Agonistic proresolutive monoclonal antibody reverses chronic inflammation.
Abstract
Resolution of inflammation is elicited by proresolving lipids, which activate GPCRs to induce neutrophil apoptosis, reduce neutrophil tissue recruitment, and promote macrophage efferocytosis. Transcriptional analyses in up to 300 patients with Inflammatory Bowel Disease (IBD) identified potential therapeutic targets mediating chronic inflammation. We found that ChemR23, a GPCR targeted by resolvin E1, is overexpressed in inflamed colon tissues of severe IBD patients unresponsive to anti-TNFα or anti-α4β7 therapies and associated with significant mucosal neutrophil accumulation. We also identified an anti-ChemR23 agonist antibody that induces receptor signaling, promotes macrophage efferocytosis, and reduces neutrophil apoptosis at the site of inflammation. This ChemR23 mAb accelerated acute inflammation resolution and triggered resolution in ongoing chronic colitis models, with a significant decrease in tissue lesions, fibrosis and inflammation-driven tumors. Our findings suggest that failure of current IBD therapies may be associated with neutrophil infiltration and that ChemR23 is a promising therapeutic target for chronic inflammation.
INTRODUCTION
Inflammatory bowel disease (IBD) such as ulcerative colitis (UC) and Crohn’s disease (CD) are chronic relapsing gastrointestinal disorders characterized by chronic intestinal inflammation, dysregulated immune responses to intestinal microbiota, and dysfunction of the epithelial barrier (1, 2). Although IBDs are associated with marked morbidity and have a major impact on quality of life and ability to work, their incidence and prevalence keep increasing worldwide (3, 4). Current conventional treatments aim to prevent inflammation by gradual administration of anti-inflammatory agents, steroids, immunosuppressive drugs or biological agents targeting tumor necrosis factor (TNF) cytokines or gut-specific α4β7 integrin, particularly for refractory and severe forms of IBD (5, 6). However, these therapies do not induce remission in over half of the patients, and relapse also occurs over time in primary responders (7), with fibrotic complications developing in a significant number of patients (8). Thus, research efforts should be invested in identifying novel mechanisms and signaling networks implicated not only in dampening or preventing inflammation but also in turning off the chronicity of inflammation and promoting tissue homeostasis.
Inflammation resolution is an active process governed in part by endogenous mediators, including members of the superfamily of lipidic specialized proresolving mediators (SPMs), as well as some proteins that promote clearance of inflammatory infiltrates, restoration of tissue integrity, and, ultimately, the return to normal function (9–15). Failed resolution occurs when excessive or unresolved protective acute inflammation response progresses to chronic inflammation, which is a universal feature in many organ-specific diseases (16). SPMs are lipids biosynthesized from omega-6 and omega-3 polyunsaturated fatty acids that act as resolution agonists by targeting distinct G protein–coupled receptors (GPCRs) expressed by immune cells, in particular, phagocytes (17–20). SPMs have been shown to stop neutrophil transmigration and trigger their apoptosis and to promote nonphlogistic phagocytosis of cellular debris (efferocytosis), including apoptotic neutrophils (18, 19). These functions are critical for inflammation resolution, as neutrophils recruited in excess or unable to exit the site of inflammation can cause unintended collateral tissue damage, which amplifies ongoing inflammation (20, 21). In IBD, delayed neutrophil apoptosis and neutrophil-mediated damage are associated with the development of UC and CD (22–26). Moreover, recruitment of activated neutrophils correlates with disease severity in CD (27), whereas complete resolution of mucosal neutrophils improves long-term clinical outcome in UC (28). SPM production is reduced in the colonic mucosa of patients with UC (29), and conversely, increased expression of SPMs in UC colon tissue is associated with remission and mucosa homeostasis (27, 28). Notably, it has been shown that exogenous treatment with resolvin E1 (RvE1) accelerates the resolution of colon inflammation in acute, spontaneously resolutive colitis mouse models (32–34). RvE1 triggers extracellular signal–regulated kinase (ERK) and Akt signaling via activation of ChemR23 GPCRs (CMKLR1 gene) expressed by myeloid cells such as macrophages, dendritic cells (DCs), neutrophils, and natural killer (NK) cells (35–37). The RvE1/ChemR23 axis has been reported to decrease inflammatory TNF-α and interleukin-12 (IL-12) secretion by DCs (33, 37), reduce neutrophil transmigration (38), increase reactive oxygen species (ROS) generation by neutrophils (39), stimulate efferocytosis (40), and promote M1 inflammatory to M2 resolutive-type macrophage conversion (35).
Despite increasing evidence that chronic inflammation in humans is associated with a deficit of SPMs, current treatments based on proresolutive drug delivery are limited to experimental validation in spontaneously resolutive acute inflammation murine models, due to the restricted bioavailability and rapid clearance of SPMs. Thus, whether this novel therapeutic approach can promote efficient resolution in nonspontaneously resolutive chronic inflammation remains unclear. Here, we report that ChemR23 receptor overexpression in the IBD colon mucosa is associated with neutrophil infiltration and unresponsiveness to anti-TNF/α4β7 therapies. Moreover, activating proresolutive ChemR23 signaling with an agonistic monoclonal antibody (mAb) efficiently accelerates resolution in acute colitis models and triggers resolution of ongoing chronic colitis in unresolved preclinical models, thereby preventing fibrosis and limiting the development of inflammation-driven colorectal tumors.
RESULTS
Increased mucosal neutrophil infiltrates and ChemR23 expression are associated with resistance to immunotherapies in IBD
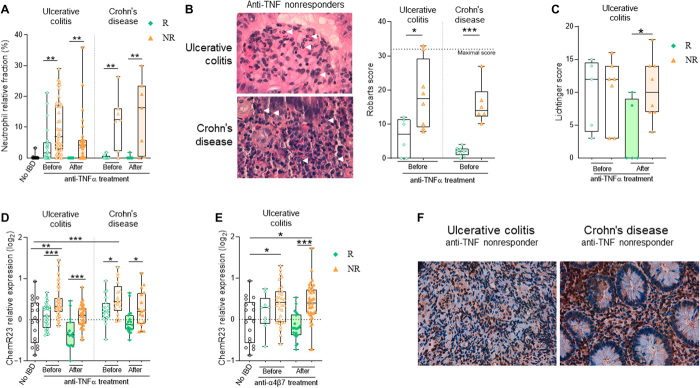
We previously reported a meta-analysis of publicly available transcriptional UC cohort datasets from colon mucosa biopsies performed before anti-TNF treatment (41). Differential analysis of all expressed genes in the meta-dataset (17,037 genes) identified only 85 genes significantly (adjusted P value < 0.05) and differentially (log2 fold change > 1) expressed between responders and nonresponders. Gene set enrichment analysis indicated overexpression of genes associated with myeloid cell activation, neutrophil activation and degranulation, leukocyte chemotaxis, and response to bacterium (fig. S1, A and B). To investigate whether local/mucosal neutrophil excessive infiltrates are associated with responsiveness to conventional immunotherapies (anti-TNF or anti-α4β7 mAbs), we first analyzed neutrophil gene expression signatures using deconvolution tools on previously published transcriptomic datasets from mucosal biopsies of IBD patients with clinical annotation. We examined colon biopsies (responders n = 28, nonresponders n = 41) from three independent UC cohort datasets [GSE16879 (42), GSE12251 (43), and GSE73661 (44)] and colon and ileal biopsies (responders n = 20, nonresponders n = 17) from one CD cohort dataset [GSE16879 (42)]. Colon biopsies were performed before the start of therapy and then 4 to 6 weeks after the first antibody infusion. Response to immunotherapy was determined by histological healing. Using two different bioinformatic deconvolution tools and validated transcriptomic signatures, we first found using the CIBERSORT transcriptomic signatures that, in both UC and CD patients, the relative fraction of neutrophils in the gut mucosa was significantly higher in nonresponder patients before and after anti-TNF treatment, when compared to responders or non-IBD controls (Fig. 1A). Similar results were obtained in patients with UC before and after anti-TNF (fig. S2A) or anti-α4β7 integrin (fig. S2B) therapy by using Quantiseq, a different deconvolution method that confirmed the CIBERSORT analysis. As previously reported (42, 43), transcriptomic signature analyses also revealed an increase in inflammatory (M1) macrophages over resolutive-type (M2) macrophages before anti-TNF treatment in colon biopsies of nonresponder patients (fig. S2C). The accumulation of neutrophils in the colon mucosa of anti-TNF nonresponder UC and CD patients was confirmed with histological sections from independent local cohorts (UC responders n = 6, UC nonresponders n = 8, CD responders n = 9, CD nonresponders n = 6 from Nantes University Hospital, France) by using pathologist blinded analysis of Robarts score, which integrates the quantity and localization of neutrophils in the tissue (Fig. 1B). No differences in disease severity (Lichtiger score) between responder and nonresponder UC patients before therapy was observed in these histological sections (Fig. 1C), further demonstrating that excessive neutrophil infiltrates in nonresponders in our local IBD cohort is not simply due to the severity of the disease.
Fig. 1. Colonic mucosal neutrophil infiltrates and ChemR23 overexpression are associated with nonresponse to anti-TNF and anti-α4β7 mAb therapies in IBD.
(A) Neutrophils transcriptomic signature fraction among immune cells using CIBERSORT analysis in intestinal biopsies of non-IBD patients (black, n = 18), responders (R) (green), and nonresponders (NR) (orange) before and after TNF treatment. Meta-analysis [datasets GSE16879 (42), GSE12251 (43), and GSE73661 (44)] of colon biopsies from three UC cohorts (responders n = 28 to 15, nonresponders n = 41 to 31) and analysis [dataset GSE16879 (42)] of colon and ileal biopsies from a CD cohort (responders n = 24 to 23, nonresponders n = 19) with histological healing as the anti-TNF response criteria. (B) Representative H&E staining (left) from colon biopsies of UC and ileal biopsies of CD patients with white arrows indicating neutrophils infiltrates. Neutrophil-associated histological Robarts score quantification (right) in colon and ileal biopsies from a local French cohort of UC and CD patients before treatment with anti-TNF: UC (responders n = 6, nonresponders n = 8) and CD (responders n = 9, nonresponders n = 6). (C) Lichtiger score of the same cohort as in (D). (Responders, green; nonresponders, orange). (D) ChemR23 transcriptomic relative expression in intestinal biopsies from the same IBD cohorts as in (A) before and after anti-TNF treatment. (E) ChemR23 transcriptomic relative expression in colon biopsies of patients with UC before and after anti-α4β7 mAb therapy: dataset GSE73661 (44), responders n = 9 to 22, nonresponders n = 30 to 37. (F) Representative staining of the ChemR23 protein expression from colon biopsies of UC and ileal biopsies of patients with CD from the local cohort in (B). *P < 0.05, **P < 0.01, ***P < 0.005 (Kruskal-Wallis and Mann-Whitney tests).
Next, we determined whether the proresolutive myeloid receptor ChemR23 (cmklr1gene) is also differentially expressed in the gut mucosa of patients with IBD in response to current immunotherapies. On the basis of the same publicly available datasets and our meta-cohort, we found that ChemR23 is significantly overexpressed in the mucosa of UC and CD patients unresponsive to anti-TNF (Fig. 1D) or anti-α4β7 (Fig. 1E) therapies, when compared to responders and non-IBD controls. Similarly, ChemR23 mucosal overexpression is detected before and after treatment in UC and CD nonresponders and significantly correlates with neutrophil transcriptomic signature and M1/M2 transcriptomic ratio (fig. S2D). Although ChemR23 expression tends to decrease after anti-TNF therapy in both responders and nonresponders, its expression remains significantly higher in nonresponders after treatment (Fig. 1, D and E). Analysis of different colon and ileal biopsy datasets from UC and CD patients after treatment with corticosteroids and/or immunosuppression showed that ChemR23 is overexpressed in gut inflamed tissues but not in proximal noninflamed segments or non-IBD controls (fig. S1, C and D). Moreover, analysis of a recent single-cell RNA sequencing (scRNA-seq) dataset from ileal tissues of patients with CD (47) confirmed that ChemR23 is mainly expressed by mucosal macrophages, particularly resident macrophages (fig. S3A). Consistent with this, scRNA-seq data from colon biopsies of patients with UC (48) show that ChemR23 is expressed by macrophages within leukocytes subsets but not by stromal or epithelial cells (fig. S4). Unfortunately, scRNA-seq technology does not permit the analysis of granulocyte subsets. However, histological ChemR23 staining on colon or ileal biopsies from a local cohort of UC and Crohn anti-TNF nonresponders showed that ChemR23 is mainly expressed by neutrophils in inflamed mucosas (Fig. 1F).
These data indicate that mucosal excessive neutrophil infiltration and ChemR23 overexpression before treatment are associated with unresponsiveness to anti-TNF and anti-α4β7 immunotherapies in UC and CD patients, suggesting that ChemR23-dependent neutrophil infiltration may be associated with refractory and severe chronic inflammation.
ChemR23 expression is regulated by inflammatory signals
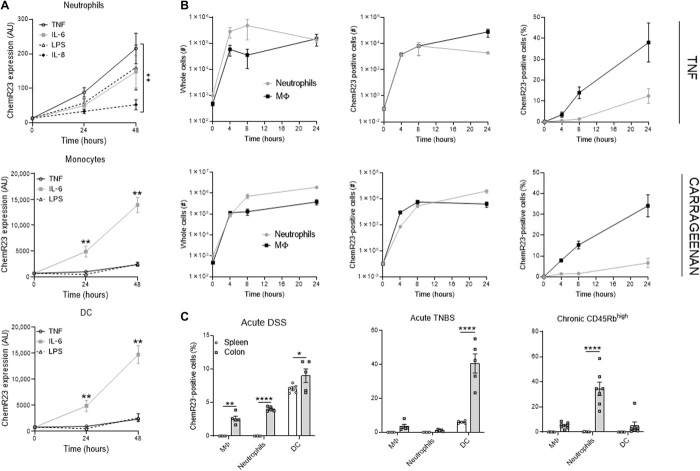
To dissect the regulatory mechanisms underpinning ChemR23 activity during inflammation, we examined ChemR23 expression in vitro in leucocyte subpopulations cultured with inflammatory signals and in vivo using murine acute inflammatory models. Whereas ChemR23 was weakly expressed on freshly isolated human neutrophils, it was significantly up-regulated in neutrophils cultured with inflammatory signals [TNFα, IL-6, lipopolysaccharide (LPS), or IL-8] (Fig. 2A and fig. S5). Similarly, IL-6 significantly increased ChemR23 expression in human monocytes and DCs over time (Fig. 2A) as previously described (32, 45–47). In naïve mice, ChemR23 expression at steady state in the spleen was significantly lower than in resident macrophages and DCs in the colon (fig. S6, A and B). Moreover, neutrophils from the spleen, bone marrow, or colon do not express ChemR23 in naïve mice. However, similar to human, mouse neutrophils or monocytes from naïve mice overexpressed ChemR23 when cultured with inflammatory mediators (IL-6 or LPS) (fig. S6C). In in vivo murine air pouch models, subcutaneous injection of TNFα or carrageenan (an algae-derived polysaccharide) induces rapid and strong local recruitment of macrophages and neutrophils expressing ChemR23 (Fig. 2B). Notably, although no ChemR23 was detected in resident macrophages and neutrophils at baseline, a significant proportion of macrophages and neutrophils recruited upon inflammation expressed ChemR23. We next analyzed ChemR23 levels in preclinical models of acute colitis induced by chemical agents [dextran sulfate sodium (DSS) or TNBS (2,4,6-trinitrobenzenesulfonic acid)] and in models of chronic colitis induced by adoptive transfer of CD4+ CD45Rbhigh T cells into Rag1 (recombination activating gene 1) knockout (KO) mice. In both these models, ChemR23 expression in macrophages and neutrophils was detected only in the inflamed colon and not in the periphery (spleen) (Fig. 2C and fig. S7). Chronic colitis models showed stronger ChemR23+ neutrophil infiltration than acute colitis models. Together, these results indicate that ChemR23 is induced by inflammatory signals and is primarily expressed in resident tissue macrophages and inflammatory neutrophils.
Fig. 2. ChemR23 expression is regulated by inflammation.
(A) ChemR23 expression (median fluorescent intensity) analyzed by flow cytometry of neutrophils (CD66b+), monocytes (CD14+), and DCs (CD11c+) after human peripheral blood leukocytes cultured with TNFα (100 U/ml), IL-6 (10 ng/ml), IL-8 (50 ng/ml), or LPS (100 ng/ml) for the indicated period of time (n = 6 to 12 per group). (B) Left: Absolute numbers of macrophages (black) and neutrophils (gray) recruitment in skin air pouch models induced by 50 ng of TNFα (top) or 1% carrageenan (bottom) injection. Middle: Absolute numbers and (right) frequency of ChemR23-positive macrophages and neutrophils recruited in these air pouch models. (C) ChemR23 expression by indicated population in spleen (white) and colon (gray) of acute DSS (day 6), acute TNBS (day 3), and chronic CD45Rbhigh transfer (day 61) colitis mouse models (n = 5 to 7 per group). *P < 0.05, **P < 0.01, ****P < 0.001 [Mann-Whitney and multiple comparisons two-way analysis of variance (ANOVA)]. AU, arbitrary units.
Agonist anti-ChemR23 mAb as new proresolving therapeutic modality
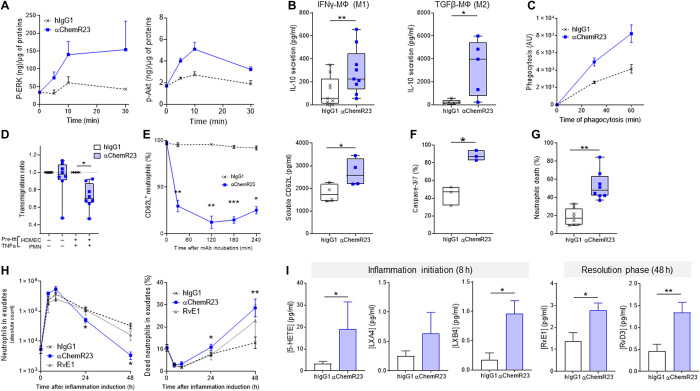
Current progress on the potential therapeutic applications of SPMs remains essentially limited to experimental validation in spontaneously resolutive acute inflammation models, due to the reduced bioavailability and rapid clearance of these molecules. To explore a different therapeutic modality for triggering efficient resolution of chronic inflammation with a long elimination half-life, we screened novel mAbs for their proresolutive agonist properties. We screened several anti-human ChemR23 mAbs and identified a unique clone with RvE1-like properties such as IL-10 secretion induction by human macrophages (fig. S8A) and selected the most efficient humanized and optimized variant of this anti-ChemR23 mAb for further exploration in vivo (fig. S8B). We found that an agonist anti-ChemR23 mAb induced rapid ERK and Akt phosphorylation (Fig. 3A), which is a well-characterized ChemR23 downstream signaling pathway typically induced by RvE1 (50). This signaling event triggered the repolarization of human macrophages into proresolving macrophages, as revealed by an increase in IL-10 secretion and decrease in CD80, CD86, and CCR7 expression in M1 interferon-γ (IFNγ)–polarized macrophages and an increase in IL-10 secretion and decrease in CD206 and CD11b expression in M2 transforming growth factor–β (TGFβ)–polarized macrophages (Fig. 3B and fig. S10A). Pretreatment of human macrophages with agonist ChemR23 mAb for 1 hour increased phagocytosis of neutrophils (Fig. 3C), consistent with previous research showing that ChemR23 proresolutive activity in macrophages induces efferocytosis of apoptotic neutrophils (51). Moreover, as previously described for RvE1 (52), agonist ChemR23 mAb significantly reduced human neutrophil transendothelial migration during inflammation of both healthy volunteers (HVs) (Fig. 3D) or patients with anti-neutrophil cytoplasmic autoantibody (ANCA) vasculitis (fig. S9), consistent with a rapid (30 min) and sustained reduction in l-selectin (CD62L) membrane expression and increase in soluble l-selectin (Fig. 3E). In addition, neutrophil cultured with the anti-ChemR23 mAb displayed significantly reduced membrane expression of both CXCR1 and CXCR2 IL-8 receptors (fig. S10B). These findings confirm that agonist ChemR23 mAb reduces neutrophil transmigration by promoting l-selectin shedding and inhibiting neutrophil chemotaxis. In addition, agonist anti-ChemR23 mAb internalization accelerated neutrophil apoptotic cell death (Fig. 3G and fig. S10C), as revealed by an increase in Caspase-3/7 activation and annexin V staining (Fig. 3F and fig. S10E). Mechanistically, this accelerated neutrophil apoptosis was associated with increased ROS production (fig. S10D). However, no significant NETosis was detected (fig. S10F).
Fig. 3. Proresolutive mechanisms of agonist ChemR23 mAb.
(A) p-ERK (Thr202/Tyr204) and p-Akt (Ser473) ChemR23 signaling measured by ELISA in protein lysate of human inflammatory M1 macrophages cultured with isotype hIgG1 or anti-ChemR23 mAb (10 μg/ml) for the indicated period of time. (B) IL-10 secretion in the supernatant of M1 IFNγ-polarized (left) and M2 TGFβ-polarized (right) human macrophages culture for 48 hours with human IgG1 isotype control or anti-ChemR23 mAb (10 μg/ml) (n = 6). (C) Efferocytosis of human neutrophils by human macrophages pretreated for 1 hour with isotype hIgG1 or anti-ChemR23 mAb (10 μg/ml) (n = 4). (D) Transmigration of human neutrophils across a monolayer of human dermal microvascular endothelial cells (HDMECs) preincubated overnight with medium or TNFα (100 U/ml). Isotype hIgG1 or anti-ChemR23 mAb was added at 10 μg/ml in a Boyden chamber (n = 8). IL-8 (50 ng/ml) was used in the lower part of the chamber as neutrophil chemoattractant for 90 min. (E) CD62L (l-selectin) expression kinetic by flow cytometry (left) and soluble CD62L concentration supernatant (right) of human neutrophils cultured with isotype hIgG1 or anti-ChemR23 mAb (10 μg/ml) for 4 hours (n = 3 to 4). (F) Percentage of Caspase-3/7 human neutrophils determined by microcopy after 10 hours of culture with isotype hIgG1 or anti-ChemR23 mAb at 10 μg/ml (n = 3). (G) Percentage of dead human neutrophils determined using Live and Dead assay after 24 hours of culture with isotype hIgG1 or anti-ChemR23 mAb at 10 μg/ml (n = 8). (H) Neutrophils absolute number (left) and percentage of dead neutrophils (right) measured in mouse exudates of TNF-induced inflammation air pouch model at indicated time points. hIgG1 control (black) or anti-ChemR23 mAb (blue) was administered twice intraperitoneally at d-1 and d0 (20 μg per injection; n = 4 to 5 per group). (I) SPMs and precursors quantification by mass spectrometry in the same exudates as in (H) 8 hours (left) and 48 hours (right) after TNF injection. *P < 0.05, **P < 0.01, ***P < 0.05 (Mann-Whitney or Wilcoxon).
Next, we identified the ChemR23 epitope recognized by the mAb using MAbTope in silico epitope mapping (fig. S11, A and B) (53) and then confirmed these results by enzyme-linked immunosorbent assay (ELISA) binding assay (fig. S11C). The antibody binds to ChemR23’s GPCR extracellular loop 3, a peptide sequence highly conserved between mouse and human, thereby suggesting cross-reactivity. Agonist anti-ChemR23 mAb triggered ERK and Akt signaling in mouse macrophages (fig. S10G) and promoted inflammation resolution in vivo. In the TNFα air pouch acute inflammation model, injection of anti-ChemR23 mAb did not reduce the initial inflammatory local recruitment of neutrophils but significantly promoted the clearance of neutrophils from the inflammatory sites (Fig. 3H). Notably, RvE1 has been previously reported to inhibit neutrophils migration in peritonitis models (52) and to trigger neutrophil apoptosis in vitro (20). In our TNF-induced air pouch model, RvE1 administration only induced a variable effect on neutrophil recruitment and cell death. Last, biochemical characterization of exudates showed that anti-ChemR23 mAb injection significantly increased local production of SPMs and their precursors at 8 hours during the inflammation phase (e.g., lipoxins) and also at 48 hours during the resolution phase (e.g., RvE1 and D3) (Fig. 3I). Together, these data show that agonist anti-ChemR23 mAb triggers inflammation resolution by reprogramming inflammatory macrophages toward proresolutive activity, thereby inducing inflammatory neutrophil clearance from inflamed sites and promoting the secretion of proresolutive molecules.
ChemR23 activation with agonist mAb triggers resolution of acute and chronic intestinal inflammation
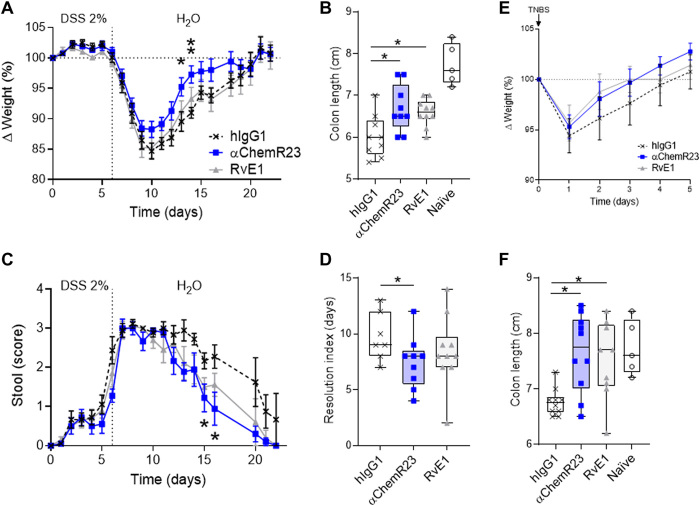
RvE1 was previously shown to accelerate the resolution of inflammation in spontaneously resolutive acute colitis mouse models (30, 31). ChemR23 mAb treatment significantly accelerated recovery from acute colitis associated with high mucosal neutrophils infiltrates (fig. S12) such as the DSS and TNBS mouse models, as shown by a reduced resolution index (Ri), acceleration of weight recovery, and normalization of stool consistency and fecal blood signs (Fig. 4). Colon retraction, a key hallmark of colonic inflammation, was also significantly restored after RvE1 or agonist ChemR23 mAb treatment, which further demonstrates their proresolutive action (Fig. 4, B and F).
Fig. 4. Agonist ChemR23 mAb accelerates the resolution of acute colon inflammation.
(A) Weight variation, (B) colon length (d22), and (C) diarrhea (stool score) of mice in the acute DSS (2%) colitis model treated intraperitoneally at d0, d2, and d4 with isotype hIgG1 (black; n = 10) or anti-ChemR23 (blue; n = 10) mAb at 1 mg/kg or RvE1 (50 μg/kg per day; gray; n = 10) daily for 5 days. (D) Resolution index of colitis based on the time to resolve 50% of the diarrhea symptoms as discussed in (90). (E) Weight variation and (F) colon length (d5) of mice in the acute intrarectal TNBS/ethanol (50%) colitis model treated intraperitoneally at d0 and d2 with isotype (control) or anti-ChemR23 mAbs (1 mg/kg) or RvE1 (50 μg/kg per day) daily for 3 days(n = 9 to 10 per group). Naïve, age-matched mice that did not receive administration of DSS or TNBS/ethanol. *P < 0.05 and **P < 0.01 (Mann-Whitney).
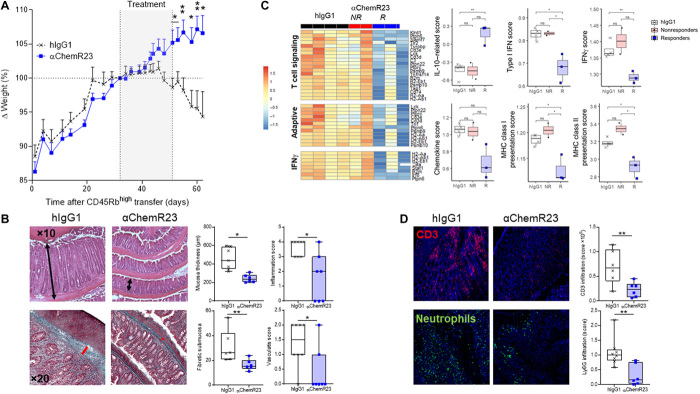
To investigate whether ChemR23 activation with a mAb effectively reduces inflammation in nonresolutive and chronic inflammatory colitis models, we conducted an adoptive transfer of CD4+ CD45Rbhigh T cells into Rag1KO mice, which induces progressive and chronic colitis over time without a resolution phase. Treatment with agonist ChemR23 mAb initiated after colitis onset [from day 32 (d32) to d51] and T lymphocyte infiltration (fig. S13A) significantly reversed ongoing chronic colitis and protected mice from fatal weight loss (Fig. 5A). Histological evaluation of the colon at sacrifice showed a significant decrease in mucosal inflammation and thickness, reduced signs of vasculitis, and nearly complete inhibition of colon fibrosis (Fig. 5B). Transcriptomic analysis (Nanostring Immunology Panel) in responding mice revealed a significant decrease in gene expression signatures associated with adaptive immune response, T lymphocyte signaling, and IFNγ pathway (Fig. 5C and fig. S13B). We also observed significant modulation of innate myeloid cell signature such as reduced antigen processing and presentation signature, IFNα and IFNβ signature, chemokine signature, and, on the opposite, increased IL-10 signaling signature (Fig. 5C). Consistent with this, histology in mice showed that agonist anti-ChemR23 mAb treatment significantly affects T cell and neutrophil infiltrates in the colon (Fig. 5D).
Fig. 5. Agonist ChemR23 mAb triggers the resolution of ongoing chronic colon inflammation.
(A) Weight variation of Rag1−/− mice reconstituted intraperitoneally with 5 × 105 CD4+ CD45Rbhigh T cells from wild-type C57Bl/6 mice and treated intraperitoneally with isotype hIgG1 (1 mg/kg; black; n = 8) or anti-ChemR23 (blue; n = 7) mAbs three times/week from days 32 to 51 after T cell transfer. (B) Representative colon Hematoxylin and eosin (H&E) (top) and Masson’s trichrome (bottom) staining and pathological quantification of colon inflammation, vasculitis, colon mucosa thickness (black bars), and fibrotic submucosa thickness (red bars) in the same model as in (A) at sacrifice (1 month after treatment initiation, control n = 6 to 8, anti-ChemR23 n = 6 to 7).(C) Representative heatmap of clustered differential gene expression and scores of transcriptomic analysis of colon in the CD4+ CD45Rbhigh adoptive transfer model using the Nanostring mouse inflammation panel. Isotype hIgG1 (black; n = 4) and anti-ChemR23 (red for nonresponders n = 2; blue for responders n = 3) mAb-treated mice. (D) Representative immunofluorescent staining of T cells (CD3+) and neutrophils (Ly6G+) and quantification in same colon. *P < 0.05 and **P < 0.01 (Mann-Whitney). MHC, major histocompatibility complex; ns, not significant.
IL-10 KO mice develop chronic colitis spontaneously over the time (54). We treated these mice for 2 weeks with the agonist ChemR23 mAb or its corresponding isotype control when significant clinical symptoms of colitis were monitored (weight decrease > 5% and/or stool score > 1). After 14 days of treatment, mice treated with anti-ChemR23 mAb displayed significantly fewer signs of diarrhea and weight loss when compared to the control group (fig. S14), showing that agonist anti-ChemR23 mAb triggers resolution of ongoing chronic colitis in different models.
Given that anti-ChemR23 mAb accelerates neutrophil apoptosis (Fig. 3, F and G) and is a human immunoglobulin G1 (hIgG1) isotype with potential cell cytotoxicity (55), we assessed its in vivo effects on the peripheral immune cell population. No significant changes were detected in immune cell populations (neutrophils, macrophages, DCs, T cells, and NK cells) in the spleen of mice treated with agonist ChemR23 mAb in three different models of induced acute or chronic colitis (fig. S15) or in the blood or bone marrow of naïve mice that received high dose of anti-ChemR23 mAb (fig. S16), in agreement with the expression pattern of ChemR23 primarily in myeloid tissue. These data further suggest that agonist anti-ChemR23 treatment does not induce peripheral immune cell depletion or neutropenia.
Agonist anti-ChemR23 mAb limits colon cancer development
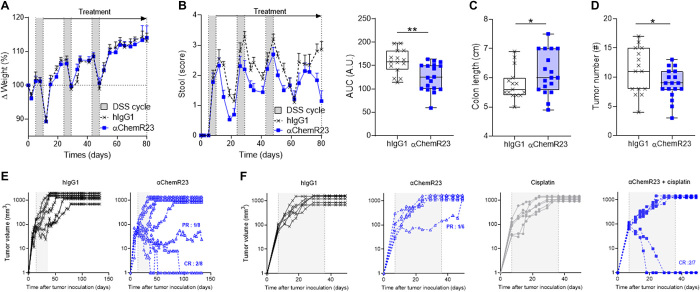
Chronic colon inflammation is an important risk factor for colorectal cancer (CRC) development (56). However, conventional therapies to inhibit inflammation can have a negative impact on the overall survival of IBD patients with CRC (57). To evaluate the potential effects of triggering inflammation resolution with agonist ChemR23 mAb on tumor development, we treated mice with chronic inflammation-mediated colon cancer receiving cycles of DSS after an initial injection of a carcinogenic agent (azoxymethane). Treatment with ChemR23 mAbs was initiated after the first cycle of DSS. Although no weight loss differences were detected (Fig. 6A), anti-ChemR23 treatment caused a significant reduction in diarrhea during the entire treatment period (3 months), as each diarrhea phase after DSS administration was shorter in mice treated with anti-ChemR23 than in controls (Fig. 6B). Colon retraction was also significantly reduced at sacrifice in mice treated with anti-ChemR23 (Fig. 6C), and macroscopic evaluation of the colon at 3 months revealed significantly fewer tumor nodules (Fig. 6D). To further investigate whether anti-ChemR23 has a protective antitumoral effect, mice implanted subcutaneously with the MC38 (Fig. 6E) or CT26 (Fig. 6F) colorectal mouse cell lines were treated with agonist anti-ChemR23 mAb in monotherapy. In CT26 mice, which are less immunogenic, we also tested a combined treatment with anti-ChemR23 and chemotherapy (cisplatin). Agonist anti-ChemR23 treatment induced partial or complete antitumor responses in MC38 and CT26 models, including with monotherapy. Notably, all mice with complete response also developed a memory immune response, as second MC38 or CT26 graft rechallenges were rejected without new treatment (0 tumor growth on four mice with previous complete response). These data suggest that activation of the proresolutive ChemR23 pathway suppresses cancer development in chronic inflammatory diseases by triggering chronic inflammation resolution.
Fig. 6. Agonist ChemR23 mAb inhibits colon tumor development.
(A) Weight variation of mice in the chronic AOM/DSS colitis–associated colorectal neoplasia model treated intraperitoneally twice/week with isotype hIgG1 (1 mg/kg; black; n = 15) or anti-ChemR23 (blue; n = 18) from day 10 (end of the first cycle of DSS) until day 80. DSS cycle is indicated by gray areas. (B) Same as in (A) for diarrhea (stool score) (left) and area under the curve (AUC) of total stool scores from day 0 to day 80 (right). (C) Colon length and (D) colon macroscopic tumors measured at day 80 in the AOM/DSS model. (E) Tumor volume after subcutaneous injection of 5 × 105 MC38 colorectal cell line injection in immunocompetent mice treated intraperitoneally three times/week for 21 days with isotype hIgG1 (1 mg/kg; black, n = 6) or anti-ChemR23 (blue, n = 8) mAbs starting when the tumor volume ranged between 50 and 100 mm3. (F) Tumor volume after subcutaneous injection of 5 × 105 CT26 colorectal cell line injection in immunocompetent mice treated intraperitoneally three times/week for 21 days with isotype hIgG1 (1 mg/kg; black, n = 6) or anti-ChemR23 (blue, empty squares, n = 7) mAbs from day 7 to day 28 and/or cisplatin (5 mg/kg) injected intraperitoneally every 5 days from d7 to day 28 (monotherapy, gray, n = 6; cisplatin + anti-ChemR23, blue solid squares, n = 7). PR, partial response; CR, complete response. *P < 0.05 and **P < 0.01 (Mann-Whitney).
DISCUSSION
Chronic inflammatory diseases, such as IBD, are currently managed with therapies that suppress the production of inflammatory mediators (i.e., inflammatory cytokines) or block immune cell recruitment or activation (i.e., integrin and costimulation). However, these strategies are not effective in a substantial proportion of patients, and significant rates of acquired resistance are observed (57). The identification of alternative therapeutic approaches targeting novel mechanisms and different disease phases is therefore urgently needed. Research in the last decade has shown that inflammation does not just peter out but is instead actively shut down by SPMs acting on receptors expressed by immune cells (13–15, 58–61). While there is growing interest in developing SPM analogs to treat inflammatory diseases (62–64), there are currently few proresolutive drugs in clinical development because stable SPM analogs have limited pharmacokinetic properties and their chemical synthesis is complex. In this study, we explored an innovative approach using an agonist mAb to activate receptors involved in inflammation resolution. Moreover, our results elucidate the mechanisms of ChemR23 regulation during inflammation and chronic inflammatory disease. Last, we show that promoting inflammation resolution via ChemR23 receptor activation with a mAb actively switches off ongoing chronic inflammation and tumor development in preclinical models.
Neutrophils recruited following infection or sterile wounding are key components of the inflammatory response. It has recently been suggested that neutrophils may have other functions in addition to providing immune protection when physical barriers are breached. For instance, neutrophils may contribute directly to inflammation resolution and recovery (65, 66) and orchestrate exacerbated adaptive responses by recruiting inflammatory macrophages and DCs that release proinflammatory cytokines associated with chronic inflammation, such as TNFα and IL-23 (67). Appropriate neutrophil apoptosis is fundamental for inducing resolution of inflammation and promoting the conversion of proinflammatory (M1) toward resolutive-type (M2) macrophages (68). In IBD, neutrophil intestinal infiltration is typically associated with severe inflammation (69) and occurrence of abscesses and/or granulomata (70), particularly in patients with UC. Our analysis of colon transcriptomic datasets from patients with IBD revealed that imbalanced M1/M2 macrophage ratios and neutrophil persistence and accumulation are strongly associated with a significant risk of resistance to anti-TNF or anti-α4β7 therapies in different cohorts of UC or CD patients. Moreover, histological analysis of an independent and local cohort of UC and CD patients showed that patients unresponsive to anti-TNF therapy have excessive neutrophil infiltrates that persist after treatment. We found that inflammatory signals (cytokines and Toll-like receptor agonist) induce ChemR23 expression in vitro, including in neutrophils, which express a very low ChemR23 level at steady state (49). In patients, ChemR23 was overexpressed in UC and CD colon biopsies from anti-TNF or anti-α4β7 nonresponders and correlated with neutrophil and M1/M2 macrophage transcriptomic signatures. scRNA-seq analysis performed on UC or CD mucosas showed that ChemR23 is predominantly expressed in myeloid cells, particularly in macrophage samples lacking granulocyte subsets. In addition, histological staining revealed that most of the neutrophils in the inflamed mucosa expressed ChemR23. These findings suggest that high ChemR23 expression in tissues is associated with excessive neutrophils and inflammatory (M1) over resolutive (M2) macrophages infiltrates, which are key features of chronic inflammation resolution deficiency.
RvE1, an endogenous lipid that binds to and activates ChemR23 receptor, has been shown to promote resolutive macrophage polarization, inhibit neutrophil recruitment, induce neutrophil apoptosis, and promote efferocytosis in resolutive-type macrophages (32, 37, 46, 69). We have identified an anti-human ChemR23 mAb with similar receptor signaling functions as RvE1, including in promoting macrophage resolutive polarization as well as inflammatory neutrophil migration and apoptosis. This dual mechanism of action identified in vitro on macrophages and neutrophils can contribute in parallel to trigger the resolution of inflammation in vivo because we observed, after treatment, reduced neutrophil infiltrates in acute and chronic inflammatory models as well as signature of repolarized macrophages, as illustrated by increased IL-10 signaling pathway and decreased antigen processing and presentation transcriptomic signatures. Moreover, the anti-ChemR23 mAb displayed proresolutive activity on overactivated neutrophils from patients with ANCA vasculitis (fig. S9) and would be interesting before clinical evaluation in IBD to confirm the effect of the antibody directly on cells or tissues isolated from patients with IBD. As expected, in vivo treatment with agonist anti-ChemR23 mAb did not affect the initiation phase of inflammation, as intensity of neutrophil recruitment, weight loss, and diarrhea was similar in acute colitis and air pouch murine models in comparison to controls. However, as previously reported for RvE1 (33, 34), agonist anti-ChemR23 mAb significantly shortened the Ri, as revealed by an accelerated clearing of leukocytes from the inflammatory site, increased apoptosis of remaining neutrophils, and faster recovery from colitis. Mass spectrometry of different resolution mediators or their precursors in exudates from TNFα air pouch model at different time points showed that agonist anti-ChemR23 mAb treatment triggers a global resolutive response, as expression of other mediators from the lipoxin or resolvin (including RvE1) families increased locally after treatment. A deficit in local and/or peripheral expression of a number of SPMs was reported not only in various chronic inflammatory diseases, including IBD (29, 72), rheumatoid arthritis (73), and severe asthma (74), but also in other inflammatory conditions such as type 2 diabetes (49), cystic fibrosis (75), or Alzheimer’s disease (76). In agreement with our findings, peripheral administration of RvE1 was shown to increase local concentration of RvE1, resolvin D, and lipoxin mediators in the hippocampus of Alzheimer’s mouse models (77), suggesting that proresolutive pathways are interconnected, and hence, proresolutive therapies may be beneficial for various types of inflammatory diseases.
Although it has been shown that RvE1 accelerates inflammation resolution in different preclinical models, such as IBD (33, 34), particularly in preclinical models with significant mucosal infiltrates of neutrophils such as acute DSS or TNBS models as described by us and others (78), periodontitis (79), and pulmonary infections (20), most of these studies used acute inflammatory models that resolved spontaneously. Furthermore, while a benefit effect of RvE1 administration has been previously reported on weight loss and colon length in the DSS acute model (34, 80), in our model, we only observed a benefit effect of RvE1 in reducing diarrhea as well as colon length. Thus, whether a potent agonist of ChemR23 may trigger nonresolutive chronic inflammation remained unknown. Our results in two different chronic colitis preclinical models characterized by aberrant T cell infiltration in the colon (81) and dysfunctional macrophages functions (83) show that treatment with agonist anti-ChemR23 mAb initiated after onset of the disease significantly reduces disease progression. Transcriptomic and histological analyses revealed significant decrease of innate immune cells and secreted cytokines, with the exception of increased IL-10 signaling pathway, and indirectly inhibition of adaptive immune responses in the colon upon ChemR23 activation, as well as reduced colon thickness and fibrosis, but without altering immune cell population frequencies in the periphery. Chronic colon inflammation is an important risk factor for CRC development in patients with IBD (56, 67). However, inhibition of inflammation with conventional anti-inflammatory drugs can have a negative impact on the overall survival of IBD patients with CRC (57). Recent preclinical studies demonstrated that various SPMs have a beneficial effect in reducing tumor development and metastasis in mouse models by promoting the elimination of apoptotic tumor cells by resolutive-type macrophages (83, 84). On the other hand, tumor-infiltrating neutrophils may play a detrimental role in the tumor microenvironment and contribute to tumor development (85, 86). We found that, besides triggering the resolution of chronic colitis, agonist ChemR23 mAb treatment also reduced colon tumor development caused by chronic inflammation and induced antitumoral immune response memory in preclinical models. Moreover, ChemR23 mAb mono- or combined therapy also significantly inhibited colon tumor growth in two CRC models.
In conclusion, our results show that ChemR23 may be overexpressed by some myeloid immune cell populations in inflammatory tissues, particularly in chronic inflammatory settings. Excessive neutrophil and proinflammatory macrophage mucosal infiltration is strongly associated with unresponsiveness to anti-TNF and anti-α4β7 therapies in patients with IBD and may contribute to chronic inflammation persistence. Moreover, we show that an agonist anti-human ChemR23 mAb is a novel proresolutive therapeutic modality that accelerates recovery from acute inflammation and triggers also chronic inflammation resolution, thereby preventing fibrosis and reducing tumor development. Together, our findings open a new therapeutic avenue for the development of a novel class of drugs to reinstate efficient resolution of chronic inflammation.
METHODS
Mice and reagents
C57Bl/6Rj and BALB/c mice were purchased from Janvier Laboratory (France) and kept in the Nantes SFR (Structure Fédérative de Recherche) Bonamy animal facility. Rag1−/− mice were provided by R.D. and IL-10−/− mice by F. Chain. Animal housing and surgical procedures were conducted according to the guidelines of the French Agriculture Ministry and were approved by the regional ethical committee [Autorisation de Projet utilisant des Animaux à des Fins Scientifiques (APAFIS) 5978, 9851, and 13136]. Colon carcinoma MC38 and CT26 cells were cultured in RPMI 1640 (Life Technologies), 10% fetal bovine serum, glutamine, and antibiotics at 37°C and 5% CO2. Cells were harvested and resuspended in phosphate-buffered saline (PBS) before inoculation into mice. The hIgG1 control mAb was purchased from Evitria (MOTA-hIgG1) and anti-RvE1 from Cayman Biochemicals. The anti-ChemR23 mAb was produced and purified by OSE Immunotherapeutics.
ChemR23 expression and PMN infiltration in colon of patients with IBD
Gene expression datasets were obtained from the Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/geo/). GSE16879 (42), GSE12251 (43), and GSE73661 (44) for colon biopsies from UC patients after treatment with immunosuppressants and/or corticosteroids; GSE16879 (42) for colon and ileal biopsies from CD patients before and 4 to 6 weeks after treatment with anti-TNF (infliximab); GSE73661 (44) for colon biopsies from UC patients before and 4 to 6 weeks after treatment with anti-TNF (infliximab), and UC patients before, 6 weeks, and 52 weeks after treatment with anti-α4β7 (vedolizumab). In all cohorts, anti-TNF or anti-α4β7 responses were defined by complete histological and endoscopic healing. Batch effect for the different cohorts was removed with Conbat. For deconvolution, the R package immunedeconv was used (https://grst.github.io/immunedeconv). To determine the fraction of neutrophils and M1/M2 macrophages in samples, we used two methods: quanTIseq and CIBERSORT. We specified that expression data were from nontumor samples (parameter tumor = FALSE) and from microarrays, instead of RNA-seq (parameters arrays = TRUE and rmgenes = “none”). Hematoxylin and eosin (H&E) colon sections from diagnosis and routine care of patients with IBD were collected from the Service of AnatomoCytoPathologie of the Nantes CHU (Centre Hospitalier Universitaire) Hospital and analyzed by a pathologist for the Robarts Histopathological Index [as previously described (87)] and ChemR23 staining (1A7 clone).
ChemR23 expression analysis
Human peripheral blood mononuclear cells (hPBMCs) isolated from the blood of HVs by Ficoll gradient and mouse bone marrow cells from male C57BL/6J mice were cultured for different times with either recombinant human or mouse TNFα at 100 U/ml (R&D Systems), IL-6 at 10 ng/ml (R&D Systems), LPS at 100 ng/ml (Sigma-Aldrich), or IL-8 at 50 ng/ml (R&D Systems). ChemR23 expression was assessed by flow cytometry with anti-human ChemR23 Ab (clone 84939) and anti-mouse ChemR23 Ab (clone 477806). To identify PMNs (Polymorphonuclear neutrophils), monocytes, and DCs, we used, respectively, anti-CD66b (clone Ms), anti-CD14 (clone M5E2), and anti-CD11c (clone Bu15) for human and anti-Ly6G (clone 1A8) and anti-CD11b (clone M1/70) for mouse.
Murine dorsal air pouch model
Dorsal air pouches were formed on male Balb/c mice (6 to 8 weeks old) by injecting subcutaneously 3 ml of sterile air in day 0 and day 3. Mice were then intraperitoneally injected with anti-ChemR23 mAb or hIgG1 control mAb at 1 mg/kg or with RvE1 (50 μg/kg) on successive days at d5 and d6. On day 6, inflammation was induced by intrapouch injection of recombinant murine TNF (50 ng) or carrageenan (1%/1 ml per pouch). Pouch lavages (PBS-EDTA 2 mM) were collected at 4, 8, 24, or 48 hours, and cells were stained for phenotyping by flow cytometry. F4/80 (BM8), Ly6G (1A8), and anti-mouse ChemR23 (clone 477806) were used to identify macrophages, PMN, and ChemR23 expression, respectively. PMN mortality was evaluated with a viability dye (Life Technologies). The extraction protocol and analysis of bioactive lipids were performed as previously described (88) and adapted according to the Ambiotis SAS (Toulouse, France) standard operating procedure. The liquid chromatography–tandem mass spectrometry experiment was performed on an ExionLC™ AD Ultra-High-Performance Liquid Chromatography system (Sciex) coupled to a QTRAP 6500+ MS (Sciex) and equipped with electrospray ionization source and performed in negative ion mode.
Macrophages polarization
Human monocytes were isolated by magnetic sorting from PBMCs of HVs (classical monocyte isolation kit, Miltenyi Biotec). Macrophages were generated with human macrophage colony-stimulating factor (M-CSF) (100 ng/ml) in RPMI 1640 (Gibco) supplemented with serum, penicillin/streptomycin, and glutamine for 5 days at 37°C and 5% CO2. Macrophages were polarized for 2 days either with human IFNγ (20 ng/ml) to obtain M1 inflammatory macrophages or with human TGFβ (50 ng/ml) to obtain M2c anti-inflammatory macrophages and then treated with coated anti-ChemR23 or hIgG1 control mAb (10 μg/ml). Mouse M1 macrophages were obtained from bone marrow after differentiation for 5 days with M-CSF (100 ng/ml), followed by polarization for 2 days with IFNγ (20 ng/ml) in complete RPMI 1640 supplemented with 50 μM β-mercaptoethanol. IL-10 secretion was assessed by ELISA (BD Biosciences) in the supernatant after the 2-day polarization, and phenotyping for CD80, CD86, CCR7, CD206, and CD11b expression was assessed by flow cytometry.
Efferocytosis assay
Freshly isolated human monocytes from HVs were incubated for 2 days at 37°C with M-CSF (100 ng/ml) to drive the differentiation into macrophages. Two-day monocytes and freshly isolated PMNs were labeled for 10 min at 37°C with cell proliferation dye (CPD) eFluor 670 and 450 (Thermo Fisher Scientific), respectively. Macrophages were incubated with the selected mAb (10 μg/ml) for 1 hour and then mixed with PMN at a 1:2 ratio for 60 min. Efferocytosis was quantified by flow cytometry. The percentage of phagocytic macrophages was calculated as the number of CPD e450+ macrophages per total number of macrophages × 100.
Human PMN isolation and related biological assays
PMN were isolated as described previously (89). Briefly, after a Ficoll gradient of blood from HVs, the pellet was collected, the PMN were separated on a dextran gradient, and red blood cells were lysed with water. Then, the PMN were incubated with coated anti-ChemR23 mAb or a control Ab at 10 μg/ml in RPMI 1640 medium. Viability of PMN was assessed using a LIVE/DEAD kit (Thermo Fisher Scientific) after a 24-hour mAb incubation and quantified by microscopy imaging (Zeiss Axiocam). NETosis was evaluated on the same images. Caspase-3/7 activity was estimated by using the CellEvent Caspase-3/7 probe (Thermo Fisher Scientific) at 2 μM after 10 hours of culture with mAbs and quantified by microscopy with a Nikon Ti2 microscope. ROS production was investigated after 5 hours of ChemR23 activation and revealed with 5 μM CM-H2DCFDA (Thermo Fisher Scientific) and 2 μM CellTracker Deep Red (Thermo Fisher Scientific) using a A1RSi confocal microscope (Nikon). All quantifications were realized using five random recorded ×20 pictures and analyzed using Fiji software. CD62L (clone DREG-56) expression was revealed by flow cytometry after 4 hours of mAb incubation, and CD62L shedding was measured by ELISA using a commercial kit (BD).
ChemR23 signaling
PMN (1.5 × 106/ml) or M1 macrophages (1.5 × 106/ml) obtained from hPBMCs or murine bone marrow were stimulated with hIgG1 control or anti-ChemR23 mAbs (10 μg/ml) for different times. Cells were lysed by adding 300 μl of radioimmunoprecipitation assay buffer 1× (Cell Signaling) with protease inhibitor cocktail (Sigma-Aldrich). ChemR23 signaling pathway was analyzed either by WB (western blot) or ELISA. Briefly, 20 μg of proteins was loaded in Mini-PROTEAN TGX gel (Bio-Rad, 10 wells, 4 to 15%) and then transferred onto a microcellulose membrane. After 2 hours of saturation in tris-buffered saline–5% bovine serum albumin, the membranes were incubated with primary antibodies: anti–P-p44/42 MAPK (T202/Y204) (1:1000) or anti–P-Akt (S473) (1:1000) purchased from Cell Signaling, for 1 hour at room temperature. Proteins were incubated with horseradish peroxidase (HRP)–coupled secondary antibodies (anti-rabbit IgG, HRP-linked antibody from Cell Signaling, 1:5000) for 1 hour at room temperature and then revealed with the chemoluminescent Super SignalTM West Femto ECL detection system. WB images and quantification were obtained with Image Lab. For some experiments, P-ERK and P-Akt were measured by ELISA using commercially available kits (R&D Systems).
PMN transmigration
Chemotaxis of PMN was evaluated in a Boyden chamber using a monolayer of human dermal microvascular endothelial cells (HDMECs) (0.2 × 106 cells per well) coated overnight in the upper chamber on a 0.1% gelatine layer. IL-8 (50 ng/ml) was used as the chemoattractant in the bottom part of the chamber, and PMN (3 × 106 cells per well) and Abs (10 μg/ml) were added in the upper part of the chamber. After a 90-min incubation at 37°C in an incubator with 5% CO2, the bottom part of the chamber was collected. Migrated cells were counted by flow cytometry using counting beads (123count eBeads, Thermo Fisher Scientific). CD62L expression (DREG-56) and shedding (R&D Systems) were analyzed by flow cytometry and ELISA, respectively, on PMN incubated with either the anti-ChemR23 or hIgG1 control mAbs at 10 μg/ml for 1 to 4 hours.
Preclinical colitis models
DSS colitis was induced in 8-week-old male C57BL/6J mice by addition of 2.5% (w/v) DSS in drinking water for 6 days. On day 6, DSS supplementation was discontinued, mice were euthanized after clinical recovery, and colon length was measured. Anti-ChemR23 or anti-hIgG1 mAbs were intraperitoneally administered at 1 mg/kg every 2 days from day 0 to day 6. RvE1 was injected intraperitoneally daily at 50 μg/kg starting on d0 for 5 days. TNBS colitis was induced by intrarectal injection of 200 μl of a solution containing 5 mg of 2,4,6-trinitrobenzenesulfonic acid (Sigma-Aldrich) in 50% ethanol (VWR International). Mice were administered with anti-ChemR23 or anti-hIgG1 mAbs intraperitoneally at 1 mg/kg on day 0 and day 2 (two injections) or RvE1 daily at 50 μg/kg for 3 days starting on d0. Adoptive transfer of 0.5 × 106 CD4+ CD45Rbhigh T cells from C57BL/6J wild-type female mice was conducted by injection intraperitoneally into Rag1KO female mice at 5 weeks old. Anti-ChemR23 mAb or hIgG1 control mAb was administered when the mice developed the disease by intraperitoneal injection three times a week for 3 weeks. IL-10 KO mice were curatively injected intraperitoneally with anti-ChemR23 or hIgG1 (1 mg/kg) three times a week for 2 weeks. The Abs were administrated when the mice lost 5% of their initial weight and/or presented a stool score > 1. Disease appearance was observed by weighting the mice three times a week and evaluating the stool score as follows: 0 > normal consistency; 1 > soft consistency but still formed; 2 > diarrhea but still formed; 3 > diarrhea; 4 > diarrhea with blood. Mice were euthanized at 25% of body weight loss. Ri was determined for each stool score as previously described (71). Tmax and T50 of stool score were measured, and Ri was calculated as follows: Tmax − T50, thus representing the required time necessary to recover 50% of maximal stool score.
Preclinical tumor models
Colorectal carcinoma MC38 cells or CT26 (0.5 × 106 cells per mouse) were injected subcutaneously in 8-week-old C57BL/6J male mice or 8-week-old Balb/c female mice, respectively. Tumor development was measured three times a week and calculated as follows: (length × width) 1.5 × 0.52. Anti-ChemR23 or hIgG1 control mAbs were administered at 1 mg/kg when the tumor volume was between 50 and 100 mm3 for MC38 and from d7 after tumor inoculation for CT26 for 3 weeks. For the azoxymethane (AOM)–DSS model, 7-week-old female C57BL/6J mice were intraperitoneally injected with azoxymethane (7.5 mg/kg, Sigma-Aldrich). Then, three 5-day cycles of 1% DSS in drinking water separated by 14-day cycle of regular drinking water were performed starting 5 days after AOM injection. Eighty days after AOM injection, the mice were euthanized, colon length was measured, and the aberrant crypt foci were counted.
Colon-, spleen-, and bone marrow–infiltrating leukocyte immunophenotyping
To collect colonic immune infiltrating cells, colon pieces were incubated in 10 ml of solution 1 [PBS, 30 mM EDTA (ethylenediaminetetraacetic acid), and 1.5 mM dithiothreitol] for 10 min at 37°C under agitation. After filtration, colon pieces were treated with 10 ml of solution 2 (PBS and EDTA 30 mM) for 10 min at 37°C under agitation and lastly filtered and incubated in 5 ml of collagenase D (Sigma-Aldrich) and 25 μl of deoxyribonuclease I (Sigma-Aldrich) solution for 15 min at 37°C under agitation. Splenocytes and bone marrow cells were isolated after lysis of red blood cells, and immune cells were immunophenotyped. Cell mortality was evaluated with a viability dye (Life Technologies). Leukocytes were stained with fluorochrome-conjugated anti-mouse antibodies: F4/80 (BM8), CD11b (M1/70), I/Ab (AF6-120.1), CD3e (500A8), CD4 (RM4-5), CD8 (53-6.7), NK1.1 (PK136), Ly6G (1A8), and CD19 (1D3), all from BD Pharmingen and anti-mouse ChemR23 (clone 477806) from R&D Systems. Fluorescence-activated cell sorting analysis was conducted using a BD Pharmingen LSR II, Canto II flow cytometers, and FlowJo software (Tree Star).
Immunohistochemistry
Colon samples from Rag1KO mice were frozen in Tissue-Tek (Thermo Fisher Scientific) and cut into 10-μm sections. Immunofluorescence was performed with an anti-mouse CD3 Ab (500A2) and an anti-mouse Ly6G (1A8). Sections were scanned and analyzed using standard fluorescence microscopy and NDPview2 imaging software (Hamamatsu). T cell and PMN infiltration was analyzed by measuring the fluorescence intensity in the different channels or manually counted and related to the tissue surface or DAPI (4′,6-diamidino-2-phenylindole)–positive cells, respectively. Tissue morphology (inflammation and vasculitis) and fibrosis were revealed by H&E staining or Masson’s trichrome staining, respectively. Submucosa and mucosa thickness was determined using NDPview2 imaging software.
Nanostring analysis
RNAs from mouse colon of DSS, TNBS, or Rag1KO preclinical models were extracted in the RLT buffer with an RNeasy Mini kit (Qiagen) supplemented by β-mercaptoethanol at 1%. Gene expression was quantified with the NanoString nCounter platform using 50 ng of total RNA in the nCounter Mouse Immunology Panel (NanoString Technologies). The code set was hybridized with the RNA overnight at 65°C. RNA transcripts were immobilized and counted using the NanoString nCounter Sprint. Normalized expression data were analyzed with the nSolver software. Standardized not log2-transformed counts were used for differential gene expression analysis with the R package DESeq2 [Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014)]. Genes were ranked in order of differential expression and P value score. Gene set enrichment analysis was performed with the GSEA software with 1000 permutations.
Statistical analysis
Continuous variables are expressed as means ± SEM unless otherwise indicated and compared with the nonparametric Mann-Whitney two-sided test or Kruskal-Wallis tests with Dunn’s ad hoc pairwise comparisons for more than two groups. Graft survival was calculated using the Kaplan-Meier method. The log-rank test was used to compare survival times between different groups. The Spearman test was used for correlation analyses. P values of <0.05 were considered statistically significant. All statistical analyses were performed on GraphPad software (GraphPad Software, San Diego, CA).
Acknowledgments
Funding: This work was supported by OSE Immunotherapeutics. We thank the MicroPICell cellular and tissular imaging core facility of Nantes University. We thank F. Chain who provided the IL10−/− mice. We thank the biological resource center for biobanking [CHU Nantes, Nantes Université, Centre de ressources biologiques (BB-0033-00040), F-44000 Nantes, France]. Author contributions: Conceived the study: N.P. and B.V. Designed and supervised the experiments: N.P., V.G., and B.V. Performed the experiments: C.T., V.G., K.B., L.B., C.M., G.T., V.T., G.C., A.P., S.P., M.N., S.B., A.G., and A.N. Analyzed data: C.T., V.G., R.D., G.R.-S., I.G., J.F.M., M.D., B.V., G.B., and NP. Wrote the paper: C.T., V.G., B.V., and N.P. Competing interests: C.T., V.G., C.M., B.V., M.D., and N.P. are inventors on a patent application related to this work filed by OSE Immunotherapeutics (no. WO 2019/193029, filed 3 April 2019, published 8 October 2019). The authors declare that they have no other competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/14/eabd1453/DC1
REFERENCES AND NOTES
- 1.Khor B., Gardet A., Xavier R. J., Genetics and pathogenesis of inflammatory bowel disease. Nature 474, 307–317 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abraham C., Cho J. H., Inflammatory bowel disease. N. Engl. J. Med. 361, 2066–2078 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Danese S., Fiocchi C., Ulcerative colitis. N. Engl. J. Med. 365, 1713–1725 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Baumgart D. C., Sandborn W. J., Crohn’s disease. Lancet 380, 1590–1605 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Ungaro R., Mehandru S., Allen P. B., Peyrin-Biroulet L., Colombel J.-F., Ulcerative colitis. Lancet 389, 1756–1770 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Torres J., Mehandru S., Colombel J.-F., Peyrin-Biroulet L., Crohn’s disease. Lancet 389, 1741–1755 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Allez M., Karmiris K., Louis E., Van Assche G., Ben-Horin S., Klein A., Van der Woude J., Baert F., Eliakim R., Katsanos K., Brynskov J., Steinwurz F., Danese S., Vermeire S., Teillaud J.-L., Lémann M., Chowers Y., Report of the ECCO pathogenesis workshop on anti-TNF therapy failures in inflammatory bowel diseases: Definitions, frequency and pharmacological aspects. J. Crohns Colitis 4, 355–366 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Singh S., George J., Boland B. S., Casteele N. V., Sandborn W. J., Primary non-response to tumor necrosis factor antagonists is associated with inferior response to second-line biologics in patients with inflammatory bowel diseases: A systematic review and meta-analysis. J. Crohns Colitis 12, 635–643 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Serhan C. N., Levy B. D., Resolvins in inflammation: Emergence of the pro-resolving superfamily of mediators. J. Clin. Invest. 128, 2657–2669 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ortega-Gómez A., Perretti M., Soehnlein O., Resolution of inflammation: An integrated view. EMBO Mol. Med. 5, 661–674 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sugimoto M. A., Sousa L. P., Pinho V., Perretti M., Teixeira M. M., Resolution of inflammation: What controls its onset? Front. Immunol. 7, 160 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fullerton J. N., Gilroy D. W., Resolution of inflammation: A new therapeutic frontier. Nat. Rev. Drug Discov. 15, 551–567 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Cash J. L., Bena S., Headland S. E., Arthur S. M., Brancaleone V., Perretti M., Chemerin15 inhibits neutrophil-mediated vascular inflammation and myocardial ischemia-reperfusion injury through ChemR23. EMBO Rep. 14, 999–1007 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cash J. L., Hart R., Russ A., Dixon J. P. C., Colledge W. H., Doran J., Hendrick A. G., Carlton M. B. L., Greaves D. R., Synthetic chemerin-derived peptides suppress inflammation through ChemR23. J. Exp. Med. 205, 767–775 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perretti M., Chiang N., La M., Fierro I. M., Marullo S., Getting S. J., Solito E., Serhan C. N., Endogenous lipid- and peptide-derived anti-inflammatory pathways generated with glucocorticoid and aspirin treatment activate the lipoxin A4 receptor. Nat. Med. 8, 1296–1302 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Serhan C. N., Yacoubian S., Yang R., Anti-inflammatory and pro-resolving lipid mediators. Annu. Rev. Pathol. 3, 279–312 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiang N., Libreros S., Norris P. C., de la Rosa X., Serhan C. N., Maresin 1 activates LGR6 receptor promoting phagocyte immunoresolvent functions. J. Clin. Invest. 129, 5294–5311 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tjonahen E., Oh S. F., Siegelman J., Elangovan S., Percarpio K. B., Hong S., Arita M., Serhan C. N., Resolvin E2: Identification and anti-inflammatory actions: Pivotal role of human 5-lipoxygenase in resolvin E series biosynthesis. Chem. Biol. 13, 1193–1202 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Dalli J., Winkler J. W., Colas R. A., Arnardottir H., Cheng C.-Y. C., Chiang N., Petasis N. A., Serhan C. N., Resolvin D3 and aspirin-triggered resolvin D3 are potent immunoresolvents. Chem. Biol. 20, 188–201 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kebir D. E., Gjorstrup P., Filep J. G., Resolvin E1 promotes phagocytosis-induced neutrophil apoptosis and accelerates resolution of pulmonary inflammation. Proc. Natl. Acad. Sci. U.S.A. 109, 14983–14988 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schett G., Neurath M. F., Resolution of chronic inflammatory disease: Universal and tissue-specific concepts. Nat. Commun. 9, 3261 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buckley C. D., Gilroy D. W., Serhan C. N., Proresolving lipid mediators and mechanisms in the resolution of acute inflammation. Immunity 40, 315–327 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koedel U., Frankenberg T., Kirschnek S., Obermaier B., Häcker H., Paul R., Häcker G., Apoptosis Is essential for neutrophil functional shutdown and determines tissue damage in experimental pneumococcal meningitis. PLOS Pathog. 5, e1000461 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kennedy A. D., DeLeo F. R., Neutrophil apoptosis and the resolution of infection. Immunol. Res. 43, 25–61 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Brannigan A. E., O’Connell P. R., Hurley H., O’Neill A., Brady H. R., Fitzpatrick J. M., Watson R. W., Neutrophil apoptosis is delayed in patients with inflammatory bowel disease. Shock 13, 361–366 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Butin-Israeli V., Bui T. M., Wiesolek H. L., Mascarenhas L., Lee J. J., Mehl L. C., Knutson K. R., Adam S. A., Goldman R. D., Beyder A., Wiesmuller L., Hanauer S. B., Sumagin R., Neutrophil-induced genomic instability impedes resolution of inflammation and wound healing. J. Clin. Invest. 129, 712–726 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Therrien A., Chapuy L., Bsat M., Rubio M., Bernard G., Arslanian E., Orlicka K., Weber A., Panzini B.-P., Dorais J., Bernard E.-J., Soucy G., Bouin M., Sarfati M., Recruitment of activated neutrophils correlates with disease severity in adult Crohn’s disease. Clin. Exp. Immunol. 195, 251–264 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pai R. K., Hartman D. J., Rivers C. R., Regueiro M., Schwartz M., Binion D. G., Pai R. K., Complete resolution of mucosal neutrophils associates with improved long-term clinical outcomes of patients with ulcerative colitis. Clin. Gastroenterol. Hepatol. 18, 2510–2517.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Mangino M. J., Brounts L., Harms B., Heise C., Lipoxin biosynthesis in inflammatory bowel disease. Prostaglandins Other Lipid Mediat. 79, 84–92 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Gobbetti T., Dalli J., Colas R. A., Canova D. F., Aursnes M., Bonnet D., Alric L., Vergnolle N., Deraison C., Hansen T. V., Serhan C. N., Perretti M., Protectin D1n-3 DPA and resolvin D5n-3 DPA are effectors of intestinal protection. Proc. Natl. Acad. Sci. U.S.A. 114, 3963–3968 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vong L., Ferraz J. G. P., Dufton N., Panaccione R., Beck P. L., Sherman P. M., Perretti M., Wallace J. L., Up-regulation of annexin-A1 and lipoxin A4 in individuals with ulcerative colitis may promote mucosal homeostasis. PLOS ONE 7, e39244 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ağış E. R., Savaş B., Melli M., Impact of colonic mucosal lipoxin A4 synthesis capacity on healing in rats with dextran sodium sulfate-induced colitis. Prostaglandins Other Lipid Mediat. 121, 63–69 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Arita M., Yoshida M., Hong S., Tjonahen E., Glickman J. N., Petasis N. A., Blumberg R. S., Serhan C. N., Resolvin E1, an endogenous lipid mediator derived from omega-3 eicosapentaenoic acid, protects against 2,4,6-trinitrobenzene sulfonic acid-induced colitis. Proc. Natl. Acad. Sci. U.S.A. 102, 7671–7676 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ishida T., Yoshida M., Arita M., Nishitani Y., Nishiumi S., Masuda A., Mizuno S., Takagawa T., Morita Y., Kutsumi H., Inokuchi H., Serhan C. N., Blumberg R. S., Azuma T., Resolvin E1, an endogenous lipid mediator derived from eicosapentaenoic acid, prevents dextran sulfate sodium induced colitis. Inflamm. Bowel Dis. 16, 87–95 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herová M., Schmid M., Gemperle C., Hersberger M., ChemR23, the receptor for chemerin and resolvin E1, is expressed and functional on M1 but not on M2 macrophages. J. Immunol. 194, 2330–2337 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Samson M., Edinger A. L., Stordeur P., Rucker J., Verhasselt V., Sharron M., Govaerts C., Mollereau C., Vassart G., Doms R. W., Parmentier M., ChemR23, a putative chemoattractant receptor, is expressed in monocyte-derived dendritic cells and macrophages and is a coreceptor for SIV and some primary HIV-1 strains. Eur. J. Immunol. 28, 1689–1700 (1998). [DOI] [PubMed] [Google Scholar]
- 37.Arita M., Bianchini F., Aliberti J., Sher A., Chiang N., Hong S., Yang R., Petasis N. A., Serhan C. N., Stereochemical assignment, antiinflammatory properties, and receptor for the omega-3 lipid mediator resolvin E1. J. Exp. Med. 201, 713–722 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Serhan C. N., Hong S., Gronert K., Colgan S. P., Devchand P. R., Mirick G., Moussignac R.-L., Resolvins: A family of bioactive products of omega-3 fatty acid transformation circuits initiated by aspirin treatment that counter proinflammation signals. J. Exp. Med. 196, 1025–1037 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Unno Y., Sato Y., Fukuda H., Ishimura K., Ikeda H., Watanabe M., Tansho-Nagakawa S., Ubagai T., Shuto S., Ono Y., Resolvin E1, but not resolvins E2 and E3, promotes fMLF-induced ROS generation in human neutrophils. FEBS Lett. 592, 2706–2715 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Ohira T., Arita M., Omori K., Recchiuti A., Van Dyke T. E., Serhan C. N., Resolvin E1 receptor activation signals phosphorylation and phagocytosis. J. Biol. Chem. 285, 3451–3461 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Belarif L., Danger R., Kermarrec L., Nerrière-Daguin V., Pengam S., Durand T., Mary C., Kerdreux E., Gauttier V., Kucik A., Thepenier V., Martin J. C., Chang C., Rahman A., Guen N. S.-L., Braudeau C., Abidi A., David G., Malard F., Takoudju C., Martinet B., Gérard N., Neveu I., Neunlist M., Coron E., Mac Donald T. T., Desreumaux P., Mai H.-L., Le Bas-Bernardet S., Mosnier J.-F., Merad M., Josien R., Brouard S., Soulillou J.-P., Blancho G., Bourreille A., Naveilhan P., Vanhove B., Poirier N., IL-7 receptor influences anti-TNF responsiveness and T cell gut homing in inflammatory bowel disease. J. Clin. Invest. 129, 1910–1925 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arijs I., De Hertogh G., Lemaire K., Quintens R., Van Lommel L., Van Steen K., Leemans P., Cleynen I., Van Assche G., Vermeire S., Geboes K., Schuit F., Rutgeerts P., Mucosal gene expression of antimicrobial peptides in inflammatory bowel disease before and after first infliximab treatment. PLOS ONE 4, e7984 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arijs I., Li K., Toedter G., Quintens R., Van Lommel L., Van Steen K., Leemans P., De Hertogh G., Lemaire K., Ferrante M., Schnitzler F., Thorrez L., Ma K., Song X.-Y. R., Marano C., Van Assche G., Vermeire S., Geboes K., Schuit F., Baribaud F., Rutgeerts P., Mucosal gene signatures to predict response to infliximab in patients with ulcerative colitis. Gut 58, 1612–1619 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Arijs I., De Hertogh G., Lemmens B., Van Lommel L., de Bruyn M., Vanhove W., Cleynen I., Machiels K., Ferrante M., Schuit F., Van Assche G., Rutgeerts P., Vermeire S., Effect of vedolizumab (anti-α4β7-integrin) therapy on histological healing and mucosal gene expression in patients with UC. Gut 67, 43–52 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Kamada N., Hisamatsu T., Okamoto S., Chinen H., Kobayashi T., Sato T., Sakuraba A., Kitazume M. T., Sugita A., Koganei K., Akagawa K. S., Hibi T., Unique CD14+ intestinal macrophages contribute to the pathogenesis of Crohn disease via IL-23/IFN-γ axis. J. Clin. Invest. 118, 2269–2280 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu H., Dasgupta S., Fu Y., Bailey B., Roy C., Lightcap E., Faustin B., Subsets of mononuclear phagocytes are enriched in the inflamed colons of patients with IBD. BMC Immunol. 20, 42 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martin J. C., Chang C., Boschetti G., Ungaro R., Giri M., Grout J. A., Gettler K., Chuang L.-S., Nayar S., Greenstein A. J., Dubinsky M., Walker L., Leader A., Fine J. S., Whitehurst C. E., Mbow M. L., Kugathasan S., Denson L. A., Hyams J. S., Friedman J. R., Desai P. T., Ko H. M., Laface I., Akturk G., Schadt E. E., Salmon H., Gnjatic S., Rahman A. H., Merad M., Cho J. H., Kenigsberg E., Single-cell analysis of Crohn’s disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy. Cell 178, 1493–1508.e20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smillie C. S., Biton M., Ordovas-Montanes J., Sullivan K. M., Burgin G., Graham D. B., Herbst R. H., Rogel N., Slyper M., Waldman J., Sud M., Andrews E., Velonias G., Haber A. L., Jagadeesh K., Vickovic S., Yao J., Stevens C., Dionne D., Nguyen L. T., Villani A.-C., Hofree M., Creasey E. A., Huang H., Rozenblatt-Rosen O., Garber J. J., Khalili H., Desch A. N., Daly M. J., Ananthakrishnan A. N., Shalek A. K., Xavier R. J., Regev A., Intra- and Inter-cellular rewiring of the human colon during ulcerative colitis. Cell 178, 714–730.e22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Freire M., Dalli J., Charles S. N., Van Dyke T. E., Neutrophil resolvin E1 receptor expression and function in type 2 diabetes. J. Immunol. 198, 718–728 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Keinan D., Leigh N. J., Nelson J. W., De Oleo L., Baker O. J., Understanding resolvin signaling pathways to improve oral health. Int. J. Mol. Sci. 14, 5501–5518 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Serhan C. N., Krishnamoorthy S., Recchiuti A., Chiang N., Novel anti-inflammatory--pro-resolving mediators and their receptors. Curr. Top. Med. Chem. 11, 629–647 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arita M., Ohira T., Sun Y.-P., Elangovan S., Chiang N., Serhan C. N., Resolvin E1 selectively interacts with leukotriene B4 receptor BLT1 and ChemR23 to regulate inflammation. J. Immunol. 178, 3912–3917 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Bourquard T., Musnier A., Puard V., Tahir S., Ayoub M. A., Jullian Y., Boulo T., Gallay N., Watier H., Bruneau G., Reiter E., Crépieux P., Poupon A., MAbTope: A method for improved epitope mapping. J. Immunol. 201, 3096–3105 (2018). [DOI] [PubMed] [Google Scholar]
- 54.Scheinin T., Butler D. M., Salway F., Scallon B., Feldmann M., Validation of the interleukin-10 knockout mouse model of colitis: Antitumour necrosis factor-antibodies suppress the progression of colitis. Clin. Exp. Immunol. 133, 38–43 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stewart R., Hammond S. A., Oberst M., Wilkinson R. W., The role of Fc gamma receptors in the activity of immunomodulatory antibodies for cancer. J. Immunother. Cancer 2, 29 (2014). [Google Scholar]
- 56.Averboukh F., Ziv Y., Kariv Y., Zmora O., Dotan I., Klausner J. M., Rabau M., Tulchinsky H., Colorectal carcinoma in inflammatory bowel disease: A comparison between Crohn’s and ulcerative colitis. Colorectal Dis. 13, 1230–1235 (2011). [DOI] [PubMed] [Google Scholar]
- 57.Luo W. Y., Bryant A. K., Stringfield S., Murphy J. D., Eisenstein S., Effects of anti-tumor necrosis factor use on colorectal cancer outcomes in patients with inflammatory bowel disease. J. Am. Coll. Surg. 227, e105 (2018). [Google Scholar]
- 58.Serhan C. N., Resolution phase of inflammation: Novel endogenous anti-inflammatory and proresolving lipid mediators and pathways. Annu. Rev. Immunol. 25, 101–137 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Serhan C. N., Chiang N., Van Dyke T. E., Resolving inflammation: Dual anti-inflammatory and pro-resolution lipid mediators. Nat. Rev. Immunol. 8, 349–361 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Serhan C. N., Pro-resolving lipid mediators are leads for resolution physiology. Nature 510, 92–101 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sugimoto M. A., Vago J. P., Perretti M., Teixeira M. M., Mediators of the resolution of the inflammatory response. Trends Immunol. 40, 212–227 (2019). [DOI] [PubMed] [Google Scholar]
- 62.de Paiva C. S., Schwartz C. E., Gjörstrup P., Pflugfelder S. C., Resolvin E1 (RX-10001) reduces corneal epithelial barrier disruption and protects against goblet cell loss in a murine model of dry eye. Cornea 31, 1299–1303 (2012). [DOI] [PubMed] [Google Scholar]
- 63.Cholkar K., Trinh H. M., Vadlapudi A. D., Wang Z., Pal D., Mitra A. K., Interaction studies of resolvin E1 analog (RX-10045) with efflux transporters. J. Ocul. Pharmacol. Ther. 31, 248–255 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Torricelli A. A. M., Santhanam A., Agrawal V., Wilson S. E., Resolvin E1 analog RX-10045 0.1% reduces corneal stromal haze in rabbits when applied topically after PRK. Mol. Vis. 20, 1710–1716 (2014). [PMC free article] [PubMed] [Google Scholar]
- 65.Fournier B. M., Parkos C. A., The role of neutrophils during intestinal inflammation. Mucosal Immunol. 5, 354–366 (2012). [DOI] [PubMed] [Google Scholar]
- 66.Kolaczkowska E., Kubes P., Neutrophil recruitment and function in health and inflammation. Nat. Rev. Immunol. 13, 159–175 (2013). [DOI] [PubMed] [Google Scholar]
- 67.Moschen A. R., Tilg H., Raine T., IL-12, IL-23 and IL-17 in IBD: Immunobiology and therapeutic targeting. Nat. Rev. Gastroenterol. Hepatol. 16, 185–196 (2019). [DOI] [PubMed] [Google Scholar]
- 68.Perretti M., Solito E., Annexin 1 and neutrophil apoptosis. Biochem. Soc. Trans. 32, 507–510 (2004). [DOI] [PubMed] [Google Scholar]
- 69.Bressenot A., Salleron J., Bastien C., Danese S., Boulagnon-Rombi C., Peyrin-Biroulet L., Comparing histological activity indexes in UC. Gut 64, 1412–1418 (2015). [DOI] [PubMed] [Google Scholar]
- 70.Naito Y., Takagi T., Yoshikawa T., Neutrophil-dependent oxidative stress in ulcerative colitis. J. Clin. Biochem. Nutr. 41, 18–26 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bannenberg G. L., Chiang N., Ariel A., Arita M., Tjonahen E., Gotlinger K. H., Hong S., Serhan C. N., Molecular circuits of resolution: Formation and actions of resolvins and protectins. J. Immunol. 174, 4345–4355 (2005). [DOI] [PubMed] [Google Scholar]
- 72.Sena A., Grishina I., Thai A., Goulart L., Macal M., Fenton A., Li J., Prindiville T., Oliani S. M., Dandekar S., Goulart L., Sankaran-Walters S., Dysregulation of anti-inflammatory annexin A1 expression in progressive crohns disease. PLOS ONE 8, e76969 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Norling L. V., Headland S. E., Dalli J., Arnardottir H. H., Haworth O., Jones H. R., Irimia D., Serhan C. N., Perretti M., Proresolving and cartilage-protective actions of resolvin D1 in inflammatory arthritis. JCI Insight 1, e85922 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Miyata J., Fukunaga K., Iwamoto R., Isobe Y., Niimi K., Takamiya R., Takihara T., Tomomatsu K., Suzuki Y., Oguma T., Sayama K., Arai H., Betsuyaku T., Arita M., Asano K., Dysregulated synthesis of protectin D1 in eosinophils from patients with severe asthma. J. Allergy Clin. Immunol. 131, 353–360.e1-2 (2013). [DOI] [PubMed] [Google Scholar]
- 75.Eickmeier O., Fussbroich D., Mueller K., Serve F., Smaczny C., Zielen S., Schubert R., Pro-resolving lipid mediator Resolvin D1 serves as a marker of lung disease in cystic fibrosis. PLOS ONE 12, e0171249 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang X., Zhu M., Hjorth E., Cortés-Toro V., Eyjolfsdottir H., Graff C., Nennesmo I., Palmblad J., Eriksdotter M., Sambamurti K., Fitzgerald J. M., Serhan C. N., Granholm A.-C., Schultzberg M., Resolution of inflammation is altered in Alzheimer’s disease. Alzheimers Dement. 11, 40–50.e2 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kantarci A., Aytan N., Palaska I., Stephens D., Crabtree L., Benincasa C., Jenkins B. G., Carreras I., Dedeoglu A., Combined administration of resolvin E1 and lipoxin A4 resolves inflammation in a murine model of Alzheimer’s disease. Exp. Neurol. 300, 111–120 (2018). [DOI] [PubMed] [Google Scholar]
- 78.Oh S. Y., Cho K.-A., Kang J. L., Kim K. H., Woo S.-Y., Comparison of experimental mouse models of inflammatory bowel disease. Int. J. Mol. Med. 33, 333–340 (2014). [DOI] [PubMed] [Google Scholar]
- 79.Lee C.-T., Teles R., Kantarci A., Chen T., Cafferty J. M., Starr J. R., Brito L. C. N., Paster B. J., Van Dyke T. E., Resolvin E1 reverses experimental periodontitis and dysbiosis. J. Immunol. 197, 2796–2806 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Campbell E. L., MacManus C. F., Kominsky D. J., Keely S., Glover L. E., Bowers B. E., Scully M., Bruyninckx W. J., Colgan S. P., Resolvin E1-induced intestinal alkaline phosphatase promotes resolution of inflammation through LPS detoxification. Proc. Natl. Acad. Sci. U.S.A. 107, 14298–14303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Basso L., Garnier L., Bessac A., Boué J., Blanpied C., Cenac N., Laffont S., Dietrich G., T-lymphocyte-derived enkephalins reduce Th1/Th17 colitis and associated pain in mice. J. Gastroenterol. 53, 215–226 (2018). [DOI] [PubMed] [Google Scholar]
- 82.Kamada N., Hisamatsu T., Okamoto S., Sato T., Matsuoka K., Arai K., Nakai T., Hasegawa A., Inoue N., Watanabe N., Akagawa K. S., Hibi T., Abnormally differentiated subsets of intestinal macrophage play a key role in Th1-dominant chronic colitis through excess production of IL-12 and IL-23 in response to bacteria. J. Immunol. 175, 6900–6908 (2005). [DOI] [PubMed] [Google Scholar]
- 83.Sulciner M. L., Serhan C. N., Gilligan M. M., Mudge D. K., Chang J., Gartung A., Lehner K. A., Bielenberg D. R., Schmidt B., Dalli J., Greene E. R., Gus-Brautbar Y., Piwowarski J., Mammoto T., Zurakowski D., Perretti M., Sukhatme V. P., Kaipainen A., Kieran M. W., Huang S., Panigrahy D., Resolvins suppress tumor growth and enhance cancer therapy. J. Exp. Med. 215, 115–140 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Panigrahy D., Gartung A., Yang J., Yang H., Gilligan M. M., Sulciner M. L., Bhasin S. S., Bielenberg D. R., Chang J., Schmidt B. A., Piwowarski J., Fishbein A., Soler-Ferran D., Sparks M. A., Staffa S. J., Sukhatme V., Hammock B. D., Kieran M. W., Huang S., Bhasin M., Serhan C. N., Sukhatme V. P., Preoperative stimulation of resolution and inflammation blockade eradicates micrometastases. J. Clin. Invest. 129, 2964–2979 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shaul M. E., Fridlender Z. G., Tumour-associated neutrophils in patients with cancer. Nat. Rev. Clin. Oncol. 16, 601–620 (2019). [DOI] [PubMed] [Google Scholar]
- 86.Gentles A. J., Newman A. M., Liu C. L., Bratman S. V., Feng W., Kim D., Nair V. S., Xu Y., Khuong A., Hoang C. D., Diehn M., West R. B., Plevritis S. K., Alizadeh A. A., The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 21, 938–945 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mosli M. H., Feagan B. G., Zou G., Sandborn W. J., D’Haens G., Khanna R., Shackelton L. M., Walker C. W., Nelson S., Vandervoort M. K., Frisbie V., Samaan M. A., Jairath V., Driman D. K., Geboes K., Valasek M. A., Pai R. K., Lauwers G. Y., Riddell R., Stitt L. W., Levesque B. G., Development and validation of a histological index for UC. Gut 66, 50–58 (2017). [DOI] [PubMed] [Google Scholar]
- 88.Le Faouder P., Baillif V., Spreadbury I., Motta J.-P., Rousset P., Chêne G., Guigné C., Tercé F., Vanner S., Vergnolle N., Bertrand-Michel J., Dubourdeau M., Cenac N., LC–MS/MS method for rapid and concomitant quantification of pro-inflammatory and pro-resolving polyunsaturated fatty acid metabolites. J. Chromatogr. B 932, 123–133 (2013). [DOI] [PubMed] [Google Scholar]
- 89.Kuhns D. B., Priel D. A. L., Chu J., Zarember K. A., Isolation and functional analysis of human neutrophils. Curr. Protoc. Immunol. 111, 7.23.1–7.23.16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Recchiuti A., Mattoscio D., Isopi E., Roles, actions, and therapeutic potential of specialized pro-resolving lipid mediators for the treatment of inflammation in cystic fibrosis. Front. Pharmacol. 10, 252 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/14/eabd1453/DC1