Abstract
Loss of RNA homeostasis underlies numerous neurodegenerative and neuroinflammatory diseases. However, the molecular mechanisms that trigger neuroinflammation are poorly understood. Viral double-stranded RNA (dsRNA) triggers innate immune responses when sensed by host pattern recognition receptors (PRRs) present in all cell types. Here, we report that human neurons intrinsically carry exceptionally high levels of immunostimulatory dsRNAs and identify long 3′UTRs as giving rise to neuronal dsRNA structures. We found that the neuron-enriched ELAVL family of genes (ELAVL2, ELAVL3, and ELAVL4) can increase (i) 3′UTR length, (ii) dsRNA load, and (iii) activation of dsRNA-sensing PRRs such as MDA5, PKR, and TLR3. In wild-type neurons, neuronal dsRNAs signaled through PRRs to induce tonic production of the antiviral type I interferon. Depleting ELAVL2 in WT neurons led to global shortening of 3′UTR length, reduced immunostimulatory dsRNA levels, and rendered WT neurons susceptible to herpes simplex virus and Zika virus infection. Neurons deficient in ADAR1, a dsRNA-editing enzyme mutated in the neuroinflammatory disorder Aicardi-Goutières syndrome, exhibited intolerably high levels of dsRNA that triggered PRR-mediated toxic inflammation and neuronal death. Depleting ELAVL2 in ADAR1 knockout neurons led to prolonged neuron survival by reducing immunostimulatory dsRNA levels. In summary, neurons are specialized cells where PRRs constantly sense “self” dsRNAs to preemptively induce protective antiviral immunity, but maintaining RNA homeostasis is paramount to prevent pathological neuroinflammation.
INTRODUCTION
Viruses generate lengthy double-stranded RNA (dsRNA) structures, and our innate immune system uses pattern recognition receptors (PRRs), expressed in all cell types, to detect this dsRNA and trigger antiviral defense. For instance, the cytosolic RIG-I–like receptors (RLRs) (i.e., RIG-I and MDA5) and the endosomal Toll-like receptor 3 (TLR3) are PRRs that sense viral dsRNA and induce expression of the antiviral cytokine type I interferon (IFN) (1). Binding of type I IFN to its cognate IFN-α/β receptor (IFNAR) induces hundreds of IFN-stimulated gene (ISG) products with antiviral and inflammatory activities. Alternatively, some PRRs, such as PKR, detect viral dsRNA and have direct antiviral function by inducing global translational shutdown to limit virus replication (1).
Aberrant PRR activation is also observed in noninfectious diseases, most prominently in many neurodegenerative diseases. For instance, an elevated IFN signature and PKR activation are widely observed in brains of patients with amyotrophic lateral sclerosis (ALS), Alzheimer’s disease, and Huntington’s disease (2-7). Systemic lupus erythematosus (SLE) is also characterized by a type I IFN signature and PKR activation, and many patients develop neuropsychiatric symptoms (8). However, the molecular events that trigger sterile inflammation in the brain are still unclear. Another stark example of sterile neuroinflammation is found in patients with Aicardi-Goutières syndrome (AGS). AGS is a type I interfer-onopathy and a rare Mendelian disorder in which spontaneous type I IFN signaling occurs without acute viral infection; hence, the pathology is a mimic of viral infection (9). The various genes mutated in patients with AGS share a common function: The encoded proteins are involved in RNA or DNA binding or processing. A substantial proportion of these mutations causes endogenous (self) dsRNAs to be mistaken as viral dsRNAs, triggering PRR activation and downstream antiviral and inflammatory responses (9-12). What is puzzling is that genes mutated in patients with AGS are all ubiquitously expressed with minimal tissue specificity, but the central nervous system (CNS) is the primary site of type I IFN production (9, 13, 14). Why the brain is especially prone to sterile inflammation is a long-standing question.
To answer this question, we postulated that the neural transcriptome may be unique in its ability to trigger an immune response. We compared dsRNA abundance and identity across different human cell types, finding that neurons carry exceptionally high levels of dsRNA that stimulates PRR-mediated inflammation. Additionally, we demonstrate that the neuron-enriched ELAVL family of genes (ELAVL2, ELAVL3, and ELAVL4) lengthen 3′ untranslated regions (UTRs), and elongated 3′UTRs serve as a major source for immunostimulatory dsRNA structures in neurons. These findings suggest that neuronal dsRNAs could serve as a major upstream trigger for inflammation in the brain.
RESULTS
Neurons have intrinsically high levels of dsRNA
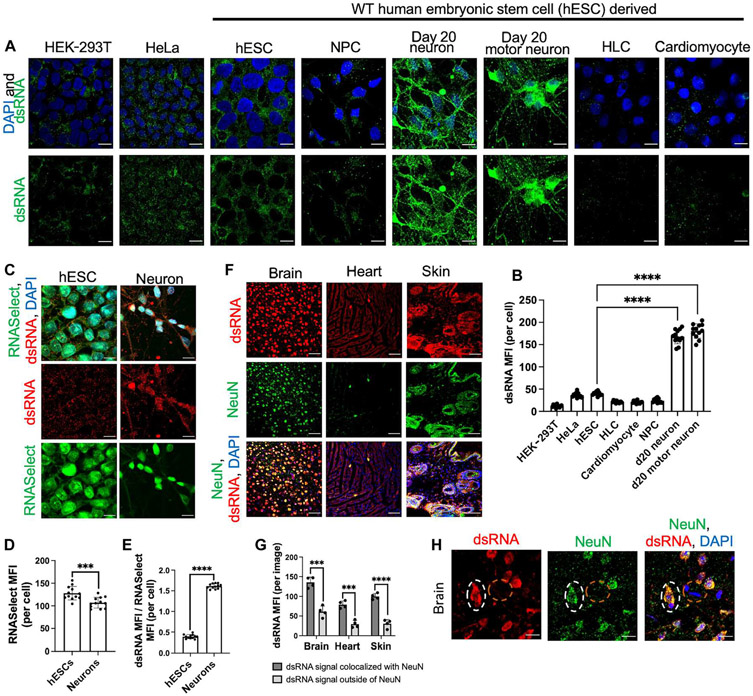
We used the J2 antibody to image dsRNAs in various human cell types (15). The J2 antibody, which binds long dsRNAs ≥40 base pairs (bp) independent of sequence, has been used for detecting both viral and endogenous dsRNAs (15-18). We focused on human cells because endogenous dsRNA levels and identity are distinct from species to species, even between mouse and human (19, 20). We differentiated wild-type (WT) HUES8 human embryonic stem cells (hESCs) to neural progenitor cells (NPCs) and neurons using well-established adherent monolayer culture methods (see Materials and Methods) (21, 22). After confirming the identity of cell types using various lineage markers (fig. S1A), we conducted J2 immunostaining. J2 antibody staining levels dynamically changed at different stages of hESC differentiation, with neurons having the noticeably strongest J2 signal (fig. S1, A and B). In parallel, we also derived hepatocyte-like cells (HLCs) from the same parental hESC line (23). HLC identity was confirmed by staining for various lineage markers (fig. S1C). HLCs had a much dimmer J2 signal than hESCs (fig. S1, C and D). Similarly, neurons derived from an independent hESC line (WA09) also had a much higher dsRNA burden than hESCs and HLCs (fig. S2, A and B).
We next compared dsRNA levels side-by-side among a broader range of cell types. We generated specialized motor neurons (fig. S1E), which are derived via embryoid bodies in cell suspension as opposed to adherent monocultures (24). We also generated cardio-myocytes (fig. S1F), a postmitotic cell type, to compare with postmitotic neurons and analyzed human embryonic kidney (HEK)–293T and HeLa cell lines as non–hESC-derived controls. Neurons had strikingly higher dsRNA levels than any other tested cell type (Fig. 1, A and B). Additionally, dsRNA subcellular localization also significantly differed by cell type. Neuronal dsRNA signal was present throughout the nucleus and cytoplasm, whereas in hESCs, dsRNA was mainly in the cytoplasm.
Fig. 1. Neurons have intrinsically high levels of dsRNA.
(A) Comparison of dsRNA levels across multiple human cell types. Cells were stained with DAPI (blue) and J2 immunostaining (green). (B) MFI quantification of (A). (C) Levels of total RNA and dsRNA compared between hESC and day 15 neurons. Total RNA (RNASelect, green) and dsRNA (J2, red). (D) Quantification of total RNA signal in (C). (E) Quantification of dsRNA signal normalized to total RNA signal in (C). (F) Immunofluorescent images of C57BL/6 mouse cortical brain, heart, and skin tissues. Tissue sections were stained with the 9D5 antibody (dsRNA, red) and neuron marker (NeuN, green). (G) Quantification of dsRNA signal in (F) colocalized with NeuN. (H) Mouse cerebral cortex stained with the 9D5 antibody (dsRNA, red) and NeuN (green). White dotted circle indicates neuron. Orange dotted circle indicates nonneuronal cells. All bar graphs are mean ± SD [n = 12 cells from biological triplicates in (B), (D), and (E); n = 4 images from biological triplicates in (G)]. All scale bars represent 10 μm, except for (F), which is 50 μm. (B) One-way ANOVA with Tukey corrected multiple comparisons. (D, E, and G) Student’s t test, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
We further confirmed J2 specificity for dsRNA and validated the above observations using different antibodies. We demonstrated that the J2 signal was sensitive to treatment with a double-stranded ribonuclease (RNase) (dsRNAse), but not a single-stranded RNase (ssRNAse) or a deoxyribonuclease (DNase) (fig. S3, A and B), and confirmed that other anti-dsRNA antibodies such as 9D5 and K1 yielded results similar to J2 (fig. S3, C and D). Additionally, by staining cells with an anti–glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody, we confirmed that cell type differences in J2 antibody staining are not due to mere differences in cell permeability to antibody. Although neurons still exhibited a significantly stronger dsRNA stain than other cell types, they did not show a higher GAPDH stain (fig. S4, A to C).
We next examined whether neurons had more dsRNA simply because they had more total RNA than other cell types. In parallel to J2, we costained total RNA using RNASelect. RNASelect staining showed that total RNA load is slightly lower in neurons than in hESCs (Fig. 1, C and D). We normalized the J2 signal (dsRNA load) to the RNASelect signal (total RNA load) from the same images and observed that normalized J2 signal is much higher in neurons than in hESCs (Fig. 1E and fig. S4, D and E). We also performed polyA FISH (fluorescence in situ hybridization) (25), which selectively stains for polyadenylated mRNA, and observed that neurons have slightly less mRNA than hESCs (fig. S3, E and F). Thus, the increase in dsRNA levels in neurons is not due to an increase in total RNA levels. Instead, the neuronal transcriptome appears to be prone to forming dsRNA structures.
Next, we examined whether neurons are enriched for dsRNAs in complex tissue. We costained tissue from 10-month-old C57BL/6 mice for dsRNA and NeuN, a pan-neuronal marker. Because the J2 antibody is a mouse monoclonal antibody, we stained dsRNA in mouse tissue with the rabbit monoclonal 9D5 antibody and confirmed that 9D5 staining in mouse tissue is sensitive to dsRNAse treatment, but not ssRNAse or DNase treatment (fig. S3G). We stained brain tissue and other innervated organs such as the heart and skin. Remarkably, we found that dsRNA signal is much stronger in cells costained with NeuN than in cells without NeuN (Fig. 1, F and G). NeuN+ cells in the periphery, such as in the skin and heart, also contain high levels of dsRNA, similar to NeuN+ cells in the brain, although further experimentation is necessary to determine whether peripheral NeuN+ cells are tissue-innervating nerves or another cell type. Additionally, in the mouse cerebral cortex, NeuN+ cells, indicative of neurons, had a higher dsRNA burden than nearby nonneuronal cells (NeuN−) (Fig. 1H). Thus, neurons are one of the most enriched cell types for dsRNA in complex tissue, and this phenotype is conserved across mouse and human cells.
Neurons are enriched for dsRNA originating from POLII
Prior studies demonstrated that mRNA noncoding regions (e.g., introns and 3′UTRs) or mitochondrial RNAs (mtRNAs) can contribute to cellular dsRNAs (16-20). We transiently treated neurons with different RNA polymerase inhibitors for 12 hours before J2 staining to determine whether RNA polymerase II (POLII) or mitochondrial RNA polymerase (POLRMT) gives rise to neuronal dsRNA. The following inhibitors were used: actinomycin D (ActD), which inhibits both POLII and POLRMT (17); α-amanitin, a POLII inhibitor; and IMT1, a POLRMT-specific inhibitor (26). We confirmed the specificity of the inhibitors in neurons (fig. S5A) and hESCs (fig. S5B): ActD down-regulated both mtRNA and mRNA levels, whereas α-amanitin and IMT1 down-regulated levels of mRNA and mtRNA, respectively. For both cell types, 12 hours of ActD treatment was sufficient to abrogate almost all dsRNA signal, revealing that cellular dsRNAs are present transiently—no longer than 12 hours. Additionally, a dual treatment with α-amanitin and IMT1 also completely reduced the dsRNA signal, suggesting that most dsRNA consists of POLII- and POLRMT-derived transcripts (fig. S6, A to D). By quantifying depletion of dsRNA upon single treatments with α-amanitin or IMT1 treatment, we calculated the proportional contribution of POLII- or POLRMT-derived dsRNAs, respectively (fig. S6E). We applied this analysis to various cell types. In HeLa cells, roughly 90% of dsRNA was derived from POLRMT (fig. S6, C and E). This is consistent with a prior study that concluded that roughly 95% of dsRNA in HeLa cells is located in mitochondria (17), validating our analysis. In both hESCs (fig. S6A) and HEK-293T cells (fig. S6D), POLRMT and POLII contribute to roughly 70% and 30% of dsRNA, respectively (fig. S6E). Conversely, in neurons (fig. S6B), POLRMT and POLII transcripts contribute to 40% and 60% of dsRNA, respectively (fig. S6E). We further validated these results by imaging colocalization of the dsRNA with the mitochondria and observed less percentage of dsRNA colocalized in the mitochondria in neurons compared with hESCs (fig. S7, A and B). In summary, mtRNA is the major component of dsRNA in HeLa, HEK-293T, and hESCs. However, in neurons, POLII-derived transcripts, presumably mRNAs, contribute to the bulk of the neuronal dsRNA load.
WT neurons constitutively produce IFN through dsRNA-sensing pathways
Past studies noted that type I IFN can be constitutively expressed at low quantities yet could have profound functions in homeostasis (27). Using single-molecule array (SIMOA), an ultrasensitive enzyme-linked immunosorbent assay (ELISA) assay, a recent study reported the intriguing observation that human neurons constitutively produce IFN-β (28). Additionally, studies using IFNAR-deficient model systems proposed a model in which constitutive type I IFN in the brain may be required to preempt viral infection, prevent neurodegeneration, and promote synaptic plasticity (28-30). These ideas align with the concept that inflammation exists on a spectrum—rather than being just active during infection or injury—and is involved in a broad range of biological processes beyond its classical role in antimicrobial defense (31).
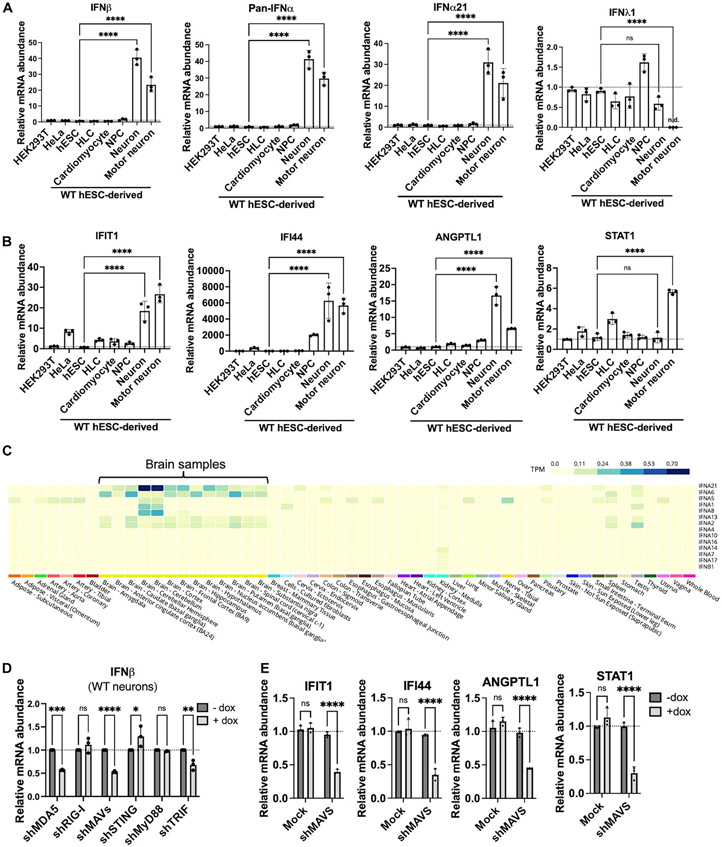
Supporting the possibility that the brain constitutively expresses “tonic” type I IFN, we observed via quantitative polymerase chain reaction (qPCR) that neurons derived from WT hESCs produced much higher levels of basal type I IFN (IFN-β and IFN-α subtypes) mRNA compared with a panel of other human cell types (Fig. 2A). Type I IFN mRNA was minimally or not detected in the other human cell types. We did not observe an up-regulation of IFN-λ1, a type III IFN (Fig. 2A). We also examined the levels of several ISGs to determine whether the tonic type I IFN in neurons was sufficient to induce ISG expression. Overall, ISGs such as IFIT1, IFI44, and ANGPTL1 were expressed highest in neurons (Fig. 2B). However, some ISGs, like STAT1, were not expressed the highest in all neurons (Fig. 2B). Because tonic type I IFN is expressed at low levels—drastically lower than viral induced type I IFN—and because many ISGs are expressed at baseline in a tissue-specific manner independently of IFN, we do not expect that neurons have the highest expression of all ISGs. Together, these data suggest that a low-grade tonic type I IFN is expressed in hESC-derived neurons.
Fig. 2. WT neurons constitutively produce tonic type I IFN through two dsRNA-sensing PRR pathways, MAVS and TRIF.
(A and B) Relative levels of type I IFN (IFN-β, pan–IFN-α, IFN-α21) and type III IFN (IFN-λ1) (A) and ISG (IFIT1, IFI44, ANGPTL1, STAT1) mRNA (B) in diverse human cell types. HLCs, cardio-myocytes, NPCs, neurons (day 15), and motor neurons (day 15) were all derived from WT hESCs (HUES8). (C) Type I IFN (IFN-β and IFN-α subtypes) expression in 54 nondiseased tissue sites analyzed by RNA-Seq. Data were derived from the GTEx database. TPM, transcript per million. (D) qPCR of IFN-β levels in WT neurons (day 20) after doxycycline (dox)–induced knockdown of the indicated immune genes using shRNA (sh). (E) qPCR of several ISGs in WT neurons (day 20) after doxycycline-induced knockdown of MAVS. For all qPCR, RPS11 was used as a housekeeping gene. All quantified data shown are mean ± SD (n = 3 biological replicates). (A and B) One-way ANOVA with Tukey corrected multiple comparisons. (D and E) Two-way ANOVA with Tukey corrected multiple comparisons, *P < 0.05, **P < 0.01, ***P < 0.001, and ****p < 0.0001.
It is unknown whether the human brain also exhibits a constitutive type I IFN signature. We analyzed the Genotype-Tissue Expression (GTEx) Portal (32) derived from nearly 1000 individuals and found that the nondiseased human brain expressed much higher levels of type I IFN compared with many other tissues (Fig. 2C). Many IFN-α subtypes were uniquely elevated in the human brain, most notably IFN-α21, which was also strongly induced in stem cell–derived neurons (Fig. 2A). In addition to the GTEx dataset, we also analyzed RNA-sequencing (RNA-Seq) datasets from the Human Protein Atlas (HPA) derived from 198 individuals (fig. S8A) (33). Analysis of the HPA RNA-Seq dataset shows that three compartments display a notable type I IFN signature: the human brain, bone marrow and lymphoid tissue, and the male epididymis. Hence, the brain is one of the compartments in the human body that constitutively expresses type I IFN in the absence of acute infection. Of note, when we analyzed publicly available single-cell RNA-Seq datasets, we could not detect significant type I IFN expression in the nondiseased human brain, likely because low-abundance transcripts such as tonic type I IFN mRNA are not effectively captured by single-cell RNA-Seq.
To investigate the mechanisms underlying constitutive type I IFN expression, we tested whether neuronal dsRNAs constantly activate PRRs to maintain tonic IFN in homeostasis. This hypothesis diverges from the classical view that PRRs distinguish self from non-self ligands. We transduced neurons with lentiviruses carrying doxycycline-inducible short hairpin RNAs (shRNAs) that target four immune adaptor proteins (STING, MAVS, MyD88, and TRIF) that lie downstream of PRRs and license transcription factors to induce transcription of type I IFN genes (34). STING is downstream of DNA-sensing cGAS, MAVS is downstream of the dsRNA-sensing RLRs MDA5 and RIG-I, MyD88 is downstream of various TLRs that sense a variety of ligands, and TRIF is downstream of the dsRNA-sensing TLR3. We confirmed successful knockdown of each adaptor protein in neurons (fig. S9A). Notably, we found that constitutive IFN-β production in WT neurons is reduced after knockdown of either MAVS or TRIF—the two adaptor proteins implicated in dsRNA sensing—but not STING or MyD88 (Fig. 2D). We also confirmed that reducing IFN-β levels by knocking down MAVS led to reduced ISG induction, suggesting that dsRNA-sensing pathways sustain basal ISG expression in neuronal homeostasis (Fig. 2E). Because tonic IFN-β production does not rely on MyD88, this suggests that single-stranded RNA (ssRNA) sensors like TLR7/8 are dispensable for tonic IFN-β production. The reduction in constitutive IFN-β after TRIF knockdown is consistent with a prior investigation, where cortical neurons derived from induced pluripotent stem cells of patients with defective TLR3 had reduced constitutive type I IFN expression (28). When comparing the two RLRs, MDA5 knockdown, but not RIG-I knockdown, significantly reduced IFN-β levels (fig. S9A and Fig. 2D).
We also asked whether there was a positive correlation between expression levels of dsRNA-sensing PRRs and constitutive IFN production. MDA5, RIG-I, and TLR3 mRNA expression was much lower in neurons compared with a panel of other human cell types (fig. S10A), suggesting that it is not high levels of PRRs that confer the ability to constitutively express IFN in neurons. Instead, high dsRNA load in neurons drives tonic type I IFN production and ISG expression via the cytoplasmic MDA5-MAVS and endosomal TLR3-TRIF signaling pathways.
Loss of ADAR1 increases dsRNA levels in diverse cell types, and neurons fail to tolerate loss of ADAR1
Next, we examined the consequences of enriched dsRNA in neurons in a disease model. AGS is an encephalopathy and genetic disorder where the brain is the primary site for inflammation (9, 13, 14). ADAR1 is a dsRNA-editing enzyme that converts adenosine (A) to inosine (I) in dsRNA and is one of the genes mutated in patients with AGS (35, 36). Past investigations demonstrated that ADAR1 deficiency in mammals leads to aberrant activation of dsRNA-sensing PRRs such as MDA5 and PKR (10, 11, 37-43). This led to the prevailing model that ADAR1 editing of self-dsRNAs introduces mismatches that disrupt dsRNA structures, thereby suppressing self-dsRNAs from triggering dysregulated PRR activation. However, this model is largely based on RNA structure prediction and in vitro biochemical studies and has yet to be conclusively demonstrated. Moreover, like other genes mutated in AGS, ADAR1 is ubiquitously expressed with low tissue specificity; thus, it was puzzling why the brain is the primary site for type I IFN production in AGS.
We reasoned that the high levels of dsRNA in neurons predispose the brain to pathological inflammation in ADAR1 deficiency. Hence, we tested whether loss of ADAR1 leads to an increase in total dsRNA levels and whether ADAR1 deficiency would induce toxic inflammation in neurons, but not other cell types with lower intrinsic dsRNA burden. ADAR1-defective mouse models exhibit multiorgan systemic inflammation, some of which also display AGS-like encephalopathy (44, 45). However, overall, it has been challenging for AGS mouse models to phenocopy neurologic disease and CNS-centric inflammation (46-48). Additionally, because mice lack Alu elements—repetitive elements prone to forming dsRNA structures that are heavily edited by human ADAR1—only ~0.004% of edited sites in human are conserved in mice (49, 50). Hence, we used ADAR1 knockout (KO) hESCs (fig. S11A) (10) to obtain NPCs and neurons. Previous attempts to obtain neurons from ADAR1 KO hESCs failed because of cell death at the NPC stage (10). However, by optimizing differentiation conditions to achieve extremely synchronized cell populations, we were able to derive ADAR1 KO NPC and neurons successfully (see Materials and Methods).
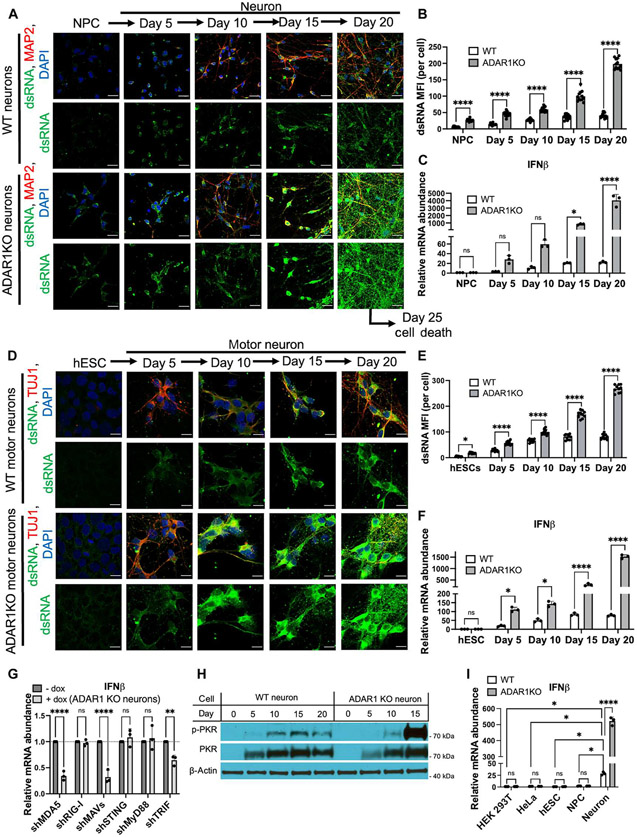
We confirmed that loss of ADAR1 did not interfere with NPC or neuron differentiation (Fig. 3A and figs. S1A and S11B). dsRNA and type I IFN production in ADAR1 KO neurons increased in an uncontrolled manner, unlike WT neurons where dsRNA and type I IFN levels are increased but plateaued at 15 to 20 days after differentiation (Fig. 3, A to C). By day 20 after differentiation, ADAR1 KO neurons had extremely high levels of dsRNA (Fig. 3, A and B)—resembling an RNA virus infection—and IFN-β production in ADAR1 KO neurons increased up to more than 100-fold compared with IFN-β levels in WT neurons (Fig. 3C). This increase in IFN-β production corresponded with an increase in the production of ISGs (fig. S12A). We also differentiated WT and ADAR1 KO hESCs to specialized motor neurons to further validate the above findings (24). ADAR1 KO hESCs had no defect in differentiating into motor neurons (Fig. 3D and fig. S13A). dsRNA and IFN-β levels were highly dysregulated in ADAR1 KO motor neurons, but not in WT motor neurons (Fig. 3, D to F).
Fig. 3. ADAR1 regulates dsRNA burden in diverse cell types and tonic IFN production in neurons.
(A to C) WT and ADAR1 KO NPCs (derived from HUES8 cells) were differentiated to neurons using adherent monolayer culture methods. Neurons were stained for dsRNA (J2, green), a neuron marker (MAP2, red), and DAPI (blue) at different stages of differentiation (A). Bar graphs quantifying dsRNA signal (B) and IFN-β mRNA levels (C). (D to F) WT and ADAR1 KO hESCs (HUES8) were differentiated to motor neurons via embryoid bodies in cell suspension. Neurons were stained for dsRNA (J2, green), a neuron marker (TUJ1, red), and DAPI (blue) at different stages of differentiation (D). Bar graphs quantifying dsRNA signal (E) and IFN-β mRNA levels (F). (G) qPCR of IFN-β levels in ADAR1 KO neurons (day 20) after doxycycline-induced knockdown of the indicated immune genes using shRNA. (H) Immunoblot measuring PKR activation in WT and ADAR1 KO neurons over time during differentiation (ADAR1 KO neurons at day 20 were too unhealthy to obtain sufficient protein). p-PKR, phosphorylated PKR. (I) Relative levels of IFN-β mRNA in WT and ADAR1 KO HEK-293T, HeLa, hESCs, NPCs, and day 15 neurons. For all qPCR, RPS11 was used as a housekeeping gene. All quantified data shown are mean ± SD [n = 3 biological replicates in (C) and (F) to (H); n = 12 cells from biological triplicates in (B) and (E)]. Scale bars, 10 μm. Two-way ANOVAwith Tukey corrected multiple comparisons, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Similar to tonic type I IFN in WT neurons, dysregulated type I IFN production in ADAR1 KO neurons also depended on MDA-MAVS and TLR3-TRIF pathways (Fig. 3G and fig. S9B). Therefore, the pathways to produce low/tonic and high/dysregulated type I IFN production are shared. Additionally, this report demonstrates that ADAR1 can suppress signaling via TRIF, an unexpected finding because ADAR1 is nuclear and cytoplasmic, and TLR3-TRIF sensing occurs in endosomes.
We observed that ADAR1 KO neurons die by day 25 after differentiation (fig. S11C) and therefore sought to understand the mechanism of cell death. We knocked down MAVS (fig. S14A) to decrease IFN-β production (fig. S14B), which resulted in a modest increase in cell survival (fig. S14, C and D), indicating that dsRNA-induced IFN-β only modestly contributes to cell death in ADAR1 KO neurons. Additionally, we knocked down PKR to observe its role in cell death. PKR is an ISG and a PRR that undergoes autophosphorylation upon binding dsRNA and inhibits translation. In addition to aberrant IFN production, ADAR1 KO neurons also exhibit hyperactivation of PKR (Fig. 3H). Knockdown of PKR (fig. S14A) did not alter IFN-β production (fig. S14B). Yet PKR depletion was able to significantly increase survival of ADAR1 KO neurons (fig. S14, C and D). Together, these data indicate that PKR activation and IFN-β production play a major and minor role in ADAR1 KO neuron death, respectively.
Finally, it is important to note that lack of ADAR1 led to elevated dsRNA levels in all cell types examined (hESCs, NPCs, and neurons) (Fig. 3, B and E), but only in neurons did ADAR1 deficiency lead to markedly increased type I IFN levels (Fig. 3, C, F, and I). Of note, previous studies into ADAR1 KO HEK-293T or HeLa cells also demonstrated that loss of ADAR1 does not induce IFN production at baseline; however, inflammation can be induced in these cells after an exogenous IFN stimulus (11, 40). In summary, these data demonstrate that ADAR1 is a global regulator of dsRNA across multiple cell types, and neurons are especially susceptible to inflammation upon loss of ADAR1.
In addition to ADAR1, another catalytically active member of the ADAR family is ADAR2. Like ADAR1, ADAR2 binds to dsRNA and performs A-to-I editing; however, ADAR2 is primarily known as a site-specific editor of coding regions (43). We examined dsRNA levels in ADAR2 KO HEK-293T cells (fig. S15A) (10). Although ADAR2 protein is expressed in HEK-293T cells, ADAR2 did not significantly alter dsRNA burden (fig. S15, B and C). Hence, ADAR1, rather than ADAR2, is the major regulator of cellular dsRNA levels.
Ectopically expressed ELAVL2, ELAVL3, and ELAVL4 (HuB, HuC, and HuD) cooperate to lengthen 3′UTRs, increase dsRNA levels, and induce inflammation
The identity of the dsRNAs that play a causal role in inflammation remains elusive. Here, we tested the hypothesis that mRNAs with elongated 3′UTRs are the main contributors to immunostimulatory dsRNAs. More than half of human genes generate alternative mRNA isoforms that differ in their 3′UTRs but encode the same protein (51, 52). Elongated 3′UTRs are thought to be versatile platforms to recruit various RNA binding proteins that can regulate transcript stability and subcellular location (52, 53). The 3′UTR is a heavily structured region in mRNA (54) and harbors many repetitive elements that can base-pair and form dsRNAs (19). The brain expresses the longest 3′UTRs of any human tissue, whereas the liver carries much shorter 3′UTRs (55, 56). Moreover, among the different CNS cell types, neurons carry the longest median 3′UTR length, even longer than glia cells (55). As expected, we observed global 3′UTR lengthening in stem cell–derived neuronal cultures (fig. S16A). 3′UTR length was determined using DaPars2 (57), a program that uses RNA sequencing data to detect alternative polyadenylation sites and produce a PDUI (percentage of distal poly-A sites usage index) value for each gene (data file S1). An increase in PDUI indicates 3′UTR lengthening, and a decrease in PDUI indicates 3′UTR shortening.
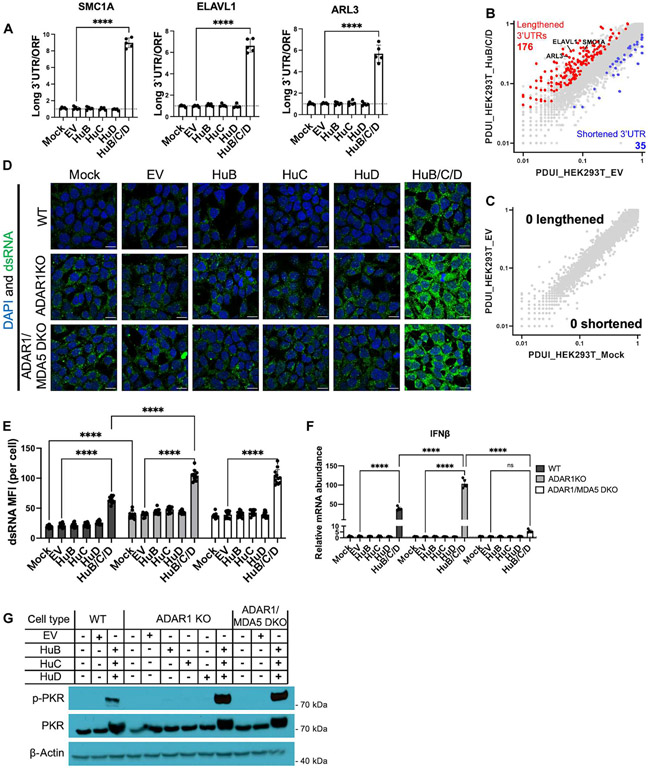
To test whether elongated 3′UTRs contribute to immunostimulatory dsRNA burden, we needed a method to force elongation of 3′UTRs; however, the neural-specific mechanism of 3′UTR lengthening is not fully understood in mammals. A recent study in Drosophila identified three genes in the ELAV/Hu family (Elav, Fne, and Rbp9) that play a role in lengthening 3′UTRs in neurons (58). The human homologs of these genes are HuB (encoded by ELAVL2), HuC (encoded by ELAVL3), and HuD (encoded by ELAVL4). HuB, HuC, and HuD are neuron-enriched RNA binding proteins that recognize AU-rich elements in the 3′UTRs and are thought to play a role in neuronal development (59-62). As expected, ELAVL2, ELAVL3, and ELAVL4 transcript expression was detected in stem cell–derived neurons but minimally detected in hESCs and HEK-293T cells (fig. S16B). We ectopically expressed FLAG-tagged HuB, HuC, or HuD in HEK-293T cells (fig. S17A) and assayed for 3′UTR lengthening of select genes. However, individual expression of HuB, HuC, or HuD failed to affect 3′UTR length (Fig. 4A). Hence, next, we tested whether combined expression of HuB, HuC, and HuD could synergistically lengthen 3′UTRs. We found that combined expression of HuB/C/D markedly induced 3′UTR lengthening of select genes (Fig. 4A and fig. S17D) and globally when DaPars2 analysis was performed (Fig. 4, B and C, and data file S1). Total transcript levels of the 3′UTR lengthened genes remained predominantly constant (fig. S17, B and C). Therefore, we demonstrated that expression of HuB/C/D can induce short-to-long 3′UTR isoform switching in a nonneural cell type such as HEK-293T cells. Because HuB-, HuC-, or HuD-deficient mice exhibit a spectrum of phenotypes ranging from various neural defects and neonatal death (60, 62, 63), this HuB/C/D ectopic expression system allowed us to decouple the neural developmental role of HuB, HuC, and HuD from their other potential roles in immunity.
Fig. 4. Neuron-enriched proteins HuB/C/D cooperate to induce 3′UTR elongation, dsRNA levels, and inflammation.
(A to C) WT HEK-293T cells were transfected with 100 ng of plasmid expressing FLAG-tagged HuB, HuC, or HuD or a combination of HuB (33 ng), HuC (33 ng), and HuD (33 ng). EV, empty vector. 3′UTR lengthening of select genes measured via qPCR (A). The long 3′UTR transcript isoform was normalized to total open reading frame (ORF)–containing transcripts. (B and C) Scatterplot of the PDUI value for genes in HEK-293T_HuB/C/D (n = 3 biological replicates) versus HEK-293T_EV (n = 3 biological replicates) (B) and HEK-293T_EV versus HEK-293T_Mock (n = 3 biological replicates) (C). PDUI values were derived via DaPars2 analysis. Significant change in 3′UTR length was defined as a change in PDUI greater than 1.5-fold. Genes with significantly lengthened (red, increase in PDUI) and shortened (blue, decrease in PDUI) 3′UTRs are colored (FDR ≤ 0.05). (D to G) WT, ADAR1 KO, or ADAR1/MDA5 double-knockout (DKO) HEK-293T cells were transfected as in (A). (D) Immunofluorescent images of dsRNA (J2, green) and DAPI (blue) in transfected cells. (E) Quantification of dsRNA in (D). (F) Relative IFN-β mRNA levels measured by qPCR. Housekeeping gene, RPS11. (G) Immunoblot measuring PKR activation. Scale bars, 10 μm. All quantified data shown are mean ± SD [n = 5 biological replicates in (A) and (F); n = 12 cells from biological triplicates in (E)]. (A) One-way ANOVA with Tukey corrected multiple comparisons. (E and F) Two-way ANOVA with Tukey corrected multiple comparisons, *P < 0.05, **P < 0.01, ***P < 0.001, and ****p < 0.0001.
With our newly developed HuB/C/D ectopic expression system, we examined how 3′UTR lengthening affected dsRNA levels and the innate immune response. At baseline, HEK-293T cells are low in dsRNA load (Fig. 1A) and express minimal IFN-β (Fig. 3I). However, triple expression of HuB/C/D led to a significant increase in dsRNA levels in both WT and ADAR1 KO HEK-293T cells compared with individual expression of each gene (Fig. 4, D and E). In all conditions, we observed higher J2 signal in ADAR1 KO HEK-293T cells than in comparable WT cells (Fig. 4, D and E), consistent with our prior conclusion that ADAR1 is a global regulator of cellular dsRNA load across cell types. Triple HuB/C/D expression also led to spontaneous IFN-β production in WT HEK-293T cells, and even higher levels of IFN-β production in ADAR1 KO cells (Fig. 4F). These effects mirror those in neurons: constitutive type I IFN production that is exacerbated by ADAR1 deficiency (Fig. 3, C and F). Additionally, lack of MDA5 did not alter total dsRNA levels as expected (Fig. 4, D and E, and fig. S18A) but did abrogate IFN-β production (Fig. 4F), suggesting that HuB/C/D-induced dsRNAs signal through MDA5 to produce type I IFN. We also checked PKR activation as another metric of spontaneous inflammation. We found that triple expression of HuB/C/D induced PKR activation in WT HEK-293T cells and ADAR1 deficiency further increased PKR activation (Fig. 4G). Together, expression of the three neuron-enriched genes HuB/C/D can cooperatively increase 3′UTR length, dsRNA burden, and cellular inflammation mediated by MDA5 and PKR. Therefore, we conclude that elongated 3′UTRs are one of the major contributors to immunostimulatory dsRNA structures.
Ectopic expression of ELAVL2, ELAVL3, and ELAVL4 (HuB, HuC, and HuD) confers antiviral activity
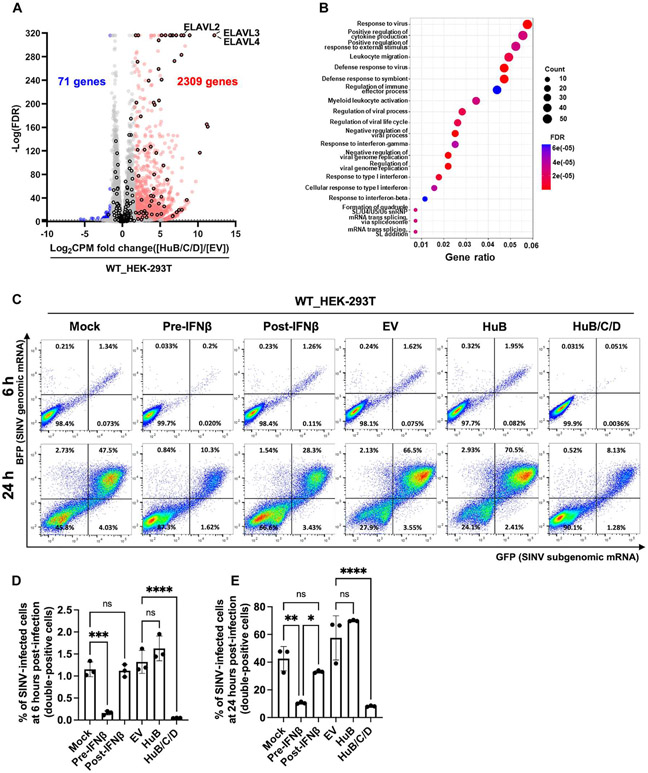
To gain a more comprehensive understanding of the consequences of HuB, HuC, and HuD expression, we conducted a transcriptome-wide analysis of genes differentially expressed upon HuB/C/D expression in WT HEK-293T cells (data file S2). Upon HuB/C/D expression, we found 2309 significantly up-regulated and 71 significantly down-regulated genes (Fig. 5A), whereas only 6 genes are up-regulated when comparing WT HEK-293T cells expressing empty vector with the mock condition (fig. S19A). Gene ontology analysis showed that up-regulated genes were predominantly enriched for antiviral defense and immune response pathways (Fig. 5B), and 64 of the up-regulated genes were ISGs (Fig. 5A), an indication that HuB/C/D expression induces an extensive antiviral and inflammatory signature.
Fig. 5. Ectopic HuB/C/D expression in HEK-293T cells protects against SINV infection.
(A and B) Effects of HuB/C/D ectopic expression on the global transcriptome. (A) Volcano plot showing differential expression of genes between WT HEK-293T_HuB/C/D (n = 3 biological replicates) and WT HEK-293T_EV cells (n = 3 biological replicates). Up-regulated genes (red; FDR ≤ 0.001, CPM fold change ≥ 3) and down-regulated genes (blue; FDR ≤ 0.001, CPM fold change ≤ 0.333) are colored. Ectopically expressed ELAVL2, ELAVL3, and ELAVL4 (HuB, HuC, and HuD) and ISGs are marked with a black border. (B) Dot plot showing gene ontology (biological process) analysis of up-regulated genes in (A). The y axis represents different pathways, and the x axis represents the ratio of the DEGs. Darker red dots represent more significant enrichment. The circle size indicates the number of genes enriched in the pathway. (C to E) EV-, HuB-, or HuB/C/D-transfected WT HEK-293T cells were infected with a SINV dual reporter (MOI = 1); BFP reports for SINV genomic mRNA, and GFP reports for SINV subgenomic mRNA. For the pre–IFN-β set, cells were pretreated with 0.05 nM IFN-β for 24 hours before infection. For the post–IFN-β set, cells were treated with 0.05 nM IFN-β for 1 hour after infection until harvest. (C) Flow cytometry analysis of SINV-infected cells (GFP and BFP double-positive cells). Representative dot plots at 6 and 24 hours after infection. (D and E) Bar graph showing the frequency (%) of SINV-infected cells at 6 hours (D) and 24 hours (E) after infection. Data are shown as mean ± SD (n = 3 biological replicates). One-way ANOVA with Tukey corrected multiple comparisons, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Because HuB/C/D expression in WT HEK-293T cells induced type I IFN and PKR activation (Fig. 4), we postulated that HuB/C/D expression emulates a pre-emptive antiviral state that protects against viral infection. To test this idea, we expressed either empty vector, HuB alone, or HuB/C/D in WT HEK-293T cells and then infected the cells with Sindbis virus (SINV) containing a dual reporter: blue fluorescent protein (BFP) reports for SINV genomic mRNA, and green fluorescent protein (GFP) reports for the SINV subgenomic mRNA. Triple expression of HuB/C/D significantly reduced the percentage of infected cells when compared with the empty vector or HuB alone at 6 and 24 hours after infection (Fig. 5, C to E). In parallel, we pretreated cells with IFN-β 24 hours before infection and found that HuB/C/D expression protects as effectively as IFN-β pretreatment (Fig. 5, C to E). Finally, we also treated cells with IFN-β 1 hour after infection. Although IFN-β after treatment had some protective effect, it was not as protective as IFN-β pretreatment or HuB/C/D expression (Fig. 5, C to E), indicating that preexisting IFN-β before viral infection has more potent antiviral activity than IFN-β induced after infection. In summary, we uncovered that HuB, HuC, and HuD proteins—classically thought to play a neurodevelopmental role—can provide cellular resistance to viral infection.
Loss of ELAVL2 and ELAVL3 (HuB and HuC) in neurons results in lower immunostimulatory dsRNA levels and enhanced susceptibility to virus infection
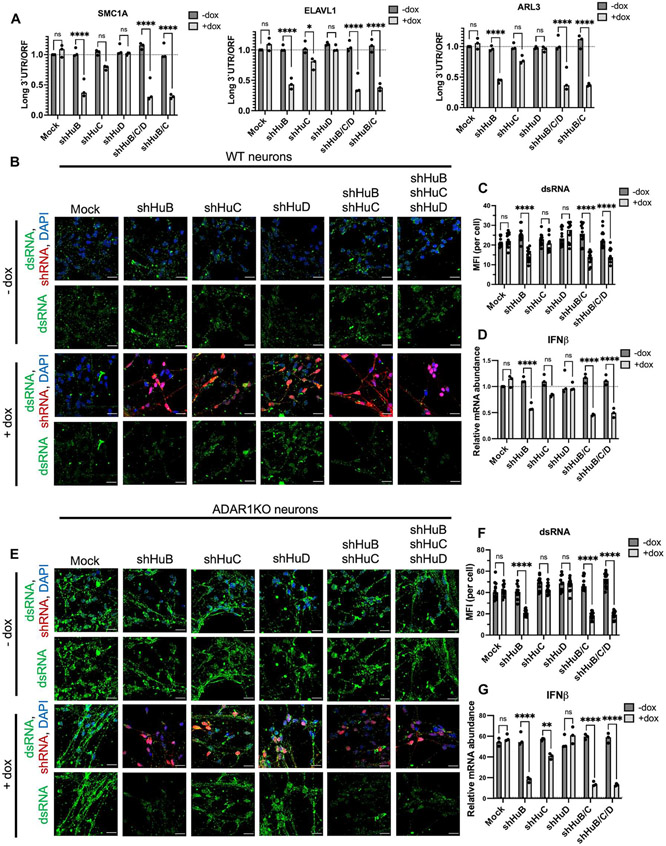
We next sought to further examine the role of HuB, HuC, and HuD proteins in a more physiologically relevant setting. Because these proteins are enriched in neurons, we used doxycycline-inducible shRNAs to down-regulate HuB, HuC, and HuD in day 20 neurons to analyze their function without potentially affecting neuron differentiation. We confirmed efficient knockdown of HuB, HuC, and HuD in neurons (fig. S20A). We found that loss of HuB greatly decreased 3′UTR length and loss of HuC showed a slight trend of decreased 3′UTR length, but loss of HuD did not alter 3′UTR length (Fig. 6A). We next investigated whether HuB, HuC, or HuD also maintained dsRNA levels. We show that loss of HuB alone, loss of HuB and HuC together, or loss of all three ELAVL proteins results in a decrease in dsRNA burden in both WT (Fig. 6, B and C) and ADAR1 KO (Fig. 6, E and F) neurons. Concordant with reduced dsRNA levels, we also observed reduced IFN-β (Fig. 6, D and G) and ISG production (fig. S20D) as well as PKR activation in both WT (fig. S20B) and ADAR1 KO (fig. S20C) neurons. Notably, we observed a striking drop in dsRNA levels in HuB-depleted ADAR1 KO neurons (Fig. 6, E and F), and down-regulating HuB, but not HuC or HuD, prolonged the survival of ADAR1 KO neurons (fig. S21A). In summary, human neurons heavily depend on HuB, and to a lesser extent on HuC, to elongate 3′UTRs, increase dsRNA levels, and induce inflammation as measured by type I IFN and PKR activation.
Fig. 6. HuB and HuC promote long 3′UTRs, dsRNA levels, and inflammation in neurons.
(A to G) Day 20 post-differentiation neurons derived from WT and ADAR1 KO hESCs were transduced with lentivirus containing the indicated doxycycline-inducible shRNA. (A) 3′UTR shortening of selected genes measured by qPCR. The long 3′UTR transcript isoform was normalized to total ORF-containing transcripts. (B) Immunofluorescent images of dsRNA (J2, green) and DAPI (blue) in transduced WT neurons. Expression of doxycycline-inducible shRNA is coupled with red fluorescent protein (RFP) expression (red). (C) Quantification of dsRNA in (B). (D) Relative IFN-β mRNA levels measured by qPCR. (E to G)Same as (B) to (D) above, except in ADAR1 KO neurons. For all qPCR, RPS11 was used as a housekeeping gene. All quantified data shown are mean ±SD [n = 3 biological replicates in (A), (D), and (G); n = 12 cells from biological triplicates in (C) and (F)]. Two-way ANOVA with Tukey corrected multiple comparisons, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
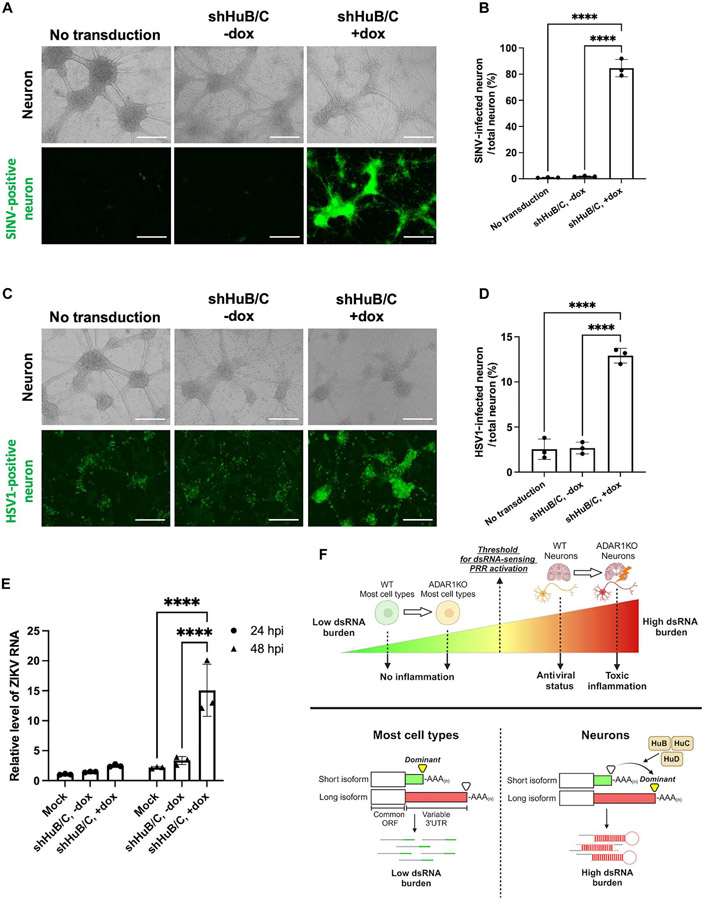
Because ELAVL proteins, especially HuB and HuC, were required to maintain tonic IFN production and PKR activation in WT neurons, we investigated whether these proteins confer resistance to viral infection. To test this, we depleted ELAVL proteins in neurons and measured infectivity with multiple viruses. We depleted both HuB and HuC simultaneously because we observed that this resulted in the greatest decrease in dsRNA and IFN production (Fig. 6). Down-regulating HuB and HuC in neurons resulted in significantly higher rates of SINV infection (Fig. 7, A and B).
Fig. 7. HuB and HuC protect neurons from SINV, HSV-1, and ZIKV infection.
(A to E) Day 20 post-differentiation WT neurons were transduced with lentivirus containing doxycycline-inducible shRNA against HuB and HuC and then were infected with various viruses. (A and B) Neurons were infected with SINV (MOI = 0.1). (A) Fluorescent and brightfield images of neurons at 24 hours after infection. (B) Quantification of GFP-positive area in (A). (C and D) Neurons were infected with an HSV1 GFP reporter virus (MOI = 1). (C) Fluorescent and brightfield images of neurons at 24 hours after infection. (D) Quantification of GFP-positive area in (C). (E) Neurons were infected with ZIKV (MOI = 0.1), and infection was measured via qPCR of Zika RNA at both 24 (circle) and 48 (triangle) hours post-infection (hpi). (F) Summary graphic showing the main findings of this study. For Zika RNA qPCR, 18S RNA was used as a housekeeping gene. All quantified data shown are mean ± SD (n = 3 experimental replicates). Scale bars, 400 μm. (B and D) One-way ANOVA with Tukey corrected multiple comparisons. (E) Two-way ANOVA with Tukey corrected multiple comparisons, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Next, we examined whether HuB/C depletion could also affect the infectivity of neurotropic viruses. Herpes simplex virus 1 (HSV-1) is a DNA virus that can cause HSV-1 encephalitis, the most common form of sporadic viral encephalitis in the Western world. Using HSV-1 with a GFP reporter (64), we found that loss of HuB and HuC significantly increased the infectivity of HSV-1 in neurons (Fig. 7, C and D). Tonic IFN has been proposed to be vital in preventing HSV-1 infection of neurons (28), and our findings serve to underline the importance of ELAVL proteins in maintaining tonic IFN production to protect from viral infection. In addition to HSV-1, we also tested Zika virus (ZIKV), which can cause severe neurological defects. Depletion of HuB/C significantly increased the infectivity of ZIKV in neurons 48 hours after infection (Fig. 7E). Together, our data suggest that HuB and HuC elongate 3′UTRs, trigger dsRNA-induced immunity, and confer an intrinsic antiviral state to neurons (Fig. 7F).
DISCUSSION
A common paradigm in innate immunity is that PRRs discriminate between self and non-self ligands. However, we show that the neuronal transcriptome is intrinsically enriched for immunostimulatory dsRNAs that are constantly sensed by PRRs, even in homeostasis (Fig. 7F). Hence, the distinction between self and non-self dsRNAs may not be as strict as previously believed, especially in neurons. One potential benefit of neuronal dsRNA may be to enhance CNS resistance to viral infections. We demonstrated that depleting dsRNAs in WT neurons dampened tonic type I IFN levels and PKR activation and led to increased susceptibility to infection by SINV, HSV-1, and ZIKV. Hence, the intrinsically high dsRNA levels in neurons may prime the activation of various PRRs (e.g., MDA5, TLR3, and PKR) to induce a preexisting and low-grade immune response that can pre-emptively counteract viral infection. This idea is in line with the emerging concept that the brain heavily relies on “constitutive immune mechanisms” for immediate control of infection that would minimize excessive cell death and inflammation that can cause irreversible damage to the brain (28, 65-67). Such pre-emptive defense mechanisms would be particularly important for postmitotic neurons that have limited capacity to regenerate after cell death or injury.
Pioneering investigations into inborn errors that cause susceptibility to HSV-1 encephalitis have uncovered mutations in various RNA-associated pathways (TLR3 signaling, RNA lariat formation, and small nucleolar RNA biogenesis) (28, 65, 66). Additionally, mutations in RNA polymerase III are also associated with severe varicella zoster virus in the CNS (68). How perturbation of RNA homeostasis leads to weakened antiviral defense in the CNS is incompletely understood. Our study sheds new light on the potential mechanistic underpinning of these observations. We speculate that mutations that alter neuronal dsRNA levels or sensing of neuronal dsRNAs by PRRs may result in weakened basal immune activation before infection, thereby lowering the threshold for viruses to successfully establish infection.
Although neuronal dsRNAs may be beneficial in antiviral defense, we show that high levels of dsRNA in neurons could also pose a risk for harmful neuroinflammation when defective ADAR1 leads to excessively high levels of dsRNA. Elevated type I IFN production in the brain has been reported since the 1980s when AGS was first diagnosed, but why the brain is especially prone to inflammation has been a long-standing question (9, 13, 14). Our study suggests that the inherently high dsRNA level in neurons could provide a molecular basis for why inflammation is most prominent in the AGS brain. We demonstrate that cell types with intrinsically lower dsRNA load tolerated the absence of ADAR1 and subsequent increase in dsRNA levels, but cell types with intrinsically higher dsRNA load such as neurons could not tolerate the absence of ADAR1 because it led to excessively high dsRNA levels that trigger uncontrolled inflammation (Fig. 7F).
High-dsRNA levels in neurons may also serve as the molecular trigger for chronic PKR activation and IFN induction observed in many neurodegenerative disorders, including Alzheimer’s disease and ALS. This is especially relevant for ALS, where perturbation of RNA binding protein dosage or expanded RNA repeat elements is a major cause of disease (2-7, 69). Additionally, regardless of the initial trigger (e.g., viral infection, self-DNA, and self-RNA) or primary source (e.g., blood and CNS) for type I IFN production (8, 70-72), once the brain is exposed to type I IFN, the high dsRNA levels in neurons could potentially pose a risk for creating a positive feedback loop for chronic brain inflammation. This is because type I IFN signaling increases PRR expression—PRRs are ISGs—that can lead to increased neuronal dsRNA sensing by PRRs, further amplifying type I IFN production. This may be why neuropsychiatric symptoms are common in patients with SLE (colloquially referred to as “lupus fog”) and why neurological symptoms are a frequent feature of post-acute infection syndrome (8, 72).
3′UTRs are best known to regulate mRNA localization, stability, and translation (52). Although the brain is understood to globally lengthen 3′UTRs, the functions of these longer 3′UTRs are incompletely understood (53). We developed a method to force 3′UTR lengthening by ectopically expressing just three neuron-enriched genes ELAVL2, ELAVL3, and ELAVL4 (HuB, HuC, and HuD) in a nonneuronal cell type. With this new experimental tool, we were able to ascribe a previously undescribed function to long 3′UTRs, a rich source for dsRNA that bolsters innate immunity and protects against viral infection. These 3′UTRs may give rise to dsRNA either by incorporating repetitive elements (e.g., Alus) prone to forming dsRNA structures or by forming complementary dsRNA with neighboring genes transcribed in opposite direction (i.e., cis-natural antisense transcripts) (73). How ELAVL proteins lengthen 3′UTRs in humans remains unclear. ELAV/Hu proteins in Drosophila are thought to lengthen 3′UTRs by binding near the proximal polyadenylation site, which mediates bypass of the proximal polyadenylation sites and promotes usage of distal polyadenylation sites (58).
From a therapeutic perspective, we envision that 3′UTRs may be lengthened to boost immune responses because increasing immunostimulatory self-dsRNA levels is remarkably effective in cancer immunotherapies (74-76) or that 3′UTRs may be shortened to reduce immune/inflammatory responses (e.g., neuroinflammatory conditions). As proof of concept, we demonstrated that targeting ELAVL2 (HuB) in ADAR1 KO neurons led to shorter 3′UTRs, reduced dsRNA levels and inflammation, and markedly enhanced cell survival of ADAR1 KO neurons. Hence, targeting dsRNAs, the upstream trigger for disease, could be an effective therapeutic strategy to treat AGS.
Future studies should examine whether glial cells (e.g., astrocytes, oligodendrocytes, and microglia) could serve as a source for immunostimulatory dsRNAs and also define their role in responding to tonic type I IFN produced by neurons. Neurons may be the major producers of dsRNA-driven type I IFN, but the major responders to type I IFN may be neighboring glial cells or endothelial cells. For instance, tonic IFN produced from neurons may increase baseline ISG levels in microglia, allowing them to quickly suppress incoming infection. Tonic type I IFN may also boost antiviral defenses by strengthening the blood-brain barrier, because type I IFN signaling can enhance tight junction integrity in endothelial cells (77). In contrast, high/dysregulated levels of type I IFN may stimulate abnormal activation of microglia and astrocytes, causing uncontrolled inflammation and neurodegeneration (78).
Together, we propose that immunostimulatory dsRNAs derived from long 3′UTRs function as a double-edged sword whereby high levels of dsRNA may activate various PRRs to induce tonic inflammation that protect the brain from virus infection; however, these dsRNAs may also predispose the brain to harmful inflammation when neural homeostasis is perturbed.
MATERIALS AND METHODS
Study design
The main objective of this study was to identify a mechanism that rendered neurons more susceptible to inflammation, in cases of both disease and health. We used hESCs to generate neurons and to allow for genetic manipulation. We assessed the level of dsRNA in cells using immunofluorescent confocal microscopy and measured inflammation via qPCR of several markers (IFN and ISGs) and Western blot of PKR activation. We used lentiviral-delivered shRNA to identify genes responsible for both increasing dsRNA levels and inducing the inflammatory response. qPCR and RNA sequencing were used to identify changes in 3′UTR length, and viral infection models were used to identify a functional significance for neural constitutive inflammation.
Cell lines
ADAR1 KO HEK-293T cells and ADAR1 KO male HUES8 hESCs and their corresponding WT control cell lines were generated as described previously (10). Female WA-09 hESCs were obtained from WiCell. WT and ADAR1 KO HeLa cells were provided by R. Cattaneo (Mayo Clinic). ADAR1/MDA5 DKO HEK-293T cells were generated using CRISPR-Cas9 and the following guide RNA sequences: ATAGCGGAAATTCTCGTCTG or TGAAGCACGAGATGAGATAG.
Cell culture reagents
HEK-293T and HeLa cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% nonessential amino acids. Both HUES8 and WA-09 hESCs were cultured in mTESR1 with its supplement (STEMCELL Technologies) and were split using ReLeSR (STEMCELL Technologies). For treatment with various polymerase inhibitors, cells were treated for 12 hours before harvest with either ActD (10 μg/ml, Sigma), α-amanitin (25 μg/ml, Sigma), or IMT1 (1 μM, MedChemExpress).
To knock down various immune proteins in neurons, lentiviruses were generated using a pTRIPZ vector (Dharmacon) that carried shRNA. At day 11 after differentiation, neurons were transduced with lentivirus carrying doxycycline-inducible shRNA targeting MDA5, RIG-I, MAVS, STING, MyD88, TRIF, PKR, HuB, HuC, and HuD. Doxycycline treatment started at day 14 after differentiation.
Ectopic expression of HuB, HuC, and HuD was done using the pFRT backbone (Addgene ID: 26360) expressing ELAVL2, ELAVL3, and ELAVL4, respectively (Addgene ID: 65758, 65759, and 65760). HEK-293T cells were transfected using Lipofectamine 2000 with either 100 ng of a single plasmid or 33.3 ng each of all three plasmids in combination. Transfections lasted for 48 hours before cells were harvested for analysis.
NPC differentiation protocol
hESCs were differentiated into NPCs using an adherent monolayer culture system (STEMCELL Technologies, STEMdiff SMADi Neural Induction Kit). hESCs were plated into Matrigel (Corning)–coated wells and grown with neural induction medium for approximately 15 days. This was followed by maintenance and expansion in neural progenitor medium. Compared with previous work (10), we increased the density of hESCs at plating (3 × 105 cells/cm2) to obtain a more homogeneous and synchronized NPC population.
Neuron differentiation protocol
NPCs were differentiated into neurons using an adherent monolayer culture system (STEMCELL Technologies, BrainPhys Neuronal Medium N2-A & SM1 Kit) (22). NPCs were plated into wells coated with poly-l-ornithine and laminin and grown in neuronal medium. The medium was changed every 3 days, and cells were harvested at the time points indicated.
HLC differentiation protocol
HLCs were generated as described previously (23). Briefly, hESCs were dissociated using Gentle Cell Dissociation Reagent (STEMCELL Technologies) and plated in Matrigel-coated wells. Endoderm differentiation was accomplished using the STEMdiff Definitive Endoderm Kit (STEMCELL Technologies). Endoderm cells were replated in Matrigel-coated wells, and HLC differentiation was started by treating with stage 1 hepatic differentiation medium for 8 days [DMEM/F-12 supplemented with 10% KnockOut Serum Replacement (KOSR), 1% nonessential amino acids, 0.8% penicillin-streptomycin, 1% glutamine, 100 ng/ml HGF (PeproTech), and 1% dimethyl sulfoxide (DMSO)]. On day 8, stage 2 hepatic differentiation medium [DMSO in stage 1 medium replaced with 0.1 μM dexamethasone (Sigma)] was added. At day 11, hepatocyte culture medium (HCM; Lonza) was added, supplemented with oncostatin-M (20 ng/ml) (R&D Systems). Cells were maintained in HCM until harvest at the indicated times (days after differentiation from endoderm).
Cardiomyocyte differentiation protocol
hESCs were differentiated into cardio-myocytes using the STEMdiff Ventricular Cardiomyocyte Differentiation Kit (STEMCELL Technologies). hESCs were dissociated using Gentle Cell Dissociation Reagent and plated into Matrigel-coated wells. Cardiomyocyte differentiation medium was added, supplemented with Matrigel. Differentiation medium was changed every 2 days for 6 days, and on day 8, cardiomyocyte maintenance medium was added. Maintenance medium was changed every 2 days until the cells were harvested at day 24.
Motor neuron differentiation protocol
Motor neurons were generated using an embryoid body–mediated protocol adapted from a previous study (24). hESCs were dissociated using accutase to generate a single-cell suspension and were plated in low-adherence culture dishes in N2B27 medium [50% DMEM/F-12 and 50% neurobasal medium (Thermo Fisher Scientific), supplemented with N2, B27, 1% penicillin-streptomycin, 1% GlutaMAX (all Thermo Fisher Scientific), and 0.1% β-mercaptoethanol (Sigma)], further supplemented with ascorbic acid (Sigma), fibroblast growth factor 2 (FGF2) (Thermo Fisher Scientific), Y-27632 (Abcam), SB431542 (Sigma), LDN 193189 (Tocris), and Chir-99021 (Tocris). At day 2 after plating, ascorbic acid, SB431542, LDN 193189, and Chir-99021 were maintained, and retinoic acid (Sigma) and smoothened agonist (SAG; Sigma) were added. At day 7 , the medium was changed, including only ascorbic acid, retinoic acid, and SAG. At day 9, in addition to the day 7 supplements, N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester (DAPT) (Sigma) was added. At day 11, brainderived neurotrophic factor (BDNF) (R&D Systems) was added, and at day 14, glial cell line–derived neurotrophic factor (GDNF) (R&D Systems) was added. On day 16, the embryoid bodies were dissociated using Accumax and plated into a monolayer in wells coated with poly-l-ornithine and laminin.
Monolayers of motor neurons were maintained in neurobasal medium supplemented with 1% GlutaMAX, 1% nonessential amino acids, 0.1% β-mercaptoethanol, N2, B27, BDNF, GDNF, CNTF (R&D Systems) and 1 μM of 1:1 uridine and fluorodeoxyuridine (Sigma). Motor neurons were harvested at the indicated time points (days after dissociation).
Immunofluorescent microscopy
Cells grown on glass coverslips were washed with phosphate-buffered saline (PBS) and fixed using 4% paraformaldehyde for 20 min at 4°C. Samples were then blocked and permeabilized in PBTG (PBS, 1% bovine serum albumin, 0.1% Triton X-100, 10% goat serum) for 1 hour at room temperature. Primary antibodies were diluted in PBTG and stained overnight at 4°C. Secondary antibodies [either goat anti-mouse immunoglobulin G (IgG) Alexa Fluor 488 or goat anti-rabbit IgG Alexa Fluor 594, Invitrogen (A11001 and A11012)] and DAPI (4′,6-diamidino-2-phenylindole) were diluted in PBTG and stained for 1 hour at room temperature before mounting. Samples were imaged on an LSM-710 confocal microscope.
The following primary antibodies were used: OCT-4A (1:2000, Cell Signaling Technology, C52G3), nestin (1:200, Neuromics, MO22183), TUJ1 (1:1000, BioLegend, 801201), MAP2 (1:100, Cell Signaling Technology, 4542S), FoxA2 (1:400, Cell Signaling Technology, 8186P), HNF4-α (1:500, Cell Signaling Technology, 3113S), albumin (1:500, Cedarlane, CL2513A), ISL1 (1:100, Developmental Studies Hybridoma Bank, 39.4D5), cardiac troponin T (1:100, Thermo Fisher Scientific, MA5-12960), J2 (1:300, ExAlpha, 10010500) (15), K1 (1:300, ExAlpha, 10020200), and 9D5 (1:250, Absolute Antibody, 00458-23.0). RNASelect stain was obtained from Thermo Fisher Scientific.
For samples treated with nucleases, after fixation, there was a permeabilization step in 0.1% Triton X-100 for 1 hour. After, samples were incubated for 30 min at 37°C with either ssRNase (RNase T1, 100 U/ml, Sigma), dsRNase (RNase III, 40 U/ml, Sigma), or DNase (RQ1, 40 U/ml, Promega) diluted in PBS containing 5 mM magnesium chloride. Samples were then stained as described above.
PolyA FISH
To image polyadenylated RNA, 30–nucleotide (nt)–long polyT probes were conjugated to Cy5 (Integrated DNA Technologies). Some samples were treated with α-amanitin as shown above. After differentiation to appropriate cell types, cells were washed with PBS and fixed using 4% paraformaldehyde for 10 min. The cells were then permeabilized in 70% ethanol overnight at −20°C. Samples were washed and hybridized overnight at 37°C in the following buffer: 125 nM probe, 2× SSC, 20% formamide, dextran sulfate (0.1 g/ml), Escherichia coli tRNA (1 mg/ml), 2 mM vanadyl ribonucleoside complex, and 0.1% Tween 20. The samples were washed with 2× SCC and 20% formamide, counter-stained with DAPI, and imaged as described above.
Tissue staining
Frozen sagittal brain sections from healthy C57BL/6 mice were obtained from Zyagen. Heart and skin tissue from healthy C57BL/6 mice were embedded in optimal cutting temperature compound (OCT) and sectioned. All tissue sections were fixed in 4% paraformaldehyde for 20 min at 4°C and permeabilized and blocked in PBTG. Primary antibodies (9D5 from Absolute Antibody, 00458-23.0 and NeuN from Abcam, ab134014) were stained as described above. Secondary antibodies [either goat anti-chicken IgG Alexa Fluor 488 or goat anti-rabbit IgG Alexa Fluor 594, Invitrogen (11039 and 11012)] and DAPI were stained as described above. Samples were imaged on an LSM-710 confocal microscope.
Mitochondria and dsRNA costaining
Cells were treated with MitoTracker Deep Red (Invitrogen) for 1 hour before fixation. Subsequently, the cells were stained with the J2 antibody as described above. Samples were imaged on an LSM-710 confocal microscope. The proportion of colocalization area was analyzed by ImageJ.
Image analysis
Images were processed and analyzed using ImageJ. Mean fluorescence intensity (MFI) of individual cells was determined by tracing cell outlines. MFI data depict 12 individual cells taken from at least three independent images. For colocalization analysis, the JACop plugin was used to measure MFI of four independent images.
Quantitative RT-PCR
RNA was extracted directly from cell samples using the Direct-zol RNA MiniPrep Kit (Zymo Research). cDNA conversion was completed using the ProtoScript II First Strand cDNA Synthesis Kit (New England Biolabs). PowerUp SYBR Green (Thermo Fisher Scientific) and QuantStudio 3 Real-Time PCR System were used for quantitative reverse transcription PCR (qRT-PCR), and all primers are listed in table S1. RPS11 or 18S ribosomal RNA (rRNA) was used as a housekeeping gene for normalization.
Western blot
Cell samples were lysed directly using NuPAGE LDS Sample Buffer (Thermo Fisher Scientific) and 400 mM dithiothreitol (Sigma). Lysates were homogenized by passing through a 26-gauge needle and were denatured by boiling for 10 min. The protein samples were run on NuPAGE 4-12% Bis-Tris Protein Gels (Thermo Fisher Scientific) and were blotted onto nitrocellulose membrane. The following proteins were probed: MDA5 (Cell Signaling Technology, 5321), RIG-I (Cell Signaling Technology, 3743S), MAVS (Cell Signaling Technology, 3993S), STING (Cell Signaling Technology, 13647S), MyD88 (Cell Signaling Technology, 4283S), TRIF (Cell Signaling Technology, 4596S), p-PKR (phospho-T451, Abcam, 227983), PKR (Abcam 32506), p-IRF3 (phospho-S386, Abcam, 76493), IRF3 (Cell Signaling Technology, 11904), ADAR1 (Cell Signaling Technology, 81284S), ADAR2 (Atlas Antibodies, 018277), FLAG (Santa Cruz Biotechnology, 166355), and β-actin (Sigma, A3854). Blots were developed using a chemiluminescent (film) method.
RNA extraction and library preparation
For the HuB/C/D ectopic expression experiment (biological triplicates of HEK-293T_Mock, HEK-293T_EV, and HEK-293T_HuB/C/D), RNA was extracted with the Direct-zol RNA MiniPrep Kit (Zymo Research). ERCC RNA Spike-In Mix (Thermo Fisher Scientific) was added to each RNA sample. rRNA was depleted using the RiboMinus Eukaryote Kit (Thermo Fisher Scientific). Libraries were prepared with the NEBNext Ultra II Directional RNA Library Prep Kit (New England Biolabs). The nine libraries were pooled and sequenced on the Illumina NovaSeq platform with a read length of 150 nt in the paired-end configuration. For analyzing global 3′UTR extension in neurons, biological triplicates of RNA samples from neurons and HEK-293T were extracted with the Direct-zol RNA MiniPrep Kit (Zymo Research). After adding ERCC RNA Spike-In Mix (Thermo Fisher Scientific), libraries were prepared with the TruSeq Stranded Total RNA with Ribo-Zero Kit (Illumina). The libraries were sequenced on the Illumina NovaSeq platform with a read length of 100 nt in paired-end configuration.
Virus infection
Viruses and infections
SINV, HSV-1 (64), and ZIKVs originated from TE5′2J, KOS-1, and MR766 strains, respectively. SINV has a dual reporter system that produces a BFP and a GFP. HSV-1 expresses GFP. After virus infection of cells at the appropriate multiplicity of infection (MOI), samples were collected for analysis at 6, 24, or 48 hours after infection. Fluorescent images were acquired using the EVOS M5000 microscope system (Invitrogen).
Flow cytometry
Cells were detached with Accumax (STEMCELL Technologies) and fixed using 2% paraformaldehyde for 20 min at 4°C. Samples were resuspended in 3% FBS solution and analyzed using a BD LSR Fortessa flow cytometer (BD Biosciences).
RNA sequencing data analysis
Preprocessing and alignment
After quality control (FastQC v0.11.5), adaptor and low-quality reads were trimmed from FASTQ files using fastp v0.20.1, and duplicated reads were removed using fastuniq v1.1. Preprocessed reads were mapped to the human genome (hg38) using STAR v2.7.3a to generate BAM files.
Alternative polyadenylation site analysis
Uniquely mapped reads in BAM files were converted to bedgraph files using bedtools v2.30.0. DaPars v2.1 took the bedgraph files as input, inferred alternative polyadenylation sites, and provided a percentage of distal polyadenylated usage index (PDUI) (57). We only used genes for which at least two PDUI values were measured in each sample for statistical analysis. Significant 3′UTR changes were defined as alterations with false discovery rate (FDR) ≤ 0.05 and ∣log2(fold change of PDUI)∣ ≥ 0.59.
Gene expression analysis
Read counts were calculated using featureCounts v1.6.4 with a GENCODE v38 primary assembly annotation file. Genes with a raw count of fewer than 10 in all sample conditions were excluded from subsequent analysis. Normalization [using the trimmed mean of M-values (TMM) method] and identification of differentially expressed genes (DEGs) were performed with the edgeR package in Bioconductor (79). Genes with FDR ≤ 0.001 and ∣log2(fold change of counts per million, CPM)∣ ≥ 1.585 are annotated as DEGs. Gene ontology (biological process) enrichment of up-regulated genes was performed with the clusterProfiler Bioconductor package (80).
HPA data
The tissue gene expression data were sourced from the HPA database, and the expression levels of type I IFN (IFN-β and IFN-α subtypes) were extracted. The resultant dataset was visualized using the heatmap function in R.
Statistical analysis
Except for RNA sequencing analysis, all statistical analysis was performed using GraphPad Prism 9 software. Where indicated, a one- or two-way analysis of variance (ANOVA) with multiple comparisons was performed with Tukey post hoc correction.
Supplementary Material
Acknowledgments:
We thank M. Gottesman, S. Goff, S. Reiner, L. Symington, and U. Basu for helpful input while preparing the manuscript; the Microbiology and Immunology Core Facility (A. Figueroa and A. Nanda), the Stem Cell Initiative (B. Corneo and M. Kissner), the Genome Center (E. Bush and K. Mejia), and the Macromolecular Bank (T. Su and X. Wang) at Columbia University Medical Center; D. Fidock for sharing laboratory equipment; C. Rice for providing us with the SINV reporter; P. Desai for providing us with the HSV-1 reporter; R. Cattaneo for ADAR1KO HeLa cells; and X. Wu for advice on generating HLCs.
Funding:
This work is supported by NIH/National Institute of Neurological Disorder and Stroke grants R01NS127802 (to H.C.) and R01NS116141 (to H.W.), NIH grants R21AI171827 (to A.-L.S.) and R35GM150778 (to A.-L.S.), NIH training grants T32AI106711 (to T.J.D.) and T32GM145440 (to J.A.G.), and Searle Scholars Program grant 20-SSP-114 (to H.C.).
Footnotes
Data and materials availability:
Raw RNA sequencing data are deposited in the GEO and can be found under accession number GSE214949. All data needed to support the conclusions of the paper are present in the paper or the Supplementary Materials. Key materials (ADAR1/MDA5 double KO HEK-293T cells) are available upon receipt of a material transfer agreement to the corresponding author. All other materials are available through Addgene, are commercially available, or were obtained from other laboratories.
Competing interests: T.J.D., H.S., and H.C. are inventors on patent number 63469895 held by Columbia University that demonstrates a method to fine-tune inflammation by altering 3′UTR length. The other authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.Chen YG, Hur S, Cellular origins of dsRNA, their recognition and consequences. Nat. Rev. Mol. Cell Biol 23, 286–301 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hugon J, Mouton-Liger F, Dumurgier J, Paquet C, PKR involvement in Alzheimer’s disease. Alzheimers Res. Ther 9, 83 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zu T, Guo S, Bardhi O, Ryskamp DA, Li J, Tusi SK, Engelbrecht A, Kiippel K, Chakrabarty P, Nguyen L, Goide TE, Sonenberg N, Ranum LPW, Metformin inhibits RAN translation through PKR pathway and mitigates disease in C9orf72 ALS/FTD mice. Proc. Natl. Acad. Sci. U.S.A 117, 18591–18599 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodriguez S,Sahin A,Schrank BR,Ai-Lawati H,Costantino i.,Benz E,Fard D,Albers AD,Cao L,Gomez AC,Evans K,Ratti E,Cudkowicz M,Frosch MP,Talkowski M,Sorger PK,Hyman BT,Albers MW,Genome-encoded cytoplasmic double-stranded RNAs, found in C9ORF72 ALS-FTD brain, propagate neuronal loss. Sci. Transl. Med 13, eaaz4699 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roy ER, Chiu G, Li S, Propson NE, Kanchi R, Wang B, Coarfa C, Zheng H, Cao W, Concerted type I interferon signaling in microglia and neural cells promotes memory impairment associated with amyloid β plaques. Immunity 55, 879–894.e6 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee H, Fenster RJ, Pineda SS,Gibbs WS, Mohammadi S,Davila-Velderrain J, Garcia FJ, Therrien M, Novis HS, Gao F, Wilkinson H, Vogt T, Kellis M, LaVoie MJ, Heiman M, Cell type-specific transcriptomics reveals that mutant huntingtin leads to mitochondrial RNA release and neuronal innate immune activation. Neuron 107, 891–908.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ochoa E, Ramirez P, Gonzalez E, De Mange J, Ray WJ, Bieniek KF, Frost B, Pathogenic tau–induced transposable element–Derived dsRNA drives neuroinflammation. Sci. Adv 9, eabq5423 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crow MK, Olferiev M, Kirou KA, Type I interferons in autoimmune disease. Annu. Rev. Pathol 14, 369–393 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Crow YJ, Stetson DB, The type I interferonopathies: 10 years on. Nat. Rev. Immunol 22, 471–483 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung H, Calis JJA, Wu X, Sun T, Yu Y, Sarbanes SL, Dao Thi VL, Shilvock AR, Hoffmann HH, Rosenberg BR, Rice CM, Human ADAR1 prevents endogenous RNA from triggering translational shutdown. Cell 172, 811–824.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahmad S, Mu X, Yang F, Greenwald E, Park JW, Jacob E, Zhang CZ, Hur S, Breaching self-tolerance to alu Duplex RNA underlies MDA5-mediated inflammation. Cell 172, 797–810.e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maharana S, Kretschmer S, Hunger S, Yan X, Kuster D, Traikov S, Zillinger T, Gentzel M, Elangovan S, Dasgupta P, Chappidi N, Lucas N, Maser KI, Maatz H, Rapp A, Marchand V, Chang YT, Motorin Y, Hubner N, Hartmann G, Hyman AA, Alberti S, Lee-Kirsch MA, SAMHD1 controls innate immunity by regulating condensation of immunogenic self RNA. Mol. Cell 82, 3712–3728.e10 (2022). [DOI] [PubMed] [Google Scholar]
- 13.Lodi L, Melki I, Bondet V, Seabra L, Rice GI, Carter E, Lepelley A, Martin-Niclós MJ, Al Adba B, Bader-Meunier B, Barth M, Blauwblomme T, Bodemer C, Boespflug-Tanguy O, Dale RC, Desguerre I, Ducrocq C, Dulieu F, Dumaine C, Ellul P, Hadchouel A, Hentgen V, Hié M, Hully M, Jeziorski E, Lévy R, Mochel F, Orcesi S, Passemard S, Pouletty M, Quartier P, Renaido F, Seidl R, Shetty J, Neven B, Blanche S, Duffy D, Crow YJ, Frémond ML, Differential expression of interferon-alpha protein provides clues to tissue specificity across type i interferonopathies. J. Clin. Immunol. 41, 603–609 (2021). [DOI] [PubMed] [Google Scholar]
- 14.Lebon P, Badouai J, Ponsot G, Goutières F, Hémeury-Cukier F, Aicardi J, Intrathecal synthesis of interferon-alpha in infants with progressive familial encephalopathy. J. Neurol. Sci 84, 201–208 (1988). [DOI] [PubMed] [Google Scholar]
- 15.Schonborn J, Oberstraß J, Breyel E, Tittgen J, Schumacher J, Lukacs N, Monoclonal antibodies to double-stranded RNA as probes of RNA structure in crude nucleic acid extracts. Nucleic Acids Res. 19, 2993–3000 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim Y, Park J, Kim S, Kim M, Kang MG, Kwak C, Kang M, Kim B, Rhee HW, Kim VN, PKR senses nuclear and mitochondrial signals by interacting with endogenous double-stranded RNAs. Mol. Cell 71, 1051–1063.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Dhir A, Dhir S, Borowski LS, Jimenez L, Teitell M, Rötig A, Crow YJ, Rice GI, Duffy D, Tamby C,, Nojima T, Munnich A, Schiff M, de Almeida CR, Rehwinkel J, Dziembowski A, Szczesny RJ, Proudfoot NJ, Mitochondrial double-stranded RNA triggers antiviral signalling in humans. Nature 560, 238–242 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao Y, Vasic R, Song Y, Teng R, Liu C, Gbyli R, Biancon G, Nelakanti R, Lobben K, Kudo E, Liu W, Ardasheva A, Fu X, Wang X, Joshi P, Lee V, Dura B, Viero G, Iwasaki A, Fan R, Xiao A, Flavell RA, Li HB, Tebaldi T, Halene S, m6A modification prevents formation of endogenous double-stranded RNAs and deleterious innate immune responses during hematopoietic development. Immunity 52, 1007–1021.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reich DP, Bass BL, Mapping the dsRNA world. Cold Spring Harb. Perspect. Biol 11, a035352 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blango MG, Bass BL, Identification of the long, edited dsRNAome of LPS-stimulated immune cells. Genome Res. 26, 852–862 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L, Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol 27, 275–280 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bardy C, Van Den Hurk M, Eames T, Marchand C, Hernandez RV, Kellogg M, Gorris M, Galet B, Palomares V, Brown J, Bang AG, Mertens J, Böhnke L, Boyer L, Simon S, Gage FH, Neuronal medium that supports basic synaptic functions and activity of human neurons in vitro. Proc. Natl. Acad. Sci. U.S.A 112, E2725–E2734 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu X, Dao Thi VL, Huang Y, Billerbeck E, Saha D, Hoffmann HH, Wang Y, Silva LAV, Sarbanes S, Sun T, Andrus L, Yu Y, Quirk C, Li M, MacDonald MR, Schneider WM, An X, Rosenberg BR, Rice CM, Intrinsic immunity shapes viral resistance of stem cells. Cell 172, 423–438.e25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maury Y, Côme J, Piskorowski RA, Salah-Mohellibi N, Chevaleyre V, Peschanski M, Martinat C,, Nedelec S, Combinatorial analysis of developmental cues efficiently converts human pluripotent stem cells into multiple neuronal subtypes. Nat. Biotechnol 33, 89–96 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Lewis YE, Moskovitz A, Mutlak M, Heineke J, Caspi LH, Kehat I, Localization of transcripts, translation, and degradation for spatiotemporal sarcomere maintenance. J. Mol. Cell. Cardiol 116, 16–28 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Bonekamp NA, Peter B, Hillen HS, Felser A, Bergbrede T, Choidas A, Horn M, Unger A, Di Lucrezia R, Atanassov I, Li X, Koch U, Menninger S, Boros J, Habenberger P, Giavalisco P, Cramer P, Denzel MS, Nussbaumer P, Klebl B, Falkenberg M, Gustafsson CM, Larsson NG, Small-molecule inhibitors of human mitochondrial DNA transcription. Nature 588, 712–716(2020). [DOI] [PubMed] [Google Scholar]
- 27.Gough DJ, Messina NL, Clarke CJ, Johnstone RW, Levy DE, Constitutive type I interferon modulates homeostatic balance through tonic signaling. Immunity 36, 166–174 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao D, Ciancanelli MJ, Zhang P, Harschnitz O, Bondet V, Hasek M, Chen J, Mu X, Itan Y, Cobat A, Sancho-Shimizu V, Bigio B, Lorenzo L, Ciceri G, McAlpine J, Anguiano E, Jouanguy E, Chaussabel D, Meyts I, Diamond MS, Abel L, Hur S, Smith GA, Notarangelo L, Duffy D, Studer L, Casanova JL, Zhang SY, TLR3 controls constitutive IFN-β antiviral immunity in human fibroblasts and cortical neurons. J. Clin. Invest 131, e134529 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ejlerskov P, Hultberg JG, Wang J, Carlsson R, Ambjorn M, Kuss M, Liu Y, Porcu G, Kolkova K, Friis Rundsten C, Ruscher K, Pakkenberg B, Goldmann T, Loreth D, Prinz M, Rubinsztein DC, Issazadeh-Navikas S, Lack of neuronal IFN-β-IFNAR causes Lewy body- and Parkinson’s disease-like dementia. Cell 163, 324–339 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hosseini S, Michaelsen-Preusse K, Grigoryan G, Chhatbar C, Kalinke U, Korte M, Type I interferon receptor signaling in astrocytes regulates hippocampal synaptic plasticity and cognitive function of the healthy CNS. Cell Rep. 31, 107666(2020). [DOI] [PubMed] [Google Scholar]
- 31.Medzhitov R, The spectrum of inflammatory responses. Science 374, 1070–1075(2021). [DOI] [PubMed] [Google Scholar]
- 32.GTEx Consortium, The Genotype-Tissue Expression (GTEx) project. Nat. Genet 45, 580–585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, Olsson IM, Edlund K, Lundberg E, Navani S, Szigyarto CAK, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, Von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, Von Heijne G, Nielsen J, Pontén F, Tissue-based map of the human proteome. Science 347, 1260419 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Tan X, Sun L, Chen J, Chen ZJ, Detection of microbial infections through innate immune sensing of nucleic acids. Annu. Rev. Microbiol 72, 447–478 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Rice GI, Kasher PR, Forte GM, Mannion NM, Greenwood SM, Szynkiewicz M, Dickerson JE, Bhaskar SS, Zampini M, Briggs TA, Jenkinson EM, Bacino CA, Battini R, Bertini E, Brogan PA, Brueton LA, Carpanelli M, De Laet C, de Lonlay P, del Toro M, Desguerre L, Fazzi E, Garcia-Cazorla A, Heiberg A, Kawaguchi M, Kumar R, Lin JP, Lourenco CM, Male AM, Marques W Jr., Mignot C, Olivieri I, Orcesi S, Prabhakar P, Rasmussen M, Robinson RA, Rozenberg F, Schmidt JL, Steindl K, Tan TY, van der Merwe WG, Vanderver A, Vassallo G, Wakeling EL, Wassmer E, Whittaker E, Livingston JH, Lebon P, Suzuki T, McLaughlin PJ, Keegan LP, O’Connell MA, Lovell SC, Crow YJ, Mutations in ADAR1 cause Aicardi-Goutières syndrome associated with a type I interferon signature. Nat. Genet 44, 1243–1248(2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bass BL, Weintraub H, An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell 55, 1089–1098 (1988). [DOI] [PubMed] [Google Scholar]
- 37.Mannion NM, Greenwood SM, Young R, Cox S, Brindle J, Read D, Nellaker C, Vesely C, Ponting CP, McLaughlin PJ, Jantsch MF, Dorin J, Adams IR, Scadden AD, Oh man M, Keegan L,P, O’Connell MA, The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep. 9, 1482–1494 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pestal K, Funk CC, Snyder JM, Price ND, Treuting PM, Stetson DB, Isoforms of RNA-editing enzyme ADAR1 independently control nucleic acid sensor MDA5-driven autoimmunity and multi-organ development. Immunity 43, 933–944 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liddicoat BJ, Piskol R, Chalk AM, Ramaswami G, Higuchi M, Hartner JC, Li JB, Seeburg PH, Walkley CR, RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science 349, 1115–1120(2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pfaller CK, Donohue RC, Nersisyan S, Brodsky L, Cattaneo R, Extensive editing of cellular and viral double-stranded RNA structures accounts for innate immunity suppression and the proviral activity of ADAR1p150. PLOS Biol. 16, e2006577 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.George CX, Ramaswami G, Li JB, Samuel CE, Editing of cellular self-RNAs by adenosine deaminase ADAR1 suppresses innate immune stress responses. J. Biol. Chem 291, 6158–6168 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Reuver R, Dierick E, Wiernicki B, Staes K, Seys L, De Meester E, Muyldermans T, Botzki A, Lambrecht BN, Van Nieuwerburgh F, Vandenabeele P, Maelfait J, ADAR1 interaction with Z-RNA promotes editing of endogenous double-stranded RNA and prevents MDA5-dependent immune activation. Cell Rep. 36, 109500(2021). [DOI] [PubMed] [Google Scholar]
- 43.Eisenberg E, Levanon EY, A-to-I RNA editing—Immune protector and transcriptome diversifier. Nat. Rev. Genet 19, 473–490(2018). [DOI] [PubMed] [Google Scholar]
- 44.Nakahama T, Kato Y, Shibuya T, Inoue M, Kim JI, Vongpipatana T, Todo H, Xing Y, Kawahara Y, Mutations in the adenosine deaminase ADAR1 that prevent endogenous Z-RNA binding induce Aicardi-Goutières-syndrome-like encephalopathy. Immunity 54, 1976–1988.e7 (2021). [DOI] [PubMed] [Google Scholar]
- 45.Guo X, Steinman RA, Sheng Y, Cao G, Wiley CA, Wang Q, An AGS-associated mutation in ADAR1 catalytic domain results in early-onset and MDA5-dependent encephalopathy with IFN pathway activation in the brain. J. Neuroinflammation 19, 285 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Behrendt R, Roers A, Mouse models for Aicardi-Goutières syndrome provide clues to the molecular pathogenesis of systemic autoimmunity. Clin. Exp. Immunol 175, 9–16 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maurano M, Snyder JM, Connelly C, Henao-Mejia J, Sidrauski C, Stetson DB, Protein kinase R and the integrated stress response drive immunopathology caused by mutations in the RNA deaminase ADAR1. Immunity 54, 1948–1960.e5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiao H, Wachsmuth L, Wolf S, Lohmann J, Nagata M, Kaya GG, Oikonomou N, Kondylis V, Rogg M, Diebold M, Tröder SE, Zevnik B, Prinz M, Schell C, Young GR, Kassiotis G,, Pasparakis M, ADAR1 averts fatal type I interferon induction by ZBP1. Nature 607, 776–783 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pinto Y, Cohen HY, Levanon EY, Mammalian conserved ADAR targets comprise only a small fragment of the human editosome. Genome Biol. 15, R5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tan MH, Li Q, Shanmugam R, Piskol R, Kohler J, Young AN, Liu KI, Zhang R, Ramaswami G, Ariyoshi K, Gupte A, Keegan LP, George CX, Ramu A, Huang N, Pollina EA, Leeman DS, Rustighi A, Goh YPS; GTEx Consortium; Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group; Statistical Methods groups–Analysis Working Group; Enhancing GTEx (eGTEx) groups; NIH Common Fund; NIH/NCI; NIH/NHGRI; NIH/NIMH; NIH/NIDA; Biospecimen Collection Source Site—NDRI; Biospecimen Collection Source Site—RPCI; Biospecimen Core Resource—VARI; Brain Bank Repository—University of Miami Brain Endowment Bank; Leidos Biomedical—Project Management; ELSI Study; Genome Browser Data Integration &Visualization—EBI; Genome Browser Data Integration &Visualization—UCSC Genomics Institute, University of California Santa Cruz, Chawla A, Del Sal G, Peltz G, Brunet A, Conrad DF, Samuel CE, O’Connell MA, Walkley CR, Nishikura K, Li JB, Dynamic landscape and regulation of RNA editing in mammals. Nature 550, 249–254(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Flerrmann CJ, Schmidt R, Kanitz A, Artimo P, Gruber AJ, Zavoian M, PolyASite 2.0: A consolidated atlas of polyadenylation sites from 3′ end sequencing. Nucleic Acids Res. 48, D149–D179 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mayr C, What are 3′ utrs doing? Cold Spring Harb. Perspect. Biot 11, a034728 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bae B, Miura P, Emerging roles for 3’ UTRs in neurons, Int. J. Mot. Sci 21, 3413 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu X, Bartel DP, Widespread influence of 3′-end structures on mammalian mRNA processing and stability. Cell 169, 905–917.e11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tushev G, Glock C, Heumuller M, Biever A, Jovanovic M, Schuman EM, Alternative 3’ UTRs modify the localization, regulatory potential, stability, and plasticity of mrnas in neuronal compartments. Neuron 98, 495–511.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Miura P, Shenker S, Andreu-Agullo C, Westholm JO, Lai EC, Widespread and extensive lengthening of 3’ UTRs in the mammalian brain. Genome Res. 23, 812–825 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li L, Huang KL, Gao Y, Cui Y, Wang G, Elrod ND, Li Y, Chen YE, Ji P, Peng F, Russell WK, Wagner EJ, Li W, An atlas of alternative polyadenylation quantitative trait loci contributing to complex trait and disease heritability. Nat. Genet 53, 994–1005 (2021). [DOI] [PubMed] [Google Scholar]
- 58.Wei L, Lee S, Majumdar S, Zhang B, Sanfilippo P, Joseph B, Miura P, Soller M, Lai EC, Overlapping activities of ELAV/Hu family RNA binding proteins specify the extended neuronal 3′ UTR landscape in drosophila. Mol. Celt 80, 140–155.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oka no HJ, Darnell RB, A hierarchy of Hu RNA binding proteins in developing and adult neurons. J. Neurosci 17, 3024–3037 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Akamatsu W, Fujihara H, Mitsuhashi T, Yano M, Shibata S, Hayakawa Y, Okano HJ, Sakakibara SI, Takano H, Takano T, Takahashi T, Noda T, Okano H, The RNA-binding protein HuD regulates neuronal cell identity and maturation. Proc. Natl. Acad. Sci. U.S.A 102, 4625–4630 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mansfield KD, Keene JD, Neuron-specific ELAV/Hu proteins suppress HuR mRNA during neuronal differentiation by alternative polyadenylation. Nucleic Acids Res. 40, 2734–2746 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ince-Dunn G, Okano HJ, Jensen KB, Park WY, Zhong R, Ule J, Mele A, Fak JJ, Yang CW, Zhang C, Yoo J, Herre M, Okano H, Noebels JL, Darnell RB, Neuronal Elavlike (Hu) proteins regulate RNA splicing and abundance to control glutamate levels and neuronal excitability. Neuron 75, 1067–1080(2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kato Y, Iwamori T, Ninomiya Y, Kohda T, Miyashita J, Sato M, Saga Y, ELAVL2-directed RNA regulatory network drives the formation of quiescent primordial follicles. EMBO Rep. 20, e48251 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Desai P, Person S, Incorporation of the green fluorescent protein into the herpes simplex virus type 1 capsid. J. Virol 72, 7563–7568 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang SY, Clark NE, Freije CA, Pauwels E, Taggart AJ, Okada S, Mandel H, Garcia P, Ciancanelli MJ, Biran A, Lafaille FG, Tsumura M, Cobat A, Luo J, Volpi S, Zimmer B, Sakata S, Dinis A, Ohara O, Garcia Reino EJ, Dobbs K, Hasek M, Holloway SP, McCammon K, Hussong SA, DeRosa N, Van Skike CE, Katolik A, Lorenzo L, Hyodo M, Faria E, Halwani R, Fukuhara R, Smith GA, Galvan V, Damha MJ, Al-Muhsen S, Itan Y, Boeke JD, Notarangelo LD, Studer L, Kobayashi M, Diogo L, Fairbrother WG, Abel L, Rosenberg BR, Hart PJ, Etzioni A, Casanova JL, Inborn errors of rna lariat metabolism in humans with brainstem viral infection. Cell 172, 952–965.e18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lafaille FG, Harschnitz O, Lee YS, Zhang P, Hasek ML, Kerner G, Itan Y, Ewaleifoh O, Rapaport F, Carlile TM, Carter-Timofte ME, Paquet D, Dobbs K, Zimmer B, Gao D, Rojas-Duran MF, Kwart D, Rattina V, Ciancanelli MJ, McAlpine JL, Lorenzo L, Boucherit S, Rozenberg F, Halwani R, Henry B, Amenzoui N, Alsum Z, Marques L, Church JA, Al-Muhsen S, Tardieu M, Bousfiha AA, Paludan SR, Mogensen TH, Quintana-Murci L, Tessier-Lavigne M, Smith GA, Notarangelo LD, Studer L, Gilbert W, Abel L, Casanova JL, Zhang SY, Human SNORA31 variations impair cortical neuronintrinsic immunity to HSV-1 and underlie herpes simplex encephalitis. Nat. Med 25, 1873–1884 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Paludan SR, Mogensen TH, Constitutive and latent immune mechanisms exert ‘silent’ control of virus infections in the central nervous system. Curr. Opin. Immunol 72, 158–166 (2021). [DOI] [PubMed] [Google Scholar]
- 68.Ogunjimi B, Zhang SY, Sorensen KB, Skipper KA, Carter-Timofte M, Kerner G, Luecke S, Prabakaran T, Cai Y, Meester J, Bartholomeus E, Bolar NA, Vandeweyer G, Claes C, Sillis Y, Lorenzo L, Fiorenza RA, Boucherit S, Dielman C, Heynderickx S, Elias G, Kurotova A, Vander Auwera A, Verstraete L, Lagae L, Verhelst H, Jansen A, Ramet J, Suls A, Smits E, Ceulemans B, Van Laer L, Wilson GP, Kreth J, Picard C, Von Bernuth H, Fluss J, Chabrier S, Abel L, Mortier G, Fribourg S, Mikkelsen JG, Casanova JL, Paludan SR, Mogensen TH, Inborn errors in RNA polymerase III underlie severe varicella zoster virus infections. J. Clin. Invest 127, 3543–3556(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Conlon EG, Manley JL, RNA-binding proteins in neurodegeneration: Mechanisms in aggregate. Genes Dev. 31, 1509–1528(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Udeochu JC, Amin S, Huang Y, Fan L, Torres ERS, Carling GK, Liu B, McGurran H, Coronas-Samano G, Kauwe G, Mousa GA, Wong MY, Ye P, Nagiri RK, Lo I, Holtzman J, Corona C, Yarahmady A, Gill MT, Raju RM, Mok SA, Gong S, Luo W, Zhao M, Tracy TE, Ratan RR, Tsai LH, Sinha SC, Gan L, Tau activation of microglial cGAS–IFN reduces MEF2C-mediated cognitive resilience. Nat. Neurosci 26, 737–750 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu CH, Davidson S, Harapas CR, Hilton JB, Mlodzianoski MJ, Laohamonthonkul P, Louis C, Low RRJ, Moecking J, De Nardo D, Balka KR, Calleja DJ, Moghaddas F, Ni E, McLean CA, Samson AL, Tyebji S, Tonkin CJ, Bye CR, Turner BJ, Pepin G, Gantier MP, Rogers KL, McArthur K, Crouch PJ, Masters SL, TDP-43 triggers mitochondrial DNA release via mPTP to activate cGAS/STING in ALS. Cell 183, 636–649.e18 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Choutka J, Jansari V, Hornig M, Iwasaki A, Unexplained post-acute infection syndromes. Nat. Med 28, 911–923 (2022). [DOI] [PubMed] [Google Scholar]
- 73.Li Q, Gloudemans MJ, Geisinger JM, Fan B, Aguet F, Sun T, Ramaswami G, Li YI, Ma JB, Pritchard JK, Montgomery SB, Li JB, RNA editing underlies genetic risk of common inflammatory diseases. Nature 608, 569–577(2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ishizuka JJ, Manguso RT, Cheruiyot CK, Bi K, Panda A, Iracheta-Vellve A, Miller BC, Du PP, Yates KB, Dubrot J, Buchumenski I, Comstock DE, Brown FD, Ayer A, Kohnle IC, Pope HW, Zimmer MD, Sen DR, Lane-Reticker SK, Robitschek EJ, Griffin G,K, Collins NB, Long AH, Doench JG, Kozono D, Levanon EY, Haining WN, Loss of ADAR1 in tumours overcomes resistance to immune checkpoint blockade. Nature 565, 43–48 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mehdipour P, Marhon SA, Ettayebi I, Chakravarthy A, Hosseini A, Wang Y, deCastro FA, LoYau H, Ishak C, Abe Ison S, O’Brien CA, De Carvalho DD, Epigenetic therapy induces transcription of inverted SINEs and ADAR1 dependency. Nature 588, 169–173 (2020). [DOI] [PubMed] [Google Scholar]
- 76.Zhang T, Yin C, Fedorov A, Qiao L, Bao H, Beknazarov N, Wang S, Gautam A, Williams RM, Crawford JC, Peri S, Studitsky V, Beg AA, Thomas PG, Walkley C, Xu Y, Poptsova M, Herbert A, Balachandran S, ADAR1 masks the cancer immunotherapeutic promise of ZBP1-driven necroptosis. Nature 606, 594–602 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Daniels BP, Klein RS, Knocking on closed doors: Host interferons dynamically regulate blood-brain barrier function during viral infections of the central nervous system. PLOS Pathog. 11, el005096 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Blank T, Prinz M, Type I interferon pathway in CNS homeostasis and neurological disorders. Glia 65, 1397–1406 (2017). [DOI] [PubMed] [Google Scholar]
- 79.Robinson MD, McCarthy DJ, Smyth GK, edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yu G, Wang LG, Han Y, He QY, ClusterProfiler: An R package for comparing biological themes among gene clusters. OM/CS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.