Structures of prostaglandin receptor EP2 revealed its agonists’ selectivity and G protein coupling mechanism.
Abstract
Selective modulation of the heterotrimeric G protein α S subunit–coupled prostaglandin E2 (PGE2) receptor EP2 subtype is a promising therapeutic strategy for osteoporosis, ocular hypertension, neurodegenerative diseases, and cardiovascular disorders. Here, we report the cryo–electron microscopy structure of the EP2-Gs complex with its endogenous agonist PGE2 and two synthesized agonists, taprenepag and evatanepag (CP-533536). These structures revealed distinct features of EP2 within the EP receptor family in terms of its unconventional receptor activation and G protein coupling mechanisms, including activation in the absence of a typical W6.48 “toggle switch” and coupling to Gs via helix 8. Moreover, inspection of the agonist-bound EP2 structures uncovered key motifs governing ligand selectivity. Our study provides important knowledge for agonist recognition and activation mechanisms of EP2 and will facilitate the rational design of drugs targeting the PGE2 signaling system.
INTRODUCTION
Prostaglandin E2 (PGE2) is the prostanoid most widely produced by cyclooxygenase (COX) enzymes. It regulates diverse physiological functions, including immune responses, bone formation, cardiovascular protection, inflammatory process, and reproductive activity, through interaction with E-prostanoid (EP) receptors. The EP receptor family comprises four G protein–coupled receptors (GPCRs), termed EP1 to EP4 (1, 2). Whereas nonsteroidal COX inhibitors, such as aspirin and indomethacin, are widely used to reduce inflammation, PGE2 has been prescribed clinically to induce childbirth and to treat ischemia or pulmonary hypertension (3). However, probably because of its nonspecific engagement with all four EP receptor family members, the side effects of PGE2, including headache, flushing, lethargy, and diarrhea, have limited its potential therapeutic uses (4, 5). To overcome this limitation, developing selective agonists for each EP receptor family member and elucidating the molecular mechanisms underlying the functional selectivity of individual EP receptor family members are highly important and clinically relevant.
EP2 is a Gs-coupled receptor with important roles in the modulation of blood pressure, bone formation, inflammatory response, neurodegeneration, tumorigenesis, and ocular hypertension 1 (3, 6–12). Selective activation of EP2 by synthetic compounds, such as taprenepag (TAP) and evatanepag (EVA), has been pursued as a promising therapeutic approach for ocular hypertension, bone fractures, and inflammatory diseases such as asthma. Although all four EP receptors are activated by the same endogenous molecule, PGE2, and recent crystallographic studies have provided molecular details regarding the inhibition of EP4 by the antagonist ONO-AE3-208 (13), as well as the binding of PGE2 and misoprostol to EP3 (14, 15), the structural basis for the interaction of selective agonists and G protein coupling mechanisms of many EP receptor members remains largely elusive. For example, EP2 does not have the conserved W6.48 toggle switch, which, in EP3, senses agonist binding and transmits the binding signal to induce conformational changes of EP2 on the cytoplasmic side. EP2 specifically favors Gs activation, whereas EP3 selectively engages with Gi. EP4 is known to couple to both Gs and Gi. These differences suggest that EP2 may accommodate the binding of PGE2 and Gs via structural features different from those of the other PGE2-binding receptors involving a distinct activation mechanism.
Here, we present the cryo–electron microscopy (cryo-EM) structures of EP2-Gs in complex with its endogenous ligand PGE2 and two highly selective agonists, EVA and TAP, at global resolutions of 2.8 Å, 2.8 Å, and 2.9 Å, respectively. Combined with the results of cellular and biochemical characterization, these structures revealed differential PGE2 binding modes within the EP receptor family and confirmed a potential conserved α chain carboxyl recognition motif shared by all prostanoid receptors. Moreover, these findings revealed the structural features responsible for the agonist selectivity of EP2, an atypical EP2 receptor activation mechanism, a different Gs protein coupling interface differing from that in other known receptor-Gs complexes, and the structural basis for Gs coupling.
RESULTS
Cryo-EM analysis and overall structure
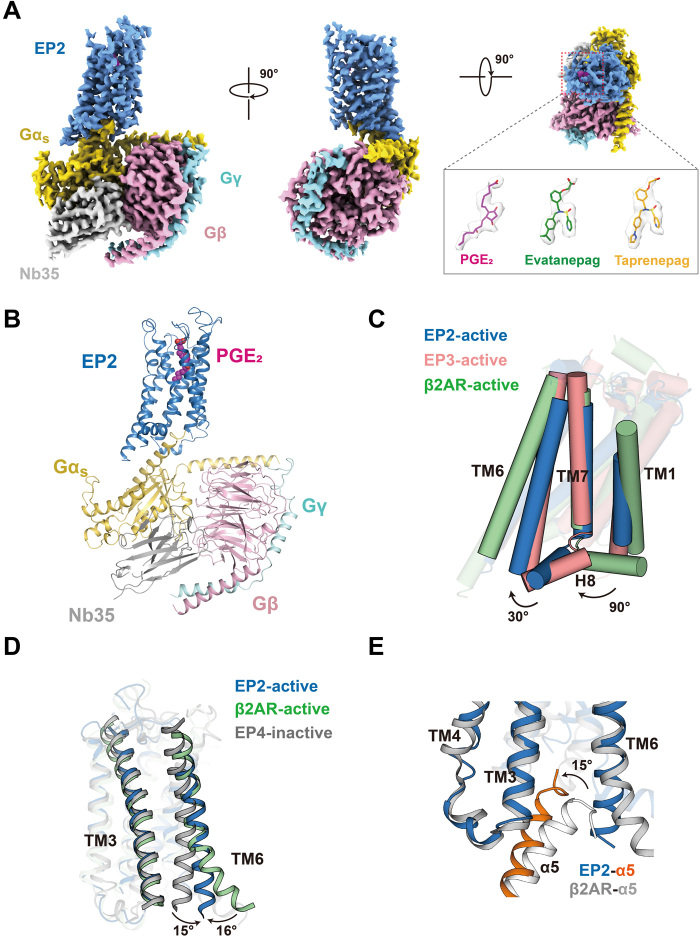
To obtain the structure of active EP2 bound to Gs complexes, we coexpressed EP2 with Gαs and Gβ1/Gγ2 in Spodoptera frugiperda (Sf9) cells, stimulated the cells with either the endogenous EP2 ligand PGE2 or a high-affinity synthetic agonist (TAP or EVA), reconstituted the complexes on the membrane, and purified them to homogeneity (fig. S1, A to C). Apyrase was used to remove guanosine diphosphate and guanosine 5′-triphosphate during complex formation to enable the formation of an agonist-bound EP2-Gs complex in a nucleotide-free state. Whereas TAP is a selective EP2 agonist undergoing a phase 2 clinical trial for the treatment of open glaucoma and ocular hypertension, EVA has been shown to improve local bone formation and fracture healing in rat models via specific EP2 binding (1, 8, 9). Cryo-imaging was carried out with a Titan Krios microscope. After two-dimensional (2D) and 3D classification, sets of 59,163, 243,913, and 238,945 particles were used to construct electron density maps for the EP2-Gs in complex with PGE2, TAP, or EVA at overall resolutions of 2.8, 2.8, and 2.9 Å, respectively (Fig. 1A, fig. S2, and table S1). In general, local Fourier shell correlation (FSC) calculations showed an overall resolution of 3.0 to 3.2 Å for the EP2 receptors and 2.7 to 2.8 Å for the G proteins. The high-resolution electron density map provided a reliable foundation for building the receptor structure containing residues 22 to 330, the three agonists, and most residues of the Gs protein trimer, except for the intracellular loop 1 (ICL1) (residues 48 to 65) and ICL3 (residues 230 to 257) domains of the receptor, the last C-terminal residue of α5 helix, and the α-helical domain of Gαs (Fig. 1, A and B, and fig. S3).
Fig. 1. Cryo-EM structures of EP2-Gs complexes.
(A) Cryo-EM density of EP2-Gs in complex with PGE2, EVA, or TAP. EP2, blue; Gαs, yellow; Gβ, pink; Gγ, cyan; Nb35, gray; PGE2, red; EVA, green; and TAP, orange. (B) Ribbon representation of the PGE2-EP2-Gs complex. EP2, blue; Gαs, yellow; Gβ, pink; Gγ, cyan; Nb35, gray; and PGE2, red. (C) Comparison of the EP2-Gs complex with the β2AR-Gs complex [Protein Data Bank (PDB) ID: 3SN6] and the active EP3 structure (PDB ID: 6AK3). The orientations of H8 of both EP2 and EP3 are significantly different from the Gs-coupled receptor β2AR. (D) Comparison of the EP2-Gs complex with the β2AR-Gs complex (PDB ID: 3SN6) and the inactive EP4 structure (PDB ID: 5YHL). The separation between TM3 and TM6 of the EP2-Gs complex was larger than the inactive EP4 structure but significantly smaller than the β2AR-Gs complex. (E) Comparison of the EP2-Gs complex structure with the β2AR-Gs complex structure (PDB ID: 3SN6). The α5 helix of the Gs in the EP2-Gs complex tilted 15° compared to that in the β2AR-Gs complex.
Although the G protein subtype selectivity differs between EP receptor subfamily members, the seven-transmembrane (7TM) bundle in the active EP2 structure is quite similar to that in the PGE2-stabilized EP3 structure, except that the orientation of the C-terminal helix 8 (H8) diverges by approximately 30° (Fig. 1C). Notably, the alignment of H8 with the 7TM bundle in both EP2 and EP3 differs notably from that in the known active GPCR structures, such as the prototype Gs-coupled receptor β2 adrenergic receptor (β2AR) (Fig. 1C). Moreover, the separation distance between transmembrane helix 6 (TM6) and TM7 and that between TM3 and TM6 in the EP2-Gs complex were significantly shorter than the corresponding distances in the β2AR-Gs complex (Fig. 1, C and D), thus leading to a 15° rotation in the insertion orientation of the Gαs α5 helix into the 7TM bundle (Fig. 1E).
Identification of a conserved α chain carboxyl recognition motif
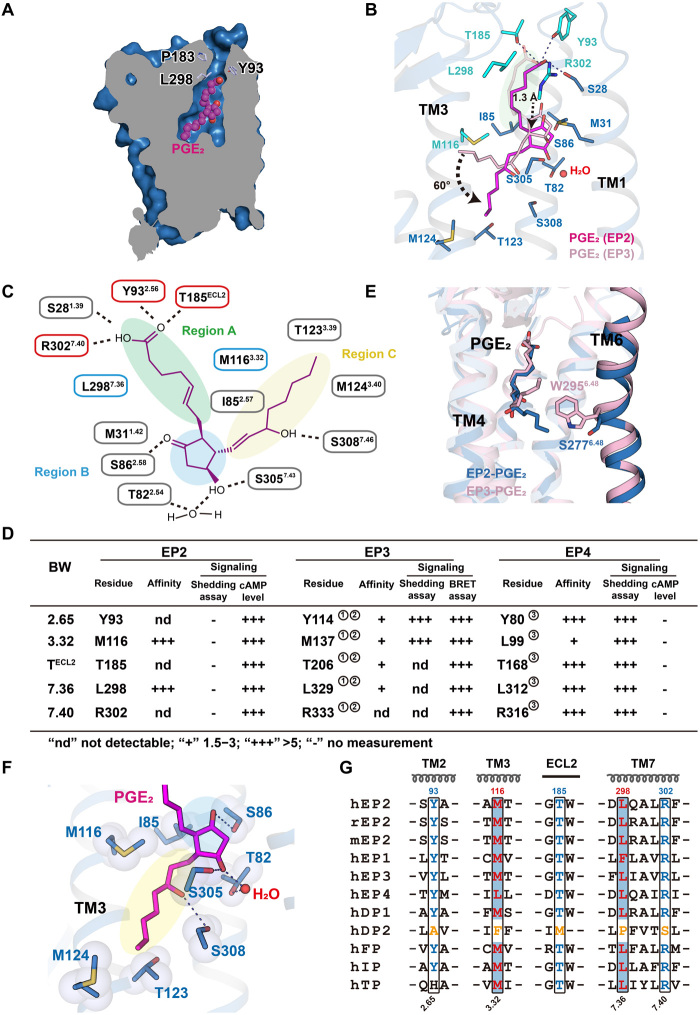
The structure of EP2 bound to the endogenous ligand PGE2 exhibited both important common characteristics and notably distinct features compared to the structure of PGE2-bound EP3 (Fig. 2, A to C and E, and fig. S4, A and B). The well-defined electron density enabled us to unambiguously assign the α chain, E ring, and ω chain of PGE2 to three separate subpockets (Fig. 2, A to C, and fig. S4C). Similar to its orientation in the PGE2-EP3 complex, the acidic group of the α chain pointed toward the outside of the plasma membrane in the PGE2-EP2 complex, with H-bonding and ionic lock interactions with the conserved Y932.65, T185ECL2, and R3027.40 residues as well as the nonconserved S281.39 residue (Fig. 2, B and C). The interactions between Y2.65, TECL2, and R7.40 with the carboxyl end of the α chain in PGE2 were also observed in the PGE2-EP3 structure 1415. Notably, this specific polar network constituting the motif Y2.65-TECL2-R7.40 and the hydrophobic residues M/L3.32-L/F7.36 along the aliphatic part of the α chain are conserved among almost all prostanoid receptor members except for prostaglandin D2 receptor (DP2), which putatively shows a distinct ligand-binding mode (Fig. 2G). The carboxyl group of DP2 is likely buried inside the ligand-binding pocket of DP2, potentially due to a different structural fold of extracellular loop 2 (ECL2) and absence of the specific polar network in the extracellular region of DP2 compared to that of other prostanoid receptors, including EP2 and EP3 (fig. S4, L to N) (16). Consistent with this observation and with sequence comparisons, mutation of these conserved residues in EP2, EP3, and EP4 significantly impaired both receptor activity and ligand binding (Fig. 2D, and fig. S4, D to H), thus highlighting the potential importance of the carboxyl α chain recognition motif containing Y2.65-TECL2-R7.40 and M/L3.32-L/F7.36 in the detection of specific endogenous prostanoid ligands.
Fig. 2. Binding of the endogenous ligand PGE2 to EP2.
(A) Vertical cut-through view showing a solvent-accessible channel in EP2 that was defined by Y932.65 and P183ECL2 (light blue). (B) Common motif composed of Y2.65, TECL2, R7.40, M/L3.32, and L/F7.36 (cyan) in the binding pocket within PGE2-EP2 and PGE2-EP3 (PDB ID: 6AK3) complex structures. PGE2 was shown in fuchsia or pink, respectively. (C) Region division in EP2-PGE2 binding pockets corresponding to the structure of PGE2. Subpockets containing H-bonds (dotted line) and hydrophobic interactions with the α chain, E ring, or ω chain of PGE2 were named region A, B, or C, respectively. (D) The effects of the common motif in EP2, EP3, and EP4 for PGE2 binding and G protein signaling. Corresponding curve of EP2-PGE2 binding and PGE2-induced Gs signaling and detailed statistical evaluation and receptor expression levels are shown in fig. S4 (D to H). Biochemical or cellular data cited here refer to ① (14), ② (15), and ③ (13). (E and F) The ω chain of PGE2 interacted with different residues in EP2 or EP3, respectively. PGE2 are shown as slate or pink sticks in EP2 or EP3 structures (E). EP2 interacts with TM3 and TM7 residues, including I852.57, M1163.32, T1233.39, M1243.40, and S3087.46 rather than TM6 (F). (G) Sequence alignment for prostanoid receptors. Notably, the specific polar network constituting motif Y2.65-TECL2-R7.40 and the hydrophobic residues M/L3.32-L/F7.36 were conserved across all prostanoid receptors except for DP2 (residues colored yellow).
Different PGE2 binding modes of the EP2 and EP3 binding pockets
Distinct features and different interaction modes were observed within the ligand binding pocket of the PGE2-EP2 complex and the PGE2-EP3 complex. Whereas the binding pocket in the PGE2-EP3 complex is totally enclosed and PGE2 assumes an L-shaped posture within the EP3 pocket, the binding pocket of the PGE2-EP2 structure contains a solvent access channel defined by Y932.65 and P183ECL2 on the extracellular side, and PGE2 assumes a more extended conformation due to a substantial downward shift in the subpocket to accommodate the ω chain (Fig. 2A and fig. S4, A and B). Compared to that in EP3, the ω chain in PGE2 is inserted more deeply and is rotated by approximately 60°, pointing toward TM3 rather than TM5 and TM6 (Fig. 2B). Notably, EP2 does not contain the conserved W6.48 toggle switch, which participates in a direct interaction with the ω chain in EP3 (Fig. 2E and fig. S4, I and J). The ω chain in PGE2 does not contact residues in TM6; instead, it interacts mainly with residues in TM3 and TM7, including I852.57, M1163.32, T1233.39, M1243.40, and S3087.46 (Fig. 2F). The downward swing of the ω chain caused the E ring in the EP2 structure to move downward by approximately 1.3 Å, which enabled the constitution of polar networks by T822.54 and S3057.43 of EP2 and the hydroxyl and carbonyl groups of the E ring (Fig. 2, A to C and F, and fig. S4K). Notably, H-bonding interactions with the E ring hydroxyl groups were hypothesized to be essential for PGE2-induced receptor activity but were not observed in the structure of the EP3-PGE2 complex (14, 15, 17).
Structural basis for selective EP2 agonism
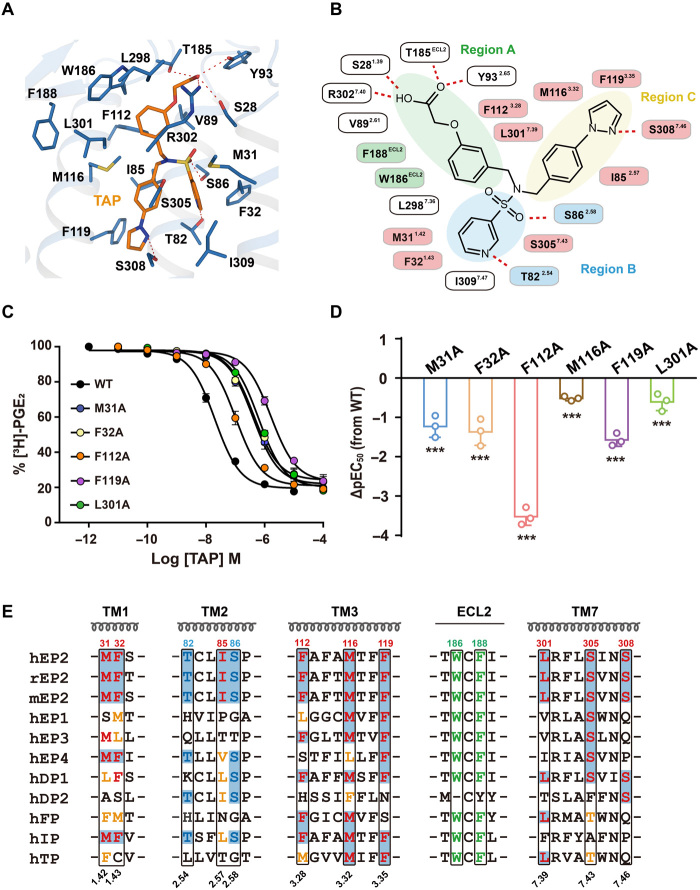
Selective activation of EP2 has therapeutic potential in many diseases, including glaucoma, bone fractures, pulmonary fibrosis, and allergy (1, 8–10). Two selective EP2 agonists, TAP and EVA, have been widely used in EP2 functional studies and are being tested in clinical trials. To understand the structural basis of the ligand selectivity of EP2 and facilitate the future design of selective EP2 agonists, we determined the cryo-EM structures of EP2-Gs in complex with TAP and EVA (Fig. 3A and fig. S5A).
Fig. 3. Specific engagement of TAP with EP2.
(A) Detail interactions of TAP (marine) with EP2 (orange) were shown. H-bonds were depicted as red dashed lines. (B) 2D representation of contacts between EP2 with the ligand TAP. The polar bonds were presented by red dotted lines. Residues F188ECL2 and W186ECL2 that were implicated in hydrophobic interactions with the region A subpocket were presented in green. T822.54 and S862.58 formed stronger H-bond engagements with the region B subpocket and were presented in blue. The hydrophobic contacts of EP2 with TAP were colored pink. (C and D) Effects of mutations of TAP contacting residues in ligand binding and Gs signaling. The competitive binding curve of TAP toward EP2 was created using 3H-PGE2 as the isotope probe (C). Bars represent the differences in calculated potency of TAP (△pEC50) for each mutation according to wild-type (WT) EP2. ***P < 0.0001 [one-way analysis of variance (ANOVA) compared with the response of WT] (D). See fig. S5 (E and F) for detailed statistical evaluation and fig. S4D for receptor expression levels. (E) Sequence alignment of prostonoid receptors. Note that the combination of F1123.28 and L3017.39 was only found in EP2, DP1, and FP; I852.57, F1193.35, S3057.43, and S3087.46exhibited great diversity among prostonoid receptor families.
Similar to the endogenous ligand PGE2, TAP and EVA bind to EP2 via a three subpocket mode but with notably more hydrophobic interactions (Fig. 3, A and B, and fig. S5, A to D and G). In the region A subpocket, whereas the carboxyl end of TAP or EVA maintains a H-bond network interaction with the Y2.65-TECL2-R7.40 motif, the phenyl linker of these two compounds forms additional interactions with hydrophobic residues contributed by TM3, TM7, and ECL2 (Fig. 3, A and B, and fig. S5, A, B and G). Among these contacting residues, W186ECL2 and F188ECL2 are conserved across all nine prostanoid receptor family members, and this interacting pattern via the combination of F1123.28 and L3017.39 was found only in EP2, DP1, and FP (Fig. 3E). Consistent with this finding, mutation of F1123.28 or L3017.39 to alanine (Ala) significantly reduced ligand binding and TAP-induced EP2 activity (Fig. 3, C and D, and fig. S5, E and F). Moreover, the F1123.28 S mutant of EP2, which substitutes F1123.28 of EP2 with an equivalent residue of S953.28 in EP4, showed no significant effects on PGE2 binding but decreased the affinity for TAP by approximately 15-fold (fig. S6). Conversely, the structurally equivalent S953.28 F mutant of EP4 decreased its binding toward its selective agonist L902688 by approximately fivefold (fig. S6). Similarly, the L3017.39V and L3017.39I mutants of EP2, which replace L3017.39 of EP2 with an equivalent residue of V3227.39 in EP3 or I3157.39 in EP4, decreased the affinity for TAP by approximately twofold (fig. S6). Moreover, the reverse mutations of V3227.39L in EP3 and I3157.39L in EP4 decreased the affinity of EP3 for sulprostone and of EP4 for L902688 by approximately twofold (fig. S6). These observations indicated that a large hydrophobic group is generally favored as a linker connecting to the carboxylic acid end in the region A subpocket of prostanoid receptors and that certain modifications at this site could improve the selectivity of specific prostanoid receptor family members.
In the region B subpocket, a pyridine ring and a sulfenic acid form stronger H-bonds with T822.54 and S862.58 in both the EVA- and TAP-bound EP2 structures than in the PGE2-bound EP2 structure (Figs. 2C and 3B and fig. S5B). Moreover, 3-pyridine-sulfonamide makes hydrophobic contacts with M311.42 and F321.43. Notably, among the nine prostanoid receptors, this pair of interacting residues was found only in EP2, EP4, and IP (Fig. 3, A, B, and E, and fig. S5, A and B). Accommodation of the tert-butyl or phenylimidazole group in EVA or TAP, respectively, is dependent on residues composing the region C subpocket in the EVA-EP2 and TAP-EP2 complexes, including I852.57, M1163.32, F1193.35, S3057.43, and S3087.46 (Fig. 3, A and B, and fig. S5, A, B and G). These residues are highly diverse among prostanoid receptor family members, allowing the design of selective agonists (Fig. 3E). The interaction of TAP with residues in the B and C regions of EP2 was supported by the results of both ligand binding and cyclic adenosine monophosphate (cAMP) accumulation assays (Fig. 3, C and D, and fig. S5, E and F). Collectively, the hydrophobic ECL2 residues and the H-bonds donated by the Y2.65-TECL2-R7.40 and T2.54-S2.58 motifs likely contribute to the high-affinity binding between EVA and TAP. In particular, the M1.42-F1.43 hydrophobic motif and the sequence diversity in the region C subpocket of EP2 could be important determinants of EVA and TAP selectivity (fig. S5G).
Structure of active EP2
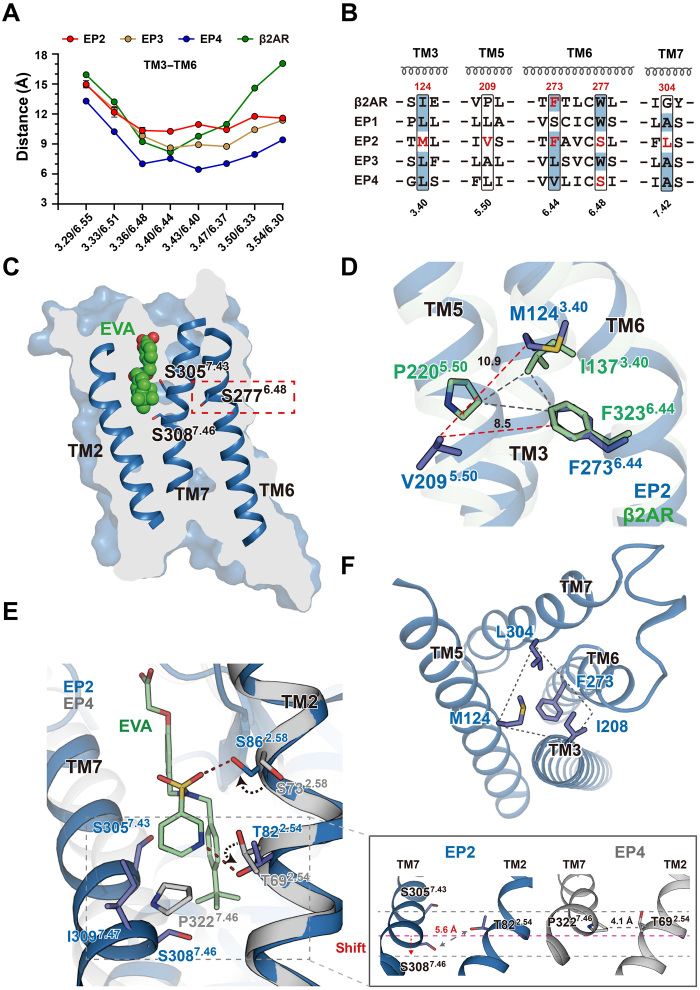
Compared to the ONO-AE3-208 (antagonist)–bound inactive structure of EP4, the three agonist-bound EP2 structures exhibit an inward shift of TM5 and TM7 toward TM3 and an outward shift of TM6 (fig. S7, A to C). Although the separation between TM3 and TM6 at the cytoplasmic end in the active structures of EP2 is smaller than that in the structure of the β2AR-Gs complex (Fig. 1D), it is significantly larger than that in the inactive structure of EP4 (17.1 versus 9.4 Å) (Fig. 4A). The larger separation of TM3 and TM6 is a hallmark of GPCR activation, indicating an active state of the EP2 structures in complex with each of the three agonists.
Fig. 4. The active structure of the EP2.
(A) Plot of Cα distances of residues between TM3 and TM6 of EP2 in PGE2, EVA and TAP-bound complex structures, and structures of the active EP3 (PDB ID: 6AK3 and PDB ID: 6M9T), active β2AR (PDB ID: 3SN6), and inactive EP4 (PDB ID: 5YHL). (B) Sequence alignment of residues involving activation in prostaglandin E receptor subfamily members (EP1 to EP4) and β2AR. The residues involving EP2 activation and their counterparts in other receptors are highlighted in red or shaded with blue backgrounds. (C) Vertical cut-through view of EVA-bound EP2 in the EP2-Gs complex structure. (D) Superimposition of EVA-bound EP2 with the active β2AR (PDB ID: 3SN6) aligned at the “IPF motif”. (E) The detailed interactions of upper parts of TM3-TM5-TM6-TM7 in the EVA-bound EP2-Gs complex. (F) Binding of EVA induced the activation of EP2 that presumably started with the rotation of side chains of S862.58 and T822.54 by approximately 150° to form polar interactions with the hydroxyl groups of the E ring of PGE2 or the pyridine ring in the EVA (left). These conformational changes and the interaction between the EVA with the S3057.43 and I3097.47 caused a downshift of the TM7 and a larger separation between the S3087.46 and the T822.54 (left). A close-up view of the structural alignment of the S/P7.46-T2.54 interactions between the EP2-EVA structure and the inactive EP4 (PDB ID: 5YHL) structure, which indicated a separation between the TM2 and TM7, as well as the overall downward shift of TM7 (right).
Notably, sequence alignment suggested that EP2 does not contain the conserved W6.48 toggle switch and that the canonical I3.40 P5.50 F6.44 (IPF) triad motif was structurally substituted with the M1243.40 V2095.50 F2736.44 motif (Fig. 4B). The toggle switch is well known for its critical role in sensing ligand binding and the subsequent structural rearrangement to tether the upper TM bundles required for receptor activation. Notably, W6.48 of EP2 was substituted with S2776.48, which does not interact with the endogenous ligand PGE2 or either of the high-affinity agonists (EVA and TAP) (Fig. 4C and fig. S7, D and E). Moreover, no tight packing occurs between the canonical motif-equivalent residues in the IPF motif (M1243.40 V2093.50 F2736.44); instead, the hydrophobic interactions between M1243.40, I2085.49, F2736.44, and L3047.42 constitute important connections for the upper TM bundles of TM3, TM5, TM6, and TM7 (Fig. 4, D and F). These observations indicate an unconventional conformation of EP2 in the active state compared to those of all known active class A GPCRs.
Inspection of all three agonist-bound EP2 structures showed that compared to the equivalent residues in antagonist-bound EP4, the side chains of both S862.58 and T822.54 are rotated approximately 150° to form polar interactions with the hydroxyl groups of the E ring in PGE2 or the pyridine ring in EVA and TAP (Fig. 4E and fig. S7, F and G). These conformational changes and the interactions between the agonists and S3057.43 and I3097.47 cause a downward shift of TM7 and increase the separation between S3087.46 and T822.54 compared to that of the equivalent residues P3227.46 and T692.54 in antagonist-bound EP4 (Fig. 4E). The overall downward shift of TM7 causes a series of structural rearrangements of TM7 and TM6, thus enabling hydrophobic packing of L3047.42 and F2736.44 with the central M1243.40, with a different interaction pattern connecting TM3-TM6-TM7 compared to that in the inactive EP4 structure (Fig. 4F and fig. S7H). In addition, a direct interaction between the ω chain and M1243.40 may further stabilize the packing of M1243.40 with L3047.42 and F2736.44 in the structure of active PGE2-bound EP2 (fig. S7I). Consistent with these observations, mutation of T822.54 and S862.58 to Ala significantly decreased both the binding affinity and the cAMP accumulation activity of EP2 (fig. S7, J to M).
On the cytoplasmic side, EP2 contains the conserved E1333.49 R1343.50 Y1353.51 (ERY) motif and the D3117.49 P3127.50 XXF/Y3157.53 motif. These motifs exhibit significant rearrangement in EP2 relative to inactive EP4 (fig. S7, N and O). In particular, Y1353.51 in the ERY motif forms an H bond with H140ICL2 to stabilize the ICL2 conformation in the structures of active EP2.
EP2-Gs coupling
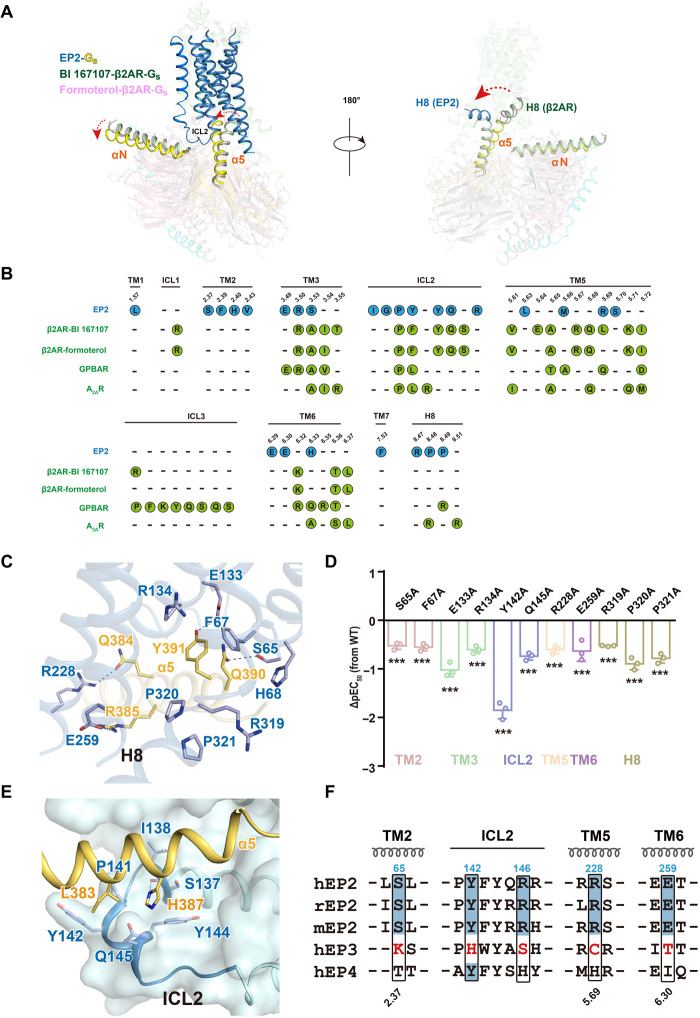
The cavity on the cytoplasmic side of the active EP2 structures is narrower than that in the structure of the β2AR-Gs complex due to the smaller separation of the TM3-TM6-TM7 bundles. This alteration triggers a 15° tilt in the α5 helix of Gαs, bringing it more perpendicular to the plasma membrane and closer to the cytoplasmic end of TM2 in the EP2-Gs complex structures than in the structures of the G-protein-coupled bile acid receptor (GPBAR)–Gs, adenosine A2A receptor (A2AR)–mini-Gs, or prototypical β2AR-Gs complexes (Fig. 5A and fig. S8A) (18–21). Unexpectedly, the C-terminal H8 helix assumes a unique orientation compared to those in reported GPCR-G protein complex structures, diverging by 90° and directly contacting the Gs protein (Figs. 1, B and C, and 5A and fig. S8A).
Fig. 5. EP2-Gs coupling and G protein selectivity.
(A) Superposition of the structures of EP2-Gs and β2AR-Gs (PDB ID: 3SN6 and 7BZ2) complex according to the transmembrane helices bundle. EP2 is in sky blue, BI 167107-β2AR is in green, and formoterol-β2AR is in light pink. Rotation of the α helices and the interaction of the Gαs with H8 were highlighted, Gαs in the EP2-Gs complex is in yellow orange, Gαs in BI 167107-β2AR-Gs is in green, and Gαs in formoterol-β2AR-Gs is in light pink. (B) Comparison of the Gs coupling interface in EP2 and known receptor-Gs complexes including β2AR-Gs (PDB ID: 3SN6 and 7BZ2), GPBAR-Gs (PDB ID: 7CFN), and A2AR–mini-Gs (PDB ID: 6GDG). Residues involved in contacting with Gs of EP2 were illustrated as blue dots, and those of β2AR, GPBAR, and A2AR were illustrated as green dots. (C) The detailed interactions of α5 helix of Gs with residues of TM2 and H8 of EP2 in the PGE2-bound EP2-Gs complex structure. Polar interactions are depicted as blue dashed lines. (D) Effects of mutations in TM2 or H8 of EP2 on PGE2-induced cAMP accumulation. Bars represent the differences of calculated potency (pEC50) between WT EP2 and its mutant. Values are means ± SD from three independent experiments performed in triplicate. ***P < 0.001; comparison between WT EP2 and its mutant. All data were analyzed by two-sided one-way ANOVA with Tukey’s test. (E) The detailed interactions of ICL2 of EP2 with α5 helix of Gs in the PGE2-bound EP2-Gs complex structure. H-bond is depicted as a dashed line. (F) Sequence alignment of residues involving Gs coupling in human, mouse, and rat EP2, which are compared with residues at corresponding positions in human EP3 and EP4.
EP2 has a buried surface (approximately 1200 Å2) for Gs, similar to that of β2AR (3SN6: 1414 Å2 and 7BZ2: 1365 Å2). The interaction interface between EP2 and Gs comprises six TM helices (TM1 to TM3 and TM5 to TM7), ICL2, and the C-terminal H8 helix (Fig. 5A). Unlike any other known receptor–G protein complex, such as GPBAR-Gs, A2AR–mini-Gs, or prototypical β2AR-Gs complexes (18–21), the C-terminal wavy hook in the Gαs α5 helix of EP2-Gs is specifically positioned on a hydrophobic surface created by the cytoplasmic end of TM1 and TM2 and the C-terminal H8 helix of EP2 (Fig. 5, B and C, and table S2). The density of the last wavy hook residue L394 of the Gαs was unclear and was not included in the final model. Both hydrophobic and polar interactions, such as interactions between the wavy hook residues in the Gαs α5 helix with S652.37, F672.39, and H682.40 in TM2 and the R3198.47 P8.48 P3218.49 motif in EP2 H8, compose this interface in the EP2-Gs-PGE2 complex (Fig. 5, B and C, and table S2). In addition, Q390 and Y391 in the α5 helix of Gαs interact extensively with the conserved E1333.49 and R1343.50 residues in the conserved Glu/Asp-Arg-Tyr (E/DRY) motif (Fig. 5C and table S2). The other helical portion of the Gαs α5 helix fits into the cavity formed by residues in TM3 and TM5-TM6 and then extends to ICL2. Mutation of most of these residues in TM2 or the H8 helix in EP2 significantly reduced ligand-induced cAMP accumulation but showed no significant effects on PGE2 binding, except for the ionic lock residue E1333.49 (Fig. 5D and fig. S8, B to D).
Interactions mediated by ICL2 in EP2 are determinants of Gs coupling and have been reported to play key roles in the mechanism underlying the G protein coupling selectivity of EP receptors (22, 23). In particular, S137, I138, P141, Y142, Y144, and Q145 in ICL2 form a hydrophobic pocket to accommodate L383 and H387 in the Gαs α5 helix (Fig. 5E). This tight hydrophobic packing between the Gαs α5 helix and the ICL2 of EP2 was not observed in A2AR–mini-Gs or GPBAR-Gs complex structures (fig. S8, E and F) (18–21). Moreover, H387 forms an additional H-bond with the backbone carbonyl of S137ICL2 (Fig. 5E). Conversely, similar to F138 in the prototypical GPCR β2AR, Y142 in EP2 fits into a well-defined pocket formed by I383G.H5.15, R380G.H5.12, C379G.H5.11, F376G.H5.08, V217G.S3.01, F219G.S3.03, and H41G.S1.02 in Gαs (fig. S8G). At the C-terminal end of ICL2, Q145 and R146 make additional contacts with Gαs. Y142ICL2 and R146ICL2 share similar properties with the corresponding residues Y125 ICL2 and H129 ICL2 in EP4, which also exhibits Gs coupling ability. However, in Gi-coupled EP3, these residues are replaced by smaller residues, namely, H163ICL2 and S167ICL2 (Fig. 5F). Consistent with these observations, mutation of most ICL2 residues involved in Gs interactions severely impaired EP2 activity but showed no significant effects on ligand binding. In particular, although Y142ICL2A reduced PGE2 binding by approximately 7-fold, it reduced PGE2-induced cAMP accumulation by nearly 50-fold, further supporting its direct role in mediating the Gs interaction (Fig. 5D and fig. S8, B to D). Moreover, previous studies have shown that replacement of the ICL2 residues in EP3 with those in EP2 switches the Gi coupling specificity to Gs specificity, whereas replacing the ICL2 residues in EP2 with those of EP3 abolishes its Gs coupling activity (22).
In addition to the ICL2 residues, residues S652.37, R2285.69, and E2596.30 participate in specific H-bonds or charge-charge interactions with Q390, D381/Q384, and R385 in the α5 helix of Gαs, respectively (Fig. 5C). Notably, Q390, Q384, and R385 in Gαs were replaced with G380, I344, and A345 in Go and with D350, I344, and K345 in Gi1; moreover, S652.37, R2285.69, and E2596.30 in EP2 were substituted with K862.37, C2605.69, and T2776.30 in EP3 (Fig. 5F and fig. S8H). Mutation of these residues diminished the ligand-induced cAMP accumulation activity of EP2, supporting the essential role of these residues in mediation of Gs coupling (Fig. 5D and fig. S8, B to D). Therefore, specific residues in TM2, TM5, and TM6 of EP2, which form interactions with the polar motif Q384RXXXXQ390 located in the α5 helix of Gαs, contribute to the Gs coupling of EP2.
DISCUSSION
Here, we present the cryo-EM structures of EP2-Gs in complex with its endogenous ligand PGE2 as well as two highly selective and potent synthesized agonists, TAP and EVA. Both the endogenous agonist PGE2 and the synthesized agonists TAP and EVA bind to EP2 in a three-subpocket mode. Whereas the conserved Y-T-R and M/L-L/F motifs are identified as important common features by which EP receptors recognize the carboxyl α chain of the endogenous ligand PGE2, the different subpockets in individual family members are shown to accommodate the ω chain. The EP receptor family is a unique group of lipid receptors that recognize the same lipid, PGE2, but transduce distinct signals by coupling to different G protein subtypes. The observed common and distinct features of recognition of PGE2 by EP2 exemplified the mechanism by which the evolution of the same subfamily of lipid receptor members promotes a diversity of receptors with different functions. By solving the structures of EP2 in complex with TAP and EVA, we demonstrate that specific combinations of hydrophobic residues in TM1 and TM3 are important structural determinants for EP receptor subtype selectivity, a finding expected to facilitate further design of EP2-selective ligands. Unlike the other EP family member EP3, EP2 does not have a conventional toggle switch to sense agonist binding. Instead, agonist binding to EP2 causes the separation of TM2 and TM7 at the ligand binding site, promotes the downward shift of one helical turn in TM7, and enables the packing of M1243.44, F2736.44, and L3047.42, an atypical activation mechanism. Last, inspection of the EP2-Gs interface and subsequent mutagenesis studies revealed key characteristics determining the Gs protein coupling of the EP2 receptor. Combined with our structure, future structural works of EP4-Gs/Gi and EP3-Gi complexes could disclose the mechanism underlying G protein coupling selectivity of the EP receptors. Collectively, our present study identifies the mechanisms underlying specific recognition of endogenous PGE2 and synthesized selective agonists by EP2, its unconventional activation, and G protein coupling mode. The structural basis of EP2 agonist selectivity elucidated from the current work is expected to greatly facilitate future design of EP2 ligands to treat various human diseases, such as osteoporosis, chronic neurodegenerative diseases, cancer, and ocular or pulmonary hypertension.
MATERIALS AND METHODS
Antibodies and reagents
The monoclonal anti-FLAG (2366), anti-Gs (HPA028386), anti-mouse IgG-Peroxidase antibody (A4416), anti-FLAG M1 affinity resin (A4596), and Apyrase (A6132-500UN) were purchased from Sigma-Aldrich. Cholesteryl hemisuccinate tris salt (CHS) (102601-49-0), lauryl maltose neopentyl glycerol (4216588), and glyco-diosgenin (GDN101) were purchased from Anatrace. Compounds including PGE2 (HY-101952), TAP (HY-14899), and EVA (HY-14839) were purchased from MedChemExpress. Protease inhibitor cocktail, EDTA-free (B14001); High-Affinity Ni-charged Resin FF (L00666-25); and X-tremGENE HP DNA Transfection were purchased from bimake, GenScript, and Roche, respectively.
Cell culture
COS-7 cells were cultured in Dulbecco’s modified Eagle’s medium (Gibco, catalog no. 11965084) with 12% fetal bovine serum (Gibco, catalog no. 16000-044) in a 37°C incubator with 5% CO2.
Constructs
The human wild-type (WT) EP2 gene was cloned into pFastBac1 vector. Native signal peptide was replaced with the hemagglutinin signal peptide (HA) to enhance receptor expression, followed by a Flag tag (DYKDDDK) to facilitate the purification of the complex. Human dominant negative Gs (DNGs), human Gβ1 with the N-terminal 6 × histidine tag, and human Gγ2 were subcloned into pFastBac vector. EP2 mutants M31A, F32A, S65A, F67A, T82A, S86A, Y93A, F112A, F112S, M116A, F119A, M124A, E133A, R134A, Y142A, Q145A, T185A, R228A, E259A, L298A, L301A, L301V, L301I, R302A, S305A, S308F, R319A, P320A, and P321A; EP3 mutant V332L; and EP4 mutants S95F and I315L were generated by using the QuikChange mutagenesis kit.
Expression of EP2 and Gs/Gβγ heterotrimer
Recombinant baculovirus was generated using the Bac-to-Bac Baculovirus expression system (Invitrogen). FuGENE HD transfection reagent (Promega) was used for baculovirus preparation as we described before (18). The Sf9 cells were grown in suspension in ESF921 medium (Expression Systems) at a density of 3 × 106 cells per milliliter, infected with EP2 baculovirus, and incubated at 27°C, 110 rpm for 48 hours. The harvested cells were collected by centrifugation, flash-frozen in liquid nitrogen, and stored at −80°C. The Gs heterotrimer was coexpressed by infected virus encoding Gs and Gβγ. The infected insect cells were cultured at 27°C, 110 rpm for 48 hours. The cells were collected by centrifugation and the cell pellets were stored at −80°C.
EP2-Gs complex formation and purification
Sf9 cell pellets transfected with EP2 and the Gs heterotrimer were resuspended in lysis buffer [20 mM Hepes (pH 7.4), 100 mM NaCl, 3 mM MgCl2, and 5 mM CaCl2], in the presence of leupeptin (2.5 mg/ml) and benzamidine (0.2 mg/ml). The EP2-Gs complex was assembled in membrane, and the mixture was incubated for 2 hours at room temperature in the presence of 10 μM EVA, PGE2, or TAP; Nb35 (10 mg/ml); and apyrase (10 mg/ml). Then, the mixture was solubilized in buffer composed of 20 mM Hepes (pH 7.4), 100 mM NaCl, 3 mM MgCl2, 5 mM CaCl2, 0.5% (w/v) lauryl maltose neopentylglycol (LMNG, Anatrace), 0.1% (w/v) CHS, leupeptin (2.5 mg/ml), and benzamidine (2.5 mg/ml) for 2 hours at 4°C. Supernatant was then collected by centrifugation at 25,000 rpm for 30 min, and the solubilized complex was immobilized on the Flag resin for 2 hours at 4°C. Complex loaded on the Flag column was washed with 20 column volumes of 20 mM Hepes (pH 7.4), 100 mM NaCl, 3 mM MgCl2, 5 mM CaCl2, 0.01% (w/v) LMNG, 0.002% (w/v) CHS, 10 μM ligand, leupeptin (2.5 mg/ml), and benzamidine (2.5 mg/ml). The EP2-Gs complex was eluted with buffer containing 20 mM Hepes (pH 7.4), 100 mM NaCl, 0.01% (w/v) LMNG, 0.002% (w/v) CHS, 10 μM ligand, 5 mM EGTA, and FLAG peptide (0.2 mg/ml). Complex was collected and concentrated, and then purified by Superdex 6 Increase 10/300 GL column (GE Healthcare). The complex fractions were concentrated with a 100-kDa molecular weight cut off (MWCO) Millipore concentrator for EM experiments.
Cryo-EM grid preparation and data collection
Three microliters of the purified EVA, TAP, and PGE2-bound EP2-Gs complexes at ~5.0 mg/ml was applied onto glow-discharged holey carbon grids (Quantifoil R1.2/1.3). Excess samples were blotted for 3.0 s with a blot force of 3 and were vitrified by plunging into liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific). Frozen grids were transferred to liquid nitrogen and stored for data acquisition. Cryo-EM imaging was performed on a Titan Krios at 300 kV in the Center of Cryo-Electron Microscopy, Zhejiang University (Hangzhou, China). Micrographs were recorded using a Gatan K2 Summit detector in counting mode with a pixel size of 1.014 Å using the SerialEM software (24). Movies were obtained at a dose rate of about 8.0 electrons per Å2 per second with a defocus ranging from −0.5 to −2.5 μm. The total exposure time was 8 s and 40 frames were recorded per micrograph. A total of 2514, 4785, and 2414 movies were collected for the EVA, TAP, and PGE2-bound EP2-Gs complexes, respectively.
Cryo-EM data processing
Cryo-EM image stacks were aligned using MotionCor2.1 (25). Contrast transfer function (CTF) parameters for each micrograph were estimated by Gctf (26). The following data processing was performed using RELION-3.0-beta2 (27).
For the EVA-bound complex, automated particle selection using Gaussian blob detection yielded 1,836,927 particle projections. The projections were subjected to reference-free 2D classification to discard poorly defined particles, producing 1,126,825 particle projections for further processing. The map of the GPBAR-Gs complex (EMD-30344) low-pass–filtered to 40 Å was used as a reference model for 3D classification, resulting in one well-defined subset with 390,854 projections. Further 3D classifications focusing the alignment on the receptor produced one good subset accounting for 238,945 particles, which were subsequently subjected to 3D refinement, CTF refinement, and Bayesian polishing. The final refinement generated a map with an indicated global resolution of 2.9 Å at an FSC of 0.143.
For the TAP-bound complex, 3,451,005 particle projections from the automated particle selection were subjected to 2D classification, producing 859,499 particle projections for further processing. The density map of the EVA-bound EP2-Gs complex low-pass–filtered to 40 Å was used as a reference model for 3D classification, resulting in one well-defined subset with 243,913 projections. The selected projections were subsequently subjected to 3D refinement, CTF refinement, and Bayesian polishing. The final refinement generated a map with an indicated global resolution of 2.8 Å.
For the PGE2-bound complex, automated particle picking yielded 1,928,972 particle projections. The projections were subjected to 2D classification, producing 627,953 particle projections for further 3D classification. Initial 3D classification using the map of EVA-bound EP2-Gs complex as reference produced two good subsets with 347,373 projections. Further 3D classifications focusing the alignment on the complex produced one high-quality subset accounting for 59,163 particles, which were subsequently subjected to 3D refinement, CTF refinement, and Bayesian polishing. The final refinement generated a map with an indicated global resolution of 2.8 Å. Local resolution was determined using the Bsoft package with half maps as input maps (28).
Model building and refinement
The initial homology model of the EP2 receptor was generated using SWISS-MODEL (29). The coordinate of the GPBAR-Gs complex (PDB ID: 7CFM) (18) was to generate the initial models of Gs and Nb35. Agonists’ (EVA, TAP, and PGE2) coordinates and geometry restraints were generated using phenix.elbow (30). Models were docked into the EM density map using UCSF Chimera (31). This initial model was then subjected to iterative rounds of manual adjustment based on the side-chain densities of bulky aromatic amino acids and automated refinement in Coot and Phenix (30, 32), respectively. The final refinement statistics were validated using the module “comprehensive validation (cryo-EM)” in PHENIX (33). The final refinement statistics are provided in table S1.
Radioligand binding assay
The plasmids encoding WT or mutant EP2 were transfected into COS-7 cells using Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific) according to the manufacturer’s instruction and cultured at 37°C with 5% CO2 for 48 hours. Transfected cells were collected by a scraper and centrifuged in ice-cold lysis buffer composed of 50 mM Hepes (pH 7.4) and 2 mM EDTA. WT or mutant EP2-expressing cell membranes were separated from other cell contents using a dounce homogenizer and then collected by centrifugation at 25,000g for 45 min. Cell membrane sediments were homogenized in ice-cold buffer containing 75 mM tris (pH 7.4), 1 mM EDTA, and 12.5 mM MgCl2. The expression of WT or mutant EP2 in COS-7 cell was determined by Western, radioligand binding, and Bradford protein assay. For saturation binding assay, the cell membranes (~100 μg) were incubated with [3H]-PGE2 (PerkinElmer) at different concentrations, shaking for 60 min at 30°C. Nonspecific binding was determined in the presence of 10 μM ligand. The same amount of membranes were used for competitive binding assay, which were incubated with 1.25 nM [3H]-PGE2 (PerkinElmer) and a range of concentrations of cold competing ligand for 60 min at 30°C shaking in binding buffer [50 mM Hepes (pH 7.4), 10 mM MgCl2, 1 mM EDTA, 0.1% protease-free BSA, and 0.01% ascorbic acid]. The cell membranes were then filtered over GF/C glass microfiber filters (Whatman) presoaked in ice-cold binding buffer supplemented with 0.5% polyethylenimine using a 96-well filtermate harvester (PerkinElmer) and syringed three times with 2 ml of ice-cold assay buffer to remove free [3H]-PGE2. Last, the filters were dried and subjected to liquid scintillation counting on a MicroBeta TriLux scintillation counter (PerkinElmer). Data were analyzed by GraphPad Prism 7 (GraphPad Software Inc.).
cAMP accumulation assay
The coding sequence of WT human EP2 followed by a hemagglutinin signal peptide and Flag tag at the N terminus was subcloned into the pcDNA3.1 vector. Point mutations of EP2 were generated using the Q5 site-Directed Mutagenesis kit (NEB). The WT and mutant EP2 constructs were transfected into COS-7 cells and harvested at 48 hours after transfection. Expression level of the WT and mutant EP2 receptors was determined by flow cytometry with an anti-Flag M2-FITC antibody (Sigma-Aldrich, F4049). cAMP level was measured using GloSensor cAMP assay (Promega) for EP2 as previously described (34). For GloSensor cAMP assay, WT and mutant EP2 and the Glosensor plasmid were cotransfected into COS-7 cells. The infected cells were plated into 96-well plates and cultured for 48 hours at 37°C with 5% CO2. A serum-free medium with 5% v/v dilution of the GloSensor cAMP reagent stock solution (Progema) was used to incubate with the EP2 plasmid transfected cells. Afterward, a range of concentrations of the EP2 ligands were added, and the luminescence signals were counted on an EnVision multimode plate reader (PerkinElmer). cAMP level was calculated according to a standard dose-response curve. EC50 (half maximal effective concentration) was calculated via nonlinear regression (curve fit) using GraphPad Prism 7 (GraphPad Software). Each measurement was performed in triplicate.
Acknowledgments
The cryo-EM data were collected at the Cryo-Electron Microscopy Center, Zhejiang University with the technical support from S. Chang and X. Zhang and support from L. Wang at the Translational Medicine Core Facility of Advanced Medical Research Institute, Shandong University. The radioactive ligand binding assays were performed at the Biomedical Isotope Research Center, Shandong University. The cAMP accumulation assays were performed at the Translational Medicine Core Facility of Advanced Medical Research Institute, Shandong University with technical assistance from Y. Yu. Funding: This project was supported by the National Key R&D Program of China (grants 2018YFC1003600 to J.-P.S. and X. Yu, and 2019YFA0904200 to J.-P.S., P.-J.Z., and P.X.), the National Science Fund for Distinguished Young Scholars (grant 81825022 to J.-P.S.), Zhejiang Province National Science Fund for Excellent Young Scholars (grant LR19H310001 to Y.Z.), the National Science Fund for Excellent Young Scholar (grant 81822008 to X. Yu), the National Natural Science Foundation of China (grants 91939301 to Z.L. and J.-P.S., 92057121 to X. Yu, 31971195 and 31700692 to P.X., 31900936 to F.Y., and 32000592 to C.Q.), the Shandong Provincial Natural Science Foundation (grant ZR2016CQ07 to P.X.), the Fundamental Research Funds of Shandong University (grant 2017JQ02 to J.-P.S.), the Shandong Key Research and Development Program (grant 2018GSF118147 to P.X.), China Postdoctoral Science Foundation (grant 2020M670062 to C.Q.), and Shandong University Youth Interdisciplinary Science Innovation Group (grant 2020QNQT002 to X. Yu). Author contributions: J.-P.S., R.-J.C., and Y.Z. conceived and supervised the overall project. R.-J.C. synthesized the large scale of compounds used and analyzed the selective mechanism. C.Q. generated the EP2 insect expression constructs. P.X., C.Q., and Y.-N.Z. performed the complex purification and prepared the samples for cryo-EM. D.-D.S. and H.Z. evaluated these samples by negative-stain EM. Y.Z., C.M., and Q.S. did all the collection of cryo-EM data and performed cryo-EM map calculation, model building, and the structure refinement. J.-P.S., Y.Z., P.X., R.-J.C., C.Q., and F.Y. analyzed these three complex structures and performed the structural-function experimental design. C.Q. and P.X. designed all the mutations for evaluating the interactions inside of the ligand binding pocket and EP2-Gs interface. C.Q., F.Y., and X.T. made all the constructions and then performed the cAMP accumulation assay. C.Q., P.X., and X. Yan analyzed the data for cellular assays. C.Q. and P.X. performed the ligand binding assay under the supervision of C.Z. and Guihua Hou. P.-J.Z. and R.-J.Z. participated in analyzing the results of ligand binding. Y.G. and J.H. prepared Nb35 for the formation of complex. X. Yu provided the COS-7 cell line and assistance for the cell culture. P.X., C.Q., C.M., F.Y., Y.-N.Z., and J.H. prepared the figures. J.-P.S., R.-J.C., and Y.-F.G. wrote the manuscript, and Y.Z., Z.L., and Guige Hou revised the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The atomic coordinates and cryo-EM density maps have been deposited in the Protein Data Bank (PDB) and Electron Microscopy Data Bank (EMDB) under accession numbers 7CX2 and EMD-30489 for the PGE2-EP2 complex, 7CX3 and EMD-30490 for the TAP-EP2 complex, and 7CX4 and EMD-30491 for the EVA-EP2 complex. Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, J.-P.S. (sunjinpeng@bjmu.edu.cn).
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/14/eabf1268/DC1
REFERENCES AND NOTES
- 1.Markovič T., Jakopin Ž., Dolenc M. S., Mlinarič-Raščan I., Structural features of subtype-selective EP receptor modulators. Drug Discov. Today 22, 57–71 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Woodward D. F., Jones R. L., Narumiya S., International union of basic and clinical pharmacology. LXXXIII: Classification of prostanoid receptors, updating 15 years of progress. Pharmacol. Rev. 63, 471–538 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Liu Q., Liang X., Wang Q., Wilson E. N., Lam R., Wang J., Kong W., Tsai C., Pan T., Larkin P. B., Shamloo M., Andreasson K. I., PGE2 signaling via the neuronal EP2 receptor increases injury in a model of cerebral ischemia. Proc. Natl. Acad. Sci. U.S.A. 116, 10019–10024 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Legler D. F., Bruckner M., Uetz-von Allmen E., Krause P., Prostaglandin E2 at new glance: Novel insights in functional diversity offer therapeutic chances. Int. J. Biochem. Cell Biol. 42, 198–201 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Breyer R. M., Bagdassarian C. K., Myers S. A., Breyer M. D., Prostanoid receptors: Subtypes and signaling. Annu. Rev. Pharmacol. Toxicol. 41, 661–690 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Säfholm J., Manson M. L., Bood J., Delin I., Orre A.-C., Bergman P., Al-Ameri M., Dahlén S.-E., Adner M., Prostaglandin E2 inhibits mast cell-dependent bronchoconstriction in human small airways through the E prostanoid subtype 2 receptor. J. Allergy Clin. Immunol. 136, 1232–1239.e1 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Sawada Y., Honda T., Nakamizo S., Nakajima S., Nonomura Y., Otsuka A., Egawa G., Yoshimoto T., Nakamura M., Narumiya S., Kabashima K., Prostaglandin E2 (PGE2)-EP2 signaling negatively regulates murine atopic dermatitis-like skin inflammation by suppressing thymic stromal lymphopoietin expression. J. Allergy Clin. Immunol. 144, 1265–1273.e9 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Cameron K. O., Lefker B. A., Ke H. Z., Li M., Zawistoski M. P., Tjoa C. M., Wright A. S., DeNinno S. L., Paralkar V. M., Owen T. A., Yu L., Thompson D. D., Discovery of CP-533536: An EP2 receptor selective prostaglandin E2 (PGE2) agonist that induces local bone formation. Bioorg. Med. Chem. Lett. 19, 2075–2078 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Plaza J., Torres R., Urbano A., Picado C., de Mora F., In vitro and in vivo validation of EP2-receptor agonism to selectively achieve inhibition of mast cell activity. Allergy Asthma Immunol. Res. 12, 712–728 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schachar R. A., Raber S., Courtney R., Zhang M., A phase 2, randomized, dose-response trial of taprenepag isopropyl (PF-04217329) versus latanoprost 0.005% in open-angle glaucoma and ocular hypertension. Curr. Eye Res. 36, 809–817 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Kennedy C. R. J., Zhang Y., Brandon S., Guan Y., Coffee K., Funk C. D., Magnuson M. A., Oates J. A., Breyer M. D., Breyer R. M., Salt-sensitive hypertension and reduced fertility in mice lacking the prostaglandin EP2 receptor. Nat. Med. 5, 217–220 (1999). [DOI] [PubMed] [Google Scholar]
- 12.Zhu S., Xue R., Zhao P., Fan F.-L., Kong X., Zheng S., Han Q., Zhu Y., Wang N., Yang J., Guan Y., Targeted disruption of the prostaglandin E2 E-prostanoid 2 receptor exacerbates vascular neointimal formation in mice. Arterioscler. Thromb. Vasc. Biol. 31, 1739–1747 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Toyoda Y., Morimoto K., Suno R., Horita S., Yamashita K., Hirata K., Sekiguchi Y., Yasuda S., Shiroishi M., Shimizu T., Urushibata Y., Kajiwara Y., Inazumi T., Hotta Y., Asada H., Nakane T., Shiimura Y., Nakagita T., Tsuge K., Yoshida S., Kuribara T., Hosoya T., Sugimoto Y., Nomura N., Sato M., Hirokawa T., Kinoshita M., Murata T., Takayama K., Yamamoto M., Narumiya S., Iwata S., Kobayashi T., Ligand binding to human prostaglandin E receptor EP4 at the lipid-bilayer interface. Nat. Chem. Biol. 15, 18–26 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Audet M., White K. L., Breton B., Zarzycka B., Han G. W., Lu Y., Gati C., Batyuk A., Popov P., Velasquez J., Manahan D., Hu H., Weierstall U., Liu W., Shui W., Katritch V., Cherezov V., Hanson M. A., Stevens R. C., Crystal structure of misoprostol bound to the labor inducer prostaglandin E2 receptor. Nat. Chem. Biol. 15, 11–17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morimoto K., Suno R., Hotta Y., Yamashita K., Hirata K., Yamamoto M., Narumiya S., Iwata S., Kobayashi T., Crystal structure of the endogenous agonist-bound prostanoid receptor EP3. Nat. Chem. Biol. 15, 8–10 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Wang L., Yao D., Deepak R. N. V. K., Liu H., Xiao Q., Fan H., Gong W., Wei Z., Zhang C., Structures of the human PGD2 receptor CRTH2 reveal novel mechanisms for ligand recognition. Mol. Cell 72, 48–59.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ungrin M. D., Carrière M.-C., Denis D., Lamontagne S., Sawyer N., Stocco R., Tremblay N., Metters K. M., Abramovitz M., Key structural features of prostaglandin E2 and prostanoid analogs involved in binding and activation of the human EP1 prostanoid receptor. Mol. Pharmacol. 59, 1446–1456 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Yang F., Mao C., Guo L., Lin J., Ming Q., Xiao P., Wu X., Shen Q., Guo S., Shen D.-D., Lu R., Zhang L., Huang S., Ping Y., Zhang C., Ma C., Zhang K., Liang X., Shen Y., Nan F., Yi F., Luca V. C., Zhou J., Jiang C., Sun J.-P., Xie X., Yu X., Zhang Y., Structural basis of GPBAR activation and bile acid recognition. Nature 587, 499–504 (2020). [DOI] [PubMed] [Google Scholar]
- 19.García-Nafría J., Lee Y., Bai X., Carpenter B., Tate C. G., Cryo-EM structure of the adenosine A2A receptor coupled to an engineered heterotrimeric G protein. eLife 7, e35946 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rasmussen S. G. F., DeVree B. T., Zou Y., Kruse A. C., Chung K. Y., Kobilka T. S., Thian F. S., Chae P. S., Pardon E., Calinski D., Mathiesen J. M., Shah S. T. A., Lyons J. A., Caffrey M., Gellman S. H., Steyaert J., Skiniotis G., Weis W. I., Sunahara R. K., Kobilka B. K., Crystal structure of the β2 adrenergic receptor–Gs protein complex. Nature 477, 549–555 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y., Yang F., Ling S., Lv P., Zhou Y., Fang W., Sun W., Zhang L., Shi P., Tian C., Single-particle cryo-EM structural studies of the β2AR–Gs complex bound with a full agonist formoterol. Cell Discov. 6, 45 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sugimoto Y., Nakato T., Kita A., Hatae N., Tabata H., Tanaka S., Ichikawa A., Functional domains essential for Gs activity in prostaglandin EP2 and EP3 receptors. Life Sci. 74, 135–141 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Yano A., Takahashi Y., Moriguchi H., Inazumi T., Koga T., Otaka A., Sugimoto Y., An aromatic amino acid within intracellular loop 2 of the prostaglandin EP2 receptor is a prerequisite for selective association and activation of Gαs. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1862, 615–622 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Mastronarde D. N., Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Zheng S. Q., Palovcak E., Armache J.-P., Verba K. A., Cheng Y., Agard D. A., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang K., Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scheres S. H. W., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heymann J. B., Guidelines for using Bsoft for high resolution reconstruction and validation of biomolecular structures from electron micrographs. Protein Sci. 27, 159–171 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arnold K., Bordoli L., Kopp J., Schwede T., The SWISS–MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Afonine P. V., Klaholz B. P., Moriarty N. W., Poon B. K., Sobolev O. V., Terwilliger T. C., Adams P. D., Urzhumtsev A., New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr. D Struct. Biol. 74, 814–840 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang H.-M., Dong J.-H., Li Q., Hu Q., Ning S.-L., Zheng W., Cui M., Chen T.-S., Xie X., Sun J.-P., Yu X., A stress response pathway in mice upregulates somatostatin level and transcription in pancreatic delta cells through Gs and β-arrestin 1. Diabetologia 57, 1899–1910 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/14/eabf1268/DC1