24 B15 B49 C3.4 C7 | DR4.7 DR13 DQA1.2 DQA3 DQB3.1 DQB6.4 |
| S9 | Infected | A3 A25.1 B7 B39 C7 C12.3 | DR11.1 DR15.1 DQA1.2 DQA5.1 DQB3.1 DQB6.2 |
Purified CD8+ cells isolated from PHA-stimulated PBMC.
Purified CD4+ cells that had been PHA-stimulated for 3 days and then infected with HIV-1SF33.
Determined by molecular methods as described in Materials and Methods.
Determined by molecular methods as described in Materials and Methods. The DR alleles shown indicate the alleles from the HLA-DRB1 locus.
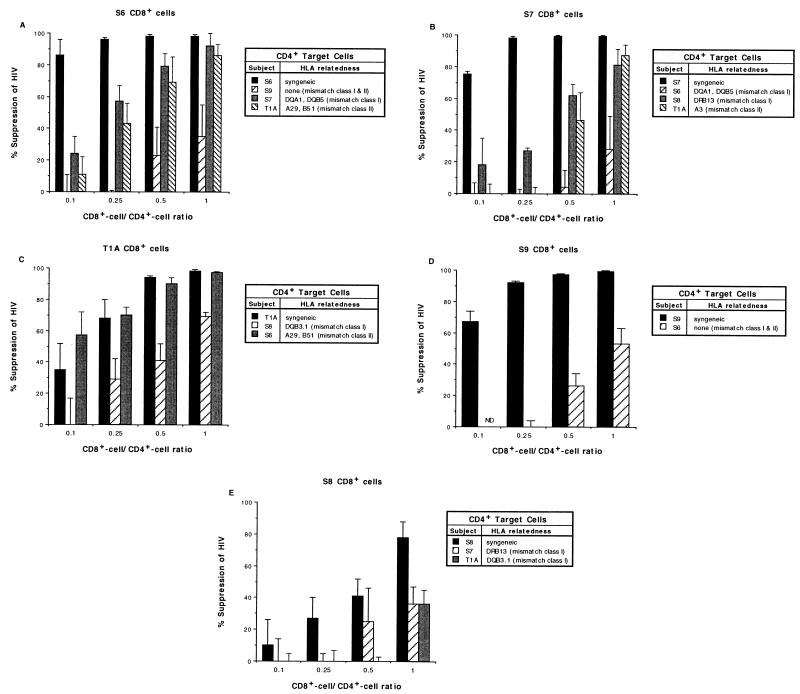
In agreement with the above-mentioned results obtained with CD4+ target cells from HIV-seronegative donors, maximal CD8+-cell-mediated suppression was observed when the effector and target cells were syngeneic (Fig. 2). For example, in the experiments with S6 CD8+ cells (Fig. 2A), >80% suppression of HIV replication was seen against autologous (syngeneic) infected CD4+ cells at the CD8+-cell/CD4+-cell ratio of 0.1. When class I-mismatched (S7 CD4+ cells with two class II DQ alleles shared) or class II-mismatched (T1A CD4+ cells with one class I A and one class I B allele shared) targets were used, ≥80% suppression was not observed until a CD8+-cell/CD4+-cell ratio of 0.5 and 1.0, respectively, was reached. The lowest extent of CD8+-cell-mediated suppression occurred when the effector and target cells were completely unrelated in both the class I and class II loci (e.g., S9 CD4+ cells). In this case, 80% suppression of HIV replication occurred at the CD8+-cell/CD4+-cell ratio of 2.0 (data not shown). These results indicated that about 20-fold more CD8+ cells were required to achieve the same amount of suppression as that seen in the syngeneic setting.
FIG. 2.
Extent of CD8+-T-cell anti-HIV activity exhibited against HIV replication in CD4+ T cells of different genetic relatedness. The anti-HIV activities of PHA-stimulated CD8+ T cells from various HIV-infected subjects were titrated against HIV-1SF33-infected CD4+ T cells from the subjects themselves (syngeneic), HLA class I-mismatched subjects, and HLA class II-mismatched subjects. The amount of antiviral activity exhibited by the CD8+ cells (% suppression of HIV) reflects the extent of reduction in RT activity in the cell culture fluid compared to the activity seen in the respective infected CD4+ T cells cultured alone. The average peak amounts of HIV replication (RT activity) in the control T1A, S6, S7, S8, and S9 CD4+ cell cultures were 174 × 103, 110 × 103, 205 × 103, 128 × 103, and 101 × 103 cpm/ml, respectively. All cultures were set up in triplicate.
Similar results were obtained with the other subjects’ CD8+ cells studied (Fig. 2B to E). Of the five data sets analyzed, the highest extent of suppression was observed in the syngeneic setting except in one case (i.e., 1 of 11 mismatches tested). In that case, the effector and target cells were mismatched in the class II locus (T1A CD8+ cells versus S6 CD4+ cells [Fig. 2C]). In general, the smallest amount of suppression occurred when the effector and target cells were class I and class II disparate or were similar at only one class II allele. No consistent difference in suppressing activity between class I- and class II-related pairings was observed. In some cases, class I mismatches showed better suppression than class II mismatches and vice versa. Because the sample number for each group was small, no statistical significance was attained. However, if all the mismatched pairings are taken as a group, then CD8+ cells showed significantly better suppressing activity in the syngeneic setting when analyzed with a stratified Wilcoxon rank sum test, the strata being the effector cell phenotypes (P = 0.035). Nevertheless, in all cases, regardless of the extent of HLA dissimilarity between the CD8+ and CD4+ cells, >50% suppression (>80% in 8 of 11 cases) could be achieved if sufficient CD8+ cells were added, i.e., to yield a CD8+-cell/CD4+-cell ratio of 1.0 or 2.0 (data not shown).
HIV replication in all target cell populations reached comparable levels (Fig. 2), and the different target cells were comparably sensitive to CD8+-cell-mediated suppression (as seen in the syngeneic setting), except, possibly, for S8 CD4+ cells. The S8 CD4+ cells showed greater sensitivity to suppression mediated by S7 and T1A CD8+ cells (both mismatched) than to that mediated by syngeneic S8 CD8+ cells. Furthermore, S8 CD8+ cells have previously been shown to have relatively low antiviral activity (unpublished observations). Together, these results suggest that the relatively weak suppressing activity of S8 CD8+ cells (Fig. 2E) is probably not due to an insensitivity of S8 CD4+ cells to CD8+-cell-mediated suppression.
Analysis of reciprocal pairings among subjects’ effector and target cells.
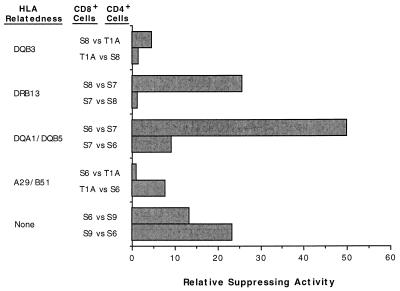
With the experimental data shown in Fig. 2, analysis of reciprocal pairings of effector and target cells (including mismatches at class I, class II, or both loci) revealed no consistent pattern of genetic regulation (Fig. 3). The CD8+-cell antiviral activity in one direction was usually markedly different than that in the opposite direction, even when the data was standardized for the intrinsic (maximal) level of an individual’s CD8+-cell antiviral activity (i.e., that seen in the syngeneic setting). This finding was made even though three pairings involved the same target cells (S6 CD4+ cells). For example, in one case, S6 CD8+ cells controlled HIV replication in the S7 mismatched CD4+ cells an average of fivefold more than S7 CD8+ cells controlled virus production in S6 CD4+ cells. In the two other reciprocal cases involving S6 cells (with T1A and S9), the opposite was true (about seven- and twofold less suppression, respectively).
FIG. 3.
Comparison of the relative CD8+-cell suppressing activities in reciprocal pairings of CD8+ and CD4+ cells. The extent of relative CD8+-cell suppressing activity was calculated for each case shown in Fig. 2 where the antiviral activity of one subject’s CD8+ cells was assessed against another subject’s infected CD4+ cells and vice versa. For a given subject’s CD8+ cells, the quotient of (RT activity in the mismatched culture)/(RT activity in the respective syngeneic culture) was obtained for the CD8+-cell/CD4+-cell ratios of 0.1, 0.25, and 0.5. These values were then averaged to yield a representative level of CD8+-cell suppressing activity relative to that measured in the respective syngeneic setting (the intrinsic CD8+-cell activity). In this fashion, the extent of antiviral activity of one subject’s CD8+ cells could be compared directly to that of another subject’s CD8+ cells without concern for the potential differences between the individuals’ intrinsic CD8+-cell functions.
DISCUSSION
CD8+ T cells from HIV-infected individuals suppress HIV replication in acutely infected CD4+ T cells (reviewed in reference 16). The present studies indicate that maximal CD8+-cell suppression of HIV replication in culture is achieved when the infected CD4+ target cells and the effector CD8+ T cells are syngeneic, and thus share the same HLA class I and II genotype. Mismatches in the class I as well as the class II locus resulted in less efficient control of HIV by antiviral CD8+ T cells, but possibly better than that in the completely mismatched setting. The results suggest that genetic relatedness between CD8+ cells and CD4+ cells influences the extent of CD8+-cell anti-HIV activity and that, unlike classic CD8+-cell cytolytic activity, this response is not class I restricted.
Of six different HIV-infected subjects’ CD8+ cells studied, nearly all suppressed HIV replication better in syngeneic infected CD4+ cells than in any of the mismatched CD4+-cell settings (Fig. 1 and 2). The increased suppressing activity in the syngeneic setting was reflected by a higher percent suppression of HIV at various CD8+-cell/CD4+-cell ratios (most prominently at the low ratios) or by a lower number of CD8+ cells required to achieve a certain level of HIV suppression (i.e., 4- to >20-fold fewer CD8+ cells needed to suppress HIV by ≥80%). This genetic compatibility effect was independent of whether the target cells used were from HIV-seropositive (Fig. 2) or -seronegative (Fig. 1) donors, and it does not appear to be due to differences in CD4+-cell HIV-replicative capacity or sensitivity to CD8+-cell-mediated suppression. While totally mismatched effector and target cells may have resulted in the smallest amount of HIV suppression, we cannot conclude that relatedness at class I or class II conferred a statistically significant increase in antiviral activity. Furthermore, no consistent difference in the extent of suppression was seen between class I-related and class II-related settings, albeit the number of shared alleles was never more than two and in several cases was only one (Fig. 1 and 2).
The similar pattern of suppressing activity seen when freshly isolated (non-exogenously stimulated) CD8+ cells were used suggests that this apparent genetic control of noncytolytic antiviral activity is not dependent on mitogen stimulation (Fig. 1C). This finding may be the consequence of the highly activated nature of CD8+ T cells in HIV-infected individuals (15).
This type of CD8+-cell noncytolytic anti-HIV response appears to result from a block in viral transcription (16, 19). The findings of this study imply that genetic compatibility in some way either enhances this mechanism for controlling HIV or provides for the activity of an additional type of antiviral mechanism that is genetically restricted. Both possibilities would be consistent with the observation that, even if the effector and target cells are totally HLA mismatched, efficient HIV suppression can be achieved if a high enough CD8+ effector cell input is used (Fig. 1 and 2). Since a β-chemokine-insensitive HIV isolate was used in the present study, involvement of β-chemokine-mediated antiviral effects (7) are not likely. Furthermore, because CD8+ T cells do not produce type 1 interferons (unpublished observations) and gamma interferon does not have anti-HIV activity in HIV-infected CD4+ T cells (20a), interferons are not likely to be responsible for the higher suppressing activity seen in HLA-matched settings.
Cytotoxic activity is an obvious antiviral mechanism worth consideration. Many studies addressing this issue provide evidence that cytotoxicity is not responsible for the control of HIV replication in the autologous (or heterologous) systems used to study CD8+ cell suppression of HIV production. Infected cells are not eliminated by the CD8+ cells (9, 14, 19, 28, 30), nor is there cytolytic release of chromium-51 from autologous target cells (22, 26). In addition, the low CD8+-cell/CD4+-cell ratios (0.25 in the syngeneic setting) required to block HIV replication (≥90% suppression) are well below that seen for polyclonal or even clonal CTL (12). Even if only 10% of the CD4+ cells were initially infected in our assays, the actual effector/target cell ratio would be 2.5 if every CD8+ cell was an effector cell, which was likely not the case. Finally, HIV-specific CD8+ CTL are exclusively class I restricted (12). The suppressing activity we observed was not genetically restricted at high CD8+-cell inputs, nor was it strictly class I restricted at lower inputs. In four of four cases in which the CD8+ cells and CD4+ cells were mismatched at class I (sharing one or two alleles at class II), a higher degree of suppression was observed relative to the completely mismatched setting (Fig. 1 and 2). Further analysis is required to determine specifically what role, if any, is played by class I or class II antigens in the increased suppressing activity seen in the syngeneic setting.
Another possible explanation for the present findings is that the lower activity seen in the HLA-mismatched cases reflects allostimulatory signals that result in either up-regulation of HIV replication in the target CD4+ cells (hence, less apparent suppression) or down-regulation of CD8+-cell antiviral activity. Allogeneic activity can involve either class I or class II antigens (24) and thus would not necessarily be restricted to either locus. The findings of Bruhl et al. (4) showing that allogeneic stimulation of CD8+ cells can induce HIV-suppressive activity would seem to argue against the latter possibility. Preliminary studies addressing the former possibility indicate that a slight enhancement of HIV replication can occur upon exposure of the infected CD4+ cells to a high input of allogeneic CD8+ cells that lack antiviral activity, but not to the extent that could explain the difference in CD8+-cell activity we have observed (unpublished observations). In addition, the results from the reciprocal pairings (Fig. 3) showing marked differences in the extent of suppression from one direction to the other are not entirely consistent with allogeneic recognition being the sole regulating factor, since in all the cases that involved nonsyngeneic matches, most or all of the HLA alleles were allogeneic.
In summary, HIV-infected individuals possess CD8+ cells that can efficiently suppress HIV replication in HLA-unrelated CD4+ lymphocytes. However, control of HIV production is maximal in a syngeneic setting regardless of whether or not the CD8+ cells are mitogen stimulated. This control does not appear to be strictly restricted to either the class I or class II locus, leaving a question as to whether a nonclassical antigen recognition process is involved. The findings suggest that many of the heterologous-cell assay systems employed to measure CD8+-T-cell-mediated noncytotoxic suppression of HIV replication are likely underestimating the relative degree of potential antiviral activity in the host. Furthermore, these studies emphasize the importance of considering the genetic relatedness between the effector and target cells when studying mechanistic issues and when quantitating and comparing noncytotoxic CD8+-T-cell antiviral activity. A better understanding of the regulation of this CD8+-T-cell anti-HIV activity could lead to improving or at least maintaining this antiviral response.
ACKNOWLEDGMENTS
These studies were funded by a grant from the NIH (RO1 AI30350) and the Histocompatibility Fund (M.R.G.). C.E.M. was supported in part by the University of California, San Francisco, AIDS Clinical Research Center (funded by the University of California Universitywide AIDS Research Program).
We thank Roland Orque and Susan Ridha for their technical assistance and Christine Beglinger and Ann Murai for help in preparation of the manuscript.
REFERENCES
- 1.Barker T D, Weissman D, Daucher J A, Roche K M, Fauci A S. Identification of multiple and distinct CD8+ T cell suppressor activities. J Immunol. 1996;156:4476–4483. [PubMed] [Google Scholar]
- 2.Blackbourn D J, Locher C P, Ramachandran B, Barnett S W, Murthy K K, Carey K D, Brasky K M, Levy J A. CD8+ cells from HIV-2-infected baboons control HIV replication. AIDS. 1997;11:737–746. doi: 10.1097/00002030-199706000-00006. [DOI] [PubMed] [Google Scholar]
- 3.Brinchmann J E, Gaudernack G, Vartdal F. CD8+ T cells inhibit HIV replication in naturally infected CD4+ T cells: evidence for a soluble inhibitor. J Immunol. 1990;144:2961–2966. [PubMed] [Google Scholar]
- 4.Bruhl P, Kerschbaum A, Zimmermann K, Eibl M M, Mannhalter J W. Allostimulated lymphocytes inhibit replication of HIV type 1. AIDS Res Hum Retroviruses. 1996;12:31–37. doi: 10.1089/aid.1996.12.31. [DOI] [PubMed] [Google Scholar]
- 5.Bunce M, Taylor C J, Welsh K W. Rapid HLA-DQB typing by eight polymerase chain reaction amplifications with sequence specific primers (PCR-SSP) Hum Immunol. 1993;37:201–206. doi: 10.1016/0198-8859(93)90502-r. [DOI] [PubMed] [Google Scholar]
- 6.Castro B A, Walker C M, Eichberg J W, Levy J A. Suppression of human immunodeficiency virus replication by CD8+ cells from infected and uninfected chimpanzees. Cell Immunol. 1991;132:246–255. doi: 10.1016/0008-8749(91)90023-5. [DOI] [PubMed] [Google Scholar]
- 7.Cocchi F, DeVico A L, Garzino-Demo A, Arya S K, Gallo R C, Lusso P. Identification of RANTES, MIP-1alpha, and MIP-1beta as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 8.Ennen J, Findeklee H, Dittmar M T, Norley S, Ernst M, Kurth R. CD8+ T lymphocytes of African green monkeys secrete an immunodeficiency virus-suppressing lymphokine. Proc Natl Acad Sci USA. 1994;91:7207–7211. doi: 10.1073/pnas.91.15.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gomez A M, Smaill F M, Rosenthal K L. Inhibition of HIV replication by CD8+ T cells correlates with CD4 counts and clinical stage of disease. Clin Exp Immunol. 1994;97:68–75. doi: 10.1111/j.1365-2249.1994.tb06581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffman A D, Banapour B, Levy J A. Characterization of the AIDS-associated retrovirus reverse transcriptase and optimal conditions for its detection in virions. Virology. 1985;147:326–335. doi: 10.1016/0042-6822(85)90135-7. [DOI] [PubMed] [Google Scholar]
- 11.Hopkins K A. Basic lymphocytotoxicity test. In: Zachary A, Teresi G A, editors. ASHI laboratory manual. 2nd ed. Vol. 195. Lenexa, Kans: American Society for Histocompatibility and Immunogenetics; 1990. p. 201. [Google Scholar]
- 12.Johnson R P, Walker B. Cytotoxic T lymphocytes in HIV infection: responses to structural proteins. Curr Opin Microbiol Immunol. 1994;189:35–63. doi: 10.1007/978-3-642-78530-6_3. [DOI] [PubMed] [Google Scholar]
- 13.Kannagi M, Chalifoux L V, Lord C I, Letvin N L. Suppression of simian immunodeficiency virus replication in vitro by CD8+ lymphocytes. J Immunol. 1988;140:2237–2242. [PubMed] [Google Scholar]
- 14.Kinter A L, Bende S M, Hardy E C, Jackson R, Fauci A S. Interleukin 2 induces CD8+ T cell-mediated suppression of human immunodeficiency virus replication in CD4+ T cells and this effect overrides its ability to stimulate virus expression. Proc Natl Acad Sci USA. 1995;92:10985–10989. doi: 10.1073/pnas.92.24.10985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Landay A L, Mackewicz C, Levy J A. An activated CD8+ T cell phenotype correlates with anti-HIV activity and asymptomatic clinical status. Clin Immunol Immunopathol. 1993;69:106–116. doi: 10.1006/clin.1993.1157. [DOI] [PubMed] [Google Scholar]
- 16.Levy J A, Mackewicz C E, Barker E. Controlling HIV pathogenesis: the role of noncytotoxic anti-HIV activity of CD8+ cells. Immunol Today. 1996;17:217–224. doi: 10.1016/0167-5699(96)10011-6. [DOI] [PubMed] [Google Scholar]
- 17.Mackewicz C, Levy J A. CD8+ cell anti-HIV activity: nonlytic suppression of virus replication. AIDS Res Hum Retroviruses. 1992;8:1039–1050. doi: 10.1089/aid.1992.8.1039. [DOI] [PubMed] [Google Scholar]
- 18.Mackewicz C E, Barker E, Greco G, Reyes-Teran G, Levy J A. Do β-chemokines have clinical relevance in HIV infection? J Clin Investig. 1997;100:921–930. doi: 10.1172/JCI119608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mackewicz C E, Blackbourn D J, Levy J A. CD8+ cells suppress human immunodeficiency virus replication by inhibiting viral transcription. Proc Natl Acad Sci USA. 1995;92:2308–2312. doi: 10.1073/pnas.92.6.2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mackewicz C E, Ortega H W, Levy J A. CD8+ cell anti-HIV activity correlates with the clinical state of the infected individual. J Clin Investig. 1991;87:1462–1466. doi: 10.1172/JCI115153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20a.Mackewicz C E, Ortega H, Levy J A. Effect of cytokines on HIV replication in CD4+ lymphocytes: lack of identity with the CD8+ cell antiviral factor. Cell Immunol. 1994;153:329–343. doi: 10.1006/cimm.1994.1032. [DOI] [PubMed] [Google Scholar]
- 21.Olerup O, Zetterquist H. HLA-DR typing by PCR amplification with sequence specific primers (PCR-SSP) in 2 hours. Tissue Antigens. 1992;39:225–236. doi: 10.1111/j.1399-0039.1992.tb01940.x. [DOI] [PubMed] [Google Scholar]
- 22.Paliard X, Lee A Y, Walker C M. RANTES, MIP-1α and MIP-1β are not involved in the inhibition of HIV-1SF33 replication mediated by CD8+ T-cell clones. AIDS. 1996;10:1317–1321. doi: 10.1097/00002030-199610000-00002. [DOI] [PubMed] [Google Scholar]
- 23.Powell J D, Yehuda-Cohen T, Villinger F, McClure H M, Sell K W, Ahmed-Ansari A. Inhibition of SIV-SMM replication in vitro by CD8+ cells from SIV/SMM infected seropositive clinically asymptomatic sooty mangabeys. J Med Primatol. 1990;19:239–249. [PubMed] [Google Scholar]
- 24.Sherman L A, Chattopadhyay S. The molecular basis of allorecognition. Annu Rev Immunol. 1993;11:385–402. doi: 10.1146/annurev.iy.11.040193.002125. [DOI] [PubMed] [Google Scholar]
- 25.Tateno M, Levy J A. MT-4 plaque formation can distinguish cytopathic subtypes of the human immunodeficiency virus (HIV) Virology. 1988;167:299–301. doi: 10.1016/0042-6822(88)90084-0. [DOI] [PubMed] [Google Scholar]
- 26.Toso J F, Chen C H, Mohr J R, Piglia L, Oei C, Ferrari G, Greenberg M L, Weinhold K J. Oligoclonal CD8 lymphocytes from persons with asymptomatic human immunodeficiency virus (HIV) type 1 infection inhibit HIV-1 replication. J Infect Dis. 1995;172:964–973. doi: 10.1093/infdis/172.4.964. [DOI] [PubMed] [Google Scholar]
- 27.Tsubota H, Lord C I, Watkins D I, Morimoto C, Letvin N L. A cytotoxic T lymphocyte inhibits acquired immunodeficiency syndrome virus replication in peripheral blood lymphocytes. J Exp Med. 1989;169:1421–1434. doi: 10.1084/jem.169.4.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walker C M, Moody D J, Stites D P, Levy J A. CD8+ lymphocytes can control HIV infection in vitro by suppressing virus replication. Science. 1986;234:1563–1566. doi: 10.1126/science.2431484. [DOI] [PubMed] [Google Scholar]
- 29.Walker C M, Thomson-Honnebier G A, Hsueh F C, Erickson A L, Pan L-Z, Levy J A. CD8+ T cells from HIV-1-infected individuals inhibit acute infection by human and primate immunodeficiency viruses. Cell Immunol. 1991;137:420–428. doi: 10.1016/0008-8749(91)90090-x. [DOI] [PubMed] [Google Scholar]
- 30.Wiviott L D, Walker C M, Levy J A. CD8+ lymphocytes suppress HIV production by autologous CD4+ cells without eliminating the infected cells from culture. Cell Immunol. 1990;128:628–634. doi: 10.1016/0008-8749(90)90054-u. [DOI] [PubMed] [Google Scholar]