Abstract
Viral hemorrhagic septicemia virus (VHSV) infections cause high losses in cultured rainbow trout in Europe. Attempts to produce a recombinant vaccine based on the transmembrane glycoprotein (G protein) have indicated that proper folding is important for the antigenicity and immunogenicity of the protein. The present study was initiated to identify the disulfide bonds and other structural aspects relevant to vaccine design. The N-terminal amino acid residue was identified as being a pyroglutamic acid, corresponding to Gln21 of the primary transcript. Peptides from endoproteinase-degraded G protein were analyzed by mass spectrometry before and after chemical reduction, and six disulfide bonds were identified: Cys29-Cys339, Cys44-Cys295, Cys90-Cys132, Cys172-Cys177, Cys195-Cys265, and Cys231-Cys236. Mass spectrometric analysis in combination with glycosidases allowed characterization of the glycan structure of the G protein. Three of four predicted N-linked oligosaccharides were found to be predominantly biantennary complex-type structures. Furthermore, an O-linked glycan near the N terminus was identified. Alignment of the VHSV G protein with five other rhabdovirus G proteins indicates that eight cysteine residues are situated at conserved positions. This finding suggests that there might be some common disulfide bonding pattern among the six rhabdoviruses.
Viral hemorrhagic septicemia virus (VHSV) is an enveloped negative-strand RNA virus belonging to the rhabdovirus family (16). It is the causative agent of viral hemorrhagic septicemia in rainbow trout (Oncorhynchus mykiss) (11). The virus results in considerable losses for European trout farming. Development of vaccines in the form of killed or attenuated virus or recombinant proteins have been attempted for many years without real success (19).
The transmembrane viral glycoprotein (G protein) is the target molecule for neutralizing antibodies (20, 23) as reported for other rhabdoviruses (9, 12, 32). Recent results with a DNA-based vaccine against VHSV (18) have demonstrated the immunostimulating and protective power of endogenously expressed G protein. However, attempts to express the G protein in Escherichia coli have generally led to improperly folded and nonimmunogenic molecules. Immunoblotting analyses further indicated that some neutralization epitopes are discontinuous and are stabilized by intramolecular disulfide bonds (1, 20, 23). Similar conformational epitopes have been identified within other rhabdoviruses, such as infectious hematopoietic necrosis virus (IHNV) and rabies virus (2, 9, 10, 27). Correct folding of the amino acid chain thus might be a prerequisite for the use of exogenously expressed G protein in a vaccine. With this in mind, it might be desirable to truncate the G protein in order to facilitate folding in vitro. Such truncation requires knowledge of the pairing of cysteine residues within the protein backbone.
The present study was undertaken in order to identify the disulfide bonds and to determine other structural aspects of potential relevance for vaccine design. The cDNA of the G protein from VHSV encodes a 507-amino-acid protein including 16 conserved cysteine residues (GenBank accession no. ACX59148 and ACX66134 [21, 30]). Twelve of these are situated in the extracellular domain of the protein and are therefore likely to be involved in the formation of six intramolecular disulfide bonds. The remaining four are positioned within the predicted transmembrane anchor. VHSV G protein was purified by immunoaffinity chromatography, using an immunobilized neutralizing monoclonal antibody (MAb). Peptides from endoproteinase-degraded G protein were analyzed by mass spectrometry before and after chemical modification or endo- and exoglycosidase digestion. The identities of the six intramolecular disulfide bonds, the position and modification of the N-terminal amino acid residue, and characteristics of the glycans of the VHSV G protein are reported.
MATERIALS AND METHODS
MAb affinity column assay.
The neutralizing MAb DK-3F1A2 used in this work recognizes a disulfide-bond-dependent, neutralizing epitope and was produced as described by Lorenzen et al. (20, 23). Hybridoma cell culture supernatant was concentrated 10 times by Amicon filtration using a YM-30 filter (Amicon, Beverly, Mass.), and mouse immunoglobulin was purified by affinity chromatography on a protein G-agarose column (gammaBind; Pharmacia Biotech, Uppsala, Sweden) following the procedures given by the supplier. Subsequently, 140 mg of purified MAb was immobilized on 56 ml of 50% divinyl sulfone-activated agarose beads (Mini Leak Low; Kem-En-Tec, Copenhagen, Denmark) as described by the manufacturer.
Virus and cells.
A low-passage field isolate of VHSV (DK-3592B [20]) was multiplied on BF-2 cells (33) in accordance with previously described procedures (20). This isolate was used for all experiments. For large-scale production of virus, BF-2 cells were grown in cell factories (CF 10 Ten Tray; Nunc, Roskilde, Denmark) until monolayers of approximately 90% confluence were obtained. Afterwards, the cells were inoculated with VHSV at a multiplicity of infection of 0.1. Three to five days later, a total cytopathic effect was attained, and cell debris was removed by centrifugation (4,182 × g for 15 min). The supernatant, containing 2 × 109 to 5 × 109 50% tissue culture infective doses, was subsequently ultracentrifugated at 86,000 × g for 2 h. The pellet was resuspended in 8 ml of TE buffer (20 mM Tris-HCl [pH 7.5], 1 mM EDTA) and kept at −80°C until used.
Preparation of the G protein sample. (i) Solubilization of virus.
The resuspended virus particles were adjusted to 1% with respect to Triton X-100 (TX-100) and incubated for 1 h at room temperature on an end-over-end mixer. Unsolubilized material was pelleted at 4,182 × g for 15 min, and the supernatant with the solubilized G protein was immediately used.
(ii) Affinity purification of the G protein.
The affinity column was pre-eluted with elution buffer (0.1 M glycine-HCl [pH 2.8], 0.05% TX-100) and pre-equilibrated with washing buffer I (TE buffer with 1% TX-100). The solubilized virus sample was diluted 25 times by the addition of TE buffer containing 1% TX-100 and was then pumped through the column. Washing of the immobilized G protein was carried out with 3 to 4 volumes of washing buffer I and then with 10 to 15 volumes of washing buffer II (TE buffer with 0.05% TX-100). Elution of the G protein was monitored at 280 nm, and the eluate was collected in 1.5-ml fractions (0.4 ml/min). All fractions were adjusted to pH 7.5 with 1 M Tris-HCl (pH 9) and examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Fractions containing pure G protein were pooled and stored at −20°C until used.
Prior to mass spectrometric analysis, the content of TX-100 was reduced by dialysis against 6 volumes of 50 mM ammonium acetate (pH 6.8) including detergent adsorber gel (Boehringer, Mannheim, Germany). The dialysis proceeded for 1 day, with a buffer change after 4 h. The dialyzed sample was concentrated to a final volume of approximately 1 ml by filtration with an ultrafiltration membrane unit (Omega Macrosep; Pall Gelman Sciences, Ann Arbor, Mich.). Subsequently, the purified G protein was adjusted to 0.1% with respect to n-octyl-β-d-glycopyranoside (Sigma, St. Louis, Mo.), a detergent known to be compatible with mass spectrometric analyses. The final concentration of the purified G protein was visually estimated from an SDS–13% polyacrylamide gel stained with Coomasie blue R-250, using step dilutions of bovine serum albumin (BSA) (fraction V; Sigma) as reference.
HPLC.
Separation of endoproteinase-generated peptides was performed by high-performance liquid chromatography (HPLC) on a Nucleosil C18 column (4 by 250 mm; 10-μm particle size; 30-nm pore size). The peptides were eluted by using a linear gradient with 0.1% trifluoroacetic acid (TFA) as buffer A and 90% acetonitrile–0.08% TFA as buffer B. The eluate was monitored at 214 nm. All fractions were collected manually and freeze-dried.
Endoproteinase digestions and chemical cleavage of the G protein.
For endoproteinase Lys-C digestion (digest A), 10 μl of 45 mM dithiothreitol (DTT) was added to 62 μg of affinity-purified G protein, and the sample was incubated at 37°C for 30 min. Then, 10 μl of 100 mM iodoacetamide was added, and the alkylation was allowed to proceed for 2 h at 37°C. After alkylation, 3 μg of endoproteinase Lys-C (Promega, Madison, Wis.) and 0.5 U of peptide-N-glycosidase (PNGase F; Boehringer) were added, and the mixture was incubated at 37°C overnight. The generated peptides were separated on the HPLC system using a linear gradient from 0 to 90% buffer B over 46 min.
Two different trypsin digestions (B and C) were performed. In the first (digest B), 62 μg of affinity-purified G protein was mixed with 3 μg of trypsin (a gift from Novo Nordisk, Bagsvard, Denmark), 0.5 U of PNGase F, and 10 mU of neuraminidase (Arthrobacter ureafaciens) (both from Boehringer), and the sample was left overnight at 37°C. In the second (digest C), 62 μg of affinity-purified G protein was mixed with 3 μg of trypsin, and the sample was left overnight at 37°C. The peptides from each digest were separated by using a linear gradient from 0 to 90% buffer B over 46 or 52 min.
PAP digestion.
Native G protein (5 to 10 μg in 50 μl of 50 mM ammonium acetate [pH 6.8]–0.1% n-octyl-β-d-glycopyranoside) was mixed with 5 μl of 14 mM DTT and 0.5 μg of pyroglutamate aminopeptidase (PAP). The Eppendorf tube was closed under argon and incubated at 37°C. After 24 h, an additional 5 μl of 14 mM DTT was added, and the tube was incubated for another 24 h. The PAP-digested protein was then subjected to Edman degradation.
PAP digestion of peptides was carried out by dissolving the peptide (10 to 100 pmol) in 10 μl of 100 mM ammonium hydrogen carbonate (pH 7.8)–14 mM DTT–5% glycerol containing 0.5 μg of PAP. The Eppendorf tube was closed under argon, and the sample was incubated at 37°C for 24 h. Then, 1 μl of 1.4 M DTT was added, and the sample was incubated for an additional 24 h. An aliquot (0.8 μl) of the digested peptide was analyzed by matrix-assisted laser desorption/ionization mass spectra (MALDI-MS), and the remaining part was sequenced by Edman degradation.
Identification of disulfide bonds.
Lyophilized peptides (30 to 50 pmol) were dissolved in 5 to 10 μl of 50 mM ammonium hydrogen carbonate (pH 8.5). In order to reduce disulfide bonds, 1 μl of 0.14 M DTT was added, and the samples were left for 30 min at 37°C. Free SH groups were alkylated by the addition of 1 μl of 0.9 M 4-vinyl pyridine (4-VP) followed by incubation at 37°C for 10 min. Aliquots of the untreated, reduced, or reduced and alkylated samples were analyzed by MALDI-MS, allowing identification of samples with inter- or intrapeptide disulfide bonds.
Glycosidase digestions of glycopeptides.
Structural characterization of the oligosaccharides was obtained by mass spectrometric analysis of glycosidase-treated peptides by using a strategy analogous to that described by Krogh et al. (13).
Characterization of N-linked oligosaccharides.
Peptides (5 to 20 pmol) expected to contain N-linked oligosaccharides were dissolved in 5 μl of 50 mM ammonium acetate (pH 5.0), and 5 mU of neuraminidase, 0.5 mU of β-galactosidase (Streptococcus pneumoniae), and 0.5 mU of N-acetyl-β-d-glucosaminidase (S. pneumoniae) (all from Boehringer) were added sequentially to the sample. Each digestion with O-glycosidase was performed overnight. PNGase F digestions were performed by dissolving the glycopeptides in 5 μl of 50 mM ammonium hydrogen carbonate (pH 7.8) containing 0.1 U of PNGase F.
Characterization of O-linked oligosaccharide.
The selected peptide (5 to 20 pmol) was analyzed by the addition of neuraminidase (as described above in the section on endoproteinase digestions) followed by the addition of 0.5 mU of O-glycosidase (S. pneumoniae; Boehringer).
After each digestion an aliquot (0.8 μl) was analyzed by MALDI-MS.
MALDI-MS.
MALDI-MS of protein and peptide samples was performed on either a Bruker Reflex MALDI-tof-MS instrument (Bruker-Franzen, Bremen, Germany) or a Voyager Elite instrument (PerSeptive Biosystems, Framingham, Mass.). Both instruments were equipped with delayed extraction, and a delay time of 250 ns was used. Mass spectra were recorded as single-shot spectra by using a UV laser at 337 nm and an acceleration voltage of 20 kV. A total of 50 to 200 single-shot spectra were averaged to give the final spectrum.
Voyager Elite spectra were externally calibrated by using human insulin. Bruker Reflex spectra were calibrated by using the α-cyano dimer (379.093 Da), which is present in each spectrum and is a constant, as described by Vorm and Mann (30).
Protein samples were prepared by mixing the G protein sample (0.1 μg in 0.8 μl) with 0.8 μl of 2% trifluoroacetic acid and 0.8 μl of matrix (sinapinic acid, 20 μg/μl in 70% acetonitrile). Peptide samples were prepared in accordance with the sandwich method (15) by applying a peptide solution (0.5 to 2 pmol in 0.8 μl) mixed with an equal amount of matrix (α-cyano-4-hydroxycinnamic acid) on a matrix thin layer (31). The sample surface was washed with 10 μl of water after drying.
Glycopeptides were analyzed by mixing the sample (0.5 to 5 pmol in 0.8 μl) with an equal volume of 2,4-dihydroxybenzoic acid in methanol (Hewlett-Packard, Palo Alto, Calif.).
Edman degradation (amino acid sequencing).
Edman degradation of peptides (5 to 50 pmol) was performed on an HP-G1000A protein sequencer, connected to an HP-1090LC HPLC system (Hewlett-Packard) for identification of phenylthiohydantoin (PTH) derivatives. Sequencing was carried out by the standard protocol provided by the manufacturer.
Programs for computer analysis.
A theoretical identification of the signal peptide and its cleavage site was performed by using the SignalP program, which is based on neural networks (http://www.cbs.dtu.dk/services/SignalP/) (26). The first 60 amino acids of the primary transcript of VHSV G protein were analyzed with the program, using the network trained on eukaryotic sequences. The entire G protein sequence was analyzed with the NetOGlyc prediction program (http://www.cbs.dtu.dk/services/NetOGlyc/) (7, 8) to predict potential O-glycosylation sites.
The enzymatic digests of the G protein were analyzed by comparing the measured molecular mass (MMmeas) of the peptides with the calculated molecular mass (MMcalc). The MMcalc was based on the translated cDNA sequence and was performed by means of the General Protein Mass Analysis for Windows program (Lighthouse Data, Odense, Denmark).
RESULTS
Purification of the G protein.
It has been demonstrated that solubilization of concentrated virus particles with TX-100 allows subsequent affinity purification of the viral G protein to a high level of purity (17). Upscaling of the immunoaffinity procedure indicated that the concentration of the solubilized G protein was critical. A concentrated sample could be eluted only if a high concentration of detergent was included in the elution buffer (0.1 M glycine-HCl [pH 2.8] with a minimum of 1% TX-100). However, if the sample was diluted 25 times in washing buffer I before its addition to the column, it was possible to elute the protein with 0.1 M glycine-HCl (pH 2.8) and 0.05% TX-100. This decrease in the TX-100 concentration made it easier to remove the detergent by dialysis. Evaluation of the immunoaffinity chromatography eluate was done by SDS-PAGE. Gels containing similar samples were either stained with silver (3) or immunoblotted and specifically stained with the same MAb DK-3F1A2 (23) as was used for affinity purification. The silver-stained gel revealed that a protein having a mass corresponding to the mass of the G protein had been purified from the crude solubilized virus solution. Immunoblotting confirmed that the purified protein could be recognized by the MAb DK-3F1A2 as well by another G-protein-specific MAb, DK-IP1H3 (22). In addition, some faint bands of high molecular mass were observed. These bands were also recognized by the G-protein-specific MAbs, indicating that a minor portion of the G protein molecules had formed di-, tri-, and multimers (not shown). The average yield of pure G protein from one cell factory was approximately 160 μg based on estimated protein concentrations with BSA as the standard.
Identification of the N-terminal residue.
The SignalP prediction program proposes Gln21 of the translated cDNA sequence to be the theoretical N-terminal residue. N-terminal Gln often converts to pyroglutamic acid (pGln), thereby preventing Edman degradation. Therefore, two samples of purified G protein were analyzed. One sample was treated with PAP, and the other was left untreated. No sequence data were obtained from the native protein sample, whereas the PAP-treated sample revealed the following sequence starting at Ile22: Ile-Xaa-Gln-Arg-Pro-Pro-Val-Glu-Xaa-Ile-Ser-Thr-Tyr-His-Ala. This confirmed that Gln21 is the modified N-terminal residue. For the remainder of this text, the amino acid residues will be numbered according to their positions in the mature protein; i.e., pGln1-Val487 corresponds to Gln21-Val507 of the translated cDNA sequence.
PTH derivates were not observed in the cycles corresponding to the positions of Thr3 and Asn10, indicating that these residues might be modified. Both residues were suspected to be glycosylated, since Asn10 is part of a potential N-glycosylation site (Asn-Xaa-Ser/Thr), and Thr3 was predicted to be a possible O-glycosylation site by NetOGlyc.
Mass determination of the intact G protein.
Four batches of purified G protein were analyzed by MALDI-MS. The spectra showed broad peaks, indicating that the protein was not homogeneous, probably due to heterogeneous glycosylation. The peaks had maximum m/z at 64,356, 64,399, 64,691, and 64,811 Da. One of the protein batches furthermore showed minor peaks at lower m/z values, indicating that this sample was contaminated with partially degraded protein. This batch was excluded from the subsequent experiments.
Enzymatic deglycosylation of the native protein was performed, but attempts to obtain a spectrum of the deglycosylated protein were unsuccessful.
Sequence verification and confirmation of the translated cDNA sequence.
Peptides were generated by LysC digestion of PNGase F-treated, reduced, and alkylated G protein (digest A) and separated by HPLC. All HPLC fractions were analyzed by MALDI-MS, and the MMmeas were compared with the MMcalc. Twenty-six peptides (L1 to L26) are theoretically generated by LysC digestion of the G protein. L1 to L20 and L23 to L24 were identified from digest A, corresponding to 80% of the protein (Table 1). A glycosylated peptide corresponding to L21 was identified from digest C. Peptides corresponding to L22 (three amino acids) and the C-terminal transmembrane region (L25 and L26) were not located in any of the digests.
TABLE 1.
Mass-based identification of VHSV G protein peptidesa
| Peptide | Amino acids | Cys position | MMmeas (Da) | MMcalc (Da) |
|---|---|---|---|---|
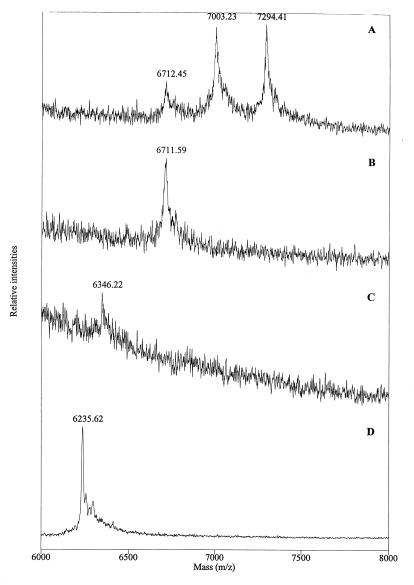
| L1c | 1–53 | C29, C44 | 6,711.5 | 6,712.3 |
| 7,002.2 | 7,003.6 | |||
| 7,293.4 | 7,294.8 | |||
| L2 | 54–61 | 798.8 | 800.0 | |
| L3 | 62–102 | C90 | 4,518.2 | 4,521.2 |
| L4 | 103–108 | 674.5 | 673.8 | |
| L5 | 109–120 | 1,335.1 | 1,334.4 | |
| L6 | 121–136 | C132 | 2,083.6 | 2,083.4 |
| L7–L9b | 137–151 | 1,859.0 | 1,858.1 | |
| L10 | 152–161 | 1,167.4 | 1,167.3 | |
| L11e | 162–192 | C172, C177 | 3,478.5 | 3,480.9 |
| L11 | 162–192 | 3,692.2 | 3,693.2 | |
| L12 | 193–216 | C195 | 2,828.4 | 2,829.2 |
| L13e | 217–242 | C231, C236 | 2,797.3 | 2,798.3 |
| L13f | 217–242 | 3,010.0 | 3,010.6 | |
| L14 | 243–259 | 1,628.8 | 1,628.8 | |
| L15–L16b | 260–264 | 586.3 | 585.7 | |
| L17 | 265–307 | C265, C295 | 4,826.6 | 4,828.3 |
| L18 | 308–315 | 894.0 | 894.1 | |
| L19 | 316–325 | 1,079.7 | 1,079.2 | |
| L20 | 326–355 | C339 | 3,494.3 | 3,493.9 |
| L21 | 356–362 | n.o.g | 861.5 | |
| L22 | 363–365 | n.o. | 419.5 | |
| L23–L24b | 366–390 | 2,856.9d | 2,839.2 | |
| L25 | 391–466 | n.o. | 8,640.9 | |
| L26 | 467–487 | n.o. | 2,318.6 |
Comparison of MMmeas and MMcalc for the peptides generated by an endoproteinase LysC digestion of PNGase F-treated, reduced, and iodoacetamide-alkylated G protein (digest A).
Two or more peptides were connected due to lack of cleavage.
An O-linked glycan was identified; masses corresponding to 0, 1, or 2 sialic acids are given.
A methione residue in the peptide was presumably oxidized, as the mass appears 16 Da higher than the theoretical mass.
An intrapeptide disulfide bond was not reduced or alkylated under the initial experimental conditions.
Additional treatment with DTT and 4-VP (as described in text) resulted in the reduction and alkylation of the bond.
n.o., not observed.
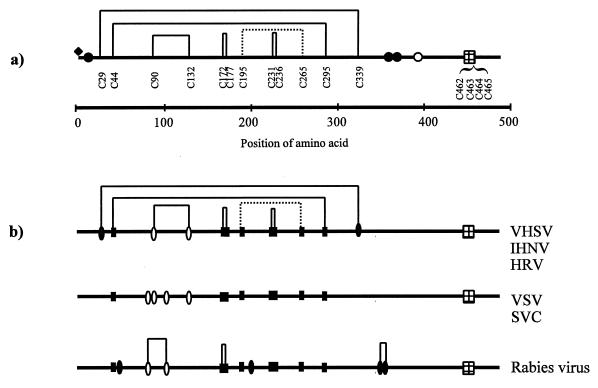
Figure 1 illustrates the deduced amino acid sequence of the VHSV G protein and the position of characterized modifications. Based on their masses, the two peptides containing double cysteines, L11 and L13, were not initially alkylated (Table 1). Additional treatment of two peptides with increased levels of DTT and 4-VP led to successful alkylation (Table 1). This need for an elevated DTT level indicated that the two disulfide bonds, Cys172-Cys177 and Cys231-Cys236, were highly stable.
FIG. 1.
The translated cDNA sequence of VHSV G protein (isolate DK-3592B; GenBank accession no. X66134). The signal peptide is indicated in italics. The sequence parts supported by the MALDI-MS analysis of the digests A through C are printed in normal type, cysteine residues are in boldface, and the unconfirmed sequence is underlined. Peptides involved in disulfide bond formation (Table 2) are underlined with dashed lines. The identified N-terminal residue, pyroglutamic acid, is abbreviated B. The filled diamond indicates the position of the characterized O-glycan, the filled circles indicate the positions of the characterized N-glycans, and the open circle indicates the uncharacterized consensus site of N-linked glycosylation.
Identification of an O-linked glycan.
One of the HPLC fractions from digest A contained a collection of three peptides showing mass variation of approximately 291 Da (Fig. 2A). The MMmeas did not correspond to any MMcalc, indicating the presence of a variable number of sialic acid (MMNeuNac = 291.26 Da) residues associated with this peptide. Aliquots of the fraction were treated sequentially with neuraminidase and O-glycosidase with intermittent analysis by MALDI-MS (Fig. 2A to C). Treatment with neuraminidase eliminated one or two sialic acid residues (MMNeuAc = 291.26 Da) and the heterogeneity. The subsequent treatment with O-glycosidase removed a Gal-NAc-Gal core (MMGalNAcGal = 365.34 Da), identifying the O-linked glycostructure as GalNac-Gal-NeuAc1–2.
FIG. 2.
MALDI mass spectra of peptide L1. Shown are spectra of the native peptide (A), L1 after digestion with neuraminidase (B), L1 after digestion with neuraminidase and O-glycosidase (C), and further digestion of the same peptide with pyroglutamate aminopeptidase (D). Note that the peaks represent MH+.
The deglycosylated peptide was further treated with PAP (Fig. 2D), resulting in the removal of a pGln residue (MMpGln = 111.14 Da) as also demonstrated on native G protein. Sequencing of the PAP-treated peptide confirmed that it is the N-terminal peptide (L1). The glycan is assumed to be on Thr3 since this is the only potential O-glycosylation site in the L1 peptide as predicted by the NetOglyc program. This was also supported by the lack of a detectable PTH derivative in cycle two of the Edman degradation of the native protein.
Assignment of disulfide bonds.
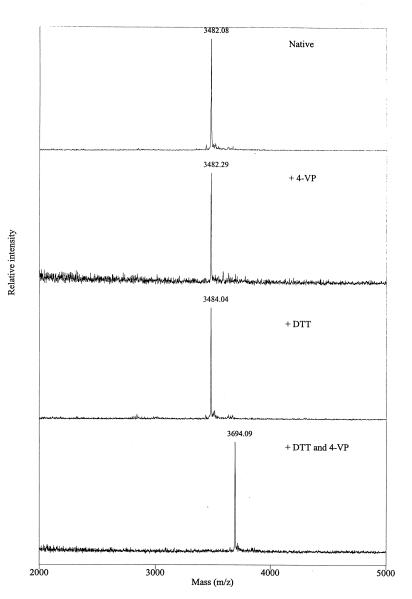
The G protein from VHSV contains 16 highly conserved cysteine residues (Cys29, Cys44, Cys90, Cys132, Cys172, Cys177, Cys195, Cys231, Cys236, Cys265, Cys295, Cys339, Cys462, Cys463, Cys464, and Cys465). Four of these are located within the presumed transmembrane region, whereas the remaining 12 are probably involved in disulfide bonds. Cleavage of the G protein between the cysteine residues was performed with trypsin on neuraminidase- and PNGase F-treated G protein (digest B). HPLC fractions which upon analysis by MALDI-MS showed molecular mass values differing from any MMcalc were subsequently reduced and alkylated to determine if a disulfide bond was present (Table 2). The identification of an intrapeptide disulfide bond between Cys172 and Cys177 in fraction B46 is illustrated in Fig. 3. Two aliquots of the fraction were treated in parallel: 4-VP was added to one aliquot, and DTT followed by 4-VP was added to the other. The molecular mass determined for the peptide was not changed upon 4-VP addition, demonstrating that no free SH groups were present. The addition of DTT caused an increase in mass of 2.0 Da, due to binding of two H+ protons as a result of the reduction of a disulfide bond. The subsequent addition of 4-VP caused an increase in mass of 210.1 Da, consistent with vinylpyridinylation of two cysteine residues (theoretical increase, 2 × 105.2 Da).
TABLE 2.
Identification of disulfide bondsa
| HPLC fraction | MMmeas (MMcalcb), Da
|
Amino acids | Disulfide bond | ||
|---|---|---|---|---|---|
| Native | + DTT | + DTT and 4-VP | |||
| L13c | 2,797.3 (2,798.3) | 2,798.9 (2,800.3) | 3,010.5 (3,010.6) | 217–242 | C231-C236 |
| B27 | 2,776.8 (2,777.0) | 1,231.4 (1,231.4) | 1,337.6 (1,336.5) | 44–53 | C44-C295 |
| 1,548.1 (1,547.7) | 1,653.4 (1,652.8) | 293–307 | |||
| B39 | 6,026.8 (6,026.5) | 3,889.9 (3,888.2) | 3,994.5 (3,994.3) | 1–30 | C29-C339 |
| 2,139.7 (2,139.3) | 2,244.5 (2,244.4) | 337–355 | |||
| B45 | 3,629.5 (3,629.1) | 1,605.3 (1,604.8) | 1,710.0 (1,709.9) | 88–102 | C90-C132 |
| 2,026.8 (2,026.3) | 2,131.6 (2,131.5) | 121–136 | |||
| B46 | 3,481.1 (3,480.9) | 3,483.0 (3,482.9) | 3,693.1 (3,693.2) | 162–192 | C172-C177 |
| B50 | 2,831.4 (2,832.2) | n.o.d (1,799.0) | n.o. (1,904.1) | 193–208 | C195-C265 |
| n.o. (1,035.1) | n.o. (1,140.4) | 265–273 | |||
PNGase F-treated G protein was digested with trypsin without prior reduction and alkylation (digest B). Peptides from the digest having molecular masses different from any possible linear sequence were reduced and alkylated (4-VP) in order to detect disulfide-linked peptides or intrachain disulfide bonds. The peptides involved in disulfide bond formation are underlined (dashed) in Fig. 1.
Disulfide bond formation is assumed.
L13 from digest A contains a highly stable intrapeptide disulfide bond (Table 1).
n.o., not observed.
FIG. 3.
MALDI mass spectra of HPLC fraction B46 revealing the presence of an intrapeptide disulfide bond. The B46 samples were treated in parallel with additions as indicated. Note that the peaks represent the MH+.
Similar analyses of fractions B27, B39, and B45 revealed that peptides linked by interpeptide disulfide bonds were present in each of these fractions (Table 2). Fraction B50 showed a peak of 2,831.4 Da, indicating linkage between the peptides 193 to 208 and 265 to 273, corresponding to an interpeptide disulfide bond between Cys195 and Cys265. No peaks at this mass were observed in any of the DTT-treated samples. Despite several attempts, it was not possible to identify these two peptides after reduction and alkylation (Table 2). Thus, five disulfide bonds (Cys29-Cys339, Cys44-Cys295, Cys90-Cys132, Cys172-Cys177, and Cys231-Cys236) were identified, and one bond (Cys195-Cys265) was tentatively identified, as shown in Fig. 4A.
FIG. 4.
Rhabdovirus G proteins. (A) A schematic overview of the disulfide bonds and posttranslational modifications of VHSV G protein. Identified disulfides are shown as solid lines, and the predicted disulfide bond is shown as a dashed line. A filled diamond indicates the position of the O-glycan (Thr3). Filled circles indicate the positions of the characterized N-linked glycans (Asn10, Asn358, and Asn369), and the open circle indicates the position of a potential but not identified N-linked glycan (Asn418). The transmembrane regions are indicated by the gridded boxes. (B) Alignment of the G proteins from VHSV (GenBank accession no. ACX66134), IHNV (GenBank accession no. ACL40874), HRV (GenBank accession no. ACU24073), SVCV (GenBank accession no. ACU18101), VSV (GenBank accession no. ACM21421), and rabies virus, ERA strain (GenBank accession no. ACJ02293). Positions of conserved cysteine residues are marked by filled boxes, cysteines common to at least two groups are marked by open ovals, and unique cysteines are marked by filled ovals. Disulfide bonds in rabies virus are marked as reported by Dietzschold and coworkers (4).
Characterization of N-linked glycans.
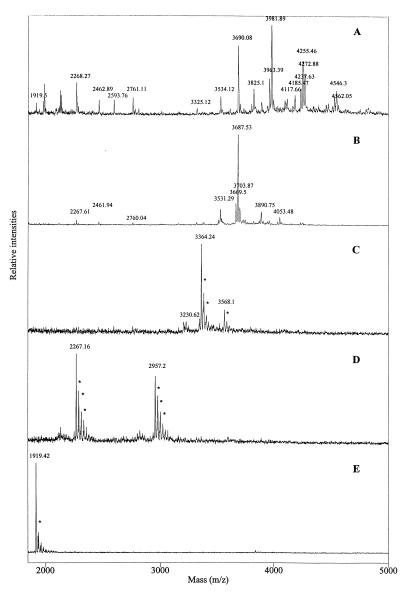
A tryptic digest on native G protein (digest C) was performed in order to locate and characterize the N-linked glycans. Three broad peaks in the HPLC chromatogram were assumed to represent heterogeneous glycopeptides. Amino acid sequencing of these fractions (C17, C31, and C39) revealed that the fractions contained peptides with predicted N-linkage sites (Asn10, Asn358, and Asn369). No PTH derivatives were observed in the cycles corresponding to the positions of the Asn residues within the N-linkage consensus sites, indicating that these residues were actually modified.
The three fractions were also analyzed by MALDI-MS before and after treatments with exo- and endoglycosidases as previously described (13). Very complex peak patterns were observed for the untreated glycopeptide (Fig. 5A), indicating highly heterogeneous glycosylation. However, dominant peaks indicated dominant glycan forms in all cases. Upon sequential glycosidase treatment, gradually reduced heterogeneity was observed (Fig. 4B to D) until only the unglycosylated peptide containing Asp instead of Asn was found (Fig. 5E). The observed peaks and the corresponding proposed glycan structures for all three glycopeptides are summarized in Table 3. The glycan structures have been proposed based on the observed masses and on the assumption that the structures are composed only of normal monosaccharide components.
FIG. 5.
MALDI mass spectra obtained by sequential glycosidase digestion of HPLC fraction C31. Spectra are shown for untreated sample (A), C31 after digestion with neuraminidase (B), C31 after sequential digestion with neuraminidase and β-galactosidase (C), C31 after digestion with neuraminidase, β-galactosidase, and N-acetyl-β-d-glucosaminidase (D), and C31 after digestion with PNGase F (E). Asterisks indicate salt adducts. Note that the peaks represent the MH+.
TABLE 3.
Estimated composition of the N- and O-linked oligosaccharidesa
| HPLC fraction | Experimental conditionsb | MMmeas (Da) | Oligosaccharide units | MMcalc (Da) | Amino acids |
|---|---|---|---|---|---|
| C17 | Native | 3,507.8 | GlcNAc4FucMan3Gal2NeuAc3 | 3,505.3 | |
| 3,419.6 | GlcNAc5FucMan3Gal2NeuAc2 | 3,417.3 | |||
| 3,218.5 | GlcNAc4FucMan3Gal2NeuAc2 | 3,214.1 | |||
| 3,128.6 | GlcNAc4FucMan2Gal2NeuAc2 | 3,126.0 | |||
| 2,925.0 | GlcNAc4FucMan3Gal2NeuAc | 2,922.8 | |||
| 2,779.7 | GlcNAc4Man3Gal2NeuAc | 2,776.7 | |||
| Neur | 2,834.9 | GlcNAc5FucMan3Gal2 | 2,834.8 | ||
| 2,631.1 | GlcNAc4FucMan3Gal2 | 2,631.3 | |||
| 2,484.8 | GlcNAc4Man3Gal2 | 2,485.4 | |||
| Neur, β-gal | 2,508.2 | GlcNAc5FucMan3 | 2,510.5 | ||
| Neur, β-gal, glcnase | 1,900.4 | GlcNAc2FucMan3 | 1,900.9 | ||
| PNGase F | 861.6 | None | 862.9 | 356–362 | |
| C31 | Native | 4,561.1 | GlcNAc4FucMan3Gal2NeuAc3 | 4,561.5 | |
| 4,271.9 | GlcNAc5FucMan3Gal2NeuAc2 | 4,270.2 | |||
| 4,184.8 | GlcNAc5FucMan3Gal2NeuAc | 4,182.1 | |||
| 3,980.9 | GlcNAc4FucMan3Gal2NeuAc | 3,978.9 | |||
| Neur | 4,052.5 | GlcNAc5FucMan3Gal3 | 4,052.0 | ||
| 3,889.8 | GlcNAc5FucMan3Gal2 | 3,890.9 | |||
| 3,686.5 | GlcNAc4FucMan3Gal2 | 3,687.4 | |||
| Neur, β-gal | 3,567.1 | GlcNAc5FucMan3 | 3,566.6 | ||
| 3,363.3 | GlcNAc4FucMan3 | 3,363.4 | |||
| Neur, β-gal, glcnase | 2,956.2 | GlcNAc2FucMan3 | 2,957.0 | ||
| PNGase F | 1,918.4 | None | 1,917.0 | 367–382 | |
| C39 | Native | N-linked glycan/O-linked glycanc | |||
| 10,520 | GlcNAc5FucMan3Gal2NeuAc2/GalNAcGalNeuAc2 | 10,529 | |||
| 10,358 | GlcNAc5FucMan3Gal2NeuAc2/GalNAcGalNeuAc | 10,368 | |||
| 10,075 | GlcNAc5FucMan3Gal2NeuAc/GalNAcGalNeuAc | 10,077 | |||
| 9,870 | GlcNAc4FucMan3Gal2NeuAc/GalNAcGalNeuAc | 9,774 | |||
| 9,785 | GlcNAc5FucMan3Gal2/GalNAcGalNeuAc | 9,785 | |||
| 9,582 | GlcNAc4FucMan3Gal2/GalNAcGalNeuAc | 9,582 | |||
| Neur | 9,499 | GlcNAc5FucMan3Gal2/GalNAcGal | 9,496 | ||
| 9,296 | GlcNAc4FucMan3Gal2/GalNAcGal | 9,292 | |||
| 8,925 | GlcNAc3Man3Gal/GalNAcGal | 8,927 | |||
| Neur, β-gal | 9,170 | GlcNAc5FucMan3/GalNAcGal | 9,171 | ||
| 8,967 | GlcNAc4FucMan3/GalNAcGal | 8,968 | |||
| Neur, β-gal, glcnase | 8,560 | GlcNAc2FucMan3/GalNAcGal | 8,561 | ||
| PNGase F | 8,104 | None/GalNAcGalNeuAc2 | 8,106 | 1–43d | |
| 7,816 | None/GalNAcGalNeuAc | 7,814 | 337–355 |
G protein was digested with trypsin under native conditions (digest C). Three HPLC fractions were analyzed by MALDI-MS analysis in combination with exo- and endoglycosidases as described in the text. It is assumed that the glycostructures are composed of normal monosaccharide components. The accuracy of the measurements of mass in these experiments is lower than that in the analysis of pure peptides due to a poor signal-to-noise ratio.
Neur, neuraminidase; β-gal, β-galactosidase; glcnase, N-acetyl-β-d-glucosaminidase.
The position of the sialic acid residues (located on either the N- or the O-linked glycan) was not identified.
Disulfide bond between the two peptides.
The masses obtained from the analysis of glycopeptides (Table 3) were not as accurate as those obtained for peptides because the molecular ion signals are distributed over several peaks, resulting in a poorer signal-to-noise ratio. Therefore, somewhat larger deviations of mass than are normally acceptable have been allowed in the interpretation. A majority of the peaks corresponding to glycosylated peptides containing sialic acid were accompanied by minor peaks 14 to 18 Da below. These minor peaks most likely represent water loss by prompt fragmentation.
The predominant oligosaccharide structures identified at site Asn369 were fucosylated biantennary complex-type structures containing various numbers of sialic acids. Peaks corresponding to a defucosylated biantennary structure as well as a bisected N-acetylglucosamine were also observed. In addition, small amounts of a triantennary complex-type structure were present. Similar oligosaccharide structures were found at sites Asn358 and Asn10, but no triantennary complex-type structures were observed. The positions of the characterized N- and O-glycostructures have been included in Fig. 1 and 4A.
DISCUSSION
To our knowledge, this is the first report of complete disulfide bonding within a rhabdovirus glycoprotein. MALDI-MS analysis allowed the identification of five of six possible extracellular disulfide bonds and strongly suggested the existence of a sixth bond between the two remaining cysteines. Furthermore, the presence of an O-linked glycan on a rhabdovirus G protein has not previously been described.
MALDI-MS analysis of alkylated, deglycosylated, and endoproteinase-digested G protein confirmed the amino acid composition in 80% of the translated cDNA sequence of the VHSV G protein (isolate DK-3592B), including the extracellular part of the protein with 12 cysteine residues. The N-terminal residue of the G protein, a pGln corresponding to Gln21 of the primary transcript, was also identified.
Based on sequential MALDI-MS analysis of nonreduced, reduced, and reduced and alkylated peptide samples from a trypsin digest of the native protein, we have identified five intramolecular disulfide bonds in the VHSV G protein: Cys29-Cys339, Cys44-Cys295, Cys90-Cys132, Cys172-Cys177, and Cys231-Cys236. Additionally, the results indicate that the two remaining cysteines, Cys195 and Cys265, form a sixth disulfide bond. We did not find indications of an alternative disulfide pattern such as described for rabies virus G protein (4) in a study performed on G protein which had not been purified by affinity chromatography. The existence of such an alternative pattern cannot be excluded, because affinity purification with a MAb recognizing a disulfide-dependent epitope might have favored a specific disulfide pairing. However, this is not likely, since no residual G protein could be detected in the solubilized virus preparation after passage through the affinity column (results not shown).
Disulfide bonds are known to be important for stabilization of the secondary and tertiary structure of proteins (25). The disulfide bonds Cys29-Cys339 and Cys44-Cys295 are formed between cysteine residues that are the most distantly positioned in the polypeptide chain, indicating a rather compact and possibly loop-like overall structure. At the same time, two highly stable disulfide bonds are formed between cysteine residues separated by only four residues, indicating that disulfide bonds also maintain structures of local importance in the VHSV G protein. Analysis of the level of conservation of cysteine positions within rhabdovirus G proteins may thus give an indication of the degree of structural similarity between these proteins.
Clustal W alignment of the VHSV G protein sequences currently available in GenBank discloses a high level of identity (>90%), including fully conserved positions of the cysteine residues. An additional alignment including the G protein sequences from VHSV, IHNV, hirame rhabdovirus (HRV), vesicular stomatitis virus (VSV), spring viremia of carp virus (SVCV), and rabies virus was performed. The overall homology of the six G proteins varies between 18 and 26%, and the identity is less than 5%. This alignment revealed that the positions of the cysteine residues can be divided into three groups: VHSV-like positions (VHSV, IHNV, and HRV); VSV-like positions (VSV and SVCV), and rabies virus-like positions (rabies virus). The numbers and positions of the cysteine residues are identical within the groups, and the sequence identity within the VHSV-like and the VSV-like proteins are 33 and 32%, respectively. As proposed by Doolittle (5), such high levels of identity (>25%) may indicate similar folding of the grouped proteins.
VHSV, IHNV, and HRV have been demonstrated to be phylogenetically closely related (14) and have recently been grouped together in a new subgenus of the rhabdoviruses assigned the name novirhabdovirus (13a). Conservation of the G-protein cysteines among these viruses is therefore less surprising. However, alignment with the VSV and rabies virus groups indicates that eight cysteines, corresponding to Cys44, Cys172, Cys177, Cys195, Cys231, Cys236, Cys265, and Cys295 in VHSV G protein, are situated similarly in all the G proteins (Fig. 4B). The disulfide bond between the distant Cys44 and Cys295 in the VHSV G protein may thus be a general feature, and interestingly the existence of such a major loop in the VSV G protein anchored by at least one disulfide bond has also been proposed by Grigera and coworkers (6). The remaining six cysteines form three bonds in the VHSV G protein situated in a region characterized by discontinuous neutralizing epitopes in VHSV, IHNV, VSV, and rabies virus (1, 2, 10, 24). This homology could reflect the presence of conserved structural features of general importance for the biological function of the rhabdovirus G protein. Presently, the information about localization of disulfides in the rabies virus and the VSV groups is not sufficient to confirm this idea, but the disulfide bonds so far identified within rabies virus (4) (Fig. 4B) support this hypothesis.
Mass-spectrometric analysis of exo- and endoglycosidase-treated peptides permitted the characterization of three N-linked oligosaccharides and one O-linked oligosaccharide. The presence of an O-linked glycan on a rhabdovirus G protein has not been previously described. It was determined to be a GalNac-Gal-NeuAc1–2 structure linked to Thr3. The identified N-glycan structures are predominantly fucosylated biantennary complex-type structures attached to Asn10, Asn358, and Asn369 positioned on the predicted Asn-Xaa-Thr/Ser consensus sites. In comparison, the VSV G protein has been demonstrated to contain tetra-antennary complex-type structures (28). The presence of a fourth glycosylation site has been proposed by Lorenzen et al. (20), but this site is positioned outside the confirmed amino acid sequence and could not be identified (Fig. 1). Attempts to locate the peptide with the fourth site by using alternative cleavage strategies with cyanogen bromide or BNPS [3-bromo-3-methyl-2-(2-nitro-phenylmercapto)-3H-indole] were not successful. However, the molecular mass determined for intact G protein by MALDI-MS indicated that the fourth consensus site (Asn418) is also glycosylated with a biantennary complex-type structure. The position and number of consensus sites for N-linked glycosylation are not conserved within the aligned rhabdovirus G protein sequences, not even within the grouped sequences, indicating that the glycans probably play more individual roles in the structure and/or function of the G proteins than the disulfide bonds. The characterized glycans represent the glycosylation pattern produced in a BF-2 cell culture. It remains to be established whether the glycan structures are the same when the virus is replicated in fish or in other cell lines.
The structural findings reported here for the VHSV G protein should contribute to a broader understanding of the structural biology of rhabdovirus G proteins, including functional aspects as well as characterization of antibody epitopes. Knowledge of disulfide bonding might further allow design of truncated G proteins in development of recombinant vaccines. Ongoing work based on the findings in this report indicates that it is possible to produce subunits of the G protein which can be recognized by conformation-dependent neutralizing antibodies.
ACKNOWLEDGMENTS
We thank H. Hermansen, L. Troels, L. Schou, I. Christiansen, and K. Rafn for excellent technical assistance.
This work was supported by the Danish Research Academy and the Danish Biotechnology Programme.
REFERENCES
- 1.Bearzotti M, Monnier A F, Vende P, Grosclaude J, de Kinkelin P, Benmansour A. The glycoprotein of viral hemorrhagic septicemia virus (VHSV): antigenicity and role in virulence. Vet Res. 1995;26:413–422. [PubMed] [Google Scholar]
- 2.Benmansour A, Leblois H, Coulon P, Tuffereau C, Gaudin Y, Flamand A, Lafay F. Antigenicity of rabies virus glycoprotein. J Virol. 1991;65:4198–4203. doi: 10.1128/jvi.65.8.4198-4203.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blum H, Beier H, Gross H J. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis. 1987;8:93–99. [Google Scholar]
- 4.Dietzschold B, Wiktor T J, Macfarlan R, Varrichio A. Antigenic structure of rabies virus glycoprotein: ordering and immunological characterization of the large CNBr cleavage fragments. J Virol. 1982;44:595–602. doi: 10.1128/jvi.44.2.595-602.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doolittle R F. Similar amino acid sequences: chance or common ancestry? Science. 1981;214:149–159. doi: 10.1126/science.7280687. [DOI] [PubMed] [Google Scholar]
- 6.Grigera P R, Keil W, Wagner R R. Disulfide-bonded discontinuous epitopes on the glycoprotein of vesicular stomatitis virus (New Jersey serotype) J Virol. 1992;66:3749–3757. doi: 10.1128/jvi.66.6.3749-3757.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hansen J E, Lund O, Engelbrecht J, Bohr H, Nielsen J O, Hansen J S, Brunak S. Prediction of O-glycosylation of mammalian proteins: specificity patterns of UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase. Biochem J. 1995;308:801–813. doi: 10.1042/bj3080801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hansen J E, Lund O, Rapacki K, Brunak S. O-GLYCBASE version 2.0: a revised database of O-glycosylated proteins. Nucleic Acids Res. 1997;25:278–282. doi: 10.1093/nar/25.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang C, Chien M, Landolt M, Winton J. Characterization of the infectious haematopoietic necrosis virus glycoprotein using neutralizing monoclonal antibodies. Dis Aquat Org. 1994;18:29–35. [Google Scholar]
- 10.Huang C, Chien M S, Landolt M, Batts W, Winton J. Mapping the neutralizing epitopes on the glycoprotein of infectious haematopoietic necrosis virus, a fish rhabdovirus. J Gen Virol. 1996;77:3033–3040. doi: 10.1099/0022-1317-77-12-3033. [DOI] [PubMed] [Google Scholar]
- 11.Jensen M H. Research on the virus of Egtved disease. Ann NY Acad Sci. 1965;126:422–426. doi: 10.1111/j.1749-6632.1965.tb14292.x. [DOI] [PubMed] [Google Scholar]
- 12.Kelly J M, Emerson S U, Wagner R R. The glycoprotein of vesicular stomatitis virus is the antigen that gives rise to and reacts with neutralizing antibody. J Virol. 1972;10:1231–1235. doi: 10.1128/jvi.10.6.1231-1235.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krogh T N, Bachmann E, Teisner B, Skjødt K, Højrup P. Glycosylation analysis and protein structure determination of murine fetal antigen 1 (mFA1)—the circulating gene product of the delta-like protein (dlk), preadipocyte factor 1 (Pref-1) and stromal-cell-derived protein 1 (SCP-1) cDNAs. Eur J Biochem. 1997;244:334–342. doi: 10.1111/j.1432-1033.1997.00334.x. [DOI] [PubMed] [Google Scholar]
- 13a.Kurath, G. Personal communication.
- 14.Kurath G, Higman K H, Björklund H V. Distribution and variation of NV genes in fish rhabdoviruses. J Gen Virol. 1997;78:113–117. doi: 10.1099/0022-1317-78-1-113. [DOI] [PubMed] [Google Scholar]
- 15.Kussmann M, Nordhoff E, Rahbek-Nielsen H, Haebel S, Rossel-Larsen M, Jakobsen L, Gobom J, Mirgorodskaya E, Kroll-Kristensen A, Palm L, Roepstorff P. Matrix-assisted laser desorption/ionization mass spectrometry sample preparation techniques designed for various peptide and protein analytes. J Mass Spectrom. 1997;32:593–601. [Google Scholar]
- 16.Lenoir G, de Kinkelin P. Fish rhabdoviruses: comparative study of protein structure. J Virol. 1975;16:259–262. doi: 10.1128/jvi.16.2.259-262.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lorenzen N. Affinity purification of the structural proteins of a fish rhabdovirus by the use of monoclonal antibodies. J Virol Methods. 1992;38:297–303. doi: 10.1016/0166-0934(92)90074-n. [DOI] [PubMed] [Google Scholar]
- 18.Lorenzen N, Lorenzen E, Einer-Jensen K, Heppell J, Wu T, Davis H. Protective immunity to VHS in rainbow trout (Oncorhunchus mykiss, Walbaum) following DNA vaccination. Fish Shellfish Immunol. 1998;8:261–270. [Google Scholar]
- 19.Lorenzen N, Olesen N J. Immunization with viral antigens: viral haemorrhagic septicaemia. Dev Biol Stand. 1997;90:201–209. [PubMed] [Google Scholar]
- 20.Lorenzen N, Olesen N J, Jørgensen P E V. Neutralization of Egtved virus pathogenicity to cell cultures and fish by monoclonal antibodies to the viral G protein. J Gen Virol. 1990;71:561–567. doi: 10.1099/0022-1317-71-3-561. [DOI] [PubMed] [Google Scholar]
- 21.Lorenzen N, Olesen N J, Jørgensen P E, Etzerodt M, Holtet T L, Thøgersen H C. Molecular cloning and expression in Escherichia coli of the glycoprotein gene of VHS virus, and immunization of rainbow trout with the recombinant protein. J Gen Virol. 1993;74:623–630. doi: 10.1099/0022-1317-74-4-623. [DOI] [PubMed] [Google Scholar]
- 22.Lorenzen N, Olesen N J, Jørgensen P E V. Production and characterization of monoclonal antibodies to four Egtved virus structural proteins. Dis Aquat Org. 1988;4:35–42. [Google Scholar]
- 23.Lorenzen, N., N. J. Olesen, and C. Koch. Immunity to VHS virus in rainbow trout. Aquaculture, in press.
- 24.Luo L, Li Y, Snyder R M, Wagner R R. Point mutations in glycoprotein gene of vesicular stomatitis virus (New Jersey serotype) selected by resistance to neutralization by epitope-specific monoclonal antibodies. Virology. 1988;163:341–348. doi: 10.1016/0042-6822(88)90274-7. [DOI] [PubMed] [Google Scholar]
- 25.Matsumura M, Signor G, Matthews B W. Substantial increase of protein stability by multiple disulphide bonds. Nature. 1989;342:291–293. doi: 10.1038/342291a0. [DOI] [PubMed] [Google Scholar]
- 26.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 27.Prehaud C, Coulon P, Lafay F, Thiers C, Flamand A. Antigenic site II of the rabies virus glycoprotein: structure and role in viral virulence. J Virol. 1988;62:1–7. doi: 10.1128/jvi.62.1.1-7.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reading C L, Penhoet E E, Ballou C E. Carbohydrate structure of vesicular stomatitis virus glycoprotein. J Biol Chem. 1978;253:5600–5612. [PubMed] [Google Scholar]
- 29.Thiry M, Lecoq-Xhonneux F, Dheur I, Renard A, de Kinkelin P. Sequence of a cDNA carrying the glycoprotein gene and part of the matrix protein M2 gene of viral haemorrhagic septicaemia virus, a fish rhabdovirus. Biochim Biophys Acta. 1991;1090:345–347. doi: 10.1016/0167-4781(91)90200-6. [DOI] [PubMed] [Google Scholar]
- 30.Vorm O, Mann M. Improved mass accuracy in matrix-assisted laser desorption/ionization time-of-flight mass spectrometry of peptides. J Am Soc Mass Spectrom. 1994;5:955–958. doi: 10.1016/1044-0305(94)80013-8. [DOI] [PubMed] [Google Scholar]
- 31.Vorm O, Roepstorff P, Mann M. Improved resolution and very high sensitivity in MALDI TOF of matrix surfaces made by fast evaporation. Anal Chem. 1994;66:3281–3287. [Google Scholar]
- 32.Wiktor T J, György E, Schlumberger H D, Sokol F, Koprowski H. Antigenic properties of rabies virus components. J Immunol. 1973;110:269–276. [PubMed] [Google Scholar]
- 33.Wolf K, Gravell M, Malsberger R G. Lymphocystis virus:isolation and propagation in centrarchid fish cell lines. Science. 1966;151:1004–1005. doi: 10.1126/science.151.3713.1004. [DOI] [PubMed] [Google Scholar]