Abstract
Background and Objectives
The aim of this study was to identify novel biomarkers for multiple sclerosis (MS) diagnosis and prognosis, addressing the critical need for specific and prognostically valuable markers in the field.
Methods
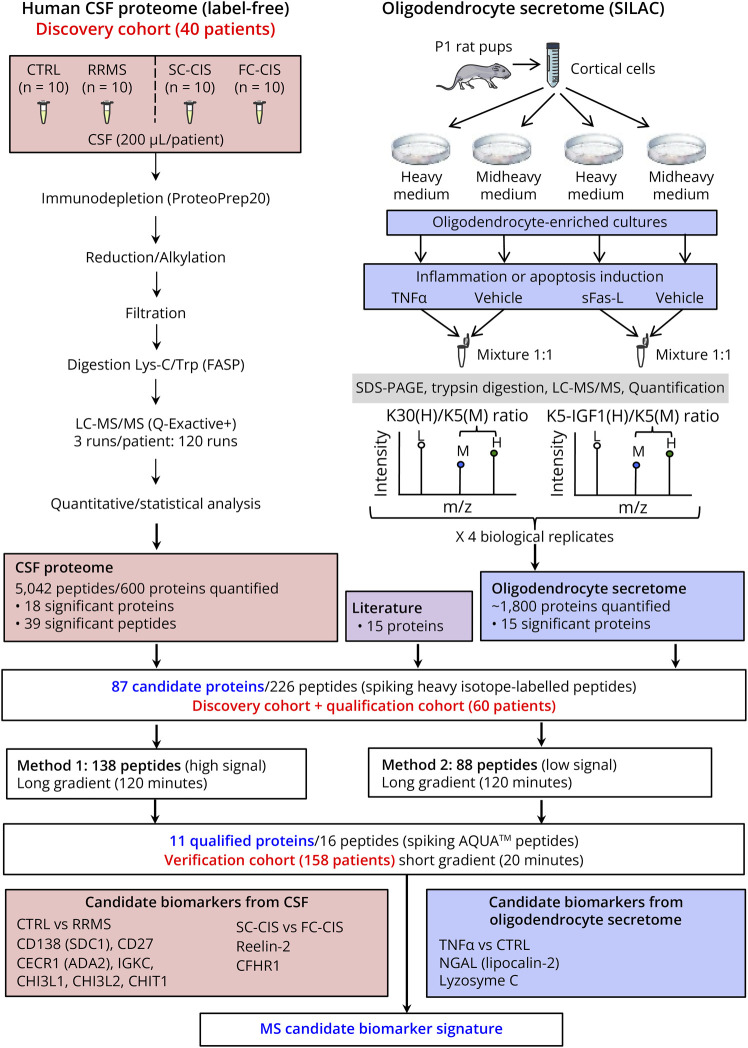
We conducted an extensive proteomic investigation, combining analysis of (1) CSF proteome from symptomatic controls, fast and slow converters after clinically isolated syndromes, and patients with relapsing-remitting MS (n = 10 per group) using label-free quantitative proteomics and (2) oligodendrocyte secretome changes under proinflammatory or proapoptotic conditions using stable isotope labeling by amino acids in cell culture. Proteins exhibiting differential abundance in both proteomic analyses were combined with other putative MS biomarkers, yielding a comprehensive list of 87 proteins that underwent quantification through parallel reaction monitoring (PRM) in a novel cohort, comprising symptomatic controls, inflammatory neurologic disease controls, and patients with MS at various disease stages (n = 10 per group). The 11 proteins that passed this qualification step were subjected to a new PRM assay within an expanded cohort comprising 158 patients with either MS at different disease stages or other inflammatory or noninflammatory neurologic disease controls.
Results
This study unveiled a promising biomarker signature for MS, including previously established candidates, such as chitinase 3-like protein 1, chitinase 3-like protein 2, chitotriosidase, immunoglobulin kappa chain region C, neutrophil gelatinase–associated lipocalin, and CD27. In addition, we identified novel markers, namely cat eye syndrome critical region protein 1 (adenosine deaminase 2, a therapeutic target in multiple sclerosis) and syndecan-1, a proteoglycan, also known as plasma cell surface marker CD138 and acting as chitinase 3–like protein 1 receptor implicated in inflammation and cancer signaling. CD138 exhibited good diagnostic accuracy in distinguishing MS from inflammatory neurologic disorders (area under the curve [AUC] = 0.85, CI 0.75–0.95). CD138 immunostaining was also observed in the brains of patients with MS and cultured oligodendrocyte precursor cells but was absent in astrocytes.
Discussion
These findings identify CD138 as a specific CSF biomarker for MS and suggest the selective activation of the chitinase 3–like protein 1/CD138 pathway within the oligodendrocyte lineage in MS. They offer promising prospects for improving MS diagnosis and prognosis by providing much-needed specificity and clinical utility.
Classification of Evidence
This study provides Class II evidence that CD138 distinguishes multiple sclerosis from other inflammatory neurologic disorders with an AUC of 0.85 (95% CI 0.75–0.95).
Introduction
Multiple sclerosis (MS) is an inflammatory autoimmune disease of the CNS that causes damage to myelin and ultimately leads to neurodegeneration. Diagnostic criteria recently evolved to better predict disease activity in patients experiencing a clinically isolated syndrome (CIS).1-3 However, the presence of active lesions in MRI and the presence of oligoclonal bands (OCBs) in the CSF only partially predict relapses and disability progression, underscoring the need for new prognostic biomarkers.4
Previous proteomic studies have identified several potential biomarkers of MS, such as chitinase-like proteins (CHI3L1 and CHI3L2) and chitotriosidase (CHIT1).5-8 However, their CSF levels are also elevated in other neurologic inflammatory and noninflammatory conditions9,10 and cannot be used for differential diagnosis of MS. More recently, neurofilament light (NfL) and heavy (NfH) chains were shown to be elevated both in the CSF and sera of patients with MS11,12 but, again, these biomarkers of neuronal/axonal damage are not specific for MS.13 Over the past years, the kappa free light chain (κFLC) index has emerged as a more specific candidate biomarker of the disease14,15 that is increasingly used in clinical practice.16-19 However, the differential diagnosis of MS remains challenging, given its common features with other neurologic inflammatory and noninflammatory conditions.20
In this study, to identify specific biomarkers of MS and/or prognostic biomarkers predicting disease evolution, we compared the CSF proteome from symptomatic controls (CTRL) and patients with relapsing-remitting multiple sclerosis (RRMS), and the CSF proteome from fast converters (FC) and slow converters (SC) after a CIS using quantitative proteomics. Moreover, to identify biomarkers reflecting oligodendrocyte damage observed in MS, we analyzed changes in the secretome of primary cultured rat oligodendrocyte precursor cells (OPCs) induced by a proinflammatory or a proapoptotic treatment to mimic the main pathologic features of the disease in vitro (Figure 1). We then complemented the resulting list of biomarker candidates with additional proteins previously involved in MS pathogenesis for validation by parallel reaction monitoring (PRM) in 2 novel cohorts comprising patients with MS at different stages of the disease and patients with inflammatory and noninflammatory diseases. This study revealed biomarker candidates that segregate MS from other inflammatory or noninflammatory neurologic disorders including the cell surface proteoglycan syndecan-1, also designated as plasma cell surface marker CD138. We then aimed at determining its accuracy to distinguish MS from other inflammatory neurologic disorders and its expression in MS brain.
Figure 1. Schematic Representation of the Workflow Used in the Study.
CTRL: symptomatic controls; CIS: clinically isolated syndrome; FC-CIS: fast conversion to MS (<1 year) after a CIS; SC-CIS: slow conversion to MS (>2 years) after CIS; RRMS: relapsing-remitting multiple sclerosis patients.
Methods
Patients
Patients were recruited in Montpellier and Nîmes University Hospitals before our study (between 2007 and 2014) and prospectively followed up for at least 2 years. We randomly included adult patients identified in our collection with a confirmed diagnosis of symptomatic control (CTRL), isolated optic neuritis (ION), CIS, RRMS, primary progressive MS (PPMS), inflammatory neurologic disease control (INDC), peripheral inflammatory neurologic disease control (PINDC), and noninflammatory neurologic disease control (NINDC). A schematic definition of CIS, RRMS, PPMS, and controls (CTRL, patients with ION, INDC, PINDC, and NINDC) is provided in eFigure 1. Conversion of CIS to RRMS was assessed using the McDonald criteria revised in 2005, after a second relapse or the apparition of new MRI lesions on follow-up scans.1 Patients with CIS with fast conversion to RRMS (<1 year after a CIS, FC-CIS) and patients with slow conversion to RRMS (>2 years after a CIS, SC-CIS) were included (eFigure 1). Demographics, CSF, and MRI characteristics of patients included in the different cohorts are described in Tables 1 and 2.
Table 1.
Characteristics of the Patients Included in the Discovery and Qualification Cohorts
| Cohort | Discovery | Qualification | ||||||||
| Diagnosis | CTRL | SC-CIS | FC-CIS | RRMS | CTRL | SC-CIS | FC-CIS | RRMS | PPMS | INDC |
| n | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 |
| Age (mean, y) | 35.7 | 37.6 | 35.3 | 36.1 | 33.4 | 34.1 | 31.6 | 34.2 | 47 | 34.9 |
| Sex (female/total ratio) | 70% | 70% | 70% | 70% | 80% | 80% | 80% | 80% | 70% | 60% |
| CSF protein level (mean, g/L) | 0.39 | 0.39 | 0.42 | 0.41 | 0.35 | 0.34 | 0.35 | 0.38 | 0.42 | 0.42 |
| Presence of OCBs | 0% | 70% | 90% | 80% | 0% | 80% | 90% | 90% | 90% | 20% |
| Elevated IgG index | 0% | 60% | 90% | 70% | 0% | 60% | 80% | 40% | 70% | 10% |
| IgG index (mean) | 0.46 | 0.87 | 1.37 | 0.97 | 0.52 | 1.02 | 1.01 | 1.40 | 0.84 | 0.58 |
| Positive CSF | 0% | 70% | 90% | 80% | 0% | 80% | 90% | 90% | 90% | 20% |
| DIS (Barkhof) | NA | 50% | 80% | 90% | NA | 100% | 50% | 80% | 80% | NA |
| DIS (Swanton) | NA | 100% | 100% | 100% | NA | 100% | 100% | 100% | 100% | NA |
| Gadolinium enhancement | NA | 30% | 50% | 50% | NA | 10% | 30% | 30% | 20% | NA |
Abbreviations: CIS = clinically isolated syndrome; CTRL = symptomatic controls; FC-CIS = fast conversion to MS (<1 y) after a CIS; INDC = inflammatory neurologic disease controls; OCB = oligoclonal bands; PPMS = patients with progressive form of multiple sclerosis without period of symptom remission; RRMS = relapsing remitting multiple sclerosis; SC-CIS = slow conversion to MS (>2 y) after a CIS.
Sample characteristics for patients included in discovery and qualification cohorts. DIS: dissemination in space of MS lesions according to Barkhof or Swanton criteria. INDCs included patients with neurolupus syndrome (n = 1), neuro-Behçet syndrome (n = 1), Susac syndrome (n = 2), neurosarcoidosis (n = 1), Neuro-Sjögren syndrome (n = 1), aseptic meningitis (n = 2), acute demyelinating encephalomyelitis (ADEM, n = 1), and isolated myelitis (n = 1).
Table 2.
Characteristics of the Patients Included in the Verification Cohort
| Diagnosis | CTRL | SC-CIS | FC-CIS | RRMS | PPMS | INDC | ION | NINDC | PINDC |
| n | 30 | 15 | 15 | 30 | 14 | 13 | 15 | 13 | 13 |
| Age (mean, y) | 38.3 | 35.2 | 33.3 | 38.2 | 46.6a | 46.4 | 31.8 | 40.8 | 56.2b |
| Sex (female/total ratio) | 80% | 73% | 93% | 77% | 43% | 23%c | 87% | 46% | 54% |
| CSF protein level (mean, g/L) | 0.36 | 0.38 | 0.35 | 0.39 | 0.51 | 0.39 | 0.36 | 0.33 | 0.41 |
| Presence of OCBs | 0% | 80% | 93% | 90% | 79% | 15% | 7% | 0% | 15% |
| Elevated IgG index | 0% | 46% | 75% | 78% | 58% | 0% | 0% | 0% | 0% |
| IgG index (mean) | 0.48 | 0.68 | 0.86 | 1.12 | 0.84 | 0.47 | 0.47 | 0.45 | 0.48 |
| Positive CSF | 0% | 80% | 93% | 90% | 79% | 15% | 7% | 0% | 15% |
| DIS (Barkhof) | NA | 40% | 87% | 80% | 79% | NA | 0% | NA | NA |
| DIS (Swanton) | NA | 100% | 100% | 100% | 100% | NA | 0% | NA | NA |
| Gadolinium enhancement | NA | 27% | 47% | 47% | 14% | NA | 0% | NA | NA |
Abbreviations: CIS = clinically isolated syndrome; CTRL = symptomatic controls; DIS = dissemination in space of white matter lesions according to Barkhof or Swanton criteria; FC-CIS = fast conversion to MS (<1 y) after a CIS; INDC = inflammatory neurologic disease controls; ION = isolated optic neuritis; NINDC = noninflammatory neurologic disease controls; OCB = presence of oligoclonal bands in the CSF; PINDC = peripheral inflammatory neurologic disease controls; PPMS = patients with progressive form of multiple sclerosis without period of symptom remission; RRMS = relapsing-remitting multiple sclerosis; SC-CIS = slow conversion to MS (>2 y) after a CIS.
Sample characteristics for patients included in the verification cohort. INDCs included patients with encephalitis (n = 4), isolated myelitis (n = 3), acute demyelinating encephalomyelitis (ADEM, n = 2), chronic lymphocytic inflammation with pontine perivascular enhancement responsive to steroids (CLIPPERS, n = 1), aseptic meningitis (n = 1), cerebral toxoplasmosis (n = 1), and sinus inflammation–related optic neuritis (n = 1); PINDCs included patients with acute inflammatory demyelinating polyneuropathy (AIDP, n = 4), chronic inflammatory demyelinating polyneuropathy (CIDP, n = 4), polyneuritis (n = 2), plexopathy (n = 2), and multiple mononeuropathy (n = 1); NINDCs included patients with TIA with small vessel disease (n = 3), frontotemporal dementia (n = 2), stroke (n = 1), generalized dystonia (n = 1), adrenoleukodystrophy (n = 1), cerebellar ataxia (n = 1), hydrocephaly (n = 1), syringomyelia (n = 1), sacral plexus compression (n = 1), and metabolic encephalitis (n = 1).
Cohorts were compared for age, sex, and CSF protein content (ANOVA followed by the Tukey-Kramer test) revealing asignificant age differences in PPMS vs FC-CIS, bsignificant age differences in PINDC vs CTRL, FC-CIS, SC-CIS, patients with RRMS, patients with ION, and NINDC, and csignificant sex ratio differences in INDC vs CTRLs, FC-CIS, patients with RRMS, and patients with ION. No differences in CSF protein content were found.
CSF samples were collected using a 25 G Whitacre-type atraumatic needle (ref 181.05, Vygon) in a 10-mL polypropylene tube (ref 62.610.201, Sarstedt) and processed according to the guidelines of the BioMS-eu network, except for sample transport and centrifugation at 4°C instead of room temperature.21
MS brain samples were obtained from a 50-year-old man who died during a fulminating MS relapse after natalizumab withdrawal and presented with extensive and active brain lesions characteristic of MS,22 a 47-year-old man with secondary progressive MS (SPMS) who died of a lung neoplasm, a 60-year-old woman with SPMS who died of an infection. Samples were rapidly (within 12 hours after death) frozen in liquid nitrogen or fixed by immersion in 10% buffered formalin and processed into liquid paraffin for histologic evaluation and immunohistochemistry (Centre des Collections Biologiques Hospitalières de Montpellier (CCBH-M), Collection tumorothèque, FINESS 340780477, Montpellier, France; and Collection Sclérose en Plaques Nantes (PFS13-003), FINESS 440000289, Nantes, France). Control brain samples were obtained from the right frontal lobe of a 62-year-old man who died after an occipital infarct and from the right frontal lobe of an 87-year-old man who died of pneumonia.
Standard Protocol Approvals, Registrations, and Patient Consents
All enrolled patients had signed informed consent for CSF biobanking after lumbar puncture prescribed for the investigation of neurologic disorders (biobank registered under DC-2008-417). The procedures were authorized by ANSM (ID RCB 2008-A01199-46) on September 12, 2008 and approved by the Comité de Protection des Personnes Sud Méditerranée IV (ethics committee) on February 10, 2009. For brain samples, we received approval from an ethical standards committee to conduct this retrospective study (IRB 2023.09.06). Operators were blinded from clinical information and reference standard results (e.g., OCBs) for all CSF analyses.
Proteomic Analysis of CSF
Label-Free Quantitative Proteomics
We compared CSF samples from 10 patients with RRMS and 10 CTRLs and CSF samples from 10 FC-CIS and 10 SC-CIS. Detailed methods used for label-free quantitative proteomics and statistical analysis of data are available in the eMethods.
Targeted Quantitative Proteomics
Targeted quantitative proteomics is detailed in the eMethods.
Selection of Additional Candidate Biomarkers From the Literature
Fifteen additional proteins that have previously been involved in MS pathogenesis (Angiotensinogen, ALCAM [CD166], Basigin, Contactin-3, Ephrin B2, Ephrin A4, GC, N-acetylglucosamine-1-phosphotransferase, Immunoglobulin lambda-like polypeptide 5, Multimerin-2, Matrix remodeling–associated protein 8, Neural cell adhesion molecule 2 [NCAM2], Neural cell adhesion molecule L1 [L1CAM], SPP1, and Tenascin-X) were analyzed by PRM to explore their potential as MS biomarkers (see Results).
Primary Cultures of Rat OPCs, Cell Treatment, and Media Conditioning
Primary cultures of rat OPCs were prepared according to the procedure described in the eMethods. After 4 days of culture in modified Bottenstein Sato medium containing heavy isotope-labeled amino acids, cells were washed 5 times with serum-free culture medium and exposed to either vehicle or TNFα (10 ng/mL) or soluble Fas ligand (sFasL) in the same medium for 24 hours. After the 24-hour secretion period, conditioned media were collected, centrifuged at 200×g for 5 minutes and then at 20,000×g for 25 minutes to remove nonadherent cells and cell debris, respectively. Immunocytochemistry in OPC cultures is detailed in the eMethods. OPC apoptosis was also determined after the treatments as previously described.23
Quantitative OPC Secretome Analysis
Methods used for OPC secretome analysis are available in the eMethods. In brief, proteins from OPC-conditioned media were concentrated by precipitation in 10% trichloroacetic acid, separated by SDS-PAGE and digested in-gel using trypsin (600 ng/band, Gold, Promega), as previously described.24 The resulting peptides were analyzed by nanoLC-MS/MS as previously described for CSF sample analysis. Data were analyzed using the MaxQuant software (version 1.4.1.2).25
ELISA
CHI3L1 concentration was determined in CSF samples from CTRLs and patients with RRMS using the MicroVue YKL-40 EIA kit (Quidel Corporation, San Diego, CA), as previously described.5 CD138 and CD27 concentrations were determined in nondiluted CSF samples from CTRLs, patients with RRMS and patients with PPMS using the R-PLEX Human Syndecan-1 (CD138) assay and the U-PLEX Human CD27 assay, respectively (Meso Scale Discovery, Rockville, MD, USA) with a MESO QuickPlex SQ 120 MM, according to the manufacturer's instructions. The CSF CD138 and CD27 lower limit of detection (LLOD) were 4.2 pg/mL and 0.3 pg/mL, respectively.
Immunohistochemistry
For horseradish peroxidase labeling, paraffin sections (4 µm thick) of fixed brains were subjected to antigen retrieval after quenching of endogenous peroxidase by immersion in citrate/EDTA buffer, pH 6, and heating (40 minutes at 100°C). After applying primary and secondary antibodies, immunoperoxidase reaction was performed using the avidin-biotin method and 3′,3′diaminobenzidine as chromogen and the ROCHE automatic immunostaining system (Benchmark ULTRA). Sections were then counterstained with hematoxylin.
For double IHC staining, 3′,3′diaminobenzidine CD138-labeled and unlabeled slices counterstained with hematoxylin were further incubated for 1 hour with primary antibodies (see eMethods). After 3 washes, slices were incubated for 30 minutes in AP one-step polymer anti-mouse/rabbit/rat (Zytomed Systems, ref ZUC068) and labeled with permanent AP red (Zytomed Systems, ref ZUC001), according to the manufacturer's instructions.
For immunofluorescence, fresh frozen brain slices on glass coverslips were incubated in PBS solution containing 10% heat-inactivated goat serum (Vector Laboratories, Ref S-100) and 0.3% Triton X-100 for 20 minutes. They were then incubated overnight at 4°C in PBS containing 3% heat inactivated goat serum, 0.1% Triton X-100, and primary antibodies (see eMethods). After 3 washes in PBS, slices were incubated for 2 hours with the Alexa Fluor 594-conjugated anti-rabbit antibody (ThermoFisher Scientific, Ref A-11037, 1:1,000 dilution) and the Alexa Fluor 680-conjugated anti-mouse antibody (ThermoFisher Scientific, Ref A-21057, 1:1,000 dilution) in PBS containing 3% heat-inactivated goat serum, 0.1% Triton X-100, and Hoechst 33342 (1 μg/mL, Sigma). After 3 washes in PBS, coverslips were mounted on superfrost ultra plus slides (ThermoFisher Scientific, Ref 10417002) using fluorescent mounting medium (Dako, Ref S3023). Immunofluorescence images were taken with the AxioImager Z1 microscope equipped with Apotome. All antibodies used in the study are detailed in the eMethods.
Data Availability
Anonymized data will be shared upon request from any qualified investigator to eric.thouvenot@chu-nimes.fr for research purposes only.
Results
Identification of MS Candidate Biomarkers by Quantitative Proteomic Analysis of the CSF
We first compared the CSF proteome of 10 CTRLs, 10 SC-CIS, 10 FC-CIS, and 10 patients with RRMS matched in age (mean age 35.3–37.6 years), sex ratio (70% in all cohorts), and CSF protein level (0.39–0.42 g/L, Table 1 and Figure 1). As expected, SC-CIS, FC-CIS, and RRMS samples showed a higher IgG index and the presence of OCBs, compared with CTRLs (Table 1). Samples from each patient were analyzed in triplicate, yielding a total of 120 LC-MS runs. Analysis of LC-MS data quality, assessed by a dispersion tree representing the protein expression in each sample after missing value imputation and data normalization, showed a regular dispersion of the data together with proximity of technical replicates (eFigure 2), thus indicating similar protein composition of all samples and reproducibility of analyses.
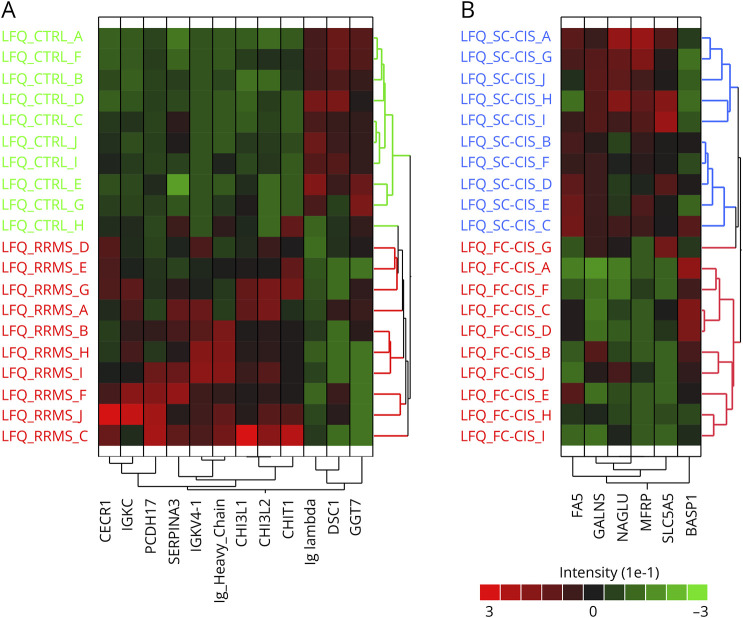
Overall, a total of 5,042 unique peptides corresponding to 600 proteins were identified and quantified after data filtering. Comparing samples at protein level revealed 12 proteins that exhibit significant difference in abundance in patients with RRMS vs CTRLs (eTable 1) and segregate both patient cohorts (Figure 2A). Likewise, 6 proteins exhibited significant differences in abundance in CSF from FC-CIS vs SC-CIS patients (eTable 1) and segregated both cohorts (Figure 2B). Analysis of data at peptide level revealed 39 additional proteins with at least 1 peptide exhibiting significant different abundance in the CSF from patients with RRMS CTRLs (eTable 2).
Figure 2. Results of Label-Free Proteomic Analysis of Human CSF Samples From the Discovery Cohort.
(A) Hierarchical clustering of the 12 proteins exhibiting difference in abundance in the CSF from patients with RRMS and CTRLs. (B) Hierarchical clustering of the 6 proteins exhibiting difference in abundance in the CSF from FC-CIS and SC-CIS.
Identification of Potential Biomarkers of Inflammation and Oligodendrocyte Apoptosis by Analysis of OPC Secretome
To identify additional candidate biomarkers reflecting 2 major pathologic features of MS (inflammation and oligodendrocyte apoptosis), we next explored the modifications of the secretome of primary cultures enriched in rat OPCs elicited by exposing cultures with either TNFα26 or sFasL27 (both at 10 ng/mL) for 24 hours, using the stable isotope labeling by amino acids in cell culture technology. As expected, exposure of OPCs to sFasL induced a significant increase in apoptotic OPCs in the cultures (46.5 ± 12.7% condensed or fragmented nuclei in sFasL-treated cell cultures vs 11.2 ± 3.0% and 13.9 ± 5.6% in vehicle-treated and TNFα-treated cultures, p < 0.0001 and p < 0.0001 (unpaired t test), respectively, eFigure 3).
A total of 2,535 proteins were identified in OPC supernatant in 4 biological replicates comparing the secretome of vehicle and TNFα or sFasL-treated cells. Of these, 36 proteins showed significant difference in abundance (assessed by significance B) in the supernatant of OPCs treated with vehicle and TNFα in at least 2 of 4 replicates, and 19 proteins showed significant difference in abundance in the supernatant of vehicle and sFasL-treated OPCs (eTable 3). Fifteen human orthologs of differentially OPC-secreted proteins were identified in our quantitative proteomic analysis of human CSF samples (eTable 3).
Qualification of Candidate Biomarkers by Targeted Quantitative Proteomics
We next combined proteins showing difference in abundance at protein (18 proteins) or peptide level (39 proteins) in patient CSF samples with 15 proteins showing different abundance in OPC secretome and 15 additional proteins linked to MS pathophysiology that we identified in human CSF (eTable 4, Figure 1). This yielded a list of 87 proteins that were further analyzed by PRM in the initial cohort and a new cohort of 60 patients (qualification cohort, Table 1) comprising CTRL, SC-CIS, FC-CIS, RRMS, PPMS, and INDC (10 patients per group). For each of these proteins, we selected a maximum of 3 proteotypic peptides providing a good signal in our label-free analyses of human CSF, yielding a list of 226 peptides that were analyzed by PRM (eTable 4, Figure 1). To improve the detection and relative quantification of peptides, digested CSF samples were spiked with a mixture of heavy isotope–labeled versions of these 226 peptides at concentrations giving signals of similar intensities to those of the endogenous peptides. We first compared label-free and PRM RRMS/CTRL ratios in the discovery cohort for the 76 proteins quantified with both approaches. As shown in eFigure 4A, a strong correlation was found (Pearson coefficient, 0.86), thus validating our PRM strategy. Comparing RRMS/CTRL PRM ratios in the discovery and qualification cohorts also indicated a good correlation (Pearson coefficient, 0.83, eFigure 4B). Proteins selected for PRM analysis include the previously described MS biomarker CHI3L1.5,6,28,29 Corroborating previous findings, CHI3L1 showed significant difference in abundance in the CSF of CTRLs and patients with RRMS both in label-free (discovery cohort, eTables 1 and 2) and PRM analyses (qualification cohort, eTable 5). Furthermore, CSF CHI3L1 concentration, determined by ELISA, was correlated with label-free and PRM CHI3L1 quantification in the discovery cohort (Pearson coefficients: 0.68 and 0.59, respectively, eFigure 4, C and D), further validating quantitative proteomics approaches used for MS biomarker discovery and verification. Of the 226 peptides analyzed, 16 peptides corresponding to 11 different proteins (see Table 3) exhibited significant PRM ratios in patients with RRMS vs CTRLs, patients with RRMS vs patients with PPMS or RRMS vs INDC comparisons. These proteins include previously identified candidate biomarkers of MS such as CHI3L1, CHI3L2, CHIT1, IGKC, and CD27 and novel candidate biomarkers of the disease such as the adenosine deaminase CECR1 (cat eye syndrome critical region protein 1) and the proteoglycan syndecan-1, also known as plasma cell surface marker CD138. None of them showed significant difference in abundance in FC-CIS vs SC-CIS.
Table 3.
Peptide Ratios Measured by PRM in the Verification Cohort
| Protein | Peptide sequence | p Value | NINDC vs CTRL | PINDC vs CTRL | INDC vs CTRL | ION vs CTRL | SC-CIS vs CTRL | FC-CIS vs CTRL | RRMS vs CTRL | PPMS vs CTRL | RRMS vs INDC | RRMS vs NINDC | RRMS vs ION | RRMS vs SC-CIS | RRMS vs FC-CIS | RRMS vs PPMS | SC-CIS vs FC-CIS |
| CD138_1a | EGEAVVLPEVEPGLTA(R) | <0.0001 | 1.00 | 1.10 | 1.13 | 1.02 | 2.02* | 2.11* | 2.70*** | 2.29* | 2.38*** | 2.70*** | 2.65*** | 1.33 | 1.28 | 1.18 | 1.04 |
| CD27_1a | HCNSGLLV(R) | <0.0001 | 1.32 | 1.35 | 3.66 | 1.19 | 5.63** | 6.61*** | 10.64*** | 5.16* | 2.91 | 8.06*** | 8.96*** | 1.89 | 1.61 | 2.06 | 1.17 |
| CECR1_1a | LLPVYELSGEHHDEEWSV(K) | <0.0001 | 1.55 | 1.50 | 2.30* | 0.85 | 2.19* | 1.70* | 2.64*** | 1.44 | 1.15 | 1.70* | 3.11*** | 1.20 | 1.55 | 1.83 * | 0.78 |
| CECR1_2a | SQVFNIL(R) | <0.0001 | 1.64 | 1.68 | 2.50* | 0.83 | 2.08 | 1.99 | 3.01*** | 1.67 | 1.21 | 1.84 | 3.61*** | 1.45 | 1.51 | 1.81 * | 0.96 |
| CHI3L2_1a | LLLTAGVSAG(R) | <0.0001 | 1.80 | 1.30 | 2.69** | 1.18 | 1.76 | 1.84* | 2.90*** | 1.80 | 1.08 | 1.61 | 2.46*** | 1.64 | 1.57 | 1.61 | 1.05 |
| CH3IL2_2a | GPSSYYNVEYAVGYWIH(K) | <0.0001 | 2.01 | 1.31 | 2.61 | 1.05 | 2.29 | 1.90 | 3.95*** | 1.98 | 1.51 | 1.96 | 3.74*** | 1.72 | 2.08 | 1.99 | 0.83 |
| CHI3L1_1a | ILGQQVPYAT(K) | <0.0001 | 2.11 | 1.91 | 2.63** | 0.92 | 1.25 | 1.26 | 2.38*** | 2.00 | 0.91 | 1.13 | 2.58*** | 1.90 | 1.89 | 1.19 | 1.01 |
| CHI3L1_3a | LVCYYTSWSQY(R) | 0.0002 | 2.60 | 1.64 | 1.62 | 0.58 | 1.15 | 1.30 | 2.46** | 1.53 | 1.52 | 0.95 | 4.22*** | 2.14 | 1.90 | 1.61 | 1.13 |
| CHIT1_1a | VGAPATGSGTPGPFT(K) | <0.0001 | 4.88 * | 2.14 | 2.29 | 1.29 | 2.98 | 6.68*** | 9.82*** | 4.92 | 4.30 | 2.01 | 7.64*** | 3.30 | 1.47 | 2.00 | 2.25 |
| CHIT1_3a | DNQWVGFDDVESF(K) | <0.0001 | 3.25 | 3.49 | 4.36 | 1.88 | 3.32 | 5.28** | 6.03*** | 4.35 | 1.38 | 1.85 | 3.20*** | 1.81 | 1.14 | 1.39 | 1.59 |
| IGKC_1a | SGTASVVCLLNNFYP(R) | <0.0001 | 1.74 | 1.30 | 1.62 | 1.71 | 3.69 | 3.81 | 4.77*** | 3.40 | 2.94* | 2.74** | 2.79** | 1.29 | 1.25 | 1.40 | 1.03 |
| IGKC_2a | VDNALQSGNSQESVTEQDS(K) | <0.0001 | 1.16 | 1.41 | 1.56 | 1.30 | 2.74** | 2.62** | 4.54*** | 2.92* | 2.92** | 3.91*** | 3.50*** | 1.66 | 1.73 | 1.55 | 0.96 |
| CFHR1_3b | INHGILYDEE(K) | 0.0650 | 1.25 | 1.89 * | 1.19 | 1.16 | 0.90 | 0.83 | 0.82 | 1.17 | 0.68 | 0.65 | 0.70 | 0.91 | 0.98 | 0.69 | 0.93 |
| RELN_2b | VIVLLPQ(K) | 0.4867 | 0.70 | 1.19 | 1.03 | 1.34 | 1.16 | 0.98 | 0.90 | 1.35 | 0.87 | 1.29 | 0.67 | 0.77 | 0.92 | 0.66 | 0.84 |
| LYZ_1c | WESGYNT(R) | <0.0001 | 1.20 | 1.24 | 2.03 | 0.58 | 0.89 | 0.83 | 1.43 | 0.84 | 0.70 | 1.19 | 2.45*** | 1.60 | 1.73 | 1.70 * | 0.93 |
| NGAL_1c | SYPGLTSYLV(R) | 0.0154 | 1.27 | 1.32 | 1.55* | 0.93 | 0.94 | 0.90 | 1.10 | 1.02 | 0.71 | 0.87 | 1.18 | 1.18 | 1.23 | 1.08 | 0.96 |
Fold changes among the different group comparisons of the 16 peptides quantified by PRM in the verification cohort. Statistical difference between the different cohorts was assessed by the Kruskal-Wallis test (p values for each peptide listed in column 3) followed by the Dunn multiple comparison test (*p value <0.05; **p-value <0.01; ***p value <0.001) using GraphPad PRISM 10.1.0. a and b indicate peptides originating from proteins identified as significant in CSF proteomic analysis comparing CTRL with RRMS and FC-CIS with SC-CIS, respectively. c indicates proteins identified as significant upon TNFα exposure in OPC secretome analysis (see also eTable 4).
Verification of Qualified Biomarkers by Targeted Quantitative Proteomics
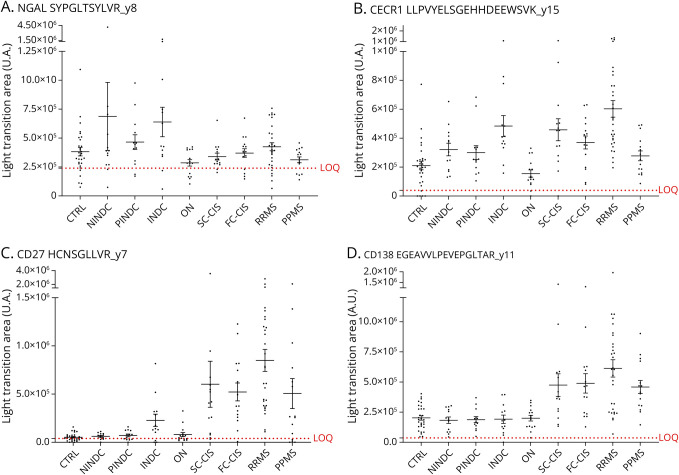
These peptides were next quantified by PRM in a new cohort of 158 patients (verification cohort), including 30 CTRLs, 13 NINDC, 13 PINDC, 13 INDC, 15 with ION, 15 SC-CIS, 15 FC-CIS, 30 with RRMS, and 14 with PPMS (Table 2). Heavy isotope–labeled, high-purity (AQUA) versions of the 16 analyzed peptides were spiked in the samples for absolute quantification and determination of LOD and LOQ. Their relative abundance between the different cohorts and their significance are summarized in Table 3. Among the 11 proteins investigated in this second PRM analysis, 7 exhibited differences in abundance between patients with RRMS and CTRLs, or between patients with MS and other inflammatory and noninflammatory neurologic diseases (Table 3). These include the previously identified MS biomarker CHI3L1, which showed an increased level not only in patients with MS (at all disease stages) but also in INDC, NINDC, and PINDC, when compared with CTRLs and patients with ION (eFigure 5). CHI3L2, CHIT1, and CECR1 showed similar differences in abundances between the different cohorts (Figure 3 and eFigure 5). Of most importance, CSF CD138 and IGKC levels were more elevated in patients with MS at all disease stages, compared with CTRL, INDC, NINDC, PINDC, and patients with ION, suggesting that these proteins could distinguish between MS at any disease stage and other diseases (Figure 3). CD27 concentration was likewise more elevated in patients with CIS and patients with MS but, contrasting with CD138 and IGKC, it was also increased in patients with INDC (Figure 3). Surprisingly, neutrophil gelatinase–associated lipocalin (NGAL, also designated as lipocalin-2) was more abundant in the CSF from patients with INDC, but not from patients with MS, compared with CTRLs (Figure 3).
Figure 3. PRM Analysis of Peptides From 4 Candidate Protein Biomarkers in the Verification Cohort.
Intensity (light transition area in arbitrary units [A.U.]) of peptides showing difference in abundance in CSF samples of the verification cohort (Table 2) is shown for NGAL (A), CECR1 (B), CD27 (C), and CD138 (D). The LOQ (limit of quantification) is indicated (red dotted line) for each peptide. Statistical analyses of group comparisons are provided in Table 3. CTRL: symptomatic controls (n = 30); CIS: clinically isolated syndrome; SC-CIS: slow converter CIS (n = 15); FC-CIS: fast converter CIS (n = 15); RRMS: relapsing-remitting multiple sclerosis (n = 30); PPMS: primary progressive multiple sclerosis (n = 14); INDC: inflammatory neurologic disease control (n = 13); ON: isolated optic neuritis (n = 15); NINDC: noninflammatory neurologic disease control (n = 13), PINDC: peripheral inflammatory neurologic disease control (n = 13).
Diagnostic Value of Verified Candidate Biomarkers
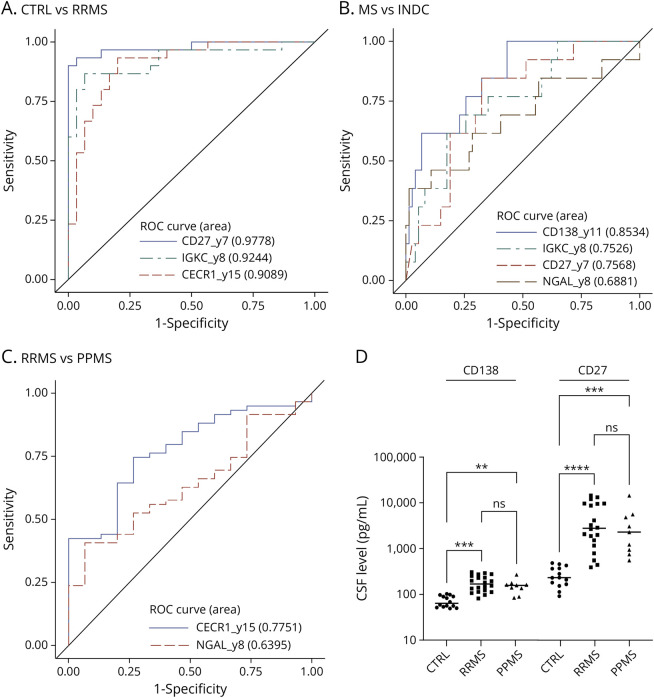
We next investigated the sensitivity and specificity of these biomarkers to discriminate patients with MS from CTRLs and other neurologic conditions. Receiver operating characteristic (ROC) curves showed that CHI3L1, CHI3L2, CHIT1, CD138, IGKC, CD27, and CECR1 discriminate CTRLs from patients with RRMS and CTRLs and patients with ION from those with any neurologic disease (SC-CIS, FC-CIS, patients with RRMS, patients with PPMS, INDC, PINDC, or NINDC; eTable 6). Of these, CD27 has higher sensitivity and specificity in discriminating CTRLs and patients with RRMS (AUC = 0.98, Figure 4A and eTable 6), while the combination of CHI3L1, CHIT1, and CD138 has higher sensitivity and specificity than each protein taken individually (AUC = 0.88, eFigure 6A and eTable 6) in discriminating CTRLs and patients with ION from those with MS (at all disease stages), INDC, PINDC, or NINDC, as assessed by multivariate analysis. On the contrary, CD27, CD138, IGKC, CHI3L2, CHIT1, and CECR1 discriminate inflammatory CNS diseases (SC-CIS, FC-CIS, RRMS, PPMS, INDC) from other neurologic diseases (PINDC or NINDC, eTable 6). Furthermore, the combination of CD138 and CD27 has higher sensitivity and specificity (AUC = 0.91, eFigure 6B and eTable 6) than each protein taken individually. CD138 (AUC = 0.85) is also more efficient than IGKC (AUC = 0.75), CD27 (AUC = 0.76), and NGAL (AUC = 0.69) to discriminate MS from INDC, and multivariate analysis showed that no combination of these markers is better than CD138 alone to discriminate these cohorts (Figure 4B and eTable 6). Finally, CECR1 (AUC = 0.78) better differentiates patients with RRMS from patients with PPMS than NGAL (AUC = 0.64), and the combination of both markers is not better than CECR1 alone to discriminate these cohorts (Figure 4C and eTable 6). Measurement of CD138 and CD27 concentration by ELISA showed that they are significantly increased in the CSF from patients with RRMS and patients with PPMS, compared with CTRLs (Figure 4D), thus validating the PRM results.
Figure 4. Diagnostic Value of Candidate MS Biomarkers.
(A) ROC curves showing the diagnostic values of CD27 (AUC = 0.98), IGKC (AUC = 0.92), and CECR1 (AUC = 0.91) to discriminate RRMS from CTRL. Multivariate analysis indicated that no combination has better diagnostic value than CD27 alone. (B) ROC curves showing the diagnostic values of CD138 (AUC = 0.85), IGKC (AUC = 0.75), CD27 (AUC = 0.76), and NGAL (AUC = 0.69) to discriminate MS from INDC. Multivariate analysis indicated that no combination has better diagnostic value than CD138 alone. (C) ROC curves showing the diagnostic values of CECR1 (AUC = 0.78) and NGAL (AUC = 0.64) to discriminate RRMS from PPMS. Multivariate analysis indicated that no combination has better diagnostic value than CECR1 alone. (D) CSF CD138 and CD27 levels measured by ELISA in CTRL (n = 14), patients with RRMS (n = 20), and patients with PPMS (n = 9) and compared with the nonparametric Mann-Whitney test. **, p value <0.01; ***, p value <0.001; ****, p value <0.0001. ROC = receiver operating characteristic.
Enhanced Expression of CD138 in MS Brain
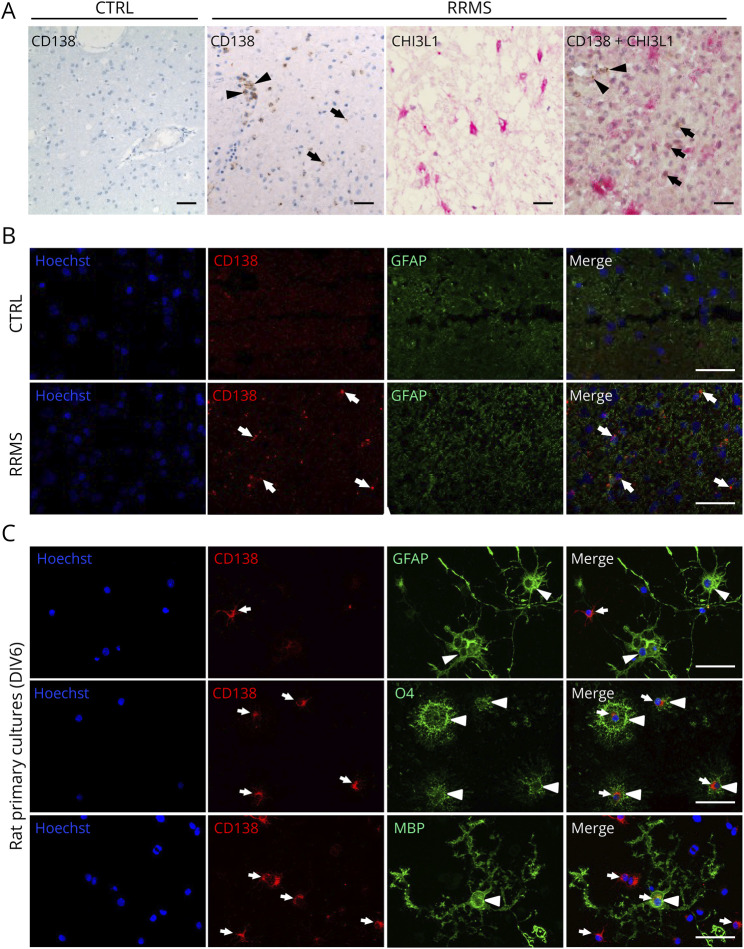
Given the potential of CD138 to discriminate MS from other CNS inflammatory and noninflammatory diseases, we investigated its expression in brain slices from a CTRL and a patient with RRMS by immunohistochemistry. Whereas CD138 immunolabeling was not detected in CTRL brain, a strong CD138 immunostaining was predominantly observed in round-shaped cells located in perivascular spaces and corresponding to plasma cells (Figure 5A). A weaker CD138 staining was also found in cells of the parenchyma, especially in the vicinity of inflamed perivascular spaces. Furthermore, CD138 staining was not colocalized with CHI3L1 immunostaining observed in reactive astrocytes of MS brain (Figure 5A) (4). Corroborating these observations, immunofluorescence staining showed that CD138 was not colocalized with GFAP in RRMS brain (Figure 5B). Likewise, CD138 was not detected in GFAP-positive astrocytes in rat primary cultures containing OPCs and astrocytes (Figure 5C). By contrast, a strong CD138 labeling was found in OPCs (O4+) and, to a lesser extent, in mature (MBP+) oligodendrocytes (Figure 5C).
Figure 5. Expression of CD138 in Control and RRMS Human Brain and Rat Primary Cultures.
(A) Immunohistochemistry of brain tissue showing predominant expression of CD138 in cells located in the perivascular spaces (arrowheads) and a sparse expression in brain parenchyma (arrows) of RRMS brain but not in the CTRL brain. CHI3L1 is strongly expressed in astrocytes (pink) from RRMS brain but shows no colocalization with CD138 (brown). Scale bar: 100 µm. (B) Immunofluorescence labeling showing a high expression of GFAP in activated astrocytes of the RRMS brain when compared with the CTRL brain. There is no colocalization of CD138 (arrows) and GFAP in the RRMS brain. Scale bar: 100 µm. (C) Immunostaining of CD138 (arrows) in rat primary cultures of OPCs at 6 DIV showing a stronger expression of CD138 in immature (O4+) than in mature (MBP+) oligodendrocytes and its absence in astrocytes (GFAP+) (arrowheads). Scale bar: 100 µm.
Discussion
Using 2 complementary proteomic strategies comparing (1) the CSF proteome of CTRLs, patients with RRMS and patients with CIS showing slow or fast conversion to RRMS and (2) the secretome of OPC cultures exposed or not to a proapoptotic or a proinflammatory treatment, we identified 72 candidate biomarkers of MS. They were combined with 15 proteins previously involved in MS pathogenesis to generate a list of 87 proteins for further quantification by PRM in a new cohort comprising patients with MS at different disease stages and control neurologic diseases. This qualified 11 proteins for a second PRM analysis in a larger cohort of 158 patients that revealed a signature of 8 biomarkers of potential interest in clinical practice. These include 5 previously described biomarkers of MS, namely CHI3L1, CHI3L2, CHIT1,29,30 IGKC, the constant region of the κFLC of immunoglobulins,18 and CD27.31,32 All exhibited increased level in the CSF of patients with MS, consistent with previous findings. The 3 other proteins that passed the verification step include NGAL, an iron-binding protein involved in innate immunity and known to inhibit remyelination in vitro,33 CD138, a cell surface proteoglycan-bearing heparan and chondroitin sulfates that links the cytoskeleton to the extracellular matrix,34 and the secreted adenosine deaminase CECR1 that binds to proteoglycans and may play a role in the regulation of cell proliferation and differentiation independently of its adenosine deaminase activity.35 NGAL was identified as a candidate biomarker of MS in the analysis of OPC secretome but not in our label-free CSF proteome analyses, underpinning the complementarities of both approaches. A decreased level of NGAL was measured in the CSF of patients with MS, compared with CTRLs, while CSF CD138 and CECR1 concentrations are increased in MS, consistent with previous findings.36,37 However, no validation of these proteins as biomarkers of MS had so far been reported. Of note, none of the 15 proteins potentially involved in MS pathogenesis selected from literature analysis were confirmed by PRM.
The 8 identified biomarkers did not arise from the comparison of the CSF proteome from SC-CIS and FC-CIS. Accordingly, they cannot be considered as biomarkers of disease activity, which remains a challenging issue. The pioneer study of the Barcelona group compared patients with CIS with normal MRI and absence of OCBs or patients with ION with patients with CIS with 3 or 4 Barkhof criteria3 and presence of OCBs (now considered as RRMS).6 Only a recent study identified homeobox protein Hox-B3 (HOXB3) as a candidate biomarker of conversion in CIS, but the results need further validation.38
The 8 identified biomarkers exhibited different sensitivities and specificities for MS but also complementary properties potentially useful for differential diagnosis of MS. Multivariate analysis indicated that a subset of them (CHI3L1, CHIT1, and CD138) discriminate CTRLs and patients with ION from any other neurologic diseases, including NINDCs. Accordingly, low CSF levels of these biomarkers might indicate the absence of any CNS disease in patients complaining about neurologic symptoms.
CD27 combined with CD138 discriminates CNS inflammatory diseases, including MS at all stages, from NINDCs and PINDCs, indicating that this combination can be considered as biomarker of central inflammation. CD27 was also the most accurate biomarker discriminating RRMS from CTRLs. MS is characterized by B-cell accumulation in the CSF. Most of them are memory B cells, with a high expression of CD27 at their surface, or short-lived plasma blasts expressing CD138 and CD38.39 The strong expression of CD138 at the surface of plasmocytes and plasma blasts present in ectopic lymphoid follicles in the meninges might contribute, at least partly, to the increased concentration of CD138 in the CSF from patients with MS.40 This could explain why our multivariate analysis revealed CD138 as the best biomarker discriminating MS (at all disease stages) from other CNS inflammatory conditions (INDC). A recent study comparing CSF CD138 level by ELISA in neuromyelitis optica (NMO), MS, and CTRLs revealed an increase in CD138 concentration in NMO, but not in MS,41 but the latter observation resulted from analysis of a small patient group (n = 12) based on low-sensitivity ELISA. High serum and CSF CD138 concentration was also found in patients with anti-N-methyl-d-aspartate receptor (NMDAR) encephalitis and was associated with poor clinical prognosis and inflammation.42 These observations indicate the need for further validation of CD138 specificity for MS, when compared with encephalitis and neuromyelitis optica spectrum disorder.
In the CNS, CD138 is also highly expressed by choroidal epithelial cells, where it reduces leukocyte recruitment to the brain across the choroid plexuses. In the myelin oligodendrocyte glycoprotein–induced experimental autoimmune encephalomyelitis (MOG-EAE) MS mouse model, CD138 knockout enhances disease severity and impairs recovery, suggesting a protective role of CD138.43 CD138 seems to play a key role in blood-brain barrier (BBB) integrity, which is altered in inflammatory brain disorders, including MS.44 On the contrary, CD138 has been identified as a receptor that binds to CHI3L1 through its heparan sulfate residues and mediates CHI3L1-operated signaling involved in inflammation and cancer.45,46 Likewise, CD138 has been suggested as an endothelial cell receptor mediating CHI3L1-induced angiogenesis.47,48 Association of CHI3L1 to CD138 promotes recruitment of integrins αvβ347 and αvβ5,48 leading to engagement of FAK and ERK1/2 signaling and VEGF expression. CHI3L1 also increases the expression of MMP9, CCL2, and CXCL2 through CD138,49 a process potentially contributing to BBB leakage in inflammation. Collectively, these results suggest that CD138 exerts both beneficial and deleterious influences on MS that depend on the binding of CHI3L1 or other growth factors and chemokines to its heparan sulfate chains.50 Further supporting the potential influence of the CHI3L1/CD138-operated pathways, sdc1 sequence variation has been associated with MS, specifically in women experiencing either PPMS or RRMS.51 We found a predominant expression of CD138 in OPCs and mature oligodendrocytes but not in astrocytes, suggesting that CD138 acts as a receptor of CHI3L1 released by activated astrocytes. This is consistent with previous data showing that the expression of CD138 and syndecan-3 is higher in the oligodendrocyte lineage cells than in astrocytes.52 Analysis of CHI3L1-CD138 interaction and associated signaling in oligodendrocytes from MS brain certainly warrants further exploration to better understand their role in the pathophysiology of MS.
Among the 8 biomarkers identified, CECR1 is also of potential interest. CSF CECR1 concentration shows a large increase in patients with RRMS, compared with CTRLs, patients with ION, and patients with PPMS. Furthermore, multivariate analysis indicated that CSF CECR1 discriminates patients with RRMS from patients with PPMS, suggesting its association with the active phase of MS. Together with adenosine deaminase 1 (ADA1), CECR1 (ADA2) plays a key role in regulating the level of adenosine. It is secreted by monocytes undergoing differentiation into macrophages or dendritic cells.53 In turn, CECR1 induces T-cell–dependent differentiation of monocytes into M2 macrophages and stimulates their proliferation through its recruitment at the cell surface through proteoglycans and adenosine receptors. In addition, CECR1 promotes the production of proangiogenic factors, inhibits Th17 differentiation, and stimulates Treg differentiation.54 CECR1 blockade by 2-chlorodeoxyadenosine (cladribine), a synthetic deoxyadenosine analog, modulates the immune responses during inflammation in specific cell types and reduces circulating T and B lymphocytes. Of interest, cladribine has been developed as a treatment for active RRMS, indicating that CECR1 might not only be a biomarker of the active phase of MS but also a therapeutic target.55,56
In conclusion, this retrospective analysis of 3 different cohorts comprising CTRLs, patients with MS, and patients with other inflammatory and noninflammatory neurologic diseases is one of the most comprehensive proteomic studies dedicated to MS biomarker discovery and validation currently available. It identified and validated CD138 as a novel biomarker that allows differential diagnosis of MS vs INDCs and confirmed that CD27 is an accurate biomarker of MS. Furthermore, it identified CECR1, a therapeutic target in MS, as a potential marker of the active phase of the disease that warrants further validation.
Glossary
- AUC
area under the curve
- BBB
blood-brain barrier
- CECR1
cat eye syndrome critical region protein 1
- CHI3L1
chitinase 3–like protein 1
- CHI3L2
chitinase 3–like protein 2
- CHIT1
chitotriosidase-1
- CIS
clinically isolated syndrome
- CTRL
symptomatic control
- EDSS
Expanded Disability Status Scale
- IGKC
immunoglobulin kappa chain region C
- ION
isolated optic neuritis
- INDC
inflammatory neurologic disease control
- MS
multiple sclerosis
- NAWM
normal-appearing white matter
- NGAL
neutrophil gelatinase–associated lipocalin
- NINDC
noninflammatory neurologic disease controls
- OCBs
oligoclonal bands
- PINDC
peripheral inflammatory neurologic disease control
- PPMS
primary progressive multiple sclerosis
- RRMS
relapsing-remitting multiple sclerosis
- WBCs
white blood cells
Appendix. Authors
| Name | Location | Contribution |
| Geoffrey Hinsinger, PhD | IGF, Université de Montpellier, CNRS, INSERM, France | Major role in the acquisition of data; analysis or interpretation of data |
| Lucile Du Trieu De Terdonck, PhD | IGF, Université de Montpellier, CNRS, INSERM, France | Major role in the acquisition of data; analysis or interpretation of data |
| Serge Urbach, PhD | IGF, Université de Montpellier, CNRS, INSERM, France | Major role in the acquisition of data; analysis or interpretation of data |
| Nicolas Salvetat, PhD | Sys2Diag, UMR 9005 CNRS / ALCEDIAG, Montpellier, France | Major role in the acquisition of data; analysis or interpretation of data |
| Manon Rival, MD, MSc | IGF, Université de Montpellier, CNRS, INSERM; Department of Neurology, Nîmes University Hospital, France | Drafting/revision of the article for content, including medical writing for content; analysis or interpretation of data |
| Manon Galoppin, PhD | IGF, Université de Montpellier, CNRS, INSERM, France | Drafting/revision of the article for content, including medical writing for content; analysis or interpretation of data |
| Chantal Ripoll, PhD | IGF, Université de Montpellier, CNRS, INSERM, France | Major role in the acquisition of data; analysis or interpretation of data |
| Renaud Cezar, MSc | IRMB, Université de Montpellier, INSERM; Department of Immunology, Nîmes University Hospital, France | Major role in the acquisition of data |
| Sabine Laurent-Chabalier, PhD | Department of Biostatistics, Clinical Epidemiology, Public Health, and Innovation in Methodology, Nîmes University Hospital, Université de Montpellier, France | Analysis or interpretation of data |
| Christophe Demattei, PhD | Department of Biostatistics, Clinical Epidemiology, Public Health, and Innovation in Methodology, Nîmes University Hospital, Université de Montpellier, France | Analysis or interpretation of data |
| Hanane Agherbi, PhD | Department of Neurology, Nîmes University Hospital, France | Major role in the acquisition of data |
| Giovanni Castelnovo, MD | Department of Neurology, Nîmes University Hospital, France | Major role in the acquisition of data |
| Sylvain Lehmann, MD, PhD | Biochemistry Department, Hôpital Saint-Eloi, Montpellier University Hospital, France | Major role in the acquisition of data |
| Valérie Rigau, MD, PhD | Department of Pathology, Montpellier University Hospital, France | Major role in the acquisition of data; analysis or interpretation of data |
| Philippe Marin, PhD | IGF, Université de Montpellier, CNRS, INSERM, France | Drafting/revision of the article for content, including medical writing for content; study concept or design; and analysis or interpretation of data |
| Eric Thouvenot, MD, PhD | IGF, Université de Montpellier, CNRS, INSERM; Department of Neurology, Nîmes University Hospital, France | Drafting/revision of the article for content, including medical writing for content; major role in the acquisition of data; study concept or design; and analysis or interpretation of data |
Footnotes
Editorial, page e200256
Study Funding
This study was supported by grants from the Fondation pour l'Aide à la Recherche sur la Sclérose en Plaques (ARSEP Foundation), the University of Montpellier, the Centre National de la Recherche Scientifique, the Institut National de la Santé et de la Recherche Médicale and la Région Languedoc-Roussillon. G. Hinsinger was recipient of fellowships from ARSEP and the Hospital of Nimes. L. du Trieu de Terdonck was a recipient of fellowships from ARSEP and la Region Languedoc-Roussillon. This investigator-sponsored study received funding from Sanofi. The funder had no influence on the study design, data analysis, or interpretation. Mass spectrometry analyses were performed using the facilities of Montpellier Proteomics Platform (PPM, BioCampus Montpellier).
Disclosure
G. Hinsinger, L. Du Trieu de Terdonck, S. Urbach, N. Salvetat, M. Rival, M. Galoppin, C. Ripoll, R. Cezar, S. Laurent-Chabalier, C. Demattei, H. Agherbi, G. Castelnovo, S. Lehmann, V. Rigau, and P. Marin have nothing to disclose; E. Thouvenot has received honoraria, travel grants, or research grants from the following pharmaceutical companies: Actelion, Biogen, BMS, Janssen-Cilag, Merck-Serono, Novartis, Roche, and Sanofi. Go to Neurology.org/NN for full disclosures.
References
- 1.Polman CH, Reingold SC, Edan G, et al. Diagnostic criteria for multiple sclerosis: 2005 revisions to the “McDonald Criteria”. Ann Neurol. 2005;58(6):840-846. doi: 10.1002/ana.20703 [DOI] [PubMed] [Google Scholar]
- 2.Polman CH, Reingold SC, Banwell B, et al. Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann Neurol. 2011;69(2):292-302. doi: 10.1002/ana.22366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thompson AJ, Banwell BL, Barkhof F, et al. Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol. 2018;17(2):162-173. doi: 10.1016/S1474-4422(17)30470-2 [DOI] [PubMed] [Google Scholar]
- 4.Tintore M, Arrambide G, Otero-Romero S, et al. The long-term outcomes of CIS patients in the Barcelona inception cohort: looking back to recognize aggressive MS. Mult Scler. 2020;26(13):1658-1669. doi: 10.1177/1352458519877810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hinsinger G, Galéotti N, Nabholz N, et al. Chitinase 3-like proteins as diagnostic and prognostic biomarkers of multiple sclerosis. Mult Scler. 2015;21(10):1251-1261. doi: 10.1177/1352458514561906 [DOI] [PubMed] [Google Scholar]
- 6.Comabella M, Fernández M, Martin R, et al. Cerebrospinal fluid chitinase 3-like 1 levels are associated with conversion to multiple sclerosis. Brain. 2010;133(Pt 4):1082-1093. doi: 10.1093/brain/awq035 [DOI] [PubMed] [Google Scholar]
- 7.Kroksveen AC, Opsahl JA, Aye TT, Ulvik RJ, Berven FS. Proteomics of human cerebrospinal fluid: discovery and verification of biomarker candidates in neurodegenerative diseases using quantitative proteomics. J Proteomics. 2011;74(4):371-388. doi: 10.1016/j.jprot.2010.11.010 [DOI] [PubMed] [Google Scholar]
- 8.Opsahl JA, Vaudel M, Guldbrandsen A, et al. Label-free analysis of human cerebrospinal fluid addressing various normalization strategies and revealing protein groups affected by multiple sclerosis. Proteomics. 2016;16(7):1154-1165. doi: 10.1002/pmic.201500284 [DOI] [PubMed] [Google Scholar]
- 9.Abu-Rumeileh S, Steinacker P, Polischi B, et al. CSF biomarkers of neuroinflammation in distinct forms and subtypes of neurodegenerative dementia. Alzheimers Res Ther. 2019;12(1):2. doi: 10.1186/s13195-019-0562-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kušnierová P, Zeman D, Hradílek P, Zapletalová O, Stejskal D. Determination of chitinase 3-like 1 in cerebrospinal fluid in multiple sclerosis and other neurological diseases. PLoS One. 2020;15(5):e0233519. doi: 10.1371/journal.pone.0233519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Disanto G, Barro C, Benkert P, et al. Serum Neurofilament light: a biomarker of neuronal damage in multiple sclerosis. Ann Neurol. 2017;81(6):857-870. doi: 10.1002/ana.24954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verberk IMW, Koel-Simmelink M, Twaalfhoven H, et al. Ultrasensitive immunoassay allows measurement of serum neurofilament heavy in multiple sclerosis. Mult Scler Relat Disord. 2021;50:102840. doi: 10.1016/j.msard.2021.102840 [DOI] [PubMed] [Google Scholar]
- 13.Khalil M, Teunissen CE, Otto M, et al. Neurofilaments as biomarkers in neurological disorders. Nat Rev Neurol. 2018;14(10):577-589. doi: 10.1038/s41582-018-0058-z [DOI] [PubMed] [Google Scholar]
- 14.Desplat-jégo S, Feuillet L, Pelletier J, Bernard D, Chérif AA, Boucraut J. Quantification of immunoglobulin free light chains in cerebrospinal fluid by nephelometry. J Clin Immunol. 2005;25(4):338-345. doi: 10.1007/s10875-005-5371-9 [DOI] [PubMed] [Google Scholar]
- 15.Presslauer S, Milosavljevic D, Brücke T, Bayer P, Hübl W. Elevated levels of kappa free light chains in CSF support the diagnosis of multiple sclerosis. J Neurol. 2008;255(10):1508-1514. doi: 10.1007/s00415-008-0954-z [DOI] [PubMed] [Google Scholar]
- 16.Presslauer S, Milosavljevic D, Huebl W, et al. Validation of kappa free light chains as a diagnostic biomarker in multiple sclerosis and clinically isolated syndrome: a multicenter study. Mult Scler. 2016;22(4):502-510. doi: 10.1177/1352458515594044 [DOI] [PubMed] [Google Scholar]
- 17.Menéndez-Valladares P, García-Sánchez MI, Adorna Martínez M, García De Veas Silva JL, Bermudo Guitarte C, Izquierdo Ayuso G. Validation and meta-analysis of kappa index biomarker in multiple sclerosis diagnosis. Autoimmun Rev. 2019;18(1):43-49. doi: 10.1016/j.autrev.2018.07.010 [DOI] [PubMed] [Google Scholar]
- 18.Leurs CE, Twaalfhoven H, Lissenberg-Witte BI, et al. Kappa free light chains is a valid tool in the diagnostics of MS: a large multicenter study. Mult Scler. 2020;26(8):912-923. doi: 10.1177/1352458519845844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levraut M, Laurent-Chabalier S, Ayrignac X, et al. Kappa free light chain biomarkers are efficient for the diagnosis of multiple sclerosis: a large multicenter cohort study. Neurol Neuroimmunol Neuroinflamm. 2023;10(1):e200049. doi: 10.1212/NXI.0000000000200049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brownlee WJ, Hardy TA, Fazekas F, Miller DH. Diagnosis of multiple sclerosis: progress and challenges. Lancet. 2017;389(10076):1336-1346. doi: 10.1016/S0140-6736(16)30959-X [DOI] [PubMed] [Google Scholar]
- 21.Teunissen CE, Petzold A, Bennett JL, et al. A consensus protocol for the standardization of cerebrospinal fluid collection and biobanking. Neurology. 2009;73(22):1914-1922. doi: 10.1212/WNL.0b013e3181c47cc2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rigau V, Mania A, Befort P, et al. Lethal multiple sclerosis relapse after natalizumab withdrawal. Neurology. 2012;79(22):2214-2216. doi: 10.1212/WNL.0b013e318275979d [DOI] [PubMed] [Google Scholar]
- 23.Lafon-Cazal M, Perez V, Bockaert J, Marin P. Akt mediates the anti-apoptotic effect of NMDA but not that induced by potassium depolarization in cultured cerebellar granule cells. Eur J Neurosci. 2002;16(4):575-583. doi: 10.1046/j.1460-9568.2002.02124.x [DOI] [PubMed] [Google Scholar]
- 24.Thouvenot E, Urbach S, Dantec C, et al. Enhanced detection of CNS cell secretome in plasma protein-depleted cerebrospinal fluid. J Proteome Res. 2008;7(10):4409-4421. doi: 10.1021/pr8003858 [DOI] [PubMed] [Google Scholar]
- 25.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367-1372. doi: 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- 26.Jana M, Pahan K. Redox regulation of cytokine-mediated inhibition of myelin gene expression in human primary oligodendrocytes. Free Radic Biol Med. 2005;39(6):823-831. doi: 10.1016/j.freeradbiomed.2005.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li W, Maeda Y, Ming X, et al. Apoptotic death following Fas activation in human oligodendrocyte hybrid cultures. J Neurosci Res. 2002;69(2):189-196. doi: 10.1002/jnr.10285 [DOI] [PubMed] [Google Scholar]
- 28.Thouvenot E, Hinsinger G, Demattei C, et al. Cerebrospinal fluid chitinase-3-like protein 1 level is not an independent predictive factor for the risk of clinical conversion in radiologically isolated syndrome. Mult Scler. 2019;25(5):669-677. doi: 10.1177/1352458518767043 [DOI] [PubMed] [Google Scholar]
- 29.Cantó E, Tintoré M, Villar LM, et al. Chitinase 3-like 1: prognostic biomarker in clinically isolated syndromes. Brain. 2015;138(Pt 4):918-931. doi: 10.1093/brain/awv017 [DOI] [PubMed] [Google Scholar]
- 30.Møllgaard M, Degn M, Sellebjerg F, Frederiksen JL, Modvig S. Cerebrospinal fluid chitinase-3-like 2 and chitotriosidase are potential prognostic biomarkers in early multiple sclerosis. Eur J Neurol. 2016;23(5):898-905. doi: 10.1111/ene.12960 [DOI] [PubMed] [Google Scholar]
- 31.Komori M, Blake A, Greenwood M, et al. Cerebrospinal fluid markers reveal intrathecal inflammation in progressive multiple sclerosis. Ann Neurol. 2015;78(1):3-20. doi: 10.1002/ana.24408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahler MR, Søndergaard HB, Buhelt S, et al. Multiplex assessment of cerebrospinal fluid biomarkers in multiple sclerosis. Mult Scler Relat Disord. 2020;45:102391. doi: 10.1016/j.msard.2020.102391 [DOI] [PubMed] [Google Scholar]
- 33.Al Nimer F, Elliott C, Bergman J, et al. Lipocalin-2 is increased in progressive multiple sclerosis and inhibits remyelination. Neurol Neuroimmunol Neuroinflamm. 2016;3(1):e191. doi: 10.1212/NXI.0000000000000191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stepp MA, Pal-Ghosh S, Tadvalkar G, Pajoohesh-Ganji A. Syndecan-1 and its expanding list of contacts. Adv Wound Care. 2015;4(4):235-249. doi: 10.1089/wound.2014.0555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zavialov AV, Yu X, Spillmann D, Lauvau G, Zavialov AV. Structural basis for the growth factor activity of human adenosine deaminase ADA2. J Biol Chem. 2010;285(16):12367-12377. doi: 10.1074/jbc.M109.083527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khalil M, Renner A, Langkammer C, et al. Cerebrospinal fluid lipocalin 2 in patients with clinically isolated syndromes and early multiple sclerosis. Mult Scler. 2016;22(12):1560-1568. doi: 10.1177/1352458515624560 [DOI] [PubMed] [Google Scholar]
- 37.Guldbrandsen A, Lereim RR, Jacobsen M, et al. Development of robust targeted proteomics assays for cerebrospinal fluid biomarkers in multiple sclerosis. Clin Proteomics. 2020;17:33. doi: 10.1186/s12014-020-09296-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Timirci-Kahraman O, Karaaslan Z, Tuzun E, et al. Identification of candidate biomarkers in converting and non-converting clinically isolated syndrome by proteomics analysis of cerebrospinal fluid. Acta Neurol Belg. 2019;119(1):101-111. doi: 10.1007/s13760-018-0954-4 [DOI] [PubMed] [Google Scholar]
- 39.Cepok S, Rosche B, Grummel V, et al. Short-lived plasma blasts are the main B cell effector subset during the course of multiple sclerosis. Brain. 2005;128(Pt 7):1667-1676. doi: 10.1093/brain/awh486 [DOI] [PubMed] [Google Scholar]
- 40.Serafini B, Rosicarelli B, Magliozzi R, Stigliano E, Aloisi F. Detection of ectopic B-cell follicles with germinal centers in the meninges of patients with secondary progressive multiple sclerosis. Brain Pathol. 2004;14(2):164-174. doi: 10.1111/j.1750-3639.2004.tb00049.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pei S, Zheng D, Wang Z, Hu X, Pan S, Wang H. Elevated soluble syndecan-1 levels in neuromyelitis optica are associated with disease severity. Cytokine. 2018;111:140-145. doi: 10.1016/j.cyto.2018.08.017 [DOI] [PubMed] [Google Scholar]
- 42.Zhu J, Li Y, Zheng D, et al. Elevated serum and cerebrospinal fluid CD138 in patients with anti-N-Methyl-d-Aspartate receptor encephalitis. Front Mol Neurosci. 2019;12:116. doi: 10.3389/fnmol.2019.00116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang X, Wu C, Song J, Götte M, Sorokin L. Syndecan-1, a cell surface proteoglycan, negatively regulates initial leukocyte recruitment to the brain across the choroid plexus in murine experimental autoimmune encephalomyelitis. J Immunol. 2013;191(9):4551-4561. doi: 10.4049/jimmunol.1300931 [DOI] [PubMed] [Google Scholar]
- 44.Zhu J, Li X, Yin J, Hu Y, Gu Y, Pan S. Glycocalyx degradation leads to blood–brain barrier dysfunction and brain edema after asphyxia cardiac arrest in rats. J Cereb Blood Flow Metab. 2018;38(11):1979-1992. doi: 10.1177/0271678X17726062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prakash M, Bodas M, Prakash D, et al. Diverse pathological implications of YKL-40: answers may lie in ‘outside-in’ signaling. Cell Signal. 2013;25(7):1567-1573. doi: 10.1016/j.cellsig.2013.03.016 [DOI] [PubMed] [Google Scholar]
- 46.Yeo IJ, Lee CK, Han SB, Yun J, Hong JT. Roles of chitinase 3-like 1 in the development of cancer, neurodegenerative diseases, and inflammatory diseases. Pharmacol Ther. 2019;203:107394. doi: 10.1016/j.pharmthera.2019.107394 [DOI] [PubMed] [Google Scholar]
- 47.Shao R, Hamel K, Petersen L, et al. YKL-40, a secreted glycoprotein, promotes tumor angiogenesis. Oncogene. 2009;28(50):4456-4468. doi: 10.1038/onc.2009.292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Francescone RA, Scully S, Faibish M, et al. Role of YKL-40 in the angiogenesis, radioresistance, and progression of glioblastoma. J Biol Chem. 2011;286(17):15332-15343. doi: 10.1074/jbc.M110.212514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kzhyshkowska J, Yin S, Liu T, Riabov V, Mitrofanova I. Role of chitinase-like proteins in cancer. Biol Chem. 2016;397(3):231-247. doi: 10.1515/hsz-2015-0269 [DOI] [PubMed] [Google Scholar]
- 50.Teixeira FCOB, Götte M. Involvement of syndecan-1 and heparanase in cancer and inflammation. In: Vlodavsky I, Sanderson RD, Ilan N, ed, Heparanase. Vol 1221. Springer International Publishing; 2020:97-135. doi: 10.1007/978-3-030-34521-1_4 [DOI] [PubMed] [Google Scholar]
- 51.Okolicsanyi RK, Bluhm J, Miller C, Griffiths LR, Haupt LM. An investigation of genetic polymorphisms in heparan sulfate proteoglycan core proteins and key modification enzymes in an Australian Caucasian multiple sclerosis population. Hum Genomics. 2020;14(1):18. doi: 10.1186/s40246-020-00264-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bansal R, Kumar M, Murray K, Pfeiffer SE. Developmental and FGF-2-mediated regulation of syndecans (1-4) and glypican in oligodendrocytes. Mol Cell Neurosci. 1996;7(4):276-288. doi: 10.1006/mcne.1996.0021 [DOI] [PubMed] [Google Scholar]
- 53.Zavialov AV, Gracia E, Glaichenhaus N, Franco R, Zavialov AV, Lauvau G. Human adenosine deaminase 2 induces differentiation of monocytes into macrophages and stimulates proliferation of T helper cells and macrophages. J Leukoc Biol. 2010;88(2):279-290. doi: 10.1189/jlb.1109764 [DOI] [PubMed] [Google Scholar]
- 54.Haskó G, Cronstein B. Regulation of inflammation by adenosine. Front Immunol. 2013;4:85. doi: 10.3389/fimmu.2013.00085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Giovannoni G, Comi G, Cook S, et al. A placebo-controlled trial of oral cladribine for relapsing multiple sclerosis. N Engl J Med. 2010;362(5):416-426. doi: 10.1056/NEJMoa0902533 [DOI] [PubMed] [Google Scholar]
- 56.Robak T, Robak P. Purine nucleoside analogs in the treatment of rarer chronic lymphoid leukemias. Curr Pharm Des. 2012;18(23):3373-3388. doi: 10.2174/138161212801227005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Anonymized data will be shared upon request from any qualified investigator to eric.thouvenot@chu-nimes.fr for research purposes only.