Abstract
Deep learning (DL) is applied in many biomedical areas. We performed a scoping review on DL in medical genetics. We first assessed 14,002 articles, of which 133 involved DL in medical genetics. DL in medical genetics increased rapidly during the studied period. In medical genetics, DL has largely been applied to small data sets of affected individuals (mean = 95, median = 29) with genetic conditions (71 different genetic conditions were studied; 24 articles studied multiple conditions). A variety of data types have been used in medical genetics, including radiologic (20%), ophthalmologic (14%), microscopy (8%), and text-based data (4%); the most common data type was patient facial photographs (46%). DL authors and research subjects overrepresent certain geographic areas (United States, Asia, and Europe). Convolutional neural networks (89%) were the most common method. Results were compared with human performance in 31% of studies. In total, 51% of articles provided data access; 16% released source code. To further explore DL in genomics, we conducted an additional analysis, the results of which highlight future opportunities for DL in medical genetics. Finally, we expect DL applications to increase in the future. To aid data curation, we evaluated a DL, random forest, and rule-based classifier at categorizing article abstracts.
Keywords: Artificial intelligence, Deep learning, Machine learning, Medical genetics, Medical genomics
Introduction
Artificial intelligence (AI) is increasingly studied and applied in a variety of biomedical contexts. Among different types of AI, deep learning (DL), a type of machine learning (ML), has shown strong potential, such as several recent, high-profile breakthroughs.1,2 In brief, ML is a type of AI that aims to enable computers to perform tasks without explicit programming to perform that task. In turn, DL is a subset of ML, in which the main objective is representation learning.3,4 Representation learning involves finding a set of features (eg, a numerical vector) that best represents a data point (eg, an X-ray image). Unlike other ML sub-branches, DL does not require domain experts (eg, a human radiologist) to handcraft these features for the raw data; instead, DL automatically learns these features using multiple processing layers (hence the term deep). Intuitively, one can think of the first few layers as learning simple localized interactions and the latter layers as learning more complex interactions of the outputs from the previous layers. For example, suppose the input is an image and the goal is to identify the image. The first layer learns the interactions of the pixels within a small contiguous region (eg, a square of 32 × 32 pixels). The second layer learns the interactions among these regions (eg, interactions among different squares of 32 × 32 pixels). The third layer learns the interactions of the outputs from the second layer, and so forth. The final layer returns a numerical vector, which can be interpreted as the feature vector representing the input image. Now, a downstream module (usually a classifier) could be applied to this vector representation to identify the correct label for the input image.3 DL can also be applied to different data types. For language data, for example, the first layer of a DL method would typically model all pair-wise interactions among the words in an input document. The subsequent layers model the relationships of these pair-wise interactions, and so forth.3,5
For the analyses we performed, we considered DL approaches as those built from architectures like convolutional neural network (CNN), long short-term memory, self-attention network (such as Transformer), or vector representation learning (such as an autoencoder). We did not confine our searches to a specific set of well-established models because many image classifiers may be used for the classification of individuals with suspected genetic conditions may be different from these well-known models, but were still built based on CNN blocks.6-9
Each biomedical discipline has nuances that affects the application of methods such as DL. The field of medical genetics involves conditions that are complex, esoteric, and individually rare, but are common in aggregate.10-12 In addition, the underlying causes for many genetic conditions are known at the molecular or cytogenomic level.13,14 By contrast, other fields of medicine more typically involve relatively common conditions affecting many people, in which the biological underpinnings or precise molecular causes of disease often remain murkier.
In addition to the earlier mentioned nuances regarding DL in medical genetics, there are reasons that DL may be especially important in this field. First, there is a severe lack of expert clinicians in medical genetics, which may mean that assistance from these types of tools may be valuable.15-17 Second, geneticists have been at the forefront of technologic breakthroughs (eg, genomic sequencing), and may be apt to embrace new approaches.18
Despite this, there has not been a systematic analysis exploring DL in medical genetics. To address this dearth, we performed a scoping review evaluating publications from 2015 to 2021. We intentionally focused on constitutional conditions because these are the conditions that clinical geneticists typically encounter in practice and research. Overlapping analyses have been performed regarding related topics, such as use of AI and ML in rare diseases19,20 or epigenetics.21 In addition to these valuable efforts, and because DL is instrumental in areas such as computer vision, we felt it would be illuminating to examine the more specific application of DL in medical genetics to determine which conditions are studied and how.22 We aimed to analyze key factors, such as study objectives, what data types and methods were used, and the origin and availability of code and data. To further explore aspects of how DL is applied in genomics, we also performed a separate analysis.
We expect DL to become an increasingly important tool in medical genetics. To support future analyses with minimal manual data curation, we present a DL, random forest (RF), and rule-based classifier to identify article abstracts on DL in medical genetics.
Materials and Methods
Data collection
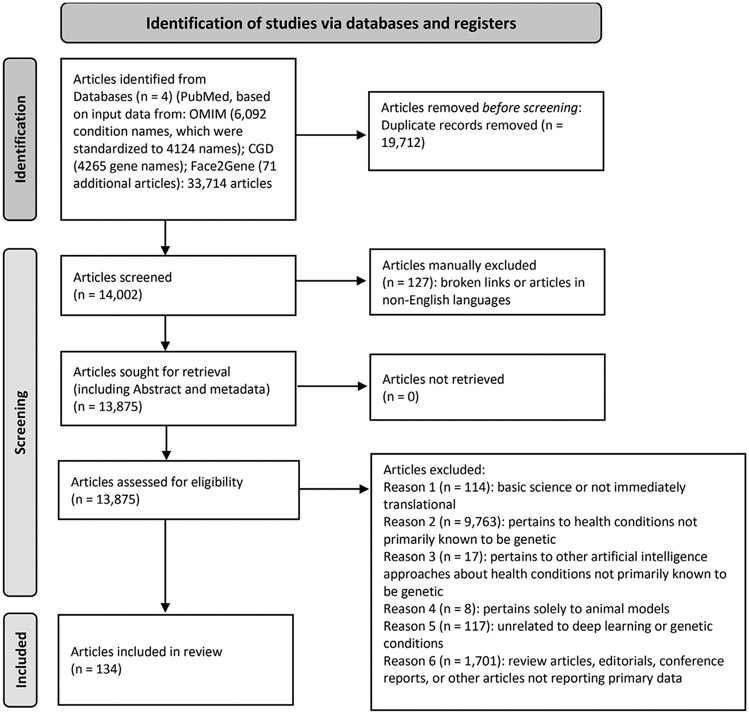
For our main analyses, we gathered articles for review based first on all monogenic genetic condition names (referred to as condition names) from OMIM (available at https://www.omim.org/). We downloaded all condition names, including synonyms, that were defined as phenotype description, molecular basis known (symbolized with # in OMIM) on June 3, 2021, and then manually standardized each condition name for syntax and to remove extraneous terms (eg, anemia, sideroblastic 1 became sideroblastic anemia). We started with 6092 condition names and synonyms, which we reduced to 4124 terms by manually removing redundant terms, such as in the case of multiple conditions with the same root name (eg, multiple instances of polycystic kidney disease). We included conditions with nonmonogenic causes (eg, cytogenomic conditions such as Williams syndrome), but excluded conditions judged to be multifactorial or involve susceptibilities without specific known individual causes. For example, we did not include studies involving susceptibilities to conditions such as diabetes mellitus except when they involved monogenic causes, such as for maturity-onset diabetes of the young. In this part of our analysis, we also excluded conditions involving somatic genetic changes related to cancer (Supplemental File 1). Second, we incorporated all gene names (referred to as gene names) related to these types of genetic conditions by downloading all genes in the Clinical Genomic Database (available at https://research.nhgri.nih.gov/CGD/)10 on June 22, 2021. We used the 4265 gene names that were then annotated in the database. We searched PubMed (June 22, 2021 for gene names, July 8, 2021 for condition names) by creating a Boolean search string of each condition name or gene name and the phrase “deep learning” (Supplemental File 2). Search results were combined into a spreadsheet and sorted by PMID. Figure 1 shows article selection and categorization on the basis of Preferred Reporting Items for Systematic Reviews and Meta-Analyses guidelines for systematic reviews.23
Figure 1. PRISMA schema for data collection and categorization.
Although we performed a scoping review, this schema was adapted from the one used for systematic reviews and is used with appropriate permission and citation as described in the guidelines.23,24 Sources used include Clinical Genomic Database (CGD), Face2Gene, OMIM, and PubMed.25-28 PRISMA, Preferred Reporting Items for Systematic Reviews and Meta-Analyses.
Article categorization
Each article in our main analyses was manually categorized into 1 of 10 categories, defined as one of the following:
(1) DL about genetic conditions (ie, pertaining to the medically-oriented application of DL in medical genetics), (2) Other AI approaches (not DL) about genetic conditions, (3) DL about genetic conditions, but judged to be primarily involving basic science or not immediately translational or used in direct clinical ways for individual patients, (4) DL about health conditions that are not necessarily primarily genetic (eg, diabetes mellitus, cancer in general), or when the cohort may have involved individuals with genetic conditions, but no analyses were performed or described to show whether individuals within the cohort actually had identified genetic etiologies, (5) Other AI approach about health conditions that are not necessarily primarily genetic (eg, diabetes mellitus, cancer in general), (6) DL applied to animal models of genetic conditions, (7) Articles about DL/AI or genetic conditions (but not a combination of these), (8) Studies unrelated to the areas of interest (eg, studies involving educational or learning theory, but not about DL), (9) Broken links/languages other than English, and (10) Review articles/editorials/conference reports/corrections that were not judged to include original data or analyses. Articles that included multiple types of information would be categorized as the lowest number category—eg, an article that could be categorized as either category 1 or 3 would be considered category 1. Duplicate articles were removed.
In an attempt to ensure inclusion of relevant articles, we also manually imported and categorized an additional 71 articles (beyond the ones already identified) reported to use the DL-based approach developed by Face2Gene (https://www.face2gene.com/) from their website (https://www.face2gene.com/publications/) on July 26, 2021. Each Face2Gene article was searched by title in PubMed to find its PMID to sort with the other articles. Articles that could not be found in the PubMed database were marked as such. These articles were categorized in the same way as other articles. To help reduce bias that may have been introduced by the inclusion of these articles, we also provide key findings in the Results section without these articles.
This combined spreadsheet had 33,714 initial rows; after categorizing and deleting uncategorized duplicates, 14,002 apparently unique rows remained. Of these, 1190 were categorized by >1 person; 341 were, owing to initial disagreement, manually adjudicated by multiple reviewers to determine final categorization. Because our main objective was to analyze the application of DL for genetic conditions, we focused our analyses on information from category 1 articles. To additionally investigate the broader use of DL in genomics, we conducted a separate review, as described later.
Additional analyses of DL in genomics
To further examine more general and other uses of DL in genomics (eg, to better capture information about DL in genome sequencing), we performed an additional PubMed-based search using the term “deep learning” with “genome,” “genome sequencing,” and “genomics.” We refer to the related analyses as DL in genomics. Unlike the main analyses described earlier, which focused on specific genetic conditions, our goal with this separate analysis on DL in genomics was to examine how DL is applied more generally and in ways not captured by the previous methods. In this search, we used the same time frame (from 2015 to July 8, 2021) as the main search. We identified 984 articles and categorized each article into 1 of 8 categories (these were intentionally different than those used in our main analyses because we hoped to explore the data set through a different lens):
(1) DL for clinically-oriented study of human genomic data (related to constitutional genetic conditions, as defined earlier); however, unlike in our main analyses, we included general methods that could be applied to data sets relevant to genetic disorders, such as methods to classify genetic variants or analyze genomic data to find answers for individuals suspected to have genetic conditions, (2) DL for clinically-oriented study of human genomic data (related to conditions that can have monogenic causes in some people and apparently multifactorial etiologies in other people; examples include autism and Alzheimer disease), (3) DL for clinically-oriented study of human genomic data (related to other health conditions or genomic analyses not involving conditions that have germline genetic causes, such as analyses on somatic cancer data), (4) DL for general analysis of human genomic data (ie, analyses that are primarily used for nonclinical, often research-related investigations, such as methods to analyze single-cell expression data), (5) DL for nonhuman genomic data sets, (6) Studies unrelated to the area of interest (a frequent example was use of DL to analyze pathology imaging data in which the potential importance of genomics was mentioned in the article but was not investigated), (7) Broken links/language other than English for the abstract, and (8) Review articles/editorials/conference reports/corrections that were not judged to include original data or analyses.
Model training
Of the 14,002 article PMIDs collected in our main analyses, 306 were excluded for the following reasons: unavailable abstracts through automated extraction methods (295); articles that were corrected or amended (6); duplicate articles cached from preprint servers (5). Abstracts for each of the remaining 13,696 articles were downloaded using the Entrez Direct Command Line Utility e-fetch.29
One-tenth (n = 1370) of the data set (n = 13,696) was chosen as the test set. The test set had 1370 articles of which 14 (1.02%) were category 1 and 1356 (99.98%) were non–category 1. With the remaining samples, we trained 9 models by using 9-fold cross validation and then tested these models on the same test set. Because the data set was severely imbalanced against category 1 (see Supplemental Figure 1; note that the numbers of articles in each category differ slightly from those shown in Supplemental Table 1, because data were not extractable for all articles for the model-building), the abstracts in each category were sampled independently to ensure equal representation in each of the 9 training folds and the test set. Each training fold had an average of 10,956 articles, of which an average of 102 (0.93%) were category 1; each validation fold had an average of 1370 articles each, of which an average of 12 (0.88%) were category 1.
Two types of ML approaches based on RF and Bidirectional Encoder Representations from Transformers (BERT) were used for the abstract classification (unpublished data: Devlin J, et al arXiv preprint:181004805. and Adhikari A, et al arXiv preprint:190408398. 2019).30 For BERT, only punctuation and stop words were removed before training. However, for RF, the words in the abstracts were stemmed and split into bigrams, which were used as training features because RF does not contain a default word tokenizer as BERT does.
Because we were primarily interested in the binary classification of category 1 abstracts vs the rest of the other categories, model performance was evaluated using binary labels, with category 1 abstracts as the positive class and all other categories as the negative class.
The RF models were trained using python sklearn.31 Each RF model was trained with a maximum depth of N/2, with N being the size of the training set (on average N = 10,956). BERT models were trained using our own modification of docBERT (unpublished data: Mulyar A, et al arXiv pre-print:191013664.). Each BERT model was trained for 200 epochs with early stopping and the same hyperparameter setting (see our code for more detail). For each of the 9 folds in the cross validation, training performance was measured on the basis of cross-entropy loss on the respective validation set.
Besides the ML predictors, we experimented with string-matching rules that predicted the label for an abstract on the basis of presence or absence of certain key phrases.32 We were interested in these simpler methods because in this context, ML acts as a statistical classifier with probability values, whereas rule-based approaches provide binary results. These rules were used in an exploratory fashion to determine how they would affect classification performance, including in combination with other predictors (Supplemental Table 5).
To handle the labeling agreement and disagreement for the abstracts among multiple classifiers, we used Snorkel to aggregate the probability predictions from each model and generate a single prediction probability.32,33
Results
Summary
The categorization of the main 14,002 articles analyzed is shown in Supplemental Table 1; articles with categorizations are included in Supplemental Table 2 (full Supplemental References for this table and for Supplemental Tables 3, 6, and 8 are available in Supplemental File 3). Although this is a scoping review, we prepared a Preferred Reporting Items for Systematic Reviews and Meta-Analyses flow diagram to depict the categorization steps (Figure 1).
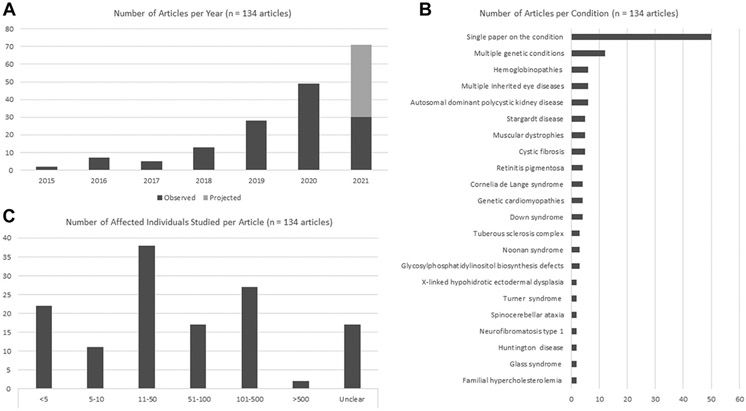
Our analyses focused on the 134 articles in category 1 (details for these articles are provided in Supplemental Table 3); these included 76 articles that did not involve Face2Gene. We note that our manual analyses initially identified 133 category 1 articles; checking the model results identified another category 1 article that had been missed. We observed an increase in the total number of articles during the selected timeframe (Figure 2A); the rate of growth, when including projected publications through the end of 2021, appears to fit an exponential curve.
Figure 2. Articles per year and characteristics of studied individuals.
A. Number of articles per year binned as category 1 (articles on deep learning [DL] applied to genetic conditions). Articles from 2021 includes observed articles as well as projected articles, the latter was calculated on the basis of the observed trend during the depicted time period (January 2015-June 2021). B. Distribution of genetic conditions studied using DL. C. Number of individuals with the studied genetic conditions included in each study. Further details are available in Supplemental Table 3.
Patients and conditions represented and data types used
The 134 articles in category 1 focused on >71 individual conditions; of these, 50 different conditions were assessed in a single article. Twenty-one conditions (or groups of conditions with heterogeneous causes, such as Noonan syndrome) were the focus of multiple papers. In total, 12 articles assessed multiple conditions, 6 papers assessed multiple inherited eye conditions, and 6 assessed hemoglobinopathies (eg, sickle cell anemia). Some articles had a more defined scope, such as several that focused on DL as applied to selected pathogenic variants related to a certain condition (Figure 2B and Supplemental Table 3).
The number of affected individuals included in the studies (for these calculations, we did not include the use of control or unaffected individuals, who might be used for training or testing purposes) ranged from single individuals, studied via application of an existing DL model, to 2400 individuals (Figure 2C). Excluding articles when data were unclear, unavailable, or when individual patients were not studied, the mean and median number of affected individuals in the remaining articles was 95 and 29, respectively. For the 76 non-Face2Gene articles, the mean and median number of affected individuals was 117 and 42, respectively. Studies involving larger numbers of individuals involved those that assessed multiple conditions and those analyzing medical records or nonfacial images (eg, radiologic or ophthalmologic images). Papers focusing on smaller numbers of individuals typically included those in which DL was used to help in the diagnostic or phenotyping processes; this was biased by the Face2Gene algorithm,6 which was used frequently by medical geneticists to assess findings in individuals with known or suspected genetic conditions (see details later in DL methods used).
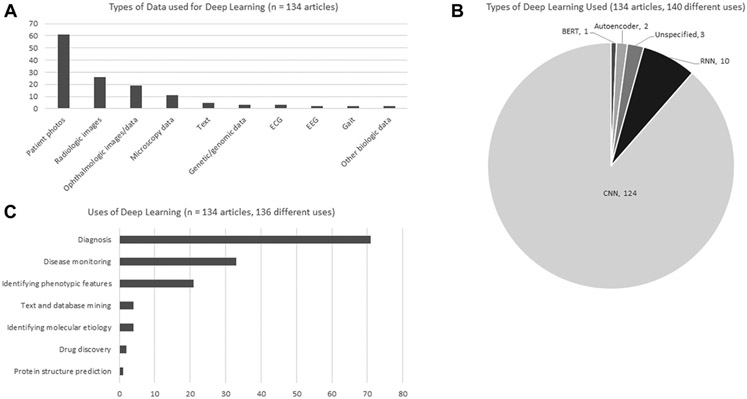
The most common data source was patient photos, used in 61 (46%) studies; the next most common data source included radiologic studies, ophthalmologic images and/or other ophthalmologic data, and microscopy data, used in 26 (19%), 19 (14%), and 11 (8%) papers, respectively. Excluding Face2Gene articles, the most common data types of the remaining 76 papers were radiologic data in 25 articles (33%), ophthalmologic images and/or other ophthalmologic data in 18 articles (24%), and microscopy data in 11 articles (14%). Of these non-Face2Gene papers, 5 (7%) used patient photos. See Figure 3A; Supplemental Figure 2 shows more details about select data types.
Figure 3. Characteristics of methods used.
A. Types of clinical input data analyzed via deep learning (DL). B. Types of DL methods used in each article. C. Categorization of the primary use of DL in each article. Further details are available in Supplemental Table 3. BERT, bidirectional encoder representations from transformers; CNN, convolutional neural network; ECG, electrocardiogram; EEG, electroencephalogram; RNN, recurrent neural network.
Purposes of studies and clinical areas
DL analysis of genetic conditions were uses in diverse ways. The most common use was for diagnosis, used in 71 (52%) of uses (we categorized some articles as using DL in multiple ways and defined 136 uses from 134 articles). The next most common use was what we termed disease monitoring (monitoring disease manifestations after diagnosis, including related to progression or treatment response), which was the main use in 33 (24%) articles. The third most common use was what we termed identifying phenotypic features, which we defined as using DL to characterize or phenotype (most commonly by examining facial features) individuals with a known diagnosis, which was used in 21 (15%) of articles. As has been shown by other reviews,20 a small number of articles focused on therapeutic approaches, such as drug discovery for genetic conditions. Among non-Face2Gene articles, and considering 78 uses among 76 articles, 33 (42%) involved diagnosis, 32 (41%) involved disease monitoring, and 4 (5%) involved text and database mining or identifying phenotypic features (Figure 3C).
Although subjective, we attempted to categorize the articles according to which patient-facing specialties the approaches were most applicable. Although this work focused on medical genetics, our methods would theoretically identify articles relevant to genetic conditions that could be diagnosed or treated in other specialty areas. For example, a genetic condition resulting in cardiac arrhythmia might be primarily diagnosed and managed by a cardiologist, whereas a neurologist might manage patients with genetic forms of epilepsy. The 3 most common areas were medical genetics, neurology, and ophthalmology, with 61 (46%), 21 (16%), and 20 (15%) articles, respectively (Supplemental Table 4).
DL methods used
We examined the types of DL used in the studies (Figure 3B). Because some articles used multiple DL types, we counted 140 uses among the 134 articles. Of these, 124 (88%) involved CNN, 10 (7%) used recurrent neural networks, 2 (1%) used autoencoders, and 1 (1%) used BERT, 3 (2%) did not supply enough data (sometimes for proprietary reasons) to enable the methodology to be accurately determined. These trends held true for non-Face2Gene articles; 65 (90%) of 72 articles in which the type of DL could be ascertained used CNN. See also Supplemental Figure 3.
We next examined whether the DL results were compared with human performance or other methods, and whether these comparisons were quantified. Of the 134 articles, 41 (31%) compared DL with a human, usually specialists in the field studied (eg, the results of DL to assess eye disease with an ophthalmologist). Of the 76 non-Face2Gene articles, 36 (49%) included comparisons with humans. Of the total articles, 83 (62%) and of the non-Face2Gene articles, 48 (63%) compared DL performance with other methods, such as another DL method or another AI approach (eg, RF). In total, 15 (11%) articles compared DL results with both human and other methods.
We were interested in code and data availability because these can affect the ability to validate, extend, and implement results. Not all papers provide both code and data, therefore we independently tallied the number of papers with code or data. Of the 134 articles, 22 (16%) stated that the code was available; of these, 18 provided an online link (however, not all links appeared functional when checked) and 3 stated that code was available on request. In total, 69 (51%) of the total articles stated that data used in the study were available; 19 of these stated that data were available only on request. Similarly, access to study code and data used in the study were provided in 19 (25%) and 30 (39%) of the non-Face2Gene articles, respectively (Supplemental Figure 4).
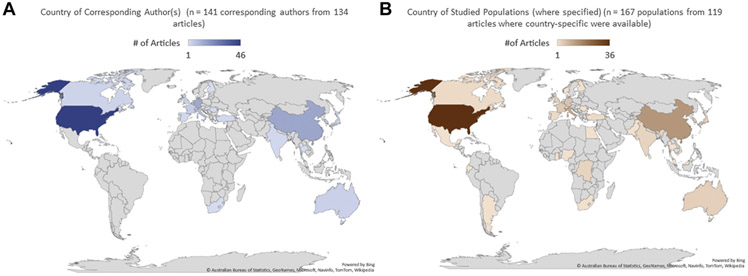
Locations of studies
To help show where research took place, we first identified the country of origin of the corresponding author. The United States accounted for most articles followed by China, Germany, Italy, and South Korea (Figure 4A). Next, we gathered information about the locations of the studied populations. If multiple countries were mentioned, all such countries were included in the tallies. For this information (excluding studies in which data were not available, or data involved multiple, unspecified countries), the United States accounted for the most articles followed by China, the Netherlands, the United Kingdom, and Singapore (Figure 4B).
Figure 4. Geographic distribution of articles.
A. Location of the corresponding author(s) for each of the 134 articles. B. Location of study populations for articles with available data.
Additional analyses of DL in genomics
The categorizations (as well as the annotations described in the following) of the 984 articles identified in this search are shown in Supplemental Table 6 and summarized in Supplemental Table 7. Of the 984 articles, 683 were identified in our previous search. For this analysis, we specifically decided not to remove these duplicates, because the goal was to examine the data set in a different way than for our main analyses.
In addition to Supplemental Tables 6 and 7, we summarize key findings of these analyses in this section. A total of 39 (4%) articles examined DL studies of human genomics data related to germline genetic conditions. We emphasize that for these categorizations, we included articles that were more general rather than specific to a certain condition. For example, we included papers on DL-based methods of analyzing genomic data or classifying genetic variants in a way that would be directly relevant to individuals with genetic conditions. Of these 39 articles, 6 (16%) involved general variant detection (including structural variants) and workflow methods, 28 (72%) involved methods of variant classification and pathogenicity annotation, and 5 (13%) involved DL-based analysis of specific conditions or classes of conditions. Of the 984 articles, 26 (3%) involved DL analyses applied to conditions with monogenic or multifactorial causes, including Alzheimer disease (8 papers), neurobehavioral conditions (10 papers), and other conditions (8 papers) (eg, susceptibility to cerebral palsy, Crohn disease). In total, 232 (24%) papers involved the use of DL to analyze data sets for conditions in which there is no known monogenic cause in at least some individuals, or when data sets relevant to nongermline conditions (eg, somatic cancer) were investigated. Of these articles, 167 (17% of the 984 total articles, and 72% of the 232 articles in this category) involved DL analyses of somatic cancer. Of note, 118 of these articles (12% of the 984 total articles and 51% of the 232 articles in this category) involved analyses of data from The Cancer Genome Atlas (TCGA) project (Supplemental Figure 5).34 Although this high proportion of TCGA-related papers was likely biased by our methods, there seems to be a correlation suggesting that the availability of this data set led to many papers on DL in cancer.
Model evaluation
The number of DL papers is expected to continue to increase (Figure 2A). To aid human curation, we built different classifiers on our abstracts; these classifiers are based on BERT and RF classifiers.
For the model evaluation, we wanted to examine how different models (alone and in combination) would perform at differentiating category 1 from non–category 1 articles. This can show how these models could be used to efficiently categorize articles. We thus focused on the true positive and false positive rate for identifying category 1 articles.
As shown in Table 1, aggregating BERT with RF and rule-based classifiers (via Snorkel) did not outperform the aggregation of BERT classifiers alone, suggesting that RF and rule-based classifiers did not add useful new information to BERT. We suspect 2 main reasons: RF may not sufficiently capture the nuances in written language in this context (eg, in situations when similar ideas may be expressed in different ways) and our string-matching rules may be overly simplistic.
Table 1.
Performance of the respective models to differentiate category 1 from non–category 1 articles on the held-out test set of articles
| Model | TPR | FPR | FNR | TNR |
|---|---|---|---|---|
| Snorkel LFs only | 0.143 | 0.378 | 0.571 | 0.469 |
| Snorkel RFs only | 0.214 | 0.000 | 0.786 | 1.000 |
| Snorkel BERTs only | 1.000 | 0.055 | 0.000 | 0.945 |
| Snorkel LFs + RFs | 0.143 | 0.378 | 0.643 | 0.469 |
| Snorkel LFs + BERTs | 0.143 | 0.378 | 0.857 | 0.622 |
| Snorkel RFs + BERTs | 1.000 | 0.055 | 0.000 | 0.945 |
| Snorkel LFs + RFs + BERTs | 0.143 | 0.378 | 0.857 | 0.622 |
Bold values show the results for the highest-perfoming models.
BERT, bidirectional encoder representations from transformers; FNR, false negative rate; FPR, false positive rate; LF, labeling functions; RF, random forest; TNR, true negative rate; TPR, true positive rate.
For the aggregated predictor based just on BERT (row 3 in Table 1), the true positivity rate was 100% (14/14 category 1 abstracts in test set correctly classified) and the false positive rate was 5.5% (75/1356 non–category 1 in test set mislabeled as category 1; see the following for more details). Because the false positive rate was 5.5%, our approach can still be helpful, including to automatically reduce papers that need to be manually assessed.
Next, we manually reviewed the 76 non–category 1 articles initially classified as false positives by this analysis; most were category 4 articles. On manual review, we noted that 62 (82%) of the articles were likely to be false positives either because of the mention of conditions that can have Mendelian forms as well as non-Mendelian forms (these false positive articles were about the latter type of condition) or because of the use of specific terms (eg, “EEG” or “OCT”) that appeared in many category 1 abstracts. Manual analysis did identify 1 article that was initially (by our manual analyses) incorrectly categorized as category 4 and should have been category 1, which has since been updated in all manuscript materials. Thus, to be accurate, 75 of the 76 results were false positives, and one was initially incorrectly assigned through our manual processes. This may be evidence of the value of automated approaches (Supplemental Table 8).
Discussion
We anticipate that DL will increasingly affect many medical fields. Applying DL to medical genetics involves specific challenges, including the rarity of many conditions.11,35,36 Condition rarity yields less data for DL training and testing. Related to this, gathering representative data from diverse individuals may be difficult, especially if ascertainment occurs in a limited geographic region.20,37,38 However, unlike many areas of medicine, the practice of medical genetics focuses on establishing precise diagnoses.10,14 The ability to accurately categorize many conditions on the basis of shared molecular causes may translate to high accuracy and applicability.38 In support of this, our analyses show that more than a third of 133 articles on DL in medical genetics focused on single rare genetic conditions. Furthermore, most studies that we identified applied methods to small numbers of individuals. However, our data were biased based on the inclusion of many articles using an established DL algorithm.6 We tried to help address this bias by providing key statistics after excluding Face2Gene articles. Overall, because AI depends on training data, inclusive data collection and recruitment are critical to ensure that methods work equitably.
The growth we see in the number of articles involving DL and medical genetics reflects a number of factors. Greater availability of high-performance computing resources, DL algorithms, and data sets have increased the adoption of DL in many venues. Our findings suggest that the early adoption of a platform for DL related to facial features helped drive the use of DL in medical genetics.6 We believe that part of the reason for this adoption was the ease-of-use of the platform. This may provide a roadmap for other tools that can be applied to other data types.
Although the growth of DL in medical genetics is impressive, it is clear from our separate analysis of DL in genomics that applications in medical genetics lag behind oncologic studies. There are undoubtedly many factors at work. One likely factor is the availability of data through projects such as TCGA.34 This suggests the potential of similarly aggregating, annotating, and sharing data relevant to medical genetics.
As shown in related analyses and reviews, our data show that DL in medical genetics concentrates on diagnosis. This is logical because the overall practice of medical genetics focuses on diagnosis (one exception is biochemical genetics, in which the work often involves patient management). Despite current diagnostic emphasis, DL may help narrow the gap between diagnosis and therapy in multiple ways, from preclinical selection of molecular testing to ensuring that a patient with a rare condition receives the right treatment quickly.37-39
This study included our own classification work. We feel that this helps highlight how data derived from manual curation can be used to enable more automated approaches. We emphasize that this work is exploratory and is not intended to maximize the prediction accuracy, but rather to show how applying and combining different techniques can affect performance. Importantly, the methods we implemented can be used for other related and unrelated data sets.
As with other scoping reviews, our analyses involve multiple limitations. First, it is unlikely that our search strategy captured all relevant articles. For example, certain forms of DL, such as related to DL-based genomic analyses,40,41 may not have been explicitly mentioned in articles in a way that was captured by our search methods. In addition, we used PubMed as our principal source of papers. PubMed emphasizes biomedical publications and limits key works from disciplines such as computer science and informatics. Second, although we tried to provide key data without certain articles, our search method likely led to bias. Third, we may have misclassified some articles and our analyses may have imperfectly appreciated details and nuances. We chose to include all articles in which DL was used; this included studies in which a DL model was trained and tested, as well as studies in which existing DL models were employed. This “lumping” could obscure differences among articles. Finally, it is likely that there are many important examples of DL in medical genetics that are not represented in the medical literature; much work occurs in biotechnology and other entities that are less likely to publish.42
Despite these limitations, we anticipate that our analyses depict key trends about DL in medical genetics. These trends include a focus on diagnostic objectives and the use of images (especially facial photos). This may point to opportunities beyond diagnosis (eg, therapy selection) or the use of less-leveraged data types.
Our analyses show that, in medical genetics, investigations tend to concentrate on a single data type, such as images or genetic variants. We found few manuscripts on the types of genetic conditions encountered by clinical geneticists that used DL to analyze both genomic (or multiomic) and phenotypic data sets. As shown in our analyses of DL in genomics, this appears to be an area in which somatic oncologic analyses are outpacing the studies related to constitutional genetic conditions. We see this as a ripe area for future study in medical genetics and suggest that examining how fields such as oncology have used data sets for these types of purposes may provide a useful model.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health. This work used the computational resources of the NIH High-Performance Computing Biowulf cluster (http://hpc.nih.gov).
Footnotes
Conflict of Interest
The authors declare no conflicts of interest.
Additional Information
The online version of this article (https://doi.org/10.1016/j.gim.2022.04.025) contains supplementary material, which is available to authorized users.
Data Availability
PMID for all categorized articles are available in Supplemental Table 2. Code is available at https://github.com/simonliu99/classify-medical-genetics-abstracts.
References
- 1.Jumper J, Evans R, Pritzel A, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583–589. 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baek M, DiMaio F, Anishchenko I, et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science. 2021;373(6557):871–876. 10.1126/science.abj8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–444. 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 4.Guo Y, Liu Y, Oerlemans A, Lao S, Wu S, Lew MS. Deep learning for visual understanding: a review. Neurocomputing. 2016;187:27–48. 10.1016/j.neucom.2015.09.116. [DOI] [Google Scholar]
- 5.Lee J, Yoon W, Kim S, et al. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. 2020;36(4):1234–1240. 10.1093/bioinformatics/btz682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gurovich Y, Hanani Y, Bar O, et al. Identifying facial phenotypes of genetic disorders using deep learning. Nat Med. 2019;25(1):60–64. 10.1038/s41591-018-0279-0. [DOI] [PubMed] [Google Scholar]
- 7.He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. Paper presented at: 2016 IEEE Conference on Computer Vision and Pattern Recognition; June 27-30, 2016; Las Vegas, Nevada, United States of America. [Google Scholar]
- 8.Szegedy C, Liu W, Jia Y, et al. Going deeper with convolutions. Paper presented at: 2015 IEEE Conference on Computer Vision and Pattern Recognition; June 7-12, 2015; Boston, Massachusetts, United States of America. [Google Scholar]
- 9.Tan M, Le QV. EfficientNet: rethinking model scaling for convolutional neural networks. Paper presented at: International Conference on Machine Learning, June 9-15, 2019; Long Beach, California, United States of America. [Google Scholar]
- 10.Solomon BD, Nguyen AD, Bear KA, Wolfsberg TG. Clinical genomic database. Proc Natl Acad Sci U S A. 2013;110(24):9851–9855. 10.1073/pnas.1302575110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferreira CR. The burden of rare diseases. Am J Med Genet A. 2019;179(6):885–892. 10.1002/ajmg.a.61124. [DOI] [PubMed] [Google Scholar]
- 12.Gonzaludo N, Belmont JW, Gainullin VG, Taft RJ. Estimating the burden and economic impact of pediatric genetic disease. Genet Med. 2019;21(8):1781–1789. Published correction appears in Genet Med. 2019;21(9):2161. 10.1038/s41436-018-0398-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bamshad MJ, Nickerson DA, Chong JX. Mendelian gene discovery: fast and furious with no end in sight. Am J Hum Genet. 2019;105(3):448–455. 10.1016/j.ajhg.2019.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katz AE, Nussbaum RL, Solomon BD, Rehm HL, Williams MS, Biesecker LG. Management of secondary genomic findings. Am J Hum Genet. 2020;107(1):3–14. 10.1016/j.ajhg.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Topol E. Deep Medicine: How Artificial Intelligence Can Make Healthcare Human Again. Basic Books; 2019. [Google Scholar]
- 16.Jenkins BD, Fischer CG, Polito CA, et al. The 2019 US medical genetics workforce: a focus on clinical genetics. Genet Med. 2021;23(8):1458–1464. 10.1038/s41436-021-01162-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Penon-Portmann M, Chang J, Cheng M, Shieh JT. Genetics workforce: distribution of genetics services and challenges to health care in California. Genet Med. 2020;22(1):227–231. 10.1038/s41436-019-0628-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kingsmore SF, Cakici JA, Clark MM, et al. A randomized, controlled trial of the analytic and diagnostic performance of singleton and trio, rapid genome and exome sequencing in ill infants. Am J Hum Genet. 2019;105(4):719–733. 10.1016/j.ajhg.2019.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brasil S, Pascoal C, Francisco R, Dos Reis Ferreira V, Videira PA, Valadao AG. Artificial intelligence (AI) in rare diseases: is the future brighter? Genes (Basel). 2019;10(12):978. 10.3390/genes10120978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schaefer J, Lehne M, Schepers J, Prasser F, Thun S. The use of machine learning in rare diseases: a scoping review. Orphanet J Rare Dis. 2020;15(1):145. 10.1186/s13023-020-01424-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brasil S, Neves CJ, Rijoff T, et al. Artificial intelligence in epigenetic studies:shedding light on rare diseases. Front Mol Biosci. 2021;8:648012. 10.3389/fmolb.2021.648012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dias R, Torkamani A. Artificial intelligence in clinical and genomic diagnostics. Genome Med. 2019;11(1):70. 10.1186/s13073-019-0689-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Page MJ, McKenzie JE, Bossuyt PM, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021;372:n71. 10.1136/bmj.n71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.PRISMA. Transparent reporting of systematic reviews and meta-analyses. PRISMA. Accessed June 3, 2021. http://www.prisma-statement.org/. [Google Scholar]
- 25.National Human Genome Research Institute. Clinical genomic database. Accessed June 22, 2021. http://research.nhgri.nih.gov/CGD/.
- 26.Publications using Face2Gene. Face2Gene. Updated February 2022. Accessed July 26, 2021. http://www.face2gene.com/publications/.
- 27.Hopkins University Johns. OMIM. Online inheritance in man. Updated May 4, 2022. Accessed June 3, 2021. http://www.omim.org/. [Google Scholar]
- 28.National Library of Medicine. National Center for Biotechnology Information. Accessed July 8, 2021. http://pubmed.ncbi.nlm.nih.gov/.
- 29.Kans J. Entrez direct: E-utilities on the Unix command line. In: Entrez Programming Utilities Help [Internet]. National Center for Biotechnology Information; 2010-. Published April 23, 2013. Updated April 18, 2022. Accessed August 14, 2021. http://www.ncbi.nlm.nih.gov/books/NBK179288/. [Google Scholar]
- 30.Vaswani A, Shazeer N, Parmar N, et al. Attention is all you need. Advances in neural information processing systems. In: Guyon I, Von Luxburg U, Bengio S, et al. , eds. Advances in Neural Information Processing Systems 30 (NIPS; 2017). [Google Scholar]
- 31.Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011;12:2825–2830. 10.5555/1953048.2078195. [DOI] [Google Scholar]
- 32.Ratner A, Bach SH, Ehrenberg H, Fries J, Wu S, Ré C. Snorkel: rapid training data creation with weak supervision. Proceedings VLDB Endowment. 2017;11(3):269–282. 10.14778/3157794.3157797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ratner A, Hancock B, Dunnmon J, Sala F, Pandey S. Ré C. Training complex models with multi-task weak supervision. Paper presented at: Proceedings of the AAAI Conference on Artificial Intelligence; 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cancer Genome Atlas Research Network, Weinstein JN, Collisson EA, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45(10):1113–1120. 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bick D, Bick SL, Dimmock DP, Fowler TA, Caulfield MJ, Scott RH. An online compendium of treatable genetic disorders. Am J Med Genet C Semin Med Genet. 2021;187(1):48–54. 10.1002/ajmg.c.31874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duong D, Waikel RL, Hu P, Tekendo-Ngongang C, Solomon BD. Neural network classifiers for images of genetic conditions with cutaneous manifestations. HGG Adv. 2021;3(1):100053. 10.1016/j.xhgg.2021.100053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muenke M, Adeyemo A, Kruszka P. An electronic atlas of human malformation syndromes in diverse populations. Genet Med. 2016;18(11):1085–1087. 10.1038/gim.2016.3. [DOI] [PubMed] [Google Scholar]
- 38.Solomon BD. Can artificial intelligence save medical genetics? Am J Med Genet A. 2022;188(2):397–399. 10.1002/ajmg.a.62538. [DOI] [PubMed] [Google Scholar]
- 39.Clark MM, Hildreth A, Batalov S, et al. Diagnosis of genetic diseases in seriously ill children by rapid whole-genome sequencing and automated phenotyping and interpretation. Sci Transl Med. 2019;11(489): eaat6177. 10.1126/scitranslmed.aat6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poplin R, Chang PC, Alexander D, et al. A universal SNP and small-indel variant caller using deep neural networks. Nat Biotechnol.2018;36(10):983–987. 10.1038/nbt4235. [DOI] [PubMed] [Google Scholar]
- 41.Jaganathan K, Kyriazopoulou Panagiotopoulou S, McRae JF, et al. Predicting splicing from primary sequence with deep learning. Cell. 2019;176(3):535–548.e24. 10.1016/j.cell.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 42.Slavotinek AM, Solomon BD. Going forward in a new world. Am J Med Genet A. 2020;182(7):1553–1554. 10.1002/ajmg.a.61715. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
PMID for all categorized articles are available in Supplemental Table 2. Code is available at https://github.com/simonliu99/classify-medical-genetics-abstracts.