Combination of functional epigenomics and single-cell CRISPR screens reveals that ER exerts its oncogenic role via downstream TFs.
Abstract
Millions of putative transcriptional regulatory elements (TREs) have been cataloged in the human genome, yet their functional relevance in specific pathophysiological settings remains to be determined. This is critical to understand how oncogenic transcription factors (TFs) engage specific TREs to impose transcriptional programs underlying malignant phenotypes. Here, we combine cutting edge CRISPR screens and epigenomic profiling to functionally survey ≈15,000 TREs engaged by estrogen receptor (ER). We show that ER exerts its oncogenic role in breast cancer by engaging TREs enriched in GATA3, TFAP2C, and H3K27Ac signal. These TREs control critical downstream TFs, among which TFAP2C plays an essential role in ER-driven cell proliferation. Together, our work reveals novel insights into a critical oncogenic transcription program and provides a framework to map regulatory networks, enabling to dissect the function of the noncoding genome of cancer cells.
INTRODUCTION
Transcription factors (TFs) impose gene expression programs tailored to a diverse range of pathophysiological states. For example, specific tumors display exquisite dependency on transcriptional regulators to maintain their malignant phenotype (1). One such example is estrogen receptor (ER)–alpha (ESR1), which is an oncogenic driver in a large fraction (≈70%) of breast tumors (2). ER binds at estrogen-responsive elements (EREs) and activates the expression of target genes to promote cell growth and survival (3–5). Over the past decade, the application of genomics contributed to elucidating the function of ER by correlating its binding sites with target genes (6, 7), and nascent RNA analysis identified ≈3000 genes regulated by ER (8). However, these studies can neither explain the mechanisms of ER-regulated enhancers nor identify functional target genes that drive the growth of cancer cells.
It became recently evident that transcriptional regulatory elements (TREs) are targeted by germline and somatic genetic alterations in cancer (9). However, the actual contribution of noncoding genetic variants to human disease remained largely open in the absence of functional assays. The advent of CRISPR-Cas9 systems filled a technological gap, and they were readily applied to study noncoding elements. In particular, CRISPR-Cas9 genetic screens tested thousands of TREs in different biological settings, such as the regulation of specific loci (10–13) or a focused set of TF binding sites (14, 15). This approach was also used to characterize regulatory interactions by coupling it with transcriptional reporters (16–19), single-cell RNA sequencing (scRNA-seq) (20, 21), and RNA–fluorescence in situ hybridization (22). However, a systematic assessment of oncogenic regulatory networks in cancer is still missing.
Here, we combine genome-scale and single-cell CRISPR screens to perform a comprehensive survey of TREs upstream and downstream of ER in breast cancer cells. We tested ≈15,000 putative TREs and identified a small subset of them controlling the proliferation of cancer cells, which are enriched in GATA binding protein 3 (GATA3), transcription factor AP-2 gamma (TFAP2C), and histone H3 lysine 27 acetylation (H3K27Ac) signal. These ER-dependent TREs control a network of downstream TFs, effectively branching out the transcriptional dependencies of ER+ breast cancer cells. We highlight the role of TFAP2C as a critical ER target gene and a potential biomarker in breast cancer. Our results provide a framework to characterize how oncogenic TFs can engage specific TREs to impose their pathogenic program.
RESULTS
High-resolution CRISPRi screen identifies critical TREs controlling ESR1 and CCND1
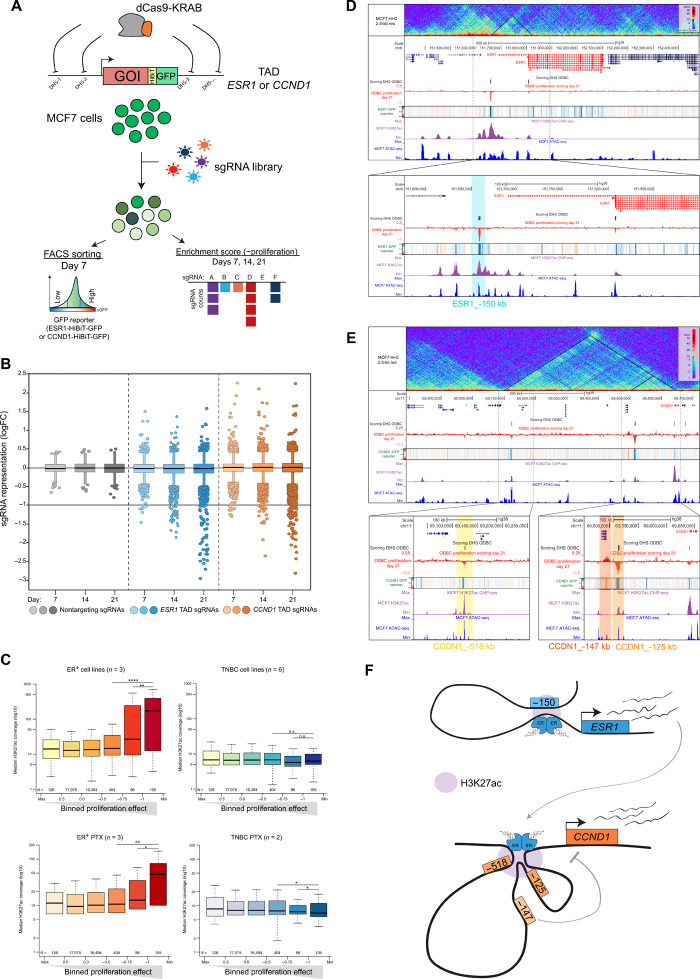
Expression of ER and its downstream target genes characterize and stratify the ER+ breast cancer subtype (23, 24). In addition, ESR1 and its target gene cyclin D1 (CCND1) are dependencies in such cancers (fig. S1A), hence critical therapeutic nodes (5, 25, 26). How ER+ breast cancer cells sustain the expression of such critical oncogenes is currently unknown. Therefore, we aimed to identify systematically TREs that control the expression of ESR1 and CCND1. We used CRISPR-Cas9 to integrate a transcriptional reporter in MCF7 cells by fusing HiBiT–green fluorescent protein (GFP) in frame with ESR1 or CCND1 to monitor gene expression changes (Fig. 1A). We observed a substantial reduction of fluorescence and luminescence signal in MCF7-ESR1-HiBiT-GFP and MCF7-CCND1-HiBiT-GFP cells upon knockdown of ESR1 and CCND1, respectively (fig. S1, B and C). This indicates that the HiBiT-GFP reporters faithfully recapitulate endogenous gene expression, and they are amenable for fluorescence-activated cell sorting (FACS)–based genetic screening. We designed a comprehensive CRISPR library to interrogate all putative TREs located within the topologically associating domains (TADs) of ESR1 and CCND1, as defined by publicly available deoxyribonuclease I hypersensitivity (i.e., DHS) (27) and Hi-C (28) data (fig. S1D). In total, we cloned 34,755 single-guide RNAs (sgRNAs) and assembled a library designated here as oncogenic drivers of breast cancer (ODBC), which contains ≈9 sgRNAs/DHS upon filtering off-targets and considering an even distribution of sgRNAs within candidate DHS (fig. S1, E and F). Of note, ≈25% of sgRNAs target putative TREs in ER+ breast cancer cells (fig. S1G), and a large fraction of these regions contain features that are indicative of promoters, enhancers, insulators, and actively transcribed regions (fig. S1H).
Fig. 1. High-resolution CRISPRi screen identifies critical TREs controlling ESR1 and CCND1.
(A) Schematic representation of the ODBC screen. GOI, gene of interest. (B) Box plot showing the representation of the ODBC library at days 7, 14, and 21. Nontargeting sgRNAs are negative controls. Data represent median logFC of three experiments. (C) Median H3K27Ac ChIP-seq signal (log10) at sgRNA target regions in ER+ and TNBC models binned by the ODBC logFC at T = 21 days (proliferation effect). TNBC, triple-negative breast cancer; PTX, primary tumor xenografts. *P < 0.05, **P < 0.01, ****P < 0.0001, and not significant (n.s.), P > 0.05. (D) Snapshot of the ESR1 locus showing Hi-C data (top track), the ODBC screening results (cell proliferation and GFP reporter), H3K27Ac ChIP-seq, and ATAC-seq in MCF7 cells. (E) Snapshot of the CCND1 locus showing Hi-C data (top track), the ODBC screening results (cell proliferation and GFP reporter), H3K27Ac ChIP-seq, and ATAC-seq in MCF7 cells. (F) Model depicting the findings of the ODBC screen.
We started by testing the functional importance of candidate TREs of ESR1 and CCND1 by performing a cell proliferation–based CRISPRi screening. We transduced MCF7-CRISPRi cells with the ODBC library and measured the representation of sgRNAs by sequencing at three different time points (Fig. 1A and table S1). We observed a substantial number of sgRNAs consistently depleted in three independent replicates, and we defined “scoring sgRNAs” based on log fold change (logFC) < −1 at day 21 (Fig. 1B and fig. S2A). Notably, scoring sgRNAs were consistently decreased in their representation, whereas nontargeting sgRNAs remained largely unchanged over time (fig. S2B). In addition, we performed a small-scale validation screen in three cell lines to validate scoring sgRNAs identified in the ODBC screens (fig. S2C). We observed that scoring ODBC sgRNAs that dropout in ER+ cells (MCF7 and T47D) have a negligible effect in MDA-MB-231 (ER−), demonstrating how our approach identifies a subset of regulatory elements that specifically drive the proliferation of ER+ breast cancer cells.
To establish a causal relationship between the proliferative effect and the regulation of ESR1 or CCND1, we performed the ODBC screen in HiBiT-GFP reporter cells and used FACS to retrieve cells based on quartiles of GFP expression at day 7 (∆GFP) (Fig. 1A). We overlaid the reporter and proliferation screening data by evaluating the enrichment score of sgRNAs in GFP-negative cells (i.e., “gene repression score”). We observed that the sgRNAs that have the strongest effect on cell proliferation are associated with decreased expression of the respective reporter gene (fig. S3, A and B). We found that the highest-scoring sgRNAs in the ESR1 TAD are associated with decreased expression of the CCND1-GFP reporter in MCF7 cells (fig. S3A). These results are in line with CCND1 being downstream of ESR1 and confirms that our screening method is suitable to identify both cis- and trans-regulatory events. In addition, scoring ODBC sgRNAs target regions that contain the highest level of H3K27Ac signal in ER+ cell lines and primary tumor xenografts (PTXs) (Fig. 1C), but not in triple-negative breast cancer (TNBC) models (Fig. 1C). This suggests that our screen can identify highly active TREs driving the expression of critical oncogenes in ER+ cells.
We then focused on scoring sgRNAs in the ESR1 TAD and identified two candidate regions that contribute to ESR1 expression and cell growth. The first corresponds to the active transcription start site (TSS) of ESR1 (ESR1_TSS) in MCF7 cells (29), and the second is a region located 150 kb upstream (ESR1_-150 kb) (Fig. 1D). To test whether the upstream element interacts with the ESR1 promoter, we performed Hi-C experiments in MCF7 cells, which produced high-resolution maps of DNA-DNA interactions and revealed significant contacts between ESR1_-150 kb and ESR1_TSS (Fig. 1D). ESR1_-150 kb is located in an accessible chromatin region that is marked by high H3K27Ac signal (Fig. 1D), which was previously described as an enhancer of ESR1 (30). Targeting ESR1_-150 kb with independent CRISPR reagents confirmed a decrease in cell growth (fig. S3C) and a concomitant down-regulation of both endogenous ESR1 expression and ESR1-HiBiT luminescence (fig. S3D). Moreover, ESR1_-150 kb activates the expression of a minimal promoter in a reporter assay (fig. S3E), further supporting its classification as an enhancer. Its transcriptional activity is stimulated by estradiol (E2), whereas small deletions in the estrogen response element (ERE) located in this region severely blunt its basal activity and stimulation by E2.
Next, we investigated the cis-regulatory network of CCND1, which is known to be regulated by several ER-bound enhancers (14, 31). In our screen, we identified three candidates that sustain CCND1 expression and cell growth: the promoter region (CCND1_TSS), a known enhancer that is bound by ER (14) (CCND1_-125 kb), and a candidate TRE located upstream of the CCND1 TSS (CCND1_-518 kb) (Fig. 1E). Using Hi-C data, we identified loops between the two candidates and the CCND1 TSS (Fig. 1E), which are indicative of long-range regulatory interactions. Alongside with CCND1_TSS, we validated that CCND1_-125 kb and CCND1_-518 kb are necessary for cell proliferation (fig. S3F), endogenous CCND1 expression, and CCND1-HiBiT luminescence (fig. S3G). We tested the transcriptional activity of CCND1_-125 kb and CCND1_-518 kb using a plasmid reporter assay, and we found that they enhance transcription in an ER-dependent manner (fig. S3H). Last, we tested the region CCND1_-147 kb, which is associated with increased CCND1 expression and cell proliferation in our screen data (Fig. 1E). We validated that inhibition of CCND1_-147 kb by CRISPRi leads to increased expression of CCND1 in MCF7 cells (fig. S3G), suggesting that this region is a repressor of CCND1. Together, our results reveal several nonredundant TREs that sustain the expression of ESR1 and CCND1 and cell proliferation in ER+ breast cancer cells (Fig. 1F).
Genome-scale CRISPRi screen identifies essential ERBS in breast cancer cells
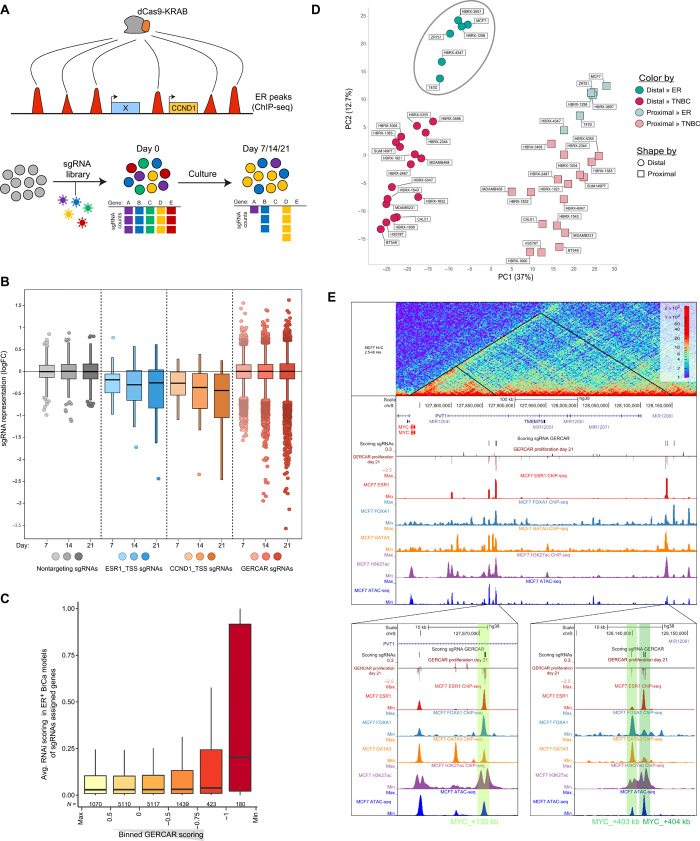
A plethora of ER binding sites (ERBS) and target genes have been described by epigenomic profiling experiments (6–8, 32). However, only few ER-bound TREs were functionally characterized, and a comprehensive assessment is still missing (14, 31, 33). To this end, we set out to functionally interrogate a consensus map of ERBS (n = 14675) based on ER chromatin immunoprecipitation sequencing (ChIP-seq) datasets (table S2) obtained from Cistrome (Fig. 2A) (34). We designed a CRISPR library, referred here as genome-wide ER CRISPR-associated repression (GERCAR), which contains an average of 5.4 sgRNAs per ER peak (fig. S4, A and B). Next, we performed a proliferation-based screen in MCF7-CRISPRi cells with the GERCAR library over a period of 21 days (Fig. 2B; fig. S4, C and D; and table S3). We observed that the representation of non-targeting control (NTC) sgRNAs remained largely unchanged, whereas positive controls and a substantial number of sgRNAs targeting ERBS were depleted over time (Fig. 2B and fig. S4E). We defined a scoring threshold based on logFC < −1 and P value of <0.05 at T = 21 days (n = 303 sgRNAs representing 242 ERBS that correspond to 1.65% of the tested regions). We validated a subset of hits by performing a small-scale validation screen of scoring sgRNAs in ER+ (MCF7 and T47D) and ER− (MDA-MB-231) cell lines. This revealed that the vast majority of scoring sgRNAs are essential only in ER+ but not in triple-negative cell models (fig. S5A), suggesting that our screen is highly specific at identifying lineage-specific TREs. We then sought to identify features correlating with sgRNA sensitivity in the GERCAR screen using three different approaches. First, we found that scoring sgRNAs are in proximity to genes deemed essential by RNA interference (RNAi) screens in ER+ breast cancer cells (Fig. 2C). This set includes several genes previously described to play a role in breast cancer biology, such as Androgen Receptor (AR) (35), Transcriptional Repressor GATA Binding 1 (TRPS1) (36), CUE Domain Containing 1 (CUEDC1) (37), and Grainyhead Like Transcription Factor 2 (GRHL2) (38). Second, we compared the ChIP-seq signal for several features with the score of the GERCAR screen (fig. S5B). We observed a significant enrichment of GATA3 signal (P = 0.032 by Wilcoxon test) at promoter-distal regions (>5 kb from annotated TSS) characterized by the highest GERCAR scoring (logFC < −1), whereas FOXA1 shows a trend for enrichment. In addition, H3K27ac signal is significantly enriched (P = 1.3 × 10−7, Wilcoxon test), whereas H3K4me1 signal is significantly depleted (p = 5.1 × 10−6, Wilcoxon test) at scoring promoter-proximal regions (<5 kb from annotated TSS). Third, we conducted a principal components analysis (PCA) of ER+ and TNBC models, focusing on the H3K27ac ChIP-seq signal at the top 1000 most variable proximal and distal sgRNA targeting regions (based on logFC at the three time points of the screen) (Fig. 2D). As expected, the PCA shows a clear separation between proximal and distal regions (PC1), while the separation between ER+ and TNBC models is more pronounced at distal regions compared to proximal ones (PC2). Collectively, our results suggest that only a small fraction of ERBS (1.65%) is required for ER-mediated cell proliferation, and, while the proximal sites display high transcriptional activity, the distal ERBS are enriched with GATA3 binding.
Fig. 2. Genome-scale CRISPRi screen identifies essential ERBS in breast cancer cells.
(A) Schematic representation of the GERCAR screen. (B) Box plot showing the representation of the GERCAR library at days 7, 14, and 21. CCND1_TSS and ESR1_TSS sgRNAs are positive controls. Nontargeting sgRNAs are negative controls. Data represent median logFC of two experiments. (C) Average RNAi score (DEPMAP-combined RNAi) of genes assigned to the closest sgRNA binned by GERCAR scoring (logFC at T = 21 days). (D) PCA of H3K27ac signal (log10) at the 2000 (union of 1000 distal and 1000 proximal) most variable sgRNA targeting regions in cell lines and primary tumor xenografts (HBRX, human breast xenograft). Distal, >5 kb from annotated TSS; proximal, <5 kb from annotated TSS. (E) Snapshot of the MYC locus showing Hi-C data (top track); the GERCAR screen results; ChIP-seq signal of ER, FOXA1, GATA3, and H3K27Ac; and ATAC-seq in MCF7 cells.
For validation experiments, we prioritized ERBS supported by multiple scoring sgRNAs and identified several within the MYC locus (Fig. 2E). MYC is a known target gene of ER (39) and is located within a ≈3-Mb TAD that contains hundreds of putative TREs predicted in different cell types (12). We observed that the three candidates (MYC_+135 kb, MYC_+403 kb, and MYC_+404 kb) are located in accessible chromatin regions marked by high levels of H3K27Ac and bound by ER, GATA3, and FOXA1 (Fig. 2E). In addition, these regions engage in long-distance interactions with the MYC promoter as measured by Hi-C, which suggests that they are putative enhancers. Validation experiments using CRISPRi reagents confirmed that targeting these regions leads to a substantial decrease of MYC expression and cell growth (fig. S5, C and D). Thereby, the GERCAR screen identifies TREs that are necessary for the ER-dependent proliferative program.
Validation of candidate TREs by single-cell CRISPR screens
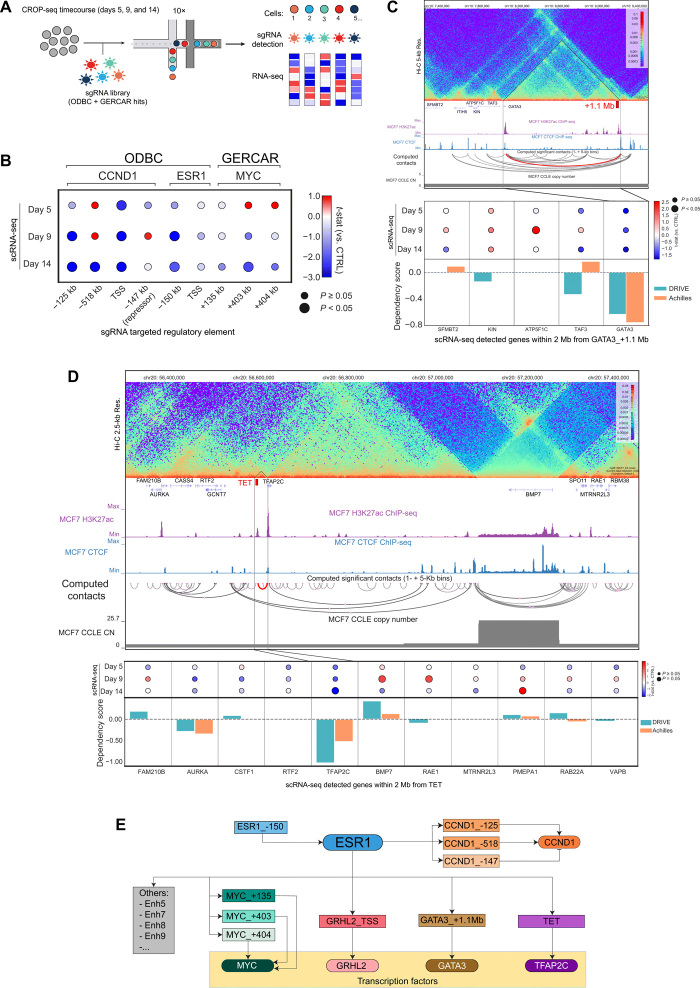
Our functional genomic screens identified a restricted number of TREs that are necessary to promote and drive the oncogenic program of ER. For characterizing the transcriptional consequences of disrupting these TREs, we used CRISPR droplet sequencing (CROP-seq) (40), which combines pooled CRISPR screens with scRNA-seq. We transduced MCF7-CRISPRi cells with a CROP-seq library, containing hits from both ODBC and GERCAR screens, and performed a proliferation-based screen using three end points (T = 5, T = 9, and T = 14 days) to capture transcriptomic changes throughout time (Fig. 3A and fig. S6A). The CROP-seq experiment yielded high-quality transcriptome profiling (fig. S6B), which enabled us to obtain >1200 cells containing a single sgRNA per time point (fig. S6C) and select TREs represented at least in 10 cells for further analysis (fig. S6D). We surveyed a 4-Mb region centered on each candidate to identify gene expression changes by scRNA-seq. Using our high-resolution Hi-C data, we identified DNA loops between the candidates and target promoters and overlaid available genetic dependency data by RNAi screens for every gene in the locus. As described earlier, we observed that TREs in the ESR1, CCND1, and MYC loci were engaged in enhancer-promoter looping (Figs. 1, D and E, and 2E). We validated by CROP-seq that their inhibition leads to specific changes in gene expression restricted to their target genes or a limited set of genes within their TAD (Fig. 3B and figs. S7, A and B, and S8A). We observed a similar pattern of specific gene expression changes when targeting a promoter-proximal ERBS near GRHL2 (fig. S8B), suggesting that this TF is downstream of ER. Of note, GRHL2 is a genetic dependency in ER+ breast cancer, and its depletion leads to altered ER binding and differential transcriptional responses to estrogen stimulation (38). Our results indicate that CROP-seq is a robust method to perturb regulatory elements and detect corresponding gene expression changes in single cells.
Fig. 3. Validation of candidate TREs by single-cell CRISPR screens.
(A) Schematic representation of the CROP-seq experiment to characterize ODBC and GERCAR scoring hits. (B) Dot plot showing gene expression of CCND1, ESR1, and MYC detected by scRNA-seq in MCF7-CRISPRi cells transduced with indicated sgRNAs. The size of each dot represents the significance of the results of differential expression (comparing target sgRNAs versus nontargeting; larger dots mean significant), and the color depicts the modulation measured as t-statistic. (C) Snapshot of the GATA3 locus showing Hi-C data, ChIP-seq signal H3K27Ac and CTCF, computed significant contacts of Hi-C (the contact between GATA3 and GATA3_+1.1 Mb is red), and copy number in MCF7 cells (top). Dot plot showing expression of selected genes by scRNA-seq in MCF7-CRISPRi cells transduced with sgRNAs targeting GATA3_+1.1 Mb (middle). Dependency score derived from DRIVE and Achilles datasets (bottom). (D) Snapshot of the TFAP2C locus showing Hi-C data, ChIP-seq signal H3K27Ac and CTCF, computed significant contacts of Hi-C (the contact between TFAP2C and TET is red), and copy number in MCF7 cells (top). Dot plot showing expression of selected genes by scRNA-seq in MCF7-CRISPRi cells transduced with sgRNAs targeting TET (middle). Dependency score derived from DRIVE and Achilles datasets (bottom). (E) Graphic summary of the ER regulatory network in breast cancer cells.
Next, we tested additional loci that were identified in the GERCAR screen. Through our approach, we identified long-range interactions between a candidate region in a gene desert (GATA3_+1.1 Mb) and the promoter of GATA3, which is located 1.1 Mb upstream (Fig. 3C). We observed that perturbing GATA3_+1.1 Mb by CRISPRi leads to the down-regulation of GATA3 expression in all time points measured by CROP-seq. In addition, we observe that GATA3_+1.1 Mb is an accessible chromatin region marked by H3K27Ac and bound by ER, FOXA1, and GATA3 (fig. S9A). GATA3 is a known cofactor of ER (33, 41) and a genetic dependency in ER+ breast cancer (42–44). We validated our findings by targeting GATA3_+1.1 Mb with individual sgRNAs, which resulted in a significant reduction of GATA3 expression (fig. S9B) and a concomitant decrease of cell growth (fig. S9C). Next, we extended our analysis to additional loci and identified regulatory interactions between Enh5 and CDK6 (fig. S10A), Enh7 and CTSD (fig. S10B), Enh8 and DPYSL4/STK32C (fig. S11A), and Enh9 and LNX2/POLR1D (fig. S11B). The CROP-seq data indicates that these candidate target genes are weakly down-regulated, and none of them is a strong hit in RNAi screens in ER+ breast cancer cells, suggesting that this set of TREs have pleiotropic transcriptional effects on a phenotypically redundant set of genes.
Last, the Hi-C data revealed that TFAP2C interacts with a candidate region [transcriptional enhancer of TFAP2C (TET)] that is located ≈30 kb upstream of its TSS (Fig. 3D). Using CROP-seq, we confirmed that TFAP2C is the only gene significantly down-regulated in this locus upon perturbing TET. In addition, data from genome-scale RNAi screens suggest that TFAP2C is a genetic dependency in MCF7 cells (Fig. 3D). TFAP2C belongs to the family of activating proteins that play a role in chromatin remodeling and accessibility (45, 46). It was reported that TFAP2C and ER overlap at putative enhancer regions (47, 48), suggesting that they cooperate to regulate gene expression. Together, our results indicate that combining Hi-C and CROP-seq data is a powerful approach to find functional enhancer-gene pairs in a high-throughput fashion (Fig. 3E).
TFAP2C is a critical target gene of ER
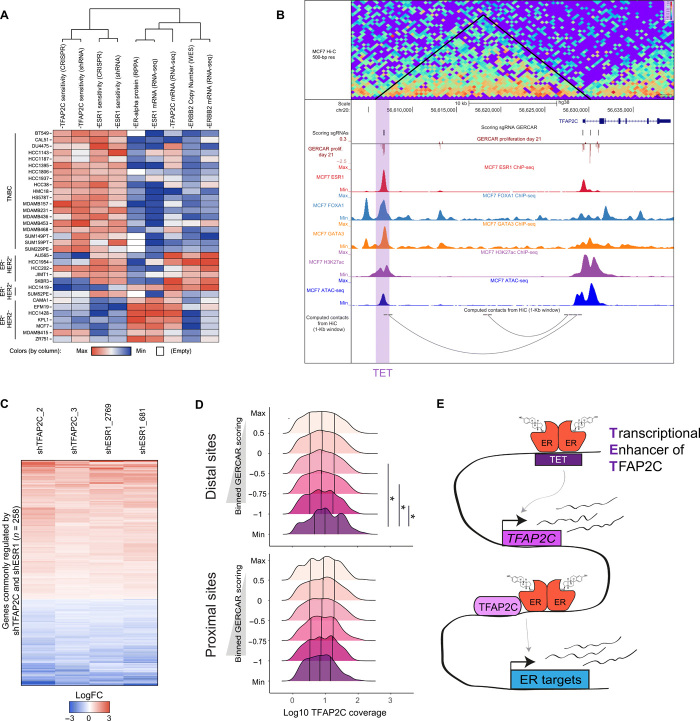
Our functional genomic approach highlights a complex network of TREs controlled by ER, ultimately impinging on master TFs that drive oncogenic phenotypes in breast cancer (Fig. 3E). Therefore, we computed core regulatory circuitry (CRC) (49) to identify critical enhancers and TFs that maintain the identity of ER+ breast cancer cells. We prioritized the top 20 candidate TFs by sensitivity to RNAi knockdown and CRISPR knockout by genome-wide screens (fig. S12A). As expected, we observe that ER, GATA3, and FOXA1 are identified by CRC and are among the most essential TFs in breast cancer (42–44). In addition, TFAP2C and SPDEF display a similar sensitivity profile and cluster together with the TF mentioned above.
We observe that the expression and sensitivity patterns of TFAP2C and ER are remarkably similar across multiple breast cancer models (Fig. 4A and fig. S12A). We identified two regions nearby TFAP2C (TFAP2C_TSS and TET) that score in the GERCAR screen (Fig. 4B). These two regions are bound by ER, GATA3, and FOXA1 and marked by high levels of H3K27Ac (Fig. 4B), indicating that they are active TREs regulated by ER. To validate these findings, we transduced MCF7-CRISPRi cells with individual sgRNAs targeting TFAP2C_TSS and TET and observed a concomitant reduction of TFAP2C expression and cell proliferation (fig. S13, A and B). These findings are in line with data from RNAi and CRISPR screens showing that TFAP2C is a genetic dependency in ER+ cells (Fig. 4A and fig. S12A). Next, we tested whether TET is able to activate the transcription of a minimal promoter in a reporter assay. We observed that the transcriptional activity of TET is stimulated by estradiol, whereas small deletions in the ERE present in this region reduce it significantly (fig. S13C), suggesting that TET is an ER-responsive TRE. We then evaluated transcriptomic changes by RNA-seq upon perturbing TFAP2C or TET. We observed a positive correlation between CRISPR reagents targeting TFAP2C and TET (fig. S13D), and 663 genes consistently modulated among these conditions (fig. S13, E to G, and table S4). In addition, we assessed the effects of down-regulating TFAP2C or ESR1 by transducing MCF7 cells with doxycycline-inducible short hairpin RNAs (shRNAs). First, we validated that the shRNAs targeting TFAP2C and ESR1 produce a robust knockdown (fig. S14, A and B), which is associated with a strong impact on cell growth (fig. S14C). Second, we performed RNA-seq and detected 258 genes that are commonly regulated by TFAP2C and ESR1 (Fig. 4C and table S4). Given our previous observations, we hypothesized that TFAP2C and ER regulate a common set of genes in breast cancer cells. To test that, we analyzed a known signature of ER target genes (50) and observed that silencing TFAP2C by different reagents attenuates their expression (fig. S14D). In addition, these genes (n = 258) are significantly associated with estrogen response and cell cycle pathways by gene set enrichment analysis (GSEA) (fig. S14E). This prompted us to evaluate the genomic occupancy of TFAP2C at the ERBS tested in the GERCAR screen, and we found that there is a poor correlation between TFAP2C and ER ChIP-seq coverage (Spearman’s = 0.26; n = 14675) (fig. S14F). However, TFAP2C signal is significantly enriched at promoter-distal ERBS that score in the GERCAR screen (Fig. 4D). These results confirm that TFAP2C contributes to the transcriptional output of the ER pathway and provide an explanation for the essentiality of this gene in breast cancer cells. Elevated expression of TFAP2C is associated with poor outcome for luminal A (ER+ and ERBB2-negative) patients (fig. S14G), which is consistent with TFAP2C playing an oncogenic role in breast cancer. Overall, our findings suggest that ER activates the expression of downstream TFs, among which TFAP2C is a critical player in modulating its oncogenic program in breast cancer cells (Fig. 4E).
Fig. 4. TFAP2C is a critical target gene of ER.
(A) Hierarchical clustering based on sensitivity of TFAP2C or ESR1 to CRISPR (AVANA dataset) and RNAi (DEPMAP Combined RNAi). ER expression, ESR1 expression, TFAP2C expression, ERBB2 copy number, and ERBB2 expression are displayed for comparison in breast cancer models. RPPA, reverse-phase protein array; WES, whole-exome sequencing. (B) Snapshot of the TFAP2C locus showing Hi-C data; the GERCAR screen results; ChIP-seq of ER, FOXA1, GATA3, and H3K27Ac; ATAC-seq; and computed contacts from Hi-C in MCF7 cells. Scoring sgRNAs were identified on the basis of logFC < −1 at T = 21 days. (C) Heatmap showing genes that have the same trend of expression and are statistically significant upon knockdown of ESR1 or TFAP2C in MCF7 cells (logFC by RNA-seq). (D) TFAP2C ChIP-seq signal at sgRNA targeting regions was binned by GERCAR scoring (logFC at T = 21 days) in MCF7 cells. The vertical lines in each ridge represent the first quartile, median, and third quartile of ChIP-seq signal. *P < 0.05. (E) Model depicting the cross-talk between ER and TFAP2C in activating downstream target genes.
DISCUSSION
To date, more than 1.3 million candidate TREs are predicted on the basis of biochemical marks in the human genome (51). This highlights the massive challenge to systematically assign candidate TREs to their bona fide target genes and test their function in specific phenotypes. The dawn of CRISPR-Cas9 technologies enabled large-scale profiling of the function and mechanisms of TREs. In our work, we systematically interrogated the ER cistrome in breast cancer cells by testing both upstream regulators and downstream effectors of this pathway. Initially, we performed gene reporter–based screens using the ODBC library to target TREs located in the TADs of ESR1 and CCND1. This approach has the advantage of disentangling the contribution of TREs for two distinct phenotypes (i.e., cell fitness and gene expression). However, there are some limitations: the discovery of functional TREs is confined to the tested TAD; testing multiple genes in a single experiment requires generating clonal cell lines with multiple knock-ins and different reporters. In the GERCAR screen, we perturbed thousands of ERBS in a genome-wide scale and evaluated their necessity for cell fitness. This approach is scalable and allows testing the contribution of TREs for a specific phenotype, yet it does not reveal the gene(s) involved in the phenotype. To tackle this issue, we used CROP-seq, which couples pooled CRISPR screenings with transcriptomic readouts by scRNA-seq. This method enables directly linking (epi)genetic perturbations to transcriptional responses in thousands of individual cells, thereby facilitating the identification of enhancer-gene pairs. The increasing throughput of single-cell transcriptomics suggests that CROP-seq and similar methods have great potential for comprehensively dissecting gene regulatory networks, although the scalability of this type of experiments can be hampered by elevated financial costs.
In the ODBC screen, we coupled gene expression and cell growth readouts, which allowed to directly correlate the TRE-mediated phenotype to the regulation of the major oncogenes ESR1 and CCND1. Our screen extends previous findings (14, 31) by systematically assessing the entire CCND1 TAD and identifying previously unknown positive and negative TREs, which are bound by ER and possibly contribute to fine-tune CCND1 expression (Fig. 1F). This TAD contains 39 different genetic haplotypes associated with human traits, including breast cancer susceptibility (52), which underscores the importance of characterizing TREs regulating CCND1. In addition, we identified ESR1_-150 kb upstream of ESR1, which is a TRE targeted by mutations that are associated with increased expression of ESR1 in human tumors (30). Several studies showed that disease-associated variants and somatic mutations are commonly found in regulatory elements (9, 53–55). A prominent example is the activation of TAL1 in T-cell acute lymphoblastic leukemia by somatic mutations that create a super-enhancer by introducing binding motifs for MYB (56). Additional work is required to determine whether the functional regions we identified are targeted by genetic variants in patient samples.
In the GERCAR screen, we observed that only 1.65% of ≈15,000 ERBS contribute to cell fitness. This observation raises the hypothesis that the vast majority of ERBS are dispensable for cell fitness, despite the well-known oncogenic function of ER in breast cancer cells. However, we cannot exclude false-negative events due to genetic compensation by redundant regulatory elements, insufficient repression by CRISPRi, low sgRNA efficiency, and exclusion of candidates for lacking biochemical marks. In addition, we focused on the proliferative phenotype exerted by ER, and it is possible that ERBS contribute to other phenotypes (e.g. differentiation, metabolic changes, epithelial-mesenchymal transition, etc.) not assessed in our genetic screens in immortalized cell lines. We also observed that the ChIP-seq GATA3 and TFAP2C signal is significantly enriched at functional TREs, in contrast with the ER and FOXA1 signal. A previous study showed that CCCTC-Binding Factor (CTCF) signal can predict the essentiality of its binding sites in breast cancer cells (15), indicating that the rules governing how different classes of TFs engage functional TREs are incompletely understood. Our analysis also indicates that high H3K27Ac ChIP-seq signal is significantly associated with functional regions. The top scoring regions of the ODBC screens are positively associated with H3K27Ac signal in breast cancer cells and PTX (Fig. 1C). In addition, we observed that ER+ models can be identified by correlating the GERCAR screen score and the H3K27Ac signal at distal ERBS (Fig. 2D), supporting the notion that this mark can predict the functionality of TREs in clinically relevant models. It was previously reported that H3K27Ac signal can be used to extrapolate phenotypic heterogeneity in tumor samples (57), suggesting that H3K27Ac genomic distribution is a potential biomarker in breast cancer.
The recent application of functional CRISPR screens coupled with scRNA-seq enables the large-scale mapping of enhancer-gene regulatory networks (20, 21). Our results indicate a significant association between scoring TREs and essential coding genes located nearby. This correlation might be partially dictated by the association rules we used, as the assignment of a TRE to the closest TSS has higher chances of success within a 50-kb window (58). However, by combining Hi-C and CROP-seq, we experimentally validated several ERBS regulating GRHL2, MYC, GATA3, and TFAP2C, which are genetic dependencies in ER+ breast cancer cells (42–44). Our results suggest that ER integrates upstream signals and, upon activation, mainly relies on downstream TFs to drive its proliferative program in cancer cells (Fig. 3E). In particular, we uncovered a regulatory network by which ER controls the expression of TFAP2C. Previously, it was reported that TFAP2C knockdown is associated with decreased response to estradiol and impaired growth of breast cancer xenografts (59). TFAP2C is known to regulate the expression of ESR1 (60), and their binding sites overlap in breast cancer cells (47, 48), suggesting a cooperative action between ER and TFAP2C. A recent study showed that therapeutic ligands promote rapid engagement and redistribution of ER binding to chromatin (50), and these binding sites contain, among others, sequence motifs of ER, GATA3, FOXA1, and TFAP2C. Moreover, the binding motif of TFAP2C is enriched in regions that gain chromatin accessibility upon resistance to ER antagonists (61), suggesting that it might play a role in therapy resistance. This hypothesis is supported by our observation that elevated TFAP2C expression is associated with poor outcome of ER+ breast cancer patients. We envision that future iterations of our method can be performed in models of drug resistance to gain mechanistic insights about ER and its cofactors in this context.
In summary, we combined epigenomic profiling with genome-scale and single-cell CRISPR screens to dissect the ER cistrome. Through this approach, we unveiled previously unknown regulatory networks between ER and its downstream TFs, such as TFAP2C. We anticipate that our approach can be applied to different models and advance our understanding of transcriptional regulation in cancer, such as identifying target genes of TFs, validating candidate TREs, and determining the function of causal regulatory variants.
MATERIALS AND METHODS
Tissue culture and cell engineering
MCF7 cells were cultured in Eagle’s minimum essential medium (EMEM) supplemented with fetal bovine serum (FBS; 10%), 2 mM l-glutamine, 1 mM sodium-pyruvate, and 10 mM Hepes. T47D cells were cultured in RPMI supplemented with FBS (10%), 2 mM l-glutamine, 1 mM sodium-pyruvate, and 10 mM Hepes. MDA-MB-231 cells were grown in Dulbecco’s modified Eagle’s medium supplemented with FBS (10%), 2 mM l-glutamine, 1 mM sodium-pyruvate, and 1% nonessential amino acids.
HiBiT-T2A-GFP reporters of ESR1, CCND1, and GATA3 were generated by cotransfecting MCF7 cells with sgRNAs and respective repair templates encompassing 800 base pairs (bp) upstream and downstream of the cleavage site. GFP-positive cells were identified and recovered by FACS using a SH800S (Sony) cell sorter. Single-cell clones derived from the bulk GFP-positive population of cells were validated by immunoblotting and knockdown experiments. Transfections of siRNAs were performed with Lipofectamine RNAi MAX (Invitrogen) according to the manufacturer’s protocol. siCtrl is AllStars negative control (QIAGEN), and siESR1 and siCCND1 were obtained from Dharmacon. HiBiT was evaluated 72 hours after transfection with siRNAs or sgRNAs. HiBiT signal was measured using the Nano-Glo HiBiT Lytic Detection System (Promega) according to the manufacturer’s recommendation.
CRISPR-Cas9 experiments were done in cells stably expressing Cas9 nuclease or dCas9-KRAB cassettes, which were delivered by lentiviral transduction and selected using blasticidin (10 μg/ml; Invitrogen). Cells expressing doxycycline-inducible shRNAs were obtained by lentiviral transduction of pLKO-TET-ON plasmids. Cells expressing constitutive sgRNA were obtained by lentiviral transduction of a modified pLKO-TET-ON plasmid. For cell growth assays, MCF7-CRISPRi cells were transduced with targeting sgRNAs (expressing mCherry) or nontargeting sgRNAs (expressing GFP). Cells containing individual lentiviral constructs were mixed (mCherry:GFP ratio of 3:1), and the fraction of cells expressing each marker was assessed by flow cytometry at the beginning of the experiment and subsequent time points. We recorded at a minimum of 2000 single cells for each condition, and the data were analyzed using FlowJo software.
Design of ODBC and GERCAR libraries
For the ODBC library, we selected DHS clusters in 95 cell types available from ENCODE (27). This enabled building a CRISPR library targeting a broad range of TREs that can be used to screen different cell types. Having designed our libraries before performing Hi-C experiments, we defined the TADs of ESR1 and CCND1 using published Hi-C data (40- and 25-kb resolution, respectively) of T47D cells available at the 3D genome browser (28). In the ESR1 TAD (≈1.16 Mb in size), we targeted 954 DHS with 7992 sgRNAs (mean of 8.66 sgRNAs per DHS). In the CCND1 TAD (≈2.56 Mb in size), we targeted 3004 DHS with 26,263 sgRNAs (mean of 9.31 sgRNAs per DHS). Overall, we have surveyed ≈21% of the total sequence of the ESR1 and CCND1 TADs (assuming that DHS are 200 bp in size). For the GERCAR library, we used seven ER ChIP-seq datasets of MCF7, T47D, and ZR-75-1 cells (table S1), available from Cistrome (34), to build a consensus set of ERBSs. We used the following rules for processing the data: For technical replicates, keep a region if it is present in both samples; for biological replicates, keep a region if it is present in two or more of the samples. We designed a library containing 80,254 sgRNAs (including positive and negative controls) to interrogate 14,675 ER peaks, with an average of 5.41 sgRNAs per peak. For both libraries, we removed overlapping sgRNAs and sgRNAs with low predicted specificity and filtered peak regions having many sgRNAs by selecting guides across the peak that were distributed as equally as possible.
The oligo pools of the ODBC (n = 34,755) and GERCAR (n = 80,254) libraries were purchased from CustomArray and Twist Bioscience, respectively. We designed 60-bp single-stranded DNA oligos containing a 20-bp sgRNA flanked by the sequences 5′-gccatccagaagacttaccg-3′ and 5′-gtttccgtcttcacgactgc-3′, which contain Bbs I restriction sites. We amplified the oligo pools by polymerase chain reaction (PCR) using matching primers for the flanking sequences, cloned the double-stranded DNA pool into a modified pLKO-TET-ON plasmid by Golden Gate, and transformed Endura electrocompetent cells (Lucigen) according to the manufacturer’s protocol. We estimated that the transformation efficiency was >500-fold over the size of the initial oligo pool, indicating that each sgRNA is highly represented in the plasmid libraries. The bacteria were expanded in LB medium for ≈16 hours [optical density at 600 nm (OD600) = 0.8], and plasmid DNA was harvested using a Genopure plasmid maxi kit (Roche). We performed a quality control of both libraries by next-generation sequencing (HiSeq2500, Illumina), which retrieved >99% of the sgRNAs present in the ODBC and GERCAR libraries.
Pooled CRISPR screenings
Proliferation-based screens
We transduced MCF7-CRISPRi cells with independent lentiviral pools [multiplicity of infection (MOI) = 0.3] of the GERCAR (n = 2 biological replicates) and small-scale validation (figs. S2C and S5A) (n = 3 biological replicates) libraries. We transduced ≈1000 cells per plasmid to ensure a correct representation of all sgRNAs in the cell population. The cells were selected using puromycin (2 μg/ml; Invitrogen) at 24 hours after transduction, after which they were expanded and harvested at indicated time points. We conducted cell proliferation–based screens up to T = 21 days to allow ≈15 doublings of the cell populations containing the CRISPR libraries and maximize the identification of scoring hits.
Gene reporter screens
In the ODBC screen, we transduced MCF7-CRISPRi-ESR1-HiBiT-GFP or MCF7-CRISPRi-CCND1-HiBiT-GFP with lentiviral pools (MOI = 0.3) of the CRISPR library. We transduced ≈1000 cells per plasmid to ensure a correct representation of all sgRNAs in the cell population. Cells were selected using puromycin (2 μg/ml; Invitrogen) at 24 hours after transduction, after which they were expanded and harvested at T = 7 days by FACS. We collected the top and bottom quartiles of GFP-expressing cells to evaluate the impact of ODBC sgRNAs on the expression of the ESR1-GFP and CCND1-GFP reporters. In parallel, we performed proliferation-based screenings (n = 3 biological replicates) by collecting unsorted pools of cells at T = 7 days, T = 14 days, and T = 21 days, which allowed measuring the impact of ODBC sgRNAs on cell fitness.
CROP-seq
The CROP-seq library was cloned in a pooled format in a modified pLKO-TET-ON plasmid by Golden Gate, which was used to transform Endura electrocompetent cells. We transduced MCF7-CRISPRi cells with lentiviral pools (MOI = 0.3) of the CROP-seq library (n = 2 biological replicates). Cells were selected using puromycin (2 μg/ml; Invitrogen) at 24 hours after transduction, after which they were expanded and harvested at defined time points (T = 5, T = 9, and T = 14 days). Single-cell suspensions were fixed in 90% methanol in Dulbecco’s phosphate-buffered saline (v/v) and stored at −80°C before rehydration and further processing. The rehydration buffer was supplemented with 1% bovine serum albumin and ribonuclease inhibitor (0.5 U/μl; Sigma-Aldrich, P/N 3335399001). The samples were further processed using the Chromium Next GEM Single-Cell 3′ Reagent Kit (10x Genomics) according to the manufacturer’s protocol (CG000184 Chromium Single Cell3 v3 Feature Barcoding CRISPR Screening UG RevB). To boost yields of the CRISPR screening library, the amplified complementary DNA was amplified for an additional 21 cycles with custom oligos (5′ CTACACGACGCTCTTCCGATCT and 5′ GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTGTGGAAAGGACGAAACACCG; Microsynth AG), using KAPA 2× HotStart HiFi ReadyMix (KAPA biosystems P/N KK2601). Libraries were sequenced on an Illumina HiSeq 2500. Raw data are deposited to the Sequence Read Archive under the BioProject accession identifier PRJNA714625 (www.ncbi.nlm.nih.gov/bioproject/PRJNA714625).
Enhancer reporter assays
DNA fragments of candidate enhancers (ESR1_-150 kb, CCND1_-125 kb, CCND1_-518 kb, and TET) were synthesized (Twist Bioscience) and cloned in pGL3 promoter (Promega). For this assay, MCF7 cells were grown in EMEM (Gibco) supplemented with 10% charcoal-striped serum (Gibco) before plasmid transfection and treatment with 10−8 M estradiol (Sigma-Aldrich) or vehicle (EtOH, Sigma-Aldrich) for 24 hours. The cells were cotransfected with a plasmid expressing Renilla, which was used to normalize the transfection efficiency (Luciferase firefly/Renilla). Reporter activity was measured 40 hours after transfection using the dual-luciferase system (Promega) according to the manufacturer’s instructions.
Gene expression analyses
For immunoblotting, cells were harvested and lysed in radioimmunoprecipitation assay buffer supplemented with protease inhibitor cocktail (Roche). Protein samples were resolved on SDS–polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes, and probed with the following antibodies: ER (Cell Signaling Technology, 13258), TFAP2C (Cell Signaling Technology, 2320), and vinculin (Sigma-Aldrich, V9131). For RNA expression analysis, total RNA was extracted from cell pellets using the RNeasy Mini Kit Plus (QIAGEN) according to the manufacturer’s instructions. Quantitative PCR was performed with QuantStudio 6 Flex (Applied Biosystems) using the iTaq Universal Probes One-step Kit (Bio-Rad). We used glyceraldehyde-3-phosphate dehydrogenase as housekeeping gene to normalize target gene expression levels.
ChIP-seq, RNA-seq, and ATAC-seq
ChIP-seq was performed as previously described (62). Briefly, cells were cross-linked in 1% formaldehyde for 10 min at room temperature after which the reaction was stopped by addition of 0.125 M glycine. Cells were lysed and harvested in ChIP buffer (100 mM tris at pH 8.6, 0.3% SDS, 1.7% Triton X-100, and 5 mM EDTA), and the chromatin was disrupted by sonication using an EpiShear sonicator (Active Motif) to obtain fragments of average 200 to 500 bp in size. Chromatin extracts were incubated for 16 hours with the following antibodies: ER (Cell Signaling Technology, 13258), FOXA1 (Cell Signaling Technology, 58613), GATA3 (Cell Signaling Technology, 5852), CTCF (Cell Signaling Technology, 2899), H3K27ac (Cell Signaling Technology, 8173), and H3K4me1 (Cell Signaling Technology, 5326). Immunoprecipitated complexes were recovered using Protein G Dynabeads (Invitrogen), and DNA was recovered by reverse cross-linking and purified using SPRIselect beads (Beckman Coulter). Libraries for ChIP-seq were generated using the Ovation Ultralow Library System V2 (NuGEN), and barcodes were added using New England Biolabs (NEB) Next Multiplex Oligos for Illumina (NEB, Index Primers Set 1) according to the manufacturer’s recommendation. For TFAP2C (GSM889425) and NCOA3 (GSM2827577), we obtained bigwig files of ChIP-seq data from Cistrome (34). RNA-seq libraries were generated using the TruSeq RNA Sample Prep Kit v2 (Illumina) according to the manufacturer’s recommendation. ATAC-seq (assay for transposase-accessible chromatin using sequencing) was performed as previously described (63). All next-generation sequencing experiments were run on a HiSeq2500 unless otherwise specified (Illumina). Data are deposited to the Dryad repository with document identifier doi: 10.5061/dryad.t1g1jwt20.
Hi-C
The Hi-C experiments were performed in MCF7 cells using the Arima-HiC Kit for mammalian cell lines according to the manufacturer’s protocol (Arima Genomics). Briefly, chromatin was cross-linked and digested using a restriction enzyme cocktail. The 5′-overhangs were filled in, causing the digested ends to be labeled with a biotinylated nucleotide. The digested ends of DNA were ligated, purified, fragmented, and finally enriched by biotin pull down. The enriched fragments were used to prepare a custom library using the Kapa HyperPrep Library Kit (Roche), and the libraries were sequenced in a NovaSeq 6000 (Illumina) using paired-end mode. Raw data are deposited to the Sequence Read Archive under the BioProject accession identifier PRJNA714625 (www.ncbi.nlm.nih.gov/bioproject/PRJNA714625).
Computational analyses
ChIP-seq and ATAC-seq data processing
Fastq files were aligned to a human reference genome (hg38) using bowtie2 v2.3.4.1 (64) and sorted using SAMtools v1.8 (65). Duplicates were marked and removed using Picard MarkDuplicates v2.18.7 (http://broadinstitute.github.io/picard), and low-quality mapped reads (below 20) were removed using SAMtools. SAMtools view was used to retain reads mapping to human chromosomes and to discard reads mapping to chrM for ATAC-seq samples. Last, peaks and their summits were called using macs2 v2.1.1 (66), with a P value threshold cutoff of 0.01. Reads per kilo base per million mapped reads (RPKM)–normalized bigwig files were created from the bam files using bamCoverage from deepTools v.3.1.0 in bin sizes of 10 with extended paired reads (67). To enable rapid downstream analyses, ChIP-seq and ATAC-seq signals were summarized at every ODBC and GERCAR guide location as follows. The center of the genomic region for each guide was obtained, and extended by 250 bp in both directions. The average signal was obtained from all 500 bp windows from the bigwig files using multiBigwigSummary from deepTools v3.1.0.
Hi-C data processing
Reads from the fastq files were mapped to ENSEMBL hg38 human genome release 97 using HiC-Pro v2.11.4 (68), the contact matrix was generated and iterative correction and eigenvector decomposition (ICE)–normalized using cooler v.0.8.7 (69) and visualized in HiGlass (70). Significant contacts were obtained from mustache (71) v.1.0.1 using the contact matrix bin sizes of 1 and 5 kb and subsequently filtered for loops having at least a false discovery rate of 0.05.
RNA-seq data processing
Reads were aligned using bowtie2 (64). Gene-level expression quantities were estimated by the Salmon algorithm (72). Differential expression analysis was performed with DESeq2 (73). GSEA was performed on the described gene intersection, considering the average logFC across conditions and using ClusterProfiler (v 3.18.1) to interrogate the gene annotation categories from MsigDB package (v 7.2.1).
Core transcription regulatory circuitry
We performed the core transcriptional regulatory circuitry analysis as previously described using CRC (74) (https://github.com/linlabcode/CRC) that required three files: a list of active genes, a list of superenhancers, and regions of open chromatin. We identified active genes as those with peaks for H3K4me3, H3K27ac, and Pol2 and without peaks for H3K27me3 and H3K9me3 within 1000 bp of a TSS. We identified superenhancers using the rank ordering of super enhancers (75) meta-algorithm (https://github.com/BradnerLab/pipeline). We identified regions of open chromatin from the ATAC-seq peaks.
Pooled CRISPR screens data analysis
ODBC and GERCAR
Sequencing reads were aligned to the sgRNA library. For each sample, sgRNA reads were counted. Results from individual samples were scaled for library size and normalized using the trimmed mean of M-values (TMM) method available in the edgeR Bioconductor package (76). The logFC in sgRNA abundance in each sample cells versus reference plasmid was calculated using the general linear model log-likelihood ratio test method in edgeR (77). For GFP reporter ODBC screening, the frequency of sgRNAs in the GFP-high or GFP-low population was compared to the one in the unsorted population. Enrichment in the two population was defined as ∆(GFPhigh/GFPlow).
Validation library
To correct for variations among samples of different time points or replicates, median counts of control sgRNAs are normalized to 1 million, and all other sgRNAs are proportionally normalized for each sample. We explicitly model the normalized counts for a given sgRNA using an exponential model. Specifically, we use the count of a given sgRNA in the library sequencing as its initial state and counts at days 5, 9, and 14 as functions of corresponding time durations. Thus, for a given sgRNA i, we have
| (1) |
where is the normalized count of sgRNA i at time t and is the normalized count in library sequencing (initial value). The solution to this equation is a single parameter: The constant αi that fully depends on lethality of the sgRNA. We refer to it as “growth rate” regardless of the sign of its value. To solve this equation, we transform it with natural log link function and get
| (2) |
We next use linear regression model from statsmodels (78) package to estimate the parameter αi for each sgRNA shown in figs. S2C and S5A.
Single-cell CRISPR screen data preprocessing
scRNA sequencing
Single-cell sequencing data were processed using Cell Ranger (version 3.0.1, 10x Genomics) and kallisto bus (79) in parallel to ensure data consistency. Human genome assembly (ENSEMBL GRCh38 release-98) was used as the reference to map mRNA reads. In addition, to map and quantify sgRNA reads for enrichment sgRNA sequencing, artificial chromosomes that represent exogenic sgRNA construct were generated by annotating sgRNA sequences in a standard genome reference format. The python-based Scanpy (80) package was used to analyze the gene expression matrices including quality control, filtering, and downstream analysis. Cells with less than 800 genes were removed, and weakly expressed genes were excluded. Quality control results are shown in fig. S6 (A and B). We found that the results from two alignment approaches (Cell Ranger and Kallisto) are comparable. Thus, Kallisto BUS output was used for downstream analysis and to generate plots shown in this manuscript.
sgRNA assignment
To assign sgRNAs, we developed a tool to model unique molecular identifier (UMI) counts of sgRNAs. Briefly, we assumed an sgRNA library of N distinct sgRNAs. In a given cell, the observed counts (oi) of sgRNA i ∈ [1, N] consist of a real signal (xi) expressed by the cell (i.e., integrated sgRNA) and a contaminating signal (ai) from ambient sgRNA. Thus, for all sgRNAs, we have
| (3) |
where are vectors of counts of integrated sgRNAs, ambient sgRNA contamination, and observations, respectively. All vectors have the dimension of the library size N. It is reasonable to assume that ambient sgRNAs are linearly correlated with their fractions in the medium, provided that the medium is the source of ambient sgRNAs. Therefore, we use as an approximation of , where represents the total ambient sgRNA counts in a cell. represents the sgRNA frequencies in empty droplets, so ei ∈ [0, 1] and . Together, we have
| (4) |
We assume that exactly one and only one sgRNA is integrated in each cell due to the low MOI used in our experiments (MOI = 0.3). Set sgRNA j is the only integrated guide, and its expression level is C ∈ ℕ. In other words
| (5), (6), (7) |
Replace with Eqs. 5 and 6, we have
| (8), (9) |
Further replace f with Eq. 7, after transformation, we get
| (10), (11) |
It is worth noting that the “pseudo-count” on the right hand side of both Eqs. 10 and 11 does not contain any unknown variables and can be calculated for all sgRNA i ∈ [1, N]. Yet, one expects for i = j and , since . As a result, the sgRNA with the highest pseudo-count, i.e., C, should indicate the true integrated sgRNA. However, assigning the highest pseudo-counts cell by cell leads to overfitting issues, and a minimum cutoff value is expected for C. Here, the cutoff value is set to maximize single-sgRNA detection rate and varies for each sample. As a result, about 40% of cells are assigned to a single sgRNA using a global cutoff as shown in fig. S6C. Knockdown groups with less than 10 cells at two or three time points were excluded, as indicated in fig. S6D.
For the analysis shown in the dot plots of Fig. 3 (B to D) and figs. S7, S8, S10, and S11, the expression of on-target genes and adjacent genes was first Z-normalized
| (9) |
We then calculated the t test of the means of knockdown groups and control group using scipy.stats.ttest_ind.
Public large-scale genomics datasets
DEPMAP (www.depmap.org)
We downloaded the RPKM-normalized mRNA expression data (version 18q3) from the Cancer Cell Line Encyclopedia (81) at https://portals.broadinstitute.org/ccle/data on 24 September 2018. We obtained the RNAi-based (https://figshare.com/articles/DEMETER2_data/6025238/2, version DEMETER2 v2) and CRISPR-based (https://figshare.com/articles/DepMap_Achilles_18Q3_public/6931364/1, version 18q3) loss-of-function screening data from the Cancer Dependency Map (DEPMAP) (44) portal at https://depmap.org/portal/ on 20 September 2018. The RNAi dataset was generated by applying the DEMETER2 algorithm (44) to the combined data from Project DRIVE (43), Project Achilles (44), and 76 additional breast cancer cell lines (Marcotte dataset) (42). The CRISPR dataset was generated as previously described (82). The sensitivity score uses all the shRNAs per gene and gives a measure of the statistical significance of the dropout of those shRNAs compared to the background of the rest of the shRNAs logFC. A detailed method to obtain the sensitivity score is described by McDonald and colleagues (43).
Acknowledgments
We thank G. Bushold, G. McAllister, F. Sigoillot, J. Reece-Hoyes, L. Morelli, G. Eliott, and L. Barys for additional technical support. We are grateful to Z. Jagani, A. Granger, and members of the Schübeler laboratory for insightful discussions and critical reading of the manuscript. Author contributions: Conceptualization: R.L. and G.G.G. Methodology: R.L., K.S., C.S., D.S., and G.G.G. Investigation: R.L., K.S., C.S., E.C.H.U., A.E.W., J.D., S.W., J.D.-M., U.Y., M.B., V.A., F.M.-M., R.K., M.E., A.V.O., P.H., J.K., W.C., R.C., A.W., M.A., and G.G.G. Supervision: R.L., K.S., U.N., J.W., A.d., A.K., G.R., D.S., and G.G.G. Writing (original draft): R.L. and G.G.G. Writing (review and editing): R.L., K.S., C.S., D.S., and G.G.G. Competing interests: R.L., K.S., C.S., E.C.H.U., A.E.W., J.D., S.W., J.D.-M., U.Y., M.B., V.A., F.M.-M., R.K., M.E., P.H., J.K., W.C., R.C., A.W., M.A., U.N., A.d., A.K., G.R., and G.G.G. are/have been employees and shareholders of Novartis Pharma. All other authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All materials can be provided by G.G.G pending scientific review and a completed material transfer agreement. Requests for the materials should be submitted to: giorgio.galli@novartis.com. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/27/eabf5733/DC1
REFERENCES AND NOTES
- 1.Bradner J. E., Hnisz D., Young R. A., Transcriptional addiction in cancer. Cell 168, 629–643 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harbeck N., Penault-Llorca F., Cortes J., Gnant M., Houssami N., Poortmans P., Ruddy K., Tsang J., Cardoso F., Breast cancer. Nat. Rev. Dis. Primers. 5, 66 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Martinez E., Givel F., Wahli W., The estrogen-responsive element as an inducible enhancer: DNA sequence requirements and conversion to a glucocorticoid-responsive element. EMBO J. 6, 3719–3727 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kato S., Endoh H., Masuhiro Y., Kitamoto T., Uchiyama S., Sasaki H., Masushige S., Gotoh Y., Nishida E., Kawashima H., Metzger D., Chambon P., Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science 270, 1491–1494 (1995). [DOI] [PubMed] [Google Scholar]
- 5.Williams C., Lin C.-Y., Oestrogen receptors in breast cancer: Basic mechanisms and clinical implications. Ecancermedicalscience 7, 370 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carroll J. S., Liu X. S., Brodsky A. S., Li W., Meyer C. A., Szary A. J., Eeckhoute J., Shao W., Hestermann E. V., Geistlinger T. R., Fox E. A., Silver P. A., Brown M., Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122, 33–43 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Carroll J. S., Meyer C. A., Song J., Li W., Geistlinger T. R., Eeckhoute J., Brodsky A. S., Keeton E. K., Fertuck K. C., Hall G. F., Wang Q., Bekiranov S., Sementchenko V., Fox E. A., Silver P. A., Gingeras T. R., Liu X. S., Brown M., Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 38, 1289–1297 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Hah N., Danko C. G., Core L., Waterfall J. J., Siepel A., Lis J. T., Kraus W. L., A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell 145, 622–634 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sur I., Taipale J., The role of enhancers in cancer. Nat. Rev. Cancer 16, 483–493 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Canver M. C., Smith E. C., Sher F., Pinello L., Sanjana N. E., Shalem O., Chen D. D., Schupp P. G., Vinjamur D. S., Garcia S. P., Luc S., Kurita R., Nakamura Y., Fujiwara Y., Maeda T., Yuan G. C., Zhang F., Orkin S. H., Bauer D. E., BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature 527, 192–197 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanjana N. E., Wright J., Zheng K., Shalem O., Fontanillas P., Joung J., Cheng C., Regev A., Zhang F., High-resolution interrogation of functional elements in the noncoding genome. Science 353, 1545–1549 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fulco C. P., Munschauer M., Anyoha R., Munson G., Grossman S. R., Perez E. M., Kane M., Cleary B., Lander E. S., Engreitz J. M., Systematic mapping of functional enhancer-promoter connections with CRISPR interference. Science 354, 769–773 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gasperini M., Findlay G. M., McKenna A., Milbank J. H., Lee C., Zhang M. D., Cusanovich D. A., Shendure J., CRISPR/Cas9-mediated scanning for regulatory elements required for HPRT1 expression via thousands of large, programmed genomic deletions. Am. J. Hum. Genet. 101, 192–205 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korkmaz G., Lopes R., Ugalde A. P., Nevedomskaya E., Han R., Myacheva K., Zwart W., Elkon R., Agami R., Functional genetic screens for enhancer elements in the human genome using CRISPR-Cas9. Nat. Biotechnol. 34, 192–198 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Fei T., Li W., Peng J., Xiao T., Chen C. H., Wu A., Huang J., Zang C., Liu X. S., Brown M., Deciphering essential cistromes using genome-wide CRISPR screens. Proc. Natl. Acad. Sci. U.S.A. 116, 25186–25195 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rajagopal N., Srinivasan S., Kooshesh K., Guo Y., Edwards M. D., Banerjee B., Syed T., Emons B. J., Gifford D. K., Sherwood R. I., High-throughput mapping of regulatory DNA. Nat. Biotechnol. 34, 167–174 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diao Y., Li B., Meng Z., Jung I., Lee A. Y., Dixon J., Maliskova L., Guan K. L., Shen Y., Ren B., A new class of temporarily phenotypic enhancers identified by CRISPR/Cas9-mediated genetic screening. Genome Res. 26, 397–405 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diao Y., Fang R., Li B., Meng Z., Yu J., Qiu Y., Lin K. C., Huang H., Liu T., Marina R. J., Jung I., Shen Y., Guan K. L., Ren B., A tiling-deletion-based genetic screen for cis-regulatory element identification in mammalian cells. Nat. Methods 14, 629–635 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klann T. S., Black J. B., Chellappan M., Safi A., Song L., Hilton I. B., Crawford G. E., Reddy T. E., Gersbach C. A., CRISPR-Cas9 epigenome editing enables high-throughput screening for functional regulatory elements in the human genome. Nat. Biotechnol. 35, 561–568 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie S., Duan J., Li B., Zhou P., Hon G. C., Multiplexed engineering and analysis of combinatorial enhancer activity in single cells. Mol. Cell 66, 285–299.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Gasperini M., Hill A. J., McFaline-Figueroa J. L., Martin B., Kim S., Zhang M. D., Jackson D., Leith A., Schreiber J., Noble W. S., Trapnell C., Ahituv N., Shendure J., A genome-wide framework for mapping gene regulation via cellular genetic screens. Cell 176, 377–390.e19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fulco C. P., Nasser J., Jones T. R., Munson G., Bergman D. T., Subramanian V., Grossman S. R., Anyoha R., Doughty B. R., Patwardhan T. A., Nguyen T. H., Kane M., Perez E. M., Durand N. C., Lareau C. A., Stamenova E. K., Aiden E. L., Lander E. S., Engreitz J. M., Activity-by-contact model of enhancer-promoter regulation from thousands of CRISPR perturbations. Nat. Genet. 51, 1664–1669 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dawson S. J., Rueda O. M., Aparicio S., Caldas C., A new genome-driven integrated classification of breast cancer and its implications. EMBO J. 32, 617–628 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ali H. R., Rueda O. M., Chin S. F., Curtis C., Dunning M. J., Aparicio S. A., Caldas C., Genome-driven integrated classification of breast cancer validated in over 7,500 samples. Genome Biol. 15, 431 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Osborne C. K., Schiff R., Mechanisms of endocrine resistance in breast cancer. Annu. Rev. Med. 62, 233–247 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arnold A., Papanikolaou A., Cyclin D1 in breast cancer pathogenesis. J. Clin. Oncol. 23, 4215–4224 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Thurman R. E., Rynes E., Humbert R., Vierstra J., Maurano M. T., Haugen E., Sheffield N. C., Stergachis A. B., Wang H., Vernot B., Garg K., John S., Sandstrom R., Bates D., Boatman L., Canfield T. K., Diegel M., Dunn D., Ebersol A. K., Frum T., Giste E., Johnson A. K., Johnson E. M., Kutyavin T., Lajoie B., Lee B. K., Lee K., London D., Lotakis D., Neph S., Neri F., Nguyen E. D., Qu H., Reynolds A. P., Roach V., Safi A., Sanchez M. E., Sanyal A., Shafer A., Simon J. M., Song L., Vong S., Weaver M., Yan Y., Zhang Z., Zhang Z., Lenhard B., Tewari M., Dorschner M. O., Hansen R. S., Navas P. A., Stamatoyannopoulos G., Iyer V. R., Lieb J. D., Sunyaev S. R., Akey J. M., Sabo P. J., Kaul R., Furey T. S., Dekker J., Crawford G. E., Stamatoyannopoulos J. A., The accessible chromatin landscape of the human genome. Nature 489, 75–82 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y., Song F., Zhang B., Zhang L., Xu J., Kuang D., Li D., Choudhary M. N. K., Li Y., Hu M., Hardison R., Wang T., Yue F., The 3D Genome Browser: A web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Genome Biol. 19, 151 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.The FANTOM Consortium and the RIKEN PMI and CLST (DGT), Forrest A. R., Kawaji H., Rehli M., Baillie J. K., de Hoon M. J., Haberle V., Lassmann T., Kulakovskiy I. V., Lizio M., Itoh M., Andersson R., Mungall C. J., Meehan T. F., Schmeier S., Bertin N., Jorgensen M., Dimont E., Arner E., Schmidl C., Schaefer U., Medvedeva Y. A., Plessy C., Vitezic M., Severin J., Semple C., Ishizu Y., Young R. S., Francescatto M., Alam I., Albanese D., Altschuler G. M., Arakawa T., Archer J. A., Arner P., Babina M., Rennie S., Balwierz P. J., Beckhouse A. G., Pradhan-Bhatt S., Blake J. A., Blumenthal A., Bodega B., Bonetti A., Briggs J., Brombacher F., Burroughs A. M., Califano A., Cannistraci C. V., Carbajo D., Chen Y., Chierici M., Ciani Y., Clevers H. C., Dalla E., Davis C. A., Detmar M., Diehl A. D., Dohi T., Drablos F., Edge A. S., Edinger M., Ekwall K., Endoh M., Enomoto H., Fagiolini M., Fairbairn L., Fang H., Farach-Carson M. C., Faulkner G. J., Favorov A. V., Fisher M. E., Frith M. C., Fujita R., Fukuda S., Furlanello C., Furino M., Furusawa J., Geijtenbeek T. B., Gibson A. P., Gingeras T., Goldowitz D., Gough J., Guhl S., Guler R., Gustincich S., Ha T. J., Hamaguchi M., Hara M., Harbers M., Harshbarger J., Hasegawa A., Hasegawa Y., Hashimoto T., Herlyn M., Hitchens K. J., Sui S. J. H., Hofmann O. M., Hoof I., Hori F., Huminiecki L., Iida K., Ikawa T., Jankovic B. R., Jia H., Joshi A., Jurman G., Kaczkowski B., Kai C., Kaida K., Kaiho A., Kajiyama K., Kanamori-Katayama M., Kasianov A. S., Kasukawa T., Katayama S., Kato S., Kawaguchi S., Kawamoto H., Kawamura Y. I., Kawashima T., Kempfle J. S., Kenna T. J., Kere J., Khachigian L. M., Kitamura T., Klinken S. P., Knox A. J., Kojima M., Kojima S., Kondo N., Koseki H., Koyasu S., Krampitz S., Kubosaki A., Kwon A. T., Laros J. F., Lee W., Lennartsson A., Li K., Lilje B., Lipovich L., Mackay-Sim A., Manabe R., Mar J. C., Marchand B., Mathelier A., Mejhert N., Meynert A., Mizuno Y., de Lima Morais D. A., Morikawa H., Morimoto M., Moro K., Motakis E., Motohashi H., Mummery C. L., Murata M., Nagao-Sato S., Nakachi Y., Nakahara F., Nakamura T., Nakamura Y., Nakazato K., van Nimwegen E., Ninomiya N., Nishiyori H., Noma S., Noma S., Noazaki T., Ogishima S., Ohkura N., Ohimiya H., Ohno H., Ohshima M., Okada-Hatakeyama M., Okazaki Y., Orlando V., Ovchinnikov D. A., Pain A., Passier R., Patrikakis M., Persson H., Piazza S., Prendergast J. G., Rackham O. J., Ramilowski J. A., Rashid M., Ravasi T., Rizzu P., Roncador M., Roy S., Rye M. B., Saijyo E., Sajantila A., Saka A., Sakaguchi S., Sakai M., Sato H., Savvi S., Saxena A., Schneider C., Schultes E. A., Schulze-Tanzil G. G., Schwegmann A., Sengstag T., Sheng G., Shimoji H., Shimoni Y., Shin J. W., Simon C., Sugiyama D., Sugiyama T., Suzuki M., Suzuki N., Swoboda R. K., t’Hoen P. A., Tagami M., Takahashi N., Takai J., Tanaka H., Tatsukawa H., Tatum Z., Thompson M., Toyodo H., Toyoda T., Valen E., van de Wetering M., van den Berg L. M., Verado R., Vijayan D., Vorontsov I. E., Wasserman W. W., Watanabe S., Wells C. A., Winteringham L. N., Wolvetang E., Wood E. J., Yamaguchi Y., Yamamoto M., Yoneda M., Yonekura Y., Yoshida S., Zabierowski S. E., Zhang P. G., Zhao X., Zucchelli S., Summers K. M., Suzuki H., Daub C. O., Kawai J., Heutink P., Hide W., Freeman T. C., Lenhard B., Bajic V. B., Taylor M. S., Makeev V. J., Sandelin A., Hume D. A., Carninci P., Hayashizaki Y., A promoter-level mammalian expression atlas. Nature 507, 462–470 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bailey S. D., Desai K., Kron K. J., Mazrooei P., Sinnott-Armstrong N. A., Treloar A. E., Dowar M., Thu K. L., Cescon D. W., Silvester J., Yang S. Y., Wu X., Pezo R. C., Haibe-Kains B., Mak T. W., Bedard P. L., Pugh T. J., Sallari R. C., Lupien M., Noncoding somatic and inherited single-nucleotide variants converge to promote ESR1 expression in breast cancer. Nat. Genet. 48, 1260–1266 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eeckhoute J., Carroll J. S., Geistlinger T. R., Torres-Arzayus M. I., Brown M., A cell-type-specific transcriptional network required for estrogen regulation of cyclin D1 and cell cycle progression in breast cancer. Genes Dev. 20, 2513–2526 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lupien M., Eeckhoute J., Meyer C. A., Wang Q., Zhang Y., Li W., Carroll J. S., Liu X. S., Brown M., FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 132, 958–970 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eeckhoute J., Keeton E. K., Lupien M., Krum S. A., Carroll J. S., Brown M., Positive cross-regulatory loop ties GATA-3 to estrogen receptor alpha expression in breast cancer. Cancer Res. 67, 6477–6483 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Liu T., Ortiz J. A., Taing L., Meyer C. A., Lee B., Zhang Y., Shin H., Wong S. S., Ma J., Lei Y., Pape U. J., Poidinger M., Chen Y., Yeung K., Brown M., Turpaz Y., Liu X. S., Cistrome: An integrative platform for transcriptional regulation studies. Genome Biol. 12, R83 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karamouzis M. V., Papavassiliou K. A., Adamopoulos C., Papavassiliou A. G., Targeting androgen/estrogen receptors crosstalk in cancer. Trends Cancer 2, 35–48 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Radvanyi L., Singh-Sandhu D., Gallichan S., Lovitt C., Pedyczak A., Mallo G., Gish K., Kwok K., Hanna W., Zubovits J., Armes J., Venter D., Hakimi J., Shortreed J., Donovan M., Parrington M., Dunn P., Oomen R., Tartaglia J., Berinstein N. L., The gene associated with trichorhinophalangeal syndrome in humans is overexpressed in breast cancer. Proc. Natl. Acad. Sci. U.S.A. 102, 11005–11010 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lopes R., Korkmaz G., Revilla S. A., van Vliet R., Nagel R., Custers L., Kim Y., van Breugel P. C., Zwart W., Moumbeini B., Manber Z., Elkon R., Agami R., CUEDC1 is a primary target of ERα essential for the growth of breast cancer cells. Cancer Lett. 436, 87–95 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Chi D., Singhal H., Li L., Xiao T., Liu W., Pun M., Jeselsohn R., He H., Lim E., Vadhi R., Rao P., Long H., Garber J., Brown M., Estrogen receptor signaling is reprogrammed during breast tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 116, 11437–11443 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang C., Mayer J. A., Mazumdar A., Fertuck K., Kim H., Brown M., Brown P. H., Estrogen induces c-myc gene expression via an upstream enhancer activated by the estrogen receptor and the AP-1 transcription factor. Mol. Endocrinol. 25, 1527–1538 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Datlinger P., Rendeiro A. F., Schmidl C., Krausgruber T., Traxler P., Klughammer J., Schuster L. C., Kuchler A., Alpar D., Bock C., Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods 14, 297–301 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Theodorou V., Stark R., Menon S., Carroll J. S., GATA3 acts upstream of FOXA1 in mediating ESR1 binding by shaping enhancer accessibility. Genome Res. 23, 12–22 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marcotte R., Sayad A., Brown K. R., Sanchez-Garcia F., Reimand J., Haider M., Virtanen C., Bradner J. E., Bader G. D., Mills G. B., Pe’er D., Moffat J., Neel B. G., Functional genomic landscape of human breast cancer drivers, vulnerabilities, and resistance. Cell 164, 293–309 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McDonald E. R. III, de Weck A., Schlabach M. R., Billy E., Mavrakis K. J., Hoffman G. R., Belur D., Castelletti D., Frias E., Gampa K., Golji J., Kao I., Li L., Megel P., Perkins T. A., Ramadan N., Ruddy D. A., Silver S. J., Sovath S., Stump M., Weber O., Widmer R., Yu J., Yu K., Yue Y., Abramowski D., Ackley E., Barrett R., Berger J., Bernard J. L., Billig R., Brachmann S. M., Buxton F., Caothien R., Caushi J. X., Chung F. S., Cortes-Cros M., deBeaumont R. S., Delaunay C., Desplat A., Duong W., Dwoske D. A., Eldridge R. S., Farsidjani A., Feng F., Feng J., Flemming D., Forrester W., Galli G. G., Gao Z., Gauter F., Gibaja V., Haas K., Hattenberger M., Hood T., Hurov K. E., Jagani Z., Jenal M., Johnson J. A., Jones M. D., Kapoor A., Korn J., Liu J., Liu Q., Liu S., Liu Y., Loo A. T., Macchi K. J., Martin T., McAllister G., Meyer A., Molle S., Pagliarini R. A., Phadke T., Repko B., Schouwey T., Shanahan F., Shen Q., Stamm C., Stephan C., Stucke V. M., Tiedt R., Varadarajan M., Venkatesan K., Vitari A. C., Wallroth M., Weiler J., Zhang J., Mickanin C., Myer V. E., Porter J. A., Lai A., Bitter H., Lees E., Keen N., Kauffmann A., Stegmeier F., Hofmann F., Schmelzle T., Sellers W. R., Project DRIVE: A compendium of cancer dependencies and synthetic lethal relationships uncovered by large-scale, deep RNAi screening. Cell 170, 577–592.e10 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Tsherniak A., Vazquez F., Montgomery P. G., Weir B. A., Kryukov G., Cowley G. S., Gill S., Harrington W. F., Pantel S., Krill-Burger J. M., Meyers R. M., Ali L., Goodale A., Lee Y., Jiang G., Hsiao J., Gerath W. F. J., Howell S., Merkel E., Ghandi M., Garraway L. A., Root D. E., Golub T. R., Boehm J. S., Hahn W. C., Defining a cancer dependency Map. Cell 170, 564–576.e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pihlajamaa P., Sahu B., Lyly L., Aittomaki V., Hautaniemi S., Janne O. A., Tissue-specific pioneer factors associate with androgen receptor cistromes and transcription programs. EMBO J. 33, 312–326 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li L., Wang Y., Torkelson J. L., Shankar G., Pattison J. M., Zhen H. H., Fang F., Duren Z., Xin J., Gaddam S., Melo S. P., Piekos S. N., Li J., Liaw E. J., Chen L., Li R., Wernig M., Wong W. H., Chang H. Y., Oro A. E., TFAP2C- and p63-dependent networks sequentially rearrange chromatin landscapes to drive human epidermal lineage commitment. Cell Stem Cell 24, 271–284.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan S. K., Lin Z. H., Chang C. W., Varang V., Chng K. R., Pan Y. F., Yong E. L., Sung W. K., Cheung E., AP-2γ regulates oestrogen receptor-mediated long-range chromatin interaction and gene transcription. EMBO J. 30, 2569–2581 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bojcsuk D., Nagy G., Balint B. L., Inducible super-enhancers are organized based on canonical signal-specific transcription factor binding elements. Nucleic Acids Res. 45, 3693–3706 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saint-Andre V., Federation A. J., Lin C. Y., Abraham B. J., Reddy J., Lee T. I., Bradner J. E., Young R. A., Models of human core transcriptional regulatory circuitries. Genome Res. 26, 385–396 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guan J., Zhou W., Hafner M., Blake R. A., Chalouni C., Chen I. P., De Bruyn T., Giltnane J. M., Hartman S. J., Heidersbach A., Houtman R., Ingalla E., Kategaya L., Kleinheinz T., Li J., Martin S. E., Modrusan Z., Nannini M., Oeh J., Ubhayakar S., Wang X., Wertz I. E., Young A., Yu M., Sampath D., Hager J. H., Friedman L. S., Daemen A., Metcalfe C., Therapeutic ligands antagonize estrogen receptor function by impairing its mobility. Cell 178, 949–963.e18 (2019). [DOI] [PubMed] [Google Scholar]
- 51.ENCODE Project Consortium, Moore J. E., Purcaro M. J., Pratt H. E., Epstein C. B., Shoresh N., Adrian J., Kawli T., Davis C. A., Dobin A., Kaul R., Halow J., Van Nostrand E. L., Freese P., Gorkin D. U., Shen Y., He Y., Mackiewicz M., Pauli-Behn F., Williams B. A., Mortazavi A., Keller C. A., Zhang X.-O., Elhajjajy S. I., Huey J., Dickel D. E., Snetkova V., Wei X., Wang X., Rivera-Mulia J. C., Rozowsky J., Zhang J., Chhetri S. B., Zhang J., Victorsen A., White K. P., Visel A., Yeo G. W., Burge C. B., Lecuyer E., Gilbert D. M., Dekker J., Rinn J., Mendenhall E. M., Ecker J. R., Kellis M., Klein R. J., Noble W. S., Kundaje A., Guigo R., Farnham P. J., Cherry J. M., Myers R. M., Ren B., Graveley B. R., Gerstein M. B., Pennacchio L. A., Snyder M. P., Bernstein B. E., Wold B., Hardison R. C., Gingeras T. R., Stamatoyannopoulos J. A., Weng Z., Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 583, 699–710 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.French J. D., Ghoussaini M., Edwards S. L., Meyer K. B., Michailidou K., Ahmed S., Khan S., Maranian M. J., O’Reilly M., Hillman K. M., Betts J. A., Carroll T., Bailey P. J., Dicks E., Beesley J., Tyrer J., Maia A. T., Beck A., Knoblauch N. W., Chen C., Kraft P., Barnes D., Gonzalez-Neira A., Alonso M. R., Herrero D., Tessier D. C., Vincent D., Bacot F., Luccarini C., Baynes C., Conroy D., Dennis J., Bolla M. K., Wang Q., Hopper J. L., Southey M. C., Schmidt M. K., Broeks A., Verhoef S., Cornelissen S., Muir K., Lophatananon A., Stewart-Brown S., Siriwanarangsan P., Fasching P. A., Loehberg C. R., Ekici A. B., Beckmann M. W., Peto J., dos Santos Silva I., Johnson N., Aitken Z., Sawyer E. J., Tomlinson I., Kerin M. J., Miller N., Marme F., Schneeweiss A., Sohn C., Burwinkel B., Guenel P., Truong T., Laurent-Puig P., Menegaux F., Bojesen S. E., Nordestgaard B. G., Nielsen S. F., Flyger H., Milne R. L., Zamora M. P., Perez J. I. A., Benitez J., Anton-Culver H., Brenner H., Muller H., Arndt V., Stegmaier C., Meindl A., Lichtner P., Schmutzler R. K., Engel C., Brauch H., Hamann U., Justenhoven C.; GENICA Network, Aaltonen K., Heikkilä P., Aittomäki K., Blomqvist C., Matsuo K., Ito H., Iwata H., Sueta A., Bogdanova N. V., Antonenkova N. N., Dörk T., Lindblom A., Margolin S., Mannermaa A., Kataja V., Kosma V. M., Hartikainen J. M.; kConFab Investigators, Wu A. H., Tseng C.-c., Van Den Berg D., Stram D. O., Lambrechts D., Peeters S., Smeets A., Floris G., Chang-Claude J., Rudolph A., Nickels S., Flesch-Janys D., Radice P., Peterlongo P., Bonanni B., Sardella D., Couch F. J., Wang X., Pankratz V. S., Lee A., Giles G. G., Severi G., Baglietto L., Haiman C. A., Henderson B. E., Schumacher F., Le Marchand L., Simard J., Goldberg M. S., Labreche F., Dumont M., Teo S. H., Yip C. H., Ng C. H., Vithana E. N., Kristensen V., Zheng W., Deming-Halverson S., Shrubsole M., Long J., Winqvist R., Pylkas K., Jukkola-Vuorinen A., Grip M., Andrulis I. L., Knight J. A., Glendon G., Mulligan A. M., Devilee P., Seynaeve C., Garcia-Closas M., Figueroa J., Chanock S. J., Lissowska J., Czene K., Klevebring D., Schoof N., Hooning M. J., Martens J. W., Collee J. M., Tilanus-Linthorst M., Hall P., Li J., Liu J., Humphreys K., Shu X. O., Lu W., Gao Y. T., Cai H., Cox A., Balasubramanian S. P., Blot W., Signorello L. B., Cai Q., Pharoah P. D., Healey C. S., Shah M., Pooley K. A., Kang D., Yoo K. Y., Noh D. Y., Hartman M., Miao H., Sng J. H., Sim X., Jakubowska A., Lubinski J., Jaworska-Bieniek K., Durda K., Sangrajrang S., Gaborieau V., McKay J., Toland A. E., Ambrosone C. B., Yannoukakos D., Godwin A. K., Shen C. Y., Hsiung C. N., Wu P. E., Chen S. T., Swerdlow A., Ashworth A., Orr N., Schoemaker M. J., Ponder B. A., Nevanlinna H., Brown M. A., Chenevix-Trench G., Easton D. F., Dunning A. M., Functional variants at the 11q13 risk locus for breast cancer regulate cyclin D1 expression through long-range enhancers. Am. J. Hum. Genet. 92, 489–503 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maurano M. T., Humbert R., Rynes E., Thurman R. E., Haugen E., Wang H., Reynolds A. P., Sandstrom R., Qu H., Brody J., Shafer A., Neri F., Lee K., Kutyavin T., Stehling-Sun S., Johnson A. K., Canfield T. K., Giste E., Diegel M., Bates D., Hansen R. S., Neph S., Sabo P. J., Heimfeld S., Raubitschek A., Ziegler S., Cotsapas C., Sotoodehnia N., Glass I., Sunyaev S. R., Kaul R., Stamatoyannopoulos J. A., Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khurana E., Fu Y., Colonna V., Mu X. J., Kang H. M., Lappalainen T., Sboner A., Lochovsky L., Chen J., Harmanci A., Das J., Abyzov A., Balasubramanian S., Beal K., Chakravarty D., Challis D., Chen Y., Clarke D., Clarke L., Cunningham F., Evani U. S., Flicek P., Fragoza R., Garrison E., Gibbs R., Gumus Z. H., Herrero J., Kitabayashi N., Kong Y., Lage K., Liluashvili V., Lipkin S. M., MacArthur D. G., Marth G., Muzny D., Pers T. H., Ritchie G. R. S., Rosenfeld J. A., Sisu C., Wei X., Wilson M., Xue Y., Yu F.; 1000 Genomes Project Consortium, Dermitzakis E. T., Yu H., Rubin M. A., Tyler-Smith C., Gerstein M., Integrative annotation of variants from 1092 humans: Application to cancer genomics. Science 342, 1235587 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weinhold N., Jacobsen A., Schultz N., Sander C., Lee W., Genome-wide analysis of noncoding regulatory mutations in cancer. Nat. Genet. 46, 1160–1165 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mansour M. R., Abraham B. J., Anders L., Berezovskaya A., Gutierrez A., Durbin A. D., Etchin J., Lawton L., Sallan S. E., Silverman L. B., Loh M. L., Hunger S. P., Sanda T., Young R. A., Look A. T., An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science 346, 1373–1377 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Patten D. K., Corleone G., Gyorffy B., Perone Y., Slaven N., Barozzi I., Erdos E., Saiakhova A., Goddard K., Vingiani A., Shousha S., Pongor L. S., Hadjiminas D. J., Schiavon G., Barry P., Palmieri C., Coombes R. C., Scacheri P., Pruneri G., Magnani L., Enhancer mapping uncovers phenotypic heterogeneity and evolution in patients with luminal breast cancer. Nat. Med. 24, 1469–1480 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chepelev I., Wei G., Wangsa D., Tang Q., Zhao K., Characterization of genome-wide enhancer-promoter interactions reveals co-expression of interacting genes and modes of higher order chromatin organization. Cell Res. 22, 490–503 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Woodfield G. W., Horan A. D., Chen Y., Weigel R. J., TFAP2C controls hormone response in breast cancer cells through multiple pathways of estrogen signaling. Cancer Res. 67, 8439–8443 (2007). [DOI] [PubMed] [Google Scholar]
- 60.McPherson L. A., Weigel R. J., AP2α and AP2γ: A comparison of binding site specificity and trans-activation of the estrogen receptor promoter and single site promoter constructs. Nucleic Acids Res. 27, 4040–4049 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cocce K. J., Jasper J. S., Desautels T. K., Everett L., Wardell S., Westerling T., Baldi R., Wright T. M., Tavares K., Yllanes A., Bae Y., Blitzer J. T., Logsdon C., Rakiec D. P., Ruddy D. A., Jiang T., Broadwater G., Hyslop T., Hall A., Laine M., Phung L., Greene G. L., Martin L.-A., Pancholi S., Dowsett M., Detre S., Marks J. R., Crawford G. E., Brown M., Norris J. D., Chang C.-Y., McDonnell D. P., The lineage determining factor GRHL2 collaborates with FOXA1 to establish a targetable pathway in endocrine therapy-resistant breast cancer. Cell Rep. 29, 889–903.e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Galli G. G., Carrara M., Yuan W. C., Valdes-Quezada C., Gurung B., Pepe-Mooney B., Zhang T., Geeven G., Gray N. S., de Laat W., Calogero R. A., Camargo F. D., YAP drives growth by controlling transcriptional pause release from dynamic enhancers. Mol. Cell 60, 328–337 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Buenrostro J. D., Giresi P. G., Zaba L. C., Chang H. Y., Greenleaf W. J., Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup , The sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nusbaum C., Myers R. M., Brown M., Li W., Liu X. S., Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ramírez F., Ryan D. P., Grüning B., Bhardwaj V., Kilpert F., Richter A. S., Heyne S., Dundar F., Manke T., deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Servant N., Varoquaux N., Lajoie B. R., Viara E., Chen C. J., Vert J. P., Heard E., Dekker J., Barillot E., HiC-Pro: An optimized and flexible pipeline for Hi-C data processing. Genome Biol. 16, 259 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Abdennur N., Mirny L. A., Cooler: Scalable storage for Hi-C data and other genomically labeled arrays. Bioinformatics 36, 311–316 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kerpedjiev P., Abdennur N., Lekschas F., McCallum C., Dinkla K., Strobelt H., Luber J. M., Ouellette S. B., Azhir A., Kumar N., Hwang J., Lee S., Alver B. H., Pfister H., Mirny L. A., Park P. J., Gehlenborg N., HiGlass: Web-based visual exploration and analysis of genome interaction maps. Genome Biol. 19, 125 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roayaei Ardakany A., Gezer H. T., Lonardi S., Ay F., Mustache: Multi-scale detection of chromatin loops from Hi-C and Micro-C maps using scale-space representation. Genome Biol. 21, 256 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Patro R., Duggal G., Love M. I., Irizarry R. A., Kingsford C., Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lin C. Y., Erkek S., Tong Y., Yin L., Federation A. J., Zapatka M., Haldipur P., Kawauchi D., Risch T., Warnatz H.-J., Worst B. C., Ju B., Orr B. A., Zeid R., Polaski D. R., Segura-Wang M., Waszak S. M., Jones D. T., Kool M., Hovestadt V., Buchhalter I., Sieber L., Johann P., Chavez L., Gröschel S., Ryzhova M., Korshunov A., Chen W., Chizhikov V. V., Millen K. J., Amstislavskiy V., Lehrach H., Yaspo M. -L., Eils R., Lichter P., Korbel J. O., Pfister S. M., Bradner J. E., Northcott P. A., Active medulloblastoma enhancers reveal subgroup-specific cellular origins. Nature 530, 57–62 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hnisz D., Abraham B. J., Lee T. I., Lau A., Saint-Andre V., Sigova A. A., Hoke H. A., Young R. A., Super-enhancers in the control of cell identity and disease. Cell 155, 934–947 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Robinson M. D., Oshlack A., A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Robinson M. D., Smyth G. K., Moderated statistical tests for assessing differences in tag abundance. Bioinformatics 23, 2881–2887 (2007). [DOI] [PubMed] [Google Scholar]
- 78.S. Seabold, J. Perktold, Statsmodels: Econometric and Statistical Modeling with Python, in Proceedings of the 9th Python in Science Confernce (SPICY, 2010). [Google Scholar]
- 79.Melsted P., Ntranos V., Pachter L., The barcode, UMI, set format and BUStools. Bioinformatics 35, 4472–4473 (2019). [DOI] [PubMed] [Google Scholar]
- 80.Wolf F. A., Angerer P., Theis F. J., SCANPY: Large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A. A., Kim S., Wilson C. J., Lehár J., Kryukov G. V., Sonkin D., Reddy A., Liu M., Murray L., Berger M. F., Monahan J. E., Morais P., Meltzer J., Korejwa A., Jane-Valbuena J., Mapa F. A., Thibault J., Bric-Furlong E., Raman P., Shipway A., Engels I. H., Cheng J., Yu G. K., Yu J., Aspesi P. Jr., de Silva M., Jagtap K., Jones M. D., Wang L., Hatton C., Palescandolo E., Gupta S., Mahan S., Sougnez C., Onofrio R. C., Liefeld T., MacConaill L., Winckler W., Reich M., Li N., Mesirov J. P., Gabriel S. B., Getz G., Ardlie K., Chan V., Myer V. E., Weber B. L., Porter J., Warmuth M., Finan P., Harris J. L., Meyerson M., Golub T. R., Morrissey M. P., Sellers W. R., Schlegel R., Garraway L. A., The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Meyers R. M., Bryan J. G., McFarland J. M., Weir B. A., Sizemore A. E., Xu H., Dharia N. V., Montgomery P. G., Cowley G. S., Pantel S., Goodale A., Lee Y., Ali L. D., Jiang G., Lubonja R., Harrington W. F., Strickland M., Wu T., Hawes D. C., Zhivich V. A., Wyatt M. R., Kalani Z., Chang J. J., Okamoto M., Stegmaier K., Golub T. R., Boehm J. S., Vazquez F., Root D. E., Hahn W. C., Tsherniak A., Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet. 49, 1779–1784 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/27/eabf5733/DC1