Abstract
A reinvestigation of cellulose degradation by Clostridium cellulolyticum in a bioreactor with pH control of the batch culture and using a defined medium was performed. Depending on cellulose concentration, the carbon flow distribution was affected, showing the high flexibility of the metabolism. With less than 6.7 g of cellulose liter−1, acetate, ethanol, H2, and CO2 were the main end products of the fermentation and cellulose degradation reached more than 85% in 5 days. The electron flow from the glycolysis was balanced by the production of H2 and ethanol, the latter increasing with increasing initial cellulose concentration. From 6.7 to 29.1 g of cellulose liter−1, the percentage of cellulose degradation declined; most of the cellulase activity remained on the cellulose fibers, the maximum cell density leveled off, and the carbon flow was reoriented from ethanol to acetate. In addition to that of previously indicated end products, lactate production rose, and, surprisingly enough, pyruvate overflow occurred. Concomitantly the molar growth yield and the energetic yield of the biomass decreased. Growth arrest may be linked to sufficiently high carbon flow, leading to the accumulation of an intracellular inhibitory compound(s), as observed on cellobiose (E. Guedon, M. Desvaux, S. Payot, and H. Petitdemange, Microbiology 145:1831–1838, 1999). These results indicated that bacterial metabolism exhibited on cellobiose was distorted compared to that exhibited on a substrate more closely related to the natural ecosystem of C. cellulolyticum. To overcome growth arrest and to improve degradation at high cellulose concentrations (29.1 g liter−1), a reinoculation mode was evaluated. This procedure resulted in an increase in the maximum dry weight of cells (2,175 mg liter−1), cellulose solubilization (95%), and end product concentrations compared to a classical batch fermentation with a final dry weight of cells of 580 mg liter−1 and 45% cellulose degradation within 18 days.
Cellulolytic clostridia play a major role in cellulose decomposition, which is a key process in carbon cycling (29). Clostridium cellulolyticum is a nonruminal cellulolytic mesophilic bacterium isolated from decayed grass and capable of degrading crystalline cellulose (36). The biotechnological exploitation of this microorganism as well as the understanding of the role it plays in its own ecosystem requires knowledge of its metabolism and of its behavior when developed on cellulose.
C. cellulolyticum is a low-G+C gram-positive anaerobe belonging to clostridial group III (39, 40); it is also placed in family 4, genus 2, in a new proposed-hierarchical structure for clostridia (7). Recent metabolic investigations with this bacterium indicated that (i) compared to a complex medium previously used, mineral salt medium clearly produced a different regulatory response and permitted better control of the carbon flow (19, 34), (ii) early growth inhibition was associated with a carbon excess (18), and (iii) carbon-limited and carbon-saturated chemostats displayed major discrepancies in the regulation of carbon flow (20). These studies were performed using cellobiose, which is considered the most important end product of the enzymatic cellulose hydrolysis (31, 44, 45), and it was assumed that growth on cellulosic material was rather difficult to monitor and that metabolism changes would be more observable with soluble sugar (14, 17).
Concerning the behavior of C. cellulolyticum towards cellulose substrate, earlier studies (i) suggested that the release of soluble sugars inhibited both cell growth and cellulase production (37) and (ii) described the growth of the bacteria on cellulose in terms of adhesion, colonization, release, and readhesion processes (13). These experiments, however, were conducted systematically in complex media without pH regulation, and many of the effects observed may have been due to a decrease in the pH of these cultures (47). In fact, compared with other low-G+C gram-positive anaerobes, particularly with bacteria defined as lactic acid bacteria, the bacteria of the clostridial type are generally considered to be restricted to a less acidic ecological niche due to their particular pattern of intracellular pH regulation (21, 41).
Bacterial growth on cellulose differs from that on cellobiose by the necessity for bacteria first to adhere on the substrate and second to degrade it into soluble sugars. Thus both substrate limitation and substrate-sufficient periods could be encountered by bacteria, and so the carbon flow on a cellulose batch culture could be somewhere between that associated with carbon limitation when bacteria are released and that associated with carbon-sufficient conditions when bacteria have adhered to cellulose fibers.
The aim of the present work was to investigate how C. cellulolyticum managed the abundance of insoluble substrate and to see whether or not the previous observations of cellobiose (15, 17–20, 34) were typical of bacterial behavior on cellulose. Taking into account previous considerations, this investigation on the metabolism and cellulolytic performance of C. cellulolyticum in batch fermentations was performed with controlled pH and using a mineral salt-based medium, which is more representative of the natural ecosystem of the bacterium.
MATERIALS AND METHODS
Chemicals.
All chemicals were of highest-purity analytical grade. Unless mentioned otherwise, commercial reagents, enzymes, and coenzymes were supplied from Sigma Chemical Co., St. Louis, Mo. All gases used were purchased from Air Liquide, Paris, France.
Organism and medium.
C. cellulolyticum ATCC 35319 was isolated from decayed grass by Petitdemange et al. (36). Stocks of spores from the original isolation, stored at 4°C, were heated to 80°C for 10 min and inoculated on cellulose medium (28, 51). Cells were subcultured once on cellobiose or cellulose before transfer and growth in a bioreactor as previously described (18). The anaerobic culture technique used was that proposed by Hungate (23) as modified by Bryant (6).
The defined medium used in all experiments was a modified CM3 medium as described previously by Guedon et al. (18). This medium contained cellobiose or cellulose MN301 (formerly MN300; Macherey-Nagel, Düren, Germany) in various amounts as specified in Results.
Growth conditions.
C. cellulolyticum was grown in batch culture with cellobiose or cellulose as the sole carbon and energy source as previously described by Guedon et al. (18). Bacteria were cultured aseptically in a 2-liter bioreactor (LSL Biolafitte, St. Germain en Laye, France) with a 1.5-liter working volume. The temperature was maintained at 34°C, and the pH was controlled at 7.2 by automatic addition of 3 N NaOH. Agitation was kept constant at 50 rpm. The inoculum was 10% by volume from an exponentially growing culture. Cellulose bioreactors were connected to a gasometer filled with a saturated NaCl-water solution and acidified at pH 1.0 with H2SO4 to prevent gas dissolution as described by Pollack (38). All tubing was made of Viton to preserve the anoxic condition of the cell culture.
Analytical procedures.
Cell growth on cellobiose was monitored spectrophotometrically at 600 nm and calibrated against cell dry weight measurement as previously described (18). On cellulose, biomass was estimated by bacterial protein measurement (33). From bacteria growing on cellobiose a cell dry weight-protein correlation was established, and this correlation was assumed to be the same for cells grown on particulate cellulose. Protein was measured by a modification of the Bradford dye method (5) as follows. A sample (30 ml) was centrifuged (8,000 × g for 15 min at 4°C) and washed twice with 0.9% (wt/vol) NaCl. The pellet was resuspended in 2 ml of 0.2 N NaOH, and this suspension was placed in a boiling water bath for 10 min (50). After cooling, the hydrolyzed sample was centrifuged as described above and the supernatant was diluted in 0.2 N NaOH as well as crystalline bovine serum albumin, which was used as the standard. The protein concentration was then estimated using the Coomassie brilliant blue reagent and reading the absorbance at 595 nm.
Cellulose concentration was determined as described by Huang and Forsberg (22). Residual cellulose was washed using acetic acid-nitric acid reagent and water to achieve removal of noncellulosic materials as described by Updegraff (46). Cellulose was then quantified by using the phenol-sulfuric acid method (8, 9) with glucose as the standard. Equivalent anhydroglucose was used for calculation.
The relative crystallinity index of the cellulose was determined as described by Shi and Weimer (43) following a technique that removed adherent microbial cells and prevented the recrystallization of the cellulose (26).
Hydrogen and carbon dioxide were analyzed on a gas chromatography unit as previously described (19). Gases dissolved in the culture medium were liberated using concentrated sulfuric acid as described by Freier et al. (12).
Culture supernatants (10,000 × g, 15 min, 4°C) were stored at −80°C until they were analyzed. The reducing sugar concentration was determined by a colorimetric ferricyanide method (32) using glucose as the standard. Glucose was assayed enzymatically using glucose oxidase and peroxidase, with o-dianisidine as a chromophore.
Acetate, ethanol, lactate, and succinate levels were determined using the appropriate enzyme kits (Boehringer Mannheim, Meylan, France).
Extracellular pyruvate was assayed enzymatically by fluorometric detection of NADH as previously described (18).
Total cellulase activity was based on the avicelase determination method described by Wood and Bhat (52). Incubation was performed at 34°C in 25 mM phosphate buffer (pH 7.2) using cellulose MN301 as the substrate. Liberation of reducing sugars was measured by the method of Miller (30) with glucose as the standard. One unit of total cellulase activity was defined as the amount of enzyme which released 1 μmol of reducing sugar per min.
All experiments were carried out in triplicate and repeated if experimental variation exceeded 10%.
Calculations.
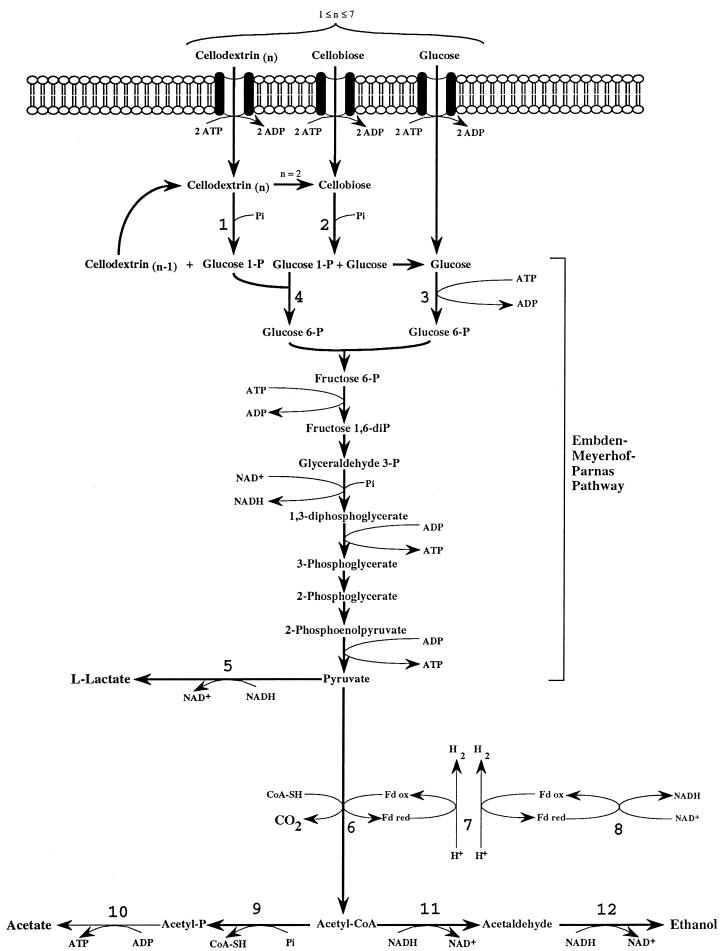
The main products of cellulose fermentation by C. cellulolyticum were acetate, ethanol, lactate, H2, and CO2 (see Results). Balance equations were established taking into account previous investigations (20, 34) and compiled in Fig. 1. Since cellodextrins are water-soluble β-1,4 oligomers of glucose with degrees of polymerization (n) between 2 and 7 (35) it was assumed that carbohydrates from glucose to celloheptaose could potentially be incorporated by bacteria. The cellulose fermented into products that can then be expressed as n hexose equivalents (hexose eq), which correspond to the glucose residue inside the cellulose chain. As specified in the scheme of cellulose catabolism (Fig. 1), it was assumed that (i) two ATP molecules were consumed for the transport system (42), (ii) one molecule of glucose and (n − 1) molecules of glucose-1-phosphate were formed from one molecule of soluble β-glucan (n) (1 ≤ n ≤ 7) according to the model of Strobel (44), (iii) the net ATP formation from glucose to pyruvate via the Embden-Meyerhof-Parnas (EMP) pathway was two molecules of ATP, (iv) a balance of three molecules of ATP was produced by the EMP pathway from glucose-1-phosphate, (v) there was production of two extra ATP molecules per hexose equivalent by acetate kinase, (vi) two NAD+ molecules per hexose eq were reduced by GAPDH (glyceraldehyde-3-phosphate dehydrogenase), (vii) two molecules of NAD+ per hexose eq were formed by the lactate dehydrogenase, and (viii) four molecules of NADH per hexose eq were reoxidized by acetaldehyde dehydrogenase and alcohol dehydrogenase. Then the cellulose conversion can be written as a general equation as follows. The equation for the conversion to acetate is
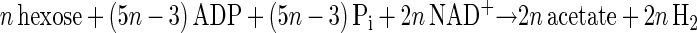 |
1 |
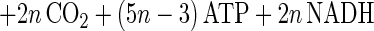 |
The equation for the conversion to ethanol is
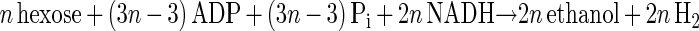 |
2 |
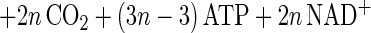 |
And the equation for the conversion to lactate is
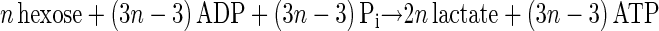 |
3 |
The energetic yield of biomass (YATP) was estimated from acetate, ethanol, and lactate concentrations. From equation 1, when a molecule of acetate was formed, (5n − 3)/2n molecules of ATP were produced. For example, if n = 1 (glucose), then 2 acetate molecules and 2 ATP molecules are produced, i.e., 1 ATP molecule per acetate molecule; if n = 2 (cellobiose), 4 acetate molecules and 7 ATP molecules are produced, i.e., 1.75 ATP molecules per acetate molecule; if n = 3 (cellotriose), 6 acetate molecules and 12 ATP molecules are produced, i.e., 2 ATP molecules per acetate molecule. From equations 2 and 3, when a molecule of ethanol or lactate was formed, (3n − 3)/2n ATP molecules were produced. For example, if n = 1, no ATP molecules and either 2 ethanol molecules or 2 lactate molecules are produced, i.e., 0 ATP molecules per ethanol or lactate molecule; if n = 2, 3 ATP molecules and either 4 ethanol molecules or 4 lactate molecules are produced, i.e., 0.75 molecules of ATP per ethanol or lactate molecule; if n = 3, 6 ATP molecules and either 6 ethanol molecules or 6 lactate molecules are produced, i.e., 1 ATP molecule per ethanol or lactate molecule. So from n = 1 to 7 averages of 1.94 ATP molecules per acetate molecule and 0.94 ATP molecules per ethanol or lactate molecule were found and YATP was estimated as follows: YATP = biomass concn/(1.94 concnacetate + 0.94 concnethanol + 0.94 concnlactate) where concn stands for concentration and YATP is expressed in grams of cells per mole of ATP produced.
FIG. 1.
Scheme of the catabolism of cellulose by C. cellulolyticum. Total cellulase activity liberated soluble β-glucans (n, the number of hexose residues inside the polymer) (i.e., 1 ≤ n ≤ 7), which were then incorporated and metabolized by bacteria. 1, cellodextrin phosphorylase (EC 2.4.1.49); 2, cellobiose phosphorylase (EC 2.4.1.20); 3, glucokinase (EC 2.7.1.2); 4, phosphoglucomutase (EC 5.4.2.2); 5, l-lactate dehydrogenase (EC 1.1.1.27); 6, pyruvate-fd oxidoreductase (EC 1.2.7.1); 7, hydrogenase (EC 1.18.99.1); 8, NADH-fd reductase (EC 1.18.1.3); 9, phosphotransacetylase (EC 2.3.1.8); 10, acetate kinase (EC 2.7.2.1); 11, acetaldehyde dehydrogenase (EC 1.2.1.10); 12, Alcohol dehydrogenase (EC 1.1.1.1). CoA-SH, coenzyme A; ox, oxidized; red, reduced. Fd, ferredoxin.
The molar growth yield, YX/S, is expressed in grams of cells per mole of hexose fermented. The qhexose is the specific rate of anhydroglucose residue fermented in millimoles per gram of cells per hour. qacetate, qethanol, qlactate, qhydrogen, and qcarbon dioxide are the specific rates of product formation in millimoles per gram of cells per hour. Extracellular qpyruvate is the specific rate of extracellular pyruvate formation in micromoles per grams of cells per hour. The specific production or utilization rates were the derivatives of the time course plots.
RESULTS
Cellulose degradation by C. cellulolyticum in batch cultures.
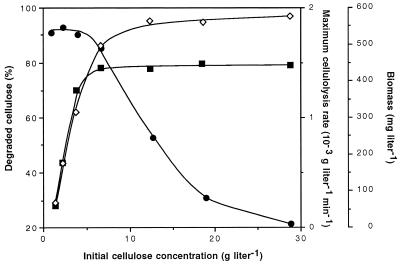
C. cellulolyticum was grown in batch culture on synthetic medium at pH 7.2 with seven concentrations of cellulose MN301: 5.6, 14.8, 24.1, 41.4, 78.4, 116.7, and 179.6 mM, expressed as hexose eq, i.e., 0.9, 2.4, 3.9, 6.7, 12.7, 18.9, and 29.1 g of cellulose liter−1, respectively. The percentage of solubilized cellulose within 120 h as a function of the initial cellulose concentration was determined (Fig. 2), and it was found that around 91% degradation was achieved with less than 3.9 g of initial cellulose liter−1, but this percentage dropped and reached 21% with the highest cellulose concentration. Such a decrease indicated that approximately the same amount of cellulose was hydrolyzed so that cellulolysis performances were close to their maximum at and above 3.9 g of cellulose added liter−1. In fact, the maximum dry weight of cells increased with initial cellulose amount, and above 6.7 g liter−1 it remained quite constant around 575 mg liter−1 (Fig. 2). The maximum rate of cellulose degradation measured in the course of each fermentation paralleled the maximum cell dry weight and reached about 1.5 × 10−3 g liter−1 min−1 at and above 6.7 g of cellulose added liter−1. The rate of cellulose hydrolysis appeared therefore to be related to biomass production, and this partly explained why the percentage of degradation was restricted above 3.9 g of cellulose added liter−1.
FIG. 2.
The percentage of solubilized cellulose ●, maximum biomass ◊, and maximum rate of cellulose degradation ■ reached within 120 h of batch fermentation of C. cellulolyticum as a function of the initial cellulose concentration.
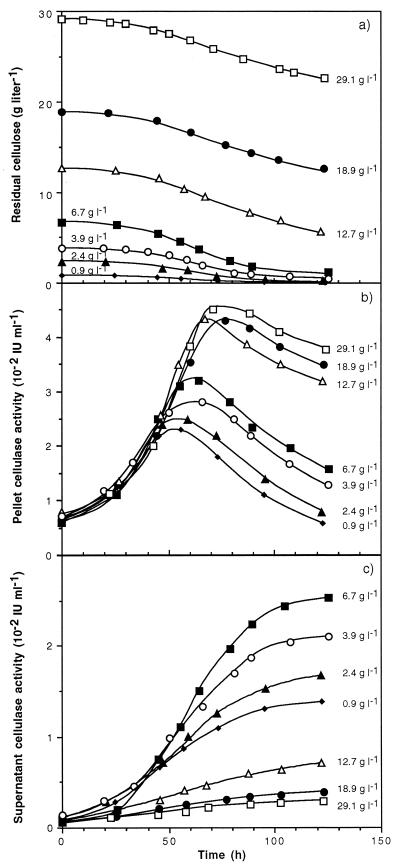
The kinetics of cellulose solubilization within 120 h of fermentation showed similar patterns whatever the initial cellulose concentration (Fig. 3a). Cellulose hydrolysis increased up to 50 to 70 h and then began to slow down. From 0.9 to 3.9 g of initial cellulose liter−1, the decrease of cellulose degradation was due to an exhaustion of the cellulose, but above 6.7 g of cellulose liter−1 this slowdown was not correlated with the cellulose depletion or a change in the crystalline structure of the cellulose. For example, at the end of the fermentation with 6.7 g of initial cellulose liter−1, the relative crystallinity index was 89.6 compared with 89.9 for the original cellulose MN301. Consequently, residual cellulose was not enriched in its crystalline content in the course of the cell culture.
FIG. 3.
Kinetics of cellulose consumption (a) and cellulase activity in the pellet (b) and in the culture supernatant (c) during batch fermentation of C. cellulolyticum on cellulose at various initial concentrations.
To understand the variation of cellulolysis, the total cellulase activity during the fermentation was then examined. With increasing initial cellulose concentration, the maximum of pellet-associated cellulase activity, including cells with cellulosome and free cellulosome adhered to cellulose fibers (2, 13, 14), rose (Fig. 3b). A peak in activity was hit between 50 and 80 h, and activity then decreased. During fermentations with initial substrate concentrations higher than 12.7 g liter−1, the maximum cellulase activity in the pellet remained quite constant around 4.4 × 10−2 IU ml−1 and the later decline was slower than with lower cellulose concentrations. In the supernatant, the cellulase activity appeared after about 24 h and reached higher values as the initial cellulose concentration increased from 0.9 to 6.7 g of cellulose liter−1 (Fig. 3c). Above 6.7 g of initial cellulose liter−1, however, the previous upward trend was reversed and little cellulase activity was measured in the supernatant. At 120 h, about 90% of the total cellulase activity was in the pellet with the highest initial substrate concentration, while with 6.7 g of cellulose liter−1 up to 70% of the total activity was in the supernatant. As the maximum degradation rate and biomass production paralleled the maximum cellulase activity in the pellet, the cellulose hydrolyzed was closely linked to the degradative activity measured. Microscopic examination at the end of the cultures revealed that most bacteria adhered to the cellulose particles, but with initial carbohydrate concentrations less than 6.7 g liter−1 many more cells were in the supernatant (data not shown). So the variations in cellulose solubilization in the course of fermentation could be attributed to a release of the cellulase system and of cellulolytic bacteria due to an excess with respect to the number of cellulose particles. These data are in agreement with the model of tight adhesion of the cellulosome as well as bacteria to cellulose fibers as the primary event required in the efficient degradation and growth on insoluble substrates (2, 3).
Accumulation of soluble sugars occurred only after growth ceased (data not shown) but remained limited. At 120 h, with 29.1 g of initial cellulose liter−1, 126 mg of reducing sugars and 15 mg of glucose liter−1, corresponding to 0.55 and 0.06% of the remaining cellulose, respectively, were detected; with 6.7 g of cellulose liter−1, the reducing sugars (77 mg liter−1) and glucose (12 mg liter−1) represented 5.83 and 0.91% of the remaining cellulose, respectively.
Metabolite production during batch cellulose fermentation.
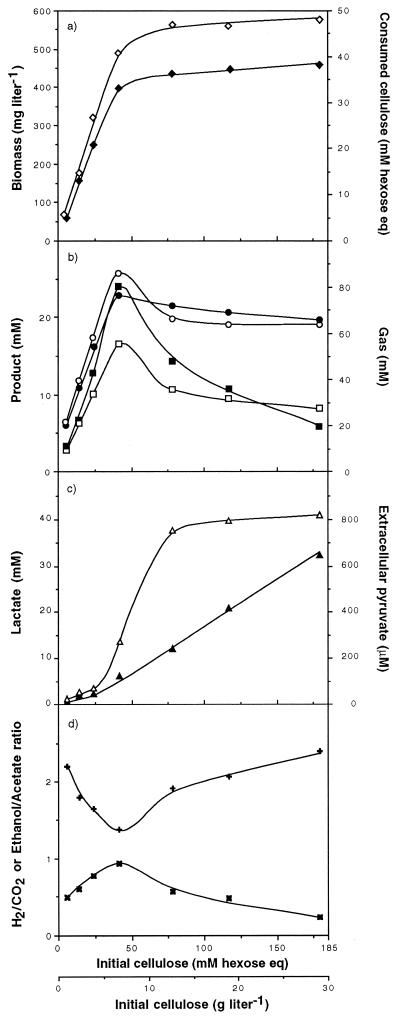
With increasing amounts of substrate and within 120 h of fermentation, the consumed cellulose concentration, expressed as millimolar hexose eq, rose (Fig. 4a). Above 41.4 mM initial cellulose (i.e., 6.7 g liter−1), the increase in the quantity of consumed cellulose slowed down and the concentration reached 36.8 mM with 179.6 mM initial cellulose (i.e., 29.1 g liter−1). The production of biomass paralleled the amount of hexose eq consumed. Substrate limitation or inhibition by fermentation products could not explain growth arrest since the same culture, when reinoculated, allowed further growth of a new cell inoculum (see Fig. 6aII).
FIG. 4.
Maximum biomass and cellulose consumption (a), metabolite concentrations (b and c), and product ratio (d) obtained within 120 h of batch culture of C. cellulolyticum as a function of the initial cellulose concentration. ◊, biomass; ⧫, consumed cellulose as hexose eq; ■, ethanol; ○, acetate; ●, H2; □, CO2; ▴, lactate; ▵, extracellular pyruvate; ✖, ethanol/acetate ratio; ✚, H2/CO2 ratio.
FIG. 6.
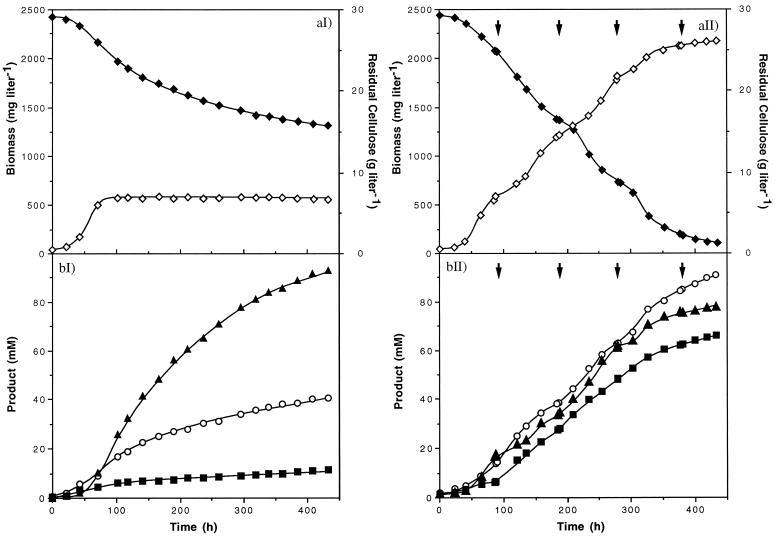
Growth and residual cellulose concentration (a) and product concentrations (b) during classical batch fermentation (I) and reinoculated culture (II) of C. cellulolyticum with 29.1 g of cellulose liter−1. Arrows, reinoculation times. ◊, biomass; ⧫, cellulose; ■, ethanol; ○, acetate; ▴, lactate.
On cellulose, as previously observed with cellobiose (18–20), acetate, ethanol, lactate, hydrogen, and carbon dioxide were the primary metabolic end products and no succinate was detected. The final levels of H2, CO2, acetate, and ethanol measured in the course of each fermentation increased linearly with cellulose added (Fig. 4b). But beyond 41.4 mM initial cellulose (i.e., 6.7 g liter−1) and up to the highest cellulose concentration, ethanol concentration decreased continuously from 24.0 to 5.6 mM. The total production of CO2 and acetate decreased sharply from 41.4 to 78.4 mM initial cellulose (i.e., 6.7 to 12.7 g liter−1, respectively) and was quite constant above 78.4 mM. H2 production decreased as well, but moderately, since it was 68.6 mM with 41.4 mM initial cellulose (i.e., 6.7 g liter−1) and 58.9 mM with 179.6 mM initial cellulose (i.e., 29.1 g of cellulose liter−1). Conversely, the final lactate concentration was low (less than 1.9 mM) but increased as soon as the initial substrate concentration was 41.4 mM (i.e., 6.7 g liter−1) and reached 32.4 mM with 179.6 mM initial cellulose (i.e., 29.1 g liter−1) (Fig. 4c). In the same way, the maximum extracellular pyruvate concentration rose sharply as soon as lactate was produced. With 41.4 mM initial cellulose (i.e., 6.7 g liter−1) the maximum extracellular pyruvate concentration was 289 μM and reached 821 μM with 179.6 mM initial cellulose (i.e., 29.1 g liter−1). From 5.6 to 41.4 mM initial cellulose (i.e., 0.9 to 6.7 g liter−1, respectively), the ethanol-to-acetate ratio increased until it reached 0.93 and then dropped with higher initial carbohydrate concentrations added (Fig. 4d). These data showed that biomass stagnation with cellulose concentrations higher than 41.4 mM (i.e., 6.7 g liter−1) was accompanied by a change in metabolic flux. Carbon flow towards acetate and ethanol dropped while lactate production rose, and a second metabolic shift arose from ethanol-to-acetate formation. The change in the upward trend of the ethanol-to-acetate ratio was also paralleled by a reverse in the downward trend of the H2-to-CO2 ratio (Fig. 4d). This ratio, always higher than 1, suggested that H2 was produced via NADH-ferredoxin (fd) reductase and hydrogenase activities in addition to the pyruvate-fd oxidoreductase and hydrogenase activities (Fig. 1) (18–20).
Kinetic analysis of fermentation products in batch culture on cellulose.
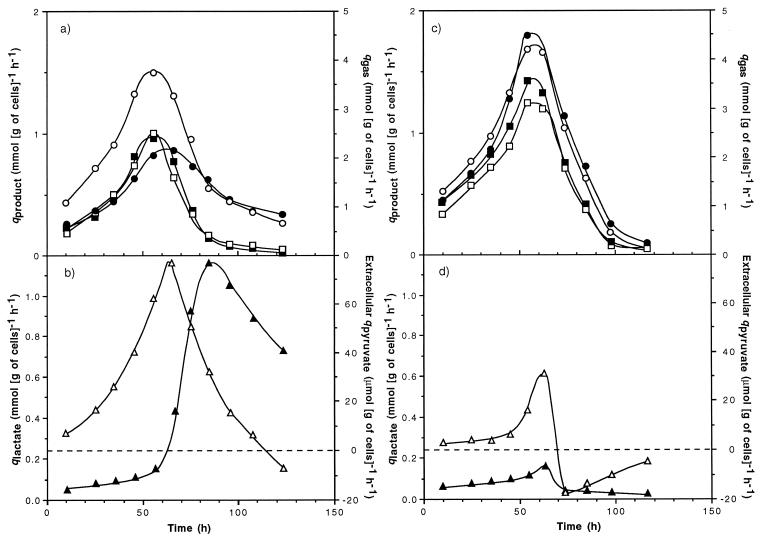
When C. cellulolyticum was grown in batch culture on a defined medium with 179.6 mM initial cellulose (i.e., 29.1 g of cellulose liter−1), the specific production rates of acetate, ethanol, CO2, and H2 accelerated during the first 60 h after inoculation, and this acceleration was followed by decreasing specific production rates (Fig. 5a). During the first 60-h period, the specific rate of lactate production was almost constant and low: around 0.07 mmol (g of cells)−1 h−1 (Fig. 5b). The peak of extracellular pyruvate formation, i.e., 78.60 μmol (g of cells)−1 h−1, coincided with the start of lactate production. The extracellular qpyruvate decreased continuously, and pyruvate was consumed about 50 h after its maximum production rate was reached. The increasing qlactate, with a maximum of 1.17 mmol (g of cells)−1 h−1, corresponded to the decrease in the specific rates of acetate, ethanol, CO2, and H2 production.
FIG. 5.
Specific rates of product formation during batch fermentations of C. cellulolyticum. The initial cellulose concentrations were 179.6 mM (i.e., 29.1 g liter−1) (a and b) and 24.1 mM (i.e., 3.9 g liter−1) (c and d). ■, ○, ▴, ●, □, and ▵, specific rates of ethanol, acetate, lactate, H2, CO2, and extracellular pyruvate formation, respectively.
With bacterial fermentation on 24.1 mM initial cellulose (i.e., 3.9 g liter−1), the specific production rates of acetate, ethanol, CO2, and H2 followed the same pattern as that previously described, but maximum specific rates attained were higher and the later decelerating phase was more abrupt (Fig. 5c). A similar increase of qlactate could be observed too (Fig. 5d). Yet after the maximum specific rate of extracellular pyruvate formation was reached, pyruvate was consumed within hours and lactate biosynthesis stopped. As a result, the specific production rates of lactate and extracellular pyruvate were much lower than with a fermentation started at 179.6 mM initial cellulose (i.e., 29.1 g liter−1) since they reached maximum values of 0.14 mmol (g of cells)−1 h−1 and 30.59 μmol (g of cells)−1 h−1, respectively.
The maximum specific growth rate (μmax) obtained on cellulose was one-third that for batch cellobiose fermentation (Table 1). Whatever the initial cellulose concentration, the μmax was around 0.056 h−1, which corresponded to a generation time of 12.4 h. The maximum qhexose obtained on cellulose was five times lower than that on cellobiose. The carbon flow entering into the cell increased with cellulose concentration added but remained constant above 41.4 mM initial cellulose (i.e., 6.7 g liter−1), whereas maximum qacetate and qethanol decreased. At and above 41.4 mM initial cellulose (i.e., 6.7 g liter−1) the maximum qlactate continuously increased, while the extracellular qpyruvate remained constant. On the average, the specific rates of acetate, ethanol, lactate, and extracellular pyruvate formation were at least twofold lower than on cellobiose. With less than 41.4 mM initial cellulose (i.e., 6.7 g liter−1), the molar growth yields remained quite constant, around 35.9 (g of cells) (mol of hexose)−1 and were comparable to that obtained on cellobiose. In this range of initial cellulose concentrations, the same comparison could be done concerning the energetic yield of biomass since YATP was about 20.6 (g of cells) (mol of ATP)−1 compared to 23.2 (g of cells) (mol of ATP)−1 on cellobiose. Above 41.4 mM initial cellulose (i.e., 6.7 g liter−1), YX/S and YATP dropped to about 25.7 (g of cells) (mol of hexose)−1 and 13.2 (g of cells) (mol of ATP)−1, respectively. The decrease in YX/S accompanied by that in YATP suggested the existence of an uncoupling growth phenomenon, namely, that some catabolized hexose and hence ATP were not associated with biomass production.
TABLE 1.
Maximum valuesa of specific rates of product formation and consumption, of molar growth yield, and of energetic yield of biomass obtained in the course of the batch fermentations of C. cellulolyticum
| Initial concn [mM hexose eq (g liter−1)] of cellulose or cellobiose |
q value (mmol [g of cells]−1 h−1) for:
|
Extracellular qpyruvate (μmol [g of cells]−1 h−1) | μmax (h−1) | YX/S ([g of cells] [mol of hexose eq]−1) | YATP ([g of cells] [mol of ATP]−1) | |||
|---|---|---|---|---|---|---|---|---|
| Hexose | Ethanol | Acetate | Lactate | |||||
| Cellulose | ||||||||
| 5.6 (0.9) | 1.42 | 0.90 | 1.20 | 0.06 | 9.68 | 0.055 | 36.8 | 18.9 |
| 14.8 (2.4) | 1.78 | 1.02 | 1.39 | 0.08 | 10.17 | 0.056 | 36.5 | 19.5 |
| 24.1 (3.9 | 1.93 | 1.43 | 1.68 | 0.14 | 30.59 | 0.057 | 35.7 | 21.2 |
| 41.4 (6.7) | 1.99 | 1.51 | 1.79 | 0.40 | 49.35 | 0.056 | 34.7 | 22.8 |
| 78.4 (12.7) | 2.06 | 1.09 | 1.61 | 0.58 | 80.38 | 0.057 | 25.5 | 12.4 |
| 116.07 (18.9) | 2.13 | 0.98 | 1.58 | 0.74 | 81.45 | 0.055 | 25.9 | 13.8 |
| 179.6 (29.1) | 2.19 | 0.97 | 1.51 | 1.17 | 78.60 | 0.056 | 25.6 | 13.3 |
| Cellobiose | ||||||||
| 46.8 (8.0) | 10.71 | 1.98 | 3.70 | 2.64 | 155.80 | 0.154 | 31.6 | 23.2 |
Values are the averages of three different experiments and were determined with an average accuracy of ±10%.
The values were the maximum ones encountered during the entire incubation time and were not calculated over the same period of time.
Cellulose degradation in a bioreactor reinoculated with C. cellulolyticum.
With substrate concentrations higher than 6.7 g of cellulose liter−1, cell growth was not limited by available cellulose sites and the growth arrest of the seeding could be explained by a carbon flow which led to an accumulation of an intracellular inhibitory compound(s) (18). Indeed the rate of cellulose catabolism apparently exceeded the rate of pyruvate consumption, since pyruvate accumulated. Taking into account this hypothesis, a reinoculation process to improve the degradation of high cellulose concentrations was attempted. By this process, when the cells entered the stationary phase (i.e., about every 96 h), a new inoculum of C. cellulolyticum was introduced into the bioreactor (Fig. 6). In this way, with 29.1 g of initial cellulose liter−1 (i.e., 179.6 mM cellulose) more than 95% degradation occurred within 18 days (i.e., four reinoculations) against 45% in classical batch fermentation. In the reinoculated culture, the final concentrations of ethanol and acetate were 65.6 and 90.5 mM, respectively (Fig. 6bII) compared to 11.4 and 40.6 mM, respectively, in classical culture (Fig. 6bI). The final lactate concentration (77.3 mM), however, was lower than that in classical batch culture (93.1 mM). A final cell dry weight of 2,175 mg liter−1 was reached with the reinoculation process compared to about 580 mg liter−1 with the classical procedure (Fig. 6aI and 6aII). In classical batch fermentation, once the stationary phase was reached, bacteria acted as resting cells (Fig. 6aI): cellulose was hydrolyzed, cell lysis did not occur within the remaining 14 days, and bacteria were catabolically active since metabolites, mainly lactate, were produced. These data indicated that acetate and ethanol production was associated with cell growth whereas lactate formation paralleled growth inhibition. With the fourth inoculum, the cellulose was not degraded further (Fig. 6aII). In fact, no growth was observed, and, at this time, the cellulose concentration was less than 2 g liter−1; the remaining cellulose was certainly saturated with bacteria coming from the preceding inocula, which were not able to initiate a new cell division and which did not allow adhesion and growth of cells from a new inoculum. Thus such a reinoculation process allowed improved maximum dry weight of cells, cellulose solubilization, and end product concentrations.
DISCUSSION
Since the early studies of cellulose hydrolysis by C. cellulolyticum, this microorganism has been considered a sluggish cellulolytic bacterium (16, 37). It was described as taking half a month to attain about 70% degradation with initial cellulose concentrations lower than 7.6 g liter−1 (16). Moreover, limitation of cellulolysis was attributed to a change in cellulose structure, such as a progressive increase in the lattice crystallinity due to the initial degradation through the cellulasic system. As a result, the kinetics of cellulose degradation could be divided into three distinct periods corresponding to three consecutive enzymatic activity levels.
In the present study, cellulolytic performance of C. cellulolyticum was improved, since with initial cellulose concentrations less than 6.7 g liter−1 more than 85% degradation occurred in 5 days. A modification of the crystalline structure of the substrate could not account for the slowdown in the cellulose degradation rate. Changes in the distribution of the cellulase activity (i.e., present on the cells or as a free cellulasic system) between the cellulose fibers and the supernatant explain the slowing down in the cellulose solubilization during the culture. Studies of the fermentation of different cellulose structures by ruminal cellulolytic bacteria indicated that the available surface area was a more important determinant of the digestion rate than the crystallinity of the cellulose (48, 49). Recent investigations of crystalline cellulose degradation by the Clostridium thermocellum cellulosome showed that the various cellulase factors acted in unison with a very efficient synergism in which the influence of digestion time on the crystallinity of the substrate was limited (4). In the present investigation, the maximum generation time was greatly reduced (i.e., 12.4 h versus the 24.0 h found by Giallo et al. [16]) and primary metabolite production, i.e., of acetate, ethanol, and lactate, evolved differently (16). These differences could be attributed to several modifications made in the present study such as the use of a synthetic medium (19, 34), the pH regulation in the course of fermentation (11), the continuous stirring of the medium (12, 31), and the fact that the cultures were carried out at atmospheric pressure without accumulation of the fermentation gases inside the bioreactor (12, 27).
With initial cellulose concentrations lower than 41.4 mM (i.e., 6.7 g liter−1), the more the cellulose was hydrolyzed the less the cellulasic system could find new adherence sites on the cellulose fibers. As the initial cellulose amount increased, the biomass production increased and in turn the total culture cellulase activity rose globally but more activity was measured in the supernatant at the end of the fermentation. Since the stationary phase occurred before cellulose was depleted, biomass limitation was more probably due to a lack of available adhesion sites on cellulose as bacteria grew (13, 14). Thus, with less than 41.4 mM cellulose (i.e., 6.7 g liter−1), cellulolysis was limited by both the accessibility of the cellulasic system to cellulose fibers and, as a result, the biomass concentration, which depends on the quantity of soluble cellodextrins released. In this range of cellulose concentrations, the ethanol-to-acetate ratio increased continuously towards 1 and concomitantly less H2 was produced. So H2 and ethanol production may regulate the intracellular NADH/NAD+ ratio as a result of a good control of the electron flow (18, 19). Likewise, carbon flow appeared to be correctly regulated since pyruvate overflow did not occur. Moreover, with increasing substrate concentration the flux, which was first mainly directed to acetate production, was partly reoriented to the ethanol pathway, while little lactate was formed. YX/S and YATP values were comparable to those obtained on cellobiose even though qhexose was at least five times lower. In these conditions, cells managed to optimize their growth on cellulose.
With higher initial cellulose concentrations, above 41.4 mM (i.e., 6.7 g liter−1), the increase of lactate production concomitant with the decrease of acetate and (mainly) ethanol biosynthesis showed the clear reorientation of the catabolism. The change was paralleled by an increase in the H2/CO2 ratio, which suggested that the intracellular NADH/NAD+ ratio was equilibrated through NADH-fd reductase and hydrogenase activities, which may compensate for the decrease of ethanol production (18–20). Moreover, cell dry weight did not increase, and most of the total cellulase activity remained on cellulose fibers. This biomass limitation could not be due to nutritional restriction or inhibitory fermentation metabolites since new cells reinoculated in the same culture were able to grow. The presence of extracellular pyruvate, though, means that the rate of cellulose catabolism exceeded the rate of pyruvate consumption. In this range of cellulose concentrations, as previously observed for cellobiose, these results suggest that the rate of hexose catabolism could exceed the rate of pyruvate consumption via pyruvate-fd oxidoreductase as well as anabolic pathways and may indicate that these enzymes are the rate-limiting step (19, 20). Values of YX/S and YATP were both lower than values obtained with less than 41.4 mM initial cellulose (i.e., 6.7 g liter−1), indicating that an uncoupling growth phenomenon had occurred. Since this decrease also came with pyruvate overflow accompanied by lactate production, growth inhibition was certainly related to an accumulation of an intracellular inhibitory compound(s) due to a deficient regulation of the entering carbon, as suggested by results with cellobiose (18). Even at high cellulose concentrations, however, lower specific rates of product formation and consumption indicated that the metabolism was not deregulated as much as it was on cellobiose (Table 1).
Whatever the initial cellulose concentrations, low soluble sugar concentrations were detected in the supernatant. These results are in agreement with the concept that the depolymerization of insoluble substrate to soluble cellodextrins limits the cellulose fermentation (33). By estimating and comparing qhexose as well as specific rates of product formation to those obtained with cellobiose batch cultures, the present investigation provides further evidence that this concept is well founded. As expected, the entering carbon flow and, as a result, the specific production rates with insoluble cellulose were lower than those with cellobiose, a soluble sugar. In addition, extracellular pyruvate was reconsumed earlier at low initial cellulose concentrations than at higher ones. These data showed that the conversion of insoluble carbohydrate to soluble cello-oligosaccharides was the rate-limiting step in cellulose fermentation, but even so, pyruvate overflow could occur.
The results from the reinoculated culture clearly indicate that the growth arrest observed in classical batch culture was not due to high end product concentrations and was not induced by signal molecules excreted by bacteria at a particular cell density or growth phase (1, 10). The reinoculation process may allow the bypassing of the cell density limitation, a priori, due to self-intoxication of the cell with time, and may then permit the further colonization of available sites on the cellulose fibers and, as a result, an increased cellulose solubilization.
Even if at high carbohydrate concentrations cellulose catabolism was similar to that observed on cellobiose in some previously described aspects, it remained clearly different especially at low cellulose concentrations. In accordance with the initial substrate concentrations used, great differences of product concentrations as well as of specific production rates were observed. In cellulose batch fermentations, the cells managed, therefore, to efficiently regulate their metabolism by reorienting the carbon and electron flows. The higher values of specific rates of product formation and consumption obtained in cellobiose batch cultures showed that metabolism was distorted compared to that for a substrate more closely related to the natural ecosystem of the bacterium.
Since the situation of a microorganism under natural conditions is most probably somewhere between the closed batch culture and the open continuous-culture system (24, 25), further investigations with chemostats should allow a better understanding of the C. cellulolyticum behavior in its ecological niche with its natural insoluble substrate, the cellulose.
ACKNOWLEDGMENTS
This work was supported by the Commission of European Communities FAIR program (contract CT95-0191 [DG 12 SSMA]) and by the Agrice program (contract 9701041).
We thank E. McRae for correcting the English and for critical reading of the manuscript.
REFERENCES
- 1.Barrow P A, Lovell M A, Barber Z. Growth suppression in early stationary-phase nutrient broth cultures of Salmonella typhimurium and Escherichia coli is genus specific and not regulated by ςs. J Bacteriol. 1996;178:3072–3076. doi: 10.1128/jb.178.11.3072-3076.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bayer E A, Shoham Y, Tormo J, Lamed R. The cellulosome: a cell surface organelle for the adhesion to and degradation of cellulose. In: Fletcher M, editor. Bacterial adhesion molecular and ecological diversity. New York, N.Y: Wiley-Liss; 1996. pp. 155–182. [Google Scholar]
- 3.Béguin P, Lemaire M. The cellulosome: an exocellular, multiprotein complex specialized in cellulose degradation. Crit Rev Biochem Mol Biol. 1996;31:201–236. doi: 10.3109/10409239609106584. [DOI] [PubMed] [Google Scholar]
- 4.Boisset C, Chanzy H, Henrissat B, Lamed R, Shoham Y, Bayer E A. Digestion of crystalline cellulose substrates by the Clostridium thermocellum cellulosome: structural and morphological aspects. Biochem J. 1999;340:829–835. [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Bryant M P. Commentary on the Hungate technique for culture of anaerobic bacteria. Am J Clin Nutr. 1972;25:1324–1328. doi: 10.1093/ajcn/25.12.1324. [DOI] [PubMed] [Google Scholar]
- 7.Collins M D, Lawson P A, Willems A, Cordoba J J, Fernandez-Garayzabal J, Garcia P, Cai J, Hippe H, Farrow J A E. The phylogeny of the genus Clostridium: proposal of five new genera and eleven species combinations. Int J Syst Bacteriol. 1994;44:812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 8.Dubois M, Gilles K, Hamilton J K, Rebers P A, Smith F. A colorimetric method for the determination of sugars. Nature. 1951;168:167. doi: 10.1038/168167a0. [DOI] [PubMed] [Google Scholar]
- 9.Dubois M, Gilles K A, Hamilton J K, Rebers P A, Smith F. Colorimetric method for determination of sugars and related substances. Anal Chem. 1956;28:350–356. [Google Scholar]
- 10.Dunny G M, Leonard B A B. Cell-cell communication in Gram positive bacteria. Annu Rev Microbiol. 1997;51:527–564. doi: 10.1146/annurev.micro.51.1.527. [DOI] [PubMed] [Google Scholar]
- 11.Duong T V C, Johnson E A, Demain A L. Thermophilic, anaerobic and cellulolytic bacteria. In: Weisman A, editor. Topics in enzyme and fermentation biotechnology. New York, N.Y: John Wiley & Sons; 1983. pp. 156–195. [Google Scholar]
- 12.Freier D, Mothershed C P, Wiegel J. Characterization of Clostridium thermocellum JW20. Appl Environ Microbiol. 1988;54:204–211. doi: 10.1128/aem.54.1.204-211.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gelhaye E, Gehin A, Petitdemange H. Colonization of crystalline cellulose by Clostridium cellulolyticum ATCC 35319. Appl Environ Microbiol. 1993;59:3154–3156. doi: 10.1128/aem.59.9.3154-3156.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gelhaye E, Petitdemange H, Gay R. Adhesion and growth rate of Clostridium cellulolyticum ATCC 35319 on crystalline cellulose. J Bacteriol. 1993;175:3452–3458. doi: 10.1128/jb.175.11.3452-3458.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giallo J. Etude physiologique d'une bactérie cellulolytique mésophile anaérobie: Clostridium cellulolyticum (ATCC n° 35319). Ph.D. thesis. Marseille, France: Université de Provence; 1984. [Google Scholar]
- 16.Giallo J, Gaudin C, Belaich J P. Metabolism and solubilization of cellulose by Clostridium cellulolyticum H10. Appl Environ Microbiol. 1985;49:1216–1221. doi: 10.1128/aem.49.5.1216-1221.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giallo J, Gaudin C, Belaich J P, Petitdemange E, Caillet-Mangin F. Metabolism of glucose and cellobiose by cellulolytic mesophilic Clostridium sp. strain H10. Appl Environ Microbiol. 1983;45:843–849. doi: 10.1128/aem.45.3.843-849.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guedon E, Desvaux M, Payot S, Petitdemange H. Growth inhibition of Clostridium cellulolyticum by an inefficiently regulated carbon flow. Microbiology. 1999;145:1831–1838. doi: 10.1099/13500872-145-8-1831. [DOI] [PubMed] [Google Scholar]
- 19.Guedon E, Payot S, Desvaux M, Petitdemange H. Carbon and electron flow in Clostridium cellulolyticum grown in chemostat culture on synthetic medium. J Bacteriol. 1999;181:3262–3269. doi: 10.1128/jb.181.10.3262-3269.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guedon E, Payot S, Desvaux M, Petitdemange H. Relationships between cellobiose catabolism, enzyme levels and metabolic intermediates in Clostridium cellulolyticum grown in a synthetic medium. Biotechnol Bioeng. 2000;67:327–335. doi: 10.1002/(sici)1097-0290(20000205)67:3<327::aid-bit9>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 21.Huang L, Gibbins L N, Forsberg C W. Transmembrane pH gradient and membrane potential in Clostridium acetobutylicum during growth under acetogenic and solventogenic conditions. Appl Environ Microbiol. 1985;50:1043–1047. doi: 10.1128/aem.50.4.1043-1047.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang L, Forsberg C W. Cellulose digestion and cellulase regulation and distribution in Fibrobacter succinogenes subsp. succinogenes S85. Appl Environ Microbiol. 1990;56:1221–1228. doi: 10.1128/aem.56.5.1221-1228.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hungate R E. A roll tube method for cultivation of strict anaerobes. Methods Microbiol. 1969;33:117–132. [Google Scholar]
- 24.Jannash H W, Egli T. Microbiol growth kinetics: a historical perspective. Antonie Leeuwenhoek. 1993;63:213–224. doi: 10.1007/BF00871219. [DOI] [PubMed] [Google Scholar]
- 25.Kovárová-Kovar K, Egli T. Growth kinetics of suspended microbial cells: from single-substrate-controlled growth to mixed-substrate kinetics. Microbiol Mol Biol Rev. 1998;62:646–666. doi: 10.1128/mmbr.62.3.646-666.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kudo H, Cheng K J, Costerton J W. Electron microscopy study of the methylcellulose-mediated detachment of cellulolytic rumen bacteria from cellulose fibers. Can J Microbiol. 1987;33:267–272. doi: 10.1139/m87-045. [DOI] [PubMed] [Google Scholar]
- 27.Lamed R J, Lobos J H, Su T M. Effect of stirring and hydrogen on fermentation products of Clostridium thermocellum. Appl Environ Microbiol. 1988;54:1216–1221. doi: 10.1128/aem.54.5.1216-1221.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lapage S P, Shelton J E, Mitchell T G, Mackenzie A R. Culture collections and the preservation of bacteria. Methods Microbiol. 1970;3A:136–228. [Google Scholar]
- 29.Leschine S B. Cellulose degradation in anaerobic environments. Annu Rev Microbiol. 1995;49:399–426. doi: 10.1146/annurev.mi.49.100195.002151. [DOI] [PubMed] [Google Scholar]
- 30.Miller G L. Use of dinitrosalicylic acid reagent for determination of reducing sugars. Anal Chem. 1959;31:426–428. [Google Scholar]
- 31.Ng T, Zeikus J G. Differential metabolism of cellobiose and glucose by Clostridium thermocellum and Clostridium thermohydrosulfuricum. J Bacteriol. 1982;150:1391–1399. doi: 10.1128/jb.150.3.1391-1399.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park J T, Johnson M J. A submicro-determination of glucose. J Biol Chem. 1949;181:149–151. [PubMed] [Google Scholar]
- 33.Pavlostathis S G, Miller T L, Wolin M J. Fermentation of insoluble cellulose by continuous cultures of Ruminococcus albus. Appl Environ Microbiol. 1988;54:2655–2659. doi: 10.1128/aem.54.11.2655-2659.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Payot S, Guedon E, Cailliez C, Gelhaye E, Petitdemange H. Metabolism of cellobiose by Clostridium cellulolyticum growing in continuous culture: evidence for decreased NADH reoxidation as a factor limiting growth. Microbiology. 1998;144:375–384. doi: 10.1099/00221287-144-2-375. [DOI] [PubMed] [Google Scholar]
- 35.Pereira A N, Mobedshashi M, Ladish M R. Preparation of cellodextrins. Methods Enzymol. 1988;160:26–45. [Google Scholar]
- 36.Petitdemange E, Caillet F, Giallo J, Gaudin C. Clostridium cellulolyticum sp. nov., a cellulolytic mesophilic species from decayed grass. Int J Syst Bacteriol. 1984;34:155–159. [Google Scholar]
- 37.Petitdemange E, Tchunden T, Valles S, Pirson H, Raval G, Gay R. Effect of carbon sources on cellulase production by Clostridium cellulolyticum. Biomass Bioenergy. 1992;3:393–402. [Google Scholar]
- 38.Pollack H. Biological waste disposal from slaughterhouses. In: Ferrero G L, Ferranti M P, Naveau H, editors. Anaerobic digestion and carbohydrate hydrolysis of waste. London, England: Elsevier Applied Science Publishers; 1984. pp. 323–330. [Google Scholar]
- 39.Rainey F A, Stackebrandt E. 16S rRNA analysis reveals phylogenetic diversity among the polysaccharolytic clostridia. FEMS Microbiol Lett. 1993;113:125–128. doi: 10.1111/j.1574-6968.1993.tb06501.x. [DOI] [PubMed] [Google Scholar]
- 40.Rainey F A, Ward N L, Morgan H W, Toalster R, Stackebrandt E. Phylogenetic analysis of anaerobic thermophilic bacteria: aid for their reclassification. J Bacteriol. 1993;175:4772–4779. doi: 10.1128/jb.175.15.4772-4779.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Russel J B, Bond D R, Cook G M. The fructose diphosphate/phosphate regulation of carbohydrate metabolism in low G+C Gram positive anaerobes. Res Microbiol. 1996;147:528–534. doi: 10.1016/0923-2508(96)84008-3. [DOI] [PubMed] [Google Scholar]
- 42.Schneider E, Hunke S. ATP-binding-cassette (ABC) transport systems: functional and structural aspects of the ATP-hydrolyzing subunits/domains. FEMS Microbiol Rev. 1998;22:1–20. doi: 10.1111/j.1574-6976.1998.tb00358.x. [DOI] [PubMed] [Google Scholar]
- 43.Shi Y, Weimer P J. Response surface analysis of the effects of pH and dilution rate on Ruminococcus flavefaciens FD-1 in cellulose-fed continuous culture. Appl Environ Microbiol. 1992;58:2583–2591. doi: 10.1128/aem.58.8.2583-2591.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strobel H J. Growth of the thermophilic bacterium Clostridium thermocellum in continuous culture. Curr Microbiol. 1995;31:210–214. [Google Scholar]
- 45.Strobel H J, Caldwell F C, Dawson K A. Carbohydrate transport by the anaerobic thermophile Clostridium thermocellum LQRI. Appl Environ Microbiol. 1995;61:4012–4015. doi: 10.1128/aem.61.11.4012-4015.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Updegraff D M. Semimicro determination of cellulose in biological materials. Anal Biochem. 1969;32:420–424. doi: 10.1016/s0003-2697(69)80009-6. [DOI] [PubMed] [Google Scholar]
- 47.Weimer P J, Zeikus J G. Fermentation of cellulose and cellobiose by Clostridium thermocellum in the absence and presence of Methanobacterium thermoautotrophicum. Appl Environ Microbiol. 1977;33:289–297. doi: 10.1128/aem.33.2.289-297.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weimer P J, French A D, Calamari T A. Differential fermentation of cellulose allomorphs by ruminal cellulolytic bacteria. Appl Environ Microbiol. 1991;57:3101–3106. doi: 10.1128/aem.57.11.3101-3106.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weimer P J, Lopez-Guisa J M, French A D. Effects of cellulose fine structure on kinetics of its digestion by mixed ruminal microorganisms in vitro. Appl Environ Microbiol. 1990;56:2421–2429. doi: 10.1128/aem.56.8.2421-2429.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wells J E, Russel J B. The effect of growth and starvation on the lysis of the ruminal cellulolytic bacterium Fibrobacter succinogenes. Appl Environ Microbiol. 1996;62:1342–1346. doi: 10.1128/aem.62.4.1342-1346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Willis A T. Techniques for the study of anaerobic spore-forming bacteria. Methods Microbiol. 1969;3B:80–115. [Google Scholar]
- 52.Wood T M, Bhat K M. Methods for measuring cellulase activities. Methods Enzymol. 1988;160:87–112. [Google Scholar]