Abstract
Genetic screens are valuable for identifying novel genes involved in the regulation of developmental processes. To identify genes associated with cell growth regulation in Drosophila melanogaster , a mutagenesis screen was performed. Undergraduate students participating in Fly-CURE phenotypically characterized the E.4.1 mutant which is associated with rough eyes and antennae overgrowth. Following complementation analysis and subsequent genomic sequencing, E.4.1 was identified as a novel mutant allele of GstE14 , a gene involved in ecdysone biosynthesis important for the timing of developmental events. The abnormal eye and antenna phenotypes observed resulting from the loss of GstE14 suggest its role in tissue growth.
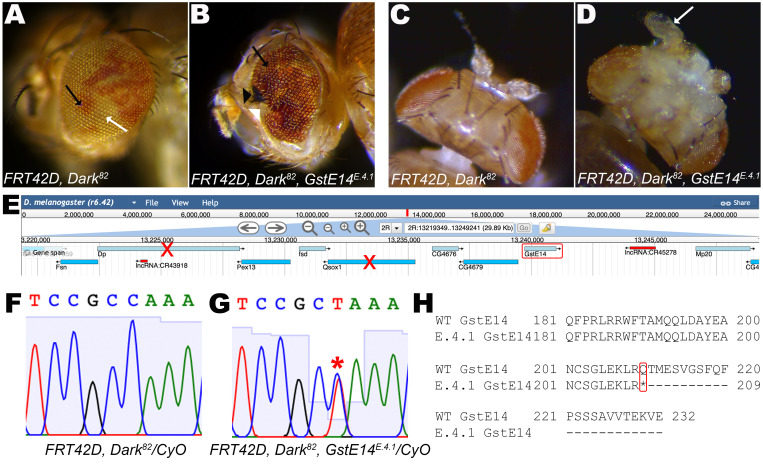
Figure 1. Characterization of the lethal GstE14 E.4.1 mutation by phenotypic analysis, complementation mapping, and genetic sequencing .
(A) FRT42D, Dark 82 control mosaic eye (B) and FRT42D, Dark 82 , GstE14 E.4.1 mutant mosaic eye showing white (wildtype, white arrow) and red (mutant, black arrow) pigmentation as a result of FRT/FLP mitotic recombination during development. (B) Overgrowth of mutant tissue in genotype FRT42D, Dark 82 , GstE14 E.4.1 is observed as clusters of red pigmentation, in addition to mutant clones displaying necrotic tissue (black arrowhead) in mosaic eye and rough eye phenotype with disorganized ommatidial arrangement (white arrowhead). (C) FRT42D, Dark 82 control fly shows wildtype antennae. (D) FRT42D, Dark 82 , GstE14 E.4.1 mutants exhibit antenna overgrowth (arrow). (E) The narrowest region in which E.4.1 failed to complement in the genomic region 2R:13,219,130..13,249,241 (Image adapted from JBrowse on FlyBase). (F-G) Sanger sequence analysis of wildtype GstE14 and mutant GstE14 E.4.1 reveals a heterozygous peak of C → T at 2R:13,240,692. (H) Alignment of amino acids of control FRT42D, Dark 82 and mutant FRT42D, Dark 82 , GstE14 E.4.1 sequence show the presence of a nonsense mutation at amino acid 210 (Gln → Stop) in the GstE14 E.4.1 mutant resulting in a truncated protein missing the last twenty two amino acids in the C-terminal region of GstE14.
Description
To identify novel genes involved in the regulation of cell growth processes in the developing Drosophila melanogaster eye, an ethyl methanesulfonate (EMS) mutagenesis screen was carried out utilizing the FLP/FRT recombination system on chromosome 2R (Kagey et al., 2012) . For this screen, the EMS concentration used was at levels that have been shown to result in an average of one lethal hit per chromosome arm. The mutations generated in this screen are homozygous lethal, so the FLP/FRT recombination system was used to generate mutant cell clones in the eye for phenotypic characterization while maintaining heterozygosity in the organism to prevent mortality. Although the Drosophila eye is not critical for survival, apoptotic pathways may be triggered in mutant cells so that an overgrowth phenotype associated with the mutation may not be observed. Therefore, apoptosis was prevented in mutant clones by the utilization of Dark 82 , a null allele of Death-associated APAF-1 related killer ( Dark ), on chromosome 2R distal to cytological site 42D (Akdemir et al., 2006; Mills et al., 2006) . Blocked apoptosis allows for an overgrowth phenotype to progress to an observable state. The Dark 82 mutant allele is due to the insertion of a mini-white P-element, allowing the identification of mutant clones by the presence of red pigmentation after mitotic recombination. Here we present the phenotypic characterization and the genetic mapping of the E.4.1 mutant line isolated in this screen.
To analyze the phenotype of E.4.1 , male flies of the mutant genotype ( FRT42D , Dark 82 , E.4.1 / CyO ) and of the control genotype ( FRT42D , Dark 82 / CyO ) were crossed with virgin females of genotype ( ey-Flp; FRT42D) . Since the eyeless ( ey ) promoter is active in the eyes, the restricted mitotic recombination leads to the generation of a mosaic eye containing homozygous E.4.1 mutant cells (red) and WT cells (white). The resulting F1 generation of the mutant cross was compared to that of the control cross to identify differences in tissue growth in the mosaic eye and morphological abnormalities. The quantified data showed an average of 56% red ( Figure 1A, black arrow ) to 44% white ( Figure 1A, white arrow ) tissue in the mosaic eyes of the control FRT42D, Dark 82 flies, with no signs of overgrowth of eyes or surrounding tissue (n=98) Figure 1A, 1C). However, FRT42D, Dark 82 , E.4.1 mutant clones ( Figure 1B, 1D) displayed overgrowth of mutant (red) eye tissue ( Figure 1B, black arrow ) and the generation of rough eyes manifesting as red clusters lacking precise ommatidial arrangement ( Figure 1B, white arrowhead ). Quantification of mosaic eye tissue resulted in an average of 95% red to 5% white tissue (n=161). Furthermore, heterozygous (orange) tissue was also present in the mutant eyes, with an average of 15% orange to 85% red (mutant) tissue (n=161). Additional abnormalities were observed, such as the presence of likely necrotic tissue on the compound eye (21% of E.4.1 mutant eyes present likely necrotic tissue, n=223) ( Figure 1B, black arrowhead ) and the antenna. We also observed enlarged antennae in E.4.1 mosaic adult fly compared to the control mosaic fly ( Figure 1D, arrow compared to Figure 1C).
In parallel, a complementation analysis was performed to narrow down the genomic location of the E.4.1 mutation and identify the gene affected by the E.4.1 mutation, using the Bloomington 2R Deficiency Kit (BDSC Df(2R) kit) with deletions of known endpoints on chromosome 2R distal to the FRT42D site (Cook et al., 2012) . Virgin females of genotype FRT42D , Dark 82 , E.4.1 / CyO were crossed with males of the genotype Df(2R)/CyO. The F1 progeny from each cross were examined for the presence or absence of straight-wing flies, where the presence of only curly-wing flies indicates a failure to complement the mutation. In the first round of mapping, E.4.1 failed to complement with deficiency lines Df(2R)CX1 and Df(2R)BSC273 (Table 1) whereas, deficiency lines Df(2R)Exel8057 and Df(2R)BSC274 complemented E.4.1. Thus, identifying the region 2R:13,219,349..13,430,464 as the putative chromosomal location for the E.4.1 mutation. Additionally, Df(2R)BSC331 failed to complement with E.4.1. However, the genomic region covered by Df(2R)BSC331 was excluded from the possible genomic location for the E.4.1 mutation since it contains Dark , and served as a positive control for the complementation mapping. A second round of complementation analysis was performed within the region 2R:13,219,349..13,430,464 to further define the genomic location of E.4.1 . Of these, Df(2R)Exel7124 and Df(2R)BSC272 failed to complement, resulting in 2R:13,219,130..13,249,241 as the smallest region that failed to complement the E.4.1 mutation ( Figure 1E ).
Next, a complementation analysis was performed for Dp and Qsox1 , the only two of the candidate genes for which homozygous lethal mutants were available at Drosophila stock centers. They both complemented with E.4.1 indicating that this mutation does not affect these two genes.
Then, to identify the gene affected by the E.4.1 mutation, we sequenced the remaining genes. To do so, genomic DNA was isolated from FRT42D , Dark 82 , E.4.1 / CyO mutant and FRT42D, Dark 82 / CyO control fly lines, and primers were designed to perform PCR amplification and Sanger sequencing. Subsequent sequence analysis showed a single nucleotide change (C→T) in the E.4.1 mutant line compared to the control at 2R:13,240,692. This mutation was independently confirmed by whole genome sequencing of the E.4.1 mutant line (Bieser, unpublished results). This mutation affects the coding region of the Glutathione S transferase E14 (GstE14) resulting in a premature stop codon at amino acid 210 (Gln → Stop) ( Figure 1H ). Overall, our data suggest that the potential loss of functional GstE14 leads to tissue overgrowth.
Following complementation and sequence analysis, we conclude that E.4.1 is a novel mutant allele of GstE14 ( GstE14 E.4.1 ) that truncates the resulting protein due to a nonsense mutation ( Figure 1H ). GstE14 , also referred to as noppera-bo ( nobo ), encodes a glutathione S-transferase, an ecdysteroidogenic enzyme that is suggested to be crucial in the biosynthesis of ecdysone (Enya et al., 2014) . Ecdysone is a major insect ecdysteroid synthesized in the prothoracic gland from exogenous sterols, such as cholesterol, and its release ensures that metamorphosis and molting occur at the appropriate time during morphogenesis (Gilbert et al., 2002; Niwa & Niwa, 2014) . GstE14 has been previously characterized as a novel Halloween gene, as its loss-of-function results in phenotypes indicative of low ecdysone production. However, functional disruption of GstE14 additionally results in atypical accumulation of cholesterol in the prothoracic gland, suggesting that its function is important in the metabolism and/or transport of cholesterol (Enya et al., 2014) .
When the biosynthesis and release of ecdysone are efficiently regulated, it directly influences the differentiation of larval imaginal disc tissues, including the eye imaginal disc, to form the adult structures, as well as signals the eradication of larval tissues that are no longer required in the adult fly through steroid-driven apoptotic pathways (Brennan et al., 1998; Cranna & Quinn, 2009) . Interferences with these mechanisms of imaginal disc differentiation or apoptotic pathways leading to the final adult structure of Drosophila melanogaster are consistent with the E.4.1 mutant phenotype observed in and around the eye.
Though GstE14 has no direct human ortholog, human glutathione S-transferases are a topic of interest in the field of oncology and chemotherapeutic treatments. Drosophila melanogaster GstE14 resides in the epsilon class of GSTs, a class unique to arthropods (Škerlová et al., 2020). Evolutionarily, this class of GSTs likely emerged in these species as an adaptation to the environment in insects specifically (Gonis et al., 2022) . However, developing an understanding of the GST protein family as a whole may be important for the treatment of many cancers. It has been found that GSTs are overexpressed in cancer cells, suggesting their potential role in metastasis (Singh & Reindl, 2021) . Since GSTs play a role in regulating redox homeostasis within the body's cells, these enzymes are able to detoxify chemotherapeutic agents, contributing to resistance against them (Shen et al., 1997) . While GstE14 's complete role in ecdysone biosynthesis and its direct substrates remains unknown, further experiments revealing these properties could aid in a greater understanding of the role of Glutathione S-transferases in human disease.
Table 1. Results of complementation analysis with deficiency lines of chromosome 2R and individual mutant alleles when scored for complementation against homozygous lethal mutant E.4.1 .
|
Bloomington Stock Center 2R Deficiency Kit | |||
|
Deficiency Stock |
BDSC Stock # |
Chromosomal Deletion |
Complementation Result with E.4.1 |
|
Df(2R)CX1 |
442 |
2R:12,700,421..14,091,140 |
Fail to complement |
|
Df(2R)Exel8057 |
7871 |
2R:13,034,847..13,219,349 |
Complement |
|
Df(2R)BSC273 |
23169 |
2R:13,159,579..13,502,150 |
Fail to complement |
|
Df(2R)BSC274 |
23170 |
2R:13,430,464..13,593,272 |
Complement |
|
Df(2R)BSC331 |
24356 |
2R:16,869,330..17,139,923 |
Fail to complement |
|
Additional Deficiency Lines | |||
|
Deficiency Stock |
BDSC Stock # |
Chromosomal Deletion |
Complementation Result with E.4.1 |
|
Df(2R)Exel7124 |
7872 |
2R:13,219,130..13,281,253 |
Fail to complement |
|
Df(2R)BSC272 |
23168 |
2R:13,219,130..13,249,241 |
Fail to complement |
|
Mutant Alleles of Individual Genes | |||
|
Gene |
BDSC Stock # |
Allele |
Complementation Result with E.4.1 |
|
5553 |
Dp 49Fk-1 |
Complement |
|
|
77650 |
Qsox1 4037-G4 |
Complement |
|
Reagents
w - ,FRT42D, Dark 82 / CyO (Akdemir et al. 2006)
w - ,FRT42D, Dark 82 , GstE14 E.4.1 / CyO (this manuscript)
w - , ey-FLP; FRT42D (BDSC 5616)
Bloomington Drosophila Stock Center 2R Deficiency Kit (Cook et al. 2012)
w 1118 ;Df(2R)Exel7124/CyO (BDSC 7872)
w 1118 ; Df(2R)BSC272/CyO (BDSC 23168)
b 1 Dp 49Fk-1 c 1 /SM5 (BDSC 5553)
w 1118 ; PBac{w +mC =IT.GAL4}Qsox 4037-G4 /CyO (BDSC 77650)
GstE14 forward primer 1: 5' AGTTACTGATCGACTTTCAAGGCGTTC 3'
GstE14 forward primer 2: 5' GCACGCAGAACGGATGAAGG 3'
GstE14 reverse primer: 5' CTGTCATGAATTTCTATTGGCGAGTCATTA 3'
Acknowledgments
Acknowledgments
Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study.
Funding Statement
Fly-CURE (JDK and KLB) is funded by a National Science Foundation IUSE Award (NSF 2021146). OD is funded by the NIH grant R15GM137236.
References
- Akdemir Fatih, Farkaš Robert, Chen Po, Juhasz Gabor, Medved'ová Lucia, Sass Miklos, Wang Lai, Wang Xiaodong, Chittaranjan Suganthi, Gorski Sharon M., Rodriguez Antony, Abrams John M. Autophagy occurs upstream or parallel to the apoptosome during histolytic cell death. Development. 2006 Apr 15;133(8):1457–1465. doi: 10.1242/dev.02332. [DOI] [PubMed] [Google Scholar]
- Brennan Catherine A., Ashburner Michael, Moses Kevin. Ecdysone pathway is required for furrow progression in the developing Drosophila eye . Development. 1998 Jul 15;125(14):2653–2664. doi: 10.1242/dev.125.14.2653. [DOI] [PubMed] [Google Scholar]
- Cook R Kimberley, Christensen Stacey J, Deal Jennifer A, Coburn Rachel A, Deal Megan E, Gresens Jill M, Kaufman Thomas C, Cook Kevin R. The generation of chromosomal deletions to provide extensive coverage and subdivision of the Drosophila melanogaster genome. Genome Biology. 2012;13(3):R21–R21. doi: 10.1186/gb-2012-13-3-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cranna Nicola, Quinn Leonie. Impact of steroid hormone signals on Drosophila cell cycle during development. Cell Division. 2009;4(1):3–3. doi: 10.1186/1747-1028-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enya Sora, Ameku Tomotsune, Igarashi Fumihiko, Iga Masatoshi, Kataoka Hiroshi, Shinoda Tetsuro, Niwa Ryusuke. A Halloween gene noppera-bo encodes a glutathione S-transferase essential for ecdysteroid biosynthesis via regulating the behaviour of cholesterol in Drosophila. Scientific Reports. 2014 Oct 10;4(1) doi: 10.1038/srep06586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert Lawrence I., Rybczynski Robert, Warren James T. Control and Biochemical Nature of the Ecdysteroidogenic Pathway. Annual Review of Entomology. 2002 Jan 1;47(1):883–916. doi: 10.1146/annurev.ento.47.091201.145302. [DOI] [PubMed] [Google Scholar]
- Gonis Elodie, Fraichard Stéphane, Chertemps Thomas, Hecker Arnaud, Schwartz Mathieu, Canon Francis, Neiers Fabrice. Expression Patterns of Drosophila Melanogaster Glutathione Transferases. Insects. 2022 Jul 7;13(7):612–612. doi: 10.3390/insects13070612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey Jacob D., Brown Jordan A., Moberg Kenneth H. Regulation of Yorkie activity in Drosophila imaginal discs by the Hedgehog receptor gene patched. Mechanisms of Development. 2012 Sep 1;129(9-12):339–349. doi: 10.1016/j.mod.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills Kathryn, Daish Tasman, Harvey Kieran F., Pfleger Cathie M., Hariharan Iswar K., Kumar Sharad. The Drosophila melanogaster Apaf-1 homologue ARK is required for most, but not all, programmed cell death . The Journal of Cell Biology. 2006 Mar 13;172(6):809–815. doi: 10.1083/jcb.200512126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa Yuko S., Niwa Ryusuke. Neural control of steroid hormone biosynthesis during development in the fruit fly <i>Drosophila melanogaster</i>. Genes & Genetic Systems. 2014;89(1):27–34. doi: 10.1266/ggs.89.27. [DOI] [PubMed] [Google Scholar]
- Shen, H, Kauvar, L, Tew, KD 1997. Importance of glutathione and associated enzymes in drug response. Oncology research. 9: 295. [PubMed]
- Singh Rahul Raj, Reindl Katie M. Glutathione S-Transferases in Cancer. Antioxidants. 2021 Apr 29;10(5):701–701. doi: 10.3390/antiox10050701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Škerlová Jana, Lindström Helena, Gonis Elodie, Sjödin Birgitta, Neiers Fabrice, Stenmark Pål, Mannervik Bengt. Structure and steroid isomerase activity of Drosophila glutathione transferase E14 essential for ecdysteroid biosynthesis . FEBS Letters. 2020 Jan 9;594(7):1187–1195. doi: 10.1002/1873-3468.13718. [DOI] [PubMed] [Google Scholar]