Cortical regions are determined by patterning transcription factors and segregated by differential cell affinity.
Abstract
Development of cortical regions with precise, sharp, and regular boundaries is essential for physiological function. However, little is known of the mechanisms ensuring these features. Here, we show that determination of the boundary between neocortex and medial entorhinal cortex (MEC), two abutting cortical regions generated from the same progenitor lineage, relies on COUP-TFI (chicken ovalbumin upstream promoter–transcription factor I), a patterning transcription factor with graded expression in cortical progenitors. In contrast with the classical paradigm, we found that increased COUP-TFI expression expands MEC, creating protrusions and disconnected ectopic tissue. We further developed a mathematical model that predicts that neuronal specification and differential cell affinity contribute to the emergence of an instability region and boundary sharpness. Correspondingly, we demonstrated that high expression of COUP-TFI induces MEC cell fate and protocadherin 19 expression. Thus, we conclude that a sharp boundary requires a subtle interplay between patterning transcription factors and differential cell affinity.
INTRODUCTION
How the brain acquires appropriate cell diversity and robust functional organization is one of the most prominent questions in the field of neural development. It is known that within the vertebrate central nervous system, some boundaries, such as the midbrain-hindbrain boundary, the zona limitans intrathalmica, or the borders between rhombomeres in the hindbrain, are established between lineage-restricted compartments and sharpened by differential cell adhesion (1). However, the neuromeric organization and clear boundaries have not been delineated in the forebrain, where the cerebral cortex arises; the only forebrain boundary that has been characterized is the pallial-subpallial boundary located between dorsal and ventral telecenphalon. The mammalian cerebral cortex consists of distinct cortical regions, including neocortex (NC), archicortex (AC), paleocortex, and transitional cortices located between these regions, each with unique functions, cytoarchitecture, patterns of gene expression, and input and output projection patterns. In this study, we sought to understand how different cortical regions segregate from each other, focusing on the sharp boundary separating the medial entorhinal cortex (MEC) and NC. Notably, the NC and MEC are two vastly distinct cortices, although both are derived from the same pool of Emx1-lineage cortical progenitors (2).
One central paradigm, the positional information theory or French flag model, posits that gradients of transcription factors (TFs) instruct newly generated neurons to adopt a specific fate (3). In line with this model, several patterning TF gradients were shown to support neuronal specification and area patterning in the NC (4–8). However, a well-known limitation of positional information theory is related to the sharpness and robustness of boundaries, where ambiguous differentiation can lead to irregular, unpredictable, or nonsharp (“salt-and-pepper”) transitions (9). Hence, various phenomena have been proposed to account for boundary regularity, including cell-sorting mechanisms (10), combined information from multiple gradients (11, 12), or cell aggregation and adhesion (13–15). To date, though, there has been no direct evidence to indicate a causal relationship between any of these processes and the regularity of boundaries between cortical regions.
In this work, we identify key molecular mediators of neuronal differentiation that distinguish the MEC and NC and also generate the border between these cortical regions. We show that the concentration of the nuclear receptor COUP-TFI (chicken ovalbumin upstream promoter–TF I; also called NR2F1) in progenitors plays a central role in determining the NC/MEC border. COUP-TFI was previously shown to be a key determinant of cortical development, as COUP-TFI mutant mice have prominent defects in neuronal specification and cortical and hippocampal patterning (16–21). Furthermore, de novo mutations in the human COUP-TFI gene cause Bosch-Boonstra-Schaaf optic atrophy syndrome, which is characterized by cortical malformation and various sensory and cognitive deficits (22–24). We confirmed that lowering the COUP-TFI expression level caudally shifted the NC/MEC border, in agreement with previous findings (18). Moreover, we demonstrated that a lack of COUP-TFI reduced the sharpness of the NC/MEC border. On the other hand, mutant mice with COUP-TFI overexpression exhibited not only an expansion of the MEC at the expense of NC but also a dislocation of the boundary and emergence of protrusions or disconnected ectopic MEC regions. Further theoretical and experimental investigations then revealed that differential cell adhesion enhances sharpness of the boundary, and excessive adhesion induces the emergence of irregular boundaries. Together, these findings demonstrate that the level of patterning TF expression can determine region-specific neuronal properties and the expression of region-specific adhesion molecules. The adhesion molecules are essential to the formation of boundaries between cortical regions, but when the differential cell adhesion is too much, the boundary may break down. Thus, the emergence of sharp boundaries in vivo requires subtle interplay between patterning TF gradients and cell adhesion.
RESULTS
Distinct gene expression profiles between NC and MEC
To study boundary formation, we focused on the border between the NC and the MEC. Although NC and MEC both consist of six layers and both are derived from the Emx1 lineage (fig. S1A), a clear cytoarchitectural border can be detected between the regions in the adult cortex (Fig. 1, A and B). Assuming that distinct cellular properties in MEC and NC segregate these two structures, we first compared the gene expression profiles in NC and MEC. We found that 2039 genes were enriched in NC and 1507 genes were enriched in MEC (Fig. 1C). These genes included some involved in nervous system development [Gene Ontology (GO):0048666], neuronal differentiation (GO:0030182), and synaptic signaling (GO:0099536) (Fig. 1D), suggesting that adult NC and MEC show substantial differences in neuronal properties. From the list of NC/MEC differentially expressed genes, we identified a battery that labels specific layers in the MEC or NC, including genes encoding TFs, such as Ctip2, Lhx2, Tbr1, Satb2, and Nurr1; cell surface molecules, such as Reelin (Rln); neurofilaments, such as Nef3; and genes involved in cellular signaling and neuronal activity, Kitl, Vglut2 (Slc17a6), Calb1 (CB), Wfs1, and Nts. Most of these factors play important roles in cortical development (26–29). More specifically, many of the genes, especially Reelin, Wfs1, and Calb1, have been shown to label specific MEC cell types (30, 31), and their differential expression patterns could be used to define the border between NC and MEC (Fig. 1E and fig. S1C).
Fig. 1. Differential gene expression in NC and MEC.
Nissl-stained sagittal sections of adult (P56) mouse cortex and its higher-magnification view around the border region between NC and MEC are shown in (A) and (B). (C) RNA sequencing (RNA-seq) analyses were performed with RNA from P56 NC and MEC. Red and blue dots in dot plots show MEC- and NC-enriched genes, respectively. (D) Top GO terms and their P values are shown for these differentially expressed genes. (E) Nissl-stained sagittal section of P7 mouse cortex shows a clear border between NC and MEC (indicated by arrowhead). Images at higher magnification around border region from RNA in situ hybridization and immunostaining reveal genes with a differential expression pattern between MEC and NC. Nissl-stained sagittal sections of embryonic (E17.5) mouse cortex and its higher magnification view around the border region between NC and AC are shown in (F) and (G). (H) RNA-seq analyses were performed with RNA from E17.5 NC and AC. Red and blue dots in dot plots show AC- and NC-enriched genes, respectively. (I) Top GO terms and their P values are shown for these differentially expressed genes. Overlap of these differentially expressed genes between P56 and E17.5 is shown in (J). (K) From the list of the differentially expressed genes, a battery of genes was identified and showed significantly differential expression between MEC/AC and NC. (L) Costaining of Satb2 (NC marker) and Nurr1 (MEC marker) reveals a sharp molecular change at the border between NC and MEC in both P7 and E17.5 cortices. FPKM, fragments per kilobase of transcript per million; FDR, false discovery rate; Hp, hippocampus. Scale bars, 500 μm (A), and 400 μm (E, F, and L).
To determine when these genetic differences between NC and MEC were established, we examined gene expression profiles in these cortical structures at embryonic stages. At embryonic day 17.5 (E17.5), a time point when most cortical neurons have been generated but before cortical neurons receive stimulation from the periphery, we could not define an apparent NC/MEC border by Nissl staining (Fig. 1, F and G). However, we did find that differences in gene expression between NC and AC (the caudal part of the dorsal telencephalon, including hippocampus and MEC) could be detected at E17.5, similar to those detected in adult (Fig. 1H). These embryonic NC/AC differentially expressed genes were enriched in nervous system development (GO:0048666), neuron differentiation (GO:0030182), cell migration (GO:0016477), and cell adhesion (GO:0007155) (Fig. 1I and fig. S1B), suggesting that distinct regional neuronal properties are established early during neurogenesis. Comparing the NC/MEC and NC/AC differentially expressed genes from adult and E17.5, respectively, we found many that were present in both lists (Fig. 1J). Most of the genes showed consistent differential expression patterns in E17.5 and adult; for example, Nurr1 and Nrp2 were enriched in the MEC, while Satb2 and RORb were enriched in the NC (Fig. 1K). However, some genes showed different patterns in E17.5 and adult, such as Lmo4 and Nef3 (Fig. 1K).
By probing NC- and MEC-specific genes, we could detect that a sharp transition at the border of the two structures is already present at E17.5 (Fig. 1L). Thus, we concluded that a molecular boundary between NC and MEC was established during embryonic development, at the position where changes in gene expression were detected.
Decreased COUP-TFI expression posteriorly shifts and degrades the border between MEC and NC
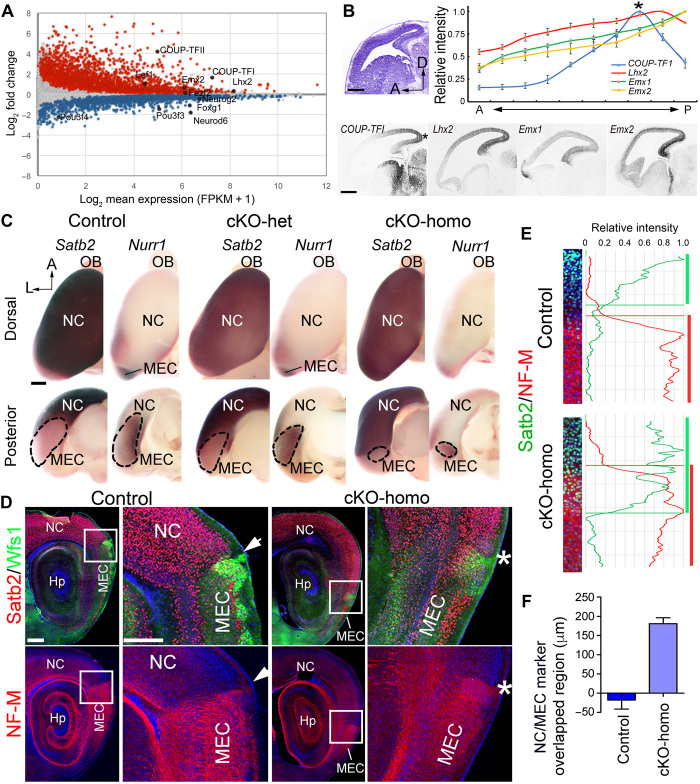
Next, we investigated how the NC/MEC border is established in the progenitors of the Emx1 lineage. Similar to our experiment with E17.5 cortices, we performed RNA sequencing (RNA-seq) analyses to compare gene expression profiles at E13.5 between the presumptive progenitors giving rise to NC and AC. We found 2112 and 1794 genes that were enriched in AC and NC progenitors, respectively (Fig. 2A). Among these differentially expressed genes, we focused on TFs that show notable differential expression in NC and AC progenitors, because TFs are known to play critical roles in specifying positional information in neuronal progenitors. AC-enriched TFs, including COUP-TFI, Emx1, Emx2, and Lhx2, could potentially regulate the formation of NC/MEC border. By quantifying their expression gradients, we unexpectedly found a unique expression pattern for COUP-TFI: It was most highly expressed by cortical progenitors located at the caudal end of the dorsal telencephalic vesicle, where MEC presumably originates (Fig. 2B and fig. S2A). Moreover, along the medial-lateral axis, the high lateral expression of COUP-TFI also correlated with the position where MEC is located relative to NC (fig. S2, B and C). On the basis of these observations, which were corroborated by the fact that conditional knockout (cKO) of COUP-TFI from cortical progenitors results in reduced MEC size (18), we hypothesized that the relatively high level of COUP-TFI expression induces the formation of MEC and therefore creates the border between MEC and NC.
Fig. 2. The expression level of COUP-TFI regulates the position of the border between MEC and NC.
(A) Dot plot of RNA-seq analyses from E13.5 NC and AC shows NC- and AC-enriched genes. (B) Nissl staining and RNA in situ hybridization of multiple patterning TFs were performed on sagittal sections of E13.5 wild-type mice. Relative expression intensities of the patterning TFs are shown along the anterior-posterior (A-P) axis. COUP-TFI showed a high-caudal–to–low-rostral gradient with its highest expression at the posterior end of the dorsal telencephalon (indicated by asterisk). (C) Dorsal and posterior view for P0 control, cKO-het (COUP-TFIf/+;Emx1-Cre), and cKO-homo (COUP-TFIf/f;Emx1-Cre) cortices. Cortices were processed for whole-mount in situ hybridization using probes for Satb2 and Nurr1 to respectively indicate NC and MEC. (D) Immunostaining for Satb2, Wfs1, and NF-M on P7 sagittal sections of control and cKO-homo cortices. Arrowhead and asterisk indicated the border between NC and MEC in the control and cKO-homo cortices, respectively. (E) Relative intensity of Satb2 (green) and NF-M (red) staining around the NC/MEC border in control (top) and cKO-homo (bottom). (F) The length of the green (Satb2+) and red (NF-M+) overlapped region is significantly increased in the cKO-homo, suggesting that the border is less clear in the cKO-homo when compared with that in control. OB, Olfactory bulb. Scale bars, 400 μm (B), 250 μm (C), 500 μm (D, left), and 300 μm (D, right).
To test how COUP-TFI expression level affects the border formation between NC and MEC, we used different conditional alleles to manipulate COUP-TFI expression levels by Emx1-Cre (2) and examined the impact on the size and relative location of MEC. We first generated cKO-het (COUP-TFIf/+;Emx1-Cre) and cKO-homo (COUP-TFIf/f;Emx1-Cre) to lower COUP-TFI expression to different levels in cortical progenitors and neurons (fig. S3A). By E15.5, COUP-TFI expression was about 50% of the control level in the cKO-het, and it was barely detectable in the cKO-homo (fig. S3, A and B). To assess the relative positions of NC and MEC within the intact cortical hemisphere, we used whole-mount in situ hybridization to detect the expression of Satb2 and Nurr1, which respectively label NC and MEC (32, 33). We found complementary Satb2 and Nurr1 expression patterns in control cortices at postnatal day 0 (P0) (Fig. 2C). Consistent with previous findings (18), the Nurr1+ domain was markedly decreased in size, with a corresponding expansion of the Satb2+ domain in the cKO-homo compared with control. Notably, we found that MEC size was regulated by COUP-TFI in a dose-dependent fashion, with the Nurr1+ domain in the cKO-het cortex being larger than that in cKO-homo cortices and smaller than that in control cortices (Fig. 2C). Thus, we showed that decreasing COUP-TFI expression level led to a caudal shift of the NC/MEC border. In addition to its position, we further examined the integrity of the NC/MEC border by examining the complementary expression of NC- and MEC-specific marker genes. We found that the border indeed became less sharp, with more overlap of NC/MEC marker gene expression in COUP-TFI cKO-homo cortices (Fig. 2, E and F). Thus, our findings suggested that COUP-TFI expression level regulates the position and sharpness of NC/MEC border.
Increased COUP-TFI induces ectopic MEC formation
In addition to the loss-of-function experiments, we also tested the impact of increasing COUP-TFI expression. We used Emx1-Cre to induce the expression of a copy of the hCOUP-TFI transgene (34) in cortical progenitors and their progeny of cTG mice (COUP-TFITG/O;Emx1-Cre or cTG-het); the hCOUP-TFI transgene expression level was about 50% of the endogenous mouse (mCOUP-TFI) expression level (fig. S3, C to E). Notably, we found that MEC was rostrally expanded in the cTG mice, as determined by the expression of MEC-enriched genes, including Nurr1, Wfs1, and VgluT2 (Fig. 3A and fig. S4A). Furthermore, ectopic domains of MEC-enriched marker gene expression were detected in the caudal NC, suggesting that ectopic MECs were generated in the NC of cTG (Fig. 3, A and B). The generation of ectopic MEC in the cTG NC was unexpected and is, to the best of our knowledge, unprecedented. We thus further carefully confirmed the properties of these ectopic MEC in the COUP-TFI cTG by comparing gene expression patterns, neuronal birthdates, and connectivity of the ectopic structures.
Fig. 3. COUP-TFI overexpression in cTG cortical progenitors leads to rostral expansion of MEC and the formation of ectopic MEC.
(A and B) Nissl staining, in situ hybridization, and immunostaining were performed for MEC-enriched genes (A) and NC-enriched genes (B) on P7 sagittal sections of control and cTG (cTG-het; COUP-TFITG/+;Emx1-Cre) cortices. The border between NC and MEC (marked by arrowhead) was rostrally shifted in the cTG cortices. Ectopic MEC domains (asterisks) were identified in the caudal NC of cTG. (C to F) Immunostaining of MEC layer markers demonstrates the similar molecular characteristics and layering structure in endogenous MEC from control cortex (C and D) and ectopic MEC from cTG (E and F). (G) DiI crystal was placed at the dorsal hippocampus in P7 control and cTG cortices. DiI-labeled neurons and neuronal processes could be detected in the dorsal MEC (arrowhead) in control and the rostrally expanded MEC (arrowhead) and ectopic MEC (asterisks) in cTG. (H) DiD was placed in the primary visual cortex (V1) in P7 control and cTG cortices, and DiD-labeled neurons and neuronal processes could be detected in the dLG in both control and cTG. DiD-labeled neuronal processes could also be detected in the perforant path (PP, arrows) in the cTG hippocampal formation (Hp). VB, ventrobasal nucleus. Scale bars, 500 μm (A, B, G, and H) and 100 μm (D and F).
Similar to the endogenous MEC (Fig. 3, A, C, and D), we found the ectopic MECs exhibited consistent layer-specific expression of Nurr1 in layers 2/3 and 5/6; Kitl and Vglut2 in layer 2/3; Calb, Lhx2, Nef3, Rln, and Wfs1 in layer 2; and Lmo4 in layer 5/6 (Fig. 3, A, E, and F, and fig. S4, A to C). Along with the rostral expansion and ectopic expression of MEC genes, we observed down-regulation of NC-enriched marker genes, including Satb2, Rorb, Id2, Er81, Fezf2, and Fosl2 (Fig. 3B and fig. S4, A to C). Furthermore, similar to that in the cTG (fig. S4D), the ectopic expression of MEC-enriched genes (e.g., Nrp2) and down-regulation of NC-enriched genes (e.g., Rorb) were also found in the D6-COUP-TFI transgenic cortices (fig. S4E), where mCOUP-TFI is overexpressed in cortical progenitors driven by the Dach1 promoter (35). The presence of ectopic domains expressing MEC-specific genes in multiple lines of COUP-TFI transgenic mice suggested that overexpression of COUP-TFI in cortical progenitors induces fate change in the NC cells, which leads to generation of ectopic MEC-like structures.
Although the sequential maturation of MEC neurons has been described (36), the pattern of neurogenesis in different MEC layers has remained uncharacterized. Therefore, we performed neuronal birthdating to compare neurogenesis programs between MEC and NC. Neurons in both structures were generated in an inside-out pattern (fig. S5, A to C). In both MEC and NC, deep-layer neurons were generated at around E11.5. However, while most NC upper-layer neurons were generated from E15.5 to E17.5, many of the layer 2/3 MEC neurons were generated at around E13.5, and no major numbers of MEC neurons were generated after E15.5 (fig. S5, A to C). The early termination of neurogenesis in MEC agrees with the fact that MEC has fewer neurons within a given cortical column than the NC (fig. S5D). We first showed that COUP-TFI overexpression in cTG does not affect neurogenesis by confirming that the number of proliferating cortical progenitors is similar in cTG and control cortices at E13.5 (fig. S5E), which is consistent with the similar cTG and control cortical sizes (fig. S3L). In the cTG, we found that most of the neurons in the ectopic MEC were generated by E13.5 and far fewer neurons were generated at E15.5 and E17.5, when compared with adjacent NC tissues (fig. S5F). Thus, our results showed that the timings of neurogenesis initiation and termination in the ectopic MEC domains were similar to those in the control endogenous MEC.
Furthermore, in line with their distinct functions, MEC and NC have different input and output projection patterns. The sensory cortices in NC have reciprocal connections with thalamus, while MEC is highly connected with hippocampus. Using DiI crystals in the dorsal hippocampus to label neurons projecting to hippocampus, we observed DiI-labeled neurons and neuronal projections in the dorsal part of the MEC in control animals. However, in the cTG hippocampus, DiI-labeled neurons and neuronal projections were rostrally shifted and could even be detected in the ectopic MEC (Fig. 3G). To further confirm the formation of ectopic MEC in the caudal NC in the cTG, we injected DiD in the P7 primary visual cortex (V1) of control and cTG cortices. While DiD labeled the reciprocal connections between V1 and thalamic dorsal lateral geniculate nucleus (dLG) but not in the hippocampus in the control cortices, we found DiD-labeled neuronal fibers in the perforant pathway in hippocampus, in addition to the dLG, in the cTG cortices (Fig. 3H).
In conclusion, on the basis of the expression of region-specific layer markers, neurogenesis patterns, and connectivity, we showed that COUP-TFI overexpression induces the formation of ectopic MEC in the caudal NC, and these ectopic MEC domains truly resembled endogenous MEC. Our results suggest that increased COUP-TFI expression is sufficient to rostrally expand MEC and induce the generation of ectopic MEC, at the expense of NC. These ectopic MEC domains were not only present transiently during development, as we found evidence for these ectopic MEC structures in the cTG caudal NC persisted to at least P120 (fig. S6).
COUP-TFI expression level regulates the number and location of ectopic MEC
To further examine whether COUP-TFI expression level determines the location of ectopic MECs, we generated cTG-homo mice (COUP-TFITG/TG;Emx1-Cre), which express two copies of hCOUP-TFI transgene, and the transgene expression level is similar to the endogenous mouse (mCOUP-TFI) (fig. S3, C to E). Notably, transgene expression did not significantly alter the expression level of endogenous mCOUP-TFI (fig. S3D). However, the expression gradient of total COUP-TFI in the cortical progenitors was altered in the transgenic cortices, with cortical progenitors in the caudal half of cTG-het and caudal three-quarters of cTG-homo cortices expressing high levels of COUP-TFI (similar or higher than the maximal COUP-TFI expression in wild type) (fig. S3, F to K). In the cTG-homo, we found an increased overall number and more rostrally located ectopic MEC domains (Satb2− and Nurr1+) when compared to cTG-het (Fig. 4, A to C). Together, these loss-of-function and gain-of-function experiments suggested that the level of COUP-TFI expression dose-dependently controls the size of MEC (fig. S3, M and N) and ectopic MEC formation.
Fig. 4. The level of COUP-TFI overexpression in cortical progenitors correlates the expansion domain of ectopic MEC.
Dorsal and posterior view for P0 cTG-het (COUP-TFITG/O;Emx1-Cre) (A) and cTG-homo (COUP-TFITG/TG;Emx1-Cre) (B) cortices. Whole-mount in situ hybridization of Satb2 and Nurr1 was used to indicate NC and MEC, respectively. (C) In situ hybridizations for Satb2 and Nurr1 on sagittal sections of P7 control, cTG-het, and cTG-homo showed ectopic Nurr1+Satb2− domains [arrowheads in (A) to (C)] in the cTG NCs. The number of ectopic Nurr1+Satb2− domains in the NC was higher, and the domains were more rostrally located in the cTG-homo compared to the cTG-het. (D) According to our new mathematical model, we can simulate the phenomenon similar to the experimentally observed phenotypes, even the formation of ectopic MEC (Figs. 2C and 4, A to C). (E) The mathematical model predicts stable homogeneous MEC and NC domains (blue regions) at high concentrations of MEC and NC patterning TFs (ρE and ρN), respectively. It also predicts Turing-like instability domain (red region) associated with ectopic domains (red) for certain ranges of 𝜌E and 𝜌N. COUP-TFI expression gradients associated with control, cKO-het, cKO-homo, cTG-het, or cTG-homo were plotted according to the data (Fig. 2B and fig. S3, F to H). Gradients of cTG-het or cTG-homo models cut through the instability region generate ectopic domains. Scale bars, 250 μm (A, B, and C).
Mathematical model predicts that cell affinity mechanisms can trigger the emergence of ectopic MEC
Previous studies of area patterning indicate that cortical progenitors adopt a specific fate depending on the level of patterning TF expression. However, changes in the expression gradient of patterning TFs lead to areal shifts (8) but not to the emergence of ectopic domains. Our observation of ectopic MEC in COUP-TFI transgenic mice thus challenges the classical conceptual frameworks of cell fate determination. Previously, self-organization models based on gene expression and diffusion were developed and shown to accurately reflect observed shifts in the boundaries between brain areas upon changes in the expression of patterning TFs (37–39). However, none of these models predict changes in boundary regularity and integrity or the emergence of regular ectopic domains like those that we observed in COUP-TFI transgenic cortices (figs. S11 and S12). We therefore revisited these classical models and developed a system of equations combining positional information and self-organization mechanisms with cell movement and differential cell affinity. Our model describes the differentiation of neural progenitors into NC or MEC cells as a result of three basic phenomena. First, external cues are provided by two gradients of patterning TF, one that promotes NC fate and an opposing gradient that promotes MEC fate, the latter mimicking in vivo expression of COUP-TFI. Second, the model emulates the competition between expression of NC and MEC genes. Eventually, the model incorporates cell movement, both isotropic, via diffusion, and directional, through aggregation terms that particularly reflect differential cell affinity. This simple model showed that mechanisms based on TF gradients and differential adhesion can support differentiation into regular regions, with a boundary location dependent on patterning TF concentrations and sufficiently steep gradients. We further observed that the boundaries may be sharpened by cell affinity levels (see fig. S13), consistent with the sharp NC/MEC border that we observed in animals (control in Fig. 2D). However, quite unexpectedly, the model also predicted that sufficiently strong differential affinity can also cause a dynamical instability akin to that observed in the celebrated Turing model (40, 41). The occurrence of such an instability yields elongated ectopic domains (typically stripes or labyrinths) or isolated aggregates depending on the expression levels of the patterning TFs (Fig. 4D and fig. S16). When patterning TF gradients do not give rise to instability (as with the fitted gradients in the control), a sharp boundary will be maintained upon changes in gradients. However, when the patterning TF gradients intersect the instability region (as is the case of COUP-TFI up-regulation; Fig. 4E), the transition between the MEC and NC becomes irregular, yielding ectopic MEC domains with topologies very similar to the experimentally identified patterns (Fig. 4D and fig. S20). Therefore, we used the model to make two testable predictions (see “Pattern formation in heterogeneous domains” section in the Supplementary Math model). First, we validated the modeling assumption that differentiation can be appropriately described as cell autonomous. To this end, we investigated whether overexpression of COUP-TFI would induce the emergence of ectopic MEC cells in the region of overexpression. Second, the instability arising in the theoretical model relies fundamentally on differential cell affinities that lead cells at the MEC/NC boundary to form clusters. We thus probed for evidence of these differential affinities experimentally, searched for the molecular pathways involved, and assessed the impact of COUP-TFI overexpression on these pathways. The model predicts that a local overexpression of COUP-TFI in the NC should induce the formation of multiple MEC clusters (Fig. 5, A and B). However, we predict that when cell affinity mechanisms are weaker, we will observe fewer clusters with lower cell density and less homogeneity (figs. S22 to S24).
Fig. 5. High level of COUP-TFI expression cell-autonomously induces the neuronal fate of MEC.
(A) Overexpression of COUP-TFI in the control parietal region leads to entering the instability region and inducing the emergence of ectopic MEC within the NC. (B) Ectopic MEC domains were visualized by the spikes (arrowheads) in expression of MEC markers (blue) and drops in expression of NC markers (orange). (C) COUP-TFITG/TG cortices were transfected with control or CAG-Cre plasmids with mCherry expression vector by in utero electroporation at E13.5. On sagittal sections of E18.5 electroporated cortices, transfected cells were labeled by mCherry, and transgene expression was detected by Myc and COUP-TFI. (D) Relative mCherry intensity was measured across the electroporation domains. The orange line indicates the mean mCherry intensity. In control cortices, mCherry+ cells show uniform distribution across the electroporation domain, while mCherry+ cells form clusters in Cre-electroporated cortices. (E) Average number of electroporated cells adjacent to a given electroporated cell (within a 15-μm radius) in Cre-electroporated cortices was significantly increased compared to the control (P < 0.0001). (F and G) Expression of NC-enriched markers (Tbr1, Ctip2, and Satb2) and MEC-enriched markers (Nrp2, Kitl, and Nurr1) among mCherry+ cells in control or Cre-electroporated cortices. Significantly fewer COUP-TFI–overexpressing cells displayed NC markers, including Tbr1 (P = 0.0235), Ctip2 (P = 0.0099), and Satb2 (P = 0.0167), while most expressed Nrp2 (P = 0.0112). Scale bars, 150 μm (C and F) and 100 μm (G).
High COUP-TFI expression cell-autonomously induces ectopic MEC
According to the theoretical prediction, increasing COUP-TFI expression levels in cells expected to adopt the NC cell fate (e.g., in parietal cortex) can move these cells into the instability region (Fig. 5A) and thereby induce cell-autonomous changes to their fate and the emergence of one or multiple isolated clusters of ectopic MEC (Fig. 5B). To experimentally test this prediction, we first investigated whether high COUP-TFI expression levels could induce MEC formation. We used in utero electroporation to transfect a control vector or a Cre expression construct (CAG-Cre) along with a mCherry expression vector (to label transfected cells) into the parietal cortex in COUP-TFITG/TG at E13.5 and analyzed the cortices at E18.5. Cre expression in COUP-TFITG/TG cortices induced the expression of hCOUP-TFI in progenitors and their progeny (Fig. 5C). Most of the transfected cells in control and COUP-TFITG/TG cortices were found in the cortical plate (CP). We found that transfected cells in the control cortices were dispersed (Fig. 5, C and D) and expressed NC markers, such as Tbr1, Ctip2, or Satb2, but not Nrp2, an MEC marker (Fig. 5, F and G). However, induction of hCOUP-TFI caused cell clustering (Fig. 5, C and D). In these clusters, significantly more transfected cells were found adjacent to a given transfected cell (Fig. 5E). Most of the COUP-TFI overexpressing cells did not express NC-enriched genes, such as Tbr1, Ctip2, Satb2, or Rorb, and instead expressed MEC-enriched genes, including Nrp2, Kitl, and Nurr1 (Fig. 5, F and G, and fig. S7, A and B). These MEC-like cell clusters were not induced when Cre expression construct was electroporated at E15.5 (fig. S7C), suggesting that there is a time window during which COUP-TFI can induce NC-to-MEC cell fate change.
Furthermore, the cell clusters expressing MEC-enriched genes induced by COUP-TFI overexpression were similar to those in the ectopic MEC domains in the cTG cortices (Fig. 3, A to F), as predicted by our mathematic model (Fig. 5B). In contrast, the model did not predict any clustering when overexpressing COUP-TFI in the MEC region. We experimentally confirmed that cell clustering was not induced when Cre was electroporated into developing MEC in COUP-TFITG/TG (fig. S7D). Thus, we provide evidence that, within a certain time window, a local increase of COUP-TFI expression in NC could induce MEC neuronal differentiation. In these COUP-TFI–induced MEC cell clusters, MEC genes are up-regulated and NC genes are repressed.
Differential affinity in MEC and NC
The model further suggested that this intriguing clustering effect relies on differential MEC and NC cell affinities. As shown in Fig. 6A, inhibiting differential cell adhesion in the model leads to a disappearance of the clusters in a region with mixed cell identity and lowers cell density in the clusters compared to the highly clustered and dense ectopic regions predicted in the presence of cell adhesion (Fig. 5A). To test this prediction, we (i) assessed whether MEC and NC cells show differentiated cell adhesion and (ii) identified the differential cell adhesion molecules enhanced by COUP-TFI. To probe whether differentiated MEC and NC cells spontaneously segregate, we performed in vitro cell aggregation assays with dye-labeled cells dissociated from E13.5 presumptive NC and MEC. After 1 hour of incubation, we assessed the propensity of cells to form aggregates in populations of red- and green-labeled NC cells [NC (red) + NC (green)] or red NC cells incubated with green MEC cells [NC (red) + MEC (green)]. We found that cells from NC or MEC were more likely to be next to cells of the same origin when compared with cells from NC (P = 0.025; Fig. 6, C and D), suggesting that MEC and NC cells spontaneously aggregate with cells of their own kind.
Fig. 6. High level of COUP-TFI expression induces Pcdh19-mediated cell clustering.
(A and B) Removing differential cell adhesion from COUP-TFI overexpression [the shaded blue region in (B)] no longer induces cell clustering and leads to the loss of instability domain (comparing with the red domain in Fig. 5A). (C and D) In vitro cell aggregation assay with dye-labeled cells from E13.5 cortices showed that mixed medial entorhinal cortical and neocortical cells (M + N) cells segregated more than neocortical cells (N + N) (P = 0.025). (E and F) COUP-TFI binds to a conserved Sp1/COUP-TFI binding site (shaded) in the Pcdh19 promoter, similarly to Rnd2 and Fabp7 promoters (Pcdh19, P = 0.00826; Rnd2, P = 0.0009; Fabp7, P = 0.0353). (G) On sagittal sections of E18.5 cortices electroporated with indicated constructs at E13.5, most of the control cells scattered in the CP. COUP-TFI overexpression formed Pcdh19- and Kitl-expressing cell clusters in the intermediate zone (IZ). (H and I) COUP-TFI overexpression repressed Satb2 (P < 0.0001) and induced Nrp2 (P = 0.0082), regardless of Pcdh19 expression (COUP-TFI + Pcdh19KD versus COUP-TFI, Satb2, P = 0.2030; Nrp2, P = 0.0871). (J) Analyses of adjacent cell number (COUP-TFI versus control, P = 0.0050; COUP-TFI + Pcdh19KD versus COUP-TFI, P = 0.0349), cell distance (COUP-TFI versus control, P < 0.0001; COUP-TFI + Pcdh19KD versus COUP-TFI, P = 0.026), and cell heterogeneity (COUP-TFI + Pcdh19KD versus COUP-TFI, P < 0.001) showed that COUP-TFI overexpression induces cell clustering and knocking down Pcdh19 in COUP-TFI–overexpressing cells reduced COUP-TFI–induced clustering. VZ, ventricular zone; SVZ, subventricular zone; IgG, immunoglobulin G; n.s., not significant. Scale bars, 10 μm (C), 300 μm (G), and 100 μm (H).
We next explored the mechanism underlying aggregation. The theoretical model predicted that the differential affinities of MEC and NC cells are key for cell clustering and boundary integrity (see Figs. 5A and 6A and “Cell Adhesion is Crucial for Ectopic Domains Formation” section in the Supplementary Math model). We therefore tested whether MEC and NC cells express different cell adhesion molecules and whether high levels of COUP-TFI could induce the expression of MEC-enriched cell adhesion molecules. On the basis of the RNA-seq analysis of NC and AC gene expression profiles at E13.5, we focused on a list of genes involved in homophilic cell adhesion via plasma membrane adhesion (GO:0007156, P = 0.00469) (fig. S8, A and B). Using the Allen Developing Mouse Brain Atlas, which includes expression patterns of about 2000 genes functionally relevant to brain development, we confirmed that several homophilic adhesion molecules have detectably differential expression along the anterior-posterior axis in the developing telencephalon at E13.5 (fig. S8C). Comparing the expression of these genes in the anterior and posterior ends of the E13.5 cortices, we found that Pcdh19 (encodes Protocadherin 19) was highly enriched in the caudal cortex (fig. S8E) and exhibited the most notable difference in relative expression levels between anterior and posterior cortices (fig. S8D) (42). Because misexpression of Pcdh19 in the developing cortex is known to cause cell clustering (43–45), we first examined whether Pcdh19 expression levels change when COUP-TFI expression is altered. We found that the high-caudal–to–low-rostral expression gradient of Pcdh19 was dose-dependently caudally shifted in the cKO (fig. S8F) and that Pcdh19 expression is significantly increased in the cTG (fig. S8, G and H).
We next demonstrated that COUP-TFI is able to directly bind to a conserved Sp1/COUP-TFI binding site in the Pcdh19 promoter region by chromatin immunoprecipitation (ChIP) (Fig. 6, E and F). Using N2a cells, in which COUP-TFI expression is relatively low, we examined the impact of the COUP-TFI–Pcdh19 genetic pathway on cell segregation. First, we showed that increasing COUP-TFI expression enhanced Pcdh19 expression (fig. S9A). Next, we performed cell aggregation assays (46), which showed that in a cluster, control cells are dispersed, but control and COUP-TFI–overexpressing cells were segregated with a higher aggregation score (fig. S9B). We then used Pcdh19 short hairpin RNA (shRNA) (fig. S9C) to knock down Pcdh19 in COUP-TFI–overexpressing cells and found that Pcdh19-knockdown COUP-TFI–expressing cells did not retain the ability to segregate from control cells (fig. S9D). These results suggested that COUP-TFI induces cell segregation via Pcdh19.
Coelectroporation of Cre and mCherry expression constructs into COUP-TFITG/TG cortices would induce COUP-TFI overexpression in the transfected progenitors and their progeny, generating more COUP-TFI–overexpressing cells than the targeted (mCherry-expressing) cells. Therefore, to test whether a high level of COUP-TFI expression indeed cell-autonomously induces Pcdh19 expression in vivo, we electroporated a CAG-COUP-TFI expression construct into wild-type parietal cortex at E13.5. As expected, we found that the high level of COUP-TFI expression induced Pcdh19 expression, and it also induced expression of other MEC-enriched genes, similar to Cre electroporation in COUP-TFITG/TG cortices (Fig. 6G and fig. S10A). Most of the COUP-TFI–overexpressing cells formed clusters in the intermediate zone (Fig. 6G) rather than the CP, as was seen in the Cre-electroporated COUP-TFITG/TG cortices (Fig. 5C). As COUP-TFI is expressed at a higher level in CAG-COUP-TFI electroporated cells than in Cre-electropored TG cells (fig. S10B), we used shRNA to knock down COUP-TFI expression in CAG-COUP-TFI transfected cells to test whether COUP-TFI overexpression level affected neuronal migration. We found that the CAG-COUP-TFI electroporated cells could form cell clusters in the CP when COUP-TFI expression was reduced (fig. S10C). This finding suggests that the COUP-TFI expression level indeed influences neuronal migration, and it must be tightly regulated during cortical development.
To directly test whether the induction of Pcdh19 expression is required for COUP-TFI–induced cell clustering, we electroporated Pcdh19 shRNA (fig. S9C) together with the CAG-COUP-TFI expression vector to knock down Pcdh19 in COUP-TFI–overexpressing cells (fig. S9E). While most of the mCherry-labeled electroporated cells in the control cortices were Satb2+ and scattered throughout the CP, most of the COUP-TFI–overexpressing cells ectopically expressed Nrp2 and Kitl but lost the expression of Satb2 (Fig. 6, H and I, and fig. S9E). We found that knocking down Pcdh19 did not change the ability of COUP-TFI to induce Nrp2 and Kitl expression or to repress Satb2 expression (Fig. 6, H and I, and fig. S9E), but it partially blocked the ability of COUP-TFI to induce cell clustering (Fig. 6J). Consistent with the theoretical predictions, clusters were significantly less dense (P = 0.026) and significantly more heterogeneous (P < 0.001) than in the presence of functional Pcdh19 (Fig. 6J). These results suggest that high levels of COUP-TFI expression induces the expression of Pcdh19, which segregates MEC cells from NC cells.
DISCUSSION
Patterning of telencephalon neuroepithelium into different progenitor regions gives rise to well-segregated functional domains that are essential to the wiring of the cerebrum and support complex cerebral functions. Hence, even minor defects in early patterning processes may be associated with serious intellectual and behavioral deficits. In this study, we discovered that the position-dependent expression gradient of COUP-TFI is a critical determinant of the cell fate decision to generate either NC or MEC. In particular, we show that mutant mice with low COUP-TFI expression have reduced MEC and expanded NC, with a less sharp caudally shifted NC/MEC border. We also found that COUP-TFI overexpression expanded MEC at the expense of NC and generated ectopic MEC domains (Figs. 2 and 3) that are not accounted for by the classical paradigm of cortical patterning. This finding then led us to further explore the determinants of boundary regularity. We theoretically examined the regularity of boundaries using a mathematical model of positional information with aggregation (such as differential adhesion). While we confirmed that cell adhesion can enhance sharpness of the boundary, we also showed that instability arises with excessive adhesion levels, leading to the emergence of irregular boundaries like those seen in our experiments. In line with the predictions from our model, we further identified Pcdh19 as a COUP-TFI–regulated factor that is responsible for MEC neuron adhesion. Our findings suggest that during cortical development, patterning TF gradients set up differential cell affinities to ensure the formation of sharp borders between cortical regions.
We demonstrated that the COUP-TFI expression level regulates neuronal fate decisions, where a high level of COUP-TFI specifies MEC neuronal fate between E10.5 (when Emx1-Cre is expressed) and E15.5 (when MEC neurogenesis terminates). These results suggest that the tight regulation of graded, position-dependent expression of COUP-TFI is crucial for cortical development. In previous work, it was proposed that graded expression of patterning TFs is established, in part, by Fgfs produced by an anterior midline signaling center, the commissural plate, and by bone morphogenetic proteins and Wnts produced by a posterior-medial signaling center, the cortical hem (8, 47). In addition, COUP-TFI protein level was shown to be regulated by fibroblast growth factor 8 via miR-21 (48), but how the expression gradient of COUP-TFI mRNA is established and maintained requires further investigation.
The observation of ectopic MEC domains in COUP-TFI–overexpressing cortices contrast with classical work that suggested that changes in patterning TF expression levels only lead to alterations in boundary locations and area sizes (8). We found that the formation of ectopic MEC formation is mediated by differential cell adhesion mechanisms involving Pcdh19. Previously, differential cell affinity was reported to sharpen and regularize boundaries (1, 49, 50), and cell type–specific combinatorial expression of adhesion molecules was recently shown to mediate cell sorting and contribute to patterning robustness (15). Our experiments and mathematical model provide further evidence that differential cell affinity plays an important role in shaping the boundaries within the cortex. Our study also shows that the effects of differential cell affinity may include shattered boundaries and heterotopias. These results identify a limit to the robustness of brain development, paving the way for future studies on the role of excessive cell adhesion in other developmental processes.
Furthermore, although our current model was built on the assumption that differential cell adhesion of MEC cells was only affected by cell-autonomous changes in COUP-TFI levels, it does not rule out a role for non–cell-autonomous interactions in boundary formation nor the involvement of differential affinity of other cell types. The facts that this model could reproduce the experimental observations and that its predictions were validated experimentally suggest that the modeled features are sufficient to explain the phenotype. Notably, the regularity of the solutions to our equations ensures that the same phenotypes will persist, even when the model is modified to include non–cell-autonomous interactions and/or differential affinity of other cell types, as they do not dominate the dynamics (see the Robustness of Phenotypes section in the Supplementary Math model). For example, although COUP-TFI–associated MEC cell adhesion is sufficient to induce the formation of an instability region, enhancing adhesion of NC cells could make the patterning more prominent (see fig. S21).
From a mathematical standpoint, these findings raise interesting questions regarding partial differential equations in heterogeneous domains. The model motivated by the cortical development problem at hand includes a spatial crossing of a Turing-like instability that was identified for a homogeneous system (i.e., with fixed levels of patterning TFs ρE and ρN). Spatial variations through opposing gradients were represented as paths on the plane (ρE, ρN). For patterns to appear at the boundary, it is necessary but not sufficient that this path intersects the instability region (Fig. 4E). In particular, the length scale (and sharpness) of the gradients is expected to play a role (see the Supplementary Math model). Identifying sufficient conditions to create a certain pattern at the boundary, as well as how the geometric properties of that pattern depend on the shape of gradients, constitutes an exciting open problem in applied mathematics, with far-reaching applications in various domains.
By showing that COUP-TFI is a key determinant of MEC development, our study advances the understanding of the topic, which is highly significant but largely underexplored. MEC plays a central role in brain function, connecting the hippocampus to other cortical regions, and it is composed of many specialized cells that are selective for speed, head direction, or localization, serving as a hub for neural correlates of spatial navigation (51–54). Because MEC is also one of the first cortical regions affected in Alzheimer’s disease, patients often experience spatial disorientation in addition to memory loss (55, 56). In addition, developmental defects in entorhinal structure, such as disruption of cortical layers, heterotopic displacement of neurons, and a paucity of neurons in superficial layers, were reportedly associated with developmental defects found in schizophrenia (57, 58). Thus, it is important to understand the unique functional properties of MEC neurons and how these properties are acquired during development. The processes that we uncovered could potentially provide a means by which to repair specific parts of the brain damaged by disease or injury. In addition, the identification of differential cell adhesion as a key determinant for precise boundary location and integrity between cortical regions has profound implications for emerging regenerative technologies and tissue engineering.
The formation of ectopic domains is reminiscent of heterotopia, a condition wherein neurons do not migrate properly during fetal development and form clusters of normal neurons in abnormal locations. These clusters are generally thought to be the result of disrupted progenitor proliferation or neuronal migration during cortical development and may be one of the causes of epilepsy in humans (59). Our findings suggest that heterotopia may also occur by the formation of ectopic cortical domains due to dysregulation of distinct region-specific cell adhesion properties. Particularly relevant to our study, heterotopia-associated cell clusters were found in the cortex of X-linked Pcdh19 heterozygous female mice, with mosaic expression of Pcdh19 in cortical neurons. These mutant mice develop epilepsy, similar to PCDH19 patients, who also show cortical abnormalities (43).
The formation of ectopic MEC in NC is also of interest from developmental and evolutionary perspectives. It has been suggested that the navigation system in birds and reptiles is located in the medial pallium (60) and that the AC contributed to the evolution of dorsomedial NC (61). Our findings show that simply changing the COUP-TFI expression level can switch the fates of NC and MEC, suggesting a close evolutionary relationship between the regions. Thus, shaping of the COUP-TFI expression gradient in cortical progenitors may have provided an intriguing molecular mechanism to regulate and fine-tune the relative sizes of functional domains in amniote cortices during evolution.
MATERIALS AND METHODS
Animals
COUP-TFI floxed and transgenic mice were provided by M.-J. Tsai. The COUP-TFI transgene consists of a human COUP-TFI gene under the control of the CAG promoter and a floxed stop cassette, in the Rosa26 locus (34). Emx1-Cre mice were provided by K. Jones. Animal care and experimental procedures were approved by and performed in accordance with guidelines provided by the Academia Sinica Institutional Animal Care and Use Committee. The day of identifying a vaginal plug and the day of birth were designated as E0.5 and P0, respectively.
Nissl staining, in situ hybridization, immunohistochemistry, and EdU labeling
Timed-pregnant mice were dissected, and embryonic cortices were fixed in 4% phosphate-buffered paraformaldehyde (PFA); postnatal brains were perfused with and postfixed in 4% PFA. For histological analyses, brains were cryoprotected with 30% sucrose in phosphate-buffered saline, embedded in Tissue-Tek OCT compound (Sakura Finetek) and cut in 20- to 25-μm sections on a cryostat (Leica). For Nissl staining, sections were stained with 0.5% cresyl violet and then dehydrated through graded alcohols. In situ hybridization on sections and whole mounts was performed as previously described (16). Antisense RNA probes were labeled with digoxigenin (DIG) using a DIG-RNA labeling kit (Roche), or with S35 radioactive isotopes. Sections were pretreated with proteinase K (5 μg/ml) at room temperature for 10 min, while whole mounts were pretreated with proteinase K (10 μg/ml) at room temperature for 30 min. The prehybridization, hybridization (for overnight), and posthybridization washes were done at 65°C, followed by incubation with anti-DIG-AP (alkaline phosphatase) (1:2000) overnight at 4°C. 4-Nitro blue tetrazolium chloride (NBT)/5-Bromo-4-chloro-3-indolyl phosphate p-toluidine salt (BCIP) (Roche) chromogenic staining was used to visualize the distribution of specific RNA transcripts. Immunohistochemistry was performed as described (62). In short, primary antibodies were incubated overnight at 4°C in blocking solution containing 3% bovine serum albumin (Sigma-Aldrich) and 0.3% Triton X-100 in phosphate buffer, followed by incubation with Alexa-conjugated secondary antibodies (Jackson ImmunoResearch) for 2 hours at room temperature. Cell nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (Vector). Primary antibodies were used at the following concentrations: Satb2 (1:500), Tbr1 (1:500), Ctip2 (1:300), Nrp2 (1:200), mCherry (1:500), COUP-TFI (1:200), Myc Tag (1:500), NF-M (1:500), Vglut2 (1:500), Wfs1 (1:500), Calbindin (1:500), and Nurr1 (1:150). Neuronal birthdating analyses were performed as described (63). Briefly, EdU (5-ethynyl-2′-deoxyuridine) (500 ng) was injected into timed-pregnant mice, and the EdU-positive cells were detected with a Click-iT EdU imaging kit (Invitrogen).
Axonal tracing
Tracing of neuronal projections was performed as described by Chou et al. (62). Small crystals of the fluorescent carbocyanide dyes, DiI or DiD (Invitrogen), were inserted into hippocampus or V1 of P7 brains. Brains were incubated for 3 to 8 weeks in 4% PFA, then embedded in 5% low-melting agarose, cut into 100-μm-thick sections on a vibratome (Leica), counterstained with DAPI (Vector), and mounted in 0.1 M phosphate buffer.
In vitro cell aggregation assay
The in vitro cell aggregation assay was performed as described with minor modifications (64). Briefly, E13.5 cortices were isolated in Hanks’ balanced salt solution (HBSS) containing 10 mM Hepes, and NCs and entorhinal cortices were dissected into small pieces and collected in separate vials on ice. Then, HBSS was replaced by dissociation buffer (Ca2+/Mg2+-free HBSS containing 10 mM EDTA) and incubated for 15 min at 37°C. Tissue pieces were then dissociated mechanically with a P200 pipette tip. After removing tissue debris with a 70-μm cell strainer, isolated cells were centrifuged for 5 min at 200g and resuspended in minimum essential media (MEM) without serum. For dye labeling, green-fluorescent (Invitrogen) and orange-fluorescent (Invitrogen) cell trackers were diluted to final concentrations of 2.5 and 10 mM in MEM, respectively. Cells were stained for 30 min at 37°C, and staining solution was subsequently washed with fresh medium. For one well of a 24-well plate, 4 × 105 dye-labeled cells were mixed in 400-μl MEM. The plate was incubated on a rotary shaker (100 rpm) at 37°C in a 5% CO2 incubator for 45 min. After shaking, mixed cells were fixed by 4% PFA and collected for further confocal imaging.
In utero electroporation
Expression constructs CAG-mCherry, CAG-COUP-TFI, and CAG-Cre were generated by subcloning mCherry, COUP-TFI, or Cre into a pCAG vector containing the cytomegalovirus early enhancer element and chicken β-actin promoter. For knockdown experiments, shRNA designed to target the coding sequence of mouse Pcdh19 RNA: TTCTGCCCTTGTCCTAATATA or shRNA designed to target the coding sequence of mouse COUP-TFI RNA: CACATCCGCATCTTTCAGGAA was cloned into the TRC2 vector (TRC2-pLKO-puro) containing the human U6 promoter. In utero electroporation was performed as previously described (65). In short, E13.5 embryos were visualized through the uterus using a fiber optic light source. Then, either CAG-Cre (1 μg/μl) or CAG-CreER (1 μg/μl) as a control was electroporated into dorsal telencephalons of embryos with paddle-type electrodes (CUY21EDIT II) in a series of five square-wave current pulses (40 V, 50-ms width, and 950-ms resting time). For mechanism exploration, a DNA solution containing CAG-COUP-TFI (1 μg/μl) and sh-Pcdh19 or sh-COUP-TFI (1 μg/μl) was electroporated, and here, CAG-Cre and sh-βgal (1 μg/μl) were used as a COUP-TFI overexpression control and a Pcdh19 knockdown control, respectively. All DNA solutions were mixed with CAG-mcherry (0.2 μg/μl) and 0.025% Fast Green (Sigma-Aldrich) and were injected with a glass capillary into the ventricle of each embryo. Electroporated embryos were allowed to develop until E18.5 and selected for further analyses by direct visualization of mCherry expression.
RNA-seq analyses
Total RNA was extracted from NC and AC manually isolated from E17.5 embryos and adult cortices with TRIzol reagent (Invitrogen). To obtain enough RNA, AC tissues from three to four embryonic brains were pooled as one sample, and three NC and AC samples were used for biological triplicates. RNA quality was assessed using a BioAnalyzer (Agilent Laboratories), and next-generation sequencing was performed with Illumina HiSeq 4000. Read sequences were then mapped to the mouse reference database (mm10) by Bowtie2 (v2.2.6). DESeq2 was used to normalize read counts by taking into account the gene length and the sequencing depth into FPKM (fragments per kilobase of transcript per million) value. Differentially expressed genes were defined by posterior probability of equal expression < 0.05. GO enrichment analysis was performed on the Gene Ontology Resource (http://geneontology.org/) (66). The heatmaps in figs. S1 and S8 were generated with relative FPKM values, where the maximal FPKM value of each gene among all samples was set as 1.
ChIP–quantitative polymerase chain reaction
Dorsal telencephalon was dissociated from E13.5 wild-type mice. The tissues were incubated with disuccinimidyl glutarate (Sigma-Aldrich) to a final concentration of 2 mM and fixed in 1% formaldehyde, followed by cross-linking with 125 mM glycine at pH 7.2. Cell lysates were sheared by sonication to generate chromatin fragments with an average length of 100 to 300 base pairs. Chromatin-protein complexes were then immunoprecipitated with mouse anti–COUP-TFI (R&D Systems) or Rabbit Gamma Globulin (Jackson ImmunoResearch) overnight at 4°C. The antibody-chromatin complexes were then incubated with Dynabeads protein G (Invitrogen) for 2 hours at 4°C. Genomic DNA fragments were purified and subjected to quantitative polymerase chain reaction (PCR) with specific primers [see table S1; primers for Fabp7 and Rnd2 were used as positive controls (67)] on real-time reverse transcription PCR (RT-PCR) using LightCycler 480 SYBR Green I Master mix (Roche).
Quantification and statistical analyses
For electroporation data, in the center of the electroporated domain, 300-μm-wide cortical columns were cropped for quantification of the cell numbers and marker intensity. The numbers of mCherry+, Ctip2+, Tbr1+, Satb2+, Nrp2+, and EdU+ cells were manually counted using ImageJ/FIJI. With a custom macro, position coordination and fluorescence intensities of selected cells were listed for further analyses of cell numbers, marker gene expression intensity, and neighboring cell distance. All analyses were performed with three or more biological replicates. The number of individual animals of the same genotype used is indicated as “n” in the text and figures. Statistical analyses were performed using GraphPad Prism 5 software. All quantitative data are presented as the means ± SEM. Minimal statistical significance was fixed at P < 0.05 for comparisons made by unpaired t test with Welch’s correction (for Figs. 1K, 2F, 5G, and 6, D, F, I, and J, and figs. S3, B, D, and E; S5D; S7C; S8, D and H; and S9, C and D); one-way analysis of variance with Bonferroni post hoc test (for fig. S9A). Significance is represented in figures as follows: *P < 0.05; **P < 0.01; ***P < 0.001.
Code availability
Custom MATLAB code used to analyze cortical imaging data and perform statistical tests on these data is available at https://github.com/Touboul-Lab/cortex_patterning. Codes were executed on MATLAB version R2019b. FIJI macro for exporting imaging numerical value is available at https://github.com/peggyscshu/Cell-grouping.
Acknowledgments
We thank M.-J. Tsai for providing the COUP-TFI floxed and transgenic allele and K. Jones for Emx1-Cre. We also thank members of the Chou laboratory for help and S.-C. P. Hsu for help on analyzing imaging data. Funding: This work was supported by Ministry of Science and Technology (MOST 108-2311-B-001-021, S.-J.C.), Academia Sinica (AS-CDA-107-L09, S.-J.C.), the Institute of Cellular and Organismic Biology of Academia Sinica (S.-J.C.), and NINDS (RO1 NS099099, J.L.R.). D.P. was partially supported by the Swartz Foundation. Author contributions: S.-J.C. designed the research. J.F., W.-H.H., C.-S.T., Z.-H.Z., H.-W.H., Y.-T.H., and S.-J.C. performed the research and analyzed data. A.F. and J.L.R. provided critical materials. D.P. and J.T performed mathematic modeling and analyzed data. J.T. and S.-J.C. wrote the paper. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/27/eabf6808/DC1
REFERENCES AND NOTES
- 1.Kiecker C., Lumsden A., Compartments and their boundaries in vertebrate brain development. Nat. Rev. Neurosci. 6, 553–564 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Gorski J. A., Talley T., Qiu M., Puelles L., Rubenstein J. L. R., Jones K. R., Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J. Neurosci. 22, 6309–6314 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolpert L., Positional information and the spatial pattern of cellular differentiation. J. Theor. Biol. 25, 1–47 (1969). [DOI] [PubMed] [Google Scholar]
- 4.Greig L. C., Woodworth M. B., Galazo M. J., Padmanabhan H., Macklis J. D., Molecular logic of neocortical projection neuron specification, development and diversity. Nat. Rev. Neurosci. 14, 755–769 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rubenstein J. L., Annual Research Review: Development of the cerebral cortex: Implications for neurodevelopmental disorders. J. Child Psychol. Psychiatry 52, 339–355 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hebert J. M., Fishell G., The genetics of early telencephalon patterning: Some assembly required. Nat. Rev. Neurosci. 9, 678–685 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alfano C., Studer M., Neocortical arealization: Evolution, mechanisms, and open questions. Dev. Neurobiol. 73, 411–447 (2013). [DOI] [PubMed] [Google Scholar]
- 8.O’Leary D. D., Chou S. J., Sahara S., Area patterning of the mammalian cortex. Neuron 56, 252–269 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Osterfield M., Kirschner M. W., Flanagan J. G., Graded positional information: Interpretation for both fate and guidance. Cell 113, 425–428 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Kay R. R., Thompson C. R., Forming patterns in development without morphogen gradients: Scattered differentiation and sorting out. Cold Spring Harb. Perspect. Biol. 1, a001503 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gregor T., Tank D. W., Wieschaus E. F., Bialek W., Probing the limits to positional information. Cell 130, 153–164 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubuis J. O., Tkacik G., Wieschaus E. F., Gregor T., Bialek W., Positional information, in bits. Proc. Natl. Acad. Sci. U.S.A. 110, 16301–16308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Godard B. G., Heisenberg C. P., Cell division and tissue mechanics. Curr. Opin. Cell Biol. 60, 114–120 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Steinberg M. S., Does differential adhesion govern self-assembly processes in histogenesis? Equilibrium configurations and emergence of a hierarchy among populations of embryonic cells. J. Exp. Zool. 173, 395–433 (1970). [DOI] [PubMed] [Google Scholar]
- 15.Tsai T. Y. C., Sikora M., Xia P., Colak-Champollion T., Knaut H., Heisenberg C.-P., Megason S. G., An adhesion code ensures robust pattern formation during tissue morphogenesis. Science 370, 113–116 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Armentano M., Chou S. J., Srubek Tomassy G., Leingärtner A., O’Leary D. D. M., Studer M., COUP-TFI regulates the balance of cortical patterning between frontal/motor and sensory areas. Nat. Neurosci. 10, 1277–1286 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Alfano C., Magrinelli E., Harb K., Hevner R. F., Studer M., Postmitotic control of sensory area specification during neocortical development. Nat. Commun. 5, 5632 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Flore G., di Ruberto G., Parisot J., Sannino S., Russo F., Illingworth E. A., Studer M., de Leonibus E., Gradient COUP-TFI expression is required for functional organization of the hippocampal septo-temporal longitudinal axis. Cereb. Cortex 27, 1629–1643 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Armentano M., Filosa A., Andolfi G., Studer M., COUP-TFI is required for the formation of commissural projections in the forebrain by regulating axonal growth. Development 133, 4151–4162 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Zhou C., Qiu Y., Pereira F. A., Crair M. C., Tsai S. Y., Tsai M. J., The nuclear orphan receptor COUP-TFI is required for differentiation of subplate neurons and guidance of thalamocortical axons. Neuron 24, 847–859 (1999). [DOI] [PubMed] [Google Scholar]
- 21.Zhou C., Tsai S. Y., Tsai M. J., COUP-TFI: An intrinsic factor for early regionalization of the neocortex. Genes Dev. 15, 2054–2059 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bosch D. G., Boonstra F. N., Gonzaga-Jauregui C., Xu M., de Ligt J., Jhangiani S., Wiszniewski W., Muzny D. M., Yntema H. G., Pfundt R., Vissers L. E., Spruijt L., Blokland E. A., Chen C. A.; Baylor-Hopkins Center for Mendelian Genomics, Lewis R. A., Tsai S. Y., Gibbs R. A., Tsai M. J., Lupski J. R., Zoghbi H. Y., Cremers F. P., de Vries B. B., Schaaf C. P., NR2F1 mutations cause optic atrophy with intellectual disability. Am. J. Hum. Genet. 94, 303–309 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen C. A., Bosch D. G. M., Cho M. T., Rosenfeld J. A., Shinawi M., Lewis R. A., Mann J., Jayakar P., Payne K., Walsh L., Moss T., Schreiber A., Schoonveld C., Monaghan K. G., Elmslie F., Douglas G., Boonstra F. N., Millan F., Cremers F. P. M., McKnight D., Richard G., Juusola J., Kendall F., Ramsey K., Anyane-Yeboa K., Malkin E., Chung W. K., Niyazov D., Pascual J. M., Walkiewicz M., Veluchamy V., Li C., Hisama F. M., de Vries B. B. A., Schaaf C., The expanding clinical phenotype of Bosch-Boonstra-Schaaf optic atrophy syndrome: 20 new cases and possible genotype-phenotype correlations. Genet. Med. 18, 1143–1150 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Bertacchi M., Romano A. L., Loubat A., Tran Mau-Them F., Willems M., Faivre L., Khau van Kien P., Perrin L., Devillard F., Sorlin A., Kuentz P., Philippe C., Garde A., Neri F., di Giaimo R., Oliviero S., Cappello S., D’Incerti L., Frassoni C., Studer M., NR2F1 regulates regional progenitor dynamics in the mouse neocortex and cortical gyrification in BBSOAS patients. EMBO J. 39, e104163 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakagawa Y., O’Leary D. D., Dynamic patterned expression of orphan nuclear receptor genes RORalpha and RORbeta in developing mouse forebrain. Dev. Neurosci. 25, 234–244 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Bulchand S., Subramanian L., Tole S., Dynamic spatiotemporal expression of LIM genes and cofactors in the embryonic and postnatal cerebral cortex. Dev. Dyn. 226, 460–469 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Nakagawa Y., Johnson J. E., O’Leary D. D., Graded and areal expression patterns of regulatory genes and cadherins in embryonic neocortex independent of thalamocortical input. J. Neurosci. 19, 10877–10885 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kashani A. H., Qiu Z., Jurata L., Lee S. K., Pfaff S., Goebbels S., Nave K. A., Ghosh A., Calcium activation of the LMO4 transcription complex and its role in the patterning of thalamocortical connections. J. Neurosci. 26, 8398–8408 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jabaudon D., Shnider S. J., Tischfield D. J., Galazo M. J., Macklis J. D., RORβ induces barrel-like neuronal clusters in the developing neocortex. Cereb. Cortex 22, 996–1006 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kitamura T., Pignatelli M., Suh J., Kohara K., Yoshiki A., Abe K., Tonegawa S., Island cells control temporal association memory. Science 343, 896–901 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ray S., Naumann R., Burgalossi A., Tang Q., Schmidt H., Brecht M., Grid-layout and theta-modulation of layer 2 pyramidal neurons in medial entorhinal cortex. Science 343, 891–896 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Saucedo-Cardenas O., Conneely O. M., Comparative distribution of NURR1 and NUR77 nuclear receptors in the mouse central nervous system. J. Mol. Neurosci. 7, 51–63 (1996). [DOI] [PubMed] [Google Scholar]
- 33.Mashiko H., Yoshida A. C., Kikuchi S. S., Niimi K., Takahashi E., Aruga J., Okano H., Shimogori T., Comparative anatomy of marmoset and mouse cortex from genomic expression. J. Neurosci. 32, 5039–5053 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu S. P., Lee D. K., Demayo F. J., Tsai S. Y., Tsai M. J., Generation of ES cells for conditional expression of nuclear receptors and coregulators in vivo. Mol. Endocrinol. 24, 1297–1304 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Faedo A., Tomassy G. S., Ruan Y., Teichmann H., Krauss S., Pleasure S. J., Tsai S. Y., Tsai M. J., Studer M., Rubenstein J. L. R., COUP-TFI coordinates cortical patterning, neurogenesis, and laminar fate and modulates MAPK/ERK, AKT, and beta-catenin signaling. Cereb. Cortex 18, 2117–2131 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Donato F., Jacobsen R. I., Moser M. B., Moser E. I., Stellate cells drive maturation of the entorhinal-hippocampal circuit. Science 355, eaai8178 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Gierer A., Meinhardt H., A theory of biological pattern formation. Kybernetik 12, 30–39 (1972). [DOI] [PubMed] [Google Scholar]
- 38.Quininao C., Prochiantz A., Touboul J., Local homeoprotein diffusion can stabilize boundaries generated by graded positional cues. Development 142, 1860–1868 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perthame B., Quininao C., Touboul J., Competition and boundary formation in heterogeneous media: Application to neuronal differentiation. Math. Models Methods Appl. Sci. 25, 2477–2502 (2015). [Google Scholar]
- 40.Turing A. M., The Chemical Basis of Morphogenesis. Philos. Trans. Roy. Soc. Lond. B Biol. Sci. 237, 37–72 (1952). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.B. t. Perthame, in Lecture Notes on Mathematical Modelling in the Life Sciences (Springer International Publishing, 2015), p. 1. [Google Scholar]
- 42.Gaitan Y., Bouchard M., Expression of the delta-protocadherin gene Pcdh19 in the developing mouse embryo. Gene Expr. Patterns 6, 893–899 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Pederick D. T., Richards K. L., Piltz S. G., Kumar R., Mincheva-Tasheva S., Mandelstam S. A., Dale R. C., Scheffer I. E., Gecz J., Petrou S., Hughes J. N., Thomas P. Q., Abnormal cell sorting underlies the unique X-linked inheritance of PCDH19 epilepsy. Neuron 97, 59–66.e55 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Hayashi S., Inoue Y., Hattori S., Kaneko M., Shioi G., Miyakawa T., Takeichi M., Loss of X-linked Protocadherin-19 differentially affects the behavior of heterozygous female and hemizygous male mice. Sci. Rep. 7, 5801 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lv X., Ren S. Q., Zhang X. J., Shen Z., Ghosh T., Xianyu A., Gao P., Li Z., Lin S., Yu Y., Zhang Q., Groszer M., Shi S. H., TBR2 coordinates neurogenesis expansion and precise microcircuit organization via Protocadherin 19 in the mammalian cortex. Nat. Commun. 10, 3946 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bisogni A. J., Ghazanfar S., Williams E. O., Marsh H. M., Yang J. Y. H., Lin D. M., Tuning of delta-protocadherin adhesion through combinatorial diversity. eLife 7, e41050 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shimogori T., Banuchi V., Ng H. Y., Strauss J. B., Grove E. A., Embryonic signaling centers expressing BMP, WNT and FGF proteins interact to pattern the cerebral cortex. Development 131, 5639–5647 (2004). [DOI] [PubMed] [Google Scholar]
- 48.Terrigno M., Bertacchi M., Pandolfini L., Baumgart M., Calvello M., Cellerino A., Studer M., Cremisi F., The microRNA miR-21 is a mediator of FGF8 action on cortical COUP-TFI translation. Stem Cell Rep 11, 756–769 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Foty R. A., Steinberg M. S., Differential adhesion in model systems. Wiley Interdiscip. Rev. Dev. Biol. 2, 631–645 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Batlle E., Wilkinson D. G., Molecular mechanisms of cell segregation and boundary formation in development and tumorigenesis. Cold Spring Harb. Perspect. Biol. 4, a008227 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O’Keefe J., Dostrovsky J., The hippocampus as a spatial map. Preliminary evidence from unit activity in the freely-moving rat. Brain Res. 34, 171–175 (1971). [DOI] [PubMed] [Google Scholar]
- 52.Hafting T., Fyhn M., Molden S., Moser M. B., Moser E. I., Microstructure of a spatial map in the entorhinal cortex. Nature 436, 801–806 (2005). [DOI] [PubMed] [Google Scholar]
- 53.Sargolini F., Fyhn M., Hafting T., McNaughton B., Witter M. P., Moser M. B., Moser E. I., Conjunctive representation of position, direction, and velocity in entorhinal cortex. Science 312, 758–762 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Witter M. P., Doan T. P., Jacobsen B., Nilssen E. S., Ohara S., Architecture of the entorhinal cortex a review of entorhinal anatomy in rodents with some comparative notes. Front. Syst. Neurosci. 11, 46 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Braak H., Braak E., Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 82, 239–259 (1991). [DOI] [PubMed] [Google Scholar]
- 56.Kunz L., Schroder T. N., Lee H., Montag C., Lachmann B., Sariyska R., Reuter M., Stirnberg R., Stocker T., Messing-Floeter P. C., Fell J., Doeller C. F., Axmacher N., Reduced grid-cell-like representations in adults at genetic risk for Alzheimer’s disease. Science 350, 430–433 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Arnold S. E., Hyman B. T., Van Hoesen G. W., Damasio A. R., Some cytoarchitectural abnormalities of the entorhinal cortex in schizophrenia. Arch. Gen. Psychiatry 48, 625–632 (1991). [DOI] [PubMed] [Google Scholar]
- 58.Harrison P. J., Weinberger D. R., Schizophrenia genes, gene expression, and neuropathology: On the matter of their convergence. Mol. Psychiatry 10, 40–68 (2005). [DOI] [PubMed] [Google Scholar]
- 59.Watrin F., Manent J. B., Cardoso C., Represa A., Causes and consequences of gray matter heterotopia. CNS Neurosci. Ther. 21, 112–122 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.E. D. Jarvis, in Encyclopedia of Neuroscience, M. D. Binder, N. Hirokawa, U. Windhorst, Eds. (Springer Berlin Heidelberg, 2009), pp. 1390–1400. [Google Scholar]
- 61.Molnár Z., Butler A. B., Neuronal changes during forebrain evolution in amniotes: An evolutionary developmental perspective. Prog. Brain Res. 136, 21–38 (2002). [DOI] [PubMed] [Google Scholar]
- 62.Chou S. J., Perez-Garcia C. G., Kroll T. T., O’Leary D. D., Lhx2 specifies regional fate in Emx1 lineage of telencephalic progenitors generating cerebral cortex. Nat. Neurosci. 12, 1381–1389 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hsing H.-W., Zhuang Z.-H., Niou Z.-X., Chou S.-J., Temporal differences in interneuron invasion of neocortex and piriform cortex during mouse cortical development. Cereb. Cortex 30, 3015–3029 (2020). [DOI] [PubMed] [Google Scholar]
- 64.Gotz M., Wizenmann A., Reinhardt S., Lumsden A., Price J., Selective adhesion of cells from different telencephalic regions. Neuron 16, 551–564 (1996). [DOI] [PubMed] [Google Scholar]
- 65.Wang C. F., Hsing H. W., Zhuang Z. H., Wen M. H., Chang W. J., Briz C. G., Nieto M., Shyu B. C., Chou S. J., Lhx2 expression in postmitotic cortical neurons initiates assembly of the thalamocortical somatosensory circuit. Cell Rep. 18, 849–856 (2017). [DOI] [PubMed] [Google Scholar]
- 66.The Gene Ontology Consortium , Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 45, D331–D338 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alfano C., Viola L., Heng J. I. T., Pirozzi M., Clarkson M., Flore G., de Maio A., Schedl A., Guillemot F., Studer M., COUP-TFI promotes radial migration and proper morphology of callosal projection neurons by repressing Rnd2 expression. Development 138, 4685–4697 (2011). [DOI] [PubMed] [Google Scholar]
- 68.B. Ermentrout, Simulating, analyzing, and animating dynamical systems: A guide to XPPAUT for researchers and students (SIAM, 2002), vol. 14.
- 69.Hecht F., New development in FreeFem++. J. Numer. Math. 20, 251–265 (2012). [Google Scholar]
- 70.Hillen T., Painter K., Global existence for a parabolic chemotaxis model with prevention of overcrowding. Adv. Appl. Math. 26, 280–301 (2001). [Google Scholar]
- 71.Hillen T., Painter K., A user’s guide to PDE models for chemotaxis. J. Math. Biol. 58, 183–217 (2009). [DOI] [PubMed] [Google Scholar]
- 72.Horstmann D., From 1970 until present: The Keller-Segel model in chemotaxis and its consequences I. Jahresber. Deutsch. Math. Verein. 105, 103–165 (2003). [Google Scholar]
- 73.Keller E. F., Segel L. A., Initiation of slime mold aggregation viewed as an instability. J. Theor. Biol. 26, 399–415 (1970). [DOI] [PubMed] [Google Scholar]
- 74.Keller E. F., Segel L. A., Model for chemotaxis. J. Theor. Biol. 30, 225–234 (1971). [DOI] [PubMed] [Google Scholar]
- 75.Wolpert L., Chapter 6 Positional information and pattern formation. Curr. Top. Dev. Biol. 6, 183–224 (1971). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/27/eabf6808/DC1