Abstract
Glioblastoma (GBM) is a common and devastating primary brain tumor, with median survival of 16-18 months after diagnosis in the setting of substantial resistance to standard-of-care and inevitable tumor recurrence. Recent work has implicated the brain microenvironment as being critical for GBM proliferation, invasion, and resistance to treatment. GBM does not operate in isolation, with neurons, astrocytes, and multiple immune populations being implicated in GBM tumor progression and invasiveness. The goal of this review article is to provide an overview of the available in vitro, ex vivo, and in vivo experimental models for assessing GBM-brain interactions, as well as discuss each model’s relative strengths and limitations. Current in vitro models discussed will include 2D and 3D co-culture platforms with various cells of the brain microenvironment, as well as spheroids, whole organoids, and models of fluid dynamics, such as interstitial flow. An overview of in vitro and ex vivo organotypic GBM brain slices is also provided. Finally, we conclude with a discussion of the various in vivo rodent models of GBM, including xenografts, syngeneic grafts, and genetically-engineered models of GBM.
Keywords: Glioblastoma, Tumor microenvironment, GBM models
Introduction:
Glioblastoma (GBM) is the most common and aggressive primary brain tumor (classified as a WHO Grade 4 primary brain tumor), with a median survival of 16-18 months after diagnosis and annual mortality of 12-14 thousand lives in the U.S. alone [32, 54, 80, 118, 128]. Conventional therapies for GBM include maximum safe surgical resection, radiation, and temozolomide chemotherapy, but these have had limited success against preventing recurrence of GBM, largely due to GBM resistance mechanisms [18, 38, 104, 144]. A long-standing frustration in the field of GBM therapeutics has been good efficacy of potential therapies in preclinical models, but subsequent failure of these therapies in clinical trials, underscoring the need for understanding and developing models that better recapitulate the genetics and phenotype of human GBM [10, 82]. In this light, there is a growing body of recent literature suggesting that GBM malignancy and resistance mechanisms are due at least in part to GBM interactions with cells of the brain microenvironment, including neurons, astrocytes, oligodendrocytes, macrophages, microglia, and other immune populations. Work by different groups have demonstrated that GBMs adapt neural mechanisms to drive malignancy and treatment resistance [65, 102, 135–138, 147]. Specifically, Monje and coworkers in 2015 demonstrated that neural activity drives glioma growth through release of the synaptic protein Neuroligin-3 [137] which was followed up by a 2019 study that demonstrated glioma integration into neural circuits at both the electrical and synaptic levels [138]. Similarly, work by Winkler and colleagues has very recently shown that GBM can hijack neuronal mechanisms to drive its invasion, including synaptic input from neurons [136] . Additional work by this group has demonstrated that GBM cells can form functional and therapy-resistant microtube networks between themselves, and that these are partially responsible for GBM invasion, proliferation, and treatment resistance [102, 147]; formation of these microtube connections is driven by TGF-β [65], a molecule known to drive tumor progression in GBM through both tumor-intrinsic and tumor-extrinsic mechanisms [7, 13, 23, 84, 92, 106, 132]. In addition to neurons, a related body of literature has also implicated astrocytes and oligodendrocytes as potential drivers of GBM proliferation, invasion, and chemoresistance, thus emphasizing the notion that GBM progression relies on the cellular brain microenvironment [15, 26, 53, 55, 67, 112, 139, 158].
In addition to interactions with neurons, astrocytes, and oligodendrocytes, it is also being increasingly recognized that GBM cells can interact with local immune cell populations in the brain tumor microenvironment (abbreviated TME), including macrophages, microglia, dendritic cells, and T cells, among other immune subsets, to facilitate GBM malignancy [3, 4, 6, 24, 31, 44, 60, 78, 100, 103, 111, 115, 119, 126, 140, 142]. Notably, GBM tumors have a substantial population of tumor-associated macrophages and microglia (collectively referred to as TAMs) which can comprise up to 50% of the cells in the brain TME [6]. In general, GBM TAMs are thought to exist on a continuum between M1 and M2 states [37, 48, 107, 145, 160, 163]. M1-like TAMs possess anti-tumor properties, including release of tumoricidal molecules, expression of proinflammatory cytokines, and direct phagocytosis and killing of tumor cells, with subsequent tumor antigen presentation to T cells to drive an adaptive immune response against GBM. In contrast, M2-like TAMs tend to be tumor-promoting via release of angiogenic and tumor growth/invasion factors and inhibition of an antitumor immune response [163]. In GBM, the majority of TAMs tend to take on more of an M2-like phenotype, with a greater M2 TAM infiltrate being associated with poorer prognosis [150, 154] . Emerging immunotherapy strategies targeting TAMs are directed towards re-programming TAMs towards an anti-tumor, M1-like, phenotype [107] and there has been much interest in repolarization of TAMs to augment immunotherapies [14, 79, 97, 105, 141], such as immune checkpoint blockade, which has not been successful as a monotherapy against GBM. Other myeloid cells in the GBM tumor microenvironment have also attracted attention, such as dendritic cells (DCs). Though there are limited circulating DCs and a prevalence of DC immune dysfunction in the GBM tumor microenvironment [2], recent studies have shown potential of certain DC subtypes to capture GBM antigens and present them to MHC-I-restricted cytotoxic T cells both in the TME and in local cervical lymph nodes (CLNs) in order to initiate a T cell response against GBM [125]. In this context, ongoing efforts are currently geared towards development of DC vaccines against GBM as a potential therapeutic option.
Collectively, the aforementioned discussion highlights the role of the complex brain microenvironment in shaping GBM malignancy and resistance to treatment. Lack of accurate preclinical models to address these interactions may have been a weakness in prior studies that could not successfully be translated to improving patient outcomes. It has become evident that understanding GBM interactions with the brain microenvironment is paramount to unveiling mechanisms of GBM progression and treatment resistance, so that the scientific community can efficiently develop and test potentially translatable therapies at the preclinical level. To rigorously interrogate these microenvironment interactions, there is a pressing need to understand, utilize, and develop preclinical models that can accurately factor these interactions in order to fuel discovery-driven research for GBM with higher probability for successful translation to the clinic. The goal of this review article is thus to provide an updated overview of the latest in vitro, ex vivo, and in vivo preclinical models of GBM with respect to their incorporation of these critical GBM-microenvironment interactions.
In Vitro Models of the Glioblastoma Microenvironment:
The in-depth exploration of glioblastoma (GBM) interactions within the brain environment has been significantly propelled by advances in in vitro models. Delving into the intricate dynamics of GBM-brain interactions and other influencing factors with these models has expanded our understanding of tumor behavior, growth, and invasion in the brain microenvironment in vivo and in humans. Among the various in vitro models of the glioblastoma microenvironment are mixed two-dimensional (2D) and three-dimensional (3D) cell co-cultures, whole organoid and spheroid cultures, and microfluidic/interstitial flow models. Each of these categories will subsequently be discussed in detail.
Mixed Two-Dimensional (2D) and Three-Dimensional (3D) Cell Co-Cultures:
GBM co-cultures typically involve directly mixing GBM cells with various cells of the GBM brain microenvironment – these can include astrocytes, oligodendrocytes, neurons, macrophages, and microglia. Among the simplest of these models are conventional 2D platforms, where GBM cells and microenvironment cells are combined in flat culture vessels (flasks, dishes, plates, etc.) and grown together in contact as a monolayer [66]. While this model has proved to be a workhorse in the GBM field in terms of drug-screening throughput and characterization of GBM cell phenotype, there are several key limitations with the 2D platform—including phenotypic and morphologic changes that occur over time, as well as greater susceptibility to genetic drift phenomenon, likely due to growth of the cells on a flat surface that does not recapitulate their growth patterns and behavior in vivo [90, 131, 152]. These drawbacks could explain the efficacy of certain drugs in 2D in vitro GBM applications, but subsequent lack of efficacy in GBM mouse models and in clinical trials. To address these limitations, 3D models have been developed to better recapitulate the growth of GBM and microenvironment cells in vivo. Generally, these 3D models have involved growth of cells in a multiprotein hydrogel (Matrigel) or scaffold to facilitate formation of three-dimensional biological tissues or cellular structures. Readers interested in a comprehensive technical evaluation of 2D and 3D cell culture platforms are encouraged to read the excellent reviews published on the topic [35, 66].
In order to distinguish the GBM cells from microenvironment cell types, GBM cells can be either labeled with a fluorescent marker such as green fluorescent protein (GFP) or modified to constitutively express a marker such as firefly luciferase (which can allow for luminescence to approximate GBM cell numbers in wells) [27, 75]. The co-culture can also be plated on culture 2D transwell inserts (transwell assay), where invasion of GBM cells can be subsequently measured. Coniglio et al. [27] optimized a protocol in which GBM cells are marked with GFP and macrophages or microglia are marked with red fluorescent protein (RFP). The GBM-macrophage/microglia mixture was then plated on matrix-coated inserts and invading GBM cells quantified with confocal microscopy 48-hours later. The authors extended this model to a 3D platform in which the GBM-macrophage/microglia mixture was seeded in a 3D matrix; the invading GBM cells were quantified by generating Z-stack series using fluorescent laser confocal microscopy, to assess the degree of invasion throughout the matrix. Interestingly, the authors noted that co-culturing GBM cells with either macrophages or microglia significantly increased GBM invasion relative to GBM cells alone with no macrophages or microglia present, in both 2D and 3D co-culture models. Other studies have used similar 2D co-culture platforms to interrogate the role of macrophage phenotype (M1 vs. M2) in the presence of GBM cells after treatment with various agents [57, 75, 151, 162]. In addition to myeloid cells, GBM-T cell co-cultures have also been employed to assess T cell phenotype in vitro in the GBM setting [52]. While many studies have focused on in vitro co-cultures of GBM with immune cells, there have been some studies that have utilized a co-culture system containing GBM cells and supporting cells of the brain in order to better understand GBM interactions with neighboring neural and glial cell types. For instance, Civita et al. [26] utilized 2D and 3D models of a GBM-astrocyte co-culture in order to understand behavior of these interacting cell types. Briefly, GBM cells and non-neoplastic astrocytes were individually labeled with Cell Trace dyes and plated in either a contact 2D co-culture or a hyaluronic acid-gelatin 3D hydrogel model; the authors found evidence in both models that astrocytes could promote GBM growth and migration, as well as form tunneling nanotube (TNT) connections with GBM cells to deliver mitochondria from astrocytes to GBM—which could determine therapeutic response to various drugs and affect GBM survival. More recently, Guyon et al. (2021) [49] optimized a GBM-neuron co-culture model in which GBM cells were cultured with patterned neurons. The authors postulated that this model allows for accurate modeling of white-matter-tract invasion and for better understanding of how neurons can modulate GBM invasion.
Spheroid and Whole Organoid Cultures:
While in vitro models involving monodispersed cells have been useful, advances have been made with both spheroid and organoid models of GBM, which better capture the heterogeneity of the brain tumor microenvironment. In general, GBM spheroids consist of aggregates of GBM and microenvironment cells either in suspension or embedded within a 3D matrix. In their simplest form, spheroids consist of either GBM cells aggregated as spheres or glioma stem cells (GSCs) grown as neurospheres [16, 17, 113, 146]. Certain culture conditions can facilitate sphere formation and maintain a phenotype and genotype similar to that of human GBM, such as growth of GSCs under serum-free conditions in Neurobasal medium supplemented with fibroblast growth factor (FGF), epidermal growth factor (EGF), and N-2 neural supplement [74, 159]. In general, the field of glioblastoma research is currently trending towards cultivating GBM cells under these stem-like conditions, as traditional serum-containing medium has been shown to induce phenotypic, morphological, and genetic alterations that alter the cellular phenotype away from that of actual human GBM, raising the question of whether serum-derived GBM lines accurately model human GBM [74]. Importantly, the aforementioned sphere aggregates contain several additional features that recapitulate GBM in vivo and in humans, such as cell-cell communication in 3D space and existence of a hypoxic core at the center of the spheres—thus providing a distinct advantage over single-cell culture models cultured in serum [17]. In the case of GSCs, the cells exhibit stem-like features such as expression of stem cell markers (specifically, CD133, nestin, and NANOG, among others), high plasticity, and possible origin from neural stem cells. Moreover, it has been found that glioma stem cell spheroids also contain other neural cell populations, including astrocyte, oligodendrocyte, and neuron precursors, implying that this model better captures GBM heterogeneity and interactions with neuronal/glial cell types [74, 83, 156].
Whole organoid models attempt to take the spheroid models a step further to account for interactions of GBM cells with other microenvironment cells of the brain, in an architecture that mimics brain organization. In general, organoids represent a 3D culture platform generated from self-organizing stem cells. Importantly, these models can better capture tumor heterogeneity and the composition of the microenvironment and native tumor organ compared to cell line cultures, as highlighted in previous reviews [64, 130]. They have been utilized broadly for applications such as modeling brain tumor formation and growth, brain microenvironment interactions with GBM, and anti-GBM drug screening [149]. In 2013, a study published in Nature [73] first reported on generation of human pluripotent stem-cell derived cerebral organoids, which spontaneously developed various brain regions such as cerebral cortex with distinct cortical neuron subtypes [73, 121]. The cerebral organoid model has more recently been carried forward in the GBM field, with several 2018 studies utilizing a variant of the model that introduced oncogenic mutations via CRISPR-Cas9 genome editing to drive tumorigenesis in the organoid [11, 99]. One year later, Linkous et al. [81] utilized a variant of this cerebral organoid model in which they used patient-derived GSCs and human embryonic stem cell-derived cerebral organoids to generate patient-specific GBMs that closely phenocopy patient GBMs (termed the GLICO model). Moreover, the authors noted that these cerebral organoid GBMs are supported by networks of tumor microtubes that facilitated GBM invasion into normal host tissue, representing an important advance in understanding microtube dynamics in 3D in vitro platforms. Recently, Fedorava et al. adapted this GLICO model for modeling glioblastoma migration [39]. Notably, new efforts have also focused on developing publicly available biobanks of GBM organoids to facilitate basic and translational research utilizing these advanced 3D in vitro platforms [61].
While spheroids and organoids have represented an intermediate platform for modelling GBM-brain microenvironment interactions (between in vitro cell cultures and in vivo mouse models), there are certain disadvantages to consider with these models. First, unlike most 2D cell culture platforms, there is no standardized protocol for generating and maintaining spheroids and organoids, implying potential difficulties in reproducibility from study-to-study between different research groups and experiment-to-experiment variability in the cellular composition of the GBM organoid. Additionally, certain features of the brain microenvironment may not be as accurately modeled with these platforms, such as the immune and vascular landscapes [108, 149].
Microfluidic and Interstitial Flow Models:
While the aforementioned model systems have proved useful in understanding interactions of GBM cells with cell types in the brain microenvironment, one key limitation is that they do not account for interstitial flow forces in the glioblastoma microenvironment. In this light, Munson and colleagues have made significant advances in our experimental understanding of these factors. In 2013, this group determined that interstitial flow was a key regulator of glioma cell invasion and that this was mediated causally by the chemokine CXCR4 [94]. The study utilized an elegant three-dimensional culture model of interstitial flow in which tumor cells were seeded in a hyaluronan gel and then placed in a cell culture insert, which was subsequently cast and subjected to a pressure head (created with culture medium) which led to a fixed average velocity through the cellular compartment. Distribution and polarization of GBM cells was evaluated in a related model in which matrix-embedded cells were added to radial flow chambers and subjected to pressure-driven flow. After fixing chambers with paraformaldehyde and immunostaining, the distribution of cells in the flow chambers was assessed with confocal microscopy. These models were extended in a follow-up publication [72], in which the group discovered that interstitial flow drives glioma stem cell invasion via CXCR4, CXCL12, and CD44. The group carried these studies forward with a paper in 2018 that reinforced the paradigm that interstitial flow enhances GBM (GL261 line) cell invasion via CXCR4 and CXCL12 using the aforementioned in vitro 3D culture model. These findings were extended to in vivo models that ultimately found that interstitial and convective flow enhanced GBM invasion in a CXCR4-dependent manner [28]. Very recently, the Munson group extended their 3D in vitro model of interstitial flow to assess how interstitial flow modulates GBM cells when they are surrounded by neighboring cells of the brain microenvironment. The study utilized a four-component 3D model of the GBM microenvironment that consisted of patient-derived glioma stem cells co-cultured with human glial cells (specifically, astrocytes and microglia) and used this model to assess invasion, stemness, and proliferation with respect to fluid forces, response to chemotherapy, and phenotype of the glial cells [29]. Importantly, the results displayed heterogeneity between different patients and could represent an important step towards precision medicine of glioblastoma patients. Another key area of research in the microfluidics space has been developing models to recapitulate the blood-brain barrier (BBB) and assess drug permeability across the barrier in the setting of GBM. In this light, Straehla et al. generated a vascularized in vitro model in a microfluidics device that consisted of self-assembled vascular networks of human endothelial cells, astrocytes, and pericytes to assess transport of nanoparticle therapeutics across the BBB and into GBM cells grown as spheroids (patient-derived GBM22 from the Mayo Clinic Brain Tumor PDX National Resource) [127]. Importantly, the research group noted concordance between in vitro and in vivo models, suggesting the utility of this approach for development of translatable nanotherapeutics. Shi et al. very recently utilized a similar in vitro model which they adapted as a screening tool for evaluating the BBB permeability and anti-glioma efficacy of traditional Chinese medicine components [124]. Evaluation of the role of complex variables such as fluid forces and BBB permeability represent important advances in GBM research towards precision medicine. Current efforts have also moved towards “glioblastoma-on-a-chip” models based on microfluidics and 3D bio-printing in an effort to develop more accurate in vitro models of the complex GBM microenvironment, as reviewed by Xie et al [148].
Consideration of Cell Types and Sources for in vitro studies:
In all of the previously discussed models, an important consideration is the source and origin of the GBM and microenvironment cells used. While many studies have employed immortalized, serum-derived human glioblastoma lines such as U251MG and U87MG, these models are falling out of favor for models that more closely recapitulate the genetics and phenotype of human GBM, such as patient-derived xenograft (PDX) lines [1, 5, 68] and glioma stem-like cells [20, 30, 143]. Importantly, these newer models can be cultivated under serum-free, stem-like conditions, which has been shown to preserve the genetics and phenotype of the cells to be more similar to that of human GBM, as previously discussed [74]. In an effort to make PDX models more accessible to researchers, the Mayo Clinic has established the Brain Tumor PDX National Resource for distribution of these lines across the world, with data on the characterization of these lines publicly available [134]. Additionally, there are multiple well-established murine GBM lines that can also be used for these studies (these models are further discussed under “in vivo glioblastoma mouse models”)
Choice of the cell types used for the microenvironment is much broader – as previously discussed, GBMs can be co-cultured with neurons, oligodendrocytes, astrocytes, microglia, and macrophages, among other cell types. To facilitate this work, there are multiple commercially available cell lines of both human and murine cell types (both primary and immortalized lines) that can be used for in vitro co-culture studies, as summarized in Table 1.
Table 1:
Overview of commercially available brain microenvironment cell types to model GBM-brain interactions in vitro
| Neurons | Species | Hyperlink to Vendor(s) | Comments |
|---|---|---|---|
| Primary Neuron Lines [101] | Human, Mouse, Rat | https://sciencellonline.com/neurons | Primary cells; not recommended for long-term culture or expansion as per the vendor’s instructions (should be used directly for experiments) |
| Oligodendrocytes/Oligodendrocyte Precursors (OPCs) | Species | Hyperlink to Vendor(s) | Comments |
| Primary OPC Lines [47] | Human, Rat |
https://sciencellonline.com/products-services/primary-cells/human/cell-types/oligodendrocytes/
https://sciencellonline.com/products-services/primary-cells/animal/cell-types/oligodendrocytes/rat/ |
Primary cells; not recommended for long-term culture or expansion as per the vendor’s instructions (should be used directly for experiments) |
| Astrocytes | Species | Hyperlink to Vendor(s) | Comments |
| Primary Human Astrocyte Lines [85] | Human | https://sciencellonline.com/products-services/primary-cells/human/cell-types/astrocytes/ | Primary Rat and Mouse lines are also available from this vendor; lines are stable for 5-15 passages post-thaw as per vendor’s instructions |
| C8-D1A (Astrocyte type I clone) [123] | Mouse | https://www.atcc.org/products/crl-2541 | Immortalized; suitable for long-term culture |
| CTX TNA2 (Astrocyte type I) [96] | Rat | https://www.atcc.org/products/crl-2006 | Immortalized; suitable for long-term culture |
| D1 TNC1 (Astrocyte type II) [63] | Rat | https://www.atcc.org/products/crl-2005 | Immortalized; suitable for long-term culture |
| SVG p12 (Astroglia) [40] | Human | https://www.atcc.org/products/crl-8621 | Immortalized; suitable for long-term culture |
| Microglia | Species | Hyperlink to Vendor(s) | Comments |
| Primary Human Microglia [161] | Human | https://sciencellonline.com/human-microglia/ | Primary Rat and Mouse lines are also available from this vendor; these lines are not suitable for long-term culture or expansion as per the vendor’s instructions (should be used directly for experiments) |
| Immortalized Human Microglia–SV40 (IMhu) [25] | Human | https://www.abmgood.com/immortalized-human-microglia-sv40.html | Immortalized; suitable for long-term culture |
| HMC3 [70] | Human | https://www.atcc.org/products/crl-3304 | Immortalized; suitable for long-term culture |
| SIM-A9 [95] | Mouse | https://www.atcc.org/products/crl-3265 | Immortalized; suitable for long-term culture |
| IMG [89] | Mouse | https://www.kerafast.com/item/1198/microglial-cell-line-img | Immortalized; suitable for long-term culture |
| Macrophages | Species | Hyperlink to Vendor(s) | Comments |
| Primary Macrophages [34] | Mouse, Rat | https://sciencellonline.com/products-services/primary-cells/animal/cell-types/macrophages/ | Primary cells; not suitable for long-term culture or expansion as per the vendor’s instructions (should be used directly for experiments) |
| J774A.1 [57] | Mouse | https://www.atcc.org/products/tib-67 | Immortalized; suitable for long-term culture |
| RAW264.7 [57] | Mouse | https://www.atcc.org/products/tib-71 | Immortalized; suitable for long-term culture |
| THP-1 [120] | Human | https://www.atcc.org/products/tib-202 | Immortalized monocytes; suitable for long-term culture; can be induced to differentiate into macrophages |
| Human M2 Macrophages – Monocyte Derived [9] | Human | https://promocell.com/product/human-m2-macrophages-m-csf-monocyte-derived/ | Can be maintained in culture for several weeks but should be used as soon as possible for experiments |
Ex Vivo and in Vitro Organotypic Brain Slice Models of Glioblastoma:
While cell co-culture models have been useful in modeling GBM and its microenvironment interactions with other cell types, as well as screening potential anti-GBM agents, one limitation of that approach is that the cell populations are dissociated and not present in their native, intact organ. To partially address this, the organotypic brain slice culture model utilizes brains harvested ex vivo from mice/rats or patient brain tissue retrieved from neurological surgeries. Briefly, slices are prepared from rodent or human brain tissue via a tissue chopper or vibratome capable of preparing 200-400 μm thick sections and cultured in a medium with a specialized composition for up to a few weeks. The model can be modified to include GBM by either generating the slices from GBM patient surgical samples, GBM-bearing mice, or by directly injecting GBM cells into regions of the slice(s) with a microinjector apparatus. The model can be used to study multiple aspects of glioma biology, including GBM invasion patterns, GBM growth/migration in the brain, screening of potential anti-GBM agents, and assessment of the brain microenvironment via a diverse array of methodologies including immunohistochemistry staining, fluorescent imaging, and confocal time-lapse microscopy [58, 62, 91, 93, 109, 114]. There have been multiple excellent studies in the literature utilizing applications of this model; in 2019, the Heiland group in Germany developed a human cortical slice system involving injection of patient-derived GBM cells into the brain slice tissue (which was derived from surgical patient samples) in order to model GBM progression [114]. The group noted that growth and invasion patterns, response to temozolomide chemotherapy, and the local cytokine milieu were similar to patterns of tumor growth seen in vivo and in humans. Furthermore, the group extensively validated this model in terms of viability/vitality of the brain tissue using multiple methods, including immunohistochemistry, electrophysiology, and RNA-sequencing. Finally, the group sought to use this GBM slice system to characterize the brain microenvironment during GBM progression in terms of astrocyte profiling using magnetic-activated cell sorting (MACS) and compared the signatures seen in this slice model to those seen in human GBMs and mouse models. The authors go on to propose that their isolation protocol will allow investigators to isolate and purify other cell types from this slice system, including neurons, oligodendrocytes, and microglia. In another study, Merz et al. [91] utilized slices prepared from human GBM surgical biopsy tissue to study the effects of irradiation, proliferation (with Ki67 staining), and temozolomide treatment. Jensen et al. [62] directly injected glioma stem cell spheroids into rat-derived brain slices to study GBM invasion patterns via confocal time-lapse microscopy and immunohistochemistry, noting that GBM invasion in brain slices closely mimicked the patterns seen in vivo. Interestingly, a report by Marques-Torrejon et al. [86] noted that the engraftment pattern of patient-derived glioma stem cells varied depending upon the specific brain slice anatomical region that was injected, with the subependymal zone being a hotspot for efficient GBM engraftment and microenvironment signaling. To evaluate the brain slice model as a GBM drug screening tool, Minami et al. [93] reported that fluorescence-based tumor imaging and immunostaining were sufficient to evaluate the effects of cisplatin, temozolomide, paclitaxel, and tranilast. Similarly, Pencheva et al. [110] utilized GBM brain slices to identify a druggable molecular pathway that could control glioma invasiveness, along with other recent reports utilizing GBM brain slices to evaluate the effects of different drugs and agents [56, 109]. The brain slice model has also been extensively used to model glioma invasion and migration patterns, with different reports describing detailed protocols and methods for generating these model systems of GBM invasion [33, 36, 133].
Organotypic brain slices confer distinct advantages over other models of GBM. First, the GBM cells are contained in a three-dimensional space where the brain spatial architecture and microenvironment is completely preserved, allowing for accurate modeling of GBM migration and invasion patterns in live neural tissue, in contrast to traditional in vitro cell culture models. Second, the brain slice approach represents a simpler, higher throughput and more cost-efficient approach to modeling GBM and its microenvironment interactions compared to in vivo mouse models. However, there are several drawbacks to this system: GBM slice cultures can usually be maintained only for a few weeks under sterile conditions and the choice of culture medium is often critical, with serum-containing medium often leading to differentiation of neural stem cells and decreased viable culture time of the slices to only a few days [86]. Moreover, age of the animal donor has also been implicated in viability of the resulting slice culture, with slices from younger animals generally resulting in more viable slice tissue and slices from older animals generally containing less viable tissue [58]. Additionally, slice cultures cannot be modified to accurately model fluidic factors such as interstitial flow, unlike with certain 3D cell culture and in vivo models. Lastly, slice cultures of GBM have not yet been adapted to accurately model the BBB and the interactions of GBM with the BBB or permeability of agents across the BBB [46, 58, 108]. Indeed, the BBB is an important variable that has not been accounted for in the studies involving drug screening on GBM brain slices.
In Vivo Glioblastoma Mouse Models:
Mouse models have proven to be critical in preclinical glioblastoma studies and in validating in vitro and ex vivo findings, and naturally include modeling of the GBM microenvironment limited only by cross-species differences. The various GBM models can broadly be divided into orthotopic and genetically engineered models, with the former involving direct implantation of GBM cells into the brains of mice using stereotactic equipment. The latter typically involves models capable of spontaneously forming GBM brain tumors secondary to targeted genetic mutation(s) in key GBM oncogenes. Orthotopic models can be further divided into xenograft and syngeneic subtypes, with xenografts involving the implantation of human GBM cells into immunodeficient mice and syngeneic grafts involving the implantation of murine GBM cells into immunocompetent mice. The key advantages of these models include the study of GBMs in living mammals (which are the closest models to humans), as well as presence of the entire, architecturally intact brain microenvironment. Thus, in vivo GBM models represent a sort of “gold standard” in evaluating potentially translatable therapeutics with respect to safety and efficacy. The various models used for in vivo GBM studies are further discussed below. Readers further interested in these models are encouraged to refer to an excellent review article by Haddad et al [50].
a). Orthotopic Xenografts.
Xenograft models involve direct implantation of human GBM cells into the brains of immunodeficient mice, such as nude and NOD/SCID mice. Key xenograft models have included both the U87 and U251 lines, both of which have been used in thousands of studies since the 1960s, when they were originally derived from GBM patients. Other human GBM models have also seen wide use, including the T98G, LN18, and LN229 lines [50, 76]. The key advantage of these models is their existence as models of human GBM, as they mimic the characteristics of human GBM histologically and genetically and allow for modeling of GBM invasion and angiogenesis. One key disadvantage, however, is that these models are grown on an immunodeficient background, which does not allow for characterization of the GBM immune landscape or evaluation of immunotherapies for GBM. There may also be major limitations in modeling human GBM-microenvironment interactions due to the placement of a human GBM in a mouse microenvironment.
Though the aforementioned models have proven to be workhorses in modeling human GBM in vivo, there have been some recent concerns with respect to just how closely they mimic the characteristics of human GBM, their susceptibility to genetic drift phenomenon when cultured in serum (which skews genotype and phenotype away from that of human GBM), and concerns of cross-contamination of the lines over time across multiple labs [131]. Thus, over time there has been decreased use of these serum-derived lines within the neuro-oncology scientific community in favor of an increased emphasis on the use of patient-derived xenograft (PDX) models of GBM. These involve the harvesting of GBM tumor tissue from patients, processing into single cell suspension, and subsequent passaging of the lines in immunodeficient mice. These PDX models have indeed been reported to better mimic characteristics of human GBM compared to the aforementioned traditional models, including more accurate assessment of GBM invasion, subtype-specific characteristics (i.e. mesenchymal vs. proneural vs. neural vs. classical), and presence of hallmark GBM histopathology, such as pseudopalisading necrosis and endovascular proliferation [134]. However, like traditional human GBM models, PDX models also suffer from the lack of an immune background. Additionally, there has been reported to be a high degree of variability between different PDX models of GBM, as each PDX line is derived from a different patient.
b). Syngeneic Grafts:
Unlike xenografts, syngeneic grafts involve implantation of murine GBM cells into the brains of immunocompetent mice. Thus, a key advantage of these models is the presence of the immune system and evaluation of various immunotherapies against GBM. Moreover, these models also allow for detailed characterization of the immune landscape in the GBM brain microenvironment. Furthermore, there should be fully intact communication between mouse GBM cells and mouse microenvironment cells and proteins. However, a key disadvantage is that the immune microenvironment seen in syngeneic models may not closely mirror that seen in human GBMs. Additionally, the genetic background in syngeneic models often does not mirror that in human GBM, and some syngeneic GBM models have been reported to have much higher mutational loads compared to that seen in human GBM [77]. Numerous syngeneic models for GBM have been established, including the GL261, CT-2A, SB28, M-005, and KR158 models, as well as the very recently established mGB/tNSC models, which are further discussed below:
GL261: the GL261 model has proved to be a workhorse among the syngeneic GBM models, having been extensively characterized and studied over the years. This model was derived in the 1970s via methylcholanthrene chemical induction in the brains of C57/Bl6 mice and passaged over time in vivo, then ultimately stably immortalized in vitro. It should be noted that GL261 has higher baseline immunogenicity and mutational burden compared to human GBMs, so much so that implanted GL261 tumors can often be rejected in vivo [69, 98, 129]. Thus, while GL261 has proved useful in evaluating potential immunotherapies at the preclinical level, some of these studies have not shown efficacy when evaluated in GBM clinical trials, such as immune checkpoint blockade, despite showing efficacy in preclinical studies [41, 43]. However, it should be noted that a subset of GL261 cells have been shown to express stem cell markers such as nestin and CD133 and have displayed the ability to grow as neurospheres in suspension when cultured with serum-free stem cell medium. This subset of GL261 neurospheres have also been shown to potentially display lower immunogenicity than traditional GL261 grown in serum, allowing for a better model of human GBM immunogenicity using the GL261 model [155].
CT-2A: Like GL261, the CT-2A line was chemically derived, rendering it genetically and immunologically distinct from human GBMs [122]. Importantly, however, the CT-2A model has been described to recapitulate several histological characteristics of human GBM, including pseudopalisading necrosis and microvascular proliferation [87]. Additionally, CT-2A can be grown as glioma stem cells under serum-free conditions, allowing an avenue to model immunocompetent GSCs [12]. It should be noted that CT-2A cells are highly tumorigenic, with mice often surviving for a shorter period of time compared to other models. In fact, there have been some accounts of CT-2A cells growing out of mouse brains due to their high degree of invasiveness.
SB28: While the GL261 and CT-2A models are older and more well-established models of GBM, the SB28 model is a newer model that more closely mimics the immunogenicity (limited CD8+ T cell infiltration and low MHC-I expression), mutational burden, and microenvironment of human GBMs and is not chemically derived [43]. Importantly, the SB28 model better recapitulates the GBM response to immunotherapy, as immune checkpoint blockade did not show efficacy against SB28, which more closely mimics the reality of human GBM [43]. This model was uniquely generated using the sleeping beauty transposon to insert constructs targeting multiple oncogenic pathways, including p53, RAS, and PDGF. Interestingly, this model grows extremely aggressively, with mice almost universally developing tumors after injection with as few as around 2000 cells [50].
M-005: The M-005 line is a newer model that was generated using Cre-loxP lentiviral vectors in immunocompetent mice. The resulting GBM cells were stem-like (expressing CD133) and were capable of forming poorly-differentiated tumorospheres that could differentiate into neurons and astrocytes [88]. The immune and molecular landscapes of this model have more recently been further characterized and, like SB28, the M-005 model seems to bear a lower degree of immunogenicity similar to that of human GBM, making it well suited for accurate evaluation of potential translatable immunotherapies [20, 69, 117]. Moreover, this model represents a non-chemically generated method of modelling glioma stem cells in immunocompetent mice and also closely mimics the genetics of mesenchymal GBM [21].
KR158: Another relatively recent model, KR158 was originally developed in 2000 as a mouse model of astrocytoma mutated in the tumor suppressor genes Nf1 and Trp53 [116]. The resulting model displayed a range of tumor histology, from low-grade astrocytoma to GBM, with the KR158B subtype mimicking the characteristics of aggressive human GBM [143]. While the detailed molecular characterization data on this line is limited, it has displayed resistance to radiation, anti-PD1 therapy and temozolomide treatment, implying that this model mimics the modes of resistance seen in human GBM, making it a good model for preclinical assessment of GBM therapeutics in the setting of treatment resistance [42, 143].
mGB/tNSC: These models were very recently developed in 2019 by Costa et al. through neural stem cell [NSC] – specific deletions in Pten and p53 [30, 143]. The resulting models recapitulated features of high-grade gliomas including histopathological characteristics (microvascular proliferation, necrosis, etc), podoplanin (PDPN) expression, and platelet aggregation. Importantly, in glioma patients, elevated expression of PDPN has been associated with a worse prognosis and correlates with platelet aggregation intratumorally, as well as an increased risk of venous thromboembolism (VTE). Thus, this model serves as an avenue to functionally interrogate the role of PDPN and platelet aggregation in vivo for preclinical GBM studies aiming to develop strategies to decrease risk of platelet aggregation and VTE in GBM patients. Costa et al. (data unpublished) have further characterized this GSC model and demonstrated transcriptomic enrichment of classical, proneural, and mesenchymal GBM subtypes, implying this model mimics human GBM heterogeneity. Furthermore, the model has been further characterized to recapitulate the immunologically “cold” GBM microenvironment, with high myeloid infiltration, but low lymphocyte infiltration into the tumor mass. Thus, the mGB and tNSC models represent latest advances and efforts in the generation of syngeneic models that more faithfully recapitulate the histopathological, molecular, and immunological characteristics of human GBM.
c). Genetically Engineered Mouse Models (GEMMs):
A key issue in multiple syngeneic models has been mutational and genetic profiles that do not closely mimic human GBM. For instance, GL261 and CT-2A, while having widespread use, have been shown to have mutational burdens higher than that of human GBM, owing to their origin as chemically-derived lines. Additionally, some concerns with grafted GBM cells include possible disruption of brain vasculature such as the BBB, as well as baseline inflammatory responses induced with the act of intracranial injections. Genetically engineered mouse models of GBM have the key advantage of spontaneously developing GBM, with no tumor cell injection required. The models themselves have been traditionally generated via genetic manipulations (global and cell-specific knockouts, Cre recombinase, viral vectors, and CRISPR-Cas9, among others) that target certain genes to drive gliomagenesis. As comprehensively reviewed by Hambardzumyan et al [51], key GEMMs have included models with mutations in key genes implicated in gliomagenesis, including PDGFR, EGFR, and NF1 that spontaneously developed glioblastoma. More recently, the M-005 line (discussed above) was also generated from a GEMM model and has seen increased use. As reviewed by Huse and Holland [59], other key GEMMs employed in glioma and brain tumor research have included 1) astrocytic glioma derived from p53 and Nf1 global mutants, 2) an Nf1/p53 double mutant in GFAP-expressing astrocytes and glial populations (cell-specific knockout), 3) a GEMM with inactivation of the retinoblastoma (Rb) pathway that was generated via expression of SV40 T antigen under the control of the GFAP promoter, leading to Rb pathway inactivation in mature astrocytes and development of astrocytomas in mice, 4) a set of GEMMs generated from expression of constitutively active RAS (V12Ras) under the GFAP promoter, 5) GEMMs derived from an avian retrovirus (RCAS) alongside its receptor, tv-a, under the nestin or GFAP promoters, with key oncogenic drivers being Akt and KRas, and PDGF-B. Tumor suppressor loss has also been employed with the RCAS/tv-a system, with key genes including Ink4a/Arf and Pten, 6) a glioma GEMM that is virally mediated and driven by a constitutively active fusion receptor tyrosine kinase (RTK), termed “FIG-ROS”. Interestingly, this GEMM usually only develops gliomas when there is another associated mutation present in the model, such as loss of Ink4a/Arf. While the aforementioned GEMMs have proved useful in modeling GBM/glioma, it should be noted that most of them require extensive cross-breeding of mice and can be time consuming to generate. Additionally, the methods used to generate these GEMMs can have unpredictable patterns with respect to genomic editing and off-target effects in other tissue types, which can cause confounding in the resulting models. To circumvent this, Kim et al [71] developed the Mosaic analysis with dual recombinase-mediated cassette exchange (MADR) system to robustly generate mosaic mouse models containing a fixed copy number and predetermined insertion sites, with limited breeding time and mouse colony maintenance required. Importantly, MADR can efficiently generate mosaic models containing a mixture of gain- and loss-of function mutations, as well as fusion proteins, thus allowing for an avenue to model diverse brain tumor types. Interestingly, the authors also utilized MADR as a personalized platform to model distinct patient GBM tumors, with models generated by MADR being similar in phenotype and heterogeneity to actual human GBMs, thus bolstering the promise of this approach as a potential preclinical drug screening pipeline for patient-specific GBMs (i.e. personalized medicine). Very recently, there has also been an effort to utilize GEMMs to assess the tumor microenvironment of brain tumors. In this light, Yao et al. [153] utilized the Mosaic Analysis with Double Markers (MADM) genetic system to delineate molecular signaling and evolution at the single-cell level within the tumor microenvironment in a spontaneous SHH-activated medulloblastoma brain tumor model. Briefly, this model involves formation of medulloblastoma via generation of Ptch1-heterozygous, p53-null, GFP-positive granule neuron progenitors through the Math1-Cre system.
The key advantages of GEMMs are that they arise spontaneously, similar to the inception of human GBM, and do not require intracranial injection of tumor cells, which could introduce baseline immunogenicity due to the injection. Moreover, these models can be developed with alterations in a single gene or pathway to investigate the effect of these genetic alterations on tumorigenesis or response to specific targeted treatments. Like syngeneic grafts, GEMMs also harbor an intact immune system, allowing for characterization of the immune landscape. Some disadvantages with GEMMs include variability in tumor formation; unlike grafted tumor cells, GEMM tumors can potentially have some variability in the location and magnitude of tumor development in the brain. These models can also require complicated breeding strategies and can be slow to form. Finally, there has been some concern with how closely GEMMs mimic the intratumoral heterogeneity seen in human GBMs, as the mechanism of tumorigenesis in GEMMs may skew the GBM microenvironment towards the molecular pathways affected by the genetic manipulations intrinsic to the model [50].
d). In utero Electroporation:
In order to study specific genetic mutations and genes in murine GBM and brain tumor models, there has been very recent progress in generation and utilization of mouse models derived from in utero electroporation. As described by Chen and LoTurco [22], this method involves simultaneous delivery of multiple plasmid DNAs into neural progenitor populations in the brains of developing murine embryos, often around embryonic day 13 or 15. Briefly, a common methodology involves pressure-injecting labeled (e.g. with GFP) plasmid DNA into the lateral ventricles of developing embryos upon exposure of the uterine horns in the mother, using pulled glass capillaries. Electric pulses are then subsequently applied with a pulse generator, after which the uterus is then placed back into the abdominal cavity. After birth, pups can be traced with tracking methods such as bioluminescent imaging to monitor transfection efficiency of the introduced plasmids or to track brain tumor development [8, 22]. In the brain tumor and GBM field, there has been additional progress in this approach by the Deneen research group [8, 19, 45, 157]. In a recent study, the group utilized in utero electroporation to overexpress the ZRfus oncogene in the brains of developing mouse embryos, which subsequently developed ependymomas and found that this mouse model was similar to human ependymomas harboring the ZRfus mutation at the transcriptomic level [8]. The group has extended this methodology in other recent papers, one of which combined in utero electroporation with CRISPR/Cas9 genome editing to generate an immunocompetent glioma mouse model bearing vascular and molecular signatures similar to that seen in human malignant gliomas [19]. A third paper by the group also utilized this methodology to understand GBM-neuron interactions, implying that this method can also be used to understand the GBM brain microenvironment [157]. To summarize, murine brain tumors derived from in utero electroporation appear to have the advantage of spontaneous development and molecular/phenotypic features that can closely mirror those seen in human GBM, including tumor heterogeneity. Moreover, brain tumors derived from in utero electroporation do not involve the same complicated and time-consuming breeding strategies needed for generation of certain brain tumor GEMMs.
Conclusions and Perspectives:
Over time, there has been a shift from focusing on tumor-intrinsic mechanisms of GBM to understanding tumor-extrinsic, or microenvironment, mechanisms of GBM progression and treatment resistance, including GBM interactions with neuronal cells of the brain as well as immune cell populations in the brain. In this vein, there have been remarkable developments in establishing more intricate models of the brain microenvironment, including whole GBM organoid models, organotypic slice models of GBM, interstitial flow in vitro 3D co-culture models, and immunocompetent mouse models mimicking the tumor mutational burden and immunogenicity of human GBM, such as SB28, M-005, and mGB/tNSC (depicted in Figure 1). With the GBM field rapidly shifting towards understanding interactions between gliomas and the brain microenvironment, it remains critical to utilize and understand the various experimental models of these interactions. As newer advancements are shifting towards more personalized medicine, it will remain both challenging and exciting to model an individual patient’s GBM brain microenvironment. Maintaining an understanding of the existing experimental tools we currently have and striving to develop more advanced models using the latest technological developments will no doubt drive more advanced innovations in the fight against this deadly brain cancer.
Fig. 1.
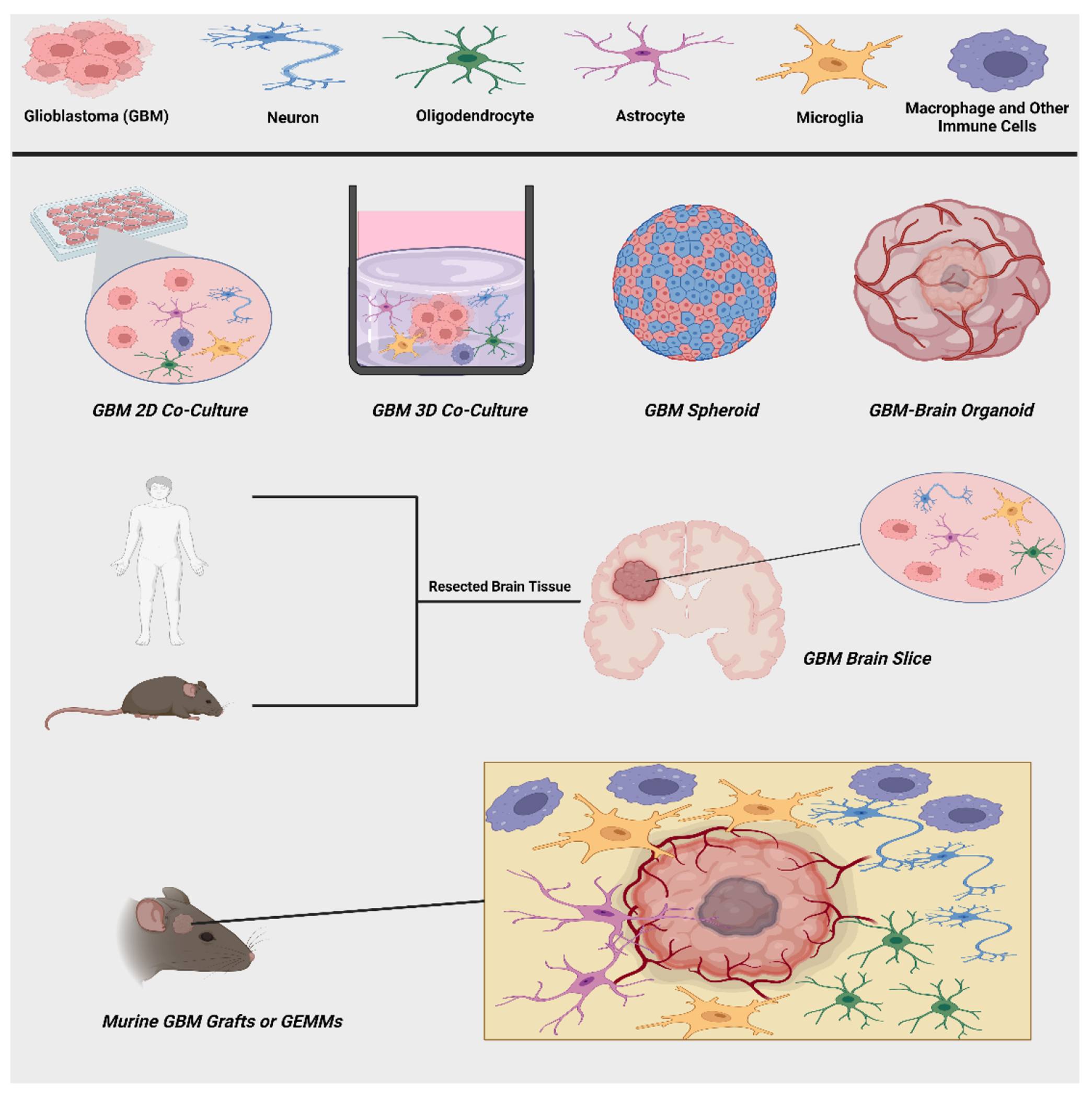
Graphical summary of in vitro GBM models (2D and 3D co-cultures, spheroids, organoids), ex vivo GBM brain slices, and in vivo GBM mouse models. These experimental models can recapitulate GBM interactions with cells of the brain microenvironment (neurons, oligodendrocytes, astrocytes, microglia, macrophages, and other immune cells) to varying extents as outlined in the text. This figure was created with BioRender (https://www.biorender.com)
Acknowledgements:
The graphical illustration made in figure 1 was created by N.Y. using BioRender (https://www.biorender.com). This work was supported by the National Institutes of Health/National Institute of Neurological Disorders and Stroke/National Cancer Institute [R01NS124787 (N.Y., B.P.), R01NS126265 (B.P.), and U54CA274499 (B.P.)]. N.Y. was supported by a Medical Scientist Training Program Grant (T32GM007267), a Cancer Training Grant (T32CA009109), and a trainee fellowship from the University of Virginia Comprehensive Cancer Center.
Footnotes
Competing Interests: Neither author has any conflicts-of-interest to disclose.
References:
- 1.Abdolahi S, Ghazvinian Z, Muhammadnejad S, et al. , Patient-derived xenograft (PDX) models, applications and challenges in cancer research. J Transl Med, 2022. 20(1): p. 206. 10.1186/s12967-022-03405-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adhikaree J, Franks HA, Televantos C, et al. , Impaired circulating myeloid CD1c+ dendritic cell function in human glioblastoma is restored by p38 inhibition - implications for the next generation of DC vaccines. Oncoimmunology, 2019. 8(7): p. 1593803. 10.1080/2162402X.2019.1593803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akkari L, Bowman RL, Tessier J, et al. , Dynamic changes in glioma macrophage populations after radiotherapy reveal CSF-1R inhibition as a strategy to overcome resistance. Sci Transl Med, 2020. 12(552). 10.1126/scitranslmed.aaw7843 [DOI] [PubMed] [Google Scholar]
- 4.Alban TJ, Bayik D, Otvos B, et al. , Glioblastoma Myeloid-Derived Suppressor Cell Subsets Express Differential Macrophage Migration Inhibitory Factor Receptor Profiles That Can Be Targeted to Reduce Immune Suppression. Front Immunol, 2020. 11: p. 1191. 10.3389/fimmu.2020.01191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alcaniz J, Winkler L, Dahlmann M, et al. , Clinically relevant glioblastoma patient-derived xenograft models to guide drug development and identify molecular signatures. Front Oncol, 2023. 13: p. 1129627. 10.3389/fonc.2023.1129627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andersen BM, Faust Akl C, Wheeler MA, et al. , Glial and myeloid heterogeneity in the brain tumour microenvironment. Nat Rev Cancer, 2021. 21(12): p. 786–802. 10.1038/s41568-021-00397-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anido J, Saez-Borderias A, Gonzalez-Junca A, et al. , TGF-beta Receptor Inhibitors Target the CD44(high)/Id1(high) Glioma-Initiating Cell Population in Human Glioblastoma. Cancer Cell, 2010. 18(6): p. 655–68. 10.1016/j.ccr.2010.10.023 [DOI] [PubMed] [Google Scholar]
- 8.Arabzade A, Zhao Y, Varadharajan S, et al. , ZFTA-RELA Dictates Oncogenic Transcriptional Programs to Drive Aggressive Supratentorial Ependymoma. Cancer Discov, 2021. 11(9): p. 2200–2215. 10.1158/2159-8290.CD-20-1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bassil R, Shields K, Granger K, et al. , Improved modeling of human AD with an automated culturing platform for iPSC neurons, astrocytes and microglia. Nat Commun, 2021. 12(1): p. 5220. 10.1038/s41467-021-25344-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bausart M, Preat V, and Malfanti A, Immunotherapy for glioblastoma: the promise of combination strategies. J Exp Clin Cancer Res, 2022. 41(1): p. 35. 10.1186/s13046-022-02251-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bian S, Repic M, Guo Z, et al. , Genetically engineered cerebral organoids model brain tumor formation. Nat Methods, 2018. 15(8): p. 631–639. 10.1038/s41592-018-0070-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Binello E, Qadeer ZA, Kothari HP, et al. , Stemness of the CT-2A Immunocompetent Mouse Brain Tumor Model: Characterization In Vitro. J Cancer, 2012. 3: p. 166–74. 10.7150/jca.4149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bodmer S, Strommer K, Frei K, et al. , Immunosuppression and transforming growth factor-beta in glioblastoma. Preferential production of transforming growth factor-beta 2. J Immunol, 1989. 143(10): p. 3222–9. [PubMed] [Google Scholar]
- 14.Bolli E, Scherger M, Arnouk SM, et al. , Targeted Repolarization of Tumor-Associated Macrophages via Imidazoquinoline-Linked Nanobodies. Adv Sci (Weinh), 2021. 8(10): p. 2004574. 10.1002/advs.202004574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brandao M, Simon T, Critchley G, et al. , Astrocytes, the rising stars of the glioblastoma microenvironment. Glia, 2019. 67(5): p. 779–790. 10.1002/glia.23520 [DOI] [PubMed] [Google Scholar]
- 16.Bruns J, Egan T, Mercier P, et al. , Glioblastoma spheroid growth and chemotherapeutic responses in single and dual-stiffness hydrogels. Acta Biomater, 2023. 163: p. 400–414. 10.1016/j.actbio.2022.05.048 [DOI] [PubMed] [Google Scholar]
- 17.Calori IR, Alves SR, Bi H, et al. , Type-I Collagen/Collagenase Modulates the 3D Structure and Behavior of Glioblastoma Spheroid Models. ACS Appl Bio Mater, 2022. 5(2): p. 723–733. 10.1021/acsabm.1c01138 [DOI] [PubMed] [Google Scholar]
- 18.Cancer Genome Atlas Research, N., Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature, 2008. 455(7216): p. 1061–8. 10.1038/nature07385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carlson JC, Cantu Gutierrez M, Lozzi B, et al. , Identification of diverse tumor endothelial cell populations in malignant glioma. Neuro Oncol, 2021. 23(6): p. 932–944. 10.1093/neuonc/noaa297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheema TA, Wakimoto H, Fecci PE, et al. , Multifaceted oncolytic virus therapy for glioblastoma in an immunocompetent cancer stem cell model. Proc Natl Acad Sci U S A, 2013. 110(29): p. 12006–11. 10.1073/pnas.1307935110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen D, Varanasi SK, Hara T, et al. , CTLA-4 blockade induces a microglia-Th1 cell partnership that stimulates microglia phagocytosis and anti-tumor function in glioblastoma. Immunity, 2023. 56(9): p. 2086–2104 e8. 10.1016/j.immuni.2023.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen F and LoTurco J, A method for stable transgenesis of radial glia lineage in rat neocortex by piggyBac mediated transposition. J Neurosci Methods, 2012. 207(2): p. 172–80. 10.1016/j.jneumeth.2012.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen ML, Pittet MJ, Gorelik L, et al. , Regulatory T cells suppress tumor-specific CD8 T cell cytotoxicity through TGF-beta signals in vivo. Proc Natl Acad Sci U S A, 2005. 102(2): p. 419–24. 10.1073/pnas.0408197102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen S, Lai SWT, Brown CE, et al. , Harnessing and Enhancing Macrophage Phagocytosis for Cancer Therapy. Front Immunol, 2021. 12: p. 635173. 10.3389/fimmu.2021.635173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiavari M, Ciotti GMP, Navarra P, et al. , Pro-Inflammatory Activation of A New Immortalized Human Microglia Cell Line. Brain Sci, 2019. 9(5). 10.3390/brainsci9050111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Civita P, M.L. D, and Pilkington GJ, Pre-Clinical Drug Testing in 2D and 3D Human In Vitro Models of Glioblastoma Incorporating Non-Neoplastic Astrocytes: Tunneling Nano Tubules and Mitochondrial Transfer Modulates Cell Behavior and Therapeutic Respons. Int J Mol Sci, 2019. 20(23). 10.3390/ijms20236017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coniglio S, Miller I, Symons M, et al. , Coculture Assays to Study Macrophage and Microglia Stimulation of Glioblastoma Invasion. J Vis Exp, 2016(116). 10.3791/53990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cornelison RC, Brennan CE, Kingsmore KM, et al. , Convective forces increase CXCR4-dependent glioblastoma cell invasion in GL261 murine model. Sci Rep, 2018. 8(1): p. 17057. 10.1038/s41598-018-35141-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cornelison RC, Yuan JX, Tate KM, et al. , A patient-designed tissue-engineered model of the infiltrative glioblastoma microenvironment. NPJ Precis Oncol, 2022. 6(1): p. 54. 10.1038/s41698-022-00290-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Costa B, Eisemann T, Strelau J, et al. , Intratumoral platelet aggregate formation in a murine preclinical glioma model depends on podoplanin expression on tumor cells. Blood Adv, 2019. 3(7): p. 1092–1102. 10.1182/bloodadvances.2018015966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cui X, Wang Q, Zhou J, et al. , Single-Cell Transcriptomics of Glioblastoma Reveals a Unique Tumor Microenvironment and Potential Immunotherapeutic Target Against Tumor-Associated Macrophage. Front Oncol, 2021. 11: p. 710695. 10.3389/fonc.2021.710695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.D’Alessio A, Proietti G, Sica G, et al. , Pathological and Molecular Features of Glioblastoma and Its Peritumoral Tissue. Cancers (Basel), 2019. 11(4). 10.3390/cancers11040469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Decotret LR, Shi R, Thomas KN, et al. , Development and validation of an advanced ex vivo brain slice invasion assay to model glioblastoma cell invasion into the complex brain microenvironment. Front Oncol, 2023. 13: p. 976945. 10.3389/fonc.2023.976945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Douvaras P, Sun B, Wang M, et al. , Directed Differentiation of Human Pluripotent Stem Cells to Microglia. Stem Cell Reports, 2017. 8(6): p. 1516–1524. 10.1016/j.stemcr.2017.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duval K, Grover H, Han LH, et al. , Modeling Physiological Events in 2D vs. 3D Cell Culture. Physiology (Bethesda), 2017. 32(4): p. 266–277. 10.1152/physiol.00036.2016 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 36.Eisemann T, Costa B, Strelau J, et al. , An advanced glioma cell invasion assay based on organotypic brain slice cultures. BMC Cancer, 2018. 18(1): p. 103. 10.1186/s12885-018-4007-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Engblom C, Pfirschke C, and Pittet MJ, The role of myeloid cells in cancer therapies. Nat Rev Cancer, 2016. 16(7): p. 447–62. 10.1038/nrc.2016.54 [DOI] [PubMed] [Google Scholar]
- 38.Eramo A, Ricci-Vitiani L, Zeuner A, et al. , Chemotherapy resistance of glioblastoma stem cells. Cell Death Differ, 2006. 13(7): p. 1238–41. 10.1038/sj.cdd.4401872 [DOI] [PubMed] [Google Scholar]
- 39.Fedorova V, Pospisilova V, Vanova T, et al. , Glioblastoma and cerebral organoids: development and analysis of an in vitro model for glioblastoma migration. Mol Oncol, 2023. 17(4): p. 647–663. 10.1002/1878-0261.13389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feng SW, Wu ZS, Chiu YL, et al. , Exploring the Functional Roles of Telomere Maintenance 2 in the Tumorigenesis of Glioblastoma Multiforme and Drug Responsiveness to Temozolomide. Int J Mol Sci, 2023. 24(11). 10.3390/ijms24119256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Filley AC, Henriquez M, and Dey M, Recurrent glioma clinical trial, CheckMate-143: the game is not over yet. Oncotarget, 2017. 8(53): p. 91779–91794. 10.18632/oncotarget.21586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flores C, Pham C, Snyder D, et al. , Novel role of hematopoietic stem cells in immunologic rejection of malignant gliomas. Oncoimmunology, 2015. 4(3): p. e994374. 10.4161/2162402X.2014.994374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Genoud V, Marinari E, Nikolaev SI, et al. , Responsiveness to anti-PD-1 and anti-CTLA-4 immune checkpoint blockade in SB28 and GL261 mouse glioma models. Oncoimmunology, 2018. 7(12): p. e1501137. 10.1080/2162402X.2018.1501137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Geribaldi-Doldan N, Fernandez-Ponce C, Quiroz RN, et al. , The Role of Microglia in Glioblastoma. Front Oncol, 2020. 10: p. 603495. 10.3389/fonc.2020.603495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Glasgow SM, Zhu W, Stolt CC, et al. , Mutual antagonism between Sox10 and NFIA regulates diversification of glial lineages and glioma subtypes. Nat Neurosci, 2014. 17(10): p. 1322–9. 10.1038/nn.3790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gomez-Oliva R, Dominguez-Garcia S, Carrascal L, et al. , Evolution of Experimental Models in the Study of Glioblastoma: Toward Finding Efficient Treatments. Front Oncol, 2020. 10: p. 614295. 10.3389/fonc.2020.614295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goudarzi S, Rivera A, Butt AM, et al. , Gas6 Promotes Oligodendrogenesis and Myelination in the Adult Central Nervous System and After Lysolecithin-Induced Demyelination. ASN Neuro, 2016. 8(5). 10.1177/1759091416668430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guillot A and Tacke F, Liver Macrophages: Old Dogmas and New Insights. Hepatol Commun, 2019. 3(6): p. 730–743. 10.1002/hep4.1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guyon J, Strale PO, Romero-Garmendia I, et al. , Co-culture of Glioblastoma Stem-like Cells on Patterned Neurons to Study Migration and Cellular Interactions. J Vis Exp, 2021(168). 10.3791/62213 [DOI] [PubMed] [Google Scholar]
- 50.Haddad AF, Young JS, Amara D, et al. , Mouse models of glioblastoma for the evaluation of novel therapeutic strategies. Neurooncol Adv, 2021. 3(1): p. vdab100. 10.1093/noajnl/vdab100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hambardzumyan D, Parada LF, Holland EC, et al. , Genetic modeling of gliomas in mice: new tools to tackle old problems. Glia, 2011. 59(8): p. 1155–68. 10.1002/glia.21142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hara T, Chanoch-Myers R, Mathewson ND, et al. , Interactions between cancer cells and immune cells drive transitions to mesenchymal-like states in glioblastoma. Cancer Cell, 2021. 39(6): p. 779–792 e11. 10.1016/j.ccell.2021.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Henrik Heiland D, Ravi VM, Behringer SP, et al. , Tumor-associated reactive astrocytes aid the evolution of immunosuppressive environment in glioblastoma. Nat Commun, 2019. 10(1): p. 2541. 10.1038/s41467-019-10493-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hess KR, Broglio KR, and Bondy ML, Adult glioma incidence trends in the United States, 1977-2000. Cancer, 2004. 101(10): p. 2293–9. 10.1002/cncr.20621 [DOI] [PubMed] [Google Scholar]
- 55.Hide T, Komohara Y, Miyasato Y, et al. , Oligodendrocyte Progenitor Cells and Macrophages/Microglia Produce Glioma Stem Cell Niches at the Tumor Border. EBioMedicine, 2018. 30: p. 94–104. 10.1016/j.ebiom.2018.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hornschemeyer J, Kirschstein T, Reichart G, et al. , Studies on Biological and Molecular Effects of Small-Molecule Kinase Inhibitors on Human Glioblastoma Cells and Organotypic Brain Slices. Life (Basel), 2022. 12(8). 10.3390/life12081258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hsu SPC, Chen YC, Chiang HC, et al. , Rapamycin and hydroxychloroquine combination alters macrophage polarization and sensitizes glioblastoma to immune checkpoint inhibitors. J Neurooncol, 2020. 146(3): p. 417–426. 10.1007/s11060-019-03360-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Humpel C, Organotypic brain slice cultures: A review. Neuroscience, 2015. 305: p. 86–98. 10.1016/j.neuroscience.2015.07.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huse JT and Holland EC, Genetically engineered mouse models of brain cancer and the promise of preclinical testing. Brain Pathol, 2009. 19(1): p. 132–43. 10.1111/j.1750-3639.2008.00234.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hutter G, Theruvath J, Graef CM, et al. , Microglia are effector cells of CD47-SIRPalpha antiphagocytic axis disruption against glioblastoma. Proc Natl Acad Sci U S A, 2019. 116(3): p. 997–1006. 10.1073/pnas.1721434116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jacob F, Salinas RD, Zhang DY, et al. , A Patient-Derived Glioblastoma Organoid Model and Biobank Recapitulates Inter- and Intra-tumoral Heterogeneity. Cell, 2020. 180(1): p. 188–204 e22. 10.1016/j.cell.2019.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jensen SS, Meyer M, Petterson SA, et al. , Establishment and Characterization of a Tumor Stem Cell-Based Glioblastoma Invasion Model. PLoS One, 2016. 11(7): p. e0159746. 10.1371/journal.pone.0159746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jimenez-Dinamarca I, Reyes-Lizana R, Lemunao-Inostroza Y, et al. , GABAergic Regulation of Astroglial Gliotransmission through Cx43 Hemichannels. Int J Mol Sci, 2022. 23(21). 10.3390/ijms232113625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jin MZ, Han RR, Qiu GZ, et al. , Organoids: An intermediate modeling platform in precision oncology. Cancer Lett, 2018. 414: p. 174–180. 10.1016/j.canlet.2017.11.021 [DOI] [PubMed] [Google Scholar]
- 65.Joseph JV, Magaut CR, Storevik S, et al. , TGF-beta promotes microtube formation in glioblastoma through thrombospondin 1. Neuro Oncol, 2022. 24(4): p. 541–553. 10.1093/neuonc/noab212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kapalczynska M, Kolenda T, Przybyla W, et al. , 2D and 3D cell cultures - a comparison of different types of cancer cell cultures. Arch Med Sci, 2018. 14(4): p. 910–919. 10.5114/aoms.2016.63743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kawashima T, Yashiro M, Kasashima H, et al. , Oligodendrocytes Up-regulate the Invasive Activity of Glioblastoma Cells via the Angiopoietin-2 Signaling Pathway. Anticancer Res, 2019. 39(2): p. 577–584. 10.21873/anticanres.13150 [DOI] [PubMed] [Google Scholar]
- 68.Kerstetter-Fogle AE, Harris PLR, Brady-Kalnay SM, et al. , Generation of Glioblastoma Patient-Derived Intracranial Xenografts for Preclinical Studies. Int J Mol Sci, 2020. 21(14). 10.3390/ijms21145113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Khalsa JK, Cheng N, Keegan J, et al. , Immune phenotyping of diverse syngeneic murine brain tumors identifies immunologically distinct types. Nat Commun, 2020. 11(1): p. 3912. 10.1038/s41467-020-17704-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Khan MI, Jeong ES, Khan MZ, et al. , Stem cells-derived exosomes alleviate neurodegeneration and Alzheimer's pathogenesis by ameliorating neuroinflamation, and regulating the associated molecular pathways. Sci Rep, 2023. 13(1): p. 15731. 10.1038/s41598-023-42485-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim GB, Rincon Fernandez Pacheco D, Saxon D, et al. , Rapid Generation of Somatic Mouse Mosaics with Locus-Specific, Stably Integrated Transgenic Elements. Cell, 2019. 179(1): p. 251–267 e24. 10.1016/j.cell.2019.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kingsmore KM, Logsdon DK, Floyd DH, et al. , Interstitial flow differentially increases patient-derived glioblastoma stem cell invasion via CXCR4, CXCL12, and CD44-mediated mechanisms. Integr Biol (Camb), 2016. 8(12): p. 1246–1260. 10.1039/c6ib00167j [DOI] [PubMed] [Google Scholar]
- 73.Lancaster MA, Renner M, Martin CA, et al. , Cerebral organoids model human brain development and microcephaly. Nature, 2013. 501(7467): p. 373–9. 10.1038/nature12517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee J, Kotliarova S, Kotliarov Y, et al. , Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell, 2006. 9(5): p. 391–403. 10.1016/j.ccr.2006.03.030 [DOI] [PubMed] [Google Scholar]
- 75.Leite DM, Zvar Baskovic B, Civita P, et al. , A human co-culture cell model incorporating microglia supports glioblastoma growth and migration, and confers resistance to cytotoxics. FASEB J, 2020. 34(1): p. 1710–1727. 10.1096/fj.201901858RR [DOI] [PubMed] [Google Scholar]
- 76.Lenting K, Verhaak R, Ter Laan M, et al. , Glioma: experimental models and reality. Acta Neuropathol, 2017. 133(2): p. 263–282. 10.1007/s00401-017-1671-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Letchuman V, Ampie L, Shah AH, et al. , Syngeneic murine glioblastoma models: reactionary immune changes and immunotherapy intervention outcomes. Neurosurg Focus, 2022. 52(2): p. E5. 10.3171/2021.11.FOCUS21556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li C, Xu X, Wei S, et al. , Tumor-associated macrophages: potential therapeutic strategies and future prospects in cancer. J Immunother Cancer, 2021. 9(1). 10.1136/jitc-2020-001341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li X, Guo X, Ling J, et al. , Nanomedicine-based cancer immunotherapies developed by reprogramming tumor-associated macrophages. Nanoscale, 2021. 13(9): p. 4705–4727. 10.1039/d0nr08050k [DOI] [PubMed] [Google Scholar]
- 80.Lim M, Xia Y, Bettegowda C, et al. , Current state of immunotherapy for glioblastoma. Nat Rev Clin Oncol, 2018. 15(7): p. 422–442. 10.1038/s41571-018-0003-5 [DOI] [PubMed] [Google Scholar]
- 81.Linkous A, Balamatsias D, Snuderl M, et al. , Modeling Patient-Derived Glioblastoma with Cerebral Organoids. Cell Rep, 2019. 26(12): p. 3203–3211 e5. 10.1016/j.celrep.2019.02.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu P, Griffiths S, Veljanoski D, et al. , Preclinical models of glioblastoma: limitations of current models and the promise of new developments. Expert Rev Mol Med, 2021. 23: p. e20. 10.1017/erm.2021.20 [DOI] [PubMed] [Google Scholar]
- 83.Loras A, Gonzalez-Bonet LG, Gutierrez-Arroyo JL, et al. , Neural Stem Cells as Potential Glioblastoma Cells of Origin. Life (Basel), 2023. 13(4). 10.3390/life13040905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lu Y, Jiang F, Zheng X, et al. , TGF-beta1 promotes motility and invasiveness of glioma cells through activation of ADAM17. Oncol Rep, 2011. 25(5): p. 1329–35. 10.3892/or.2011.1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Maas RR, Soukup K, Fournier N, et al. , The local microenvironment drives activation of neutrophils in human brain tumors. Cell, 2023. 10.1016/j.cell.2023.08.043 [DOI] [PubMed] [Google Scholar]
- 86.Marques-Torrejon MA, Gangoso E, and Pollard SM, Modelling glioblastoma tumour-host cell interactions using adult brain organotypic slice co-culture. Dis Model Mech, 2018. 11(2). 10.1242/dmm.031435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Martinez-Murillo R and Martinez A, Standardization of an orthotopic mouse brain tumor model following transplantation of CT-2A astrocytoma cells. Histol Histopathol, 2007. 22(12): p. 1309–26. 10.14670/HH-22.1309 [DOI] [PubMed] [Google Scholar]
- 88.Marumoto T, Tashiro A, Friedmann-Morvinski D, et al. , Development of a novel mouse glioma model using lentiviral vectors. Nat Med, 2009. 15(1): p. 110–6. 10.1038/nm.1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McCarthy RC, Lu DY, Alkhateeb A, et al. , Characterization of a novel adult murine immortalized microglial cell line and its activation by amyloid-beta. J Neuroinflammation, 2016. 13: p. 21. 10.1186/s12974-016-0484-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Melissaridou S, Wiechec E, Magan M, et al. , The effect of 2D and 3D cell cultures on treatment response, EMT profile and stem cell features in head and neck cancer. Cancer Cell Int, 2019. 19: p. 16. 10.1186/s12935-019-0733-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Merz F, Gaunitz F, Dehghani F, et al. , Organotypic slice cultures of human glioblastoma reveal different susceptibilities to treatments. Neuro Oncol, 2013. 15(6): p. 670–81. 10.1093/neuonc/not003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Merzak A, McCrea S, Koocheckpour S, et al. , Control of human glioma cell growth, migration and invasion in vitro by transforming growth factor beta 1. Br J Cancer, 1994. 70(2): p. 199–203. 10.1038/bjc.1994.280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Minami N, Maeda Y, Shibao S, et al. , Organotypic brain explant culture as a drug evaluation system for malignant brain tumors. Cancer Med, 2017. 6(11): p. 2635–2645. 10.1002/cam4.1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Munson JM, Bellamkonda RV, and Swartz MA, Interstitial flow in a 3D microenvironment increases glioma invasion by a CXCR4-dependent mechanism. Cancer Res, 2013. 73(5): p. 1536–46. 10.1158/0008-5472.CAN-12-2838 [DOI] [PubMed] [Google Scholar]
- 95.Nagamoto-Combs K, Kulas J, and Combs CK, A novel cell line from spontaneously immortalized murine microglia. J Neurosci Methods, 2014. 233: p. 187–98. 10.1016/j.jneumeth.2014.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Narvaez-Perez LF, Paz-Bermudez F, Avalos-Fuentes JA, et al. , CRISPR/sgRNA-directed synergistic activation mediator (SAM) as a therapeutic tool for Parkinson s disease. Gene Ther, 2023. 10.1038/s41434-023-00414-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nishida-Aoki N and Gujral TS, Polypharmacologic Reprogramming of Tumor-Associated Macrophages toward an Inflammatory Phenotype. Cancer Res, 2022. 82(3): p. 433–446. 10.1158/0008-5472.CAN-21-1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Noffsinger B, Witter A, Sheybani N, et al. , Technical choices significantly alter the adaptive immune response against immunocompetent murine gliomas in a model-dependent manner. J Neurooncol, 2021. 154(2): p. 145–157. 10.1007/s11060-021-03822-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ogawa J, Pao GM, Shokhirev MN, et al. , Glioblastoma Model Using Human Cerebral Organoids. Cell Rep, 2018. 23(4): p. 1220–1229. 10.1016/j.celrep.2018.03.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Okonogi N, Suzuki Y, Sato H, et al. , Combination Therapy of Intravenously Injected Microglia and Radiation Therapy Prolongs Survival in a Rat Model of Spontaneous Malignant Glioma. Int J Radiat Oncol Biol Phys, 2018. 102(3): p. 601–608. 10.1016/j.ijrobp.2018.06.018 [DOI] [PubMed] [Google Scholar]
- 101.Olsson M, Hultman K, Dunoyer-Geindre S, et al. , Epigenetic Regulation of Tissue-Type Plasminogen Activator in Human Brain Tissue and Brain-Derived Cells. Gene Regul Syst Bio, 2016. 10: p. 9–13. 10.4137/GRSB.S30241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Osswald M, Jung E, Sahm F, et al. , Brain tumour cells interconnect to a functional and resistant network. Nature, 2015. 528(7580): p. 93–8. 10.1038/nature16071 [DOI] [PubMed] [Google Scholar]
- 103.Ott M, Prins RM, and Heimberger AB, The immune landscape of common CNS malignancies: implications for immunotherapy. Nat Rev Clin Oncol, 2021. 18(11): p. 729–744. 10.1038/s41571-021-00518-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ou A, Yung WKA, and Majd N, Molecular Mechanisms of Treatment Resistance in Glioblastoma. Int J Mol Sci, 2020. 22(1). 10.3390/ijms22010351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Oyarce C, Vizcaino-Castro A, Chen S, et al. , Re-polarization of immunosuppressive macrophages to tumor-cytotoxic macrophages by repurposed metabolic drugs. Oncoimmunology, 2021. 10(1): p. 1898753. 10.1080/2162402X.2021.1898753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Padua D and Massague J, Roles of TGFbeta in metastasis. Cell Res, 2009. 19(1): p. 89–102. 10.1038/cr.2008.316 [DOI] [PubMed] [Google Scholar]
- 107.Pan Y, Yu Y, Wang X, et al. , Tumor-Associated Macrophages in Tumor Immunity. Front Immunol, 2020. 11: p. 583084. 10.3389/fimmu.2020.583084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Paolillo M, Comincini S, and Schinelli S, In Vitro Glioblastoma Models: A Journey into the Third Dimension. Cancers (Basel), 2021. 13(10). 10.3390/cancers13102449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Parker JJ, Lizarraga M, Waziri A, et al. , A Human Glioblastoma Organotypic Slice Culture Model for Study of Tumor Cell Migration and Patient-specific Effects of Anti-Invasive Drugs. J Vis Exp, 2017(125). 10.3791/53557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pencheva N, de Gooijer MC, Vis DJ, et al. , Identification of a Druggable Pathway Controlling Glioblastoma Invasiveness. Cell Rep, 2017. 20(1): p. 48–60. 10.1016/j.celrep.2017.06.036 [DOI] [PubMed] [Google Scholar]
- 111.Prinz M, Erny D, and Hagemeyer N, Ontogeny and homeostasis of CNS myeloid cells. Nat Immunol, 2017. 18(4): p. 385–392. 10.1038/ni.3703 [DOI] [PubMed] [Google Scholar]
- 112.Pustchi SE, Avci NG, Akay YM, et al. , Astrocytes Decreased the Sensitivity of Glioblastoma Cells to Temozolomide and Bay 11-7082. Int J Mol Sci, 2020. 21(19). 10.3390/ijms21197154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Quereda V, Hou S, Madoux F, et al. , A Cytotoxic Three-Dimensional-Spheroid, High-Throughput Assay Using Patient-Derived Glioma Stem Cells. SLAS Discov, 2018. 23(8): p. 842–849. 10.1177/2472555218775055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ravi VM, Joseph K, Wurm J, et al. , Human organotypic brain slice culture: a novel framework for environmental research in neuro-oncology. Life Sci Alliance, 2019. 2(4). 10.26508/lsa.201900305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ravi VM, Neidert N, Will P, et al. , T-cell dysfunction in the glioblastoma microenvironment is mediated by myeloid cells releasing interleukin-10. Nat Commun, 2022. 13(1): p. 925. 10.1038/s41467-022-28523-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Reilly KM, Loisel DA, Bronson RT, et al. , Nf1;Trp53 mutant mice develop glioblastoma with evidence of strain-specific effects. Nat Genet, 2000. 26(1): p. 109–13. 10.1038/79075 [DOI] [PubMed] [Google Scholar]
- 117.Saha D, Martuza RL, and Rabkin SD, Macrophage Polarization Contributes to Glioblastoma Eradication by Combination Immunovirotherapy and Immune Checkpoint Blockade. Cancer Cell, 2017. 32(2): p. 253–267 e5. 10.1016/j.ccell.2017.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sampson JH, Gunn MD, Fecci PE, et al. , Brain immunology and immunotherapy in brain tumours. Nat Rev Cancer, 2020. 20(1): p. 12–25. 10.1038/s41568-019-0224-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sarkar S, Yang R, Mirzaei R, et al. , Control of brain tumor growth by reactivating myeloid cells with niacin. Sci Transl Med, 2020. 12(537). 10.1126/scitranslmed.aay9924 [DOI] [PubMed] [Google Scholar]
- 120.Scobie MR, Abood A, and Rice CD, Differential Transcriptome Responses in Human THP-1 Macrophages Following Exposure to T98G and LN-18 Human Glioblastoma Secretions: A Simplified Bioinformatics Approach to Understanding Patient-Glioma-Specific Effects on Tumor-Associated Macrophages. Int J Mol Sci, 2023. 24(6). 10.3390/ijms24065115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Setia H and Muotri AR, Brain organoids as a model system for human neurodevelopment and disease. Semin Cell Dev Biol, 2019. 95: p. 93–97. 10.1016/j.semcdb.2019.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Seyfried TN, el-Abbadi M, and Roy ML, Ganglioside distribution in murine neural tumors. Mol Chem Neuropathol, 1992. 17(2): p. 147–67. 10.1007/BF03159989 [DOI] [PubMed] [Google Scholar]
- 123.Shahab SW, Roggeveen CM, Sun J, et al. , The LIN28B-let-7-PBK pathway is essential for group 3 medulloblastoma tumor growth and survival. Mol Oncol, 2023. 17(9): p. 1784–1802. 10.1002/1878-0261.13477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shi Y, He X, Wang H, et al. , Construction of a novel blood brain barrier-glioma microfluidic chip model: Applications in the evaluation of permeability and anti-glioma activity of traditional Chinese medicine components. Talanta, 2023. 253: p. 123971. 10.1016/j.talanta.2022.123971 [DOI] [PubMed] [Google Scholar]
- 125.Srivastava S, Jackson C, Kim T, et al. , A Characterization of Dendritic Cells and Their Role in Immunotherapy in Glioblastoma: From Preclinical Studies to Clinical Trials. Cancers (Basel), 2019. 11(4). 10.3390/cancers11040537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Stessin AM, Clausi MG, Zhao Z, et al. , Repolarized macrophages, induced by intermediate stereotactic dose radiotherapy and immune checkpoint blockade, contribute to long-term survival in glioma-bearing mice. J Neurooncol, 2020. 147(3): p. 547–555. 10.1007/s11060-020-03459-y [DOI] [PubMed] [Google Scholar]
- 127.Straehla JP, Hajal C, Safford HC, et al. , A predictive microfluidic model of human glioblastoma to assess trafficking of blood-brain barrier-penetrant nanoparticles. Proc Natl Acad Sci U S A, 2022. 119(23): p. e2118697119. 10.1073/pnas.2118697119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Stupp R, Mason WP, van den Bent MJ, et al. , Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med, 2005. 352(10): p. 987–96. 10.1056/NEJMoa043330 [DOI] [PubMed] [Google Scholar]
- 129.Szatmari T, Lumniczky K, Desaknai S, et al. , Detailed characterization of the mouse glioma 261 tumor model for experimental glioblastoma therapy. Cancer Sci, 2006. 97(6): p. 546–53. 10.1111/j.1349-7006.2006.00208.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Takahashi T, Organoids for Drug Discovery and Personalized Medicine. Annu Rev Pharmacol Toxicol, 2019. 59: p. 447–462. 10.1146/annurev-pharmtox-010818-021108 [DOI] [PubMed] [Google Scholar]
- 131.Torsvik A, Stieber D, Enger PO, et al. , U-251 revisited: genetic drift and phenotypic consequences of long-term cultures of glioblastoma cells. Cancer Med, 2014. 3(4): p. 812–24. 10.1002/cam4.219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Uhl M, Aulwurm S, Wischhusen J, et al. , SD-208, a novel transforming growth factor beta receptor I kinase inhibitor, inhibits growth and invasiveness and enhances immunogenicity of murine and human glioma cells in vitro and in vivo. Cancer Res, 2004. 64(21): p. 7954–61. 10.1158/0008-5472.CAN-04-1013 [DOI] [PubMed] [Google Scholar]
- 133.van Asperen JV, van Bodegraven EJ, Robe P, et al. , Determining glioma cell invasion and proliferation in ex vivo organotypic mouse brain slices using whole-mount immunostaining and tissue clearing. STAR Protoc, 2022. 3(4): p. 101703. 10.1016/j.xpro.2022.101703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Vaubel RA, Tian S, Remonde D, et al. , Genomic and Phenotypic Characterization of a Broad Panel of Patient-Derived Xenografts Reflects the Diversity of Glioblastoma. Clin Cancer Res, 2020. 26(5): p. 1094–1104. 10.1158/1078-0432.CCR-19-0909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Venkataramani V, Tanev DI, Strahle C, et al. , Glutamatergic synaptic input to glioma cells drives brain tumour progression. Nature, 2019. 573(7775): p. 532–538. 10.1038/s41586-019-1564-x [DOI] [PubMed] [Google Scholar]
- 136.Venkataramani V, Yang Y, Schubert MC, et al. , Glioblastoma hijacks neuronal mechanisms for brain invasion. Cell, 2022. 185(16): p. 2899–2917 e31. 10.1016/j.cell.2022.06.054 [DOI] [PubMed] [Google Scholar]
- 137.Venkatesh HS, Johung TB, Caretti V, et al. , Neuronal Activity Promotes Glioma Growth through Neuroligin-3 Secretion. Cell, 2015. 161(4): p. 803–16. 10.1016/j.cell.2015.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Venkatesh HS, Morishita W, Geraghty AC, et al. , Electrical and synaptic integration of glioma into neural circuits. Nature, 2019. 573(7775): p. 539–545. 10.1038/s41586-019-1563-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Watson DC, Bayik D, Storevik S, et al. , GAP43-dependent mitochondria transfer from astrocytes enhances glioblastoma tumorigenicity. Nat Cancer, 2023. 4(5): p. 648–664. 10.1038/s43018-023-00556-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wei J, Marisetty A, Schrand B, et al. , Osteopontin mediates glioblastoma-associated macrophage infiltration and is a potential therapeutic target. J Clin Invest, 2019. 129(1): p. 137–149. 10.1172/JCI121266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wei Z, Zhang X, Yong T, et al. , Boosting anti-PD-1 therapy with metformin-loaded macrophage-derived microparticles. Nat Commun, 2021. 12(1): p. 440. 10.1038/s41467-020-20723-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Woolf Z, Swanson MEV, Smyth LC, et al. , Single-cell image analysis reveals a protective role for microglia in glioblastoma. Neurooncol Adv, 2021. 3(1): p. vdab031. 10.1093/noajnl/vdab031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wouters R, Bevers S, Riva M, et al. , Immunocompetent Mouse Models in the Search for Effective Immunotherapy in Glioblastoma. Cancers (Basel), 2020. 13(1). 10.3390/cancers13010019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wu W, Klockow JL, Zhang M, et al. , Glioblastoma multiforme (GBM): An overview of current therapies and mechanisms of resistance. Pharmacol Res, 2021. 171: p. 105780. 10.1016/j.phrs.2021.105780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Xiang X, Wang J, Lu D, et al. , Targeting tumor-associated macrophages to synergize tumor immunotherapy. Signal Transduct Target Ther, 2021. 6(1): p. 75. 10.1038/s41392-021-00484-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Xiao A, Brenneman B, Floyd D, et al. , Statins affect human glioblastoma and other cancers through TGF-beta inhibition. Oncotarget, 2019. 10(18): p. 1716–1728. 10.18632/oncotarget.26733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Xie R, Kessler T, Grosch J, et al. , Tumor cell network integration in glioma represents a stemness feature. Neuro Oncol, 2021. 23(5): p. 757–769. 10.1093/neuonc/noaa275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Xie Z, Chen M, Lian J, et al. , Glioblastoma-on-a-chip construction and therapeutic applications. Front Oncol, 2023. 13: p. 1183059. 10.3389/fonc.2023.1183059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Xu C, Yuan X, Hou P, et al. , Development of glioblastoma organoids and their applications in personalized therapy. Cancer Biol Med, 2023. 20(5): p. 353–68. 10.20892/j.issn.2095-3941.2023.0061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Xu J, Zhang J, Zhang Z, et al. , Hypoxic glioma-derived exosomes promote M2-like macrophage polarization by enhancing autophagy induction. Cell Death Dis, 2021. 12(4): p. 373. 10.1038/s41419-021-03664-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Xue N, Zhou Q, Ji M, et al. , Chlorogenic acid inhibits glioblastoma growth through repolarizating macrophage from M2 to M1 phenotype. Sci Rep, 2017. 7: p. 39011. 10.1038/srep39011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Yang YK, Ogando CR, Wang See C, et al. , Changes in phenotype and differentiation potential of human mesenchymal stem cells aging in vitro. Stem Cell Res Ther, 2018. 9(1): p. 131. 10.1186/s13287-018-0876-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yao M, Ventura PB, Jiang Y, et al. , Astrocytic trans-Differentiation Completes a Multicellular Paracrine Feedback Loop Required for Medulloblastoma Tumor Growth. Cell, 2020. 180(3): p. 502–520 e19. 10.1016/j.cell.2019.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ye XZ, Xu SL, Xin YH, et al. , Tumor-associated microglia/macrophages enhance the invasion of glioma stem-like cells via TGF-beta1 signaling pathway. J Immunol, 2012. 189(1): p. 444–53. 10.4049/jimmunol.1103248 [DOI] [PubMed] [Google Scholar]
- 155.Yi L, Zhou C, Wang B, et al. , Implantation of GL261 neurospheres into C57/BL6 mice: a more reliable syngeneic graft model for research on glioma-initiating cells. Int J Oncol, 2013. 43(2): p. 477–84. 10.3892/ijo.2013.1962 [DOI] [PubMed] [Google Scholar]
- 156.Yi Y, Hsieh IY, Huang X, et al. , Glioblastoma Stem-Like Cells: Characteristics, Microenvironment, and Therapy. Front Pharmacol, 2016. 7: p. 477. 10.3389/fphar.2016.00477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Yu K, Lin CJ, Hatcher A, et al. , PIK3CA variants selectively initiate brain hyperactivity during gliomagenesis. Nature, 2020. 578(7793): p. 166–171. 10.1038/s41586-020-1952-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhang H, Zhou Y, Cui B, et al. , Novel insights into astrocyte-mediated signaling of proliferation, invasion and tumor immune microenvironment in glioblastoma. Biomed Pharmacother, 2020. 126: p. 110086. 10.1016/j.biopha.2020.110086 [DOI] [PubMed] [Google Scholar]
- 159.Zhang L, Yu H, Yuan Y, et al. , The necessity for standardization of glioma stem cell culture: a systematic review. Stem Cell Res Ther, 2020. 11(1): p. 84. 10.1186/s13287-020-01589-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhang M, Hutter G, Kahn SA, et al. , Anti-CD47 Treatment Stimulates Phagocytosis of Glioblastoma by M1 and M2 Polarized Macrophages and Promotes M1 Polarized Macrophages In Vivo. PLoS One, 2016. 11(4): p. e0153550. 10.1371/journal.pone.0153550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zhou C, Zhang Y, Dai J, et al. , Pygo2 functions as a prognostic factor for glioma due to its up-regulation of H3K4me3 and promotion of MLL1/MLL2 complex recruitment. Sci Rep, 2016. 6: p. 22066. 10.1038/srep22066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zhou F, Shi Q, Fan X, et al. , Diverse Macrophages Constituted the Glioma Microenvironment and Influenced by PTEN Status. Front Immunol, 2022. 13: p. 841404. 10.3389/fimmu.2022.841404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zhou J, Tang Z, Gao S, et al. , Tumor-Associated Macrophages: Recent Insights and Therapies. Front Oncol, 2020. 10: p. 188. 10.3389/fonc.2020.00188 [DOI] [PMC free article] [PubMed] [Google Scholar]


