Abstract
Background
Streptococcus pneumoniae is one of the leading causes of mortality and morbidity worldwide. The dramatic increase in in-vitro resistance of antimicrobial agents, particularly beta-lactams and macrolides, makes pneumococcal infections difficult to treat. The aim of this study was to describe the drug resistance rate, assess the prevalence of macrolide-resistant genes and review the clinical complications of pneumococcal infections among patients presented to Hospital Universiti Sains Malaysia (HUSM), Kelantan, Malaysia.
Methods
This is a descriptive cross-sectional study. All S. pneumoniae isolates collected from clinical specimens within a 1-year period were subjected to selected antimicrobial susceptibility testing using E-test strips. Polymerase chain reaction (PCR) analysis was conducted to detect macrolide-resistant determinants. The patient’s clinical data were obtained from clinical notes.
Results
A total of 113 patients with a positive growth of S. pneumoniae were included in the study. The most common predisposing factors among them were bronchopulmonary diseases (15.9%). The penicillin-resistant rate was 7.1%, with minimal inhibitory concentration (MIC) ranging between 0.012 μg/mL and >32 μg/mL, and the erythromycin-resistant rate was 26.5%, with a MIC range of 0.03 μg/mL–> 256 μg/mL. Most of the erythromycin-resistant isolates were found to have the mef(A) gene (50.4%) and the erm(B) gene (20%); 16.7% had a combination of genes mef(A) and erm(B), and 13.3% had none of the two genes. Community-acquired pneumonia is the predominant type of pneumococcal infection. There was no significant association between the presence of macrolide resistance determinants and mortality (P = 0.837) or complications (P > 0.999 for empyema and cardiac complication; P = 0.135 for subdural abscess).
Conclusion
The majority of erythromycin-resistant isolates were found to have the mef(A) gene, followed by the erm(B) gene and a combination of genes mef(A) and erm(B).
Keywords: macrolide, resistant, genes, Streptococcus pneumoniae, susceptibility
Introduction
Streptococcus pneumoniae is one of the leading causes of mortality and morbidity worldwide, posing a major public health problem. It causes more serious infections in infants under 2 years old (1) and adults older than 65 years old (2). S. pneumoniae is a Gram-positive bacteria spread by airborne droplets, and it is a primary cause of bacterial pneumonia, acute otitis media, sinusitis, bacteremia and meningitis. The World Health Organization (WHO) estimates that, in the Asia Pacific region, 49 out of 98 cases of pneumonia deaths in children are due to pneumococcal pneumonia. Several studies reported that pneumonia is the most common pneumococcal disease in Malaysia (3, 4). Additionally, about 4% of the 7,000 deaths among children less than 5 years old were thought to be caused by S. pneumoniae (5).
The dramatic increase in the prevalence of antibiotic resistance, especially against first-line antibiotics such as penicillin and macrolide, makes pneumococcal infections difficult to treat, especially in children and the elderly (6). In some Asian countries, the reported prevalence of penicillin and macrolide resistance was the highest in the world (7, 8). Previous surveillance studies showed that more than 60% of pneumococcal isolates from Asian countries were resistant to erythromycin and 22.7% were penicillin-resistant (9). In China, erythromycin resistance rates for S. pneumoniae increased from 35% to 53% in 1996–1999, to over 75% by 2001, and South Korean resistance rates against erythromycin remained high from 1996 to 2001 (75%–85%). While for Malaysia, 32% were resistant to erythromycin in the year 2012 (9).
Ribosomal methylation encoded by erm(B) gene was the most common mechanism of erythromycin resistance in China, Taiwan, Sri Lanka and Korea (7). While in Hong Kong, Singapore, Thailand and Malaysia, an efflux pump encoded by the mef(A) gene was more common. Erm(B) gene was found in more than 50% of pneumococcal isolates either alone or in combination with mef(A) gene in most Asian countries except Hong Kong, Malaysia and Singapore (7). Although antibiotic resistance among S. pneumoniae has been increasingly noted worldwide, the clinical significance of in-vitro resistance is not widely studied (10, 11). The aim of this study is to describe the antibiotic susceptibility pattern and the distribution of macrolide-resistance determinants of S. pneumoniae isolated from patients presented to Hospital Universiti Sains Malaysia (HUSM), Kelantan, Malaysia.
Methods
Pneumococcal Isolates
A total of 113 non-duplicate isolates of S. pneumoniae were collected from patients admitted to HUSM from June 2014 to December 2015. Pneumococcal isolates were collected from various clinical specimens including sputum, throat swab, endotracheal secretion, nasopharyngeal swab/aspirate, blood, cerebrospinal fluid (CSF), pus swab, high vaginal swab (HVS) and other body fluids. The isolates were presumptively identified as S. pneumoniae by their colony morphology, Gram-stain results and susceptibility to 5 μg optochin disc (ethyl hydrocuprein hydrochloride, Becton, Dickinson and Company, USA). The isolates were stored at –70 oC in brain heart infusion broth plus 20% glycerol in the laboratory until further testing.
Antimicrobial Susceptibility Test
The minimal inhibitory concentrations (MICs) of six antimicrobial agents: i) penicillin, ii) erythromycin, iii) azithromycin, iv) amoxicillin-clavulanic acid, v) trimethoprim-sulphamethoxazole and vi) vancomycin were determined by using E-test strips (BioMerieux SA, France) according to manufacturer’s recommendations. The MIC results are interpreted according to Clinical and Laboratory Standard Institute (CLSI) guidelines, 2015. S. pneumoniae ATCC 49619 was used as the control strain.
Detection of Macrolide-Resistant Genes; erm(B) and mef(A) Genes
Out of 113 isolates of S. pneumoniae, 108 were subjected to polymerase chain reaction (PCR) analysis to determine the presence of macrolide-resistant genes. Three of the isolates were non-viable and the other two isolates were contaminated with other bacteria. According to the manufacturer’s instructions, DNA extractions were performed using a DNA extraction kit (DNeasy Blood & Tissue Kit, Qiagen, USA). The PCR mixture consists of a 20 μL total volume containing extracted DNA template, DreamTaq DNA polymerase (Thermo Scientific, Malaysia), 10X DreamTaq buffer which contains 20 mM MgCl2, 2 mM dNTP Mix, nuclease-free water and primer mixture. The primer sequences for erm(B) gene are 5′-CGTACCTTGGATATTCACCG-3′ and 5′-GTAAACAGTTGACGATATTCTCG-3′ whilst for mef(A) gene are 5′-CTGTATGGAGCTACCTGTCTGG-3′ and 5′-CCCAGCTTAGGTATACGTAC-3′ (12). The primer mixture was prepared by mixing an equal concentration of genes erm(B) and mef(A) primers (20 pmol/μL) for each forward and reverse primer. The amplification was performed with a thermal cycler (MJ Research Peltier Thermal Cycler, GMI, USA). The positive control strains; S. pneumoniae ATCC 700673 (Hungary19A-6) for erm(B) gene and S. pneumoniae ATCC 51916 (Tennessee23F-4) for mef(A) gene (13) and a negative control were included in each run. Following amplification, 2 μL of the PCR products were electrophoresed on 1% agarose gel and visualised using a UV light transilluminator (Syngene, USA) with GeneSnap image analysis software.
Statistical Analysis
Statistical analysis was performed using SPSS, version 22.0. Fisher’s exact test or X2 test was applied to determine the association between the presence of macrolide resistance genes with mortality and complications.
Results
Distribution of Clinical Specimens of S. pneumoniae Isolates
A total of 113 non-duplicate isolates of S. pneumoniae from various clinical specimens were included in this study. The majority of the isolates were from sputum (33.6%), followed by endotracheal tube secretion (ETT) (29%), eye swab (14%), blood (10.6%), high vaginal swab (HVS) (2.7%), ear swab (1.8%) and corneal scrapping (1.8%). Other specimens (12.4%) included CSF, broncho-alveolar lavage (BAL) fluid, vitreous fluid, bone tissue, bile, nasal swab and pus swab.
Demographic and Clinical Characteristics of Patients with S. pneumoniae Infection
Among the 113 patients, 22 (19.5%) were children aged 5 years old or below and 16 (14.2%) were elderly patients (≥ 65 years old). The median age of the patients was 33 years old. Of these patients, 73 (64.6%) were males and 40 (35.4%) were females. Community-acquired pneumonia was the most common type of infection (59.3%), followed by conjunctivitis (8.8%) and bacteremia (7.1%). About 9.9% of the patients had S. pneumoniae (the majority were from ETT specimens). The most common predisposing factors among patients with culture-positive S. pneumoniae were bronchopulmonary diseases (15.9%), underlying malignancy (6.2%) and chronic renal disease (4.4%).
In vitro Susceptibility Pattern of S. pneumoniae Isolates
The susceptibility pattern of 113 S. pneumoniae isolates was shown in Table 1 and the MIC distribution of macrolide antibiotic based on the presence of macrolide resistance determinants was shown in Table 3.
Table 1.
Susceptibility rates of S. pneumoniae isolate for six antimicrobial agents (n = 113)
| Antibiotics | MIC50 (μg/mL) | MIC90 (μg/mL) | MIC range (μg/mL) | MIC interpretive criteria (μg/mL) | Antimicrobial susceptibility | ||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| S | I | R | S n (%) | I n (%) | R n (%) | ||||
| Penicillin parenteral (nonmeningitis) | 0.032 | 1 | 0.012–> 32 | ≤ 2 | 4 | ≥ 8 | 110 (97.3) | 2 (1.8) | 1 (0.9) |
| Penicillin parenteral (meningitis) | 0.032 | 1 | 0.012–> 32 | ≤ 0.06 | - | ≥ 0.12 | 88 (77.9) | 0 (0.0) | 25 (22.1) |
| Penicillin (oral) | 0.032 | 1 | 0.012–> 32 | ≤ 0.06 | 0.12–1 | ≥ 2 | 88 (77.9) | 17 (15.0) | 8 (7.1) |
| Erythromycin | 0.125 | 32 | 0.03–> 256 | ≤ 0.25 | 0.5 | ≥ 1 | 83 (73.5) | 0 (0.0) | 30 (26.5) |
| Azithromycin | 1 | 96 | 0.10–> 256 | ≤ 0.5 | 1 | ≥ 2 | 22 (19.5) | 57 (50.4) | 34 (30.1) |
| Amoxicillin-clavulanic acid | 0.03 | 1 | 0.015–8 | ≤ 2/1 | 4/2 | ≥ 8/4 | 110 (97.3) | 0 (0.0) | 3 (2.7) |
| Trimethoprim-sulfamethoxazole | 0.25 | > 32 | 0.064–> 32 | ≥ 0.5/9.5 | 1/19–2/38 | ≥ 4/76 | 77 (68.1) | 16 (14.2) | 20 (17.7) |
| Vancomycin | 0.5 | 0.5 | 0.12–1 | ≤ 1 | - | - | 113 (100.0) | 0 (0.0) | 0 (0.0) |
Notes: MIC50/90 = MIC at which 50% or 90% of the isolates are inhibited; S = susceptible; I = intermediate; R= resistant
Table 3.
MIC distribution of macrolide antibiotic based on the presence of macrolide resistance determinant (n = 113)
| Antimicrobial agent | MIC (μg/mL) distribution | MIC range | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||||||||
| 0.032 | 0.125 | 0.25 | 0.5 | 0.75 | 1 | 2 | 3 | 4 | 6 | 8 | 12 | 16 | 24 | 32 | 48 | 96 | > 256 | ||
| Erythromycina | |||||||||||||||||||
| erm(B) | 1 | 5 | 16–> 256 | ||||||||||||||||
| mef(A) | 1 | 3 | 1 | 1 | 2 | 1 | 3 | 5 | 1 | 1 | 1 | 0.032–> 32 | |||||||
| erm(B) + mef(A) | 1 | 4 | 2–> 256 | ||||||||||||||||
| Azithromycinb | |||||||||||||||||||
| erm(B) | 1 | 5 | 1–> 256 | ||||||||||||||||
| mef(A) | 1 | 1 | 2 | 1 | 1 | 2 | 2 | 1 | 2 | 1 | 1 | 1 | 3 | 1 | 0.125–> 256 | ||||
| erm(B) + mef(A) | 1 | 4 | 8–> 256 | ||||||||||||||||
Notes:
Resistance to erythromycin was defined as MIC ≥ 1 μg/mL;
Resistance to azithromycin was defined as MIC ≥ 2 μg/mL
Distribution of erm(B) and mef(A) Genes
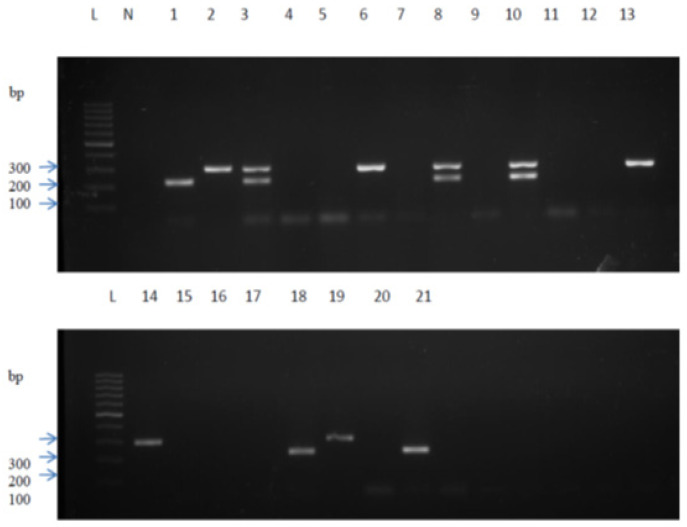
Out of 108 isolates subjected to PCR analysis, erm(B) gene, mef(A) gene or a combination of erm(B) and mef(A) genes were identified in 31 (27.4%) isolates. Of these isolates, 20 (64.5%) had mef(A) gene alone and 6 (19.4%) had erm(B) gene alone, whereas 5 (16.15) contained both mef(A) and erm(B) genes. The PCR gel image showed amplified DNA fragments, as demonstrated in Figure 1.
Figure 1.
Scanned image of gel showing results of amplified DNA fragments of positive control (lanes 1–3) and 18 test isolates (lanes 4–21). PCR amplicons of mef(A) (298 bp) gene were shown in lanes 6, 13, 14 and 19, erm(B) (224 bp) gene in lanes 18 and 21 and a combination of mef(A) and erm(B) gene was shown in lane 8 and 10. A lane with no band indicates the absence of both genes
Notes: Lane L = 100 bp DNA ladder; Lane N = negative control (DNase-free water); Lane 1 = positive control (S. pneumoniae ATCC 700673 (Hungary19A-6) - for erm(B) gene; Lane 2 = positive control (S. pneumoniae ATCC 51916) (Tennessee23F-4) - for mef(A) gene; Lane 3 = positive control (S. pneumoniae ATCC 700673 (Humgary19A-6) + S. pneumoniae ATCC 51916 (Tennessee23F-4) - for erm(B) gene + mef(A) gene combination; Lane 4–21 = S. pneumoniae isolates
Among 30 erythromycin-resistant S. pneumoniae, mef(A) and erm(B) genes were detected in 15 (50.0%) and 6 (20.0%) isolates, respectively. Both mef(A) and erm(B) genes were detected in 5 (16.7 %) isolates and 4 (13.3%) did not harbour either of the genes. Five (6.4%) of the erythromycin-sensitive isolates were detected to have have the mef(A) gene. Among the 34 azithromycin-resistant isolates, 15 (44.1%) and 5 (14.7%) had mef(A) gene and erm(B) gene, respectively, and 5 (14.7%) had both genes. Nine (26.5%) did not contain any of the genes. Two azithromycin-sensitive S. pneumoniae (10.0%) had mef(A) gene (Table 2). Isolates that harbour erm(B) gene alone or both erm(B) and mef(A) genes showed higher MICs ranging from 2 μg/mL to > 256 μg/mL and 8 μg/mL to > 256 μg/mL for erythromycin-resistant S. pneumoniae and azithromycin-resistant S. pneumoniae, respectively, as compared to isolates carrying mef(A) gene alone, with MICs ranging from 1 μg/mL to 32 μg/mL and 3 μg/mL to > 256 μg/mL for erythromycin-resistant S. pneumoniae and azithromycin-resistant S. pneumoniae, respectively (Table 3).
Table 2.
Distribution of macrolide resistance determinants according to the susceptibility of macrolide antibiotics (n = 108)
| Susceptibility to | No. of isolates with macrolide resistance determinant | Total n (%) |
|||
|---|---|---|---|---|---|
|
| |||||
|
mef(A) n (%) |
erm(B) n (%) |
mef(A) + erm(B) n (%) |
None n (%) |
||
| Erythromycin | |||||
| Sensitive | 5 (6.4) | 0 (0.0) | 0 (0.0) | 73 (93.6) | 78 (100.0) |
| Intermediate | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Resistant | 15 (50.0) | 6 (20.0) | 5 (16.7) | 4 (13.3) | 30 (100.0) |
| Azithromycin | |||||
| Sensitive | 2 (10.0) | 0 (0.0) | 0 (0.0) | 18 (90.0) | 20 (100.0) |
| Intermediate | 3 (5.6) | 1 (1.9) | 0 (0.0) | 50 (92.6) | 54 (100.0) |
| Resistant | 15 (44.1) | 5 (14.7) | 5 (14.7) | 9 (26.5) | 34 (100.0) |
Impact of Resistance to Mortality and Complications
Six patients (5.3%) died in the hospital while receiving treatment. One patient (0.9%) had empyema-related complications and another had subdural abscess-related complications. Two (1.8%) developed arrhythmia. The results (Tables 4 and 5) showed that there is no significant association between mortality rate and the development of complications between patients harbouring macrolide-resistance determinants with those without macrolide-resistance determinants.
Table 4.
Association between macrolide resistance determinants with clinical outcome (n = 111)
| Macrolide resistance determinant | Outcome | P-valuea | |
|---|---|---|---|
|
| |||
| Alive n (%) |
Death n (%) |
||
| erm(B) | 6 (100) | 0 (0) | 0.837 |
| mef(A) | 18 (90) | 2 (10) | |
| erm(B) + mef(A) | 4 (100) | 0 (0) | |
| None | 72 (94.7) | 4 (5.3) | |
| Non-viable/contaminated | 5 (100) | 0 (0) | |
Note:
Fisher’s exact test was applied
Table 5.
Association between macrolide resistance determinants with complications (n = 111)
| Complication | Genes | P-valuea | |||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| erm(B) | mef(A) | erm(B) + mef(A) | None | NV/C | |||
| Empyema | Absent | 6 (100) | 20 (100) | 4 (100) | 75 (98.7) | 5 (100) | > 0.999 |
| Present | 0 (0) | 0 (0) | 0 (0) | 1 (1.3) | 0 (0) | ||
| Subdural abscess | Absent | 5 (83.3) | 20 (100) | 4 (100) | 76 (100) | 5 (100) | 0.135 |
| Present | 1 (16.7) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ||
| Cardiac | Absent | 6 (100) | 20 (100) | 4 (100) | 74 (97.4) | 5 (100) | > 0.999 |
| Present | 0 (0) | 0 (0) | 0 (0) | 2 (2.6) | 0 (0) | ||
Notes:
Fisher’s exact test was applied; NV/C= non-viable/contaminated
Discussion
In Malaysia, the prevalence of macrolide-resistant S. pneumoniae appeared to have increased over the years from 36.8% in 2001 (14) to 46.2% in 2005, then drastically increased to 62.2% in the year 2010 (15, 16). In our study, the rates of susceptibility towards macrolide antibiotics were not much different to those of a previous study (16), where the resistance rates to erythromycin and azithromycin were 26.5% and 30.1%, respectively. However, we reported a decrease in erythromycin resistance compared to more recent studies. However, the resistance rate towards erythromycin in 2010, which was more than 60%, was higher (14, 15).
The level of macrolide resistance has increased remarkably, with very high MIC90s value (64 μg/mL to ≥ 128 μg/mL) in Asian countries, including Malaysia (9). In this study, we found that isolates which are resistant to erythromycin or azithromycin also have high MIC values. The majority of them have an MIC of > 256 μg/mL, with MIC90 values of 32 μg/mL and 128 μg/mL for erythromycin and azithromycin, respectively. Compared with previous studies that investigated S. pneumoniae isolates in the years 2008–2010, the MIC90s of erythromycin are higher, with MIC values of ≥ 256 μg/mL (14, 15). In this study, 96.7% of erythromycin-resistant isolates were also resistant to azithromycin, which is consistent with previous studies where 94.4%–97% of the erythromycin-resistant pneumococcal isolates had concomitant erythromycin and azithromycin resistance (6, 15).
The erm(B) gene-mediated resistance in pneumococci gives high-level resistance (MLSB phenotype), with MIC values typically ≥ 64 μg/mL. In contrast, the efflux pump (M phenotype) encoded by the mef(A) gene confers a low-level resistance, with MIC usually 1 μg/mL–32 μg/mL (9, 17, 18). Similarly, in this study, among the 30 erythromycin-resistant pneumococci, isolates that carry the erm(B) gene alone or both erm(B) and mef(A) genes showed higher MICs, ranging from 2 μg/mL to > 256 μg/mL, with MIC90 values for erm(B) gene being > 256 μg/mL. This is in contrast to isolates carrying mef(A) gene alone, which have much lower MICs ranging from 1 μg/mL to 32 μg/mL and a MIC90 of 4 μg/mL for mef(A) gene. These findings were in accordance with the other study that reported erythromycin-resistant S. pneumoniae isolates with both erm(B) and mef(A) genes showed very high MICs ≥ 256 μg/mL (14).
Ones risk of contracting pneumococcal infections varies depending on the age and gender of the individual. The prevalence of pneumococcal disease is substantially higher in children and the elderly above 65 years old than in young adults (2). The study’s findings also emphasise on the preponderance of pneumococcal disease among males, which might be linked to the higher incidence of underlying factors such as smoking and exposure to outdoor air pollution among males. Deficiencies in the non-specific or specific defence mechanisms against colonisation, aspiration or invasion of S. pneumoniae increase the likelihood of pneumococcal disease among this group of patients.
The clinical significance of the in-vitro resistance of macrolide antibiotics is still controversial (19). The failure of macrolide therapy caused by erythromycin-resistant strain in patients with pneumococcal diseases has been reported in previous studies (18, 20). A study investigating the mortality rates associated with invasive pneumococcal diseases reported almost similar findings with patients with erythromycin-resistant (18%) or erythromycin-susceptible strains (14%) (21). This study also had similar findings, where the mortality rate among patients with macrolide-resistant strains was not significantly different from that of patients with macrolide-susceptible strains (P > 0.999 for erythromycin and P = 0.667 for azithromycin). In this study, we also specifically assessed the relationship between the macrolide resistance determinants and the mortality and complications in patients with pneumococcal diseases since these resistant determinants are the main cause of macrolide resistance. This revealed a similar result where mortality and complication in patients carrying the resistance determinants did not differ from those in patients without resistance determinants. These findings were supported by another study by the Asian Network for Surveillance of Resistant Pathogens (ANSORP) group that reported that pneumococcal infection with erythromycin-resistant strains was not significantly associated with more severe illness or higher mortality (22).
The prevalence of the macrolide-resistant determinant gene of S. pneumoniae can provide the management teams in hospitals with crucial insights for making appropriate decisions on antimicrobial usage in patients’ treatment. By knowing its prevalence, healthcare professionals can understand the likelihood of resistance and choose alternative antibiotics for patients with macrolide-resistant infections, improving treatment efficacy. They may avoid unnecessary macrolide prescriptions in areas with high resistance rates, reducing the risk of further resistance development and preserving the effectiveness of these antibiotics. Furthermore, they can implement targeted infection control measures to limit the spread of macrolide-resistant infections within healthcare facilities.
This study has several limitations. The identification of S. pneumoniae isolates based on colony morphology, Gram-stain results and susceptibility to the optochin disc reflects the use of classical microbiological methods that have been historically relied upon for bacterial identification. These methods offer a cost-effective and relatively simple way to distinguish between different streptococcal species which is often encountered in clinical settings. However, the challenge arises because closely related species like S. pseudopneumoniae and S. mitis share similar characteristics and can be mistaken for S. pneumoniae using these conventional techniques. The similarity in optochin susceptibility and bile solubility between these species can lead to misclassification. As advancements in molecular techniques and genetic analysis have emerged, it has become apparent that incorporating more precise and specific methods, such as DNA sequencing and PCR-based assays, can enhance the accuracy of S. pneumoniae identification. However, due to limited resources and a small amount of grant money allocation, the molecular identification method could not be used.
Another limitation was the inadequacy of clinical data collected from medical records that largely depend on the clinician’s observation and interpretation. Incomplete documentation of the medical records will largely influence our data, especially on the predisposing factors and outcome of the diseases. Finally, since the sources of the isolates were collected only from the state of Kelantan, data from this study may not be representative of the Malaysian population.
Conclusion
The majority of erythromycin-resistant isolates were found to have a mef(A) gene, an erm(B) gene or a combination of mef(A) and erm(B) genes. Overall, the isolates showed good susceptibility towards all antibiotics tested except for azithromycin. The outcome and complications of pneumococcal diseases were not significantly different between macrolide-resistant groups and macrolide-susceptible groups. They also were not affected by the presence of macrolide-resistant determinants in the pneumococcal isolates. Current data regarding the in-vitro susceptibility patterns of S. pneumoniae and the prevalence of the macrolide-resistant determinants gene could help the management team make appropriate decisions regarding antimicrobial usage in treating patients. Continuous surveillance of antibiotic resistance at the national level is very important to monitor the changing trends in antimicrobial resistance in Malaysia.
Acknowledgements
We thank the Department of Medical Microbiology and Parasitology, School of Medical Sciences, Universiti Sains Malaysia for the facility provided.
Footnotes
Ethics of Study: This study was approved by the Human Research Ethics Committee, Universiti Sains Malaysia, Kubang Kerian, Kelantan, Malaysia (JEPeM code: USM/JEPeM/15010025) prior to conducting the study.
Conflict of Interest: None.
Funds: The study was funded by Universiti Sains Malaysia, Short Term Grant, (304/PPSP/61311069) and RU Grant (1001.PPSP.812064).
Authors’ Contributions: Conception and design: NI
Analysis and interpretation of the data: WNWM, SAH
Drafting of the article: WNWM
Critical revision of the article for important intellectual content: NI, SAH, ZAH, WNAWAW
Final approval of the article: NI, SAH, ZAH, WNAWAW
Provision of study materials or patients: NI
Statistical expertise: WNWM
Obtaining of funding: NI
References
- 1.Thadchanamoorthy V, Dayasiri K. Review on pneumococcal infection in children. Cureus. 2021;13(5):e14913. doi: 10.7759/cureus.14913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martinez-Vega R, Jauneikaite E, Thoon KC, Chua HY, Huishi Chua A, Khong WX, et al. Risk factor profiles and clinical outcomes for children and adults with pneumococcal infections in Singapore: a need to expand vaccination policy? PLoS ONE. 2019;14(10):e0220951. doi: 10.1371/journal.pone.0220951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shafie AA, Ahmad N, Naidoo J, Foo CY, Wong C, Pugh S, et al. Estimating the population health and economic impacts of introducing a pneumococcal conjugate vaccine in Malaysia—an economic evaluation. Hum Vaccin Immunother. 2020;16(7):1719–1727. doi: 10.1080/21645515.2019.1701911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arushothy R, Ahmad N, Amran F, Hashim R, Samsudin N, Azih CRC. Pneumococcal serotype distribution and antibiotic susceptibility in Malaysia: a four-year study (2014–2017) on invasive paediatric isolates. Int J Infect Dis. 2019;80:129–133. doi: 10.1016/j.ijid.2018.12.009. [DOI] [PubMed] [Google Scholar]
- 5.Yasin RM, Zin NM, Hussin A, Nawi SH, Hanapiah SM, Wahab ZA, et al. Current trend of pneumococcal serotypes distribution and antibiotic susceptibility pattern in Malaysian hospitals. Vaccine. 2011;29(34):5688–5693. doi: 10.1016/j.vaccine.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 6.Ktari S, Ben Ayed N, Ben Rbeh I, Garbi N, Maalej S, Mnif B, et al. Antibiotic resistance pattern, capsular types, and molecular characterization of invasive isolates of Streptococcus pneumoniae in the south of Tunisia from 2012 to 2018. BMC Microbiol. 2023;23(1):36. doi: 10.1186/s12866-023-02784-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang CI, Song JH. Antimicrobial resistance in Asia: current epidemiology and clinical implications. Infect Chemother. 2013;45(1):22–31. doi: 10.3947/ic.2013.45.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lister AJJ, Le CF, Cheah ESG, Desa MNM, Cleary DW, Clarke SC. Serotype distribution of invasive, non-invasive and carried Streptococcus pneumoniae in Malaysia: a meta-analysis. Pneumonia. 2021;13:9. doi: 10.1186/s41479-021-00086-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim SH, Song JH, Chung DR, Thamlikitkul V, Yang Y, Wang H, et al. Changing trends in antimicrobial resistance and serotypes of Streptococcus pneumoniae isolates in Asian countries: an Asian Network for Surveillance of Resistant Pathogens (ANSORP) study. Antimicrob Agents Chemother. 2012;56(3):1418–1426. doi: 10.1128/AAC.05658-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karcic E, Aljicevic M, Bektas S, Karcic B. Antimicrobial susceptibility/resistance of Streptococcus pneumoniae. Mater Sociomed. 2015;27(3):180–184. doi: 10.5455/msm.2015.27.180-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larsson M, Nguyen HQ, Olson L, Tran TK, Nguyen TV, Nguyen CTK. Multi-drug resistance in Streptococcus pneumoniae among children in rural Vietnam more than doubled from 1999 to 2014. Acta Paediatr. 2021;110(6):1916–1923. doi: 10.1111/apa.15795. [DOI] [PubMed] [Google Scholar]
- 12.Nagai K, Shibasaki Y, Hasegawa K, Davies TA, Jacobs MR, Ubukata K, Appelbaum PC. Evaluation of PCR primers to screen for Streptococcus pneumoniae isolates and β-lactam resistance, and to detect common macrolide resistance determinants. J Antimicrob Chemother. 2001;48:915–918. doi: 10.1093/jac/48.6.915. [DOI] [PubMed] [Google Scholar]
- 13.Sirekbasan L, Gönüllü N, Sirekbasan S, Kuşkucu M, Midilli K. Phenotypes and genotypes of macrolide-resistant Streptococcus pneumoniae. Balkan Med J. 2015;32:84. doi: 10.5152/balkanmedj.2015.15169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nathan JJ, Mohd Taib N, Mohd Desa MN, Masri SN, Md Yasin R, Farida Jamal F, et al. Prevalence of macrolide resistance and in vitro activities of six antimicrobial agents against clinical isolates of Streptococcus pneumoniae from a multi-center surveillance in Malaysia. Med J Malaysia. 2013;68(2):119–124. [PubMed] [Google Scholar]
- 15.Arushothy R, Ahmad N, Md Yassin R. Evolution of erythromycin resistance among Streptococcus pneumoniae isolates in Malaysia from 2005 and 2010. J Biosci Med. 2016;4:116–122. doi: 10.4236/jbm.2016.45012. [DOI] [Google Scholar]
- 16.Rohani MY, Zin NM, Hussin A, Nawi SH, Hanapiah SM, Wahab ZA, et al. Current trend of pneumococcal serotypes distribution and antibiotic susceptibility pattern in Malaysian hospitals. Vaccine. 2011;29:5688–5693. doi: 10.1016/j.vaccine.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 17.Zhou XZ, Liu JH, Zhang ZJ, Cui B, Wang YL, Zhang Y, et al. Characterization of Streptococcus pneumoniae macrolide resistance and its mechanism in Northeast China over a 20-year period. Microbiol Spectr. 2022;10(5):1–13. doi: 10.1128/spectrum.00546-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McFarlane A, Sligl W. The value of macrolide-based regimens for community-acquired pneumonia. Curr Infect Dis Rep. 2015;17:50. doi: 10.1007/s11908-015-0507-4. [DOI] [PubMed] [Google Scholar]
- 19.Joseph P, Lynch III, Martinez FJ. Clinical relevance of macrolide-resistant Streptococcus pneumoniae for community acquired pneumonia. Clin Infect Dis. 2002;34:S27–S46. doi: 10.1086/324527. [DOI] [PubMed] [Google Scholar]
- 20.Cao B, Huang Y, She DY, Cheng QJ, Fan H, Tian XL, et al. Diagnosis and treatment of community-acquired pneumonia in adults: 2016 clinical practice guidelines by the Chinese Thoracic Society, Chinese Medical Association. Clin Respir J. 2018;12:1320–1360. doi: 10.1111/crj.12674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cilloniz C, Albert RK, Liapikou A, Gabarrus A, Rangel E, Bello S, et al. The effect of macrolide resistance on the presentation and outcome of patients hospitalized for Streptococcus pneumoniae. Am J Respir Crit Care Med. 2015;191(11):1265–1272. doi: 10.1164/rccm.201502-0212OC. [DOI] [PubMed] [Google Scholar]
- 22.Song JH, Lee NY, Ichiyama S, Yoshida R, Hirakata Y, Fu W, et al. Spread of drug-resistant Streptococcus pneumoniae in Asian countries: Asian Network for Surveillance of Resistant Pathogens (ANSORP) study. Clin Infect Dis. 1999;28:1206–1211. doi: 10.1086/514783. [DOI] [PubMed] [Google Scholar]