Artificial transmembrane proteins can be used to dissect virus entry and provide new insights into cell biology.
Abstract
Retromer, a cellular protein trafficking complex, sorts human papillomaviruses (HPVs) into the retrograde pathway for transport of HPV to the nucleus during virus entry. Here, we conducted a protein modulation screen to isolate four artificial transmembrane proteins called traptamers that inhibit different steps of HPV entry. By analyzing cells expressing pairs of traptamers, we ordered the trafficking steps during entry into a coherent pathway. One traptamer stimulates ubiquitination of the L2 capsid protein or associated proteins and diverts incoming virus to the lysosome, whereas the others act downstream by preventing sequential passage of the virus through retrograde compartments. Complex genetic interactions between traptamers revealed that a cell-penetrating peptide (CPP) on L2 mediates transient insertion of L2 into the endosome membrane, which is stabilized by retromer-L2 binding. These results define the retrograde entry route taken by HPV and show that retromer can play a role in CPP-mediated membrane insertion.
INTRODUCTION
Human papillomavirus (HPV) infections are widespread and responsible for 5% of human cancer, including essentially all cervical cancer (1). Despite the existence of effective prophylactic vaccines, HPV infection and the cancers it causes will remain a major public health problem for decades because vaccine uptake is poor in some populations, current vaccines do not target many pathogenic HPV types, and vaccination does not clear established infections. There are no specific drugs that inhibit HPV. Better understanding of HPV infection will not only provide a rational basis to develop antiviral drugs but also provide important insights into fundamental cell biology, as has been historically the case for many viruses (2).
HPV are nonenveloped viruses containing an ~8000–base pair DNA genome encased in a capsid consisting of 360 molecules of the L1 major capsid protein and up to 72 copies of the L2 minor capsid protein (3). The L1 protein is responsible for HPV binding to the cell surface, and the L2 protein plays an important role in trafficking of the viral genome to the nucleus where viral gene expression and DNA replication occur. Similar to other nonenveloped DNA viruses, HPV must navigate to the nucleus while crossing membranes that impede its progress. HPV has evolved a unique entry strategy, traveling to the nucleus inside a series of retrograde transport compartments, which appears to protect it from cytoplasmic innate immune sensors (4–8).
We conducted a genome-wide RNA interference screen that revealed that the cytoplasmic protein trafficking complex named retromer plays a key role in mediating delivery of HPV into the retrograde pathway and transport to the trans-Golgi network (TGN) and downstream locations (5). Retromer is composed of three core subunits (VPS26, VPS29, and VPS35) that in cooperation with late endosomal small guanosine triphosphatase (GTPase) and Rab7 binds to the cytoplasmic segment of cellular transmembrane (TM) proteins in the endosome membrane and initiates their transport to the TGN or the cell surface (9, 10). Retromer action is thought to be confined to binding to the cytoplasmic segment of TM cargo proteins, recruiting other sorting factors, and possibly contributing to the formation of the protein coat around nascent vesicles for transport.
A nonenveloped virus, HPV lacks classic TM proteins, but a conserved motif in the C terminus of the L2 protein nevertheless binds directly to retromer (11). To access retromer, a cell-penetrating peptide (CPP) sequence near the C terminus of all papillomavirus L2 proteins mediates protrusion of L2 through the endosomal membrane into the cytoplasm where it can bind retromer and other cytoplasmic entry factors such as SNX17 and SNX27 (11–14). γ-Secretase and its adaptor protein p120-catenin promote insertion of L2 into the endosome membrane and proper HPV trafficking during entry (7, 15, 16). A protein complex related to retromer known as retriever also binds the C-terminal segment of L2 and plays a poorly understood role in HPV entry (17, 18). After the L2 protein protrudes into the cytoplasm, it may be anchored in the membrane by a TM domain in its N terminus (19, 20), thus being an “inducible TM protein” [reviewed in (21)]. These studies reveal that CPPs can transfer proteins from one intracellular compartment to another and possibly convert soluble proteins into TM ones, but the mechanism of CPP action remains obscure.
HPV is handled differently from known cellular retromer cargos. Retromer knockdown, mutation of the retromer binding sites or CPP on L2, or titration of retromer away from incoming HPV by cytoplasmic delivery of a peptide containing a retromer binding site causes HPV to accumulate in the endosome without reaching the TGN (5, 11, 12). In contrast, impairment of retromer function typically causes cellular retromer cargos such as Wntless (22), β2-adrenergic receptor (23), glucose transporter type 1, monocarboxylate transporter 1, and ATPase copper transporting alpha (24) to be missorted to the lysosome for degradation [reviewed in (9, 25)]. In addition, HPV trafficking requires Rab7 cycling, whereas Rab7–guanosine triphosphate (GTP) is sufficient for trafficking of divalent metal transporter 1-II and cation independent mannose 6-phosphate receptor (26). These differences suggest that further analysis of the role of retromer during HPV infection may provide new insight into the action of this conserved cellular trafficking complex.
Through gene knockdown and biochemical studies, a variety of cellular proteins in addition to retromer and γ-secretase have been shown to play a role in HPV entry (13, 14, 18, 27–35). However, these studies typically focus on a single protein, so a comprehensive picture of the HPV entry pathway has not emerged, and the factors that determine the fate of incoming HPV are not known.
To further characterize HPV entry, we developed a functional genetics screen to isolate artificial TM proteins that modulate the activity of natural TM proteins (36, 37). In this approach, we construct libraries that express millions of proteins containing a 15- to 26-residue stretch of randomized hydrophobic amino acids and screen these libraries in cells to select rare proteins that by chance bind to the TM domain of a cellular or viral protein and modulate its activity. We name these proteins “traptamers” (TM protein aptamers). To select traptamers that inhibit HPV infection, we introduced a traptamer library into HeLa cervical cancer cells and then infected them with a HPV16 pseudovirus (PsV) that expresses the bovine papillomavirus E2 transcription factor (HPV16-BE2) (Fig. 1A). Expression of E2 represses the HPV E6 and E7 oncogenes in these cells and causes them to undergo growth arrest (38–40). We recovered a traptamer named JX2 from cells that continued to proliferate despite exposure to HPV16-BE2 (26). JX2 associates with and inhibits TBC1 domain family member 5 (TBC1D5), a GTPase activating protein (GAP) that modulates retromer function by regulating Rab7 activity, resulting in increased levels of Rab7 in the GTP-bound form, prevention of retromer dissociation from HPV, and accumulation of HPV in the endosome without trafficking to the TGN. Expression of constitutively active or dominant-negative Rab7 also causes incoming HPV to accumulate in the endosome, confirming that Rab7 must cycle between GTP- and GDP-bound forms to support the role of retromer in HPV entry. TBC1D5 was not identified in previous screens for HPV entry factors, confirming the utility of traptamer screening.
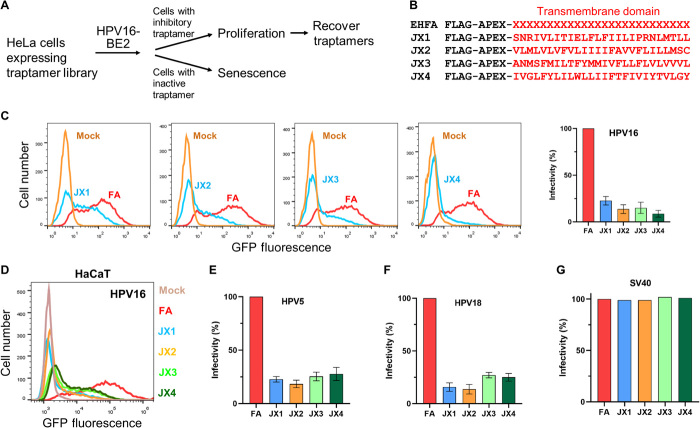
Fig. 1. Isolation of traptamers that inhibit HPV infection.
(A) Overview of scheme to isolate traptamers that inhibit HPV infection. See the main text for details. (B) Sequences of traptamers that inhibit HPV infection. In the EHFA library (top line), FLAG is the epitope tag, APEX is ascorbate peroxidase segment, and red “Xs” represent randomized hydrophobic positions. The red letters in other lines show the sequences of the hydrophobic segments of the four active traptamers in the single-letter amino acid representation. (C) Clonal HeLa-tTA cells stably expressing control FA (FLAG-APEX) (red) or JX1, JX2, JX3, or JX4 (blue) were infected at multiplicity of infection (MOI) of 2 with HPV16 PsV containing the green fluorescent protein (GFP) reporter plasmid (HPV16-GFP). Two days later, GFP fluorescence was measured by flow cytometry to assess infectivity (histograms shown in left). Similar results were graphed in three independent cell lines for each traptamer, showing means and SD. (D) HaCaT-tTA cells expressing FA or the indicated traptamer were infected with HPV16-GFP PsV-GFP. Cells expressing FA were also mock-infected (brown). Two days later, GFP fluorescence was measured by flow cytometry and displayed as histograms. (E and F) Cells as in (C) were infected at MOI of 2 with HPV5-HcRed (E) or HPV18-HcRed (F) PsV. Two days later, HcRed fluorescence was measured by flow cytometry in three independent experiments for each PsV and graphed as in (C). (G) Traptamers do not inhibit SV40. HeLa-tTA cells expressing FA or the indicated traptamer were mock-infected or infected with SV40. Two days later, cells were stained with an antibody recognizing SV40 large T antigen and analyzed by flow cytometry. In all bar graphs, the results are graphed as % infectivity in cells expressing a traptamer normalized to cells expressing FA.
Here, we describe the isolation and characterization of three additional traptamers that block HPV entry. All four traptamers block entry at different steps. The existence of multiple traptamers with different effects on HPV infection allowed us to deploy them as tools to dissect HPV entry in unprecedented detail. By studying HPV entry in cells expressing these traptamers separately and in combination, we ordered the steps of HPV entry into a coherent sequential pathway and showed that ubiquitination of L2 or of proteins associated with L2 is accompanied by diversion of L2 and the encapsidated plasmid genome from the retrograde transport pathway to the lysosome. We also found that the L2 CPP mediates transient insertion of L2 into the endosome membrane and that L2-retromer binding stabilizes membrane insertion. These results define the HPV retrograde entry pathway and identify a previously unknown role of retromer in CPP-mediated membrane insertion.
RESULTS
Isolation of four traptamers that inhibit HPV infection
To isolate traptamers that inhibit HPV16 infection, we used the EHFA retrovirus library, which encodes millions of different traptamers containing an N-terminal FLAG tag, the APEX2 ascorbic peroxidase, and a 26-residue randomized, hydrophobic segment intended to act as a TM domain (Fig. 1, A and B) (26). After introducing EHFA into HeLa S3 cells, we selected cells that were resistant to HPV16 PsV infection and recovered traptamer genes from the genomic DNA of these cells as reported previously (26).
We cloned a traptamer gene from four independent cell cultures into the Retro-X Tet-Off inducible retrovirus vector, pTight (pT), which drives high-level transgene expression in cells expressing the tetracycline transactivator (tTA) in the absence of doxycycline. The four recovered traptamers (designated JX1, JX2, JX3, and JX4) each contained a hydrophobic sequence unrelated to each other or to any known protein (Fig. 1B). HeLa S3 cells expressing tTA (HeLa-tTA cells) were infected with a retrovirus expressing a recovered traptamer or with a control retrovirus, T-FA, expressing the FLAG and APEX2 segment but lacking a randomized hydrophobic segment. Puromycin-resistant cells were infected with HPV16 PsV expressing green fluorescent protein (GFP) from an encapsidated reporter plasmid (HPV16-GFP), and 2 days later, GFP fluorescence was measured by flow cytometry to assess infection. Each traptamer markedly reduced GFP fluorescence compared to infected control FA cells, showing that they inhibited HPV16-GFP PsV infection (Fig. 1C). For all four traptamers, doxycycline addition repressed traptamer expression and abrogated the inhibitory effect, demonstrating that HPV inhibition was due to traptamer expression (fig. S1, A and B) (26). Removal of the APEX2 segment from the traptamers did not affect inhibitory activity (fig. S1C).
We also tested whether continuous expression of the traptamers was required for inhibition. First, we used immunoblotting to show that 1 day of doxycycline treatment caused marked inhibition of traptamer expression (fig. S1D). We then infected duplicate plates of cells expressing traptamers in the absence of doxycycline for 8 hours. We then added doxycycline to one plate of each and measured infectivity 3 days later. As shown in fig. S1E, doxycycline treatment had no significant effect on the inhibition of infection by any of the traptamers. This result shows that 8 hours of full traptamer expression is sufficient to impose a substantial block to infection, which is not reversed by traptamer repression after this time.
HPV16 PsV infection of human HaCaT skin keratinocytes was also markedly inhibited by all four traptamers (Fig. 1D), as was infection of HeLa cells by HPV5 and HPV18 PsV, other carcinogenic HPV types that infect skin and genital mucosa, respectively (Fig. 1, E and F). In contrast, the traptamers did not inhibit SV40 (Fig. 1G), an unrelated, nonenveloped DNA virus. We previously reported that JX2 inhibited the Rab7 GAP, TBC1D5 (26). Here, we report the analysis of cells expressing the other active traptamers, additional analysis of cells expressing JX2, and analysis of cells expressing pairs of traptamers.
Active traptamers are TM proteins that localize differently
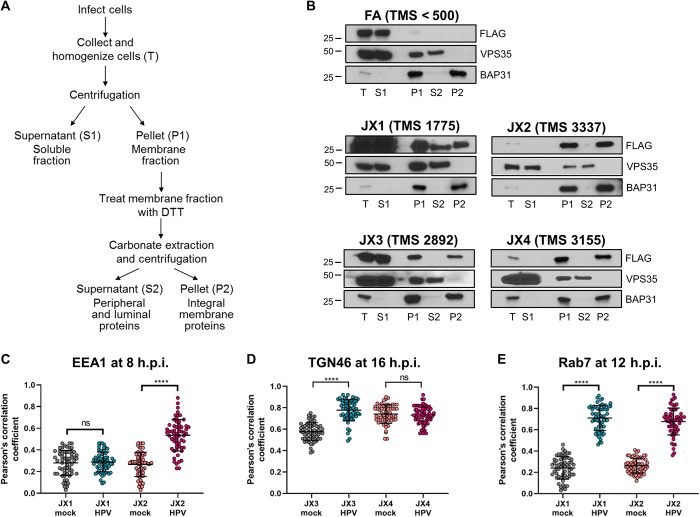
To test whether the traptamers were TM proteins, we performed a carbonate extraction assay, in which uninfected cells expressing traptamers were mechanically homogenized, and proteins were separated by centrifugation into a soluble (S1) and a membrane (P1) fraction (Fig. 2A). The membrane fraction was resuspended, treated with alkaline carbonate solution, and subjected to ultracentrifugation. Peripheral membrane-associated proteins (such as VPS35) and vesicle luminal proteins [such as protein disulfide isomerase (PDI)] were extracted by carbonate into the S2 fraction, while bona fide TM proteins [such as B-cell receptor associated protein 31 (BAP31)] were present in the P2 pellet fraction (Fig. 2B) (15, 41). Membrane-associated JX2, JX3, and JX4 were present exclusively in the P2 fraction, indicating that they were integral membrane proteins. Most of JX1 was in the soluble S1 fraction, and the remainder partitioned equally in S2 and P2 fractions, suggesting that it was anchored less strongly in the membrane than the other traptamers. The TM scores (TMS) from the TM helix prediction program TMpred were consistent with the results of the extraction experiment, with JX2 and JX4 having the highest scores and JX1 the lowest (Fig. 2B). In contrast, FA, which lacks a randomized hydrophobic segment, was exclusively in the S1 fraction and had a TMS inconsistent with TM status, showing that the hydrophobic segment of the active traptamers was responsible for their membrane insertion.
Fig. 2. Characterization of traptamer membrane association and localization.
(A) Schematic of carbonate extraction protocol. (B) JX1, JX2, JX3, and JX4 are TM proteins. HeLa-tTA cells expressing FA, JX1, JX2, JX3, or JX4 were osmotically swollen and mechanically homogenized. The cell lysates (T) were fractionated into soluble (S1) and membrane (P1) fractions by centrifugation. The membrane fraction was extracted with 0.1 M sodium carbonate and again separated into supernatant (S2) and integral membrane pellet (P2) fractions by ultracentrifugation. All fractions were subjected to SDS–polyacrylamide gel electrophoresis (PAGE) and immunoblotted with antibodies recognizing the FLAG epitope tag on FA and the four traptamers, VPS35 (a peripheral membrane protein), or BAP31 (a TM protein), as markers of successful fractionization. Filters were exposed for different lengths of time so that the signals in the P2 and/or S2 lanes were comparable to facilitate comparison. TMS, maximum TMS predicted by TMpred. (C) Pearson’s correlation coefficient for costaining between EEA1 and JX1 or JX2 in mock-infected and HPV16 PsV-infected cells at 8 h.p.i. (at least 60 cells for each condition). Each dot represents an individual cell. ns, not significant; ****P < 0.0001. Representative primary data are shown in fig. S2A. (D) Pearson’s correlation coefficient for costaining between TGN46 and JX3 or JX4 at 16 h.p.i. as in (C). Representative primary data are shown in fig. S2B. (E) Pearson’s correlation coefficient for costaining between Rab7 and JX1 or JX2 at 12 h.p.i. as in (C). Representative primary data are shown in fig. S2C.
Immunofluorescence staining with anti-FLAG antibody was performed to determine the subcellular localization of traptamers. FA was widely distributed in the cell, its localization did not change in response to HPV infection, and it did not colocalize with early endosome antigen 1 (EEA1) or TGN46, markers of the early endosome and TGN, respectively (fig. S2, A and B). On the basis of the virus localization studies described in the next section, we assessed colocalization of JX1 and JX2 with EEA1 and colocalization of JX3 and JX4 with TGN46. JX1 did not colocalize with EEA1 in uninfected or HPV-infected cells, while as reported previously, JX2 did not colocalize with EEA1 in uninfected cells but displayed strong colocalization with EEA1 in HPV-infected cells at 8 hours postinfection (h.p.i.) (Fig. 2C and fig. S2A) (26). In uninfected cells, JX3 colocalized to a moderate extent with TGN46, which was increased by 16 h.p.i., while JX4 and TGN46 highly colocalized in both uninfected and infected cells (Fig. 2D and fig. S2B). Thus, each traptamer displayed a different localization pattern, and HPV infection caused JX2 and JX3 to localize to compartments through which HPV traffics during entry. None of the traptamers caused an obvious redistribution of the cellular markers tested.
Traptamers block HPV entry at different steps
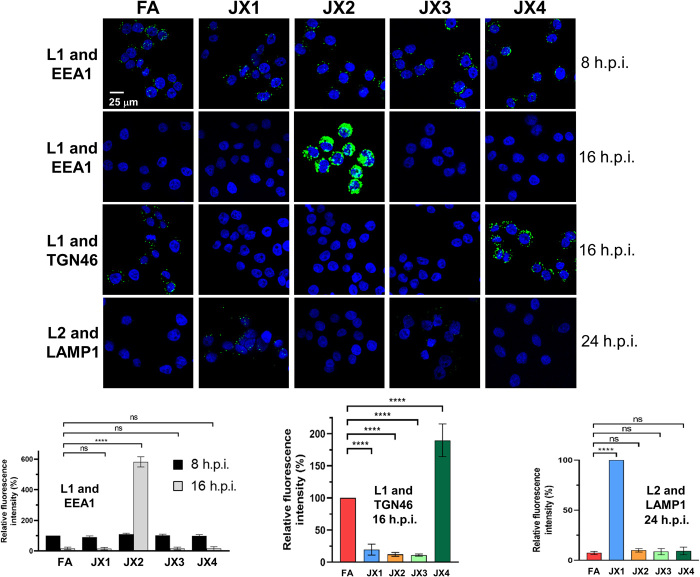
The different localization of the traptamers suggested that they interfered with HPV infection at different steps. To test this, we performed proximity ligation assay (PLA) to examine HPV trafficking in cells expressing each of the traptamers. In PLA, a fluorescent signal is generated only if the two proteins of interest are within 40 nm. We first used an antibody recognizing HPV16 L1 in combination with an antibody recognizing EEA1 or TGN46 (Fig. 3 and fig. S3, A and B), because HPV traffics from the endosome to the TGN during entry. At 8 h.p.i., there were similar levels of L1-EEA1 PLA signal in cells expressing FA or any of the traptamers, showing that the traptamers did not interfere with virus binding to cells, internalization, or localization to the endosome. At 16 h.p.i. in control FA cells, there was little L1-EEA1 PLA signal but abundant L1-TGN46 PLA signal, due to the departure of HPV from the endosome and arrival at the TGN by this time (11). In cells expressing JX2, at 16 h.p.i., there was increased L1-EEA1 PLA signal and no detectable L1-TGN46 PLA signal due to inhibition of endosome exit, as reported previously (26). In cells expressing JX1 or JX3, there was neither L1-EEA1 nor L1-TGN PLA signal at 16 h.p.i., indicating that HPV did not accumulate in the endosome and failed to arrive at the TGN. In contrast, in cells expressing JX4, the L1-TGN46 PLA signal at 16 h.p.i. was increased compared to cells expressing FA or any of the other traptamers. Similarly, in cells expressing JX4 there was increased PLA signal for L1 and the cis-Golgi marker, GM130 (fig. S3C). Thus, JX4 appears to arrest HPV at the TGN and Golgi apparatus in a nonproductive state.
Fig. 3. Traptamers block HPV entry at different steps.
HeLa-tTA cells expressing FA, JX1, JX2, JX3, or JX4 were mock-infected or infected with wild-type HPV16-HcRed PsV or PsV containing HA-tagged L2 at MOI of 150. At 8, 16, or 24 h.p.i., PLA was performed with an antibody recognizing L1 or HA and an antibody recognizing EEA1, TGN46, or LAMP1, as indicated. (Top) PLA signal is green; nuclei are stained blue with 4′,6-diamidino-2-phenylindole (DAPI). Representative primary data and mock-infected controls are shown in fig. S3. (Bottom) Multiple images obtained in each experiment were processed by using BlobFinder software to quantify the fluorescence intensity per cell (at least 100 cells for each condition). The graphs show mean results and SD (three independent experiments), in which the results for the L1-EEA1 samples and L1-TGN46 samples were normalized to those of infected FA cells at 8 and 16 h.p.i., respectively, set at 100%. L2-LAMP1 samples were normalized to JX1 cells at 24 h.p.i., set at 100%. ****P < 0.0001.
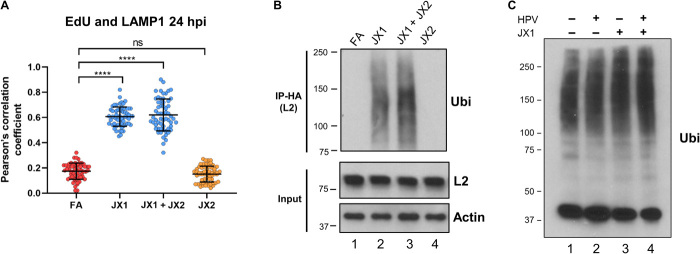
Because missorted retromer cargo is often directed to the lysosome for degradation (25), we also assessed virus localization in the lysosome. For this experiment, we determined localization of L2, because some L1 enters the lysosome upon capsid disassembly during normal infection (42, 43). We conducted PLA at 24 h.p.i. for hemagglutinin (HA)–tagged L2 and lysosome-associated membrane protein 1 (LAMP1), a lysosome marker. In infected cells expressing FA, JX2, JX3, or JX4, there was no L2-LAMP1 PLA signal. In contrast, cells expressing JX1 displayed a clear L2-LAMP1 PLA signal at 24 h.p.i. but not at 16 h.p.i. (Fig. 3 and fig. S3, D and E). To test whether lysosomal localization is accompanied by accelerated degradation of L2, we infected cells expressing FA or JX1 with HPV16-HcRed PsV containing FLAG-tagged L2 and performed immunoblotting at 12 and 24 h.p.i. with an antibody recognizing FLAG. As shown in fig. S3F, there is slightly less L2 expressed at 12 h.p.i. in cells expressing JX1 compared to control cells and a marked fourfold lower L2 expression at 24 h.p.i. Thus, JX1 expression diverts L2 to the lysosome and causes L2 to be removed from the cell.
We also determined the localization of PsV DNA in cells expressing traptamers. After infection with PsVs containing DNA substituted with the nucleoside analog 5-ethynyl-2′-deoxyuridine (EdU), plasmid DNA was localized in infected cells with a fluorescent azide probe that reacts with EdU. We observed strong nuclear EdU signal that overlapped with promyelocytic leukemia protein (PML) staining at 24 h.p.i. in control FA cells, demonstrating that PsV DNA arrived at the nucleus (fig. S4A). At this time, abundant EdU signal was evident in cells expressing each of the traptamers at this time, indicating that the PsV DNA persisted in the cells, but the EdU signal was not nuclear. Thus, none of the traptamers blocked HPV internalization or caused rapid degradation of viral DNA, and all blocked nuclear entry. Costaining for EdU and cellular marker proteins in cells expressing the traptamers was consistent with the localization of HPV L1 and L2 described in the preceding paragraphs (fig. S4, B to E). These virus localization experiments show that each of the four traptamers blocks HPV infection at a different step. JX1 directs HPV to the lysosome, JX2 causes it to accumulate in the endosome, JX4 causes it to accumulate in the TGN, and JX3 did not cause accumulation in any of these compartments.
Delineating the retrograde pathway of HPV intracellular trafficking
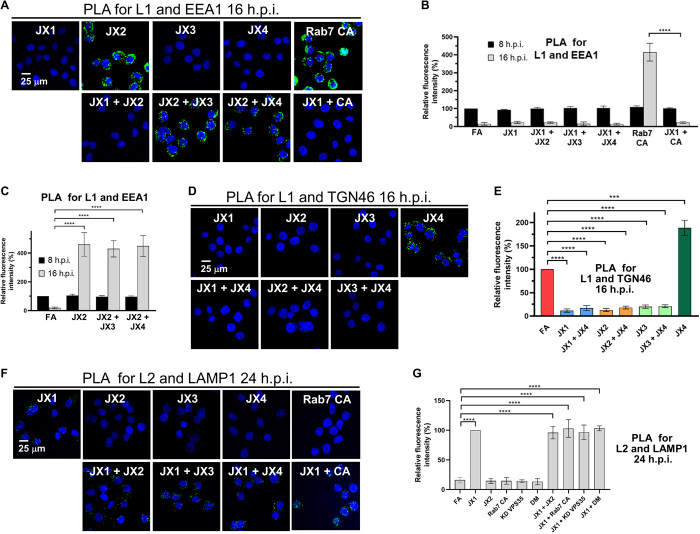
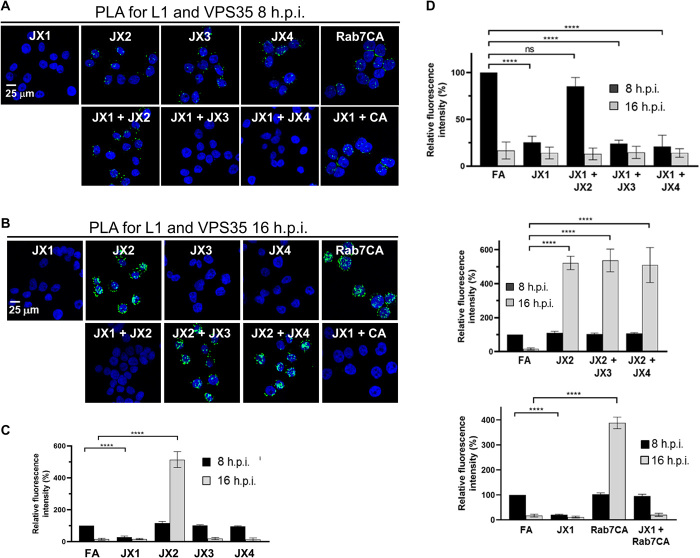
Because the traptamers block different steps in HPV intracellular trafficking, we assayed the effect of traptamers expressed in pairwise combinations. These types of studies are similar to formal genetic epistasis analyses of mutants and can reveal information not ascertainable by studying individual factors. Because only JX2 caused endosome accumulation, we first conducted PLA for L1 and EEA1 at 16 h.p.i. in cells coexpressing JX2 with each of the other traptamers (Fig. 4, A to C, and fig. S5, A and B). In cells coexpressing JX1 and JX2, the lack of endosome accumulation caused by JX1 masked the endosome accumulation phenotype caused by JX2. In contrast, HPV accumulated in the endosome in cells coexpressing JX2 and JX3 or JX2 and JX4. These results imply that JX1 acts upstream of JX2 and that JX2 acts upstream of JX3 and JX4. We previously showed that JX2 affected HPV entry by causing accumulation of Rab7-GTP (26). Accordingly, JX2 and constitutively active Rab7 (Rab7CA), which also increases Rab7-GTP, should display the same epistatic relationships. Consistent with this prediction, there was endosome accumulation in cells expressing Rab7CA alone but not in cells coexpressing JX1 and Rab7CA (Fig. 4, A and B).
Fig. 4. Traptamers inhibit HPV trafficking.
(A) HeLa-tTA cells expressing the indicated protein(s) were infected at MOI of 150 with HPV16-HcRed PsV. At 16 h.p.i., PLA was performed with anti-HPV L1 and anti-EEA1 antibodies. PLA signal is green; nuclei are blue. Several images in the top row are also in Fig. 3. Additional controls and primary data are shown in figs. S3A and S5. (B and C) At least 100 cells in each condition were imaged and processed as described in Fig. 3. Quantitation of images as in (A) shows the mean PLA signal per cell and SD at 8 and 16 h.p.i., normalized to the signal of infected FA cells at 8 h.p.i. (D) HeLa-tTA cells were infected as in (A). At 16 h.p.i., PLA was performed with anti-HPV L1 and anti-TGN46 antibodies. Images were processed as in (A). The image showing JX4 cells is also in Fig. 3. Additional controls and primary data are shown in figs. S3B and S5C. (E) Quantitation of images as in (D) shows the mean PLA signal per cell and SD, normalized to the signal of infected FA cells. (F) HeLa-tTA cells were infected as in (A), except the PsV contained HA-tagged L2. At 24 h.p.i., PLA was performed with anti-HA and anti-LAMP1 antibodies. Images were processed as in (A). Several images in the top row are also in Fig. 3. Additional controls and primary data are shown in figs. S3D and S5D. (G) Quantitation of images as in (F) shows the mean PLA signal per cell and SD at 24 h.p.i., normalized to the signal of infected JX1 cells. ***P < 0.001 and ****P < 0.0001.
We also performed PLA for L1 and TGN46 at 16 h.p.i. in cells coexpressing JX4 with each of the other traptamers (Fig. 4, D and E, and fig. S5C). In all three combinations, the TGN accumulation phenotype of JX4 was masked. In addition to providing additional evidence that JX1 and JX2 act upstream of JX4, this result implies that JX3 acts upstream of JX4.
PLA for HA-tagged L2 and LAMP1 at 24 h.p.i. showed lysosomal HPV in cells coexpressing JX1 and each of the other traptamers or Rab7CA (Fig. 4, F and G, and fig. S5D), providing further evidence that JX1 acts upstream of the other traptamers and Rab7-GTP. Similarly, viral DNA colocalized with LAMP1 in cells coexpressing JX1 and JX2 at 24 h.p.i. (Fig. 5A and fig. S4E). In addition, L2 was directed to the lysosome when retromer is knocked down in cells expressing JX1 or when cells expressing JX1 are infected with PsV containing an L2 mutant [designated the double mutant (DM)] that cannot bind retromer because of mutations in the retromer binding site (Fig. 4G and fig. S5D) (11), showing that L2-retromer binding is not required for JX1-induced lysosomal localization.
Fig. 5. HPV L2 ubiquitination correlates with virus trafficking to lysosome.
(A) HeLa-tTA cells expressing FA, JX1, JX2, or JX1 plus JX2 were infected at MOI of 150 with HPV16-HcRed PsV containing EdU-substituted DNA. At 24 h.p.i., cells were treated with click chemistry to label EdU-substituted DNA and permeabilized and stained with an antibody recognizing LAMP1. Graph shows Pearson’s correlation coefficient for costaining between EdU and LAMP1 in images as in fig. S4E, as described in Fig. 2C. ns, not significant; ****P < 0.0001. (B) HeLa-tTA cells expressing FA, JX1, JX2, or JX1 plus JX2 were infected at MOI of 50 with HPV16-HcRed PsV containing HA-tagged L2. At 8 h.p.i., extracts were prepared, immunoprecipitated (IP) with anti-HA antibody, and blotted with an antibody recognizing ubiquitin (Ubi). Bottom panels show immunoblotting for L2 and actin as a loading control in the absence of immunoprecipitation. (C) HeLa-tTA cells expressing FA or JX1 were mock-infected or infected as in (B). At 8 h.p.i., extracts were prepared, subjected to SDS-PAGE in the absence of immunoprecipitation, and immunoblotted with an antibody recognizing ubiquitin.
Ubiquitination of the cytoplasmic domain of endosomal TM proteins can target them to the lysosome (44, 45). To test whether L2 or associated proteins was ubiquitinated during HPV entry, we infected HeLa cells with HPV16 PsV containing HA-tagged L2, immunoprecipitated L2 with anti-HA antibody, and blotted with anti-ubiquitin antibody. As shown in Fig. 5B, at 8 h.p.i. in infected control cells and cells expressing JX2, there was no detectable ubiquitination. In contrast, there was prominent smear of ubiquitinated proteins in cells expressing JX1 alone or coexpressing JX1 and JX2. The two conditions that increased ubiquitination also induced lysosomal localization of L2 and PsV DNA (Figs. 4, F and G, and 5A, and fig. S4E), suggesting that ubiquitination of L2 or associated proteins directs incoming virus to the lysosome. JX1 or HPV infection did not cause a global increase in ubiquitination (Fig. 5C).
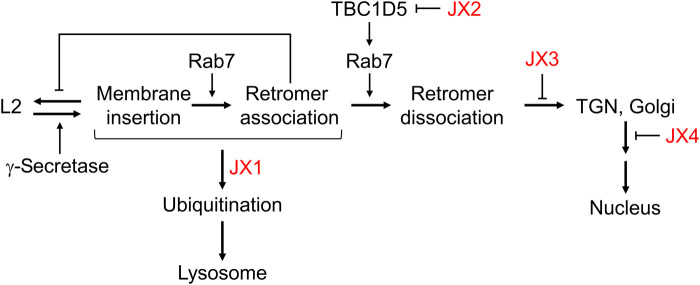
These experiments delineate the sequential passage of HPV through a retrograde pathway in which each step perturbed by a traptamer is dependent on the prior upstream step (Fig. 6). In the presence of JX1, L2 is diverted to the lysosome and does not accumulate in retrograde compartments. In the absence of JX1, the virus travels via the retrograde transport pathway in steps controlled by JX2, JX3, and JX4 in that order, consistent with the localization of the traptamers themselves. Infection is aborted by interruption of any of these steps, showing their essential nature for HPV entry and establishing that HPV cannot efficiently enter cells via an auxiliary pathway when any of these steps is inhibited.
Fig. 6. Model of HPV retrograde trafficking.
The model shows the sites of action of the four traptamers (red), the role of γ-secretase in promoting insertion of L2 into the endosome membrane, the reversible nature of L2 membrane insertion, and the stabilization of L2 membrane insertion by retromer-L2 binding.
JX1 inhibits HPV-retromer association and L2 membrane insertion and is antagonized by JX2
In addition to examining intracellular trafficking of viral proteins and DNA, we also examined the interaction of HPV and retromer and the insertion of L2 into the membrane. We previously reported that JX2 prevents dissociation of HPV from retromer (26). To test the effect of the other traptamers, we conducted PLA for L1 and retromer subunit VPS35 at 8 and 16 h.p.i. in cells expressing each of the traptamers [Fig. 7, A and B (top rows of images) and C, and fig. S6, A and B]. As reported earlier (11), L1-VPS35 PLA signal was readily apparent in infected FA cells at 8 h.p.i. but not at 16 h.p.i., reflecting HPV-retromer association at the earlier time point, followed by retromer dissociation. At 8 h.p.i., there is little or no detectable L1-VPS35 PLA signal in cells expressing JX1 but clear PLA signal in cells expressing JX2, JX3, or JX4, suggesting that JX1 prevents HPV-retromer association. At 16 h.p.i., there was a strong L1-VPS35 PLA signal in cells expressing JX2 or Rab7CA (Fig. 7B and fig. S6B), as reported earlier (26), but little or no L1-VPS35 PLA signal in cells expressing any of the other traptamers (fig. S6, A and B).
Fig. 7. Traptamers regulate HPV-retromer interaction.
(A) HeLa-tTA cells expressing the indicated protein(s) were infected at MOI of 150 with HPV16-HcRed PsV. At 8 h.p.i., PLA was performed with antibodies recognizing HPV L1 and VPS35. PLA signal is green, and nuclei were stained blue with DAPI. Representative primary data including additional controls are shown in fig. S6. (B) As in (A), except PLA was performed at 16 h.p.i. (C) At least 100 cells expressing each traptamer as in (A) and (B) in three independent experiments were imaged and processed as described in Fig. 3. The graph shows the mean L1-VPS35 PLA signal per cell and SD at 8 or 16 h.p.i., normalized to the PLA signal of FA cells at 8 h.p.i., set at 100%. (D) Graphs show L1-VPS35 PLA signal as in (C) for cells expressing JX1 and JX2, JX3, or JX4 (top); JX2 and JX3 or JX4 (middle); or JX1, Rab7CA, or JX1 and Rab7CA (bottom). ****P < 0.0001.
Because JX1 and JX2 affect the retromer-HPV interaction, we used immunofluorescence to assess their colocalization with the late endosome marker Rab7, which recruits retromer to the endosome membrane. Neither traptamer colocalized with Rab7 in uninfected cells, but HPV infection caused significant colocalization of JX1 and JX2 with Rab7 at 12 h.p.i. (Fig. 2E and fig. S2C). Thus, HPV recruits both JX1 and JX2 to the late endosome.
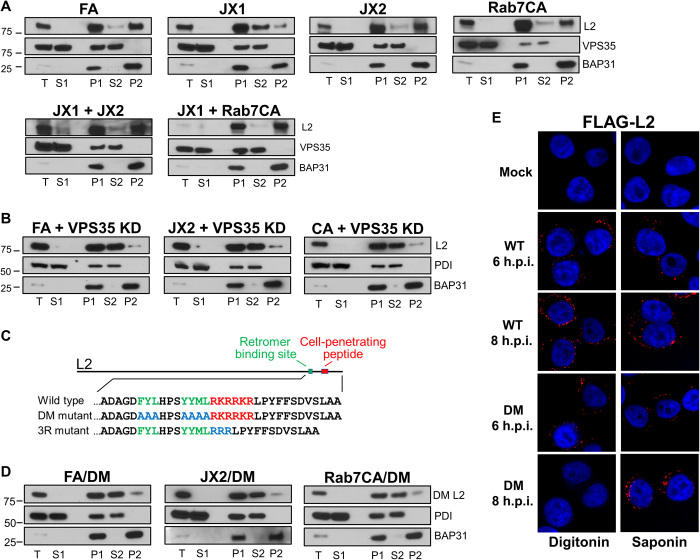
To determine if JX1 or JX2 affected whether L2 is an integral TM protein, extracts of cells expressing FA, JX1, JX2, or Rab7CA were prepared at 12 h.p.i. and subjected to carbonate extraction and Western blotting for FLAG-tagged L2. In control FA cells and cells expressing JX2 or Rab7CA, most L2 was in the P2 fraction at 12 h.p.i., demonstrating its integral membrane nature at this time (Fig. 8A). In contrast, in cells expressing JX1, most L2 was in the S2 fraction, showing that JX1 caused a membrane insertion defect at 12 h.p.i. The inhibition of stable membrane insertion and HPV-retromer association by JX1 is consistent with it acting earlier in the entry process than the other traptamers.
Fig. 8. Retromer is required for HPV L2 membrane insertion.
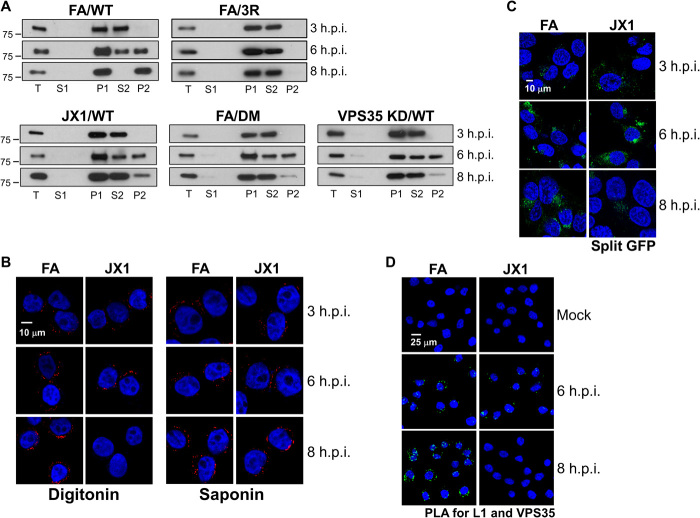
(A) HeLa-tTA cells expressing the indicated protein(s) were infected at MOI of 50 with HPV16-HcRed PsV containing FLAG-tagged L2. At 12 h.p.i., the cells were lysed, fractionated, and extracted with sodium carbonate as outlined in Fig. 2A. Fractions were subjected to SDS-PAGE and immunoblotted for FLAG (recognizing L2), VPS35, and BAP31. Lanes are labeled as in Fig. 2B. (B) HeLa-tTA cells expressing FA, JX2, or Rab7CA were transfected with small interfering RNA (siRNA) targeting VPS35. Forty-eight hours after transfection, cells were infected and processed as in (A), except PDI (a luminal protein) was used as a fractionation marker instead of VPS35. KD, knockdown. (C) Schematic diagram of the HPV16 L2 protein. Top shows L2 protein and positions of the retromer binding sites and the CPP. Bottom shows the wild-type and mutant C-terminal L2 sequence with the retromer binding site shown in green and the CPP shown in red. Mutant sequences are shown in blue. (D) Cells expressing FA, JX2, or Rab7CA were infected with FLAG-tagged HPV16 DM PsV containing the number of encapsidated HcRed reporter plasmids that corresponds to wild-type PsV at MOI of 50. At 12 h.p.i., cells were processed as in (B). (E) HeLa S3 cells were mock-infected or infected at MOI of 150 with HPV16-HcRed PsV containing FLAG-tagged wild-type or DM L2. At 6 and 8 h.p.i., cells were permeabilized with digitonin (2 μg/ml) (left) or 1% saponin (right) and then stained with anti-FLAG antibody (red). Nuclei are stained blue. Additional primary data including controls and quantitation are shown in fig. S7 (A and B).
We next examined retromer-HPV association and L2 membrane insertion in cells expressing pairs of traptamers. Because JX2 blocks retromer-HPV dissociation at 16 h.p.i., we first conducted PLA for VPS35 and L1 at this time point. As shown in Fig. 7 (B and D) and fig. S6 (B to D), JX1 masked the HPV-retromer dissociation defect phenotype caused by JX2 or Rab7CA, and the dissociation defect caused by JX2 persisted despite coexpression of JX3 or JX4. We also conducted PLA at 8 h.p.i., a time when JX1 inhibited retromer-HPV association (Fig. 7, A and D, and fig. S6, B to D). There was little VPS35-L1 PLA signal in cells coexpressing JX1 and either JX3 or JX4. Unexpectedly, at 8 h.p.i. in cells coexpressing JX1 and either JX2 or Rab7CA, we observed substantial VPS35-L1 PLA signal, the phenotype caused by JX2 expression, not the phenotype caused by JX1. We also used carbonate extraction to assess membrane insertion of L2. In cells coexpressing JX1 and either JX2 or Rab7CA, L2 was largely in the P2 fraction at 12 h.p.i., the phenotype displayed by cells expressing JX2 or Rab7CA alone, not the JX1 phenotype (Fig. 8A). These show that JX2 and Rab7CA masked the JX1 phenotypes of reduced retromer association at 8 h.p.i. and reduced L2 membrane insertion at 12 h.p.i. Thus, JX1 and JX2 engage in complex genetic interactions. JX1 is upstream of JX2 in terms of virus trafficking and is dominant in terms of inducing ubiquitination, but JX2 is dominant in terms of retromer association at 8 h.p.i. but not at 16 h.p.i. and in terms of membrane insertion at 12 h.p.i.
Retromer is required for stable membrane insertion
The experiments in the previous section showed that L2 stably inserts into membranes and associates with retromer in cells coexpressing JX1 and JX2, in contrast to cells expressing JX1 alone, which inhibits these activities. Because JX1 prevents membrane insertion and JX2 acts by regulating retromer, this result raised the intriguing possibility that retromer may affect the behavior of L2 even when retromer is nominally on the opposite side of the endosome membrane from L2 before insertion. To test whether retromer plays a role in inserting L2 into the membrane, we conducted carbonate extraction experiments in retromer knockdown cells. Knockdown of VPS35 reduced the partitioning of L2 into the integral membrane pellet at 12 h.p.i., whereas L2 is primarily an integral membrane protein in control FA cells (Fig. 8, A and B). This unexpected result indicates that retromer is essential for membrane insertion of L2. To test whether this membrane insertion defect is due to the lack of retromer-L2 binding or to an indirect, global effect of retromer knockdown, we also assayed membrane insertion in cells infected with PsV containing the L2 DM mutant defective for retromer binding (Fig. 8C). As shown in Fig. 8D, the mutant L2 protein is also depleted from the integral membrane fraction in infected cells expressing FA. These results show that retromer-L2 binding is required for insertion of the L2 protein into the endosome membrane. Moreover, little membrane insertion occurred if VPS35 knockdown or the L2 mutant was combined with expression of JX2 or Rab7CA (Fig. 8, B and D), implying that retromer plays a more direct role in L2 membrane insertion than does JX1, because the insertion defect caused by JX1 can be bypassed by JX2 or Rab7CA (Fig. 8A).
We used antibody staining to test whether retromer was also required for L2 protrusion into the cytoplasm. When the plasma membrane was selectively permeabilized with digitonin (2 μg/ml) at 8 h.p.i., a condition that does not allow antibody access to the endosome lumen (20), the anti-FLAG antibody did not stain the FLAG tag at the C terminus of L2 in cells knocked down for VPS35 or infected with a PsV containing the L2 DM, whereas L2 was stained in control-infected cells (Fig. 8E and fig. S7, A and B). L1 displayed minimal staining under these digitonin conditions, consistent with the impaired ability of the antibody to access luminal contents, whereas both L1 and L2 were stained if the cells were permeabilized with saponin, which allows antibody access to luminal proteins. These results show that retromer-L2 binding is required for insertion of L2 into the membrane and exposure of the L2 C terminus in the cytoplasm.
Transient membrane insertion is stabilized by retromer
How can insertion of L2 from the luminal side of the membrane be affected by retromer, which is cytoplasmic? We hypothesized that insertion of L2 is a dynamic process that occurs transiently even without L2-retromer binding, providing a window for retromer to bind L2 and trap it in the inserted state. To test this, we performed carbonate extraction at times before 12 h.p.i. As shown in Fig. 9A, in FA cells infected with wild-type HPV16 PsV, there was little L2 insertion at 3 h.p.i., L2 partitioned equally between P2 and S2 fractions at 6 h.p.i., and L2 was exclusively in P2 by 8 h.p.i. In contrast, in cells infected with a PsV containing an L2 mutant with a defective CPP (the 3R mutant; Fig. 8C) (12), L2 was not inserted at any time (Fig. 9A). Thus, CPP activity is absolutely required for L2 insertion into the membrane. In addition, L1 was not an integral membrane protein at any time examined (fig. S7C), suggesting that the carbonate resistance of L2 is an intrinsic property of the L2 protein and not a consequence of the behavior of the intact capsid. Notably, if retromer was knocked down or if the cells were infected with the L2-retromer binding mutant, then L2 was inserted into membranes at 6 h.p.i. but not at 8 h.p.i. (Fig. 9A). Similarly, FLAG staining was observed at 6 h.p.i. in digitonin-permeabilized cells even if L2 binding to retromer was disrupted by VPS35 knockdown or by mutations in the retromer binding site (Fig. 8E and fig. S7A). Together, these results show that in the absence of L2-retromer binding, L2 inserts transiently into the membrane and protrudes into the cytoplasm, but CPP-mediated membrane insertion and protrusion of L2 are reversible unless L2-retromer binding stabilizes the inserted state.
Fig. 9. L2 transiently inserts into the endosome membrane.
(A) HeLa-tTA cells expressing FA were transfected with siRNA targeting VPS35. Forty-eight hours after transfection, knockdown cells or cells expressing FA or JX1 were infected at MOI of 50 with FLAG-tagged HPV16-HcRed PsV. In addition, cells expressing FA were infected with FLAG-tagged HPV16 DM or HA-tagged HPV16 3R mutant PsV containing the same number of encapsidated HcRed reporter plasmids as in the wild-type (WT) infections. At 3, 6, or 8 h.p.i., cells were lysed, fractionated, and extracted with sodium carbonate as described in Fig. 2A. Fractions were subjected to SDS-PAGE and immunoblotted with anti-FLAG or anti-HA antibody, as appropriate. Proper fractionation was confirmed by immunoblotting for marker proteins. (B) HeLa-tTA cells expressing FA or JX1 were infected at MOI of 150 with HPV16-HcRed PsV containing HA-tagged L2. At 3, 6, and 8 h.p.i., cells were permeabilized with digitonin (2 μg/ml) (left) or 1% saponin (right) and then immunostained with anti-HA antibody (red). Nuclei are blue. Additional primary data including controls and quantitation are shown in fig. S8 (A and B). (C) HaCaT–GFP1-10NES cells expressing FA or JX1 were infected at MOI of 1200 with HPV16 PsV containing GFP11 fused to the C terminus of L2. Cells were imaged by confocal microscopy at 3, 6, and 8 h.p.i. Reconstituted GFP is green; nuclei are blue. Additional controls and quantitation are shown in fig. S8 (C and D). (D) HeLa-tTA cells expressing FA or JX1 were mock-infected or infected at MOI of 150 with HPV16-HcRed PsV. At 6 and 8 h.p.i., PLA was performed with anti-HPV L1 and anti-VPS35 antibodies. PLA signal is green; nuclei are stained blue with DAPI. Quantitation is shown in fig. S8E.
We also examined the kinetics of L2 insertion and protrusion in cells expressing JX1. L2 displayed little integral membrane nature at 8 h.p.i. in cells expressing JX1 but partitioned equally in P2 and S2 at 6 h.p.i. (Fig. 9A), indicating that L2 membrane insertion is also transient in JX1 cells. We performed three additional experiments to confirm transient membrane insertion in these cells. First, in digitonin-permeabilized infected cells, antibody staining for the HA tag at the C terminus of L2 was evident at 6 but not 8 h.p.i. in cells expressing JX1 but at both time points in control cells, implying that the L2 C terminus is cytoplasmic at the earlier but not the later time point in JX1 cells (Fig. 9B and fig. S8, A and B). In contrast, L1 and L2 staining progressively increased with time in saponin-permeabilized JX1 cells. Second, we performed a split GFP assay in HaCaT cells in which fluorescence is reconstituted only if the C terminus of L2 fused to GFP11 associates with GFP1-10 in the cytoplasm. In this experiment, fluorescence peaked at 6 h.p.i. in cells expressing JX1 and then declined, whereas it progressively increased in cells expressing FA (Fig. 9C and fig. S8, C and D). Last, PLA showed HPV-VPS35 association at 6 h.p.i in cells expressing JX1 but not at 8 h.p.i., whereas the PLA signal increased at 8 h.p.i. in control FA cells (Fig. 9D and fig. S8E). These experiments show that when L2 carries a CPP, L2 transiently inserts into the membrane and protrudes into the cytoplasm, but stable insertion and protrusion require L2-retromer binding and the absence of JX1.
DISCUSSION
Traptamer screening generated a set of tools to dissect HPV entry. By analyzing the localization of viral proteins and DNA in cells expressing individual traptamers or pairs of traptamers, we placed viral trafficking steps during HPV entry in a defined order and implicated ubiquitination as a signal that diverts the incoming virus from its productive trafficking pathway to the lysosome. Overall, these results define retrograde trafficking steps essential for HPV entry, identify inhibitors of each of these steps, and show that there are unlikely to be major alternative HPV entry pathways.
While analyzing the phenotype of cells expressing pairs of traptamers, we found that insertion of L2 into membranes is initially transient in the absence of L2 binding to retromer and that insertion is stabilized by retromer binding. Thus, CPP-mediated transfer of the C terminus of L2 through the endosome membrane into the cytoplasm is reversible, and retromer action is not restricted to its canonical role of binding and sorting proteins that are already stably inserted in membranes. Binding of retromer to L2 may directly anchor L2 in the cytoplasm, although more complex mechanisms related to the sorting activity of retromer are also possible. The ability of retromer to stabilize membrane insertion was not recognized previously because all other known retromer cargos are stably inserted into membranes before encountering retromer.
The presence of HPV inside retrograde transport vesicles during entry protects it from cytoplasmic immune sensors to support successful infection (8). HPV evolved a unique strategy to enter the retrograde pathway, namely, to insert a segment of a viral capsid protein across the endosome membrane into the cytoplasm where it can bind retromer and other sorting factors. To transit the membrane, the virus uses a short segment of the L2 protein that acts as a CPP, as well as retromer expression and the nearby retromer binding site on L2 to stabilize L2 in the membrane. Thus, HPV not only exploits the protein-sorting activity of retromer but also relies on retromer to present L2 in a suitable form to use this activity. This is an economical solution to the presence of a membrane barrier between the incoming virus and essential entry factors. In this way, HPV expends minimal genetic resources to engage complex cellular machinery to travel to the nucleus. By inserting transiently into the membrane in the absence of retromer binding, the exposure of viral proteins to the cytoplasm is minimized until conditions are conducive to proper trafficking, thereby reducing the likelihood of virus elimination by classic cytoplasmic immune sensors or by diversion of the virus to the lysosome. The existence of multiple copies of L2 in the capsid may provide numerous opportunities for the capsid to engage retromer and enter the retrograde transport pathway.
On the basis of our results, we propose a model for HPV trafficking during entry (Fig. 6). In cells lacking traptamers, the C terminus of L2 inserts into the endosome membrane soon after endocytosis and samples the cytoplasm for retromer. Insertion is transient unless it is stabilized by retromer binding. If L2 binds retromer in the presence of Rab7 cycling, then HPV dissociates from retromer and is sorted into the retrograde transport pathway for normal trafficking. In contrast, if HPV does not bind retromer or if HPV binds but does not dissociate from retromer, as is the case when Rab7 cycling is blocked, then trafficking is arrested, and HPV accumulates in the endosome.
This pathway is perturbed by the traptamers. By inhibiting TBC1D5, JX2 prevents Rab7 cycling and retromer dissociation from L2, causing L2 and viral DNA to accumulate in the endosome (26). JX3 acts downstream of JX2, but the fate of the virus in cells expressing JX3 is not known. Analysis of cells expressing pairs of traptamers indicates that JX3 acts after virus departure from the endosome but before its exit from the TGN. The increased localization of JX3 to the TGN and Golgi apparatus caused by virus infection implies that JX3’s site of action is at the TGN, but we have not detected L2 in this compartment, possibly because JX3 interferes with docking of HPV-containing vesicles with the TGN. Localization of HPV components in cells expressing JX3 and identification of proteins bound by JX3 may elucidate unknown aspects of the endosome to TGN transition and the fate of cargo when this transition in blocked. JX4 acts downstream of JX3, causing HPV to accumulate in the TGN and Golgi apparatus, a phenotype similar to that of an L2 mutant reported by the Sapp laboratory (46). Identification of cellular proteins bound by JX4 and analysis of the trafficking of wild-type virus in cells expressing JX4 and of the mutant virus in cells lacking a traptamer may provide new insights into Golgi exit during HPV trafficking.
In cells expressing JX1 alone, L2 transiently inserts into the endosome membrane, but by 8 h.p.i., L2 no longer is resistant to carbonate extraction and does not protrude into the cytoplasm or associate with retromer, whereas in cells coexpressing JX1 and JX2, L2 stably inserts into the membrane and associates with retromer in the cytoplasm. In both situations, JX1 diverts the virus to the lysosome by 24 h.p.i. Thus, stable membrane insertion and retromer binding do not prevent JX1-mediated diversion to the lysosome. These findings may be explained by a lag before diversion, during which time JX1 causes progressive modification of L2, regardless of retromer binding or expression of the other traptamers. Once a certain threshold of L2 modification occurs, it is sensed by the molecular machinery that diverts the virus to the lysosome. Our data suggest that L2 ubiquitination may play a critical role in HPV trafficking. When trafficking of cellular retromer cargos is disrupted, they frequently undergo ubiquitination, which targets them to the lysosome where they are degraded (25). After the C terminus of L2 protrudes into the cytoplasm during normal entry, we propose that it is ubiquitinated by cytoplasmic ubiquitinases, possibly at the C-terminal lysines in the CPP. In the absence of JX1, L2 ubiquitination is opposed by deubiquitination, and ubiquitin does not accumulate on L2 even if HPV is arrested in the endosome, whereas JX1 shifts the balance of ubiquitination and deubiquitination, allowing ubiquitin to accumulate on L2, which eventually diverts HPV to the lysosome for degradation. Further experiments are required to determine the mechanism of L2 ubiquitination and its role in HPV trafficking and to determine whether lysosomal HPV diversion involves known cellular mechanisms or unknown mechanisms consistent with the unconventional nature of L2 as an inducible TM protein. For example, do proteins and mechanisms that mediate ER-associated degradation, a process that extracts cargo through membranes for ubiquitination and proteasome degradation, help remove HPV from the endosome or are the membrane-associated RING-CH-type finger E3 ubiquitin ligases, which are themselves TM proteins (47), involved.
Our findings highlight the value of studying a collection of inhibitory proteins, which allowed us to arrange entry steps in order and revealed phenotypes caused by pairs of traptamer that were not readily predicable from the phenotypes of the individual traptamers. Because the first four traptamers we isolated inhibit entry at different steps, it is likely that we can isolate additional inhibitory traptamers with other sites of action. It should be possible to design other selection strategies to isolate traptamers that perturb a wide variety of cellular processes, including entry of other viruses (37). Traptamer screening may allow the interrogation of proteins required for cell viability, which are difficult to identify by knockdown/knockout screening, because a traptamer might modulate an activity of a cellular protein required for the studied process without disrupting other essential activities of the protein. In addition, functional genomic screens for the same process often identify nonoverlapping hits, so it seems likely that traptamer screening, which is based on a radically different protein modulation strategy, will identify an even more diverse set of proteins.
Last, we note that a fundamental problem in biology is the mechanism of insertion of proteins into membranes. Most proteins are inserted into the endoplasmic reticulum membrane cotranslationally, although other mechanisms exist, e.g., insertion of fusion proteins of enveloped viruses into host cell membranes during virus entry. Our results suggest the existence of a previously unrecognized mechanism of protein membrane insertion involving CPPs that is reversible unless it is stabilized by binding proteins on the distal side of the membrane. Other proteins may use this mechanism to enter cells, move from one intracellular compartment to another, or convert from a soluble existence into a TM one.
MATERIALS AND METHODS
Producing HPV PsV
HPV PsVs were produced by using polyethyleneimine to cotransfect 293TT cells with pCAG-HcRed (Addgene #11152), pCINeo-GFP [obtained from C. Buck, National Institutes of Health (NIH)], or pCAG-BE2-IRES-GFP (26), together with wild-type p16sheLL, p5sheLL, p18sheLL (48), p16sheLL.L2F or p16sheLL-L2HA (7), p16sheLL-DM expressing FLAG- or HA-tagged L2 lacking retromer binding sites (11), or p16sheLL-3R containing a mutation that inactivates the L2 CPP (12). Packaged PsVs were purified by density gradient centrifugation in OptiPrep as described (48). The quality of the PsV stocks was assessed by Coomassie brilliant blue staining for L1 and L2 levels after SDS–polyacrylamide gel electrophoresis (PAGE). Wild-type PsV stocks were titered by flow cytometry for fluorescence 2 days after infection of HeLa S3 cells. For experiments comparing wild-type and noninfectious HPV16 PsVs, encapsidated reporter plasmids were quantified by quantitative polymerase chain reaction (PCR) as described (12), and wild-type and mutant PsV stocks containing the same number of encapsidated reporter plasmids were used to infect cells. Reporter plasmids in PsV stocks used in the same experiment were quantified in parallel.
Selection and recovery of traptamers that block HPV16-BE2 infection
The EHFA retroviral library was generated by using Lipofectamine 2000 (Invitrogen) to cotransfect 293T cells with EHFA DNA in MSCVpuro, pCL-Eco, and pVSV-G plasmids (26). To generate stable cells expressing EHFA traptamer library, 1 million HeLa S3 cells in 10-cm2 plates were infected with EHFA retrovirus at multiplicity of infection (MOI) of ~0.2 followed with puromycin selection. To initiate the screen, 1 million cells stably transduced with EHFA were seeded per plate in 10 15-cm2 plates. After 16 hours, the cells were infected with HPV16-BE2 PsV at MOI of 20. HPV16-BE2 is an HPV16 PsV expressing bovine papillomavirus E2 (26). One day later, the HPV16-BE2 infection was repeated, and the growth medium was changed every 2 days. Fourteen days after the first infection, individual colonies of proliferating cells were isolated using cloning cylinders and were expanded. Genomic DNA was isolated from the expanded cells by using the DNeasy Blood and Tissue Kit (Qiagen).
Retroviral inserts in genomic DNA from the expanded cells were amplified by using short primers specific to fixed sequences in the library, flanking the randomized segment (forward short and reverse short in table S2) [PCR protocol as in (26)]. PCR products were purified, digested with Bam HI and Eco RI, ligated into pMSCVpuro-FA vector to generate full-length traptamers, and transformed into Escherichia coli. FA consists of the FLAG tag and APEX2 segment without a randomized hydrophobic sequence (26).
Inducible system for traptamer expression
To construct pT-FA, pT-JX1, pT-JX2, pT-JX3, and pT-JX4, the corresponding fragments were amplified from pMSCVpuro constructs or directly from cellular DNA and cloned into the pT-WOB variant of pRetroX-Tightpuro (pTpuro, Clontech #632104) (26). pRetroX-Tet-Off Advanced plasmid (Tet-Off) (Clontech #632105) was used to produce retrovirus expressing the tetracycline (Tet)–controlled transactivator protein tTA. Polyclonal HeLa-tTA or HaCaT-tTA cells were generated by infection with tTA retrovirus followed by neomycin selection. Cells expressing FA, JX1, JX2, JX3, JX4, or Rab7CA were produced by transduction in HeLa-tTA or HaCaT-tTA cells with the corresponding pT retrovirus followed by puromycin selection. To construct cells expressing pairs of proteins, one gene was selected with puromycin and the second was cloned into pRetroX-Tighthygro and selected with hygromycin. Unless specified otherwise, all experiments were performed in the absence of doxycycline to achieve maximal traptamer expression.
HPV and SV40 infectivity
To measure the effect of traptamers on HPV infection, 1 × 105 HeLa-tTA or HaCaT-tTA cells expressing FA or a traptamer in 12-well plates were incubated with wild-type HPV16-GFP, HPV5 HcRed, or HPV18 HcRed PsV at infectious MOI of approximately two. Fivefold more virus was used to attain a comparable level of infectivity in HaCaT cells. Flow cytometry was performed at 48 h.p.i. to measure GFP or HcRed fluorescence to determine reporter gene expression. To show that HPV inhibition was due to traptamer expression, clonal HeLa-tTA cells expressing FA, JX1, JX2, JX3, or JX4 from pT were treated with various concentrations of doxycycline for 2 days to repress traptamer expression or left untreated. Traptamer expression was determined by immunoblotting for the FLAG tag on the traptamer. The cells were then infected with HPV16-GFP PsV at MOI of 2 and maintained in the absence or presence of doxycycline. Two days after infection, GFP fluorescence was measured by flow cytometry. To determine whether the inhibitory effects of the traptamers were reversible, 5 × 104 HeLa-tTA cells expressing FA or a traptamer in 12-well plates were infected at MOI of 2 with wild-type HPV16-GFP PsV. At 8 h.p.i., the cells were incubated with doxycycline (10 ng/ml) or left untreated. Three days later, GFP fluorescence was measured by flow cytometry.
SV40 was produced in CV-1 cells. SV40 infection was performed in HeLa-tTA cells expressing FA or a traptamer, as previously described (26, 49). To measure infectivity, cells were permeabilized with cold methanol for 30 min and stained for intracellular SV40 large T antigen with a 1:50 dilution of Sc-147 antibody (Santa Cruz Biotechnology Inc.) at 48 h.p.i., and flow cytometry was performed.
Effect of JX1 on L2 levels
Clonal HeLa-tTA cells (5 × 104) expressing FA or JX1 were seeded in 24-well plates. The cells were then mock-infected or infected at MOI of 20 with HPV16 PsV containing FLAG-tagged L2. At 12 h.p.i., cells were harvested immediately or washed twice with phosphate-buffered saline (PBS), and the culture medium was replaced with fresh medium. At 12 and 24 h.p.i., cells were washed with PBS (pH 10.7), and lysates were prepared in Laemmli sample buffer. After SDS-PAGE, samples were probed with antibodies recognizing FLAG (Sigma-Aldrich, #A8592) and actin.
TM domain prediction and carbonate extraction assay
The full-length sequence of FA and each of the traptamers was analyzed by the TM domain prediction program TMpred (https://embnet.vital-it.ch/software/TMPRED_form.html). For all traptamers, the protein segment with the highest TMS was the C-terminal randomized segment.
To assess the TM behavior of FA and the traptamers, 3.6 × 106 HeLa-tTA cells expressing FA or a traptamer in the absence of HPV infection were plated in one 6-cm2 plate for each condition. After overnight incubation, cells were harvested and resuspended in 500 μl of swelling buffer [10 mM Hepes (pH 7.5), 1.5 mM MgCl2, 10 mM KCl, and 0.5 mM dithiothreitol (DTT)] for 30 min on ice with occasional vortex mixing and then lysed by using a 2-ml Dounce homogenizer as previously reported (26). The cell homogenate was centrifuged at 16,100g for 10 min at 4°C to remove intact cells and nuclei. The resulting supernatant (fraction T) was further centrifuged at 100,000g in a SW55Ti rotor for 30 min at 4°C. The supernatant was concentrated with Amicon ultracentrifugal filters (3 kDa of molecular weight cutoff) and saved as fraction S1. The pellet containing membranes was then gently washed with HN buffer [50 mM HEPES (pH 7.5) and 150 mM NaCl] and centrifuged at 100,000g in a SW55Ti rotor for 10 min at 4°C. The rinsed pellet (P1) was resuspended in 50 μl of buffer containing 10 mM tris (pH 7.5), 5 mM DTT, 150 mM NaCl, and 2 mM MgCl2 for 15 min on ice, mixed with 500 μl of freshly-made solution of 0.1 M Na2CO3 (pH ~11.7) for 30 min on ice with occasional vortex mixing, and then centrifuged at 100,000g in a SW55Ti rotor for 30 min at 4°C. The supernatant (fraction S2) was concentrated with Amicon ultracentrifugal filters (3 kDa of molecular weight cutoff). The pellet was washed in cold HN buffer and recentrifuged as above to generate pellet fraction P2, which was dissolved in 60 μl of Laemmli loading buffer. All fractions were analyzed by immunoblotting with anti-FLAG antibody to detect traptamer and with antibodies recognizing marker proteins to assess fractionization.
To assess membrane insertion of the HPV16 L2 protein, 2.4 × 106 HeLa-tTA cells expressing FA, a traptamer, or Rab7CA separately or in combination were plated in one 6-cm2 plate for each condition, incubated overnight, and infected with wild-type HPV16-HcRed PsV at MOI of 100 or with HPV16 DM or HPV16 3R PsV containing the same number of encapsidated HcRed reporter plasmids, all with a FLAG or HA tag at the C terminus of L2. At various time points, cells were harvested, fractionated, and extracted as descripted above. Fractions were analyzed by immunoblotting with anti-FLAG or anti-HA antibody to detect tagged L2 or antibody recognizing HPV L1 (BD, 554171) and with antibodies to assess fractionization. Where indicated, cells were transfected with VPS35 small interfering RNA (siRNA) before infection.
Immunofluorescence microscopy
HeLa-tTA cells (3.5 × 104) expressing FA or a traptamer were seeded on glass cover slips in 24-well plates, incubated overnight, and mock-infected or infected with wild-type HPV16-HcRed PsV at MOI of 150. At various h.p.i., cells were fixed for 15 min at room temperature with 4% paraformaldehyde, permeabilized with 1% saponin for 30 min, and incubated with 1:400 anti-FLAG mouse antibody (Sigma-Aldrich, F3165), 1:100 anti-EEA1 rabbit antibody [Cell Signaling Technology (CST), 2411], 1:75 anti-Rab7 rabbit antibody (CST, 9367), 1:300 anti-TGN46 rabbit antibody (Abcam, ab50595), or 1:100 anti-LAMP1 rabbit antibody (Abcam, ab24170) at 4°C overnight. Cells were then incubated at room temperature for 1 hour with 1:200 Alexa Fluor–conjugated secondary antibody (Life Technologies). The slides were mounted in mounting solution with 4′,6-diamidino-2-phenylindole (DAPI), and images were captured using a Leica SP5 confocal microscope. To quantify colocalization, Pearson’s correlation coefficient between channels was obtained in ImageJ by using the coloc2 plugin (NIH; https://imagej.net/Coloc_2).
Proximity ligation assay
Clonal HeLa-tTA cells (3.5 × 104) expressing FA, a traptamer, or Rab7CA separately or in combination on glass coverslips in 24-well plates were mock-infected or infected at MOI of 150 with HPV16-HcRed PsV. For PLA to detect L2, HPV16 PsV containing HA-tagged L2 was used. At various h.p.i., cells were fixed in 4% formaldehyde and permeabilized by 1% saponin. Cells were then incubated with pairs of antibodies (one from mouse and one from rabbit) recognizing HPV L1 or HA-L2 and a cellular protein (source and dilutions of antibodies are listed in table S1). PLA probes and reagents (Duolink) were used according to the manufacturer’s directions as described (12, 50). Cells were imaged with a Leica SP5 inverted confocal microscope. Images were processed by Fiji and quantitatively analyzed by BlobFinder software to measure total fluorescence intensity in each sample. The average fluorescence intensity per cell in each sample was normalized to the appropriate control sample as indicated in each experiment.
Staining for EdU-substituted HPV16 PsV DNA
To produce HPV16 PsV containing reporter plasmids substituted with EdU, 100 μM EdU (Thermo Fisher Scientific, C10337) was added to the 293TT cell growth medium at 6 hours after transfection of pSheLL16 L2 and the HcRed reporter plasmid. Cells were harvested and PsV was purified as described above. HeLa cells were mock-infected or infected at MOI of 150 with EdU-labeled HPV16 PsV. At various h.p.i., the cells were fixed in 4% paraformaldehyde and permeabilized with 1% saponin. The cells were incubated with the Click-iT reaction mixture prepared according to the protocol provided using the Alexa Fluor 488 Imaging Kit (Thermo Fisher Scientific, C10425). The cells were then incubated with antibodies recognizing cellular proteins [1:100 anti-EEA1 mouse antibody (BD Biosciences, #610457), 1:300 anti-TGN46 rabbit antibody (Abcam, ab50595), 1:100 anti-LAMP1 rabbit antibody (Abcam, ab24170), or 1:50 anti-PML mouse antibody (Santa Cruz Biotechnology, sc-966)] and the appropriate secondary antibodies provided by the manufacturer. Cells were imaged with a Leica SP8 gated STED 3X super-resolution microscope. The images were deconvolved by Huygens software (Scientific Volume Imaging) and then processed by LAS X software (Leica Microsystems).
Detection of ubiquitination
Clonal HeLa-tTA cells (1.2 × 106) expressing FA, JX1, JX2, or JX1 plus JX2 were seeded in 6-cm2 plates. The cells were then mock-infected or infected with HPV16 PsV containing HA-tagged L2 at MOI of 100. At 8 h.p.i., cells were washed twice with cold PBS and lysed with 400 μl of ice-cold lysis buffer [20 mM Hepes (pH 7.4), 50 mM NaCl, 5 mM MgCl2, 1 mM DTT, and 0.125% Triton X-100] supplemented with 1× Halt protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific). The lysate was clarified by centrifugation at 14,000 rpm at 4°C for 20 min. Immunoprecipitation was performed by adding 1:80 anti-HA mouse antibody (BioLegend, 901513) to the clarified supernatant for 3 hours at 4°C, followed by protein A/G PLUS-agarose beads (Santa Cruz Biotechnology) at 4°C for an additional 2 hours, followed by four washes with cold lysis buffer. Protein complexes were eluted from the beads by heating to 100°C in 2× SDS sample buffer. Eluates and the input were normalized for total extracted protein as determined by bicinchoninic acid protein assay and analyzed by SDS-PAGE followed by immunoblotting with rabbit antibody recognizing ubiquitin (Abcam, ab7780). Input L2 and actin were also detected in the samples in the absence of immunoprecipitation. Total ubiquitin in mock-infected or infected FA or JX1 cells was also detected by immunoblotting in the absence of immunoprecipitation.
Selective permeabilization experiments
We modified an approach used by the Sapp lab to selectively permeabilize the plasma membrane while sparing the endosome membrane (20). HeLa-tTA cells (3.5 × 104) expressing FA or JX1 were seeded on glass cover slips in 24-well plates, incubated overnight, and mock-infected or infected at MOI of 150 with wild-type HPV16-HcRed PsV containing HA-tagged L2. At 3, 6, or 8 h.p.i., cells were treated with PBS at pH 10.7, fixed for 15 min at room temperature with 4% paraformaldehyde, and washed with PBS. Cells were then permeabilized with freshly made 1% (w/v) saponin in PBS for 30 min at room temperature or selectively permeabilized with freshly made digitonin (2 μg/ml) in PBS for 10 min at room temperature, washed with PBS, and blocked with Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum for 1 hour at room temperature. Cells were incubated with 1:100 anti-L1 mouse antibody (BD, 554171) or 1:100 anti-HA rabbit antibody (CST, 3724) at 4°C overnight. Cells were then incubated at room temperature for 1 hour with 1:200 Alexa Fluor–conjugated secondary antibodies (Life Technologies). The slides were mounted in mounting solution with DAPI, and images were captured using a Leica SP5 confocal microscope. In experiments in cells lacking FA or JX1 expression, HeLa S3 cells were infected with HPV16 PsV containing FLAG-tagged wild-type or DM L2. In some cases, cells were transfected with siRNA targeting VPS35 48 hours before infection. In these experiments, L2 was detected with 1:300 rabbit anti-FLAG antibody (CST, 14793S) following digitonin or saponin permeabilization as above.
Split GFP assay for cytoplasmic protrusion of L2
To conduct split GFP assays, we first cloned the GFP1-10NES gene into the doxycycline-regulated pTightpuro vector to generate pT–GFP1-10NES (12, 26). Retroviruses were produced by cotransfecting 293T cells with pT–GFP1-10NES, pCL-Eco, and pVSV-G plasmids. Two days later, the retroviral supernatant was harvested, filtered, and stored at −80°C for later use. Cells stably expressing GFP1-10NES were constructed by infecting HaCaT cells with GFP1-10NES retrovirus and selection with puromycin (1 μg/ml) for 2 days. To minimize potential cell toxicity, polyclonal, puromycin-resistant cell lines were maintained in the presence of doxycycline (50 ng/ml) to repress expression of GFP1-10NES. pThygro-FA and pThygro-JX1 retrovirus produced in 293T cells was used to infect HaCaT–GFP1-10NES cells in the presence of doxycycline, and cells were selected with hygromycin. To assess cytoplasmic protrusion of L2 during PsV infection, cells were cultured in medium without doxycycline for 2 days and then seeded on eight-chambered glass slides with 1.8 × 104 cells in each chamber. After overnight culture, the cells were incubated for 3, 6, or 8 hours at MOI of 1200 with HPV16 containing an HcRed reporter plasmid and wild-type L2 or L2 containing GFP11 inserted at the C terminus of the protein (HPV16-CPP-GFP11) (12). Nuclei were stained with Hoescht 33342, and live cells were analyzed with a Leica SP5 confocal microscope.
Quantitation and statistical analysis
Quantitation of PLA assay
To quantify PLA signals, BlobFinder was used to perform a single-cell analysis and quantify the fluorescence signal intensity per cell for each sample (51). The PLA fluorescence signal intensity of at least 100 cells for each condition in each experiment was averaged. The average intensity for L1-EEA1 and L1-VPS35 samples was normalized to control FA cells infected with wild-type HPV16 PsV at 8 h.p.i., the average intensity for L1-TGN46 samples was normalized to control FA cells infected with wild-type HPV16 PsV at 16 h.p.i., the average intensity for L2-LAMP1 samples was normalized to JX1 cells infected with wild-type HPV16 PsV at 24 h.p.i, and the average intensity for L1-GM130 samples was normalized to control FA cells infected with wild-type HPV16 PsV at 16 h.p.i. The normalized fluorescence intensity of three individual experiments were averaged and plotted as means and SD. Statistical analysis was performed using Prism 7 (GraphPad) software. Comparisons between control and experimental groups were made using one-way analysis of variance (ANOVA) or unpaired two-tailed Student’s t tests. P values < 0.05 were considered to indicate statistical significance.
Quantitation of immunofluorescence and EdU staining
To quantify colocalization, Pearson’s correlation coefficient between the respective channels was obtained in ImageJ by using the coloc2 plugin. A total of at least 60 cells from three independent experiments were analyzed for each condition. Statistical analysis was performed using Prism 7 software. Results are presented as means and SD, with each cell represented by a single symbol. Comparisons between control and experimental groups were made using one-way ANOVA tests. P values < 0.05 were considered to indicate statistical significance.
Quantitation of selective permeabilization and split GFP assays
Quantitation of fluorescence intensity in selective permeabilization assay and reconstituted GFP signal in cells were performed using Fiji software. An outline was drawn around each cell; area, integrate density, and mean fluorescence were measured, along with several adjacent background readings. The corrected total cellular fluorescence (CTCF) = Integrated density − (Area of selected × Mean fluorescence of background readings) was calculated. The CTCF of 60 cells in selective permeabilization assays and 100 cells in split GFP assays for each condition was quantified and averaged. Statistical analysis was performed using GraphPad Prism 7 software. Data are presented as means and SD. Comparisons between control and experimental groups were made using one-way ANOVA or Student’s t tests. P values < 0.05 were considered to indicate statistical significance.
Acknowledgments
We thank A. Edwards for assistance in characterizing the doxycycline-regulated HaCaT GFP1-10NES cell line. We also thank C. Burd for helpful discussions and J. Zulkeski for assistance in preparing this manuscript. Flow cytometry was conducted in the Yale Cancer Center Flow Cytometry Shared Resource. Funding: M.C. was supported by a predoctoral training grant from the NIH and an NSF-GRFP fellowship (T32 AI055403 and DGE-1752134). This work was supported by grants from the NIH to D.D. (R35 CA242462 and R01 AI150897). Author contributions: Conceptualization: J.X. and D.D. Data curation: J.X., P.Z., M.C., and C.V.L. Funding acquisition: D.D. Investigation: J.X., P.Z., M.C., and C.V.L. Methodology: J.X., P.Z., and M.C. Project administration: D.D. Supervision: D.D. Writing (original draft): J.X. and D.D. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/27/eabh4276/DC1
REFERENCES AND NOTES
- 1.Farmer E., Cheng M. A., Hung C. F., Wu T. C., Vaccination strategies for the control and treatment of HPV infection and HPV-associated cancer. Recent Results Cancer Res. 217, 157–195 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DiMaio D., Small size, big impact: How studies of small DNA tumour viruses revolutionized biology. Philos. Trans. R. Soc. Lond. B Biol. Sci. 374, 20180300 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buck C. B., Cheng N., Thompson C. D., Lowy D. R., Steven A. C., Schiller J. T., Trus B. L., Arrangement of L2 within the papillomavirus capsid. J. Virol. 82, 5190–5197 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Day P. M., Thompson C. D., Schowalter R. M., Lowy D. R., Schiller J. T., Identification of a role for the trans-Golgi network in human papillomavirus 16 pseudovirus infection. J. Virol. 87, 3862–3870 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lipovsky A., Popa A., Pimienta G., Wyler M., Bhan A., Kuruvilla L., Guie M. A., Poffenberger A. C., Nelson C. D. S., Atwood W. J., DiMaio D., Genome-wide siRNA screen identifies the retromer as a cellular entry factor for human papillomavirus. Proc. Natl. Acad. Sci. U.S.A. 110, 7452–7457 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie J., Zhang P., Crite M., DiMaio D., Papillomaviruses Go retro. Pathogens 9, 267 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang W., Kazakov T., Popa A., DiMaio D., Vesicular trafficking of incoming human papillomavirus 16 to the Golgi apparatus and endoplasmic reticulum requires γ-secretase activity. MBio 5, e01777-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uhlorn B. L., Jackson R., Li S., Bratton S. M., van Doorslaer K., Campos S. K., Vesicular trafficking permits evasion of cGAS/STING surveillance during initial human papillomavirus infection. PLOS Pathog. 16, e1009028 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burd C., Cullen P. J., Retromer: A master conductor of endosome sorting. Cold Spring Harb. Perspect. Biol. 6, a016774 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seaman M. N. J., The retromer complex: From genesis to revelations. Trends Biochem. Sci., (2021). [DOI] [PubMed] [Google Scholar]
- 11.Popa A., Zhang W., Harrison M. S., Goodner K., Kazakov T., Goodwin E. C., Lipovsky A., Burd C. G., DiMaio D., Direct binding of retromer to human papillomavirus type 16 minor capsid protein L2 mediates endosome exit during viral infection. PLOS Pathog. 11, e1004699 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang P., Monteiro da Silva G., Deatherage C., Burd C., DiMaio D., Cell-penetrating peptide mediates intracellular membrane passage of human papillomavirus L2 protein to trigger retrograde trafficking. Cell 175, 1465–1476.e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pim D., Broniarczyk J., Bergant M., Playford M. P., Banks L., A novel PDZ domain interaction mediates the binding between human papillomavirus 16 L2 and sorting nexin 27 and modulates virion trafficking. J. Virol. 89, 10145–10155 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bergant Marusic M., Ozbun M. A., Campos S. K., Myers M. P., Banks L., Human papillomavirus L2 facilitates viral escape from late endosomes via sorting nexin 17. Traffic 13, 455–467 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inoue T., Zhang P., Zhang W., Goodner-Bingham K., Dupzyk A., Maio D. D., Tsai B., γ-secretase promotes membrane insertion of the human papillomavirus L2 capsid protein during virus infection. J. Cell Biol. 217, 3545–3559 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harwood M. C., Dupzyk A. J., Inoue T., DiMaio D., Tsai B., p120 catenin recruits HPV to γ-secretase to promote virus infection. PLOS Pathog. 16, e1008946 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McNally K. E., Faulkner R., Steinberg F., Gallon M., Ghai R., Pim D., Langton P., Pearson N., Danson C. M., Nägele H., Morris L. L., Singla A., Overlee B. L., Heesom K. J., Sessions R., Banks L., Collins B. M., Berger I., Billadeau D. D., Burstein E., Cullen P. J., Retriever is a multiprotein complex for retromer-independent endosomal cargo recycling. Nat. Cell Biol. 19, 1214–1225 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pim D., Broniarczyk J., Siddiqa A., Massimi P., Banks L., Human papillomavirus 16 L2 recruits both retromer and retriever complexes during retrograde trafficking of the viral genome to the cell nucleus. J. Virol. 95, e02068-20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bronnimann M. P., Chapman J. A., Park C. K., Campos S. K., A transmembrane domain and GxxxG motifs within L2 are essential for papillomavirus infection. J. Virol. 87, 464–473 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DiGiuseppe S., Keiffer T. R., Bienkowska-Haba M., Luszczek W., Guion L. G. M., Müller M., Sapp M., Topography of the human papillomavirus minor capsid protein L2 during vesicular trafficking of infectious entry. J. Virol. 89, 10442–10452 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campos S. K., Subcellular trafficking of the papillomavirus genome during initial infection: The remarkable abilities of minor capsid protein L2. Viruses 9, 370 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harterink M., Port F., Lorenowicz M. J., McGough I. J., Silhankova M., Betist M. C., van Weering J. R. T., van Heesbeen R. G. H. P., Middelkoop T. C., Basler K., Cullen P. J., Korswagen H. C., A SNX3-dependent retromer pathway mediates retrograde transport of the Wnt sorting receptor Wntless and is required for Wnt secretion. Nat. Cell Biol. 13, 914–923 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Temkin P., Lauffer B., Jäger S., Cimermancic P., Krogan N. J., von Zastrow M., SNX27 mediates retromer tubule entry and endosome-to-plasma membrane trafficking of signalling receptors. Nat. Cell Biol. 13, 715–721 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steinberg F., Gallon M., Winfield M., Thomas E. C., Bell A. J., Heesom K. J., Tavaré J. M., Cullen P. J., A global analysis of SNX27-retromer assembly and cargo specificity reveals a function in glucose and metal ion transport. Nat. Cell Biol. 15, 461–471 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cullen P. J., Steinberg F., To degrade or not to degrade: Mechanisms and significance of endocytic recycling. Nat. Rev. Mol. Cell Biol. 19, 679–696 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Xie J., Heim E. N., Crite M., DiMaio D., TBC1D5-catalyzed cycling of Rab7 is required for retromer-mediated human papillomavirus trafficking during virus entry. Cell Rep. 31, 107750 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishii Y., Nakahara T., Kataoka M., Kusumoto-Matsuo R., Mori S., Takeuchi T., Kukimoto I., Identification of TRAPPC8 as a host factor required for human papillomavirus cell entry. PLOS ONE 8, e80297 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Broniarczyk J., Bergant M., Gozdzicka-Jozefiak A., Banks L., Human papillomavirus infection requires the TSG101 component of the ESCRT machinery. Virology 460-461, 83–90 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Broniarczyk J., Pim D., Massimi P., Bergant M., Goździcka-Józefiak A., Crump C., Banks L., The VPS4 component of the ESCRT machinery plays an essential role in HPV infectious entry and capsid disassembly. Sci. Rep. 7, 45159 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dziduszko A., Ozbun M. A., Annexin A2 and S100A10 regulate human papillomavirus type 16 entry and intracellular trafficking in human keratinocytes. J. Virol. 87, 7502–7515 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang H. S., Buck C. B., Lambert P. F., Inhibition of gamma secretase blocks HPV infection. Virology 407, 391–396 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karanam B., Peng S., Li T., Buck C., Day P. M., Roden R. B. S., Papillomavirus infection requires gamma secretase. J. Virol. 84, 10661–10670 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor J. R., Fernandez D. J., Thornton S. M., Skeate J. G., Lühen K. P., da Silva D. M., Langen R., Kast W. M., Heterotetrameric annexin A2/S100A10 (A2t) is essential for oncogenic human papillomavirus trafficking and capsid disassembly, and protects virions from lysosomal degradation. Sci. Rep. 8, 11642 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wüstenhagen E., Hampe L., Boukhallouk F., Schneider M. A., Spoden G. A., Negwer I., Koynov K., Kast W. M., Florin L., The cytoskeletal adaptor obscurin-like 1 interacts with the human papillomavirus 16 (HPV16) capsid protein L2 and is required for HPV16 endocytosis. J. Virol. 90, 10629–10641 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aksoy P., Meneses P. I., The role of DCT in HPV16 infection of HaCaTs. PLOS ONE 12, e0170158 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Freeman-Cook L. L., DiMaio D., Modulation of cell function by small transmembrane proteins modeled on the bovine papillomavirus E5 protein. Oncogene 24, 7756–7762 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Xie J., DiMaio D., Traptamer screening: A new functional genomics approach to study virus entry and other cellular processes. FEBS J. 10.1111/febs.15775 , (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goodwin E. C., DiMaio D., Repression of human papillomavirus oncogenes in HeLa cervical carcinoma cells causes the orderly reactivation of dormant tumor suppressor pathways. Proc. Natl. Acad. Sci. U.S.A. 97, 12513–12518 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goodwin E. C., Yang E., Lee C. J., Lee H. W., DiMaio D., Hwang E. S., Rapid induction of senescence in human cervical carcinoma cells. Proc. Natl. Acad. Sci. U.S.A. 97, 10978–10983 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hwang E.-S., Naeger L. K., DiMaio D., Activation of the endogenous p53 growth inhibitory pathway in HeLa cervical carcinoma cells by expression of the bovine papillomavirus E2 gene. Oncogene 12, 795–803 (1996). [PubMed] [Google Scholar]
- 41.Fujiki Y., Hubbard A. L., Fowler S., Lazarow P. B., Isolation of intracellular membranes by means of sodium carbonate treatment: Application to endoplasmic reticulum. J. Cell Biol. 93, 97–102 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siddiqa A., Broniarczyk J., Banks L., Papillomaviruses and endocytic trafficking. Int. J. Mol. Sci. 19, 2619 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gottschalk E. Y., Meneses P. I., A dual role for the nonreceptor tyrosine kinase Pyk2 during the intracellular trafficking of human papillomavirus 16. J. Virol. 89, 9103–9114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clague M. J., Liu H., Urbe S., Governance of endocytic trafficking and signaling by reversible ubiquitylation. Dev. Cell 23, 457–467 (2012). [DOI] [PubMed] [Google Scholar]
- 45.Steinberg F., Heesom K. J., Bass M. D., Cullen P. J., SNX17 protects integrins from degradation by sorting between lysosomal and recycling pathways. J. Cell Biol. 197, 219–230 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DiGiuseppe S., Bienkowska-Haba M., Hilbig L., Sapp M., The nuclear retention signal of HPV16 L2 protein is essential for incoming viral genome to transverse the trans-Golgi network. Virology 458-459, 93–105 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin H., Li S., Shu H.-B., The membrane-associated MARCH E3 ligase family: Emerging roles in immune regulation. Front. Immunol. 10, 1751 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buck C. B., Pastrana D. V., Lowy D. R., Schiller J. T., Generation of HPV pseudovirions using transfection and their use in neutralization assays. Methods Mol. Med. 119, 445–462 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Goodwin E. C., Naeger L. K., Breiding D. E., Androphy E. J., DiMaio D., Transactivation-competent bovine papillomavirus E2 protein is specifically required for efficient repression of human papillomavirus oncogene expression and for acute growth inhibition of cervical carcinoma cell lines. J. Virol. 72, 3925–3934 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lipovsky A., Zhang W., Iwasaki A., DiMaio D., Application of the proximity-dependent assay and fluorescence imaging approaches to study viral entry pathways. Methods Mol. Biol. 1270, 437–451 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Allalou A., Wahlby C., BlobFinder, a tool for fluorescence microscopy image cytometry. Comput. Methods Programs Biomed. 94, 58–65 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/27/eabh4276/DC1