Phylogenomic analyses reveal the origin and evolution of anaerobic alkane metabolism in the domain Archaea.
Abstract
Methanogens are considered as one of the earliest life forms on Earth, and together with anaerobic methane-oxidizing archaea, they have crucial effects on climate stability. However, the origin and evolution of anaerobic alkane metabolism in the domain Archaea remain controversial. Here, we present evidence that methylotrophic methanogenesis was the ancestral form of this metabolism. Carbon dioxide–reducing methanogenesis developed later through the evolution of tetrahydromethanopterin S-methyltransferase, which linked methanogenesis to the Wood-Ljungdahl pathway for energy conservation. Anaerobic multicarbon alkane metabolisms in Archaea also originated early, with genes coding for the activation of short-chain or even long-chain alkanes likely evolving from an ethane-metabolizing ancestor. These genes were likely horizontally transferred to multiple archaeal clades including Candidatus (Ca.) Bathyarchaeia, Ca. Lokiarchaeia, Ca. Hadarchaeia, and the methanogenic Ca. Methanoliparia.
INTRODUCTION
Methanogenesis, one of the most ancient biochemical pathways on Earth, also plays a critical role in global climate change, as this process largely controls the formation of methane, a strong greenhouse gas. Methanogenesis is exclusively found in the domain Archaea, and changes in its activity may have caused severe fluctuations of Earth’s surface temperature and subsequent life mass extinction events, e.g., the mass extinction potentially caused by the methanogenic burst in the end-Permian (1). By contrast, microbial methanogenesis might have prevented early snowball Earth scenarios during the Hadean and Archean eons by producing methane to keep a warm atmosphere in the “faint young Sun” period of Earth history (2, 3). However, methane can be also consumed in anoxic habitats by anaerobic methane-oxidizing archaea (ANME) using a reverse methanogenesis pathway (4, 5). It has been estimated that the anaerobic oxidation of methane removes ~80% of the methane produced by methanogenesis in the modern ocean, therefore keeping the atmospheric methane at a relatively low concentration, avoiding potential global warming effect (6).
Both geological evidence and molecular dating indicated that methanogenesis originated very early (2, 7), and it has been suggested that methanogens may represent one of the earliest life forms (8). The methyl–coenzyme M reductase (MCR) is the key enzyme of anaerobic methane metabolism, while the related alkyl–coenzyme M reductases (ACRs) catalyze the oxidation of multicarbon alkanes (4, 5, 9, 10). Most of the cultured methanogens reduce carbon dioxide with electron donors such as hydrogen. These organisms contain the enzymes of the Wood-Ljungdahl pathway to reduce carbon dioxide to tetrahydromethanopterin-bound methyl groups (11). Other methanogens thrive on the acetoclastic reaction or disproportionation of methylated substrates such as methanol to methane and carbon dioxide (11). ANME and the recently found anaerobic short-chain alkane-oxidizing archaea use the Wood-Ljungdahl pathway and MCR/ACR in an oxidative direction (9, 10, 12). The enzyme interconnecting the Wood-Ljungdahl pathway with MCR is the tetrahydromethanopterin S-methyltransferase (MTR) that transfers the methyl groups between the cofactors tetrahydromethanopterin and coenzyme M (11, 13). However, hydrogen-dependent methylotrophic archaea of the order Methanomassiliicoccales have been cultured, which lack both the Wood-Ljungdahl pathway and MTR and consequently require methylated compounds and hydrogen as electron acceptor and donor, respectively (14, 15). In recent years, environmental genomics has revealed many previously unknown lineages of potential methanogens across the archaeal species tree (16–22) [reviewed in (5)]. Phylogenomic and comparative genomic analyses of archaeal diversity have supported the hypothesis that the last common ancestor of Euryarchaeota and TACK (an archaeal superphylum including Thaumarchaeota, Aigarchaeota, Crenarchaeota, Korarchaeota, Geoarchaeota, and Bathyarchaeota, among other phyla) archaea might have been a methanogen (19–22). Some studies suggested that the first methanogens were carbon dioxide reducers using the Wood-Ljungdahl pathway (19). Others also pointed to the possibility that the ancestral methanogen was likely a hydrogen-dependent methylotroph (20). The finding of anaerobic multicarbon alkane-oxidizing archaea from different archaeal phyla suggested a more complex evolutionary history of ACR, potentially involving multiple horizontal gene transfers (HGTs) of ACR-encoding genes (20–22). Clearly elucidating the origin and evolution of methanogenesis and anaerobic alkane metabolisms requires better genomic sampling of early-diverging members of the Euryarchaeota and TACK archaea and mapping of mcr/acr and mtr gene family origins onto the archaeal species tree.
RESULTS AND DISCUSSION
Here, we studied the origin and diversification of methanogens and anaerobic multicarbon alkane-oxidizing archaea. To do so, we retrieved MCR/ACR-containing metagenome-assembled genomes (MAGs) from eight metagenome datasets collected from Tengchong hot spring in China and Captain Aryutinov mud volcano, two public metagenome datasets, and the Genomes from Earth’s Microbiomes (GEM) catalog (23). The data analyzed here [Table 1 and data file S1 (table S1)] comprise 24 MAGs binned from our own sequencing data, two MAGs binned from public metagenomic data, and 30 published MAGs from the GEM project (23). All of the data originally generated by other laboratories (including MAGs from GEM) were used with their permission, and we thank these colleagues for their generosity and willingness to share their data. The provenance of all data used is indicated in table S1 as well as the Acknowledgements for further details. The MAGs assigned to the class I (Methanobacteriales, Methanococcales, and Methanopyrales) and class II (Methanosarcinales, Methanomicrobiales, and Methanocellales) methanogens are well represented by genomes from cultured strains and, hence, were not included in subsequent analyses. A total of 56 MAGs of MCR/ACR-containing organisms mostly with completeness >80% (39 MAGs) and contamination below 5% (49 MAGs) were retained for further analyses [Table 1 and data file S1 (table S1)]. In the following discussion, we use the Genome Taxonomy Database (GTDB) to label and refer to MAGs (24), but in the interest of clarity, we retain the conventional names, i.e., Euryarchaeota, TACK, Asgard, and DPANN (an archaeal superphylum currently including Diapherotrites, Parvarchaeota, Aenigmarchaeota, Nanoarchaeota, Nanohaloarchaeota, and several other phyla), for higher taxonomic description. These MAGs and reference genomes of the domain Archaea from the National Center for Biotechnology Information (NCBI) prokaryotes database were used to construct a phylogenomic tree on the basis of a concatenated set of 37 marker genes [fig. S1 and data file S1 (tables S2 to S3)] (25). All MAGs with MCR/ACR affiliated with the Euryarchaeota, TACK, and Asgard archaea. Despite intensive search for additional MCR/ACR sequences on both NCBI prokaryotes and metagenomic databases (July 2019), no MCR/ACR-encoding gene was found in genomes assigned to the DPANN archaea or the domain Bacteria, indicating that our analysis covered the diversity of MCR/ACR-encoding archaeal lineages.
Table 1. MAGs described in this study and their main potential metabolic features.
| Classification | MAGs |
Metabolic
potentials |
Database
source |
|||
| Superphylum | Phylum | Class | Order or family | |||
| Euryarchaeota | Methanobacteriota | Thermococci | Ca. Nuwarchaeales | 6 | CH3-reducing methanogen |
Present study, PRJNA357337, and Nayfach et al. (23) |
| Thermoplasmatota | Thermoplasmata | Methanomassiliicoccales | 1 | CH3-reducing methanogen or C2H6 metabolism |
PRJNA443587 and Reiss et al. (75) |
|
| Halobacteriota | Ca. Methanoliparia | Ca. Methanoliparales | 1 | CO2-reducing methanogen with CnH2n+2 metabolism |
Nayfach et al. (23) | |
| Archaeoglobi | Archaeoglobales | 4 | CO2-reducing methanogen or CH4 oxidation |
Present study & Nayfach et al. (23) |
||
| Ca. Polytropaceales | 1 | CnH2n+2 oxidation | Nayfach et al. (23) | |||
| Methanosarcinia | Ca. Ethanoperedenaceae | 1 | C2H6 oxidation | Nayfach et al. (23) | ||
| Ca. Syntropharchaeia | Ca. Santabarbaracales | 1 | CnH2n+2 oxidation (n > 2) |
Hawley et al. (31, 32) and Nayfach et al. (23) |
||
| Ca. Alkanophagales | 1 | Dombrowski et al. (30) | ||||
| Ca. Hadarchaeota | Ca. Hadarchaeia | Ca. Hadarchaeales | 3 | CnH2n+2 oxidation | Nayfach et al. (23) | |
| TACK | Thermoproteota | Ca. Nezhaarchaeia | Ca. Nezhaarchaeales | 5 | CO2-reducing methanogen |
Present study, Nayfach et al. (23), and Dombrowski et al. (30) |
|
Ca. Methanomethylicia |
Ca. Methanomethylicales | 30 | CH3-reducing methanogen or C2H6 metabolism |
Present study and Nayfach et al. (23) |
||
| Ca. Korarchaeia | Ca. Korarchaeales | 1 | CH3-reducing methanogen | Nayfach et al. (23) | ||
| Asgard | Asgardarchaeota | Ca. Lokiarchaeia | Ca. Helarchaeales | 4 | CnH2n+2 oxidation | Present study |
Vertical evolution of MCR-encoding genes in Euryarchaeota and TACK
This and other recent metagenome-based studies (5, 16–22, 26) remarkably expanded the known distribution of MCR/ACR-encoding genes in the domain Archaea. In most of these archaea, the MCR alpha subunit (McrA)–encoding genes are well conserved and can be used as a phylogenetic marker, in the sense that their evolution follows the species tree (27). Consistent with previous reports (15), topologies of trees for McrA sequences and species trees in the current study are highly congruent for the class I and II methanogens in Euryarchaeota (Fig. 1, A and B, and fig. S1). McrA-encoding genes are found less frequently in TACK archaea, but the McrA-encoding genes that have been identified [in Nitrososphaeria, Candidatus (Ca.) Methanomethylicia, Ca. Korarchaeia, and Ca. Nezhaarchaeia] form a monophyletic group whose internal branching order is congruent with the inferred TACK species tree (Fig. 1, A and B, and figs. S1 and S2), potentially consistent with vertical inheritance of McrA from their common ancestor. However, the branching position of Ca. Korarchaeia varied when using different combinations of conserved genes and phylogenetic algorithm, placing Ca. Korarchaeia either close to Ca. Bathyarchaeia and Nitrososphaeria or to Thermoproteia and Ca. Methanomethylicia (20–22, 26). Nevertheless, with substitution model (LG+C60+F+G) of a better Bayesian information criterion (BIC) score, the phylogenomic tree of anaerobic alkane-metabolizing archaea with 37 concatenated conserved protein sequences shows identical tree topology with their McrA phylogenetic tree (Fig. 1, A and B, and fig. S2).
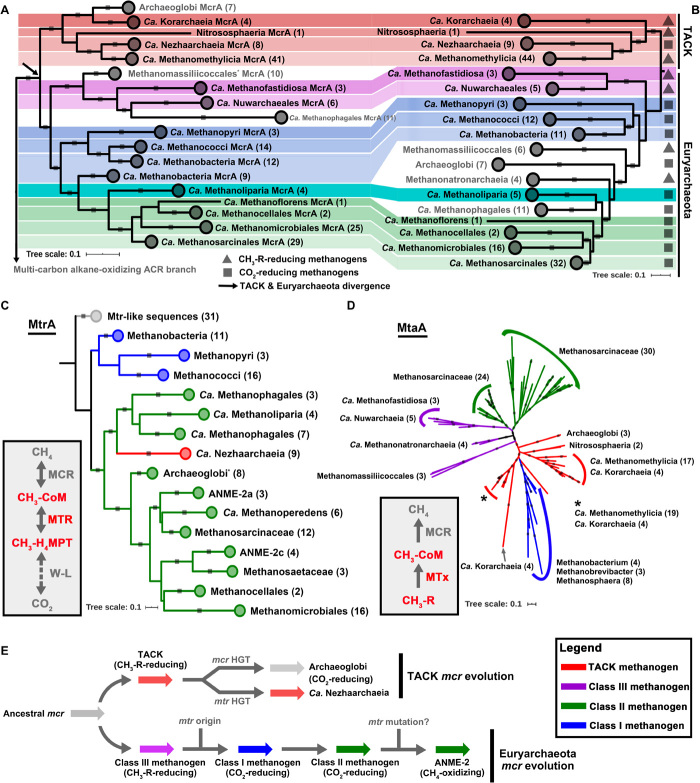
Fig. 1. Phylogenetic analyses of enzymes in archaeal methane metabolism.
(A) The McrA/AcrA phylogenetic tree is constructed on the basis of the alignments of 259 McrA/AcrA sequences with 472 aligned positions. Only the McrA branches are showed here. (B) Phylogenomic affiliation of 177 MAGs is based on 37 conserved protein sequences using representative methane metabolism archaea. Background colors: TACK, red shaded; class I methanogen, blue shaded; class II methanogen, green shaded; class III methanogen, pink shaded. (C) MtrA phylogenetic tree shows the classification of these sequences from different archaeal lineages. The phylogenetic tree is constructed on the basis of the alignments of 152 MtrA sequences with 148 aligned positions. Other phylogenetic trees are presented in fig. S5, which are in agreement with Liu et al. (73). The MtrA and H-like subunits are possibly either HGT from methanogens or originated before class I methanogens, as also described in Wang et al. (74). (D) One type of methyltransferase (MtaA, alpha subunit of methanol-corrinoid protein:coenzyme M methyltransferase) phylogenetic tree (134 sequences with 309 aligned positions) shows that methyltransferases are likely vertically transferred and might originate before the divergence of TACK and Euryarchaeota. Some class I methanogens that contain MtaA might be HGT from the TACK methanogens. Other methyltransferase phylogenetic trees are displayed in fig. S7. All conserved protein phylogenomic and McrA, MtrA, and MtaA phylogenetic alignments are based on MAFFT and then filtered with trimAl, and the trees that were built by the IQ-Tree method with model LG+C60+F+G using SH approximate likelihood ratio test implemented with 1000 bootstrap replicates with bootstrap higher than 0.8 are shown with gray squares on tree branches. (E) Evolutionary history of MCR-based methane metabolism.
In addition, we found that 13 MAGs assembled here (Table 1) and in previous studies (18, 20–22, 26) branch near the base of Euryarchaeota and TACK in both the MCR and species trees, helping provide insight into the early evolution of methanogenesis (Fig. 1, A and B, and fig. S1). Two early diverging lineages from Euryarchaeota branch between the TACK and Euryarchaeota class I methanogens: Ca. Methanofastidiosales and one group branching next to Ca. Methanofastidiosales (including a previously reported MAG, NM3) (20) belonging to a new order-level lineage, which we here propose as Ca. Nuwarchaeales (Nuwa, the mother of earth in the Chinese myth; fig. S3), a potential hydrogen-dependent methylotrophic methanogen without the Wood-Ljungdahl pathway as also described in (20). Ca. Methanofastidiosales and Ca. Nuwarchaeales branch together with Thermococci and Ca. Theionarchaea and are close to the divergence of Euryarchaeota and TACK on the species tree, which is known as a superclass Ca. Acherontia and has been reported previously (28) (fig. S1), such that comparisons of their gene content with other Euryarchaeota may elucidate early metabolic transitions within the clade. Their MCR sequences also form a monophyletic cluster on the MCR/ACR phylogenetic tree, and therefore, we here classified these MAGs as the class III methanogens (Fig. 1, A and B). Within the class I and II methanogens, the previously reported Ca. Methanoliparia (20, 29) branches between these two groups of methanogens. This position is supported by both MCR and genome trees, and the branching position of Ca. Methanoliparia in both the MCR and species trees strengthens the existing case (19–22) for vertical evolution of MCR-based metabolism within the class I and II methanogen clades, with lineage-specific losses giving rise to the metabolisms of methanogenesis and carbon fixation (Fig. 1, A and B, and fig. S1). Together, the results here suggest that the MCR-encoding genes of the TACK and Euryarchaeota methanogens have descended vertically from the common ancestor of the two superphyla, as suggested previously (19–22).
Although most methanogen lineages showed vertical evolution, we identified and confirmed some incongruences with the species tree, in accordance with a previous report (20) (Fig. 1, A and B, and fig. S1) (20). Within Euryarchaeota, McrA-encoding genes belonging to the class I/II (carbon dioxide–reducing) and class III (methyl-reducing) methanogens form reciprocally monophyletic lineages with high bootstrap support (100 and 93%, respectively); however, some methyl-reducing methanogens such as Methanomassiliicoccales and Methanonatronarchaeia have the class III MCR-encoding genes but branch within the class I/II methanogens on species trees (fig. S1). This pattern might reflect the sorting out of a putatively duplicated ancestral euryarchaeotal MCR-encoding gene through parallel gene losses or, alternatively, gene transfer of a single ancestral MCR-encoding gene during the diversification of Euryarchaeota (Fig. 1, A and B, and figs. S1 and S4). In the first scenario, ancestral Euryarchaeota should have both the class I and III MCR but gradually lost one during their evolution, and only one type of MCR-encoding gene was kept within their genomes. In the latter scenario, the MCR-encoding genes of the Methanomassiliicoccales/Methanonatronarchaeia clades would have been acquired from an ancient class III methanogen ancestral to Ca. Methanofastidiosales and Ca. Nuwarchaeales, and the deeper MCR lineage branching outside Methanomassiliicoccales on this cluster is still somewhere that was not sampled in this study. The phylogeny indicates that Ca. Methanophagales (previously called ANME-1), a lineage of methane oxidizers, may also have acquired MCR-encoding genes from a relative of Ca. Nuwarchaeales by HGT, as suggested by Borrel et al. (20). Ca. Methanophagales also contains genes for beta-oxidation, which would not be required for methane metabolism; however, this organism emerges from within a clade of archaea capable of multicarbon alkane metabolism and may have retained these beta-oxidation genes by vertical inheritance (fig. S3). All the neighboring branches of Ca. Methanophagales, including Ca. Syntrophoarchaeales, the Ca. Alkanophagales proposed here (previously called ANME-1 B39_G2) (30), and Ca. Santabarbaracales (from Santa Barbara Coal Oil Point) (31, 32), contain ACR and beta-oxidation enzyme-encoding genes for anaerobic multicarbon alkane metabolisms (fig. S3). Therefore, the Ca. Methanophagales ancestor, such as those archaea described above, may originally metabolize multicarbon alkanes rather than methane.
The ancestral methanogen likely reduced methylated compounds
The congruence between the topologies of the McrA phylogenetic tree and the methanogen species tree suggests that the common ancestor of Euryarchaeota and TACK archaea already contained methanogenic MCR-encoding genes (Fig. 1, A and B). Previous studies pointed out the possibility that both Euryarchaeota and TACK archaea combine the MCR and Wood-Ljungdahl pathways via the MTR complex and, therefore, that the ancestral methanogen might have been able to reduce carbon dioxide to methane (19). However, on the basis of our current knowledge, the combination of MCR, MTR, and the Wood-Ljungdahl pathway is restricted to the class I and II methanogens in Euryarchaeota, as well as Ca. Nezhaarchaeia in TACK. The other early-diverging lineages of methanogens within Euryarchaeota and TACK lack the MTR complex (Fig. 1, A to C, and fig. S3). To resolve the evolutionary history of MTR-encoding genes, we inferred phylogenetic trees of MTR subunits (Fig. 1C). For the Euryarchaeota class I and II methanogens, the MTR-encoding genes have very similar topologies as the corresponding phylogenomic trees. However, MTR-encoding genes from Ca. Nezhaarchaeia clusters with those of Ca. Methanophagales and Ca. Methanoliparia and an approximately unbiased (AU) test (33) for the catalytic subunit MtrA rejected the monophyly of Ca. Nezhaarchaeia MtrA cluster with other TACK archaea. This suggests a horizontal transfer of MTR-encoding genes from an ancestral lineage of Ca. Methanophagales/Ca. Methanoliparia to Ca. Nezhaarchaeia and, consequently, an origin of the MTR well within Euryarchaeota, likely in the class I methanogens (see MTR phylogenetic tree; Fig. 1C and fig. S5). Some archaea from TACK and Asgard contain MtrA or H-like subunits; however, these appear to be only distantly related to the enzymes of the MTR complex in methanogens and thus may have alternative functions (16, 22). To evaluate the origination points of the McrA- and MtrA-encoding gene families statistically, we used probabilistic gene tree–species tree reconciliation [the amalgamated likelihood estimation (ALE) method (34)] to estimate the probability that these gene families were present at each internal node of their rooted species tree. ALE reconciliations account for gene duplication, transfer, and loss by estimating the rates of these events for the data, and analysis of empirical data and simulations suggest that ALE is an accurate method for rooting gene trees (35), particularly in the presence of HGT (36). The results suggested that MtrA evolved later than McrA during archaeal evolution. The nodes in the species tree to which McrA could be mapped with high confidence [presence probability (PP) > 0.95] are the common ancestor of class I and II methanogens, Ca. Methanofastidiosales/Ca. Nuwarchaeales in Euryarchaeota, as well as Ca. Methanomethylicia, Ca. Korarchaeia, and Ca. Nezhaarchaeia in TACK (fig. S6). By contrast, the earliest node at which MtrA is present with high confidence (PP = 0.94 and PP > 0.95 on descendant nodes) is only within the class I and II methanogens following the divergence of Archaeoglobales (fig. S6). This is consistent with the hypothesis that MtrA evolved later than McrA in archaeal evolution. This late origin of MTR is consistent with a methylotrophic origin of methanogenesis and the subsequent evolution of carbon dioxide–reducing methanogenesis within Euryarchaeota. By contrast, in addition to MCR, both the Euryarchaeota and TACK methanogens contain a variety of methyltransferase genes such as methanol-corrinoid protein:coenzyme M methyltransferase (Mta), methylamine-corrinoid protein:coenzyme M methyltransferase (Mtb), and methylated-thiol-corrinoid protein:coenzyme M methyltransferase (Mts). These methyltransferases enable methanogens to use methyl compounds such as methanol, methylamine, and methanethiol. The phylogenetic analyses of these methyltransferases also indicate that they were likely vertically inherited and that they might be present in the common ancestor of Euryarchaeota and TACK archaea (Fig. 1D and fig. S7).
Hydrogen-dependent methyl-group reduction as the original metabolic mode of methanogens is consistent with the conditions of the early Earth. Our planet was anoxic but likely rich in methylated compounds and hydrogen, as well as other simple organic compounds (37, 38). These environments should have been suitable for hydrogen-dependent methyl-reducing methanogenesis. This finding does not contradict the hypothesis that both MCR and the Wood-Ljungdahl pathway were early metabolic traits of archaea (4, 8, 15, 28, 36, 39) because methane production via the methylotrophic methanogenesis pathway and carbon dioxide fixation through Wood-Ljungdahl pathway can work separately. Ancestral class I methanogens likely developed the MTR complex shortly after the divergence of the TACK and Euryarchaeota methanogens. The MTR connected the MCR with the Wood-Ljungdahl pathway and enabled energy generation by carbon dioxide reduction to methane. After that, some class II methanogens acquired the capability for cytochrome c synthesis that allows for more efficient methanogenic growth (11, 40). The class II methanogens are now the most successful on the modern Earth environments; members of this group thrive on carbon dioxide reduction, disproportion of methylated substrates, and acetate (11). With the accumulation of electron acceptors such as sulfate on Earth in the late Archean (41), several methanogenic lineages started to reverse the methanogenesis pathway and turned into anaerobic methane oxidizer coupling with sulfate-reducing bacteria or nitrate or metal oxides (4, 5). All known members of this group have cytochromes and are suggested to have crucial roles in direct interspecies electron transfer (42, 43).
Multicarbon alkane metabolism in Ca. Methanomethylicia and Methanomassiliicoccales
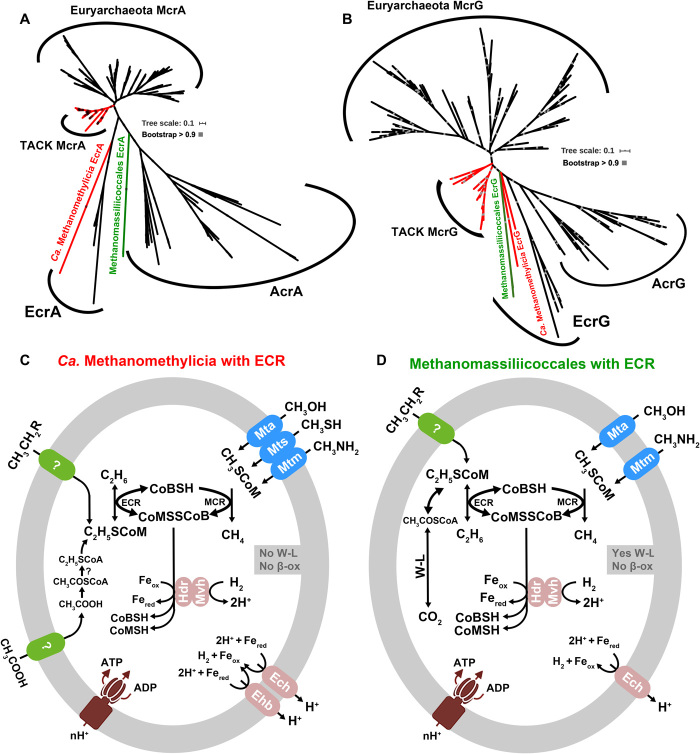
Experimentally characterized ACRs catalyze the anaerobic oxidation of multicarbon alkanes including ethane, propane, and n-butane, similar to the biochemical process in anaerobic methane oxidation, i.e., activating alkanes to alkyl–coenzyme M (9, 10). ACR-encoding genes are found in at least seven euryarchaeotal lineages (5), as well as in Ca. Bathyarchaeia from the TACK superphylum (16) and Ca. Lokiarchaeia from the Asgard superphylum (44). Among the ACR-containing MAGs assembled in the present study (Table 1), we identified five Ca. Methanomethylicia MAGs from five independent hot spring sediment samples that code both an MCR and an ACR (Fig. 2, A to C, and figs. S4 and S8). The contigs coding for these genes appears to be endogenous to Ca. Methanomethylicia because these genes show the highest amino acid identities (~87.2% on average) to sequences of other Ca. Methanomethylicia MAGs (fig. S9, A and B). One mcr gene clusters with those of other Ca. Methanomethylicia, and this gene is arranged with other genes of the methanogenesis pathway (fig. S9C). On the MCR/ACR phylogenetic tree, the ACR sequences of Ca. Methanomethylicia cluster closely but as a basal lineage with the ACR from Ca. Argoarchaeum (10) and Ca. Ethanoperedens (45). These organisms activate ethane as ethyl–coenzyme M; hence, we propose to assign these ACR sequences to the subgroup of ethyl–coenzyme M reductases (ECRs). The Ca. Methanomethylicia ECR-encoding genes are surrounded by genes coding for cobalamin-binding protein and carboxylesterase (fig. S9C), enzymes that might help metabolize reactants or products of ethane metabolism (Fig. 2C). In addition to the ECR-containing Ca. Methanomethylicia, we assembled one MAG from Methanomassiliicoccales in Euryarchaeota from peatland samples that also contain both MCR- and ECR-encoding genes as well as bacterial-type Wood-Ljungdahl pathway with tetrahydrofolate as C1 carrier (Fig. 2, A, B, and D). Both MCR/ECR-containing Ca. Methanomethylicia and Methanomassiliicoccales have several methyltransferases such as Mta, Mtb, and Mts that enable activation of methylated compounds such as methanol, methylamines, and methyl sulfides (Fig. 2, C and D, and fig. S3). Both organisms also contain genes coding for heterodisulfide reductase/F420-nonreducing hydrogenase complex and Ech hydrogenase (ECH) for heterodisulfide (CoB-SS-CoM) cycling and energy conservation. However, in contrast to Ca. Argoarchaeum, Ca. Ethanoperedens, and Methanomassiliicoccales, the ECR-containing Ca. Methanomethylicia lack the gene coding for the Wood-Ljungdahl pathway (the completeness of one MAG approaches 100% with 0.93% contamination); it therefore lacks an obvious pathway to oxidize coenzyme M–bound ethyl groups to carbon dioxide. In contrast, Ca. Methanomethylicia may rather reduce C2 compounds such as acetate or ethanol to ethane using hydrogen as an electron donor (Fig. 2, C and D). Biogenic ethanogenesis is thermodynamically feasible even at low hydrogen and acetate or ethanol concentrations, and this process has been postulated to occur in the subsurface and may account for ethanogenesis in anoxic sediments (46). Ethanogenic organisms are not yet cultured, but on the basis of their coded pathways, ECR-containing Ca. Methanomethylicia might perform such reactions; however, at this stage, we cannot rule out the possibility of oxidation of ethane or propane, etc., using unknown pathways.
Fig. 2. Phylogeny of MCR/ECR/ACR subunits and metabolic models of studied organisms.
(A) Phylogenetic analyses of McrA/EcrA/AcrA sequences. (B) Phylogenetic analyses of McrG/EcrG/AcrG sequences. McrAG/EcrAG/AcrAG phylogenetic alignments are based on MAFFT and then filtered with trimAl, and the trees were built by the IQ-Tree method with model LG+C60+F+G using SH approximate likelihood ratio test implemented with 1000 bootstrap replicates. (C) Metabolic model of MCR- and ECR-containing Ca. Methanomethylicia, one MAG with the completeness of 100% and contamination of 0.93%. (D) Metabolic model of MCR- and ECR-containing Methanomassiliicoccales, with the completeness of 83.24% and contamination of 6.45%. They contain both MCR and ECR, as well as methyltransferases and hydrogenases, and therefore have the potential to use methylated compounds to produce methane. MAGs from Ca. Methanomethylicia lack Wood-Ljungdahl (W-L) and beta-oxidation (β-ox) pathways, while the MAG from Methanomassiliicoccales has a bacterial-type Wood-Ljungdahl pathway but lacks a beta-oxidation pathway. Their ECR may enable them to gain energy from the reduction of acetate or ethyl compounds to produce ethane or oxidize ethane or propane, etc., using unknown pathways.
Origin of anaerobic multicarbon alkane metabolism in Archaea
Genes coding for putative anaerobic multicarbon alkane metabolism are widespread within three archaeal superphyla (5). MCR- and ACR-encoding genes are part of the same broader gene family. On the basis of the congruence of the MCR tree with their species tree (Fig. 1, A and B), MCR- and ACR-encoding genes likely originated from an ancient gene divergence that took place before the radiation of the Euryarchaeota and TACK archaea. However, while the MCR subfamily has evolved largely vertically during archaeal evolution, ACR-encoding genes appear to have been subjected to frequent horizontal transfer (Fig. 2, A and B). Reconciliation of the MCR and ACR subtrees of the MCR/ACR gene family independently against the archaeal species tree suggested that the MCR subfamily is the older of the two, appearing coincident with the family as a whole in the common ancestor of class I and II methanogens at the latest (PP = 0.95). By contrast, other ACR genes can only be mapped confidently (PP > 0.95) to relatively shallow nodes within the Euryarchaeota and TACK archaea. Together, these analyses are consistent with a root for the ACR/MCR tree on, or within, the MCR subfamily (fig. S6). The exclusion of the root from the ACR clade indicates that the ACR sequences that bind longer-chain alkanes evolved from within a paraphyletic grade of ECR sequences, including those newly discovered from Ca. Methanomethylicia and Methanomassiliicoccales. This suggests that the enzymes that potentially metabolize longer-chain alkanes, such as the ACRs in Ca. Lokiarchaeia in Asgard, Ca. Bathyarchaeia in TACK, and Ca. Hadarchaeia and Ca. Methanoliparia, evolved or transferred from those involved in potential ethane metabolism. By extension, it is tempting to speculate that the genes of potential ethane metabolism might have evolved from within the MCR clade (i.e., that the root lies on either the TACK MCR or Euryarchaeota MCR stem branches, two of the three root positions that are not rejected by the ALE analysis). Alternatively, they might have evolved from an ancestral gene coding for methyl-binding enzyme at the root of the MCR/ACR family. However, since there are still not enough ECR/ACR-containing MAGs that have been sampled up to now, our calculation only showed a strong statistic earliest node within the Euryarchaeota, perhaps also as a result of high rates of gene transfers and losses for this family (Fig. 3, A and B). Comparison of sequence identity among the MCR-, ECR-, and ACR-encoding genes (fig. S10) indicates that ECR-encoding genes show greater sequence identities to MCR-encoding genes generally and, at least for EcrG, TACK MCR-encoding genes specifically, than to other ACR-encoding genes. Speculatively, this higher sequence identity might indicate a specific evolutionary relationship between ECR-encoding genes and TACK MCR-encoding genes, as might be expected if TACK MCR and all ACRs form a clade on the rooted gene tree. However, this pattern might also reflect lower rates of sequence evolution in TACK MCR- and ECR-encoding genes compared to other ACR family members.
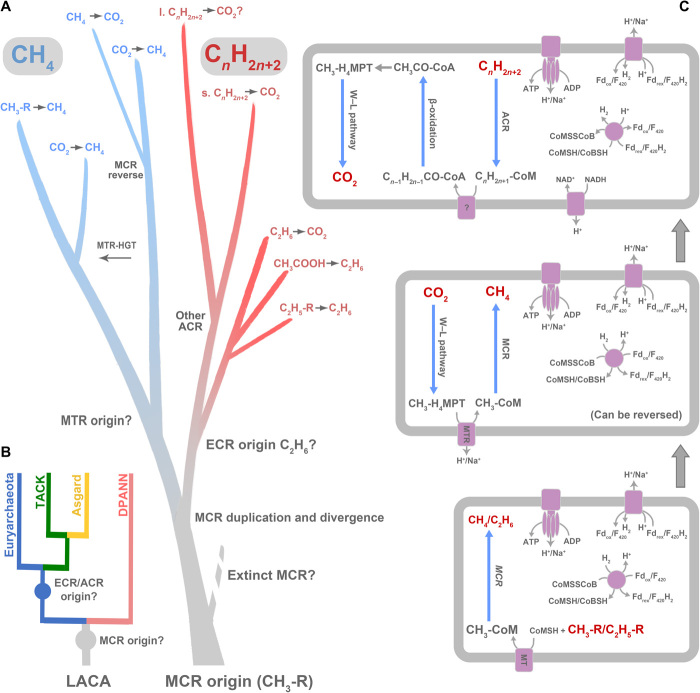
Fig. 3. Evolutionary history of MCR/ECR/ACR-based anaerobic alkane metabolisms.
(A) A scenario for the evolutionary history of MCR/ECR/ACR-encoding genes. After the emergence of an ancestral MCR that reduced methylated compounds, in the common ancestor of Euryarchaeota, TACK, and Asgard, MCR was inherited by each of the two lineages. ECR/ACR evolved before the radiation of extant Euryarchaeota on the euryarchaeotal or TACK stem or potentially earlier. MTR originated in Euryarchaeota and was subsequently horizontally transferred to Ca. Nezhaarchaeia in TACK. Before the radiation of Euryarchaeota, a putative duplication of MCR-encoding genes created the ECR-encoding genes, which later evolved to metabolize longer-chain alkanes and spread by both vertical descent and horizontal transfer in the domain Archaea. “l.” refers to long chain, and “s.” refers to short chain. (B) A cartoon phylogenetic tree for archaeal evolution and MCR/ECR/ACR divergence. MCR-encoding gene might have originated by the time of the last archaea common ancestor (LACA), assuming that DPANN had lost their MCR-encoding gene during evolution. ECR/ACR might have been present in LACA or the stem lineage of Euryarchaeota, TACK, and Asgard but underwent multiple HGT processes. (C) Predicted ancestral MCR/ECR/ACR-based anaerobic alkane metabolism evolution in Archaea. First, a methyl-reducing methanogen or an ethyl-metabolizing archaea appeared. Second, the origin of MTR complex created the carbon dioxide-reducing methanogen; then, the introduction of beta-oxidation pathway enabled the anaerobic multicarbon alkane-metabolizing archaea.
A root for the gene tree on the ACR stem would imply that in addition to MCR, at least one ACR-encoding gene was present at one time in a lineage ancestral to extant TACK and Asgard archaea, either in the common ancestor of TACK and Euryarchaeota. However, this ACR clade appears to be absent from the described genomes of extant TACK, with the possible exception of the new sequences from Ca. Methanomethylicia. Trees of Mcr/Ecr/AcrA and G suggest that the new Ca. Methanomethylicia and Methanomassiliicoccales sequences branch either within the ECR clade or between MCRs and all ACRs. Thus, one possibility is that these sequences represent a vestige of this otherwise extinct, or unsampled, ACR lineage (Fig. 2, A and B). Exploration of MAGs from other extreme environments such as hot springs or hydrothermal vents will help determine whether other TACK archaea encode relatives of this deep-branching ACR lineage. Alternatively, the Ca. Methanomethylicia sequences might have been acquired by gene transfer from an unsampled deep-branching euryarchaeotal lineage.
The second scenario focuses on metabolic features of the ACR-containing archaea. All those deep-branching organisms with ACR such as Ca. Bathyarchaeia, Ca. Lokiarchaeia, and Ca. Hadarchaeia encode the Wood-Ljungdahl and beta-oxidation pathways and, hence, could potentially oxidize long-chain alkanes. Notably, they do not contain cytochromes and hence likely interact with partner organisms via interspecies hydrogen or formate transfer (47). Considering the low sulfate levels in the Archean ocean (41), at that time, the partners of the alkane oxidizers might have been rather methanogens than sulfate reducers, or they might use a methanogenic alkane degradation pathway, which requires a chain length of at least six carbons (hexane) (48). In contrast, the more modern lineages of Ca. Syntrophoarchaeia have cytochromes that allow direct electron transfer with cytochrome-containing partners such as Ca. Desulfofervidus (49). The phylogenetic positions of these ACR sequences, from one Archaeoglobi lineage and members in the Euryarchaeota class Ca. Syntrophoarchaeia, match with their species tree (fig. S11). This suggests that these ACR-encoding genes might be vertically inherited from their last common ancestor.
Dating the origin and subsequent evolution of anaerobic alkane metabolisms
The acquisition of SMC (structural maintenance of chromosomes) protein-encoding genes by the cyanobacterial ancestor from an archaeal lineage in Euryarchaeota, most likely within Methanomicrobia or Halobacteria, has been recently used to date the origin of euryarchaeal methanogens before 3.51 billion years (Ga) ago (2). We here combined the temporal constraint implied by this HGT event to estimate the time at which methanogenesis first evolved and the divergence times of carbon dioxide–reducing methanogens. Because the euryarchaeotal SMC donor lineage must be older than the common ancestor of Cyanobacteria, we can import an absolute time constraint from Cyanobacteria [which have fossil records (50, 51)] to date events within Archaea, for which no unambiguous fossil record is available. The SMC gene partition provides a fossil calibration for the archaeal tree at the point at which the cyanobacterial clade branches inside Euryarchaeota (fig. S12). The SMC sequences of Cyanobacteria cluster with Halobacteria and Methanomicrobia in Euryarchaeota, and the topology within cyanobacterial clade for the SMC phylogenetic tree shows high congruence with the phylogenomic tree of nonphotosynthetic and photosynthetic Cyanobacteria, indicating that HGT of SMC-encoding genes occurred even long before the origin of photosynthetic Cyanobacteria lineages (fig. S12). On the basis of the timing constraints of Cyanobacteria fossils (50, 51) and the predicated divergence of archaea and bacteria (52), we predict that the common ancestor of methanogenic archaea originated at ~3.8 to 4.1 Ga ago, at the border between the Hadean and the Archean eon (Fig. 3, A and B, and figs. S12 and S13). The age of the common ancestor of class I and II methanogens, that is, a conservative estimate for the origin of MCR genes based on the earliest node to which MCR could be mapped with high confidence in the ALE analysis, was 3.66 ± 0.20 Ga. The posterior age estimates for the key nodes in the analysis of the tree built by the conserved ribosome protein plus SMC sequence alignment from Archaea and Cyanobacteria were similar even in an analysis that did not include the sequence alignment, e.g., age of the most recent common ancestor of Euryarchaeota, TACK, and Asgard at 3.89 ± 0.21 Ga (fig. S13), suggesting that the tree topology and calibrations are providing much of the information for this analysis, as has been noted previously for estimates of ancestral methanogen age (2, 53, 54). This is earlier than the recent prediction for the origin of methanogenesis (55), and the authors also stated that the discovery of non-euryarchaeal methanogens would result in an even earlier origin of methanogenesis. Our analyses indicate that the first methanogen was a hydrogen-dependent methylotroph (Fig. 3, A to C), a possibility that has been raised previously (20). It is not clear whether methanogenesis is at the root of all archaea, as, so far, MCR-encoding genes have not been found in the DPANN superphylum. This might be because the DPANN genomes have undergone reductive evolution (56), as it remains difficult to determine whether the absence of MCR-encoding genes is ancestral to the DPANN clade or has resulted from gene loss. If so, the archaeal ancestor would be capable of anaerobic alkane metabolism. Following the divergence of Euryarchaeota and TACK, some Euryarchaeota developed the MTR complex that provided a link between the MCR and the Wood-Ljungdahl pathway. The latter pathway might have first been developed or acquired to allow an autotrophic lifestyle. With this, the class I carbon dioxide–reducing methanogens emerged, possibly before ~3.66 Ga ago (Fig. 3, A to C), supporting the geological evidence that methane produced via carbon dioxide reduction as characterized by highly depleted isotope signatures was dated back to 3.46 Ga ago (7). In TACK, the ancestral methyl-reducing methanogenesis pathway persisted, although some members such as Ca. Nezhaarchaeia might have acquired the MTR complex from Euryarchaeota (Fig. 1C).
Together, our results indicate that methanogenesis developed soon after the divergence of Bacteria and Archaea, possibly in the late Hadean period. The first methanogen was likely a hydrogen-dependent methylotrophic archaeon. Life likely originated at hydrothermal vents or serpentinization sites that provided ideal conditions for such methanogens (fig. S14) (8, 57, 58). The temperatures were elevated compared to the otherwise possibly cold planet owing to the low illumination from the young Sun, and large amounts of molecular hydrogen and simple organic compounds such as methanol and acetate would provide separated carbon and energy sources (59). Meanwhile, microorganisms may have developed the Wood-Ljungdahl pathway either for carbon fixation or energy conservation from acetate metabolism. The later evolution of MTR could have allowed methanogenesis from carbon dioxide reduction, which would have strongly increased methane production on Earth. In the anoxic Archean atmosphere, methane remained ~1000 times longer than today (60), and hence, methanogenesis may have had a crucial impact on the climate of the early Earth. The accumulating methane would enhance an early greenhouse effect and retain the radiation from the young Sun, which would increase the surface temperature on Earth, providing suitable habitats for other life to evolve.
MATERIALS AND METHODS
Raw sequencing reads treatment and genome binning
Six sediment samples from Tengchong hot spring in China and two methane-enriched samples from Captain Aryutinov mud volcano were collected, and DNA were extracted and sequenced on Illumina NovaSeq platform (Novogene Co.). Two metagenomic datasets from NCBI Sequence Read Archive (SRA) were downloaded [data file S1 (table S1)], and GEM catalog was downloaded from the website (https://genome.jgi.doe.gov/portal/GEMs/GEMs.home.html) (23). The raw sequencing reads were trimmed using the Sickle algorithm version 1.33 (https://github.com/najoshi/sickle). The trimmed reads were assembled using MEGAHIT version 1.0.6-hotfix1 (61) with step 4 and/or SPAdes-3.13.1 (62) with k-step 4. We then also downloaded the McrA/AcrA sequences from the NCBI protein nr database and built a local McrA/AcrA database by DIAMOND version 0.8.28.90 (http://github.com/bbuchfink/diamond). A DIAMOND search of potential McrA/AcrA sequences was carried out for the assembled metagenomic datasets, and McrA/AcrA sequences with best hits to the recently published alkane-metabolizing McrA/AcrA sequences or McrA/AcrA with low identities or without the known sequences were selected [data file S1 (table S1)]. Then, each assembled contig coverage was determined by mapping the trimmed reads back to the contigs using Bowtie version 2.2.8 (63) with parameter --very-sensitive. The assembled metagenomic sequences were binned on the basis of MaxBin version 2.2.4 (64) with the run_MaxBin.pl script and on the basis of abundance and tetranucleotide frequency using MetaBAT version 2.12.1 (65) with 1 kb (or 1.5 kb) and 3 kb as contig length cutoffs. The 4-mer and 5-mer frequencies of each contig were calculated, and dimensionality was reduced by t-sne clustering algorithm (https://github.com/lejon/T-SNE-Java/tree/master/tsne-core) (66). The selected MAGs were then checked with the mmgenome package version 0.6.3. Final completeness and contamination of each MAG [data file S1 (table S1)] were assessed with CheckM version 1.0.7 (67) using lineage-specific (149 to 228) marker genes, and the MAGs with higher completeness and lower contamination were selected from each dataset for further analyses. Taxonomic classification was calculated using GTDB-Tk (24) with slight modification based on the potential metabolic differences. Open reading frames (ORFs) of these MAGs were predicted with Prodigal version 2.6.3 (https://github.com/hyattpd/Prodigal). The predicted ORFs were searched against the NCBI nr protein database (July 2019) and eggnog (68) database with the BLASTP algorithm (e values < 1 × 10−10) to check their protein identities to the most closely related sequences using DIAMOND (data file S2). For metabolic pathway analyses, we used the web portal GhostKOALA on the Kyoto Encyclopedia of Genes and Genomes website (www.kegg.jp/ghostkoala/).
Phylogenetic analyses based on conserved proteins and Mcr/Ecr/Acr, SMC, Mtr, Mts, Mtb, and Mta protein sequences
For phylogenomic analysis, a total of 337 representative archaea reference genomes [data files S1 (table S2)] from the superphyla Euryarchaeota, TACK, Asgard, and DPANN were downloaded from the NCBI prokaryote genome database (www.ncbi.nlm.nih.gov/assembly/). These reference genomes and the MAGs from this study were used to construct a phylogenomic tree on the basis of a concatenated alignment of a set of 37 marker genes as suggested by (25) [data file S1 (table S3)]. Specifically, each of the 37 marker protein sequences from the reference genomes and the MAGs was aligned using the MAFFT algorithm version 7.313 (https://mafft.cbrc.jp/alignment/software/) with parameters --ep 0 –genafpair --maxiterate 1000 and filtered with trimAl version 1.4.rev2 (https://sourceforge.net/projects/trimal) with parameter -automated1. Then, all 37 marker genes were concatenated into a single alignment, and phylogenetic trees were built using IQ-Tree version 1.6.6 (69) with model LG+C60+F+G with bootstrap value of 1000. The tree was rooted at DPANN, as suggested by Williams et al. (36). For the phylogenetic analysis of functional marker proteins (Mcr/Ecr/Acr, SMC, Mtr, Mts, Mtb, and Mta; data file S3), the protein sequences were retrieved from the MAGs and the reference genomes in the present study by a DIAMOND search of reference protein sequences with parameters of coverage >75% (e values < 1 × 10−20). Alignment and filtering were carried out with the same programs described above, and phylogenetic trees were built using IQ-Tree version 1.6.6 with the fitting model chosen according to the BIC criterion (LG+C60+F+G) with 1000 ultrafast bootstraps. The ALE analyses were performed using the maximum likelihood implementation of the undated ALE algorithm using a sample of 1000 ultrafast bootstrap trees for each gene family (McrA/EcrA/AcrA and MtrA) and their species trees to estimate conditional clade probabilities.
Divergence time estimation
Molecular timing analyses were conducted by three methods, i.e., Bayesian analysis of molecular sequences using Markov chain Monte Carlo (MCMC) BEAST version 1.10.4 (70) with Yela relaxed model, Bayesian estimation of species divergence times using soft fossil constraints MCMCTree version 4.9c (71) with JC69 model, and treePL (72) with thorough option, which will continue iterating until convergence. Two different age constraints were considered for the divergence time estimation. One is the fossil evidence of the cyanobacterial clades Nostocales and Stigonematales (1.2 to 2.0 Ga, fossil resting cells similar to the Nostocales and Stigonematales from Franceville Group of Gabon) (2, 50, 51). The other is the predicted differentiation time of bacterial and archaeal lineages, i.e., 4.26 Ga (52). The SMC phylogenetic tree calculating with model LG+C60+F+G by IQ-Tree were used for BEAST, MCMCTree, and treePL configurations. The 37 conserved protein sequences plus SMC from archaea in the present study and Cyanobacteria were firstly concatenated, and then, the phylogenomic tree was also constructed with model LG+C60+F+G by IQ-Tree. The time tree was calculated by BEAST version 1.10.4 with cyanobacterial clades Nostocales and Stigonematales (1.2 to 2.0 Ga) and the time inferred from (2).
Acknowledgments
We wish to clarify an unusual situation regarding the work presented here. An earlier version of this paper unintentionally included data that had been deposited in the NCBI SRA database but was under embargo in another database; to protect the interests of the original data generators, we retracted that version of the paper. In this updated and reworked paper, all of the data generated by other laboratories that we analyzed here were used with the explicit permission of the original data generators whom we thank for generously sharing the data with us. In particular, we thank Z. Du, R. Reiss, E. Lilleskov, C. Schadt, B. Hedlund, S. Caffrey, K. McMahon, R. Stepanauskas, R. Kelly, M. Hess, I. N. Sierra Garcia, and B. J. Baker for help, as well as the GEM project. We are grateful to the researchers who published their sequence data on NCBI (www.ncbi.nlm.nih.gov/) and the U.S. Department of Energy (DOE) Joint Genome Institute (JGI) (https://jgi.doe.gov/), a DOE Office of Science User Facility, supported under contract no. DE-AC02-05CH11231. We also thank the scientists who analyzed the public datasets and assembled MAGs that were reassembled in the current study. The computations here were run on the π 2.0 cluster supported by the Center for High Performance Computing at Shanghai Jiao Tong University and with help from C. Li and J. Wang, and we thank W. Liu for the MAGs submission to eLMSG (eLibrary of Microbial Systematics and Genomics; www.biosino.org/elmsg/index). We also thank E. Lilleskov for valuable suggestions for this manuscript and J. Wang and X. Feng for useful discussion. Funding: We thank the following sources for funding: the National Key Research and Development Program of China (grant nos. 2018YFC0309800 and 2016YFA0601102), COMRA project (DY135-B2-12), the National Natural Science Foundation of China (grant nos. 41525011, 41902313, 91751205, and 92051116), the Natural Science Foundation of Shanghai (20ZR1428000), the Senior User Project of RV KEXUE (KEXUE2019GZ06), and Shanghai Jiao Tong University interdisciplinary grant (20CX-01). G.W. was supported by the DFG Cluster of Excellence, The Ocean Floor-Earth’s Uncharted Interface (EXC-2077-390741603) at MARUM, University of Bremen. T.A.W. is supported by a Royal Society University Research Fellowship (UF140626). Author contributions: Y.W. designed the research, performed the analyses, and wrote the paper. G.W. provided knowledge on metabolism of archaea and wrote the paper. T.A.W. provided guidance on inferring and interpreting phylogenies, performed gene tree–species tree analyses, and wrote the paper. R.X. performed the analyses of evolutionary time calculation. J.H. performed double-blind assessments of the MAGs. C.T. and Y.Z. provide important MAGs in the present study. F.W. and X.X. performed the analyses and wrote the paper. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All scripts and analyses necessary to perform metagenome processing can be accessed from GitHub (https://github.com/) or the websites provided in the original research articles. The specific links to the custom software are listed below: DIAMOND version 0.8.28.90, http://ab.inf.uni-tuebingen.de/software/diamond/; Sickle version 1.33, https://github.com/najoshi/sickle; MEGAHIT version 1.0.6-hotfix1, https://hku-bal.github.io/megabox/; Bowtie version 2.2.8, http://bowtie-bio.sourceforge.net/bowtie2/index.shtml; Prodigal version 2.6.3, https://github.com/hyattpd/Prodigal; MaxBin version 2.2.4, http://sourceforge.net/projects/maxbin/; MetaBAT version 2.12.1, https://bitbucket.org/berkeleylab/metabat; CheckM version 1.0.7, http://ecogenomics.github.io/CheckM; compareM version 0.0.23, https://github.com/dparks1134/CompareM; MAFFT version 7.313, https://mafft.cbrc.jp/alignment/software/; trimAl version 1.4.rev2, http://trimal.cgenomics.org; IQ-Tree version 1.6.6, www.cibiv.at/software/iqtree; t-SNE version 2.4, https://github.com/lejon/T-SNE-Java/tree/master/tsne-core; treePL, https://github.com/blackrim/treePL; BEAST, http://beast.community/; and MCMCTree, http://abacus.gene.ucl.ac.uk/software/MCMCtree.Tutorials.pdf. The public datasets analyzed during the current study are available in NCBI (www.ncbi.nlm.nih.gov/) and JGI (https://img.jgi.doe.gov/). The genomes from the GEM project can be downloaded at (https://genome.jgi.doe.gov/portal/GEMs/GEMs.home.html). The genomes from the current study have been deposited in eLMSG (www.biosino.org/elmsg/index) under accession numbers LMSG_G000001525.1 to LMSG_G000001550.1. Reprint and permission information is available online.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/27/eabj1453/DC1
REFERENCES AND NOTES
- 1.Rothman D. H., Fournier G. P., French K. L., Alm E. J., Boyle E. A., Cao C., Summons R. E., Methanogenic burst in the end-Permian carbon cycle. Proc. Natl. Acad. Sci. U.S.A. 111, 5462–5467 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolfe J. M., Fournier G. P., Horizontal gene transfer constrains the timing of methanogen evolution. Nat. Ecol. Evol. 2, 897–903 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Sauterey B., Charnay B., Affholder A., Mazevet S., Ferrière R., Coevolution of primitive methane-cycling ecosystems and early Earth’s atmosphere and climate. Nat. Commun. 11, 2705 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Evans P. N., Boyd J. A., Leu A. O., Woodcroft B. J., Parks D. H., Hugenholtz P., Tyson G. W., An evolving view of methane metabolism in the Archaea. Nat. Rev. Microbiol. 17, 219–232 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Wang Y., Wegener G., Ruff S. E., Wang F., Methyl/alkyl-coenzyme M reductase-based anaerobic alkane oxidation in archaea. Environ. Microbiol. 23, 530–541 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Knittel K., Boetius A., Anaerobic oxidation of methane: Progress with an unknown process. Annu. Rev. Microbiol. 63, 311–334 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Ueno Y., Yamada K., Yoshida N., Maruyama S., Isozaki Y., Evidence from fluid inclusions for microbial methanogenesis in the early Archaean era. Nature 440, 516–519 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Martin W. F., Sousa F. L., Early microbial evolution: The age of anaerobes. Cold Spring Harb. Perspect. Biol. 8, a018127 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laso-Pérez R., Wegener G., Knittel K., Widdel F., Harding K. J., Krukenberg V., Meier D. V., Richter M., Thermophilic archaea activate butane via alkyl-coenzyme M formation. Nature 539, 396–401 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Chen S. C., Musat N., Lechtenfeld O. J., Paschke H., Schmidt M., Said N., Popp D., Calabrese F., Stryhanyuk H., Jaekel U., Zhu Y. G., Joye S. B., Richnow H.-H., Widdel F., Musat F., Anaerobic oxidation of ethane by archaea from a marine hydrocarbon seep. Nature 568, 108–111 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Thauer R. K., Kaster A. K., Seedorf H., Buckel W., Hedderich R., Methanogenic archaea: Ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 6, 579–591 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Hallam S. J., Putnam N., Preston C. M., Detter J. C., Rokhsar D., Richardson P. M., DeLong E. F., Reverse methanogenesis: Testing the hypothesis with environmental genomics. Science 305, 1457–1462 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Gottschalk G., Thauer R. K., The Na+-translocating methyltransferase complex from methanogenic archaea. BBA-Bioenergetics 1505, 28–36 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Borrel G., O’Toole P. W., Harris H. M., Peyret P., Brugere J. F., Gribaldo S., Phylogenomic data support a seventh order of methylotrophic methanogens and provide insights into the evolution of methanogenesis. Genome Biol. Evol. 5, 1769–1780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borrel G., Adam P. S., Gribaldo S., Methanogenesis and the Wood-Ljungdahl pathway: An ancient, versatile, and fragile association. Genome Biol. Evol. 8, 1706–1711 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evans P. N., Parks D. H., Chadwick G. L., Robbins S. J., Orphan V. J., Golding S. D., Tyson G. W., Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics. Science 350, 434–438 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Vanwonterghem I., Evans P. N., Parks D. H., Jensen P. D., Woodcroft B. J., Hugenholtz P., Tyson G. W., Methylotrophic methanogenesis discovered in the archaeal phylum Verstraetearchaeota. Nat. Microbiol. 1, 16170 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Nobu M. K., Narihiro T., Kuroda K., Mei R., Liu W. T., Chasing the elusive Euryarchaeota class WSA2: Genomes reveal a uniquely fastidious methyl-reducing methanogen. ISME J. 10, 2478–2487 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berghuis B. A., Yu F. B., Schulz F., Blainey P. C., Woyke T., Quake S. R., Hydrogenotrophic methanogenesis in archaeal phylum Verstraetearchaeota reveals the shared ancestry of all methanogens. Proc. Natl. Acad. Sci. U.S.A. 116, 5037–5044 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borrel G., Adam P. S., McKay L. J., Chen L. X., Sierra-García I. N., Sieber C. M., Letourneur Q., Ghozlane A., Andersen G. L., Li W. J., Hallam S. J., Muyzer G., de Oliveira V. M., Inskeep W. P., Banfield J. F., Gribaldo S., Wide diversity of methane and short-chain alkane metabolisms in uncultured archaea. Nat. Microbiol. 4, 603–613 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y., Wegener G., Hou J., Wang F., Xiao X., Expanding anaerobic alkane metabolism in the domain of Archaea. Nat. Microbiol. 4, 595–602 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Hua Z. S., Wang Y. L., Evans P. N., Qu Y. N., Goh K. M., Rao Y. Z., Qi Y. L., Li Y. X., Huang M. J., Jiao J. Y., Chen Y. T., Insights into the ecological roles and evolution of methyl-coenzyme M reductase-containing hot spring Archaea. Nat. Commun. 10, 4574 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nayfach S., Roux S., Seshadri R., Udwary D., Varghese N., Schulz F., Wu D., Paez-Espino D., Chen I. M., Huntemann M., Palaniappan K., Ladau J., Mukherjee S., Reddy T. B. K., Nielsen T., Kirton E., Faria J. P., Edirisinghe J. N., Henry C. S., Jungbluth S. P., Chivian D., Dehal P., Wood-Charlson E. M., Arkin A. P., Tringe S. G., Visel A.; IMG/M Data Consortium, Woyke T., Mouncey N. J., Ivanova N. N., Kyrpides N. C., Eloe-Fadrosh E. A., A genomic catalog of Earth’s microbiomes. Nat. Biotechnol. 39, 499–509 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chaumeil P.-A., Mussig A. J., Hugenholtz P., Parks D. H., GTDB-Tk: A toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 36, 1925–1927 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hug L. A., Baker B. J., Anantharaman K., Brown C. T., Probst A. J., Castelle C. J., Butterfield C. N., Hernsdorf A. W., Amano Y., Ise K., Suzuki Y., A new view of the tree of life. Nat. Microbiol. 1, 16048 (2016). [DOI] [PubMed] [Google Scholar]
- 26.McKay L. J., Dlakić M., Fields M. W., Delmont T. O., Eren A. M., Jay Z. J., Klingelsmith K. B., Rusch D. B., Inskeep W. P., Co-occurring genomic capacity for anaerobic methane and dissimilatory sulfur metabolisms discovered in the Korarchaeota. Nat. Microbiol. 4, 614–622 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Friedrich M. W., Methyl-coenzyme M reductase genes: Unique functional markers for methanogenic and anaerobic methane-oxidizing Archaea. Method Enzymol. 397, 428–442 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Adam P. S., Borrel G., Brochier-Armanet C., Gribaldo S., The growing tree of Archaea: New perspectives on their diversity, evolution, and ecology. ISME J. 11, 2407–2425 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laso-Pérez R., Hahn C., van Vliet D. M., Tegetmeyer H. E., Schubotz F., Smit N. T., Pape T., Sahling H., Bohrmann G., Boetius A., Knittel K., Wegener G., Anaerobic degradation of nonmethane alkanes by “Candidatus Methanoliparia” in hydrocarbon seeps of the Gulf of Mexico. MBio 10, e01814-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dombrowski N., Teske A. P., Baker B. J., Expansive microbial metabolic versatility and biodiversity in dynamic Guaymas Basin hydrothermal sediments. Nat. Commun. 9, 4999 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hawley E. R., Piao H., Scott N. M., Malfatti S., Pagani I., Huntemann M., Chen A., del Rio T. G., Foster B., Copeland A., Jansson J., Pati A., Tringe S., Gilbert J. A., Lorenson T. D., Hess M., Metagenomic analysis of microbial consortium from natural crude oil that seeps into the marine ecosystem offshore Southern California. Stand Genomic Sci. 9, 1259–1274 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hawley E. R., Malfatti S. A., Pagani I., Huntemann M., Chen A., Foster B., Copeland A., del Rio T. G., Pati A., Jansson J. R., Gilbert J. A., Tringe S. G., Lorenson T. D., Hess M., Metagenomes from two microbial consortia associated with Santa Barbara seep oil. Mar. Genomics 18, 97–99 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Shimodaira H., An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 51, 492–508 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Szöllősi G. J., Rosikiewicz W., Boussau B., Tannier E., Daubin V., Efficient exploration of the space of reconciled gene trees. Syst. Biol. 62, 901–912 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coleman G. A., Davín A. A., Mahendrarajah T. A., Szánthó L. L., Spang A., Hugenholtz P., Szöllősi G. J., Williams T. A., A rooted phylogeny resolves early bacterial evolution. Science 372, eabe0511 (2021). [DOI] [PubMed] [Google Scholar]
- 36.Williams T. A., Szöllősi G. J., Spang A., Foster P. G., Heaps S. E., Boussau B., Ettema T. J., Embley T. M., Integrative modeling of gene and genome evolution roots the archaeal tree of life. Proc. Natl. Acad. Sci. U.S.A. 114, E4602–E4611 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chyba C. F., Thomas P. J., Brookshaw L., Sagan C., Cometary delivery of organic molecules to the early Earth. Science 249, 366–373 (1990). [DOI] [PubMed] [Google Scholar]
- 38.Tian F., Toon O. B., Pavlov A. A., de Sterck H., A hydrogen-rich early Earth atmosphere. Science 308, 1014–1017 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Adam P. S., Borrel G., Gribaldo S., An archaeal origin of the Wood-Ljungdahl H4MPT branch and the emergence of bacterial methylotrophy. Nat. Microbiol. 4, 2155–2163 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Lyu Z., Shao N., Akinyemi T., Whitman W. B., Methanogenesis. Curr. Biol. 28, R727–R732 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Habicht K. S., Gade M., Thamdrup B., Berg P., Canfield D. E., Calibration of sulfate levels in the Archean ocean. Science 298, 2372–2374 (2002). [DOI] [PubMed] [Google Scholar]
- 42.McGlynn S. E., Chadwick G. L., Kempes C. P., Orphan V. J., Single cell activity reveals direct electron transfer in methanotrophic consortia. Nature 526, 531–535 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Wegener G., Krukenberg V., Riedel D., Tegetmeyer H. E., Boetius A., Intercellular wiring enables electron transfer between methanotrophic archaea and bacteria. Nature 526, 587–590 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Seitz K. W., Dombrowski N., Eme L., Spang A., Lombard J., Sieber J. R., Teske A. P., Ettema T. J., Baker B. J., Asgard archaea capable of anaerobic hydrocarbon cycling. Nat. Commun. 10, 1822 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hahn C. J., Laso-Pérez R., Vulcano F., Vaziourakis K. M., Stokke R., Steen I. H., Teske A., Boetius A., Liebeke M., Amann R., Knittel K., Wegener G., “Candidatus Ethanoperedens,” a thermophilic genus of archaea mediating the anaerobic oxidation of ethane. MBio 11, e00600-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hinrichs K. U., Hayes J. M., Bach W., Spivack A. J., Hmelo L. R., Holm N. G., Johnson C. G., Sylva S. P., Biological formation of ethane and propane in the deep marine subsurface. Proc. Natl. Acad. Sci. U.S.A. 103, 14684–14689 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schink B., Stams A. J., Syntrophism among prokaryotes. The Prokaryotes 2, 309–335 (2006). [Google Scholar]
- 48.Jones D. M., Head I. M., Gray N. D., Adams J. J., Rowan A. K., Aitken C. M., Bennett B., Huang H., Brown A., Bowler B. F. J., Oldenburg T., Erdmann M., Larter S. R., Crude-oil biodegradation via methanogenesis in subsurface petroleum reservoirs. Nature 451, 176–180 (2008). [DOI] [PubMed] [Google Scholar]
- 49.Krukenberg V., Harding K., Richter M., Glöckner F. O., Gruber-Vodicka H. R., Adam B., Berg J. S., Knittel K., Tegetmeyer H. E., Boetius A., Wegener G., Candidatus Desulfofervidus auxilii, a hydrogenotrophic sulfate-reducing bacterium involved in the thermophilic anaerobic oxidation of methane. Environ. Microbiol. 18, 3073–3091 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Amard B., Bertrand-Sarfati J., Microfossils in 2000 Ma old cherty stromatolites of the Franceville Group, Gabon. Precambrian Res. 81, 197–221 (1997). [Google Scholar]
- 51.Shih P. M., Hemp J., Ward L. M., Matzke N. J., Fischer W. W., Crown group Oxyphotobacteria postdate the rise of oxygen. Geobiology 15, 19–29 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Sheridan P. P., Freeman K. H., Brenchley J. E., Estimated minimal divergence times of the major bacterial and archaeal phyla. Geomicrobiol. J. 20, 1–14 (2003). [Google Scholar]
- 53.Roger A. J., Susko E., Molecular clocks provide little information to date methanogenic Archaea. Nat. Ecol. Evol. 2, 1676–1677 (2018). [DOI] [PubMed] [Google Scholar]
- 54.Wolfe J. M., Fournier G. P., Reply to ‘Molecular clocks provide little information to date methanogenic Archaea’. Nat. Ecol. Evol. 2, 1678 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Betts H. C., Puttick M. N., Clark J. W., Williams T. A., Donoghue P. C., Pisani D., Integrated genomic and fossil evidence illuminates life’s early evolution and eukaryote origin. Nat. Ecol. Evol. 2, 1556–1562 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dombrowski N., Lee J. H., Williams T. A., Offre P., Spang A., Genomic diversity, lifestyles, and evolutionary origins of DPANN archaea. FEMS Microbiol. Lett. 366, fnz008 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Russell M. J., Hall A. J., Martin W., Serpentinization as a source of energy at the origin of life. Geobiology 8, 355–371 (2010). [DOI] [PubMed] [Google Scholar]
- 58.Westall F., Hickman-Lewis K., Hinman N., Gautret P., Campbell K. A., Bréhéret J. G., Foucher F., Hubert A., Sorieul S., Dass A. V., Kee T. P., Georgelin T., Brack A., A hydrothermal-sedimentary context for the origin of life. Astrobiology 18, 259–293 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goldblatt C., Zahnle K. J., Faint young Sun paradox remains. Nature 474, E1 (2011). [DOI] [PubMed] [Google Scholar]
- 60.Kasting J. F., Siefert J. L., Life and the evolution of Earth’s atmosphere. Science 296, 1066–1068 (2002). [DOI] [PubMed] [Google Scholar]
- 61.Li D., Liu C. M., Luo R., Sadakane K., Lam T. W., MEGAHIT: An ultrafast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676 (2015). [DOI] [PubMed] [Google Scholar]
- 62.Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., Lesin V. M., Nikolenko S. I., Pham S., Prjibelski A. D., Pyshkin A. V., Sirotkin A. V., Vyahhi N., Tesler G., Alekseyev M. A., Pevzner P. A., SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu Y. W., Simmons B. A., Singer S. W., MaxBin 2.0: An automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 32, 605–607 (2016). [DOI] [PubMed] [Google Scholar]
- 65.Kang D. D., Froula J., Egan R., Wang Z., MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ 3, e1165 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Maaten L. V. D., Hinton G., Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008). [Google Scholar]
- 67.Parks D. H., Imelfort M., Skennerton C. T., Hugenholtz P., Tyson G. W., CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huerta-Cepas J., Forslund K., Coelho L. P., Szklarczyk D., Jensen L. J., Von Mering C., Bork P., Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol. 34, 2115–2122 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nguyen L. T., Schmidt H. A., Von Haeseler A., Minh B. Q., IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Suchard M. A., Lemey P., Baele G., Ayres D. L., Drummond A. J., Rambaut A., Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 4, vey016 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.dos Reis M., Notes on the birth–death prior with fossil calibrations for Bayesian estimation of species divergence times. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371, 20150128 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smith S. A., treePL: Divergence time estimation using penalized likelihood for large phylogenies. Bioinformatics 28, 2689–2690 (2012). [DOI] [PubMed] [Google Scholar]
- 73.Liu Y. F., Chen J., Zaramela L. S., Wang L. Y., Mbadinga S. M., Hou Z. W., Wu X. L., Gu J. D., Zengler K., Mu B. Z., Genomic and transcriptomic evidence supports methane metabolism in Archaeoglobi. mSystems 5, e00651-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang S., Chen Y., Cao Q., Lou H., Long-lasting gene conversion shapes the convergent evolution of the critical methanogenesis genes. G3 5, 2475–2486 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reiss R. A., Guerra P., Makhnin O., Metagenome phylogenetic profiling of microbial community evolution in a tetrachloroethene-contaminated aquifer responding to enhanced reductive dechlorination protocols. Stand Genomic Sci. 11, 88 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/27/eabj1453/DC1